Abstract
Although there is no standard treatment protocol for inflammatory breast cancer (IBC), multi-modality treatment has improved survival. In this study we profiled the NCI approved oncology drug set in a qHTS format to identify those that are efficacious in basal type and ErbB2 overexpressing IBC models. Further, we characterized the sensitivity of an acquired therapeutic resistance model to the oncology drugs. We observed that lapatinib-induced acquired resistance in SUM149 cells led to cross-resistance to other targeted- and chemotherapeutic drugs. Removal of the primary drug to which the model was developed led to re-sensitization to multiple drugs to a degree comparable to the parental cell line; this coincided with the cells regaining the ability to accumulate ROS and reduced expression of anti-apoptotic factors and the antioxidant SOD2. We suggest that our findings provide a unique IBC model system for gaining an understanding of acquired therapeutic resistance and the effect of redox adaptation on anti-cancer drug efficacy.
Keywords: qHTS, SUM149, SUM190, lapatinib, XIAP, redox adaptation
1. Introduction
Cancer is a complex and heterogeneous disease, and increasingly large scale profiling efforts have been undertaken at both the genetic and protein levels to help enhance our understanding of this disease and the variability in molecular phenotype, including for example, identifying genes critical to the growth of breast cancer subtypes [1]. More recently, chemical biology or systems pharmacology [2; 3] approaches have been undertaken to link drug treatment and tumor cell responses with the goal of identifying signatures that predict drug sensitivity or clinical responses in patients [4; 5; 6; 7]. Drug profiling on a collection of breast cancer subtypes revealed that therapeutic compounds showed significant subtype specificity [8], and efficacy profiling was able to identify relationships between compounds that were not apparent from gene expression profiling [9].
Inflammatory breast cancer (IBC), an understudied and relatively rare subtype of breast cancer, is a distinct and highly aggressive form of locally advanced breast cancer (LABC). Current data from the National Cancer Institute (NCI) Surveillance, Epidemiology and End Results (SEER) program suggests that IBC accounts for 1-5% of all newly diagnosed breast cancer in the United States annually, although many believe it is more common but difficult to diagnose [10]. IBC is clinically distinguished by rapid onset of primary skin changes and progression to Stage IIIb/IV within 6 months. IBC carries a guarded prognosis with 5-&10-yr disease-free survival rates approaching 45% and 20% respectively compared to around 90% (5-yr) for non-IBC [11; 12; 13].
Despite the improvements gained from multidisciplinary management and anthracycline- and taxane-based polychemotherapy regimens [14], women with IBC continue to have worse survival outcomes compared with those with non-IBC LABC [15]. IBC has been designated by the FDA as an orphan cancer and is a health disparity as it disproportionately affects minority populations globally including African American and American Indian women, and is more prevalent in young women of child bearing age. There shows no standard IBC-specific treatment for patients with advanced disease, and enrollment in clinical trials has been strongly recommended [16].
For IBC tumors with over-expression of epidermal growth factor receptors (ErbB1/EGFR and ErbB2/HER2), ErbB1/2 targeting agents including trastuzumab (HER2 targeting) and lapatinib (a dual ErbB1/2 tyrosine kinase inhibitor) have been approved for treatment. Although the clinical response (CR) rate of patients receiving lapatinib monotherapy has been remarkable (50% CR rate in ErbB2-overexpressing IBC patients [17]), a significant number of patients do not respond to lapatinib, and acquired resistance is frequent [18; 19]. A recent study indicates that expression of surrogate markers ER, PR, and HER2 is significantly associated with overall survival and loco-regional relapse outcomes in patients with IBC of the triple negative (TN) subtype, consistently predicting the worst outcomes [20]. Since endocrine and anti–HER2 therapies are ineffective in TN IBC, EGFR inhibitors (gefitinib, erlotinib), an anti-EGFR antibody (cetuximab), and a c-KIT inhibitor (imatinib) are in clinical trials; however, recent trial results with cetuximab have been disappointing in TNBC patients with EGFR overexpression [21].
A major unmet clinical challenge in IBC is the rapid development of chemo- and radiotherapy resistance. The efficacy of chemo- and radio-therapeutics is largely influenced by optimal levels of cellular reactive oxygen species (ROS) which act on redox sensitive oncogenic signaling pathways and induce cell death response to therapy in the cancer cells [22]. We identified that IBC cells with acquired therapeutic resistance to lapatinib have adapted to oxidative stress induced by lapatinib, resulting in insignificant accumulation of ROS [23]. This results in inhibition of apoptosis and enhanced cancer cell survival. Further, we discovered that the therapeutically resistant IBC cells overexpressed X-linked inhibitor of apoptosis protein (XIAP), the most potent mammalian anti-apoptotic/caspase inhibitor [24; 25]. XIAP overexpression in these cells correlated with redox adaptation, in particular increased expression of superoxide dismutases (SOD1/2), leading to enhanced ROS detoxification activity [26].
Few profiling studies have looked more specifically at linking tumor cell drug responses to drug resistance. In this study, we employed a high throughput (HT) approach to profile the NCI Developmental Therapeutics Program (NCI-DTP) Approved Oncology Drug Set II in inflammatory breast cancer models (SUM149, basal type and SUM190, ErbB2 overexpressing) established from IBC patient tumors [27]. To obtain pharmacological information from the screening data, we employed a titration-based screening paradigm, quantitative HTS (qHTS) where compounds were screened at multiple concentrations. The primary objective of the current study was to characterize whether the development of resistance to lapatinib also affects sensitivity to other chemo- and targeted drugs and secondly, whether removal of lapatinib would have an impact on sensitivity to lapatinib and other anti-cancer agents. Therefore, we also characterized efficacy of this drug set in rSUM149, an isogenic derived line from SUM149 wherein a clonal population of cells was selected for acquired resistance to lapatinib [25]. In addition, another isogenic derivative was generated through removal of lapatinib from rSUM149 for an extended period of time (rrSUM149/resistance reversal line), and we present data that this cell line regains sensitivity to lapatinib and other anti-cancer drugs. Analysis of the parental and isogenic derived IBC cell lines provides a unique model system for gaining an understanding of acquired therapeutic resistance and the effect of redox adaptation on anti-cancer drug efficacy.
2. Materials and methods
2.1 Cell lines and reagents
SUM149PT and SUM190PT cells were obtained from Asterand Inc. (Detroit, MI). SUM cell lines were cultured as per the manufacturer’s instructions and as described previously [24; 25; 27]. SUM149 cells were routinely maintained in Ham’s F-12 supplemented with 5% fetal bovine serum (FBS), insulin (5 μg/ml) and hydrocortisone (1 μg/ml). SUM190 cells were routinely maintained in Ham’s F-12 supplemented with 0.5 g/L bovine serum albumin (BSA), insulin (5 μg/ml), hydrocortisone (1 μg/ml), transferrin (5 μg/ml) and triido thyronine (10 nM) as previously described [24; 27]. The lapatinib-resistant SUM149-derived cell line (rSUM149) was generated as described previously [25]. Briefly, rSUM149 cells were established by chronic exposure of parental SUM149 cells to increasing concentrations of the lapatinib analog GW583340 (up to 20 μM) for >3 months. The rSUM149 cells are routinely grown in the regular SUM149 media described above supplemented with 7.5 μM GW583340. In order to generate the rrSUM149 cells (resistance reversal model), rSUM149 cells were transferred to regular SUM149 growth media without GW583340, and cells were maintained in GW583340-free media for an extended period (~2 months) prior to use in this study. Ham’s F-12 media and Hoechst-33342 dye was from Life Technologies (Grand Island, NY). FBS was from Thermo Fisher Scientific (Hyclone Labs., Logan, UT). Insulin, hydrocortisone, GW583340 and 3-[4, 5-dimethylthiazol-2yl]-2, 5 diphenyl tetrazolium bromide (MTT) were from Sigma (St. Louis, MI).
2.2 Oncology drug library
The Approved Oncology Set II was obtained from the NCI-DTP (www.dtp.nci.nih.gov). This set consists of 89 drugs provided in 96-well plate format (Supplemental Table S1) as 10 mM stock concentrations in DMSO. Compounds from this set were also resourced from NCI-DTP as dry powders (5 mg) and dissolved in DMSO to give 10 mM stock concentrations for retesting. Compound plates are stored at −20°C. All drugs were registered in ActivityBase (IDBS Inc., Guildford UK) with associated SD file data.
2.3 Dose response and intermediate drug plates
All drugs were serially diluted in DMSO in 384-well Greiner V-bottom polypropylene plates (Greiner Bio-One, Monroe, NC). Briefly, a 10-point dose range was generated for each compound from the Approved Oncology Drug Set II (10 mM stock concentration in DMSO) by 2-fold serial dilution in 100% DMSO (5 μL final volume in each well) using a Biomek® NX robotics workstation (Beckman Coulter Inc., Fullerton, CA) and P20 tips. Each dose response plate contained 32 compounds in 10-point serial dilution and sufficient volume to generate 8 replicate intermediate drug plates.
To generate intermediate drug plates, 0.5 μL of each compound serial dilution from a dose response plate was spotted into columns 3 - 22 of dry Costar clear flat bottom 384-well assay plates (Corning Corp., Corning, NY) using the Biomek® NX. Unless otherwise stated, columns 1, 2, 23 and 24 were spotted with 0.5 μL of DMSO for use as controls. Unless otherwise stated, 120 μL of Ham’s F-12 culture media was added to each well resulting in 10 final concentrations for each compound spanning 40 μM to 0.08 μM.
