Summary
NusG is a conserved regulatory protein interacting with RNA polymerase (RNAP) and other proteins to form multi-component complexes that modulate transcription. The crystal structure of Thermotoga maritima NusG (TmNusG) shows a three-domain architecture, comprising well conserved amino-terminal (NTD) and carboxy-terminal (CTD) domains with an additional, species-specific domain inserted into the NTD. NTD and CTD directly contact each other, occluding a surface of the NTD for binding to RNAP and a surface on the CTD interacting either with transcription termination factor Rho or transcription anti-termination factor NusE. NMR spectroscopy confirmed the intra-molecular NTD-CTD interaction up to the optimal growth temperature of Thermotoga maritima. The domain interaction involves a dynamic equilibrium between open and closed states and contributes significantly to the overall fold stability of the protein. Wild type TmNusG and deletion variants could not replace for endogenous Escherichia coli NusG, suggesting that the NTD-CTD interaction of TmNusG represents an auto-inhibited state.
Introduction
Regulatory proteins are frequently composed of several domains, each of which typically sustains interactions to different binding partners (Pawson and Nash, 2000; Pawson and Nash, 2003). Multiple interaction modules can form the basis for assembling and regulating multi-component complexes. Some of these multi-domain proteins are known to be regulated by an auto-inhibitory mechanism, in which intra-molecular interactions block binding sites for other proteins on the interacting domains (Burmann, et al., 2012; Mackereth, et al., 2011; Pufall and Graves, 2002). The release of auto-inhibition requires large scale conformational changes that in turn critically depend on the specific intra-molecular dynamics of the system (Li, et al., 2008). Knowledge of thermodynamics and kinetics of auto-inhibition release helps to understand the physical principles of affinity control. However, detection of lowly populated high-energy states is necessary, which often represents experimental challenges (Cho, et al., 2011).
NusG is a general regulator of bacterial transcription that exerts diverse effects on RNA polymerase (RNAP) in a context-dependent manner. It is the only universally conserved transcription factor in all three domains of life (Werner, 2012). Escherichia coli (Ec) NusG increases the elongation rate of RNAP in vivo and in vitro (Artsimovitch and Landick, 2000; Burns and Richardson, 1995; Burova, et al., 1995) by suppressing transcriptional pauses that involve backtracking of RNAP (Artsimovitch and Landick, 2000; Pasman and von Hippel, 2000). In conjunction with several other Nus-factors, NusG is part of anti-termination complexes that resist pausing and termination (Torres, et al., 2004; Zhou, et al., 2002). These complexes are necessary for bacteriophage λ growth (Li, et al., 1992; Sullivan, et al., 1992) and for the efficient transcription of ribosomal RNA operons (Squires, et al., 1993). At the same time, EcNusG directly interacts with transcription termination factor Rho (Burmann, et al., 2010; Pasman and von Hippel, 2000) and enhances Rho-dependent termination (Sullivan and Gottesman, 1992). EcNusG also couples transcription and translation (Burmann, et al., 2010).
EcNusG consists of two domains (amino-terminal domain – NTD; carboxy-terminal domain – CTD) connected by a flexible linker (Mooney, et al., 2009). In some bacteria an additional domain (domain II) is inserted in the NTD (Supplemental figure S1) (Knowlton, et al., 2003; Liao, et al., 1996; Steiner, et al., 2002) and a similar domain expansion is found in the functional NusG analog, Spt5, of archaea and eukaryotes (Guo, et al., 2008; Wenzel, et al., 2009). The NusG NTD consists of a four-stranded anti-parallel β-sheet flanked by two anti-parallel helices on one side and by a third, C-terminal helix on the other side. The CTD forms a barrel-type anti-parallel β-sheet with an embedded KOW motif (Kyrpides, et al., 1996), a known RNA-binding element.
The multi-domain architecture of NusG enables it to act as an adaptor protein that links other proteins bound to these domains in multi-component complexes. A crystal structure of the NusG-like NTD (NGN) of transcription factor Spt5 from Pyroccous furiosus (Pf) in complex with transcription factor Spt4 and the clamp domain of RNAP suggested that a large hydrophobic patch between the C-terminal helix, and the β-sheet of the NusG NTD represents a binding region for the conserved clamp region of RNAP (Martinez-Rucobo, et al., 2011). Furthermore, NMR analyses have demonstrated that the CTD of EcNusG forms mutually exclusive complexes with Rho and NusE (equivalent to ribosomal protein S10) (Burmann, et al., 2010). These multiple, domain-wise interactions of NusG allow it to integrate the activities of the transcriptional and translational machineries in E. coli (Burmann, et al., 2010).
Here, we present evidence that in some organisms NusG might be regulated by auto-inhibition. The crystal structure of NusG from the marine hyperthermophilic bacterium Thermotoga maritima (TmNusG) and the solution structure of a deletion variant thereof reveal a direct interaction between the conserved NTD and CTD that is incompatible with other known protein contacts of these domains. In addition, thermodynamic parameters as well as interconversion dynamics between open and closed states determined by NMR spectroscopy point to an autoregulatory mechanism.
Results and discussion
Crystal structure analysis of TmNusG
Based on sequence analysis, TmNusG possesses an insertion (domain II) in its NTD not found in most other bacteria (Liao, et al., 1996; Steiner, et al., 2002). To investigate the fold and domain architecture of TmNusG, we crystallized the full-length recombinant protein, produced in E. coli. Since molecular replacement with known NusG NTD and CTD structures as the sole search models failed, and well diffracting crystals of full-length TmNusG were poorly reproducible, we separately produced and crystallized the Thermotoga-specific domain II (residues 42 to 233) and solved its structure by selenomethionine (SeMet) single anomalous dispersion (SAD) at 1.9 Å resolution (Table 1). The structure of domain II together with known NusG NTD and CTD structures allowed us to solve the crystal structure of the full-length protein by molecular replacement at 2.4 Å resolution. Both structures were refined to acceptable R-factors with good stereo-chemistry (Table 1).
Table 1.
