Abstract
A collection of 238 eggplant breeding lines, heritage varieties and selections within local landraces provenanced from Asia and the Mediterranean Basin was phenotyped with respect to key plant and fruit traits, and genotyped using 24 microsatellite loci distributed uniformly throughout the genome. STRUCTURE analysis based on the genotypic data identified two major sub-groups, which to a large extent mirrored the provenance of the entries. With the goal to identify true-breeding types, 38 of the entries were discarded on the basis of microsatellite-based residual heterozygosity, along with a further nine which were not phenotypically uniform. The remaining 191 entries were scored for a set of 19 fruit and plant traits in a replicated experimental field trial. The phenotypic data were subjected to principal component and hierarchical principal component analyses, allowing three major morphological groups to be identified. All three morphological groups were represented in both the “Occidental” and the “Oriental” germplasm, so the correlation between the phenotypic and the genotypic data sets was quite weak. The relevance of these results for evolutionary studies and the further improvement of eggplant are discussed. The population structure of the core set of germplasm shows that it can be used as a basis for an association mapping approach.
Introduction
Eggplant (Solanum melongena L.) belongs to the large Solanaceae family, which also includes a number of other significant crop species, in particular tomato, potato, sweet and hot peppers and tobacco. Unlike all of the latter, eggplant is an Old World species. Lester et al. [1] have suggested that the eggplant’s pre-domestication ancestor was the subtropical species S. incanum, a native of north Africa and West Asia which is being used in eggplant breeding programs as a source of variation for phenolics content and resistance to drought [2]; others have postulated that the ancestor was rather S. undatum [3], [4]. However, recent morphological and molecular work has shown that species-level differences exist between S. incanum and S. melongena while, on the basis of a new nomenclature, S. undatum and S.cumingii have been reclassified as S. insanum. The latter, distributed from India to SE Asia, and found also in Madagascar and Mauritius, is fully inter-fertile with S. melongena and is considered almost certainly its wild progenitor [2]. Sanskrit documents have revealed that the domestication of eggplant was achieved around 100–300 BCE and archaeological records, based on the analysis of microfossils starch grains, suggest that eggplant was present in the diet of inhabitants of the Indus valley during Harappan civilisation, thus Rajasthan may have been an area of domestication [5]. On the other hand the use of eggplant as a vegetable crop was described in Chinese literature dating to 59 BCE [6]. The crop spread westwards to Persia, was unknown by the ancient Greeks and Romans, and was introduced to the Mediterranean Basin by Muslim invaders in the 7th to 8th century CE [7].
The global production of eggplant is estimated to be around 46 Mt (http://faostat.fao.org); it represents an economically and nutritionally important crop in Asia and southern Europe. The bulk of production is concentrated in China, India, Iran, Egypt and Turkey, with Italy representing the most important European producer (http://faostat.fao.org). Eggplant is highly regarded as a source of antioxidants [8], in particular flavonoids and the phenolic chlorogenic acid [9], [10]. These compounds are present in both the fruit’s flesh and skin [11] and their content and profile are developmentally regulated during fruit ripening [12]. Fruit extracts have been shown to have anti-oxidant [13], hepato-protective [14], anti-carcinoma [15], anti-microbial, anti-LDL. anti-viral [16]–[18] and cardio-protective properties [19].
Selection and breeding over some hundreds of years has resulted in the elaboration of a large number of eggplant varieties. These are conventionally grouped as “Occidental” (preferred – grown in North Africa, Europe and the Americas) and “Oriental” eggplants (East and Southeast Asia). They vary from one another both with respect to their overall plant morphology and physiology, with their fruit size, color and shape being particularly distinctive. Fruit color can be cream, green, red, reddish-purple, dark purple or black, and some varieties produce fruit which is striped. Global trade is concentrated on an diminishing number of elite varieties [20]. These include F1 hybrids [7] which through their expression of heterosis for yield and their unique genetic status, have become extremely attractive for seed suppliers and breeders [21], [22]. As a result of the growing dominance of commercial hybrids, the genetic diversity of material in cultivation finds itself under some pressure; the conservation and characterization of germplasm is therefore becoming a priority, since this is exactly where the genetic variation necessary for future varietal improvement and for addressing future breeding challenges will be found [23].
A number of investigations aiming to characterize the phenotypic and genetic diversity of local collections of eggplant germplasm have been published in recent years [20], [24]–[27]. Hurtado et al. [28] have described both the phenotypic and DNA-based diversity present in a collection of entries sampled from three geographically well separated centers of diversity (China, Spain and Sri Lanka); their conclusion was that a combination of six plant traits was sufficient to assign the geographical origin of each entry, but that a similar level of discrimination was not possible using a set of 12 microsatellites; rather, the genotypic data suggested a measure of gene flow between the three centers of diversity. Furthermore, Meyer et al. [4], through historic and morphologic and molecular data based on nrITS sequences and AFLPs, made assumptions on phylogeographic relationships among candidate progenitors and Asian eggplant landraces and suggested a minimum of two domestications events which occurred in India and Southern China/SE Asia.
Here we describe a combined marker-based and morphological characterization of a wide set of “Occidental” and “Oriental” breeding lines, heritage varieties and selections from landraces. The objective was to assess the extent of genetic diversity that they contain, to illuminate the genetic relationship between “Occidental” and “Oriental” germplasm, and to provide criteria for the identification of a core germplasm collection. The genotypic data was represented by microsatellites, a class of genetic marker which thanks to its informativeness, reproducibility and co-dominant nature, has been widely employed for the analysis of plant genetic resources in many crops, including eggplant [22], [23], [25], [29].
Our results are of interest for conservation of genetic resources, their use in breeding programs, and contribute to the understanding of the evolutionary history of the species. Furthermore, in the context of our own research program, this data set sets the scene for an intended genotype/phenotype association study.
Methods
Permission
No specific permits were required for the described field studies, which took place in two experimental fields at the CRA-ORL in Montanaso Lombardo and CRA-ORA in Monsampolo del Tronto (Italy). These field plots were used by the authors of this paper affiliated to the fore mentioned institution (FC, LT, NA, TC, TS and GLR) for phenotypic characterization of the eggplant collection.
Germplasm and Genotyping
The set of 238 entries was composed of 94 “Oriental” (Eastern - EA) types, hailing from China, Indo-China (specifying the region when known i.e. Thailand or Myanmar), Indonesia, India and Japan, and 139 “Occidental” (Western - WE) ones from Italy, France, Spain, Turkey and North Africa (Table 1). Genomic DNA was extracted from 2 g fresh young leaf harvested from three randomly chosen plants of each entry, using an E.Z.N.A.T.M. Plant DNA mini kit (OMEGA bio-tek) according to the manufacturer’s protocol. The quality of each DNA sample was monitored by 0.8% agarose gel electrophoresis and its DNA concentration estimated spectrophotometrically (Beckman Coulter®, DU730). Each entry was then genotyped using a set of 24 microsatellite markers of known map location [30] and uniformly distributed across all 12 eggplant chromosomes (Table 2). Twenty-two were genomic SSRs [31]–[33]; while two (e.g. ecm001 and ecm023) were EST-SSRs [31]. PCR amplification was performed according to [29], and successful amplicons were separated by denaturing 6% polyacrylamide gel electrophoresis on a LI-COR Gene ReadIR 4200 device, as described by Barchi et al. [30].
Table 1. The set of germplasm used for genotypic and phenotypic characterization.