2.4 Automated qHTS cell proliferation assay
An automated method was used to measure cell proliferation by MTT [28] adapted from a manual version of the assay previously described by us [29] in which cells were seeded into a 96-well plates and all reagent additions made utilizing multichannel pipettors. For our fully automated procedure, SUM149 or SUM190 cells were seeded at a density of 800 or 1000 cells per well respectively (40 μL) in columns 3-24 of clear 384-well tissue culture plates using a Multidrop 384 (Thermo Fisher, Waltham, MA) bulk dispenser sterilized for cell culture studies and cells allowed to adhere for 24 h. For qHTS drug screening studies, media alone was added to columns 1 and 2 as minimum signal controls. All subsequent aspiration or drug reagent addition steps were carried out using the Biomek NX workstation which was equipped with a plate stacker, bar code reader, and wash station. Intermediate dose response compound plates were generated separately as in Section 2.3 and then added to the cell plates allowing dilution of compounds directly in 100% DMSO prior to adding media and avoiding potential solubility issues with carrying out serial dilutions in aqueous cell culture media. Culture medium was aspirated from the cell plates and the drugs from the intermediate drug plate added (20 μL) to the cell plate. Cells were then incubated for a further 72 h unless otherwise stated. To facilitate multiplexing of IBC cell lines, the same cell culture media was used for the 72 h incubation of drugs with cells.
For our automated procedure, after treatment with drugs for 72 h, culture media was aspirated using the Biomek NX and 8 μL MTT (5 mg/ml) added to each well using the Biomek NX. Cells were incubated at 37°C for 4 h to allow formation of the colored formazan product. The reagent was aspirated using the Biomek NX and 40 μl of DMSO was added in each well using the Multidrop. Plates were incubated for a further 2 h and then absorbance was read at 550 nm in a SpectraMax Plus 384 plate reader (Molecular Devices, Sunnyvale, CA). Absorbance well data was saved in Microsoft Excel.
2.5 High content image cytometry assay
For high content analysis of cell counts, live/dead and morphology, SUM149 cells were seeded at 200 cells per well (40 μL) in clear 384-well tissue culture plates using the Multidrop 384 and cells allowed to adhere for 24 h. Dose response plates were generated as described in Section 2.3. Compounds were then added from the dose response plates directly to the cell plates using a Biomek NX equipped with a 384-well Pintool head with 50 nL pins (VP Scientific, San Diego, CA). Two 50 nL dips were used from the 10mM stock of drug giving a top dose of 25 μM. The plates were incubated for a further 72 h. After the incubation period, Hoechst-33342 dye was added using the Biomek NX to the cell plates to a final concentration of 10 μg/ml to stain the nuclei using a protocol adapted from Abraham et al. [30]. All cell plates were centrifuged at 800 rpm for 1 min and then entire wells were imaged using a BD Pathway 855 high-content imaging system (BD Biosciences, San Jose, CA) with an Olympus UPlanASpo 10X/0.40 objective lens. Entire-well image montages were analyzed using a customized pipeline based on the open source CellProfiler® software [31] to automatically identify all cells. The whole plate was imaged classifying nuclear morphology (nuclei count, size and intensity). After tabulation using the CellProfiler software, data is exported to a relational SQL database for data analysis.
2.6 Measurement of reactive oxygen species
Cells were cultured in 6-well plates (Corning) in regular growth media until reaching 70–80% confluence and then treated with GW583340 for 24 h. Cells were harvested and incubated for 30 min with dyes to detect reactive oxygen species (ROS) [10 μM dihydroethidium (DHE) and 10 μM carboxy-H2-DCFDA, Molecular Probes, Carlsbad, CA]. Cells were washed twice with 1% BSA/PBS and analyzed for fluorescence by flow cytometry. At least 25,000 events were collected on a FACScalibur flow cytometer (Beckton Dickinson, Rockville, MD) and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). High fluorescence was calculated by setting a gate on the control cells where the peak reached a minimum, and all experimental samples were compared to this control gate.
2.7 Western immunoblotting
Western immunoblot analysis was conducted as described previously [24]. Cells were lysed, run on a 10 or 4-20% gel (BioRad, Hercules, CA), and transferred to a PVDF membrane. Membranes were incubated with primary antibodies against XIAP (BD Bioscience, San Jose, CA), cIAP1, cIAP2, survivin, Smac, SOD2, AMPK, p-AMPK (Cell Signaling Technologies, Danvers, MA), catalase (Abcam, Cambridge, MA), Bcl-2, actin, or glyceraldehyde–3–phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology, Dallas, TX), overnight at 4°C.
2.8 Data analysis
To calculate percent relative cell proliferation, absorbance (Abs) values from the MTT assay were normalized using the in-plate vehicle (DMSO + cells, columns 23 and 24) and no cells (columns 1 and 2) control wells for maximum and minimum signal calculations respectively. For each drug concentration, percent inhibition values were calculated using the following relationship: relative cell proliferation (%) = {1- [(Abs - Avgmin)/(Avgmax-Avgmin)]} * 100. Where Abs is the compound well absorbance signal, and Avgmin and Avgmax are the plate-averages of the minimum and maximum signal controls respectively. IC50 (concentration of half-maximal inhibition) values determined using a four-parameter dose-response (variable slope) equation in GraphPad Prism 5 (GraphPad Software, San Diego, CA). Heat maps using percent inhibition values were generated in Microsoft Excel using conditional formatting. For the high content Hoechst assay, nuclei count data was normalized to in-plate controls and IC50 values determined in GraphPad Prism 5. Z’ factor was calculated using the method of Zhang et al. [32].
3. Results
3.1 Validation of automated proliferation assay
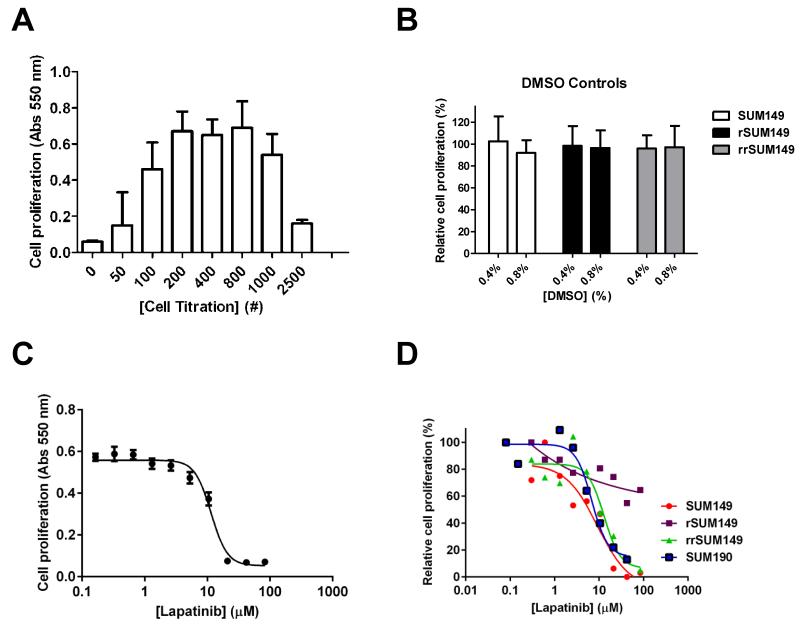
For efficacy drug profiling, we have adapted and validated in 384-multiwell format an automated proliferation screen based on the MTT assay [28] utilizing our previously described manual procedures [24; 33; 34]. The automated cell proliferation assay was first optimized for cell number and DMSO tolerance. SUM149 cells have a relatively short doubling time of 14 h [25], making them amenable to this type of profiling [6]. For SUM149 cells, a density of 800 cells per well for the 384-well format was chosen based on cell titration experiments (Fig. 1A). Doubling times for the lapatinib-resistant (rSUM149) [25] and lapatinib resistance reversal (rrSUM149) [26] isogenic SUM149-derived cell lines were comparable, and so the same 800 cells per well seeding density was used for these isogenic cell lines. For the slower growing SUM190 [24], a seeding density of 1000 cells per well was used. As all drugs to be screened were dissolved in DMSO and then diluted into cells with the highest concentration of DMSO cellular exposure being 0.8%, we assessed the tolerance of the cell lines to DMSO and observed no loss of proliferation up to concentrations of 0.8% DMSO for the three SUM149-derived cell lines (Fig. 1B) and the SUM190 cell line (data not shown). To validate the proliferation assay for screening in dose response, replicate dose response curves were performed using lapatinib over multiple experiments with the results for SUM149 shown in Fig. 1C. These assays were highly reproducible with an average IC50 value of 11.6 ± 1.06 μM for lapatinib on SUM149 cells (Table 1). To assess the robustness and reproducibility of the assay, Z’-factors [32] were calculated for two independent experiments and determination of the values as >0.5 indicate an acceptable assay for HTS (Table 1).
Fig. 1.
Optimization and validation of 384-well format proliferation assay. SUM149 cells were seeded in 384 well plates and the MTT assay was carried out after 72 h as described in Material and Methods. (A) Cell number titration. SUM149 cells were seeded at the indicated cell numbers. Data from triplicate samples. (B) DMSO tolerance. Cells were seeded (800 cells/well) with the indicated concentration of DMSO included in the media and MTT assay carried out for 72 h. For these experiments, in plate controls for minimum signal (Avgmin) in columns 1-2 were no cells and no DMSO and for maximum signal (Avgmax) in columns 23-24 were +cells and no DMSO. The relative proliferation (%) was calculated using the same relationship as in Materials and Methods. Mean and SD based on a minimum of three independent experiments. (C) Sixteen replicate 10-point dose response curves for lapatinib. SUM149 cells (800 cells/well) were incubated with the indicated concentrations of lapatinib for 72 h. (D) Representative qHTS dose response curves for lapatinib on parental SUM149, isogenic rSUM149, rrSUM149 and SUM190 cells. Data was analyzed in GraphPad Prism 5.0.