Crystallographic data
| Data Collection | Full-length | Domain II (native) | Domain II (SeMet) |
|---|---|---|---|
| Wavelength (Å) | 1.05 | 0.97985 | 0.97893 |
| Temperature (K) | 100 | 100 | 100 |
| Space group | P4322 | C2 | C2 |
| Unit cell parameters | |||
| a, b, c (Å) | 60.0, 60.0, 311.5 | 136.3, 42.2, 88.3 | 134.9, 42.0, 89.1 |
| α, β,γ (°) | 90, 90, 90 | 90, 106.3, 90 | 90, 106.3, 90 |
| Resolution (Å)a | 50.0 – 2.4 (2.44 – 2.40) | 50.0 – 1.9 (1.97 – 1.90) | 50.0 – 2.0 (2.07 – 2.00) |
| Reflections | |||
| Unique | 23,437 (1,082) | 37,677 (1,846) | 29,850 (1,495) |
| Completeness (%) | 99.2 (99.6) | 99.9 (72.7) | 89.9 (45.7) |
| Redundancy | 4.9 (4.7) | 3.8 (3.7) | 6.2 (3.0) |
| Mean I/σ(I) | 16.6 (2.0) | 13.3 (1.8) | 14.6 (2.1) |
| Rsym(I)b | 7.4 (56.7) | 9.4 (78.8) | 8.6 (39.2) |
|
| |||
| Phasing | |||
|
| |||
| Resolution (Å) | 35.0 – 2.0 | ||
| Heavy atom sites | 4 | ||
| SHELXD CC/CCweakc | 56.5 / 26.9 | ||
| SHARP overall figure of merit | |||
| acentric / centric | 0.29 / 0.22 | ||
|
| |||
| Refinement | |||
|
| |||
| Resolution (Å) | 50.0 – 2.4 | 50.0 – 1.9 | |
| Reflections | |||
| Number | 23,437 | 35,358 | |
| Completeness (%) | 95.4 | 92.7 | |
| Test set (%) | 5 | 5 | |
| Rworkd (%) | 22.0 | 18.1 | |
| Rfreed (%) | 27.4 | 23.0 | |
| ESU (Å)e | 0.31 | 0.25 | |
| Contents of A.U.f | |||
| Protein molecules / residues / atoms | 1 / 341/ 2763 | 2 / 386/ 3220 | |
| Water oxygens | 27 | 204 | |
| Acetate ions | - | 6 | |
| Mean B-factors (Å2) | |||
| Wilson | 56.1 | 22.5 | |
| Protein | 76.0 | 33.4 | |
| Water | 60.1 | 40.4 | |
| Ramachandran plotg | |||
| Preferred (%) | 94.1 | 99.2 | |
| Allowed (%) | 4.1 | 0.8 | |
| Disallowed (%) | 1.8 | 0 | |
| RMSDh from target geometry | |||
| Bond lengths (Å) | 0.006 | 0.009 | |
| Bond angles (°) | 1.036 | 1.140 | |
| Chirality (Å) | 0.067 | 0.087 | |
| Dihedral angles (°) | 16.01 | 13.31 | |
| PDB ID | 2XHC | 2XHA | |
Data for the highest resolution shell in parentheses.
Rsym(I) = ΣhklΣi|Ii(hkl) − <I(hkl)> | / ΣhklΣi|Ii(hkl) |; for n independent reflections i observations of a given reflection; <I(hkl)> – average intensity of the i observations.
CC = [ΣwEoEcΣw −ΣwEoΣwEc]/{[ΣwEo2Σw−(ΣwEc)2] [ΣwEc2Σw−(ΣwEc)2]}½; w – weight (see http://shelx.uni-ac.gwdg.de/SHELX/shelx_de.pdf for full definitions).
R = Σhkl||Fobs| − |Fcalc|| / Σhkl|Fobs|; Rwork − hkl ∉ T; Rfree − hkl ∈ T; Rall – all reflections; T – test set.
ESU – estimated overall coordinate error based on maximum likelihood.
A.U. – asymmetric unit
According to (Lovell, et al., 2003)
RMSD – root-mean-square deviation
Domain II (TmNusGDII) crystallized in space group C2 with two molecules per asymmetric unit, which are structurally very similar (root-mean-square deviation [RMSD] 1.37 Å for 187 superimposed Cα atoms). The two molecules differed slightly in some surface loops and at the domain termini (Figure 1A,B). All TmNusG residues contained in the domain II construct could be traced for both independent copies of the molecule in the electron density. The regularly structured part of domain II encompasses residues 52–224 and is composed of two sub-domains (Figure 1A). The composite N-terminal sub-domain (residues 52–80 and 200–224) adopts a β-sandwich fold made up of two anti-parallel four- and three-stranded β-sheets. The continuous C-terminal sub-domain (residues 83–197) exhibits a dumbbell-like structure whose central scaffold is formed by a four-stranded anti-parallel β-sheet. Short, anti-parallel peptides (residues 81–82 and 198–199) connect the sub-domains. Comparison with entries in the Protein Data Bank (www.pdb.org) showed that the overall fold of domain II is unique with respect to the spatial organization and the connectivity of the secondary structure elements. PDBeFold/SSM (Krissinel and Henrick, 2004) yielded no matches using the default threshold value (lowest acceptable match 70 % for query and target). Lowering the threshold value did not yield hits with RMSDs below 4 Å or Q-scores above 0.06, confirming the novelty of the fold.
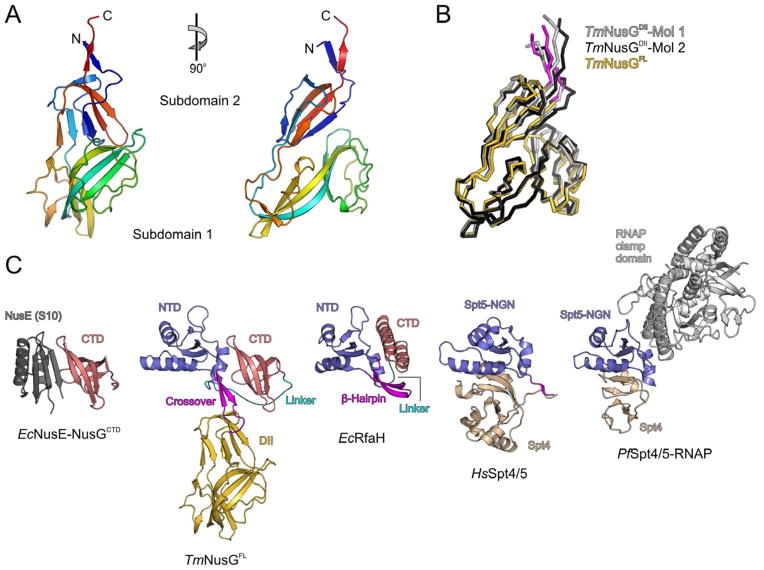
Figure 1.
Crystal structure of TmNusG. A) Orthogonal ribbon plots of the Thermotoga-specific domain II insertion comprising two sub-domains. The protein is colored blue to red from N-terminus to C-terminus to illustrate the chain trace. B) Superimposed ribbon plots of the two crystallographically independent molecules of the TmNusGDII crystal structure (light and dark gray) and of the domain II portion of the full-length protein (domain II – gold; crossovers to the NTD – magenta). Minor differences are seen in surface loops and at the domain termini. C) Overall structure of TmNusGFL (panel 2) in comparison with an EcNusE-NusGCTD complex (panel 1), EcRfaH (panel 3), HsSpt4-Spt5 complex (panel 4) and PfSpt4-Spt5-RNAP clamp domain complex (panel 5). The NusG CTD (panels 1 and 2) or the equivalents of the NusG NTD (panels 2–5) are shown in the same orientation as the corresponding domains in the structure of TmNusG (panel 2). The orientation of TmNusGFL is the same as for TmNusGDII in the left panel of A). NTD – blue; domain II – gold; CTD – light red; NusE – dark gray; Spt4 – beige; RNAP – gray; NTD-CTD linker – cyan; NTD-domain II crossovers or topologically equivalent regions – magenta.