| ID | Accession Name | Origin | Areal | Structure subpopulations | Morphological grups |
| AM_001 | Dadali | Indonesia | EA | B | 1 |
| AM_004 | Cima viola | Italy | WE | A | 1 |
| AM_005 | Bianca ovale | Italy | WE | B | 2 |
| AM_010 | 1F5 (9) | Breeding line | WE | A | 2 |
| AM_011 | Bianca Sicilia | Italy | WE | B | 2 |
| AM_013 | CCR3 | Breeding line | WE | A | 1 |
| AM_014 | Mel | Italy | WE | B | 2 |
| AM_015 | Luga 063 | Italy | WE | A | 1 |
| AM_016 | Prosperosa | Italy | WE | B | 3 |
| AM_018 | Lunga Violetta Cinese | China | EA | B | 1 |
| AM_021 | Tal 1/1 | Italy | WE | A | 1 |
| AM_022 | Angiò 4 | China | EA | B | 1 |
| AM_023 | BLK 1269 | Breeding line | WE | Admixtured | 2 |
| AM_024 | GIC/27-9 | Breeding line | WE | A | 2 |
| AM_025 | Tina | Italy | WE | A | 1 |
| AM_026 | DR2 | Italy | WE | A | 1 |
| AM_027 | TBE80 D | Breeding line | WE | - | - |
| AM_028 | TBE84 D | Breeding line | WE | A | 2 |
| AM_029 | FanE13 D | Breeding line | WE | A | 2 |
| AM_030 | FanE27 D | Breeding line | WE | A | 2 |
| AM_031 | FanE63 D | Breeding line | WE | A | 2 |
| AM_032 | SNL 534-11 | India | EA | B | 3 |
| AM_033 | SNL 533-8 | India | EA | B | 3 |
| AM_034 | SNL 600-1 | India | EA | B | 2 |
| AM_035 | Cin 01/24-6 | China | EA | B | 2 |
| AM_036 | Viola Cin-A-1 | China | EA | B | 2 |
| AM_037 | Violetta di toscana | Italy | WE | B | 3 |
| AM_038 | Bellezza nera | Italy | WE | A | 2 |
| AM_040 | Violetta di Metaponto | Italy | WE | B | 3 |
| AM_041 | 28-08/3 (23-09) | Breeding line | WE | B | 3 |
| AM_042 | 31-08/4 (25-09) | Breeding line | WE | B | 3 |
| AM_043 | 51-08/4 (29-09) | Breeding line | WE | B | 3 |
| AM_044 | 52-08/4 (30-09) | Breeding line | WE | B | 3 |
| AM_045 | 55-08/5 (31-09) | Breeding line | WE | B | 3 |
| AM_046 | 16-set | Breeding line | WE | B | 3 |
| AM_047 | P621-08 | Breeding line | WE | B | 3 |
| AM_048 | P623-08 | Breeding line | WE | B | 3 |
| AM_049 | P645-08 | Breeding line | WE | B | 3 |
| AM_050 | P649-08 | Breeding line | WE | B | 3 |
| AM_051 | P612-08 | Breeding line | WExEA | B | 3 |
| AM_052 | P390 | Breeding line | WExEA | B | 3 |
| AM_053 | P328 | Breeding line | WExEA | B | 3 |
| AM_054 | P656-08 | Breeding line | WE | B | 3 |
| AM_055 | msp 73-08 | Breeding line | WE | A | 2 |
| AM_056 | S 1052-08 | Breeding line | WE | A | 1 |
| AM_057 | LI324/06 | Italy | WE | A | 1 |
| AM_058 | msp 36-08 | Italy | WE | A | 1 |
| AM_059 | msp 42-08 | Italy | WE | A | 1 |
| AM_060 | msp 30-08 | Italy | WE | A | 1 |
| AM_062 | msp 55-08 | Italy | WE | A | 1 |
| AM_063 | L422-08 | Italy | WE | A | 1 |
| AM_064 | L717-289 | Italy | WE | A | 1 |
| AM_067 | Uga | Italy | WE | A | 2 |
| AM_068 | Tana | Italy | WE | A | 1 |
| AM_069 | Bin 6 | Italy | WE | A | 2 |
| AM_070 | Floralba | Italy | WE | A | 2 |
| AM_071 | Ind Min | India | EA | B | 1 |
| AM_072 | SM5/2 | Breeding line | WE | A | 2 |
| AM_073 | SM5/13 | Breeding line | WE | A | 2 |
| AM_074 | SM5/22 | Breeding line | WE | A | 1 |
| AM_076 | S.Nicandro | Italy | WE | Admixtured | 2 |
| AM_085 | LS96 | Japan | EA | - | - |
| AM_086 | LS 3805 minden | Japan | EA | B | 2 |
| AM_098 | CIN6 | China | EA | B | 3 |
| AM_099 | CIN5 | China | EA | B | 3 |
| AM_100 | CIN7 | China | EA | B | 3 |
| AM_102 | CIN9 | China | EA | B | 3 |
| AM_103 | LS611 | Japan | EA | B | 2 |
| AM_106 | Naga-Ungu | Indonesia | EA | B | 1 |
| AM_107 | N 286-1 | India | EA | - | - |
| AM_109 | N 285-B | India | EA | - | - |
| AM_110 | N 24-6 | India | EA | - | - |
| AM_111 | N 243-3 | India | EA | - | - |
| AM_112 | N 286-A | India | EA | - | - |
| AM_113 | N 321-14 | India | EA | - | - |
| AM_114 | N 258-4 | India | EA | B | 1 |
| AM_116 | N 220-A | India | EA | - | - |
| AM_119 | N 324-A | India | EA | - | - |
| AM_121 | Indom melanz | Indonesia | EA | B | 2 |
| AM_123 | Pusa kranti | India | EA | - | - |
| AM_124 | PI17 | Italy | WE | A | 2 |
| AM_126 | Almagro | Spain | WE | Admixtured | 2 |
| AM_127 | Larga negra | Spain | WE | A | 1 |
| AM_128 | Listada | Spain | WE | A | 2 |
| AM_129 | Tolga | Algeria | WE | A | 2 |
| AM_133 | Black Beauty | Italy | WE | A | 2 |
| AM_134 | Viserba | Italy | WE | A | 1 |
| AM_135 | Black Beauty | Italy | WE | A | 2 |
| AM_136 | Tonda Violetta Firenze | Italy | WE | B | 2 |
| AM_137 | Violetta Lunga Romagna | Italy | WE | A | 1 |
| AM_138 | Barbentane | France | WE | A | 1 |
| AM_139 | Lunga Marina | Italy | WE | A | 1 |
| AM_140 | Tonda di Valence | France | WE | B | 3 |
| AM_141 | Lunga Violetta Scura Cannellina | Italy | WE | A | 1 |
| AM_142 | Tonda Black Beauty | Italy | WE | A | 2 |
| AM_143 | Bellezza Nera | Italy | WE | A | 2 |
| AM_144 | Lunga Violetta Napoli | Italy | WE | A | 1 |
| AM_145 | Bianca Ovale | Italy | WE | - | - |
| AM_146 | Black Beauty | Italy | WE | A | 2 |
| AM_147 | Violetta Mostruosa NY | Italy | WE | A | 2 |
| AM_148 | Slim Jim | India | EA | B | 1 |
| AM_149 | Tonda Violetta Scura Valence | France | WE | Admixtured | 3 |
| AM_150 | Grossissima Violetta Firenze | Italy | WE | B | 2 |
| AM_151 | Violetta Lunga | Italy | WE | A | 1 |
| AM_152 | Tonda Bianca | Italy | WE | B | 2 |
| AM_153 | Prospera | Italy | WE | A | 3 |
| AM_155 | Daejang | China | EA | B | 1 |
| AM_156 | Buia | Italy | WE | A | 2 |
| AM_157 | Baffa | Italy | WE | A | 2 |
| AM_158 | Ank2 | India | EA | A | 2 |
| AM_159 | CN2 | China | EA | B | 3 |
| AM_160 | Dourga | France | WE | A | 2 |
| AM_162 | Tunisia Baharia | Italy | WE | B | 3 |
| AM_163 | Pusa Purple Cluster | India | EA | B | 1 |
| AM_167 | Angio 3 | China | EA | B | 1 |
| AM_168 | Angio 5 | China | EA | B | 2 |
| AM_169 | Bianca striata verde | Italy | WE | B | 2 |
| AM_170 | SM19/14 | Breeding line | WE | A | 2 |
| AM_171 | Palermitana | Italy | WE | B | 3 |
| AM_172 | 97-3 Drago volante | China | EA | - | - |
| AM_173 | Pusa Purple Long | India | EA | B | 1 |
| AM_174 | JM (Slim Jim) | India | EA | B | 1 |
| AM_175 | Cannellina Sarnense | Italy | WE | A | 1 |
| AM_176 | Sita | Italy | WE | B | 2 |
| AM_177 | FiL withe | Turchia | WE | B | 2 |
| AM_178 | Lunga napoli | Italy | WE | A | 1 |
| AM_179 | 1237/06 | Italy | WE | A | 1 |
| AM_180 | Listada Tacconi | Italy | WE | A | 2 |
| AM_181 | Suraj(143) | India | EA | Admixtured | 2 |
| AM_182 | Pusa Round | India | EA | B | 2 |
| AM_183 | Chaojiuye Yuanquie | China | EA | B | 3 |
| AM_184 | He Shanwang | China | EA | B | 3 |
| AM_185 | Pp | Indochinese Region | EA | B | 3 |
| AM_186 | Zf | Italy | WE | - | - |
| AM_187 | Naveen | India | EA | Admixtured | 2 |
| AM_188 | TAI 444 | Indochinese Region | EA | B | 2 |
| AM_189 | TAI 445 | Indochinese Region | EA | B | 1 |
| AM_190 | TAI 446 | Indochinese Region | EA | Admixtured | 1 |
| AM_191 | TAI 449 | China | EA | B | 3 |
| AM_192 | TAI 452 | Indochinese Region | EA | - | - |
| AM_193 | TAI 453 | Indochinese Region | EA | A | 2 |
| AM_194 | TAI 455 | Thailand | EA | B | 2 |
| AM_195 | TAI 456 | Myanmar | EA | B | 2 |
| AM_196 | TAI 457 | India | EA | Admixtured | 2 |
| AM_197 | TH 4438 Yad thip | Thailand | EA | B | 2 |