Table 1.
Assay validation statistics.
| Expt. | Lapatinib IC50 ± SD (μM) |
Z’-factor |
|---|---|---|
| 1 | 11.5 ± 1.06 | 0.53 |
| 2 | 11.7 ± 1.26 | 0.53 |
IC50 value determination was performed on 16 replicates for SUM149 cells in two independent experiments.
Representative lapatinib dose response curves for the parental and isogenic SUM149 and the SUM190 cell lines are shown in Fig. 1D, giving the expected lapatinib sensitivity profile for the parental and lapatinib-resistant cells comparable to previous reports [23; 24; 25; 26]. Further, the lapatinib efficacy data in rrSUM149 demonstrate that maintenance of the lapatinib-resistant rSUM149 cells for an extended period of time in lapatinib-free media causes reversal of lapatinib resistance.
3.2 qHTS of NCI approved oncology drug library: identification of potent drugs with efficacy on SUM149 and SUM190 cells
We first assessed the sensitivity of the parental SUM149 and SUM190 cells to the 89 drugs in the NCI Approved Oncology set. Drugs in this set include both chemo- and targeted therapeutics. Based on literature [6; 9; 35; 36; 37; 38], we grouped the drug Set into 12 drug classes (Supplemental Table 2), including antimetabolites, alkylating agents, antineoplastic antibiotics, platinum-based agents, alkaloids, taxanes, and kinase inhibitors; mechanism of action was included if known. For our qHTS format, cells were incubated with the drugs in 10-point dose response for 72 h under standard culture conditions, and effects on cell proliferation assessed by an automated MTT assay. Dose response curves were generated, and IC50 values determined.
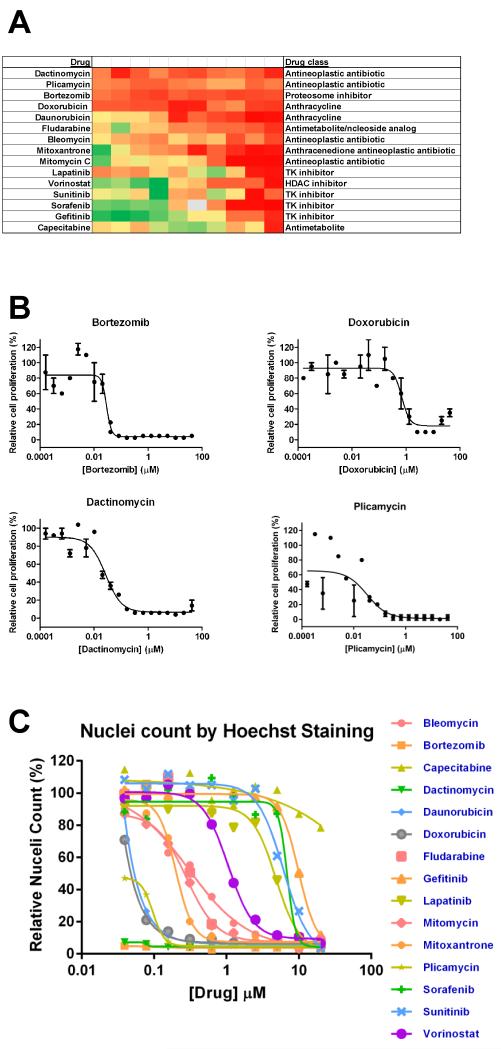
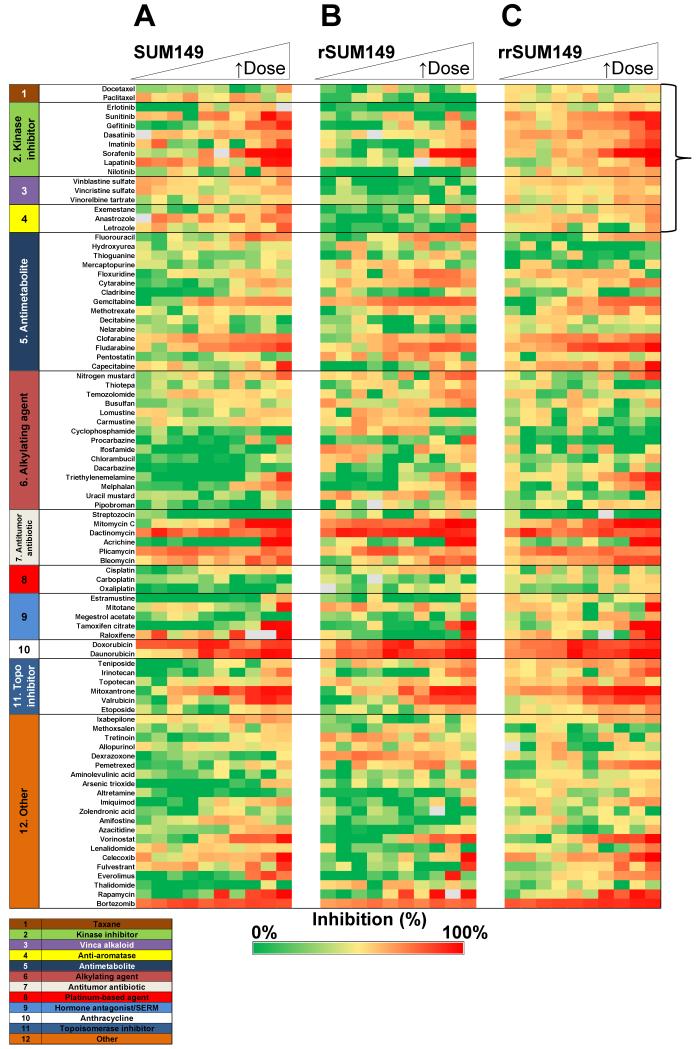
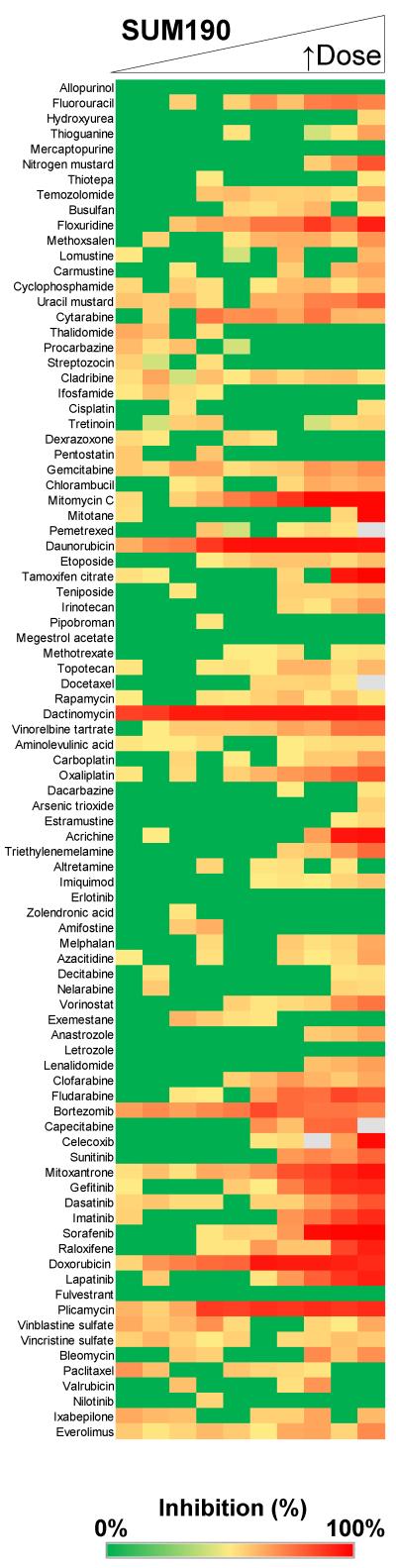
This qHTS strategy was first used to identify potent drugs that consistently demonstrated IC50 values <15 μM for inhibition of proliferation in SUM149 (Table 2) and SUM190 (Table 3). The heat map profile for the dose response of the top 15 drugs on SUM149 is shown in Fig. 2A. To generate the heat map, proliferation data was normalized to in-plate controls. In addition to lapatinib, the 15 top drugs included antimetabolites, antineoplastic antibiotics, anthracyclines, kinase inhibitors, an HDAC inhibitor, and a proteasome inhibitor (Fig. 2A, Table 2). A representative proliferation heat map profile of the complete 89 drug set screened in dose response against the SUM149 cells is shown in Fig. 3A. Dose response data in the heat map was grouped based on the drug classes/mechanism of action (from supplemental 2). Data from the heat map profile of all 89 drugs on SUM149 cells (Fig. 3A), reveals that alkylating (Fig. 3A, group 6) and platinum-based agents (Fig. 3A, group 8) were generally ineffective at reducing SUM149 proliferation. Although not in the top 15 drugs for SUM149 cells, gemcitabine, the vinca alkaloids and mTOR inhibitors rapamycin and everolimus also demonstrated modest micromolar activity. A representative heat map profile for all 89 drugs also tested in SUM190 cells is shown in Fig. 4. From a comparison of the top drugs (Tables 2 and 3) and heat map profiles (Fig. 3A and Fig. 4), SUM190 cells were generally less sensitive to this set of drugs when compared to SUM149, with the more potent drugs that act on both cell lines having ~10-fold lower potency in SUM190 cells.
Table 2.