Crystals of full-length TmNusG (TmNusGFL) belonged to space group P4322 with one molecule per asymmetric unit. The crystal structure of TmNusGFL exhibits a “T”-shape (Figure 1C, panel 2), in which the NTD (residues 3–40 and 234–281) and CTD (residues 299–352) form the bar of the “T” while the stem is formed by domain II that is inserted within the NTD. Both NTD and CTD adopt very similar folds as in other NusG proteins (compared to Aquifex aeolicus (Aa) NusG, PDB ID 1M1G: RMSD NTD 1.65 Å for 84 superimposed Cα common atoms, CTD 0.77 Å for 53 superimposed Cα atoms). The NTD comprises an anti-parallel four-stranded β-sheet flanked by two α-helices on one side and an additional α-helix on the other. The CTD consists of a five-stranded, anti-parallel β-barrel. The two terminal domains are connected by a long, flexible linker (residues 282–298) whose central part (residues 288–296) lacked well defined electron density (dashed cyan line in Figure 1C, panel 2).
In TmNusGFL, domain II adopts the same structure as in the two copies of the isolated domain construct (RMSD 1.20/1.66 Å for 184/179 superimposed Cα atoms; Figure 1B). It is connected to the N- and C-terminal portions of the NTD via two peptides (residues 41–51 and 225–233) arranged as two anti-parallel β-strands (Figure 1C, panel 2). The bulk of domain II lacks direct contacts to either NTD or CTD. Previously, an additional domain has also been found inserted at the corresponding site in the NTD of AaNusG (Knowlton, et al., 2003; Steiner, et al., 2002). Although the AaNusG insertion exhibits a β-sandwich fold reminiscent of the first sub-domain of TmNusG domain II, it folds with a different topology. In EcNusG, domain II is replaced by an elongated loop (Burmann, et al., 2011; Mooney, et al., 2009).
NTD-CTD contacts in TmNusG are reminiscent of inter-domain interactions in the NusG paralog, RfaH
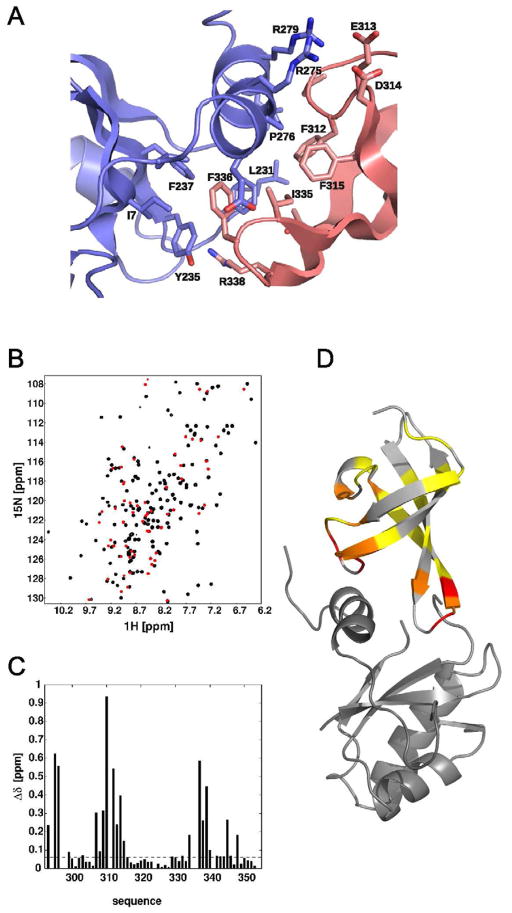
In the crystal structure of TmNusG, the NTD and CTD directly contact each other, burying 1544 Å2 of combined surface area at their interface (contribution of NTD and CTD 686 Å2 and 858 Å2, respectively). In the NTD, residues from strands β1 (Ile7), β3 (Tyr235, Phe237) and its preceding loop (Leu231, Phe232, Pro233), as well as residues from helix α3 (Pro276, Leu280) and its preceding loop (Val269) form a hydrophobic surface patch (Figure 2A). The CTD associates with this surface patch via two loops, between strands β1′ and β2′ (Pro311, Phe312) and between strands β3′ and β4′ (Ile335, Phe336), forming extensive, hydrophobic inter-domain contacts. In addition, two salt bridges (Arg275-Asp314, Arg279-Glu313) and a hydrogen bond (between the side chain of Arg338 and backbone of Pro233) are sustained between the domains (Figure 2).
Figure 2.
Domain interaction of TmNusG. A) Expanded view of the domain interface between NTD (blue) and CTD (light red). Side chains with buried surfaces compared to isolated domains are shown in stick representation. B) Superposition of 1H, 15N HSQC spectra of TmNusGNTD-CTD (black) and TmNusGCTD (red). C) Normalized chemical shift differences of backbone amide resonances between TmNusGNTD-CTD and TmNusGCTD as a function of sequence. D) Mapping of chemical shift differences onto the crystal structure of TmNusG. The inserted domain II is omitted for clarity. Color coding from yellow to red indicates increasing chemical shift differences.
In E. coli the specialized transcription factor RfaH, a paralog of NusG, exhibits a NusG-like NTD and a differently folded, α-helical CTD that are tightly associated (Belogurov, et al., 2007). The NTD-CTD interactions in TmNusG and RfaH make use of an equivalent hydrophobic patch on the respective NTDs (Figure 1C, panel 3). In contrast to TmNusG, RfaH-CTD adopts a helical conformation in the closed state. Upon domain opening RfaH-CTD refolds into the all-β-sheet conformation similar to NusG-CTD (Burmann, et al., 2012). The domain interaction in RfaH has important physiological consequences. RfaH requires a specific DNA sequence, ops, to unlock its domains and to use the hydrophobic patch on the liberated NTD for binding to RNAP (Belogurov, et al., 2007). Thus, the tight domain interaction in RfaH represents an auto-inhibited state that restricts the use of the transcription factor to genes bearing an ops sequence. These observations suggest that similar auto-inhibitory mechanisms may be exploited to regulate the functions of TmNusG. The requirement of a certain factor for opening of TmNusG is unknown. It might also be possible that the affinity to RNAP is sufficient to open TmNusG without an additional factor.
NTD and CTD contacts are mutually exclusive with NusG-RNAP, NusG-NusE, and NusG-Rho interactions
The surfaces of the NTD and the CTD involved in intra-molecular interaction are congruent with regions of these domains known to bind other protein partners (Figure 1C). In the complex of the EcNusG CTD with NusE (Burmann, et al., 2010) the two loops of the CTD which sustain inter-domain contacts in TmNusG are involved in the protein-protein interaction (Figure 1C, panel 1). Moreover, the identical CTD surface contacts transcription termination factor Rho (Burmann, et al., 2010).
While presently no structure is available of bacterial NusG in complex with RNAP, the high structural similarity of the NusG NTD with the Spt5 NGN domain of archaea and eukaryotes (Figure 1C, panels 4 and 5) allows the prediction of the RNAP-binding site on NusG based on the crystal structure of an archaeal Spt4-Spt5-RNAP clamp domain complex (Martinez-Rucobo, et al., 2011). Again, the region of the NusG NTD expected to bind the clamp domain of RNAP partly overlaps with the area of the NTD involved in inter-domain contacts in TmNusG (Figure 1C, panel 5). These observations suggest that intra-molecular domain interactions observed in TmNusG would interfere with other functional interactions of the NTD and CTD, further supporting the notion that the present conformation of TmNusG represents an auto-inhibited state.