| AM_198 | TH 6413 Raos | Indonesia | EA | B | 1 |
| AM_199 | TAI 470 | Thailand | EA | B | 2 |
| AM_200 | TAI 475 | Thailand | EA | B | 2 |
| AM_201 | TAI 477 | Thailand | EA | Admixtured | 2 |
| AM_202 | TAI 480 | India | EA | B | 2 |
| AM_203 | TAI 481 | China | EA | B | 1 |
| AM_204 | TAI 483 | India | EA | Admixtured | 1 |
| AM_205 | TAI 484 | India | EA | Admixtured | 1 |
| AM_206 | 7 CN | China | EA | B | 3 |
| AM_207 | 9 CN | China | EA | B | 3 |
| AM_208 | 17 CN | China | EA | B | 1 |
| AM_209 | 19 CN | China | EA | Admixtured | - |
| AM_210 | 67-3 | Breeding line | WExEA | B | 3 |
| AM_211 | 305 E40 | Breeding line | WE | A | 1 |
| AM_212 | CGN17464 (PI 176759) | Turkey | WE | A | 2 |
| AM_213 | CGN23345 (PI 169641) | Turkey | WE | A | 2 |
| AM_214 | CGN18783 (Croisette) | France | WE | B | 1 |
| AM_215 | CGN18531 (Patchem) | Turkey | WE | B | 1 |
| AM_216 | CGN18510 (Bostan selection; PI 169666) | Turkey | WE | A | - |
| AM_217 | CGN17449 (Topak; PI 175917) | Turkey | WE | A | 2 |
| AM_218 | CGN17451 (Dolmalik; PI 176758) | Turkey | WE | A | 2 |
| AM_219 | CGN17571 (PI 169641) | Turkey | WE | - | - |
| AM_220 | CGN17574 (PI 169643) | Turkey | WE | - | - |
| AM_221 | CGN17579 (PI 169648) | Turkey | WE | A | - |
| AM_222 | CGN23346 (Topatan; PI 169649) | Turkey | WE | Admixtured | 2 |
| AM_223 | CGN23347 (PI 169650) | Turkey | WE | - | - |
| AM_224 | CGN17581 (PI 169651) | Turkey | WE | A | 1 |
| AM_225 | CGN17564 (PI 166994) | Turkey | WE | A | - |
| AM_226 | CGN23341 (Kemer Patlican; PI 167101) | Turkey | WE | A | - |
| AM_227 | CGN23342 (Patlican; PI 167209) | Turkey | WE | A | - |
| AM_228 | CGN23343 (PI 167328) | Turkey | WE | A | 1 |
| AM_229 | CGN17568 (Yuvorlak Patlican; PI 167373) | Turkey | WE | - | - |
| AM_230 | CGN23344 (Bostan; PI 169639) | Turkey | WE | A | 2 |
| AM_231 | CGN18591 (PI 171847) | Turkey | WE | A | 2 |
| AM_232 | CGN18595 (PI 171852) | Turkey | WE | A | 1 |
| AM_233 | CGN18779 (De Barbentane) | France | WE | A | 1 |
| AM_234 | CGN23309 (Dolg; PI 358232) | Macedonia | WE | A | 1 |
| AM_235 | CGN18484 (Morska Pata; PI 358242) | Macedonia | WE | B | 1 |
| AM_236 | CGN18782 (Violette Longue Hative) | France | WE | A | 1 |
| AM_237 | CGN18512 (Violette Noire d`Orient) | France | WE | - | - |
| AM_238 | CGN17453 (Yesilkoy 27) | Turkey | WE | A | 1 |
| AM_239 | CGN17582 (Alacali; PI 169652) | Turkey | WE | - | - |
| AM_240 | CGN18578 (Kemer; PI 169655) | Turkey | WE | Admixtured | - |
| AM_241 | CGN23348 (PI 169658) | Turkey | WE | A | 1 |
| AM_242 | CGN18581 (PI 169660) | Turkey | WE | Admixtured | - |
| AM_243 | CGN18585 (PI 169663) | Turkey | WE | A | 1 |
| AM_244 | CGN23349 (PI 169663) | Turkey | WE | - | - |
| AM_245 | CGN18601 (PI 173106) | Turkey | WE | - | - |
| AM_246 | CGN18602 (PI 173107) | Turkey | WE | - | - |
| AM_247 | CGN18605 (PI 173807) | Turkey | WE | - | - |
| AM_248 | CGN18610 (Bostan; PI 174361) | Turkey | WE | Admixtured | - |
| AM_249 | CGN23351 (PI 174362) | Turkey | WE | A | 2 |
| AM_250 | CGN23352 (PI 174367) | Turkey | WE | - | - |
| AM_251 | CGN24467 (Berenjena Listada) | Spain | WE | A | 2 |
| AM_252 | CGN18505 (Berenjena Redonda) | Spain | WE | Admixtured | 3 |
| AM_253 | CGN24468 (Caminal) | France | WE | A | 1 |
| AM_254 | CGN17472 (Redonda Negra Lisa) | Spain | WE | - | - |
| AM_255 | CGN23318 (Larga Negra) | Spain | WE | - | - |
| AM_256 | CGN18511 (Indonesische Aubergine) | Indonesia | EA | - | - |
| AM_257 | CGN18776 (Longue Hative) | France | WE | A | 1 |
| AM_258 | CGN17456 (Monda) | France | WE | Admixtured | 2 |
| AM_259 | CGN23315 (Ronde de Valence) | France | WE | Admixtured | 3 |
| AM_260 | CGN17479 (Semiredonda Jaspeada) | Spain | WE | A | 2 |
| AM_261 | CGN23323 (PI 120770) | Turkey | WE | - | - |
| AM_262 | CGN23772 | Nigeria | WE | Admixtured | 2 |
| AM_263 | Violetta precoce | Italy | WE | Admixtured | - |
| AM_264 | Mezza Lunga Violetta | Italy | WE | A | 1 |
| AM_265 | Lunghissima Precoce Violetta | Italy | WE | A | 1 |
| AM_266 | Dingaras | China | EA | B | 1 |
| AM_267 | Bioleta | Spain | WE | - | - |
| AM_268 | L 129 | Indonesia | EA | B | 1 |
| AM_269 | Talindo | Indonesia | EA | B | 1 |
| AM_271 | DS1 | Breeding line | WE | A | 2 |
| AM_273 | DS2 | Breeding line | WE | A | 2 |
| AM_274 | DS4 | Breeding line | WE | A | 2 |
| AM_275 | 1 CAAS | China | EA | B | 3 |
| AM_276 | 2 CAAS | China | EA | - | - |
| AM_277 | 3 CAAS | China | EA | - | - |
| AM_278 | 4 CAAS | China | EA | B | 3 |
| AM_279 | 5 CAAS | China | EA | B | 3 |
| AM_280 | 6 CAAS | China | EA | - | - |
| AM_281 | 7 CAAS | China | EA | - | - |
| AM_282 | 8 CAAS | China | EA | - | - |
| AM_283 | 9 CAAS | China | EA | - | - |
| AM_284 | 10 CAAS | China | EA | A | 1 |
| AM_285 | 11 CAAS | China | EA | B | 1 |
| AM_287 | 13 CAAS | China | EA | - | - |
| AM_288 | 14 CAAS | China | EA | A | 2 |
| AM_289 | 15 CAAS | China | EA | B | 3 |
| AM_290 | 16 CAAS | China | EA | B | 3 |
| AM_291 | 17 CAAS | China | EA | B | 3 |
| AM_292 | 18 CAAS | China | EA | B | 1 |
| AM_293 | 19 CAAS | China | EA | B | 1 |
Those shown in italics refer to entries retaining a level of heterozygosity >10%, and those shown underlined produced off-types with respect to plant and/or fruit type. Retained entries are shown in bold.
Table 2. The 24 microsatellite loci used for genotyping.
| Marker | Chromosome | Position (cM) | Alleles | Rare alleles | PIC |
| CSM 31 | E01 | 107.4 | 12 | 2 | 0.83 |
| ecm001 | E01 | 77.7 | 7 | 2 | 0.73 |
| emh21J12 | E01 | 91.8 | 11 | 5 | 0.76 |
| emf01G17 | E02 | 35.4 | 10 | 5 | 0.65 |
| EM 133 | E02 | 11.6 | 6 | 4 | 0.24 |
| emg11I03 | E03 | 6.0 | 6 | 2 | 0.77 |
| emj03A17 | E03 | 34.3 | 3 | 1 | 0.38 |
| emf01K16 | E04 | 0.0 | 4 | 0 | 0.63 |
| EM 117 | E04 | 47.6 | 6 | 1 | 0.76 |
| emf01A06 | E05 | 64.3 | 4 | 1 | 0.45 |
| EM 146 | E05 | 50.6 | 7 | 2 | 0.68 |
| CSM 7 | E06 | 35.6 | 3 | 0 | 0.48 |
| CSM 19 | E07 | 0.0 | 4 | 1 | 0.55 |
| CSM 69 | E07 | 73.4 | 2 | 0 | 0.48 |
| ecm023 | E08 | 13.9 | 2 | 0 | 0.35 |
| emi03M03 | E08 | 13.5 | 3 | 0 | 0.45 |
| CSM 54 | E09 | 16.1 | 7 | 1 | 0.66 |
| eme03B08 | E10 | 46.5 | 6 | 0 | 0.74 |
| emf11F07 | E10 | 53.7 | 7 | 1 | 0.75 |
| emf21K08 | E11 | 0.0 | 9 | 3 | 0.63 |
| EM 080 | E11 | 25.6 | 2 | 0 | 0.35 |
| CSM 29 | E12 | 82.3 | 6 | 1 | 0.67 |
| CSM 73 | E12 | 0.0 | 5 | 1 | 0.69 |
| emb01O01 | E12 | 43.9 | 8 | 1 | 0.77 |
| total | 140 | 34 | 0.60 |
Phenotypic Characterization
The entries were each scored for 19 plant, leaf, flower and fruit traits (Table 3), included among the European Cooperative Program for Plant Genetic Resource Solanaceae and/or the International Board for Plant Genetic Resources eggplant descriptors. Peel color was measured using a Chroma-meter Minolta CR-400 on the basis of the three Hunter color coordinates (L*, a* and b*), and represented the average of three randomly chosen portions of each fruit. The measurements were reduced to a single variate by calculating the Euclidean distance from white (L* = 100, a* = 0, b* = 0), following Prohens et al. [34].