IC50 values for oncology drugs tested on SUM149 cells in qHTS and confirmation MTT and Hoechst assays.
| qHTS MTTa IC50 (μM) |
Confirmation MTTb IC50 (μM) |
Hoechstc IC50 (μM) |
Drug classd | |
|---|---|---|---|---|
| Dactinomycin | <0.02 | 0.027 | <0.02 | Antineoplastic antibiotic |
| Plicamycin | <0.02 | 0.028 | 0.12 r | Antineoplastic antibiotic |
| Bortezomib | <0.02 | 0.029 | <0.02 | Proteosome inhibitor |
| Doxorubicin | <0.02 | 0.70 | <0.05 | Anthracycline |
| Daunorubicin | <0.02 | 0.25 | 0.04 | Anthracycline |
| Fludarabine | 2.1 ± 1.1 | 0.48 | 0.45 | Antimetabolite |
| Mitoxantrone | 2.5 ± 0.6 | 3.08 | 0.20 | Anthracenedione antineoplastic antibiotic |
| Sorafenib | 4.5 ± 0.4 | 3.93 | 8.69 | Kinase inhibitor |
| Sunitinib | 5.0 ± 0.8 | 5.55 | 6.10 | Kinase inhibitor |
| Gefitinib | 6.2 ± 2.3 | 7.27 | 14.10 | Kinase inhibitor |
| Vorinostat | 8.9 ± 4.9 | 2.44 | 2.02 | HDAC inhibitor |
| Lapatinib | 9.0 ± 0.3 | 10.85 | 6.73 | Kinase inhibitor |
| Mitomycin | 9.1 ± 0.4 | 1.61 | 0.53 | Antineoplastic antibiotic |
| Bleomycin | 13.8 ± 2.0 | 0.82 | 0.30 | Antineoplastic antibiotic |
| Capecitabine | 15.0 ± 9.5 | 17.51 | >20 | Antimetabolite |
aIC50 values derived from qHTS screen of complete approved oncology drug set (89 drugs originally obtained from NCI DTP in plate format). Average and SD values derived from minimum of three independent experiments. The approved oncology drugs were resourced from the NCI DTP as dry powders and retested in MTTb and Hoechstc assays as described in Materials and Methods. dSee supplementary Table 2 for full listing of drug class and mechanism of action for the approved oncology drug set.
Table 3.
IC50 values for oncology drugs tested on SUM190 cells in qHTS.
| qHTS MTT IC50 (μM) |
Drug class | |
|---|---|---|
| Dactinomycin | 0.2 | Antineoplastic antibiotic |
| Doxorubicin | 0.26 | Anthracycline |
| Plicamycin | 0.42 | Antineoplastic antibiotic |
| Daunorubicin | 0.47 | Anthracycline |
| Fludarabine | 0.9 | Antimetabolite |
| Mitomycin | 1.6 | Antineoplastic antibiotic |
| Sorafenib | 3.5 | Kinase inhibitor |
| Lapatinib | 6.1 | Kinase inhibitor |
Fig. 2.
Identification of top oncology drugs affecting proliferation of SUM149 cells. (A) Heat map profile for proliferation inhibitory effects of the top 15 drugs from the NCI approved oncology set on parental SUM149 cells. SUM149 cells were incubated with drugs in dose response as a 10-point two-fold dilution series for 72 h and cell proliferation assessed by MTT assay as described in Materials and Methods. For each concentration, percent inhibition values were calculated and data normalized to vehicle and no cell control wells. The heat map key indicates red for 100% inhibition through to green for 0% inhibition. Data from a representative qHTS experiment is shown. (B) 20-point dose response curves for four most potent drugs. Data from duplicate samples. (C) High content Hoechst staining assay for nuclei count. Nuclei count data was normalized and dose response curves for top 15 drugs on SUM149 cells generated by non-linear regression using GraphPad Prism 5.
Fig. 3.
Comparative efficacy profiling of the 89 oncology drugs against the parental SUM149 and isogenic rSUM149 and rrSUM149 cells. Heat map profile for proliferation inhibitory effects of the NCI approved oncology set. SUM149 (A), rSUM149 (B) and rrSUM149 (C). Cells were incubated with drugs in dose response as a 10-point two-fold dilution series for 72 h and cell proliferation assessed by MTT assay as described in Materials and Methods. Each row represents dose response data for a single drug. For each concentration, percent inhibition values were calculated and data normalized to vehicle and no cell control wells. The heat map key indicates red for 100% inhibition through to green for 0% inhibition. Outlier values are shown as grey. Drug names are listed on the vertical axis and grouped for drug class as detailed in Supplemental Table 2. A representative experiment is shown. Each cell line was profiled versus the NCI Oncology set for a minimum of two independent screens. Dose response curves were generated using GraphPad Prism 5.0.
Fig. 4.
Efficacy profiling of the 89 oncology drugs against the SUM190 cells. SUM190 cells (1000 cells per well) were incubated with drugs in dose response as a 10-point two-fold dilution series for 72h and cell proliferation assessed by MTT assay as described in Materials and Methods. Each row represents dose response data for a single drug. For each concentration, percent inhibition values were calculated and data normalized to vehicle and no cell control wells. The heat map key indicates red for 100% inhibition through to green for 0% inhibition. Drugs are listed in plating order and a representative experiment is shown. Cells were profiled versus the NCI Oncology set for a minimum of two independent screens.
3.3 Confirmation of oncology drugs identified as having anti-proliferative activity
A number of approaches were undertaken to confirm the fifteen anti-cancer drugs identified from our primary qHTS. Chemical interference by reducing compounds with tetrazolium substrates has been reported ([39; 40] and references therein), resulting in the potential for false positives. To rule out that the 15 drugs we identified as reducing SUM149 proliferation were not directly interfering with MTT formation, parallel cell-free screens were carried out, and in these we did not observe any conversion to a colored product (data not shown).
Compounds stored in DMSO can be subject to degradation, especially if they undergo multiple freeze-thaw cycles. Hence, the approved oncology drugs were resourced from the NCI-DTP as dry powders, fresh 10 mM drug stocks prepared in 100% DMSO, and the drugs retested in the confirmation MTT assay (Table 2). In general, IC50 values were comparable (<3-fold difference) between the primary qHTS MTT using the original DMSO drug plates from NCI-DTP and the confirmation MTT assay using dry powders resourced from NCI-DTP. One exception was bleomycin, with a ~15-fold more potent IC50 value observed from the resourced dry powder, suggesting that the concentration in the DMSO plate may have been significantly less than the supposed 10 mM stock value whether due to changes in solubility or stability.
In the primary qHTS screens, four compounds including doxorubicin, bortezomib, dactinomycin and plicamycin inhibited proliferation of SUM149 cells at all concentrations tested (Fig. 2A). For the confirmation MTT assay, we retested these drugs over an extensive 20-point dose response range so as to observe full dose response curves (Fig. 2B) and determine IC50 values (Table 2). Dactinomycin, plicamycin and bortezomib all had IC50 values in the 20-30 nM range and doxorubicin had an IC50 value of 700 nM. Together, these studies confirmed and strengthened our previous findings from the qHTS system.
3.4 Secondary screen for drugs
As another assessment of the qHTS data, we also screened the 15 top drugs in an alternative secondary assay using high-content imaging of nuclear count following Hoechst staining [30; 41] as an alternative measure of cell proliferation [30]. Dose response data in Fig. 2C and Table 2 show comparable potencies for the drugs in both assay formats, thus confirming our initial qHTS screening results.
3.5 Lapatinib-resistant IBC cells are cross-resistant to multiple oncology drugs and regain sensitivity in the resistance reversal model
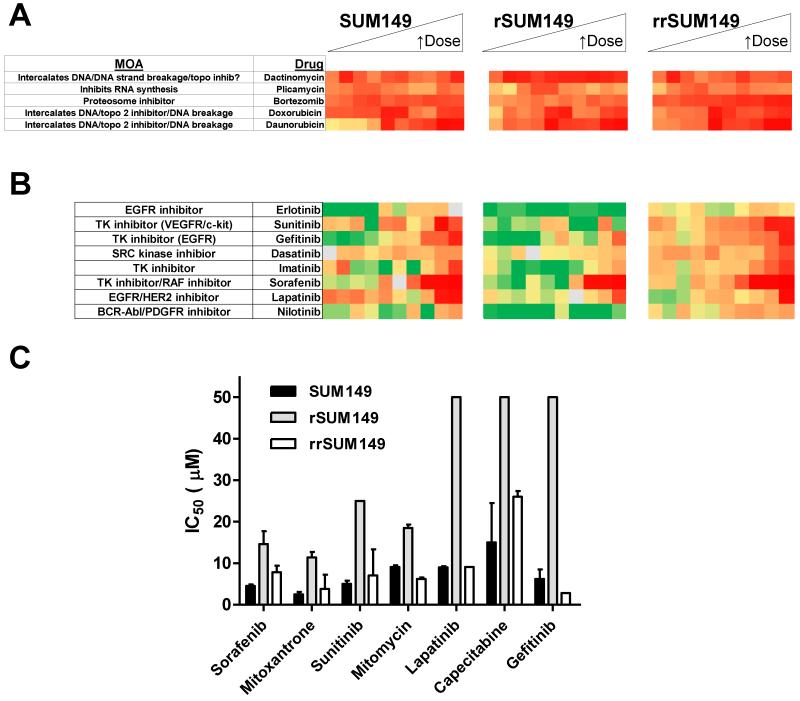
We then sought to profile the drug panel in the resistant rSUM149 and in a resistance reversal (rrSUM149) cell line. rrSUM149 is a unique isogenic cell line which was generated by removing lapatinib from the rSUM149 culture conditions for a period of >2 months, which allowed for selection of a population of cells that were resensitized to lapatinib treatment (resistance reversal model). A comparative profile for all 89 drugs on parental SUM149 and the two isogenic cell lines rSUM149 and rrSUM149 is shown in heat map format in Fig. 3.