NTD-CTD contacts are maintained in solution
Previous structural analyses of other NusG orthologs have failed to disclose stable NTD-CTD interactions (Burmann, et al., 2011; Mooney, et al., 2009; Reay, et al., 2004; Steiner, et al., 2002). Interestingly, however, regions identical to the NTD-CTD contact areas in TmNusG are involved in lattice contacts between neighboring molecules in crystal structures of AaNusG (Knowlton, et al., 2003; Steiner, et al., 2002) and have been suggested to reflect a functional intra-molecular interaction in solution (Knowlton, et al., 2003). We therefore asked whether the NTD-CTD interactions observed in the TmNusG crystal structure prevail in solution. In order to address these questions using NMR spectroscopy, we generated a shorted variant of TmNusG, TmNusGNTD-CTD, in which we replaced a region (residues 43–230) encompassing domain II with the corresponding loop of EcNusG (residues 52–61). The replaced region did not exhibit any direct interactions with the NTD or CTD in the crystal structure.
The 1H, 15N HSQC spectrum of TmNusGNTD-CTD showed the characteristic signal dispersion of well folded globular proteins. Almost all resonances could be assigned by applying standard hetero-nuclear through-bond correlations. Secondary chemical shifts correlated well with the crystal structure. Expression of the isolated CTD also resulted in a well folded globular protein whose 1H, 15N correlations were assigned using 3D 15N-edited NOESY and 15N-edited TOCSY spectra. However, the chemical shifts of the isolated CTD displayed remarkable differences in their magnitudes compared to the spectrum of TmNusGNTD-CTD (Figure 2B–D). Such differences are typically found upon complex formation of protein complexes. Mapping chemical shift changes onto the structure revealed that both loops of the CTD participating in the interaction with the NTD in the crystal structure of TmNusG are strongly affected by the presence of the NTD in solution. EcNusG, in contrast, did not show any chemical shift differences between isolated domains and the full-length protein (Burmann, et al., 2011). These observations indicate that NTD and CTD of TmNusGNTD-CTD mutually interact in solution, presumably in a similar manner as observed in the TmNusG crystal structure.
Spin relaxation experiments support similar NTD-CTD interactions in solution as in the crystal
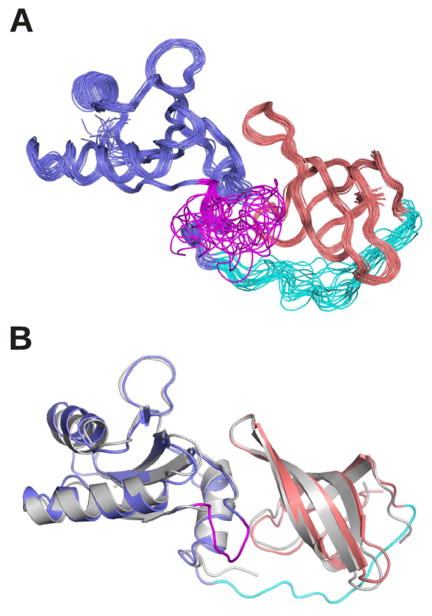
To further confirm the similar association of NTD and CTD in TmNusG in solution as in the crystal, we conducted spin relaxation experiments, a powerful method to determine the overall tumbling of proteins in solution. Defined domain interactions in a multi-domain protein result in a uniform rotational tumbling of the entire protein. In this case, the stochastic rotation can be described in a single frame represented by a unique rotational diffusion tensor for both domains. In the case of a multi-domain protein with non-interacting domains, the relative domain movement requires an individual description of the rotational tumbling for each domain. The ratio of the transverse (R2) and the longitudinal (R1) relaxation rates is a key parameter for characterizing overall tumbling of a protein because contributions of fast internal dynamics are cancelled out to a large extend. The R2/R1 ratio of TmNusGNTD-CTD showed a mono-modal distribution (Figure 3) with an effective isotropic correlation time of 7.4 ns at 50 °C, indicating coupling of the two domains on the timescale of the molecular rotation. The situation is different in EcNusG, which displayed different R2/R1 distributions for its NTD and CTD, consistent with domain decoupling in that protein (Burmann, et al., 2011). The overall shape of a protein is reflected in the relaxation rates of its nuclear spins due to the orientation of the corresponding inter-nuclear vectors relative to the principal axis of the rotational diffusion tensor. We derived a rotational diffusion tensor of TmNusGNTD-CTD using the 15N relaxation data (Supplemental Table 1) from TmNusGNTD-CTD and the coordinates of full length TmNusG. The tumbling of TmNusGNTD-CTD was very well described by a prolate, axial symmetric diffusion tensor with a tumbling time of 7.44 ns and an axial ratio of 1.42. These observations showed that both domains tumble as a rigid unit with an overall elongated shape, exhibiting a stable domain interaction at least on the time scale of the molecular rotation. The derived values perfectly fit the coordinates of NTD and CTD of the TmNusG crystal structure, indicating that TmNusGNTD-CTD in solution adopts an identical overall structure as in the crystal. In particular, NTD and CTD strongly interact in solution. We directly verified the above conclusions by determining the solution structure of TmNusGNTD-CTD, using inter-domain NOEs detected in isotope-edited NOESY spectra, and residual dipolar coupling, determined in two liquid crystalline media (pf1 phages (5 mg/ml) and 3 % C6E12/Hexanol) (Figure 4, Table 2). The resulting structural ensemble shows good coordinate precision and reasonable stereo-chemical properties. The average solution structure superimposes very well with the crystal structure (backbone RMSD of 1.1 Å, Figure 5B).
Figure 3.
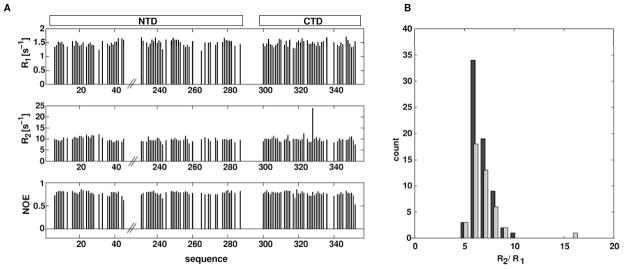
15N spin relaxation data of TmNusGNTD-CTD at 14.1 T magnetic field strength and 323 K. A) Longitudinal (R1, top), transverse (R2, middle) relaxation rates, and {1H}15N steady state NOE (bottom) as a function of amino acid sequence. B) Distributions of R2/R1 ratios. Data from residues of the NTD are shown black, data from residues of the CTD are in light gray. The similar distributions of both domains indicate an uniform overall tumbling of the protein.
Figure 4.
Solution structure of TmNusGNTD-CTD. A) Superposition of 20 accepted structures. B) Comparison of the crystal structure (gray) with the solution structure. The inserted domain II of the crystal structure is omitted for clarity. NTD, blue; CTD, light red; NTD-CTD, linker cyan; NTD-domain II crossover, magenta.
Table 2.
