Abstract
With age, muscle mass and integrity are progressively lost leaving the elderly frail, weak and unable to independently care for themselves. Defined as sarcopenia, this age-related muscle atrophy appears to be multifactorial but its definite cause is still unknown. Mitochondrial dysfunction has been implicated in this process. Using a novel transgenic mouse model of mitochondrial DNA (mtDNA) double-strand breaks (DSBs) that presents a premature aging-like phenotype, we studied the role of mtDNA damage in muscle wasting. We caused DSBs in mtDNA of adult mice using a ubiquitously expressed mitochondrial-targeted endonuclease, mito-PstI. We found that a short, transient systemic mtDNA damage led to muscle wasting and a decline in locomotor activity later in life. We found a significant decline in muscle satellite cells, which decreases the muscle's capacity to regenerate and repair during aging. This phenotype was associated with impairment in acetylcholinesterase (AChE) activity and assembly at the neuromuscular junction (NMJ), also associated with muscle aging. Our data suggests that systemic mitochondrial dysfunction plays important roles in age-related muscle wasting by preferentially affecting the myosatellite cell pool.
INTRODUCTION
Sarcopenia is the age-associated loss of muscle mass and strength causing frailty in the elderly. Healthy individuals over 25 years of age begin to lose an estimated 0.5–1% of muscle mass per year (1,2). The molecular basis for sarcopenia is unknown, but it has been suggested that mitochondrial dysfunction, protein synthesis/degradation dysregulation, systemic inflammation and loss of supporting cell types such as satellite cells and motor neurons may be causal or secondary factors contributing to age-related muscle loss (3–7).
The skeletal muscle is an aerobic tissue highly reliant on mitochondria (8). Aerobic capacity, mitochondrial proteins and ATP production decline with age (9). Transcriptional gene arrays on different mammalian species also revealed that the expression of mitochondrial genes declined in aging muscle (10,11). Mitochondrial DNA (mtDNA) is a 16.5 kb genome, which is transcribed and translated inside the mitochondria, contributing subunits to the oxidative phosphorylation (OXPHOS) pathway to produce ATP (12). Low levels of mtDNA deletions, in a heteroplasmic context, have been observed in aged tissues, including muscle (13). However, individual skeletal muscle fibers can harbor mtDNA deletion loads between ∼60 and 80%. These high levels of mutated mtDNA correlated with an OXPHOS defect at the single-fiber level (14,15). Because the percentage of such defective fibers is low (1–5%), their role in sarcopenia is unclear.
Motor neuron damage and inactivity are also major contributors to myofiber integrity and muscle wasting. Mitochondrial dysfunction in motor neuron leads to deterioration and subsequent muscle atrophy in mouse models of amyotrophic lateral sclerosis (ALS) and Charcot Marie Tooth 2A disease (CMT2) (16–18). Nevertheless, the role of mtDNA damage in the progressive decline of motor neurons during normal aging is still not clear (4). The sedentary lifestyle in the elderly may also contribute to exacerbated losses in muscle quantity and quality (19,20). We and several others have previously shown that exercise provides benefits to help prevent muscle atrophy in aging and multiple mouse models where mitochondrial dysfunction occurs in the muscle, systemically, or in motor neurons (21–24). In addition, we found that expression of the peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1α), which links mitochondrial function with muscle integrity and acetylcholinesterase (AChE) expression during aging, can impact the function of aged muscle (25).
The role of mitochondrial function in the aging muscle remains unclear. To better understand this role, we examined how systemic and transient mtDNA damage and mitochondrial dysfunction contribute to muscle wasting in a novel mouse model.