We first assessed how the 15 compounds identified as having efficacy on the parental SUM149 cells (Table 2, Fig. 3A) profiled on the resistant rSUM149 and resistance-reversal rrSUM149 cells (Fig. 3B and 3C). Table 4 lists the most potent drugs identified from the NCI-DTP oncology set affecting proliferation of SUM149 cells and the corresponding IC50 values in the rSUM149 and rrSUM149 cells. The five compounds that we previously identified as inhibiting proliferation in SUM149 cells at all the qHTS concentrations tested (doxorubicin, bortezomib, dactinomycin, plicamycin and daunorubicin) were as effective on the rSUM149 and rrSUM149 cells (Table 4, Fig. 5A). In addition to those drugs, fludarabine and vorinostat were highly potent even in the acquired resistance model as well (Table 4).
Table 4.
MTT assay IC50 values derived for qHTS screen data for oncology drugs on parental and isogenic SUM149 cells.
| Primary IC50 ± SD (μM) | |||
|---|---|---|---|
| SUM149 | rSUM149 | rrSUM149 | |
| Fludarabine | 2.1 ± 1.1 | 1.7 ± 0.8 | 1.4 ± 0.9 |
| Mitoxantrone | 2.5 ± 0.6 | 11.4 ± 1.3 | 3.8 ± 3.4 |
| Sorafenib | 4.5 ± 0.4 | 14.6 ± 3.1 | 78 ± 16 |
| Sunitinib | 5.0 ± 0.8 | >25 | 7.05 ± 6.3 |
| Gefitinib | 6.2 ± 2.3 | >50 | 28 |
| Vorinostat | 8.9 ± 4.9 | 6.2 ± 0.6 | 2.75 ± 2.1 |
| Lapatinib | 9.0 ± 0.3 | >50 | 9.1 |
| Mitomycin | 9.1 ± 0.4 | 18.5 ± 0.8 | 6.2 ± 0.4 |
| Bleomycin | 13.8 ± 2.0 | >50 | 9.5 |
| Capecitabine | 15.0 ± 9.5 | >50 | 26 ± 1.4 |
Doxorubicin, bortezomib, dactinomycin, plicamycin and daunorubicin all had IC50 values <0.2 μM on all three isogenic cell lines.
Fig. 5.
Lapatinib-resistant IBC cells are cross-resistant to multiple oncology drugs. (A) Heat map proliferation profile of five most potent drugs identified for SUM149 and corresponding profile on rSUM149 and rrSUM149 cells. (B) Comparative heat map proliferation profile of tyrosine kinase drugs on parental SUM149 and isogenic rSUM149 and rrSUM149 cells. (C) Comparative IC50 values for drugs demonstrating cross-resistance and cross-resensitization. Mean (bars) and SD (error bars) with IC50 values determined using GraphPad Prism 5.0.
Compared to the parental lapatinib-sensitive SUM149 cells, the lapatinib-resistant rSUM149 cells were also cross-resistant to multiple targeted- and chemo-therapeutic agents amongst the top 15 potent drugs identified in SUM149 and some other lower potency drugs. These were predominantly drugs from four of the classes (highlighted by bracket in Fig. 3), including the taxanes (Fig. 3, group 1), a subset of the kinase inhibitors (group 2), vinca alkaloids (group 3), and anti-aromatases (group 4). Several drugs showed shifts in their IC50 values from the parental to resistant SUM149 cells. From the initial top 15 drugs identified for efficacy in parental SUM149 cells, RTK inhibitors, capecitabine, mitomycin, mitoxantrone and bleomycin showed dramatically reduced potencies in rSUM149 in addition to previously published lapatinib resistance (Table 4, Fig. 5B and 5C).
Interestingly, the data in Table 4 and Figure 5B heat maps show that the rrSUM149 regained sensitivity and potency comparable to the parental SUM149 cells for not only lapatinib, but also for other targeted- and chemo-therapy agents in the top 15 panel (Fig. 5C).
Comparative efficacy profiling of the approved oncology drug set in SUM149 parental and isogenic models of acquired resistance and re-sensitization demonstrates that lapatinib-resistant IBC cells are cross-resistant to multiple oncology drugs.
3.6 Mechanism of resistance reversal in the rrSUM149 cell line
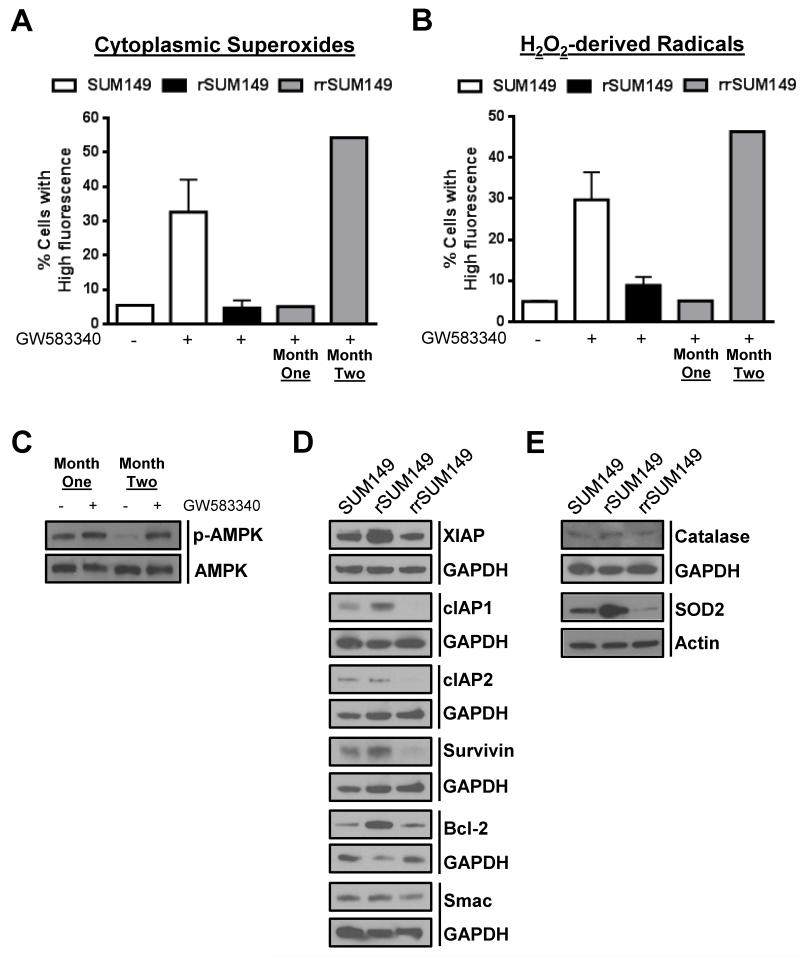
To further characterize the resistance reversal model and understand their cross-resensitization, we compared the rrSUM149 cells to rSUM149 and SUM149 cells using several important factors associated with acquired resistance in the rSUM149 model. Measurement of cytoplasmic superoxides and hydrogen peroxide-derived radicals at two different time points during the generation of the resistance reversal model showed that after one month of growth in GW583340 (research grade lapatinib analog)-free media, the rrSUM149 cells (Fig. 6A and 6B respectively, “one month” gray bars) remained insensitive to lapatinib-induced accumulation of ROS, similar to the rSUM149 model (Fig. 6A and 6B, black bars) which is growing in lapatinib. However, following two months of growth in lapatinib-free media, the rrSUM149 cells (Fig. 6A and 6B, “two months” gray bars) had regained their ability to accumulate ROS at levels similar to the parental SUM149 cells (Fig. 6A and 6B respectively, white bars) following challenge with lapatinib.
Fig. 6.
Reversal of resistance in rrSUM149 is mediated by enhanced sensitivity toward ROS-mediated cell death. (A) Cytoplasmic superoxides as measured by flow cytometric staining with DHE of (left to right) SUM149 parental cells (white bars) untreated and treated with 7.5 μM lapatinib, rSUM149 cells (black bars) growing in 7.5 μM lapatinib, and rrSUM149 cells following removal of drug for one or two months (gray bars) treated with 7.5 μM lapatinib. (B) Hydrogen peroxide-derived radicals as measured by flow cytometric staining with H2DCFDA in SUM149 parental cells (white bars) untreated and treated with 7.5 μM lapatinib, rSUM149 cells (black bars) growing in 7.5 μM lapatinib, and rrSUM149 cells following removal of drug for one or two months (gray bars) treated with 7.5 μM lapatinib. (C) Western immunoblot analysis of AMPK expression and phosphorylation in rrSUM149 cells treated with 7.5 μM lapatinib following one or two months of growth in lapatinib-free media. (D) Western immunoblot analysis of basal XIAP, cIAP1, cIAP2, survivin, Bcl-2, and Smac expression in untreated SUM149, rSUM149, and rrSUM149 cells. GAPDH was used as a loading control. (E) Western immunoblot analysis of basal catalase and SOD2 expression in untreated SUM149, rSUM149, and rrSUM149 cells. GAPDH and β-actin were used as loading controls.
We previously showed that SUM149 cells treated with lapatinib have increased phosphorylation of 5–AMP-activated protein kinase (AMPK), a marker of oxidative stress, but that rSUM149 cells, which are growing in lapatinib, show no increased p-AMPK compared to SUM149 cells, indicating a partial loss of stress responsive signaling to lapatinib [26]. Thus, we were interested to examine the phosphorylation status of AMPK in the resistant reversal cell line rrSUM149 following lapatinib treatment. Following one month of growth in lapatinib-free media, at which time the cells had not displayed any evidence of resistance reversal, the cells displayed very minimal increase in p-AMPK levels in response to lapatinib treatment (Fig. 6C, Month One). However, after two months of growth in lapatinib-free media, there was an approximately two-fold increase in AMPK phosphorylation in the rrSUM149 cells following lapatinib treatment, indicating that the cells are undergoing significant oxidative stress (Fig. 6C, Month Two).