Solution structure statistics
| Experimental derived restraints | |
|---|---|
| distance restraints | |
| NOE | 1908 |
| intraresidual | 679 |
| sequential | 480 |
| medium range | 224 |
| long range | 525 |
| hydrogen bonds | 73 |
| dihedral restraints | 302 |
| residual dipolar couplings | 173 |
|
| |
| restraint violation | |
|
| |
| average distance restraint violation (Å) | 0.007 +/− 0.001 |
| maximum distance restraint violation (Å) | 0.18 |
| average dihedral restraint violation (°) | 0.93 +/− 0.2 |
| maximum dihedral restraint violation (°) | 14 |
| average rdc restraint violation (Hz) | 0.59 +/− 0.05 |
| maximum rdc restraint violation (Hz) | 3.2 |
| deviation from ideal geometry | |
| bond length (Å) | 0.00075 +/− 0.00005 |
| bond angle (°) | 0.16 +/− 0.008 |
|
| |
| coordinate precisiona,b | |
|
| |
| backbone heavy atoms (Å) | 0.49 |
| all heavy atoms (Å) | 0.99 |
|
| |
| Ramachandran plot statisticsc (%) | 84.1/12.4/2.5/1.0 |
The precision of the coordinates is defined as the average atomic root mean square difference between the accepted simulated annealing structures and the corresponding mean structure calculated for the given sequence regions.
calculated for residues Lys5-Pro43, Lys230-Glu287, Phe299-Ile352 (numbering according to full length tmNusG)
Ramachandran plot statistics are determined by PROCHECK and noted by most favored/additionally allowed/generously allowed/disallowed.
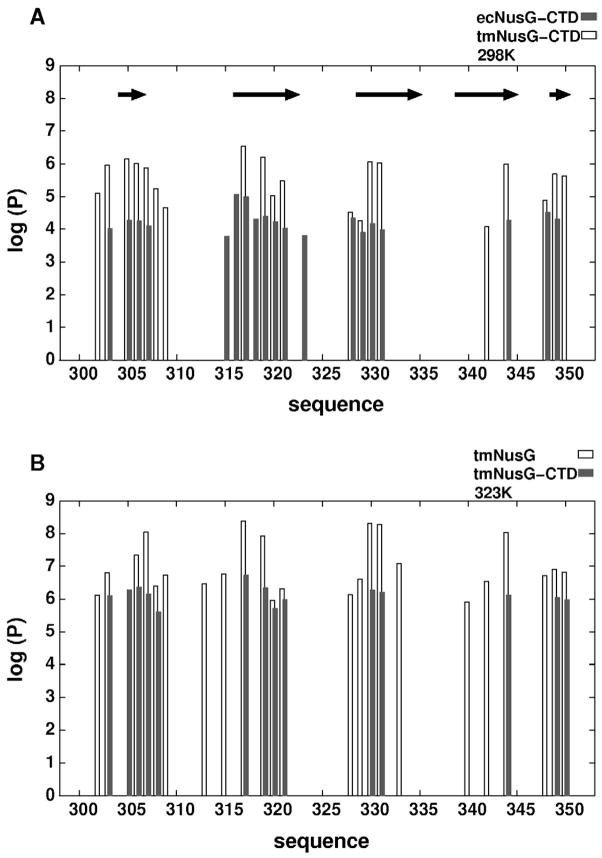
Figure 5.
Amide protection factors determined by H/D exchange. A) Protection factors of TmNusGCTD (dark gray) and EcNusGCTD (white) at 298 K as a function of sequence. Protection factors are expressed as the ratio of the intrinsic exchange rate and the experimentally determined exchange rate. Residues Phe299-Ile352 of TmNusG correspond to residues Phe128-Ala181 of EcNusG. The arrows indicate the location of the β-strands. B) Protection factors of the CTD from TmNusGNTD-CTD (white) and isolated TmNusGCTD at 323K.
NTD-CTD contacts persist at high temperatures
The analyses described above demonstrate that in solution TmNusGNTD-CTD exhibits a defined domain interaction at least at 50 °C, a low growth temperature for Thermotoga maritima. The solution structure closely resembles the domain interaction in the crystal. To test whether this domain interaction also persists at temperatures closer to the optimal growth conditions for Thermotoga maritima, a series of HSQC spectra with increasing sample temperatures up to 80 °C were recorded. The prominent chemical shift differences between TmNusGNTD-CTD and TmNusGCTD were still observed at 80 °C, revealing the presence of intra-molecular domain interactions even at this temperature. However, sample stability was significantly reduced under these conditions and the protein precipitated after roughly two hours.
Contributions of the inter-domain interaction to protein stability
The optimal growth temperature of 84 °C for Thermotoga maritima presents particular challenges for the integrity of protein folds. To estimate the contribution of the domain interaction to protein fold stability, we conducted hydrogen/deuterium (H/D) exchange experiments followed by NMR spectroscopy of TmNusGNTD-CTD, an isolated CTD of TmNusG (TmNusGCTD), and an isolated CTD of EcNusG (EcNusGCTD). H/D exchange was monitored via the decay of signal intensities in a series of 1H, 15N HSQC experiments recorded at different time points after dissolving lyophilized proteins in D2O at 298 K or 323 K.
On average, the exchange rates of TmNusGNTD-CTD and TmNusGCTD were 30 times faster at 323 K than at 298 K. At 323 K, the H/D exchange of EcNusGCTD was too fast to detect, although the protein still showed the well dispersed 1H, 15N HSQC spectrum of a folded domain. At 298 K, the exchange rates of EcNusGCTD were 65 times faster than for TmNusGCTD. All slowly exchanging amide protons were found in regular secondary structure elements, and the patterns of exchanging amide protons were similar for all three proteins, suggesting similar opening dynamics for all three constructs.
To characterize the stabilization effect due to NTD-CTD interaction, the amide proton exchange was expressed as Protection Factor (PF; Figure 5). From the PF, the free energy of hydrogen exchange was calculated and the highest ΔGHX was assumed to be equal to the free energy of folding. For EcNusGCTD, the maximum PF (Asn145) was 8.8·103 which translates into a ΔGHX(298 K) of 28.8 kJ/mol. For TmNusGCTD the maximum PF (Gly317) was 3.4·106 corresponding to a ΔGHX(298 K) of 37.3 kJ/mol. At 323 K, the maximum PF (Gly317) of TmNusGCTD was 5.2·106 corresponding to a ΔGHX(323 K) of 41.5 kJ/mol. ΔGHX of TmNusGCTD is lower at 298 K than at 323 K indicating that the measurements were performed below the optimal temperature for stabilization, which is expected to be close to the optimal growth temperature of Thermotoga maritima (84 °C). Thus, compared to the mesophilic EcNusG, at 298 K the thermophilic TmNusGCTD is stabilized by about 8.5 kJ/mol. Comparison of the exchange rates of amide protons of TmNusGNTD-CTD and of TmNusGCTD shows that the protection is increased 40–50 fold (PF = 2.4·108, ΔGHX(323 K) = 51.8 kJ/mol for Gly317 in TmNusGNTD-CTD). This corresponds to an additional stabilization of 10.3 kJ/mol, which is due to the presence of the NTD. This additional stabilization can be attributed to the interaction between NTD and CTD. Converting the portion of the free energy of folding that is caused by the domain interaction into the equilibrium between open and closed states yielded a fraction of 2 % of the TmNusGNTD-CTD molecules in the open conformation at 323 K. As a consequence, the closing rate is about 50 times faster than the opening rate.