Table 3. Traits analysed to generate the phenotypic data set.
| Trait | Code | Evaluation method |
| Peel color | pcol | L*a*b* color coordinates |
| Peel glossiness | pglo | Scale from 0 (high opacity) to 3 (high glossiness) |
| Fruit curvature | fcur | Scale: 1 (no curvature), 5 (curved), 9 (U shaped) |
| Fruit weight | fw | Grams |
| Fruit length | fl | Centimeters (from the base of the calyx to the tip of the fruit) |
| Fruit diameter max | fdmax | Centimeters |
| Fruit diameter max position | fdmaxp | Scale from 1 (close to the calyx) to 8 (close to the apex) |
| Fruit shape | fs | fl/frdmax |
| Flesh fruit firmness | firm | Scale from 1 (very loose) to 9 (very dense) |
| Leaf hairiness | lha | Scale from 0 (no hairiness) to 5 (highly hairiness) |
| Adaxial leaf lamina anthocyanin | adlan | Scale from 0 (green) to 5 (complete purple coloration) |
| Stem anthocyanin | stean | Scale from 0 (green) to 5 (complete purple coloration) |
| Calyx coverage of the fruit | cacov | Scale from 1 (<10% of the fruit length) to 5 (>50%) |
| Fruit calix prickliness | fcpri | Scale from 0 (no prickles) to 9 (high prickliness) |
| Leaf prickliness | lepri | Scale from 0 (no prickles) to 5 (high prickliness) |
| Plant growth habit | hab | Scale from 1 (upright ) to 9 (prostrate) |
| Inflorescence flowers | inflw | Number of flowers for inflorescence |
| Flowering abundance | flwab | Number of flowers on the plant, scale from 1 (very few) to 5 (many) |
| Flowering time | flwt | Number of days from seedling emergence after sowing when at least 50% of the plants have its first flower opened |
The germplasm was grown in two locations: Montanaso Lombardo [ML]: 45 20′N, 9 26′E, and Monsampolo del Tronto [MT]: 42 53′N; 13 47′E in each of 2010 and 2011. For each field experiment, six plants per entry were planted in two completely randomized blocks with a 1 m inter-row and a 0.8 m inter-plant within row distance. Standard crop management practices were applied.
Analysis of Marker Data
The scoring of microsatellite data was imported into Past 2.08 software [35]. and pair-wise similarity coefficients [36] were computed. Alleles occurring at a frequency ≤1% were considered as rare. A principal co-ordinate (PCO) analysis was carried out to display the multi-dimensional relationships between entries. The polymorphic information content (PIC) of each microsatellite locus was evaluated by applying the following equation, as suggested by Anderson [37]: PIC = 1-∑ P 2 ij, where P ij represented the frequency of the j th allele at the i th microsatellite locus and the summation was extended over n alleles. The Bayesian-based model procedure implemented by STRUCTURE 2.3 software [38] was used to determine population structure; K values from 1 to 15 were tested. A burn-in period of 50,000 and 100,000 rounds from ten independent simulations were used to assess the population structure. The most likely number of sub-groups present was based on minimizing ΔK [39]. Population structure was also characterized using the fixation index statistics provided within the STRUCTURE 2.3 package. To identify the minimum number of entries required to retain 100% of the allelic diversity present in the full germplasm set, the M strategy suggested by Schoen and Brown [40], as implemented in the MSTRAT software [41], was used. The number of iterations per MSTRAT run was 30, and the number of repetitions for core sampling was 20. The entries most frequently represented across the 30 replicates formed the core collection. The efficiency of the strategy was assessed by comparing the total number of alleles captured using MSTRAT in samples of increasing size to the number of alleles captured in randomly chosen collections of equal size (ten independent samplings).
Analysis of Morphological Data
The morphological data were treated as adjusted entry means (best linear unbiased predictors, BLUPs). The variance components were determined using the restricted maximum likehood (REML) method applying the mixed linear model pijsb = lj+ys+gi+rbjr+eijs, where pijsb was the phenotypic value of the bth replicate of ith entry at the jth location in the sth year, lj the contribution of the jth location, ys the contribution of the sth year, gi the contribution of the ith entry, rbjs the contribution of the bth replicate within the jth location in the sth year, and eijs the residual error. A principal component analysis (PCA) was carried out to determine which traits acted as the prime discriminators between entries. Common components coefficients, eigenvalues and the proportion of the total variance expressed by each single trait were calculated. The Scree plot was used to select the components most relevant for the ordination analysis. Correlations between traits and each principal component were calculated, and those ones having an absolute value >0.5 were considered relevant for the trait’s determination [42]. An hierarchical clustering on principal components (HCPC) analysis was performed to define a set of clusters based on phenotypic traits. The cluster analysis was performed only on the most significant PCA components, with the remaining minor ones considered to represent noise [43]. Only dimensions having an eigenvalue >1 (Kaiser’s method) were considered. The hierarchical clustering was performed according to the Ward criterion, based on variance evaluation (inertia) as well as on the principal component method. In order to define the appropriate number of clusters, both the overall shape of the tree and the bar plot of the gain in inertia were considered. The presence of a difference between the clusters for each trait was tested using a Kruskal-Wallis analysis of variance, and a Nemenyi post hoc test was performed on traits displaying differences to identify which groups were involved. The above analyses were implemented with R software [44]. A co-phenetic correlation between the genotypic and phenotypic data matrices was calculated, and tested using the Mantel [45] method, including 5,000 permutation as implemented in Past 2.08 software [35].
Results
Microsatellite Diversity
Across the set of 238 eggplant entries, 140 alleles were identified at the 24 microsatellite loci (average 5.8 per locus) (Table 2), and each entry had a distinct genotype. The loci varied in terms of the number of alleles present from two (EM 080, ecm023 and CSM 69) to 12 (CSM 31), while their PICs ranged from 0.24 (EM 133) to 0.83 (CSM 31), with a mean of 0.60. There were 34 rare alleles, of which 14 were only found in the “Oriental” germplasm and the other 20 only in the “Occidental” germplasm. A residual level of heterozygosity >10% was present in 38 entries, and as a result, these entries were not considered for phenotyping (Table 1). The average Dice similarity coefficient for the 200 fixed lines was 0.32.
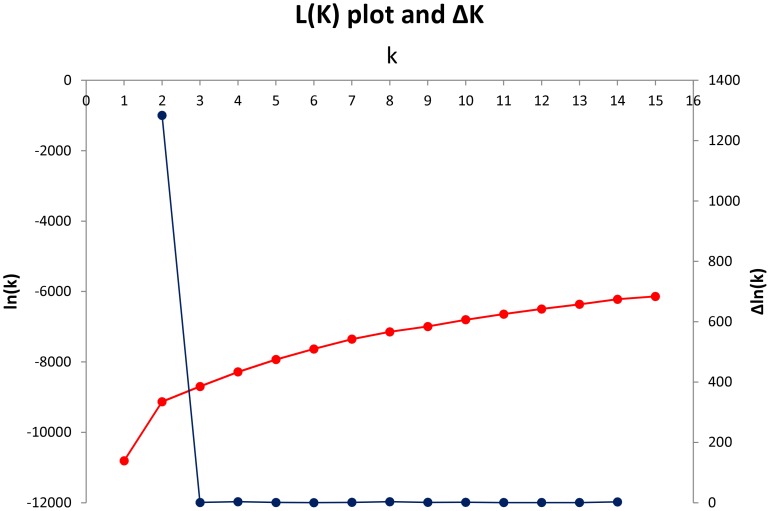
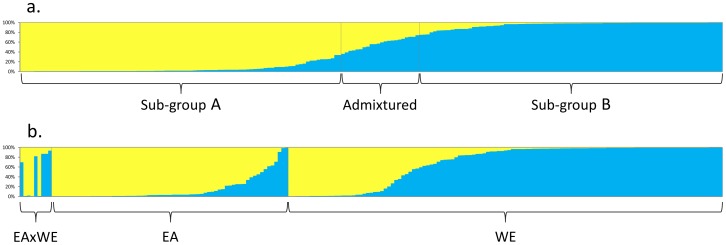
STRUCTURE analysis with different K-levels (1–15) were assayed and K value for 2 was optimal (Figure 1). According to output of structure analysis (Figure 2a) each accession was assigned to a sub-group (A or B) when its level of membership was higher than 70% (Table 1). Sub-group A comprised 89 entries and sub-group B 90 entries, with the remaining 21 defined as admixed. The fixation index was 0.30 for sub-group A and 0.18 for sub-group B, indicating that a certain amount of structuration was still present within each of them. Applying the M method showed that the minimal set sufficient to capture all 106 non-rare alleles was 16 (“sub-16”), while the size of set required to capture all 140 alleles was 48 (“sub-48”). Random sampling was less efficient at retaining alleles, since randomly chosen sets of 16 entries captured only 96.5 alleles on average, and randomly chosen sets of 48 only 109.3 alleles.
Figure 1. STRUCTURE analysis.
(K) and ΔK plots derived from the genotypic data. The germplasm set forms two distinct sub-groups, with a small number of entries being intermediate.
Figure 2. STRUCTURE output at K = 2.
Each entry is represented by a horizontal line representing subgroup 1 (yellow) and subgroup 2 (blue). a) Entries ordered according to their subgroup membership. b) Entries ordered according to their geographical origin: WE: “Occidental”, EA: “Oriental”.