Acquired resistance in rSUM149 was previously correlated with upregulation of XIAP compared to parental SUM149 cells [25]. To further investigate the mechanisms behind the reversal of resistance in rrSUM149 cells, we investigated the levels of XIAP as well as several other anti-apoptotic proteins and Smac/DIABLO, a negative regulator of XIAP, in the panel of SUM149, rSUM149, and rrSUM149. Data in Fig. 6D show that expression of XIAP was upregulated approximately two-fold in rSUM149, similar to previous results, and interestingly, XIAP expression in the rrSUM149 cells was reduced to levels comparable to the parental SUM149 cells. Additionally, cIAP1 and Bcl-2 were both increased to some degree in rSUM149 cells, further supporting their high degree of apoptotic dysregulation. cIAP2 and survivin levels were unchanged in rSUM149 cells compared to SUM149, but there was a decrease in all anti-apoptotic proteins examined in the rrSUM149 cells, either returning to parental SUM149 levels or in some cases even lower (cIAP1/2, survivin). Levels of Smac/DIABLO were unchanged across the panel of cell lines.
As there is a distinct difference in ROS accumulation across the cell lines (Fig. 6A and 6B) and we have previously reported increased antioxidant expression in the rSUM149 cell line [26], we examined the expression of SOD2 and catalase, two important intracellular antioxidants. Catalase expression did not show any significant changes across the cell lines (Fig. 6E). In contrast, SOD2 expression was significantly upregulated in the rSUM149 cells, and expression was markedly decreased in the rrSUM149 resistance reversal model (Fig. 6E).
Together, these data indicate that rrSUM149 cells are more susceptible to ROS-mediated cell death than the rSUM149 cells.
4. Discussion
IBC is characterized by rapid progression from onset of disease, therefore an early and aggressive multimodal therapy is essential to improve outcome. IBC tumors are frequently ErbB2- and EGFR-positive, and the use of trastuzumab in combination with lapatinib has seen clinical success; however, de novo and acquired resistance to these agents due to interplay between different members of the ErbB family is a significant challenge. In addition, it is well recognized that there is a paucity of IBC cellular models. In the present study we have characterized a novel isogenic-derived progression model of drug resistance and reversal. A set of IBC cell lines (SUM149, rSUM149, rrSUM149 and SUM190) representing either basal type or HER2 overexpression were profiled in a qHTS format to identify approved drugs from the NCI-DTP 89 approved oncology drug panel that can act as potent inhibitors of IBC cell proliferation.
High throughput profiling approaches have utilized a range of phenotypic endpoints such as cytotoxicity, viability [9], survival [42], growth inhibition [2], and proliferation [8]. We chose to use the MTT cell proliferation assay as the basis for the high throughput assay due to its ease of automation, widespread use, and economical cost per well. Cell-based high throughput assays screening compounds at single dose are typically highly variable and can be prone to high false positive rates. To reduce this possibility, we chose to screen a dose response of all compounds in the primary assay. Screening compounds in dose response allowed us to minimize the false-positive and false-negative rates typically observed when screening compounds in a cell-based assay at a single concentration [43]. Further, we used an alternative secondary assay based on bioimaging of Hoechst stained cells as an orthogonal measure of cell proliferation [30; 41], and observed significant agreement between both assay formats for inhibition values. Using this qHTS approach, we were able to rapidly identify and confirm a subset of drugs with efficacy in these models.
From our profiling efforts we have identified drug classes including antimetabolites, antineoplastic antibiotics, anthracyclines, and a subset of kinase inhibitors to be potent in the SUM149 cell line, a well-established model of basal type, EGFR-activated IBC [44; 45; 46; 47; 48]. In the ErbB2 overexpression SUM190 model, the top chemotherapeutics included dactinomycin, doxorubicin, plicamycin and daunorubicin. Anthracyclines such as doxorubicin are recommended for initial treatment of IBC patients [14]. SUM190 cells were relatively less sensitive to the panel when compared to SUM149, with those drugs that act on both cell lines having ~10-fold lower potency on SUM190 cells. It is known that SUM190 cells proliferate at a much slower rate than the SUM149 cells (doubling times of approximately 42 and 14-21 hours respectively) [49], which would make them inherently less sensitive towards chemotherapeutic agents that target dividing cells such as the anti-metabolites, alkylating agents, and topoisomerase inhibitors.
Additionally, we showed previously that XIAP, a chemoresistance factor, is an important determinant of lapatinib sensitivity in SUM190 and SUM149 cells [25], and that XIAP determined TRAIL sensitivity, with targeting of XIAP by the small molecule inhibitor embelin or shRNA increasing the sensitivity of rSUM149 cells to TRAIL-induced apoptosis [23]. Unpublished data from the same studies showed that SUM190, which expresses XIAP at higher levels than SUM149 [24; 25], was highly resistant to TRAIL treatment, mimicking its relative insensitivity to the drugs in this study compared to the SUM149-derived lines. Similarly, Foster et al. [50] also showed that depletion of XIAP could overcome intrinsic TRAIL resistance in two breast cancer lines BT20 and MDA-MB-468. Others have also shown that XIAP can function as a chemoresistance factor for gemcitabine [51; 52], cisplatin [53; 54], 5- fluorouracil [51], mitomycin C [54], and docetaxel [55] in a variety of cancers. Thus, our data here support XIAP’s role as an important therapeutic resistance factor which could contribute to the relative insensitivity of SUM190 cells observed in this study.
A subset of the drugs tested were highly potent even in the acquired lapatinib-resistant rSUM149 cell model and included dactinomycin, plicamycin, bortezomib, doxorubicin, daunorubucin, fludarabine and vorinostat. Table 5 summarizes those drugs showing efficacy on the resistant SUM149 cells along with corresponding mechanism of action. In contrast, rSUM149 cells showed cross-resistance to a number of the drugs previously shown to act on the parental cells, including RTK inhibitors (sorafenib, sunitinib and gefitinib), capecitabine, mitoxantrone, and bleomycin. Our results profiling oncology drugs on parental and resistant SUM149 cells suggests a mechanism of lapatinib-induced acquired resistance whereby the cells become cross-resistant to other tyrosine kinase inhibitors.
Table 5.
Top Drugs from NCI Approved Oncology Set with efficacy on lapatinib-resistant rSUM149 cells
| Drug Name | Type | Mechanism of action |
|---|---|---|
| Dactinomycin | Chromopeptide antineoplastic antibiotic |
Intercalates DNA; causes single- strand DNA breaks, possibly via a free-radical intermediate or an interaction with topoisomerase II. |
| Plicamycin | Antibiotic | Intercalates DNA |
| Bortezomib | Dipeptide boronic acid analogue |
Proteasome inhibitor |
| Doxorubicin | Anthracycline | Intercalates DNA; inhibits topoisomerase II; forms oxygen free radicals |
| Daunorubicin | Anthracycline | Inhibits topoisomerase II |
| Fludarabine | Fluorinated nucleotide antimetabolite analog |
DNA synthesis inhibitor |
| Vorinostat | Synthetic hydroxamic acid |
HDAC inhibitor |
| Mitoxantrone | Anthracenedione antibiotic |
Intercalates DNA; inhibits topoisomerase II; |
| Mitomycin C | Antineoplastic antibiotic |
Generates oxygen radicals, alkylates DNA, and produces interstrand DNA cross-links |
The present study shows that long term removal of lapatinib, the primary drug against which resistance was developed in rSUM149 cells, led to isolation of a population of cells (rrSUM149) that are resensitized to multiple drugs (sorafenib, sunitinib, gefitinib, capecitabine, mitoxantrone and bleomycin, Fig. 5C), behaving in a manner comparable to the parental SUM149 cell line. Resensitization was accompanied by a decrease in expression of anti-apoptotic proteins including XIAP, cIAP1, cIAP2, survivin, and Bcl-2 (Fig. 6D) as well as the antioxidant SOD2 (Fig. 6E) and increased ROS accumulating abilities (Fig. 6A and 6B).
Additionally, the rrSUM149 cells regained the ability to activate AMPK, a cellular metabolic regulator that also acts as a sensor for oxidative stress [56], in response to lapatinib. Thus, the mechanistic phenotype of redox adaptation in the rSUM149 cells (Fig. 7) supports our current observations of cross-resistance to multiple drugs, which is also commonly seen in patients; further, decreases in survival factors and antioxidant mechanisms restored ROS accumulating capabilities in the rrSUM149 cell line, resulting in resensitization of the cells to a wide variety of treatments (Fig. 7).
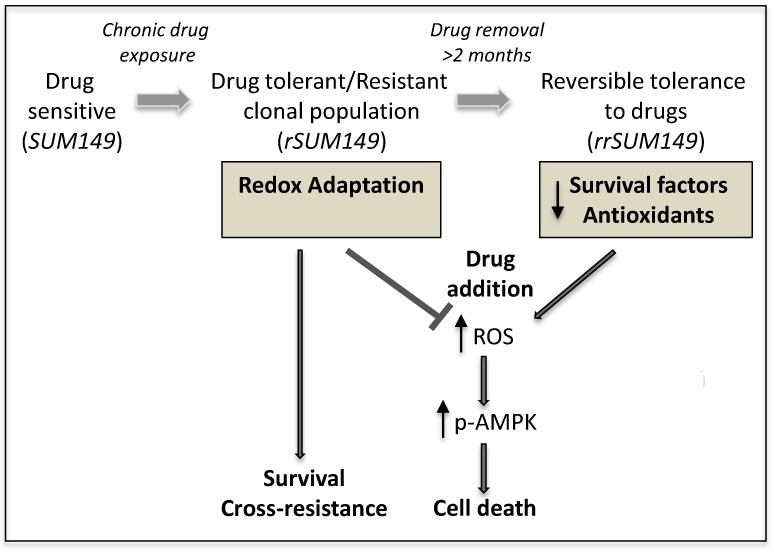
Fig. 7.