Dynamics of the inter-domain interactions
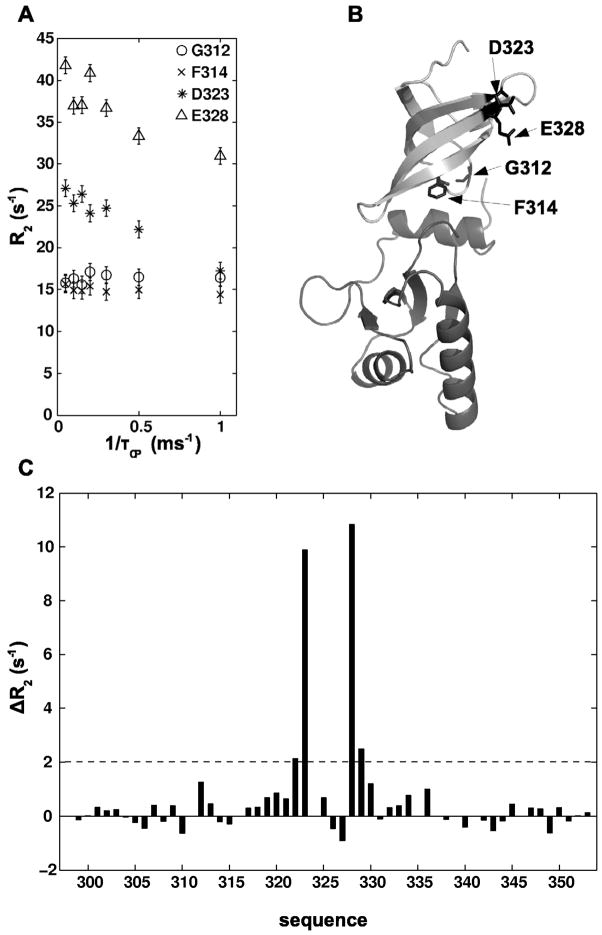
To test the presence of μs-ms dynamics in the domain interface, 15N relaxation dispersion experiments were performed with TmNusGNTD-CTD. By this method we did not observe any contribution of chemical exchange to the transverse relaxation rates of nitrogen spins for any of the residues of the CTD which are located at the domain interface and exhibited significant chemical shift differences between TmNusGNTD-CTD and TmNusGCTD. Only for two residues of the CTD (Asp323 and Glu328), relaxation dispersion curves showed a contribution of chemical exchange to the transverse relaxation (Figure 6). These residues are located face to face at the end of two β-strands, far away from the domain interface, and their observed dynamics reflect local conformational flexibility. Therefore, the exchange rate for domain opening/closing must either be in the fast exchange regime with kex ≫ 2πΔνmax (Δνmax being the largest 15N resonance frequency change, ca. 370 Hz for Phe312) or, less likely, in the slow exchange limit for all residues (kex < 1 s−1). The slow exchange situation would result in observable signals for the open state in a 1H, 15N HSQC spectrum with sufficient signal/noise ratio. In a spectrum with signal/noise > 200:1 for characteristic residues in the domain interface, no signals of the open state could be observed.
Figure 6.
15N-relaxation-dispersion of TmNusGNTD-CTD. A) Selected 15N-relaxation-dispersion curves of residues from the domain interface (Gly312, Phe314), and Asp323 and Glu328. B) Structure of TmNusGNTD-CTD. Gly312, Phe314, Asp323, and Glu328 are shown in stick representation. C) Differences between transverse relaxation (R2) at the maiximum and minimum applied effective field for residues of the CTD. The residues located at the domain interface show no dependence of the inter-pulse delay during CPMG pulse trains, indicating the lack of dynamics on the μs-ms scale causing an exchange contributing to the transverse relaxation.
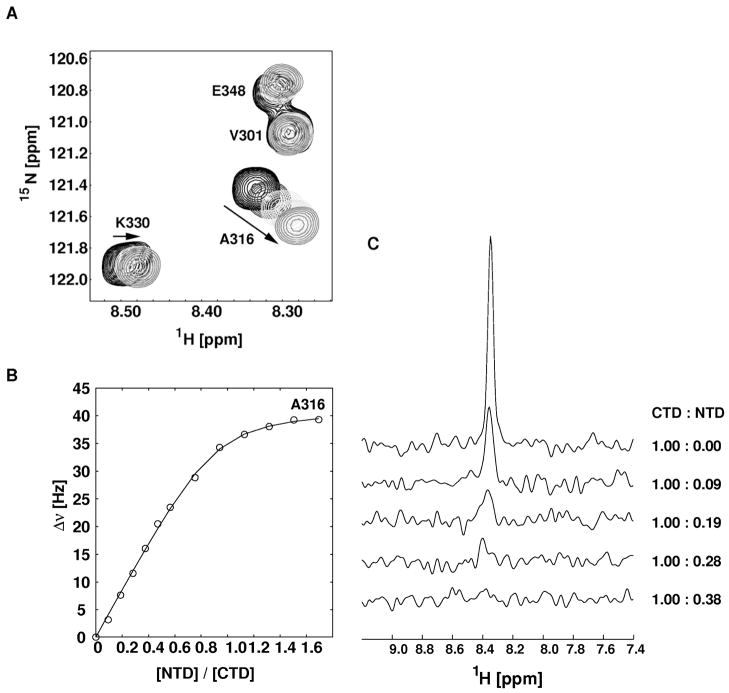
The exchange contribution in TmNusGNTD-CTD is determined by the rate for closing (kex = kopen + kclose = (1/50 + 1) kclose, due to KD = kopen/kclose = 1:50). In a titration using separate domains, the exchange rate is given by kex = koff(1+pEL/pE) with a dissociation constant KD,binary = kon/koff, in which pEL, pE are the molar fractions of bound (pEL) and free (pE) 15N-labeled domains. Using a sample composition with known fractions of free and bound (separated) domains, koff can be estimated from the coalescence of signals in the spectra. During the titration of 15N-labeled TmNusGCTD with unlabeled TmNusGNTD, residues with only small (< 60 Hz) chemical shift differences between TmNusGNTD-CTD and isolated TmNusGCTD showed continuous chemical shift changes characteristic for exchange behavior in the fast exchange regime (Figure 7). From the chemical shift changes, a KD,binary of 5 μM could be determined. Resonances with chemical shift differences of about 200 Hz either in the proton or nitrogen dimension disappeared beyond pEL/pE > 0.2. This situation corresponds to koff ≈ 1000 s−1 and kon ≈ 2*108 M−1s−1, a value typical for a diffusion-limited association. Under the assumption that the dissociation rate of the binary complex is similar to the opening rate of the full-length protein, a closing rate of 50,000 s−1 can be estimated. This rate is faster than the effective rate of association in the binary complex at typical concentrations ([NTD] = 0.1 – 1 mM), given by kon * [NTD], reflecting the tethered protein domains in the full-length protein.
Figure 7.
Titration of isolated 15N-labeled TmNusGCTD with unlabeled TmNusGNTD. A) Expanded region of superimposed 1H, 15N HSQC spectra of TmNusG-CTD with increasing concentrations of TmNusGNTD. The arrows mark the direction of chemical shift changes during titration. B) Chemical shift changes (expressed as Δν = sqrt((Δν1H)2+(Δν15N)2) of Ala316 during the titration. Fitting the titration curve to a two-state binding model resulted in a dissociation constant (KD) of 5 μM. C) One dimensional traces of the 1H, 15N HSQC spectra with given ratios of CTD:NTD showing the dissappearing signal of Gly314. The signal disappearing at a CTD:NTD ratio of 0.3 corresponds to a dissociation rate of approximately 1000 s−1.