Morphological Variation
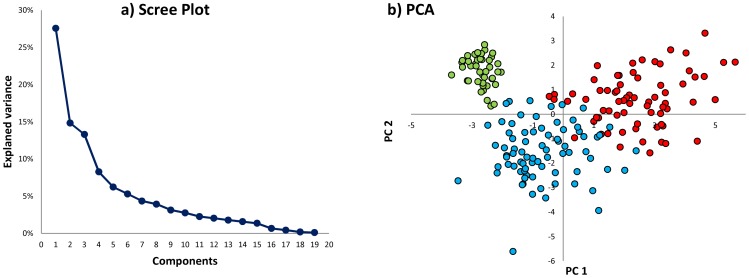
Among the 200 fixed lines, off types with respect to plant and/or fruit traits were present in nine, so the full phenotyping set was further reduced to 191 entries (Table 1). The phenotypic performance of these entries is reported in Table 4. The most variable traits were fruit size, weight, shape and curvature, along with peel color. The PCA scree plot showed that 55.7% of the overall phenotypic variation was captured by the first three principal components (PC’s) (Figure 3a). The correlation coefficients for each trait with each of these three PC’s, along with the associated eigenvalues and proportions of the total variance explained, are detailed in Table 5. The first PC explained 27.6% of the variance and was positively correlated with fruit length (+0.89), shape (+0.92) and curvature (+0.89), as well as the distance of the widest part of the fruit from the fruit apex (+0.76); it was simultaneously negatively correlated with the maximum diameter of the fruit (−0.91), fruit weight (−0.76) and flesh firmness (−0.74). PC2 explained 14.8% of the variance, and was positively correlated with the anthocyanin content of the stem (+0.86) and leaf (+0.76), and with the intensity of the peel color (+0.52). PC3 explained 13.3% of the variance, and was positively correlated with late flowering (+0.55) and negatively with flowering abundance (−0.71) and the presence of a prostrate growth habit (−0.51). The subsequent HCPC analysis was based on the leading six PC’s (with eigenvalues >1), which together explained 75.4% of the variance. Three main morphological groups were identified (Figure 3b) and the differences between these groups are detailed in Table 4. Entries belonging to the group 1 (Figure 3b, area I/II) produced long, light (average weight ∼150 g) and curved fruits, the flesh of which was of only limited firmness and the peel was purple; the anthocyanin content of both the leaves and stems was intermediate, plant habit was erect and the plants formed many flowers per inflorescence. The entries within group 2 (Figure 3b, area II/III) produced oblong-shaped fruits of average weight of ∼250 g; peel color was white, green or light violet, the plants were semi-erect and the leaves and stems contained little anthocyanin. Finally, group 3 entries (Figure 3b, area IV) produced rounded, heavy (average weight ∼400 g) and dark purple colored fruits; calyx and leaf prickliness was largely absent, the anthocyanin content of both the leaves and stems was high and the number of flowers per inflorescence was low. Examples of fruits belonging to the three morphological groups are reported in Figure 4.
Table 4. The distribution of trait-by-trait performance across the 191entries phenotyped (the ones not showing residual heterozygosity as well as phenotypic variation), and the statistical significance of the three morphology-based groups identified.
| Trait code | Average | St. Dev. | ANOVAbetween groups | Post hoc test | Group average | ||||
| 1 vs 2 | 1 vs 3 | 2 vs 3 | 1 | 2 | 3 | ||||
| pcol | 74.57 | 15.60 | *** | NS | NS | *** | 79.89 | 69.21 | 81.61 |
| pglo | 2.37 | 0.73 | *** | NS | ** | *** | 2.36 | 2.19 | 2.76 |
| fcur | 2.08 | 1.38 | *** | *** | *** | NS | 3.95 | 1.61 | 1.03 |
| fw | 256.97 | 122.26 | *** | *** | *** | *** | 148.28 | 243.99 | 395.11 |
| fl | 14.39 | 5.53 | *** | *** | *** | ** | 21.43 | 13.16 | 9.32 |
| fdmax | 7.39 | 2.59 | *** | *** | *** | *** | 4.53 | 7.28 | 10.74 |
| fdmaxp | 5.84 | 0.72 | *** | ** | *** | *** | 6.48 | 5.84 | 5.05 |
| fs | 2.49 | 1.88 | *** | *** | *** | *** | 5.03 | 1.95 | 0.88 |
| firm | 5.33 | 1.62 | *** | ** | *** | *** | 3.58 | 5.72 | 6.38 |
| lha | 1.91 | 1.15 | NS | NS | NS | NS | 1.58 | 2.05 | 1.98 |
| adlan | 1.37 | 1.49 | *** | *** | ** | *** | 1.79 | 0.42 | 3.13 |
| stean | 2.80 | 1.84 | *** | *** | ** | *** | 3.59 | 1.53 | 4.95 |
| cacov | 2.44 | 0.74 | NS | NS | NS | NS | 2.29 | 2.52 | 2.28 |
| fcpri | 1.65 | 1.83 | NS | NS | NS | NS | 1.35 | 1.88 | 1.16 |
| lepri | 0.43 | 0.65 | *** | NS | * | * | 0.46 | 0.51 | 0.20 |
| hab | 3.83 | 1.62 | *** | *** | NS | *** | 2.94 | 4.47 | 3.41 |
| inflw | 1.94 | 0.95 | *** | NS | * | *** | 2.20 | 1.99 | 1.52 |
| flwab | 2.24 | 0.76 | NS | NS | NS | NS | 2.25 | 2.25 | 2.32 |
| flwt | 86.43 | 5.97 | NS | NS | NS | NS | 85.90 | 86.48 | 85.98 |
P<0.05
P<0.01
P<0.001
Figure 3. HCPC analysis, based on the leading six PC’s (eigenvalues >1).
a) Scree plot showing the proportion of variance explained by each PC. b) PCA based on the leading two PC’s. Entries belonging to each morphological group marked by a different color (red: group 1, blue: group 2, green: group 3).
Table 5. Correlation coefficients between each trait and the leading three PC’s, along with the associated eigenvalues and proportions of the overall variance explained.
| Trait code | Common principal component coefficient | ||
| First | Second | Third | |
| pcol | −0.01 | 0.52 | 0.45 |
| pglo | −0.14 | 0.34 | 0.49 |
| fcur | 0.89 | 0.20 | 0.12 |
| fw | −0.76 | 0.09 | 0.44 |
| fl | 0.89 | 0.06 | 0.30 |
| fdmax | −0.91 | 0.11 | 0.23 |
| fdmaxp | 0.76 | −0.19 | 0.20 |
| fs | 0.92 | 0.15 | 0.08 |
| firm | −0.74 | −0.10 | −0.09 |
| lha | −0.28 | −0.20 | 0.04 |
| adlan | −0.22 | 0.76 | −0.05 |
| stean | −0.14 | 0.86 | 0.03 |
| cacov | −0.05 | −0.44 | 0.42 |
| fcpri | −0.09 | −0.50 | 0.42 |
| lepri | 0.04 | −0.48 | 0.30 |
| hab | −0.24 | −0.44 | −0.51 |
| inflw | 0.20 | −0.09 | −0.39 |
| flwab | 0.01 | 0.15 | −0.71 |
| flwt | −0.05 | −0.16 | 0.55 |
| Eigenvalue | 5.24 | 2.82 | 2.53 |
| Variability % | 27.6% | 14.8% | 13.3% |
| Accumulated variability % | 27.6% | 42.4% | 55.7% |
Figure 4. Fruits of accessions belonging to the three main morphological groups.
Group 1∶1a = AM 269-Talindo; 1b = AM 026-Dr2; Group 2∶2a = AM 168-Angio 5; 2b = AM 031-FantE63D; 2c = AM 160-Dourga; Group 3∶3a = AM 037-Violetta di Toscana; 3b = AM 291∶17CAAS; 3c = AM 210-67/3.