Schematic representation of drug sensitivity and tolerance in SUM149-derived models. Cancer cells have inherently high levels of ROS due to oncogenic signals and altered metabolism compared to normal cells. Presence of therapeutic insults/cellular stress can cause further increase in ROS, which in the absence of adaptive mechanism is associated with drug sensitivity and induction of cell death (SUM149). However, chronic stress leads to redox adaptive mechanisms such as increases in survival factors and antioxidants/ROS scavenging systems. These cells can suppress therapy-mediated ROS induction and/or rapidly clear ROS, which selects for a phenotypically distinct drug tolerant subpopulation of cells (rSUM149) that are drug tolerant due to their ROS protective mechanisms and contributes to multidrug resistance. Drug tolerance in this population of cells can be reversed by removal of the therapeutic stress for an extended period; loss of their redox adaptive mechanisms renders the resistant reversal cells (rrSUM149) highly therapy-sensitive.
From these studies, it appears that non-mutational mechanism(s) involving the engagement of pro-survival pathways and redox adaption play a role in the observed acquired resistance to lapatinib and concomitant cross-resistance to other drugs. The reversibility of this state back to a re-sensitized state after removal of drug is intriguing mechanistically. Recently the idea of a drug-tolerant sub-population of cells (persisters) termed DTPs has been proposed from other observations of acquired drug resistance [57]. The suggestion is that within a heterogeneous cell population there exists a subpopulation of reversibly drug tolerant cells that are dormant until dosed with high concentrations of drugs, after which a subset of DTPS tolerate growing in the presence of drug indefinitely giving rise to drug-tolerant expanded persisters (DTEPs) [57] – perhaps analogous to the rSUM149 cells in our isogenic model. There is some evidence that these drug tolerant clones exist de novo and may share some properties with cancer stem cells [57]. SUM149 cells show significant ALDH positivity [58; 59; 60], a characteristic associated with cancer stem cells, and thus may also contain a subpopulation of DTPs. Hence, it may be possible to eliminate the emergence of drug-tolerant cells in our SUM149 isogenic model by treatment of the parental cells with lapatinib in combination with other drugs targeting the adaptive mechanisms described here. These results strengthen the need for novel strategies to modulate cellular redox to prevent and overcome drug resistance in IBC.
In conclusion, we report the first description of drug efficacy profiles in IBC cells with differential phenotypes by screening approved drugs that can act as potent inhibitors of IBC cell proliferation. These studies have the potential to identify combinations and facilitate their inclusion in clinical trials immediately. Our findings in addition to the currently available database of IBC gene signatures facilitate a significant step toward better treatment protocols for IBC, even if potential side effects and dosing regimens still need to be identified for combination strategies.
Supplementary Material
Acknowledgements
This study was supported in part by NIH grant CA137844 (K. P. Williams), American Cancer Society grant RSG-08-290-01-CCE (G. R. Devi), Duke Comprehensive Cancer Institute Core Grant Pilot Project (G.R. Devi), NIH Grant T32CA009111 (J.L. Allensworth), with additional funding from the Golden LEAF Foundation and the BIOIMPACT Initiative of the State of North Carolina.
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
Appendix A. Supplementary data.
Supplementary data associated with this article can be found in the online version, at http://dx.doi.org/10/1016/j.canlet.2013.05.017.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I, Costa-Cabral S, Rafiq R, Ahmad AS, Cerone MA. Functional Viability Profiles of Breast Cancer. Cancer Discov. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kuo WL, Das D, Ziyad S, Bhattacharya S, Gibb WJ, Heiser LM, Sadanandam A, Fontenay GV, Hu Z, Wang NJ. A systems analysis of the chemosensitivity of breast cancer cells to the polyamine analogue PG-11047. BMC Med. 2009;7:77. doi: 10.1186/1741-7015-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yang R, Niepel M, Mitchison T, Sorger P. Dissecting variability in responses to cancer chemotherapy through systems pharmacology. Clin. Pharmacol. Ther. 2010;88:34–38. doi: 10.1038/clpt.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Benes C, Settleman J. Integrating complex genomic datasets and tumour cell sensitivity profiles to address a‘simple’question: which patients should get this drug? BMC Med. 2009;7:78. doi: 10.1186/1741-7015-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lee JK, Havaleshko DM, Cho HJ, Weinstein JN, Kaldjian EP, Karpovich J, Grimshaw A, Theodorescu D. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc. Natl. Acad. Sci. USA. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- [7].Smith SC, Baras AS, Lee JK, Theodorescu D. The COXEN principle: translating signatures of in vitro chemosensitivity into tools for clinical outcome prediction and drug discovery in cancer. Cancer Res. 2010;70:1753–1758. doi: 10.1158/0008-5472.CAN-09-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wolpaw AJ, Shimada K, Skouta R, Welsch ME, Akavia UD, Pe’er D, Shaik F, Bulinski JC, Stockwell BR. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc. Natl. Acad. Sci. USA. 2011;108:E771–780. doi: 10.1073/pnas.1106149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dawood S, Cristofanilli M. Inflammatory Breast Cancer: What Progress Have We Made? Oncology. 2011;25:264–270. [PubMed] [Google Scholar]
- [11].Gonzalez-Angulo A, Hennessy B, Broglio K, Meric-Bernstam F, Cristofanilli M, Giordano S, Buchholz T, Sahin A, Singletary S, Buzdar A. Trends for inflammatory breast cancer: is survival improving? The Oncologist. 2007;12:904–912. doi: 10.1634/theoncologist.12-8-904. [DOI] [PubMed] [Google Scholar]
- [12].Kleer CG, Van Golen KL, Merajver SD. Molecular biology of breast cancer metastasis. Inflammatory breast cancer: clinical syndrome and molecular determinants. Breast Cancer Res. 2000;2:423–429. doi: 10.1186/bcr89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Woodward WA, Cristofanilli M. Inflammatory breast cancer. Semin. Radiat. Oncol. 2009;19:256–265. doi: 10.1016/j.semradonc.2009.05.008. [DOI] [PubMed] [Google Scholar]
- [14].Goldfarb JM, Pippen JE. Inflammatory breast cancer: the experience of Baylor University Medical Center at Dallas. Proceedings (Baylor University. Medical Center) 2011;24:86–88. doi: 10.1080/08998280.2011.11928688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dawood S, Broglio K, Buzdar AU, Hortobagyi GN, Giordano SH. Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J. Clin. Oncol. 2010;28:92–98. doi: 10.1200/JCO.2008.19.9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, Krishnamurthy S, Le-Petross H, Bidaut L, Player AN, Barsky SH, Woodward WA, Buchholz T, Lucci A, Ueno N, Cristofanilli M. Inflammatory Breast Cancer: The Disease, the Biology, the Treatment. CA. Cancer J. Clin. 2010;60:351–375. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- [17].Johnston S, Trudeau M, Kaufman B, Boussen H, Blackwell K, LoRusso P, Lombardi DP, Ben Ahmed S, Citrin DL, DeSilvio ML. Phase II study of predictive biomarker profiles for response targeting human epidermal growth factor receptor 2 (HER-2) in advanced inflammatory breast cancer with lapatinib monotherapy. J. Clin. Oncol. 2008;26:1066–1072. doi: 10.1200/JCO.2007.13.9949. [DOI] [PubMed] [Google Scholar]
- [18].Burris HA, III, Hurwitz HI, Dees EC, Dowlati A, Blackwell KL, O’Neil B, Marcom PK, Ellis MJ, Overmoyer B, Jones SF. Phase I safety, pharmacokinetics, and clinical activity study of lapatinib (GW572016), a reversible dual inhibitor of epidermal growth factor receptor tyrosine kinases, in heavily pretreated patients with metastatic carcinomas. J. Clin. Oncol. 2005;23:5305–5313. doi: 10.1200/JCO.2005.16.584. [DOI] [PubMed] [Google Scholar]
- [19].Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin. Cancer Res. 2008;14:6730–6734. doi: 10.1158/1078-0432.CCR-08-0581. [DOI] [PubMed] [Google Scholar]
- [20].Li J, Gonzalez-Angulo AM, Allen PK, Tse KY, Woodward WA, Ueno NT, Lucci A, Krishnamurthy S, Gong Y, Bondy ML. Triple-Negative Subtype Predicts Poor Overall Survival and High Locoregional Relapse in Inflammatory Breast Cancer. The Oncologist. 2011;16:1675–1683. doi: 10.1634/theoncologist.2011-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Petrelli F, Cabiddu M, Ghilardi M, Barni S. Current data of targeted therapies for the treatment of triple-negative advanced breast cancer: empiricism or evidence-based? Expert Opin. Investig. Drugs. 2009;18:1467–1477. doi: 10.1517/13543780903222268. [DOI] [PubMed] [Google Scholar]