These results show that TmNusGNTD-CTD exists in a dynamic equilibrium between the major closed conformation and the minor open conformation. The fast opening with a rate of approximately 1000 s−1 is not expected to be a kinetic barrier during formation of protein complexes required for transciptional processes.
Inter-domain salt bridges contribute little to the domain association
The crystal structure of TmNusG identified two salt bridges (Arg275-Asp314 and Arg279-Glu313) between NTD and CTD. An increased number of salt bridges is frequently observed in proteins from thermophilic organisms compared to their mesophilic counterparts (Kumar, et al., 2000). Sequence comparison of TmNusG with NusG from other bacteria suggests that these salt bridges are a unique feature of TmNusG; no other NusG ortholog shows amino acid combinations at these positions suitable for analogous ionic interactions.
To test whether these salt bridges are essential for the NTD-CTD interaction in TmNusG, two single point mutants (Arg275Ala, Arg279Ala) and the double mutant (Arg275Ala+Arg279Ala) were studied. 1H, 15N HSQC spectra of TmNusGNTD-CTD(Arg275Ala) and TmNusGNTD-CTD showed no significant chemical shift differences. Only small changes were observed for residues sequentially and spatially close to the mutated site. Specifically, in comparison to the isolated TmNusGCTD the characteristic large chemical shift differences were still present, showing that TmNusGNTD-CTD(Arg275Ala) maintains the closed conformation.
Compared to TmNusGNTD-CTD, TmNusGNTD-CTD(Arg279Ala) and the double mutant TmNusGNTD-CTD(Arg275Ala+Arg279Ala), showed larger chemical shifts for resonances of both, NTD and CTD. Especially in the carboxy-terminal helix of the NTD, where the two arginines are located, it was no longer possible to assign the residues unambiguously by HQSC comparisons. Moreover, these mutants exhibited a significantly lower solubility (< 100 μM), rendering NMR experiments for more detailed information (15N relaxation, assignment by triple-resonance NMR) impossible. As the signals from the CTD in TmNusGNTD-CTD still showed remarkable differences to those of the isolated CTD, we conclude that a significant fraction of both mutant proteins stayed in the closed conformation. Compared to the parent TmNusGNTD-CTD, the large chemical shift changes may have resulted from minor differences in the relative domain orientations, which are due to the lack of the salt bridges. Together, these results suggest that additional interactions are essential and predominantly responsible for the stable intra-molecular domain interaction. This interpretation would also explain that the solubility of TmNusGNTD is much lower than that of EcNusGNTD. Similarly to the NTD of EcRfaH the hydrophobic surface exposed to the solvent is much larger in TmNusGNTD.
TmNusG or TmNusGNTD-CTD do not complement a NusG deletion in E. coli
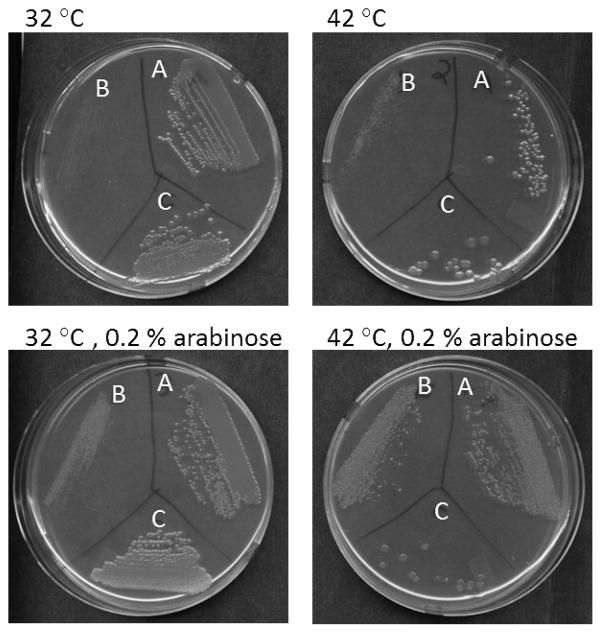
The genome of T. maritima does not encode for another NusG-like protein (suggesting that T. maritima lacks an RfaH ortholog) but contains genes for putative NusA, NusB, NusE/S10 and Rho proteins (Nelson, et al., 1999). Notably, TmNusA also contains a different domain arrangement compared to EcNusA (Shin, et al., 2003; Worbs, et al., 2001), in this case lacking two C-terminal double helix-hairpin-helix domains(Bonin, et al., 2004; Eisenmann, et al., 2005), which in E. coli serve an autoinhibitory function (Mah, et al., 2000; Schweimer, et al., 2011). The conservation of NusG in all bacteria and higher organisms, the lack of an additional NusG-like open reading frame in T. maritima and the observation that also other presumed T. maritima transcription factors have different domain compositions compared to E. coli orthologs suggest that despite the DII insertion and the NTD-CTD interaction, TmNusG should maintain similar functions as known for E. coli. We assumed that the preferred closed state of TmNusG at 298 K and 323 K might interfere with the functions of this protein as a transcription factor at elevated temperatures. Therefore, we tested whether TmNusG or TmNusGNTD-CTD could rescue an E. coli nusG deletion. The corresponding genes were cloned into plasmid pBAD/HisA to allow induction by arabinose. The E. coli strain W3102 bearing a chromosomal nusG::kan insert and a temperature-sensitive plasmid that expresses EcNusG was used for gene expression (Sullivan and Gottesman, 1992) (Figure 8). At 42 °C the ecnusG plasmid is lost and cells are not viable in the absence of an alternative source of active NusG. After the addition of arabinose, full length TmNusG (or TmNusGNTD-CTD, data not shown) expressed from the pBAD/HisA plasmid was not able to support cell growth at 42 °C. In agreement with this finding, previous analyses have also shown that TmNusG or a domain II deletion variant of the protein were unable to substitute for E. coli NusG (Liao, et al., 1996).
Figure 8.
Test for complementation of EcNusG by TmNusG. The growth of the following E. coli strains was tested at the growth conditions indicated: A) 9388, which carries the temperature-sensitive plasmid ecnusG and plasmid pBAD-aanusG, B) 9391, which was derived from 9388 at 42 °C in the presence of arabinose and has lost the ecnusG plasmid, and C) 11480, which carries the temperature-sensitive plasmid ecnusG and plasmid pBAD-tmnusG.
In contrast, cells supplemented with an Aquifex aeolicus nusG gene on the same plasmid grew with 100 % efficiency at 42 °C in the presence of arabinose. Even though both NusG proteins originate from hyperthermophilic organisms, AaNusG, unlike TmNusG, does not exhibit a stable NTD-CTD interaction (Knowlton, et al., 2003; Steiner, et al., 2002). These results indicate that TmNusG is not functional in E. coli. This is possibly due to its preference for a stable closed conformation at E. coli growth temperatures. Expression of TmNusG in cells producing EcNusG induces a small colony phenotype at 37 °C and kills at 42 °C (data not shown), suggesting that TmNusG interacts with and inhibits some E. coli transcription reaction, possibly Rho-dependent transcription termination.