The Relationship between Phenotype, Genotype and Geographical Origin
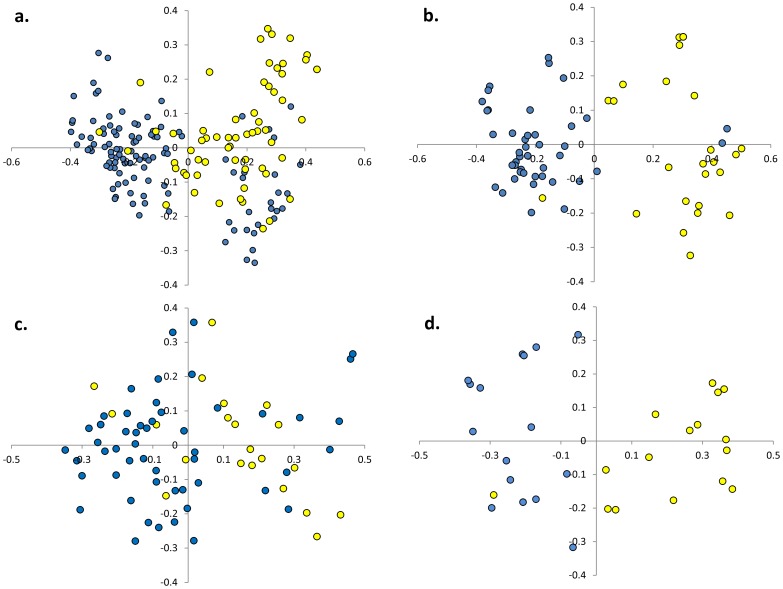
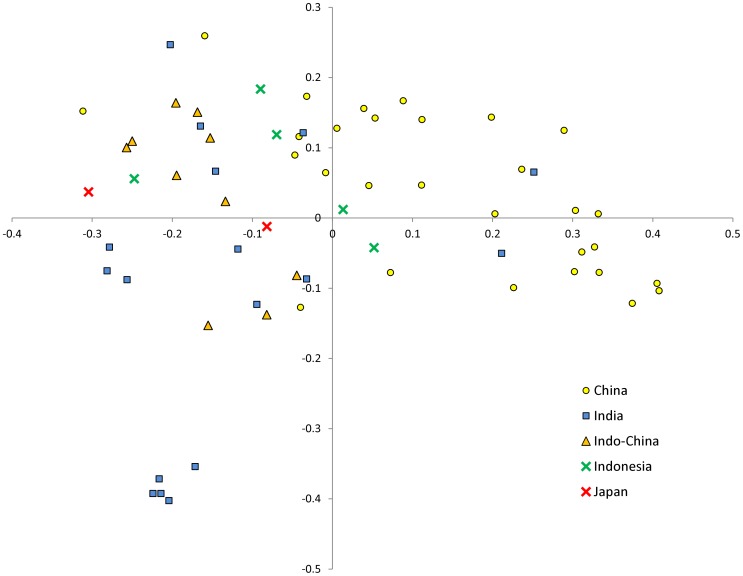
All three morphological groups were represented in both the “Occidental” and the “Oriental” germplasm (Table 1). Group 1 types comprised 39% of the “Occidental” set, group 2 types comprised 45% and group 3 types comprised 16%, while the respective proportions for the “Oriental” germplasm were 35%, 30% and 35%. According to a Mantel test, there was only a weak correlation (0.23) between the phenotypic and the genotypic data sets. A PCO analysis of the microsatellite data showed that entries belonging to each of the three morphological groups were scattered across the whole PC space (not shown). However, there was a perceptible relationship between genotype and geographical origin, since the PCO analysis showed that most of the “Oriental” entries mapped to the right hand section of the PC plane and the most of the “Occidental” ones to the left hand section (Figure 5a). A similar relationship was revealed by STRUCTURE analysis, once the entries were grouped according to their geographical provenance (Figure 2b). Some 65% of the “Oriental” entries were captured by sub-group A, as were 96% of the “Occidental” entries by sub-group B. The average pair-wise genetic similarity between the “Oriental” and “Occidental” entries was just 0.31, highlighting the extent of genetic differentiation between these two sets of germplasm. In contrast, the average pair-wise genetic similarity between entries within a geographical group was 0.44 (“Oriental”) and 0.46 (“Occidental”); although the entries within these groups were more similar to one another than were the entries between the groups, there still remains a considerable amount of within group genetic variation in both regions. When the PCO was applied to entries sorted by morphological group, the “Occidental” vs “Oriental” distinction was retained (Figure 5b–d), although the relationship was weakest for the group 2 types (Figure 5c). A PCO analysis of the genotypic data performed within each of the two areas showed a clustering of Chinese germplasm within the “Oriental” germplasm (right hand section of Figure 6), and similarly of India/Burma entries (left hand side). No equivalent clustering was evident in the “Occidental” germplasm (data not shown).
Figure 5. PCA based on geographical origin (blue: “Occidental”; yellow: “Oriental”).
a) The full germplasm set, and entries within b) morphological group 1, c) morphological group 2, d) morphological group 3.
Figure 6. PCA based on geographical origin showing the clustering of the “Oriental” entries with their country of origin.
The accessions from Myanmar and Thailand were classified as Indochinese region.
Discussion
Eggplant varieties/landraces are morphologically, physiologically and biochemically highly variable, but the progressive dominance of elite F1 hybrids in commercial cultivation presents a threat of genetic erosion, which in the longer term may well have negative implications by narrowing the source of useful genes exploitable in breeding programmes [26]. Previous attempts to characterize diversity have been restricted to a limited number of local varieties/landraces; [1], [24], [31], [33], [46]–[51]. Two recent studies have focused on 52 accessions identified from three secondary centers of origin of the crop [23] or 115 genotypes from Asian landraces and some wild relatives [4]. Here we have presented a phenotypic (19 traits) and genotypic (24 microsatellite loci) survey of a large germplasm collection originating from both Asia and the Mediterranean Basin, and representing a mixture of breeding lines, heritage and current varieties and landrace selections.
S. melongena is a largely autogamous species, so that the expectation is that most heritage and commercial varieties should be highly homozygous. The microsatellite-based genotyping uncovered some residual heterozygosity in the germplasm set, which led to the discarding of some 16% of the entries. A further 4% produced phenotypic off-types, presumably also reflecting the presence of residual heterozygosity (although it may also reflect admixture), leaving a panel of 191 true-breeding, largely homozygous entries. There was ample variation with respect to both plant and fruit traits within both “Oriental” and “Occidental” entries, and it was possible to derive a set of just three morphology-based PC’s to explain over half of the phenotypic variance displayed by the full set of 19 traits (Figure 3a). Both the leading two PC’s were correlated with fruit shape and dimension, as well as with anthocyanin content, as has previously been reported for a set of Spanish varieties [24]. As for many other crops [52], the fruit has been a major target of anthropogenic selection. Anthocyanin content, a trait acquired during domestication (since the eggplant’s putative ancestor S. insanum produces green fruit [1]), may have been under both indirect selection, based on its involvement in tolerance to a number of environmental stresses, and direct selection, due to cultural preferences towards pigmented fruits [53], [54].
The HCPC analysis identified three main groups (Figure 3b). The first one included genotypes producing elongated fruits, with a mean fs (fruit length/fruit maximum diameter) around 5.05 (Table 4). This group corresponds to the one previously detected within the eggplant Spanish germplasm (fs >2) [55], [56] as well as to the fruit typology defined var. serpentinum (long and slender fruit) identified by Choudhury [57]within the Indian germplasm. The second and the third morphological groups, with a mean fs of 1.95 and 0.98 respectively, are classified together in the fruit typology var. esculenta (round or egg-shaped fruit) identified by Choudhury [57], while they are separately identified as genotypes bearing semi-long fruits (with a fs >1.2 and <2) and round fruits (with a fs ∼1) by Prohens et al. [56] and Nuez et al. [55].The three morphological groups cut across the “Oriental” vs “Occidental” divide. In contrast, the conclusion of Hurtado et al. [23], based on an analysis of entries originating from China, Spain and Sri Lanka, was that a number of traits could be associated with the geographical origin of the material. The apparent discrepancy can be explained by either the difference in size of the two germplasm sets (52 vs 191) and/or by the somewhat different set of traits assessed in the two studies. Germplasm sets which capture a wide range of phenotypic variation tend to form many clusters when many traits are scored and few when only few traits are scored [58], [59]. The present HCPC analysis identified three distinct and robust groups, based on variation in 14 out of the full set of 19 traits recorded. Nevertheless, there was only a weak correlation between phenotype and molecular fingerprinting, an experience also recorded by Hurtado et al. [23]; in contrast, both the Munoz-Falcon et al. [22] and Prohens et al. [24] studies showed a reasonable level of phenotype/genotype correlation, probably because both focused on germplasm of rather limited diversity. The relationship between rates of phenotypic evolution and genetic change has been a matter of debate, but the rate of molecular evolution has been by many authors considered to be not strictly associated to the rate of morphological change, as only a tiny portion of the genome is directly responsible for the measurable phenotypic changes [60]. The two types of markers follow different evolutionary paths and provide complementary information contributing in understanding both evolutionary history and identifying the most suitable strategy for germplasm management [61].
When the STRUCTURE analysis was based on geographical provenance (Figure 2b), most of the “Oriental” entries fell into one cluster and most of the “Occidental” ones into another. The PCO analysis of the microsatellite data also differentiated clearly between the two provenances. A clustering in relation to provenance was also detected when PCO analysis was separately performed within each of the three main morphological groups (Figure 5b, 4c, 4d). This highlights that a molecular differentiation is detectable also between Oriental and Occidental entries with similar phenotypic traits.
When the PCO analysis was applied to just the “Occidental” entries, no evidence of any correlation between provenance and genetic relatedness was found (data not shown), suggesting that this gene pool has experienced extensive exchange of breeding materials. The picture is rather different for the “Oriental” gene pool (figure 6), in which a trend of clustering was detected and most of the genotypes from the Indian, Indo-Chinese and Indonesian regions grouped together and separately from the Chinese ones. Recent studies highlight that the modern eggplant evolved from the species S. insanum [2], and it has been generally assumed that it was domesticated in Indian subcontinent[3], ,possibly in Rajasthan region [5]. The distinct genetic content of Chinese germplasm uncovered in the present analysis supports the alternate idea proposed by Wang et al. [6], Ali et al. [20] and Meyer et al. [4], that a secondary site of domestication also developed in China. Multiple, rather than single, domestication events seem to apply for a number of crops [66]. The introduction of the eggplant to the Mediterranean Basin by the Arabs would have generated a temporary bottleneck in genetic diversity [54] but still maintaining a rather large share of variability [67] and which was alleviated by subsequent selection, de novo mutations and recombination events as well as adaptation to different environments [68].
This, despite some movement of germplasm across the Asian and Mediterranean countries occurred over time, justify the genetic differentiation we detected between genotypes from the two geographical areas.
Plant germplasm management is pivotal for providing the plant scientist with sufficient genetically, well-characterized material for research and crop improvement. To this purpose the development of genetic core collections helps to provide a reduced set of accessions, in terms of entry number but not in terms of allelic coverage, that are feasible to study and handle. A critical examination of the various methods used to evaluate the quality of core collections suggests a lack of consensus regarding the optimal selection criteria to be applied [69]. Here, the retention of about 25% of the collection was required to capture all the microsatellite alleles present in the full set; the need for such a large proportion is a consequence of the species’ high level of homozygosity, since a heterozygote by definition harbors two alleles, whereas a homozygote only harbors one. Similar proportions have to be retained in both Arabidopsis thaliana (18%, [70]) and Medicago truncatula (31% [71]), while a heterozygous species, such as grapevine, required a retention level as low as 4% [72].