- [22].Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- [23].Allensworth JL, Aird KM, Aldrich AJ, Batinic-Haberle I, Devi GR. XIAP Inhibition and Generation of Reactive Oxygen Species Enhances TRAIL Sensitivity in Inflammatory Breast Cancer Cells. Mol. Cancer Ther. 2012;11:1518–1527. doi: 10.1158/1535-7163.MCT-11-0787. [DOI] [PubMed] [Google Scholar]
- [24].Aird KM, Ding X, Baras A, Wei J, Morse MA, Clay T, Lyerly HK, Devi GR. Trastuzumab signaling in ErbB2-overexpressing inflammatory breast cancer correlates with X-linked inhibitor of apoptosis protein expression. Mol. Cancer Ther. 2008;7:38, 47. doi: 10.1158/1535-7163.MCT-07-0370. [DOI] [PubMed] [Google Scholar]
- [25].Aird KM, Ghanayem RB, Peplinski S, Lyerly HK, Devi GR. X-Linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol. Cancer Ther. 2010;9:1431–1442. doi: 10.1158/1535-7163.MCT-10-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Aird KM, Allensworth JL, Batinic-Haberle I, Lyerly HK, Dewhirst MW, Devi GR. ErbB1/2 tyrosine kinase inhibitor mediates oxidative stress-induced apoptosis in inflammatory breast cancer cells. Breast Cancer Res. Treat. 2012;132:109–119. doi: 10.1007/s10549-011-1568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi O-P, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br. J. Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- [29].Amantana A, London CA, Iversen PL, Devi GR. X-linked inhibitor of apoptosis protein inhibition induces apoptosis and enhances chemotherapy sensitivity in human prostate cancer cells. Mol. Cancer Ther. 2004;3:699–707. [PubMed] [Google Scholar]
- [30].Abraham VC, Towne DL, Waring JF, Warrior U, Burns DJ. Application of a high-content multiparameter cytotoxicity assay to prioritize compounds based on toxicity potential in humans. J. Biomol. Screen. 2008;13:527–537. doi: 10.1177/1087057108318428. [DOI] [PubMed] [Google Scholar]
- [31].Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang J, Chung T, Oldenburg K. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [33].Rangwala F, Williams KP, Smith GR, Allensworth J, Thomas Z, Lyerly HK, Diehl AM, Morse MA, Devi GR. Differential effects of arsenic trioxide on chemosensitization in human hepatic tumor and stellate cell lines. BMC Cancer. 2012;12:402. doi: 10.1186/1471-2407-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thomas Z, Gibson W, Sexton J, Aird K, Ingram S, Aldrich A, Lyerly H, Devi G, Williams K. Targeting GLI1 expression in human inflammatory breast cancer cells enhances apoptosis and attenuates migration. Br. J. Cancer. 2011;104:1575–1586. doi: 10.1038/bjc.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brunton LL, Lazo JS, Parker KL. Goodman & Gilman’s the pharmacological basis of therapeutics. McGraw-Hill New York: 2006. [Google Scholar]
- [36].DeVita VT, Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68:8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- [37].Gheeya JS, Chen QR, Benjamin CD, Cheuk AT, Tsang P, Chung JY, Metaferia BB, Badgett TC, Johansson P, Wei JS. Screening a panel of drugs with diverse mechanisms of action yields potential therapeutic agents against neuroblastoma. Cancer Biol. Ther. 2009;8:2386–2395. doi: 10.4161/cbt.8.24.10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Labay E, Efimova E, Quarshie B, Golden D, Weichselbaum R, Kron S. Ionizing radiation-induced foci persistence screen to discover enhancers of accelerated senescence. Int. J. High Throughput Screen. 2011;2:1–13. doi: 10.2147/IJHTS.S17076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ulukaya E, Colakogullari M, Wood EJ. Interference by anti-cancer chemotherapeutic agents in the MTT-tumor chemosensitivity assay. Chemotherapy. 2004;50:43–50. doi: 10.1159/000077285. [DOI] [PubMed] [Google Scholar]
- [40].Riss TL, Moravec RA, Niles AL. Assay Development for Cell Viability and Apoptosis for High-Throughput Screening. In: Chen T, editor. A Practical Guide to Assay Development and High-Throughput Screening in Drug Discovery. CRC Press; 2009. pp. 99–122. [Google Scholar]
- [41].Sexton J, Williams K. Evaluating the Peroxisomal Phenotype in High-Content Toxicity Profiling. In: Steinberg P, editor. High-Throughput Screening Methods in Toxicity Testing. Wiley, New Jersey: 2013. pp. 501–518. [Google Scholar]
- [42].Valiathan C, McFaline JL, Samson LD. A rapid survival assay to measure drug-induced cytotoxicity and cell cycle effects. DNA Repair. 2011;11:92–98. doi: 10.1016/j.dnarep.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- [44].Dong HM, Liu G, Hou YF, Wu J, Lu JS, Luo JM, Shen ZZ, Shao ZM. Dominant-negative E-cadherin inhibits the invasiveness of inflammatory breast cancer cells in vitro. J. Cancer Res. Clin. Oncol. 2007;133:83–92. doi: 10.1007/s00432-006-0140-6. [DOI] [PubMed] [Google Scholar]
- [45].Hoffmeyer MR, Wall KM, Dharmawardhane SF. In vitro analysis of the invasive phenotype of SUM 149, an inflammatory breast cancer cell line. Cancer Cell Int. 2005;5:11. doi: 10.1186/1475-2867-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ignatoski K, Ethier S. Constitutive activation of pp125fak in newly isolated human breast cancer cell lines. Breast Cancer Res. Treat. 1999;54:173–182. doi: 10.1023/a:1006135331912. [DOI] [PubMed] [Google Scholar]
- [47].Singh B, Irving L, Tai K, Lucci A. Overexpression of COX-2 in Celecoxib-resistant breast cancer cell lines. J. Surg. Res. 2010;163:235–243. doi: 10.1016/j.jss.2010.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wu M, Wu Z, Rosenthal D, Rhee E, Merajver S. Characterization of the roles of RHOC and RHOA GTPases in invasion, motility, and matrix adhesion in inflammatory and aggressive breast cancers. Cancer. 2010;116:2768–2782. doi: 10.1002/cncr.25181. [DOI] [PubMed] [Google Scholar]
- [49].Robertson FM, Chu K, Fernandez SV, Mu Z, Zhang X, Liu H, Boley KM, Alpaugh RK, Ye Z, Wright MC, Luo AZ, Oraes R, Wu H, Zook M, Barsky SH, Krishnamurthy S, Cristofanilli M. Genomic Profiling of Pre-Clinical Models of Inflammatory Breast Cancer Identifies a Signature of Epithelial Plasticity and Suppression of TGFß Signaling. J. Clin. Exp. Path. 2012;119 [Google Scholar]
- [50].Foster FM, Owens TW, Tanianis-Hughes J, Clarke RB, Brennan K, Bundred NJ, Streuli CH. Targeting inhibitor of apoptosis proteins in combination with ErbB antagonists in breast cancer. Breast Cancer Res. 2009;11:R41. doi: 10.1186/bcr2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li Y, Jian Z, Xia K, Li X, Lv X, Pei H, Chen Z, Li J. XIAP is related to the chemoresistance and inhibited its expression by RNA interference sensitize pancreatic carcinoma cells to chemotherapeutics. Pancreas. 2006;32:288–296. doi: 10.1097/01.mpa.0000218314.67111.fb. [DOI] [PubMed] [Google Scholar]
- [52].Shrikhande SV, Kleeff J, Kayed H, Keleg S, Reiser C, Giese T, Buchler MW, Esposito I, Friess H. Silencing of X-linked inhibitor of apoptosis (XIAP) decreases gemcitabine resistance of pancreatic cancer cells. Anticancer Res. 2006;26:3265–3273. [PubMed] [Google Scholar]
- [53].He X, Khurana A, Maguire JL, Chien J, Shridhar V. HtrA1 sensitizes ovarian cancer cells to cisplatin induced cytotoxicity by targeting XIAP for degradation. Int. J. Cancer. 2011;130:1029–1035. doi: 10.1002/ijc.26044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tong Q-S, Zheng L-D, Wang L, Zeng F-Q, Chen F-M, Dong J-H, Lu G-C. Downregulation of XIAP expression induces apoptosis and enhances chemotherapeutic sensitivity in human gastric cancer cells. Cancer Gene Ther. 2005;12:509–514. doi: 10.1038/sj.cgt.7700813. [DOI] [PubMed] [Google Scholar]
- [55].Sapi E, Alvero AB, Chen W, O’Malley D, Hao X-Y, Dwipoyono B, Garg M, Kamsteeg M, Rutherford T, Mor G. Resistance of ovarian carcinoma cells to docetaxel is XIAP dependent and reversible by phenoxodiol. Oncol. Res. 2004;14:11–12. doi: 10.3727/0965040042707943. [DOI] [PubMed] [Google Scholar]
- [56].Cardaci S, Filomeni G, Ciriolo MR. Redox implications of AMPK-mediated signal transduction beyond energetic clues. J. Cell Sci. 2012;125:2115–2125. doi: 10.1242/jcs.095216. [DOI] [PubMed] [Google Scholar]
- [57].Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra J-M, Bertucci F, Jacquemier J. Aldehyde dehydrogenase 1-Positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Debeb BG, Cohen EN, Boley K, Freiter EM, Li L, Robertson FM, Reuben JM, Cristofanilli M, Buchholz TA, Woodward WA. Pre-clinical studies of Notch signaling inhibitor RO4929097 in inflammatory breast cancer cells. Breast Cancer Res. Treat. 2012;134:495–510. doi: 10.1007/s10549-012-2075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rosenthal DT, Zhang J, Bao L, Zhu L, Wu Z, Toy K, Kleer CG, Merajver SD. RhoC Impacts the Metastatic Potential and Abundance of Breast Cancer Stem Cells. PLoS One. 2012;7:e40979. doi: 10.1371/journal.pone.0040979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.