Conclusions
We have shown by crystal and solution NMR structural analyses that the transcription factor NusG of the hyperthermophilic bacterium Thermotoga maritima exhibits a stable interaction between its NTD and CTD. The interaction is subject to fast dynamics on the NMR time scale with ca. 98 % of the molecules existing in the closed conformation at 323 K. H/D exchange studies revealed that this interaction contributes significantly to the overall fold stability of the protein. Thus, it may represent a safeguard against unfolding or aggregation at high temperature. However, NusG proteins from other hyperthermophilic organisms, such as Aquifex aeolicus, which exhibit a very similar domain architecture as TmNusG, do not show a stable NTD-CTD domain interaction. Therefore, the NTD-CTD interaction does not represent a mandatory adaption to life at high temperatures.
We showed that the closed state of TmNusG is incompatible with other functional interactions of the NTD (with RNAP) and CTD (with NusE or Rho). In contrast to TmNusG, which predominantly adopts a closed conformation, NusG from Aquifex aeolicus, which does not exhibit a stable NTD-CTD interaction, fully complemented a NusG deletion in E. coli. Therefore, we conclude that in some organisms, e.g. Thermotoga maritima, NusG is subject to auto-inhibition via the characterized domain interaction. Auto-inhibition may prevent NusG from interacting prematurely with other components of the transcription complex or may preclude non-specific interactions of NusG with other cellular components. Release from auto-inhibition may be achieved by the presentation of NusG binding sites with sufficiently high affinity on preformed transcription complexes. Alternatively, it may require interaction of NusG with a specific component of the transcription complex, such as the participating nucleic acids. An auto-inhibited state in the isolated protein that is released in the presence of a specific DNA signal sequence, ops, has been described for the NusG paralog RfaH (Belogurov, et al., 2007; Burmann, et al., 2012). In support of this notion, TmNusG has been found to strongly interact with various nucleic acids (Liao, et al., 1996) unlike the mesophilic EcNusG, in which no comparable auto-inhibited conformation was detected.
Experimental Procedures
Experimental procedures including details of cloning and of protein production, modification, purification, crystallographic and NMR spectroscopic analysis, structure calculation, and complementation test in E. coli are described in Supplemental Experimental Procedures.
Data deposition
The structure coordinates were deposited in the Protein Data Bank (www.pdb.org) under accession codes 2XHC (crystal structure of TmNusGFL), 2XHA (crystal structure of TmNusGDII) and 2LQ8 (solution structure of TmNusGNTD-CTD). Chemical shift assignments of TmNusGNTD-CTD were deposited in the BioMagResBank (BMRB) under the accession code 18298.
Supplementary Material
Highlights.
Thermotoga maritima NusG shows a dynamic intramolecular NTD-CTD interaction.
The NTD-CTD interaction hides the binding surfaces for RNA polymerase, S10 and Rho.
Domain interaction contributes to thermostability.
Thermotoga maritima NusG does not complement a NusG-deficient E. coli strain.
Acknowledgments
We are grateful to Elke Penka and Ramona Heiβmann for technical support and Homa Ghalei for help in diffraction data collection. We thank Gleb Bourenkov and Hans Bartunik for support at beamline BW6 (DESY, Hamburg, Germany). This work was supported by grants Ro 617/17-1 (PR) and WA1126/5-1 (MCW) from the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural Basis for Converting a General Transcription Factor into an Operon-Specific Virulence Regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin I, Muhlberger R, Bourenkov GP, Huber R, Bacher A, Richter G, Wahl MC. Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda. Proc Natl Acad Sci U S A. 2004;101:13762–13767. doi: 10.1073/pnas.0405883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Knauer SH, Sevostyana A, Schweimer K, Mooney RA, Landick R, Artsimovitch I, Rösch P. An α-helix to β-barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rösch P. A NusE:NusG Complex Links Transcription and Translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Scheckenhofer U, Rösch P. Domain interactions of the transcription:translation coupling factor E.coli NusG are intermolecular and transient. Biochem J. 2011;435:783–789. doi: 10.1042/BJ20101679. [DOI] [PubMed] [Google Scholar]
- Burns CM, Richardson JP. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci U S A. 1995;92:4738–4742. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burova E, Hung S, Sagitov V, Stitt B, Gottesman M. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Muralidharan V, Vila-Perello M, Raleigh DP, Muir TW, Palmer AG., 3rd Tuning protein autoinhibition by domain destabilization. Nat Struct Mol Biol. 2011;18:550–555. doi: 10.1038/nsmb.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann A, Schwarz S, Prasch S, Schweimer K, Rosch P. The E. coli NusA carboxy-terminal domains are structurally similar and show specific RNAP- and lambdaN interaction. Protein Sci. 2005;14:2018–29. doi: 10.1110/ps.051372205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Xu F, Yamada J, Egelhofer T, Gao Y, Hartzog GA, Teng M, Niu L. Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation. Structure. 2008;16:1649–1658. doi: 10.1016/j.str.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton JR, Bubunenko M, Andrykovitch M, Guo W, Routzahn KM, Waugh DS, Court DL, Ji X. A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by Site-Directed Mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Chrystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tsai CJ, Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- Li P, Martins IR, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat Struct Mol Biol. 2008;15:613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Lurz R, Dobrinski B, Dennis PP. A NusG-like protein from Thermotoga maritima binds to DNA and RNA. J Bacteriol. 1996;178:4089–4098. doi: 10.1128/jb.178.14.4089-4098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–75. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rösch P, Gottesman ME, Landick R. Two Structurally Independent Domains of E. coli NusG Create Regulatory Plasticity via Distinct Interactions with RNA Polymerase and Regulators. J Mol Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Pasman Z, von Hippel PH. Regulation of Rho-Dependent Transcription Termination by NusG Is Specific to the Escherichia coli Elongation Complex†. Biochemistry (N Y ) 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- Reay P, Yamasaki K, Terada T, Kuramitsu S, Shirouzu M, Yokoyama S. Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus. Proteins. 2004;56:40–51. doi: 10.1002/prot.20054. [DOI] [PubMed] [Google Scholar]
- Schweimer K, Prasch S, Santhanam SP, Bubunenko M, Gottesman ME, Rösch P. NusA interaction with the α-subunit of E. coli RNA Polymerase is via the UP-element site and releases autoinhibition. Structure. 2011;19:945–954. doi: 10.1016/j.str.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Nguyen HH, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain. Biochemistry. 2003;42:13429–37. doi: 10.1021/bi035118h. [DOI] [PubMed] [Google Scholar]
- Squires CL, Greenblatt J, Li J, Condon C, Squires CL. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci U S A. 1993;90:970–4. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Ward DF, Gottesman ME. Effect of Escherichia coli nusG function on lambda N-mediated transcription antitermination. J Bacteriol. 1992;174:1339–1344. doi: 10.1128/jb.174.4.1339-1344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M, Balada JM, Zellars M, Squires C, Squires CL. In vivo effect of NusB and NusG on rRNA transcription antitermination. J Bacteriol. 2004;186:1304–10. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel S, Martins BM, Rosch P, Wohrl BM. Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface. Biochem J. 2009;425:373–380. doi: 10.1042/BJ20091422. [DOI] [PubMed] [Google Scholar]
- Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Filter JJ, Court DL, Gottesman ME, Friedman DI. Requirement for NusG for transcription antitermination in vivo by the lambda N protein. J Bacteriol. 2002;184:3416–8. doi: 10.1128/JB.184.12.3416-3418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.