Some of the phenotypic diversity identified in the present germplasm would doubtless be of interest to conventional eggplant improvement programs. However, the application of more efficient selection programs requires the understanding of the genetic basis of key agronomic traits, via the development of linkage maps and quantitative trait locus (QTL) analysis. Thus, for example, Miyatake et al. [73] have defined the genetic control of parthenocarpy, while Barchi et al. [74] were able to identify a number of QTL underlying anthocyanin pigmentation. The association mapping approach has been proposed as an alternative platform to conventional linkage analysis for QTL detection [75]. The concept relies on analyzing a large set of germplasm in which there is a substantial level of morphological and genetic diversity built up by a history of recombination and re-assortment and whose population structure has been carefully assessed. One of the intentions of the present study is to identify such a population in eggplant, and the present analysis has provided important information regarding both the potential diversity available in the species and the likely sources of population structure. The data set as a whole contributes significantly to the knowledge base regarding the level and distribution of genetic diversity in the “Occidental” and “Oriental” eggplant gene pool, and sets the scene for a well-founded association mapping exercise to derive genotype-phenotype relationships.
Acknowledgments
The authors thank the institutions and company which kindly supplied seeds material: UPV Universitat Politècnica de València Spain (from AM_126 to AM_128); East West Seed (AM_197 and AM_198) Institute of Vegetables and Flowers CAAS Beijing China (from AM_206 to AM_209 and from AM_275 to AM_293 ); The Centre for Genetic Resources, the Netherlands (CGN) Wageningen (from AM_212 to AM_262).
Funding Statement
This research was partially supported by the Italian Ministry of Agricultural Alimentary and Forest Politics in the framework of “ESPLORA”project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding were received for this study.
References
- 1.Lester (1991) Origin and domestication of the brinjal eggplant, Solanum melongena, from S. incanum, in Africa and Asia. Solanaceae III: taxonomy, chemistry, evolution. London: The Linnean Society of London. pp. 369–387.
- 2. Knapp S, Vorontsova MS, Prohens J (2013) Wild Relatives of the Eggplant (Solanum melongena L.: Solanaceae): New Understanding of Species Names in a Complex Group. PLoS ONE 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weese TL, Bohs L (2010) Eggplant origins: Out of Africa, into the Orient. Taxon 59: 49–56. [Google Scholar]
- 4. Meyer RS, Karol KG, Little DP, Nee MH, Litt A (2012) Phylogeographic relationships among Asian eggplants and new perspectives on eggplant domestication. Molecular Phylogenetics and Evolution 63: 685–701. [DOI] [PubMed] [Google Scholar]
- 5.Kashyap AASW (2010) Starch Grains from Farmana Give New Insights into Harappan Plant Use. Antiquity Volume 084 Issue 326.
- 6. Wang J, Gao T, Knapp S (2008) Ancient Chinese Literature Reveals Pathways of Eggplant Domestication. Annals of Botany 102: 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daunay (2008) Eggplant. Prohens J, Nuez F, editors. Handbook of Plant Breeding - Vegetables II. New York: Springer. 163–220.
- 8. Cao GH, Sofic E, Prior RL (1996) Antioxidant capacity of tea and common vegetables. Journal of Agricultural and Food Chemistry 44: 3426–3431. [Google Scholar]
- 9. Mennella G, Rotino GL, Fibiani M, D’Alessandro A, Francese G, et al. (2010) Characterization of Health-Related Compounds in Eggplant (Solanum melongena L.) Lines Derived from Introgression of Allied Species. Journal of Agricultural and Food Chemistry 58: 7597–7603. [DOI] [PubMed] [Google Scholar]
- 10. Stommell JR, Whitaker BD (2003) Phenolic acid content and composition of eggplant fruit in a germplasm core subset. Journal of the American Society for Horticultural Science 128: 704–710. [Google Scholar]
- 11. Huang HY, Chang CK, Tso TK, Huang JJ, Chang WW, et al. (2004) Antioxidant activities of various fruits and vegetables produced in Taiwan. International Journal of Food Sciences and Nutrition 55: 423–429. [DOI] [PubMed] [Google Scholar]
- 12. Mennella G, Lo Scalzo R, Fibiani M, D’Alessandro A, Francese G, et al. (2012) Chemical and Bioactive Quality Traits During Fruit Ripening in Eggplant (S. melongena L.) and Allied Species. Journal of Agricultural and Food Chemistry 60: 11821–11831. [DOI] [PubMed] [Google Scholar]
- 13. Lo Scalzo R, Fibiani M, Mennella G, Rotino GL, Dal Sasso M, et al. (2010) Thermal Treatment of Eggplant (Solanum melongena L.) Increases the Antioxidant Content and the Inhibitory Effect on Human Neutrophil Burst. Journal of Agricultural and Food Chemistry 58: 3371–3379. [DOI] [PubMed] [Google Scholar]
- 14. Akanitapichat P, Phraibung K, Nuchklang K, Prompitakkul S (2010) Antioxidant and hepatoprotective activities of five eggplant varieties. Food and Chemical Toxicology 48: 3017–3021. [DOI] [PubMed] [Google Scholar]
- 15. Eleveld-Trancikova D, Triantis V, Moulin V, Looman MWG, Wijers M, et al. (2005) The dendritic cell-derived protein DC-STAMP is highly conserved and localizes to the endoplasmic reticulum. Journal of Leukocyte Biology 77: 337–343. [DOI] [PubMed] [Google Scholar]
- 16. Sudheesh S, Presannakumar G, Vijayakumar S, Vijayalakshmi NR (1997) Hypolipidemic effect of flavonoids from Solanum melongena . Plant Foods for Human Nutrition 51: 321–330. [DOI] [PubMed] [Google Scholar]
- 17. Matsubara K, Kaneyuki T, Miyake T, Mori M (2005) Antiangiogenic activity of nasunin, an antioxidant anthocyanin, in eggplant peels. Journal of Agricultural and Food Chemistry 53: 6272–6275. [DOI] [PubMed] [Google Scholar]
- 18. Han SW, Tae J, Kim JA, Kim DK, Seo GS, et al. (2003) The aqueous extract of Solanum melongena inhibits PAR2 agonist-induced inflammation. Clinica Chimica Acta 328: 39–44. [DOI] [PubMed] [Google Scholar]
- 19. Das S, Raychaudhuri U, Falchi M, Bertelli A, Braga PC, et al. (2011) Cardioprotective properties of raw and cooked eggplant (Solanum melongena L). Food & Function 2: 395–399. [DOI] [PubMed] [Google Scholar]
- 20. Ali Z, Xu ZL, Zhang DY, He XL, Bahadur S, et al. (2011) Molecular diversity analysis of eggplant (Solanum melongena) genetic resources. Genetics and Molecular Research 10: 1141–1155. [DOI] [PubMed] [Google Scholar]
- 21. Rodriguez-Burrruezo A, Prohens J, Nuez F (2008) Performance of hybrids between local varieties of eggplant (Solanum melongena) and its relation to the mean of parents and to morphological and genetic distances among parents. European Journal of Horticultural Science 73: 76–83. [Google Scholar]
- 22. Munoz-Falcon JE, Prohens J, Vilanova S, Ribas F, Castro A, et al. (2009) Distinguishing a protected geographical indication vegetable (Almagro eggplant) from closely related varieties with selected morphological traits and molecular markers. Journal of the Science of Food and Agriculture 89: 320–328. [Google Scholar]
- 23. Hurtado M, Vilanova S, Plazas M, Gramazio P, Fonseka HH, et al. (2012) Diversity and Relationships of Eggplants from Three Geographically Distant Secondary Centers of Diversity. PLoS ONE 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prohens J, Blanca JM, Nuez F (2005) Morphological and molecular variation in a collection of eggplants from a secondary center of diversity: Implications for conservation and breeding. Journal of the American Society for Horticultural Science 130: 54–63. [Google Scholar]
- 25. Demir K, Bakir M, Sarikamis G, Acunalp S (2010) Genetic diversity of eggplant (Solanum melongena) germplasm from Turkey assessed by SSR and RAPD markers. Genetics and Molecular Research 9: 1568–1576. [DOI] [PubMed] [Google Scholar]
- 26. Muñoz-Falcón J, Vilanova S, Plazas M, Prohens J (2011) Diversity, relationships, and genetic fingerprinting of the Listada de Gandía eggplant landrace using genomic SSRs and EST-SSRs. Scientia Horticulturae 129: 238–246. [Google Scholar]
- 27. Behera TK, Sharma P, Singh BK, Kumar G, Kumar R, et al. (2006) Assessment of genetic diversity and species relationships in eggplant (Solanum melongena L.) using STMS markers. Scientia Horticulturae 107: 352–357. [Google Scholar]
- 28. Hurtado MA, Romero C, Vilanova S, Abbott AG, Llacer G, et al. (2002) Genetic linkage maps of two apricot cultivars (Prunus armeniaca L.), and mapping of PPV (sharka) resistance. Theoretical and Applied Genetics 105: 182–191. [DOI] [PubMed] [Google Scholar]
- 29. Stàgel A, Portis E, Toppino L, Rotino GL, Lanteri S (2008) Gene-based microsatellite development for mapping and phylogeny studies in eggplant. BMC Genomics 9: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barchi L, Lanteri S, Portis E, Stagel A, Vale G, et al. (2010) Segregation distortion and linkage analysis in eggplant (Solanum melongena L.). Genome 53: 805–815. [DOI] [PubMed] [Google Scholar]
- 31. Nunome T, Negoro S, Kono I, Kanamori H, Miyatake K, et al. (2009) Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theoretical and Applied Genetics 119: 1143–1153. [DOI] [PubMed] [Google Scholar]
- 32. Vilanova S, Manzur JP, Prohens J (2012) Development and characterization of genomic simple sequence repeat markers in eggplant and their application to the study of diversity and relationships in a collection of different cultivar types and origins. Molecular Breeding 30: 647–660. [Google Scholar]
- 33. Nunome T, Suwabe K, Iketani H, Hirai M (2003) Identification and characterization of microsatellites in eggplant. Plant Breeding 122: 256–262. [Google Scholar]
- 34. Prohens J, Rodriguez-Burruezo A, Raigon MD, Nuez F (2007) Total phenolic concentration and browning susceptibility in a collection of different varietal types and hybrids of eggplant: Implications for breeding for higher nutritional quality and reduced browning. Journal of the American Society for Horticultural Science 132: 638–646. [Google Scholar]
- 35. Hammer Ø, Harper DAT, Ryan PD (2001) Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4(1): 9pp. [Google Scholar]
- 36. Dice LR (1945) Measures of the Amount of Ecologic Association Between Species. Ecology 26: 297–302. [Google Scholar]
- 37. Anderson J, Churcill G, Autrique J, Tanksley S, Sorrels M (1992) Optimizing parental selection for genetic linkage maps. Genome 36: 181–186. [DOI] [PubMed] [Google Scholar]
- 38. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 40. Schoen DJ, Brown AHD (1991) intraspecific variation in population gene diversity and effective population-size correlates with the mating system in plants. Proceedings of the National Academy of Sciences of the United States of America 88: 4494–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gouesnard B, Bataillon TM, Decoux G, Rozale C, Schoen DJ, et al. (2001) MSTRAT: An algorithm for building germ plasm core collections by maximizing allelic or phenotypic richness. Journal of Heredity 92: 93–94. [DOI] [PubMed] [Google Scholar]
- 42. Matus (1996) Evaluation of phenotypic variation in a Chilean collection of garlic (Allium sativum L.) clones using multivariate analysis. In: Matus GG, A. del Poso, editor. Plant Genet. Res. Newsl. 117: 31–36. [Google Scholar]
- 43.Husson F (2010) Principal component methods - hierarchical clustering - partitional clustering: why would we need to choose for visualizing data? Technical Report – Agrocampus, Applied Mathematics Department.
- 44.R Development Core Team (2006) R: a language and environment for statistical computing. http://www.r-project.org/Accessed 2013 Agoust 1.
- 45.Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Research. pp. 209–220. [PubMed]
- 46. Nunome T, Ishiguro K, Yoshida T, Hirai M (2001) Mapping of fruit shape and color development traits in eggplant (Solanum melongena L.) based on RAPD and AFLP markers. Breeding Science 51: 19–26. [Google Scholar]
- 47. Isshiki S, Suzuki S, Yamashita K (2003) RFLP analysis of mithocondrial DNA in eggplant and related Solanum species. Genetic Research and Crop Evolution 50: 133–137. [Google Scholar]
- 48. Mace ES, Lester RN, Gebhardt CG (1999) AFLP analysis of genetic relationships among the cultivated eggplant, Solanum melongena L., and wild relatives (Solanaceae). Theoretical and Applied Genetics 99: 626–633. [DOI] [PubMed] [Google Scholar]
- 49. Karihaloo J, Brauner S, Gottlieb L (1995) Random amplified polymorphic DNA variation in the eggplant, Solanum melongena L. (Solanaceae). Theoretical and Applied Genetics 90: 767–770. [DOI] [PubMed] [Google Scholar]
- 50. Karihaloo JL, Kaur M, Singh S (2002) Seed protein diversity in Solanum melongena L. and its wild and weedy relatives. Genetic Resources and Crop Evolution 49: 533–539. [Google Scholar]
- 51. Isshiki S, Okubo H, Fujieda K (1994) phylogeny of eggplant and related solanum species constructed by allozyme variation. Scientia Horticulturae 59: 171–176. [Google Scholar]
- 52.Harlan (1992) Crops and man. 2nd ed.Amer.Soc.Agron.,Madison,Wis.
- 53. Chalker-Scott (1999) Characterization and typification of spanish eggplant landraces. 2nd ed.Amer.Soc.Agron.,Madison,Wis. : PlantAmnesty Newsletter, 11(1): 4–5. [Google Scholar]
- 54. Atanassova B, Daskalov S, Shtereva L, Balatcheva E (2001) Anthocyanin mutations improving tomato and pepper tolerance to adverse climatic conditions. Euphytica 120: 357–365. [Google Scholar]
- 55.Nuez FJ, Prohens JV, Valcárcel JV, Fernández de Córdova P (2002) Collection of eggplant seeds from the Centro de Conservación y Mejora de la Agrodiversidad Valenciana (in Spanish). Ministerio de Ciencia y Tecnología, Madrid.
- 56. Prohens J, Valcárcel JV, Fernandez de Cordova P, Nuez F (2003) Characterization and typification of Spanish eggplant landraces. Capsicum Eggplant Nwsl. 22: 135–138. [Google Scholar]
- 57.Choudhury B. (1976) Vegetables. 4th Revised Edition National Book Trust, New Delhi, India. 214 pp.
- 58.Parker PF (1986) The classification of cultivated plants - problems and prospects. B.T.Styles (ed.). Infraspecific classification of wild and cultivated plants. Claredon Press, Oxford U.K.
- 59. Spooner DM HW, Van Den Berg RG, Brandenburg WA (2003) Plant nomenclature and taxonomy: an horticoltural and agronomic perspective. Hort. Rev. 28: 1–60. [Google Scholar]
- 60. Bromham L, Woolfit M, Lee MSY, Rambaut A (2002) Testing the relationship between morphological and molecular rates of change along phylogenies. Evolution 56: 1921–1930. [DOI] [PubMed] [Google Scholar]
- 61. Bretting PK, Widrlechner MP (1995) Genetic markers and horticultural germplasm management. Hortscience 30: 1349–1356. [Google Scholar]
- 62. Wu FN, Eannetta NT, Xu YM, Tanksley SD (2009) A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theoretical and Applied Genetics 118: 927–935. [DOI] [PubMed] [Google Scholar]
- 63. Polignano G, Uggenti P, Bisignano V, Della Gatta C (2010) Genetic divergence analysis in eggplant (Solanum melongena L.) and allied species. Genetic Resources and Crop Evolution 57: 171–181. [Google Scholar]
- 64. Daunay (2007) History and iconography of eggplant. In: Janick, editor. Chronica Hort 47(3): 16–21. [Google Scholar]
- 65.Sękara (2007) Cultivated eggplants–origin, breeding objectives and genetic resources, a review. In: A. Sękara SC, E Kunicki, editor. Folia Hort., 19 (2007), pp. 97–114.
- 66. Olsen KM, Gross BL (2008) Detecting multiple origins of domesticated crops. Proceedings of the National Academy of Sciences of the United States of America 105: 13701–13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hufford MB, Martinez-Meyer E, Gaut BS, Eguiarte LE, Tenaillon MI (2012) Inferences from the Historical Distribution of Wild and Domesticated Maize Provide Ecological and Evolutionary Insight. PLoS ONE 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prohens J, Nuez F (2001) Spanish traditional varieties of eggplant (in Spanish). Vida Rural 130: 46–50. [Google Scholar]
- 69. Odong TL, Jansen J, van Eeuwijk FA, van Hintum TJL (2013) Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theoretical and Applied Genetics 126: 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McKhann HI, Camilleri C, Berard A, Bataillon T, David JL, et al. (2004) Nested core collections maximizing genetic diversity in Arabidopsis thaliana . Plant Journal 38: 193–202. [DOI] [PubMed] [Google Scholar]
- 71. Ellwood SR, D’Souza NK, Kamphuis LG, Burgess TI, Nair RM, et al. (2006) SSR analysis of the Medicago truncatula SARDI core collection reveals substantial diversity and unusual genotype dispersal throughout the Mediterranean basin. Theoretical and Applied Genetics 112: 977–983. [DOI] [PubMed] [Google Scholar]
- 72. Le Cunff L, Fournier-Level A, Laucou V, Vezzulli S, Lacombe T, et al. (2008) Construction of nested genetic core collections to optimize the exploitation of natural diversity in Vitis vinifera L. subsp sativa . BMC Plant Biology 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miyatake K, Saito T, Negoro S, Yamaguchi H, Nunome T, et al. (2012) Development of selective markers linked to a major QTL for parthenocarpy in eggplant (Solanum melongena L.). Theoretical and Applied Genetics 124: 1403–1413. [DOI] [PubMed] [Google Scholar]
- 74. Barchi L, Lanteri S, Portis E, Vale G, Volante A, et al. (2012) A RAD Tag Derived Marker Based Eggplant Linkage Map and the Location of QTLs Determining Anthocyanin Pigmentation. PLoS ONE 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mackay I, Powell W (2007) Methods for linkage disequilibrium mapping in crops. Trends in Plant Science 12: 57–63. [DOI] [PubMed] [Google Scholar]