RESULTS
Systemic inducible mito-PstI mouse model
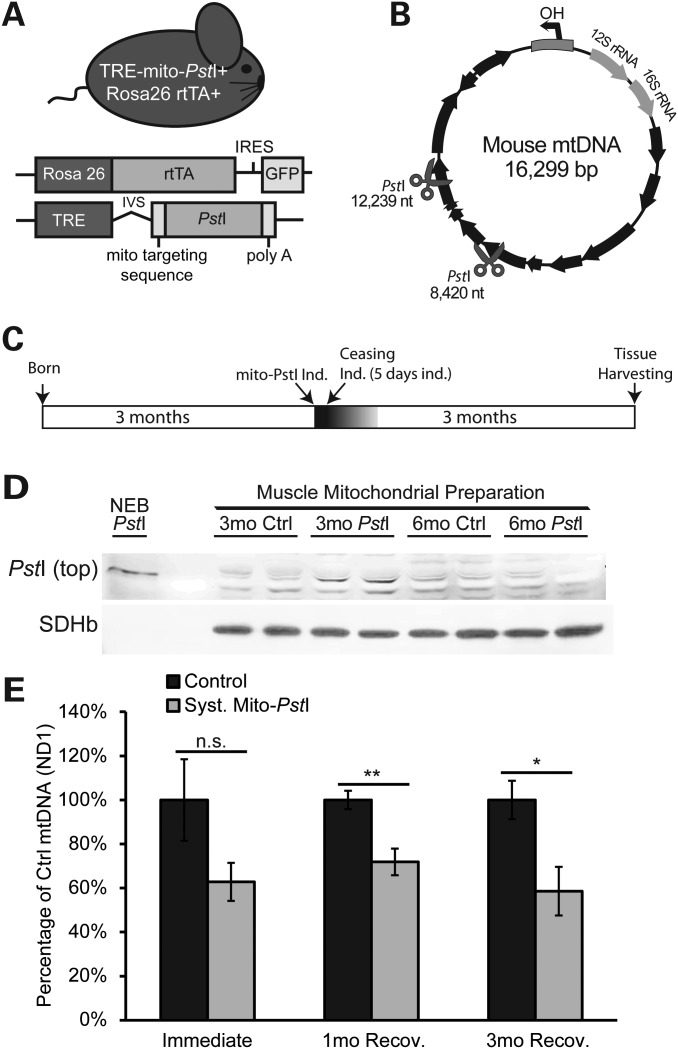
During aging, every organ system is exposed to mtDNA damage. Therefore, to better model the effect of mtDNA damage on the aging muscle, we reasoned that targeting exclusively myotubes would not fully reflect the aging process. With this concern in mind, we created a transgenic mouse model expressing a mitochondrial-targeted PstI (mito-PstI) under a tetracycline response element promoter (26). We mated these mito-PstI mice with a mouse line that expresses a reverse tetracycline transactivator (rtTA) protein under the control of the ubiquitous Rosa26 promoter (27). Experimental animals harboring both alleles are hereafter referred to as ‘systemic mito-PstI’ mice (mito-PstI(+)/Rosa26-rtTA(+)), where mito-PstI expression could be induced not only in the muscle, but also in all tissues when supplying mice with a doxycycline (DOX) diet (Fig. 1A). Siblings, which were positive only for one of the transgenes (mito-PstI(−)/Rosa26-rtTA(+), or mito-PstI(+)/Rosa26-rtTA(−)), did not express mito-PstI protein and were used as controls.
Figure 1.
Characterization of expression of mito-PstI in skeletal muscle of transgenic mice. (A) Schematic representation of the systemic mito-PstI mouse and the transgenic constructs. (B) MtDNA map showing the targeted sites of PstI (scissors). Black arrows denote protein-coding genes. (C) Schematic representation of mito-PstI induction and experimental paradigm: 5-day induction at 3 months of age, and up to 3 months follow-up after the induction. (D) Western blotting using an anti-PstI antibody to detect mito-PstI expression in mitochondrial extracts of gastrocnemius muscle from systemic mito-PstI mice after a 5-day induction at 3 months of age, and at 6 months of age. NEB (New England Biolabs) PstI restriction enzyme was used as a molecular weight control. Antibody against SDHβ was used as a loading control. (E) Real-time PCR quantification of mtDNA/nuclear DNA ratios of quadriceps from the systemic mito-PstI mice and age-matched controls after 5-day induction, 1 and 3 months recovery (n = 5 per group for ‘immediate’ and ‘3mo’ time points, and n = 4 per group for ‘1mo’ time point). An unpaired t-test was performed for each time point. Values are presented as mean ± SEM (*P< 0.05, **P< 0.01, ***P< 0.001).
Mito-PstI has previously been shown to localize exclusively in mitochondria, and to cause double-strand breaks (DSBs) in the mouse mtDNA at nucleotide (nt) positions 8420 and 12 238 (Fig. 1B). These DSBs result in mostly a depletion of mtDNA, but also in the formation of low levels of mtDNA deletions (26,28,29). We transiently induced the systemic mito-PstI mice at 3 months, by substituting a standard diet with DOX chow for 5 days. After this brief induction period, mice were allowed to live without induction for three additional months (Fig. 1C). Mouse behavior and tissue biochemistry were analyzed at this point (6 months of life or 3 months after the brief mito-PstI induction; Fig. 1C). At the end of induction, PstI expression was detected in different organs of systemic mito-PstI mice, but it was below detection limits in muscle homogenates (data not shown). However, PstI was detected when mitochondrial preparations were analyzed (Fig. 1D). This expression was only observed in muscle mitochondrial preparations immediately after DOX induction, and not in those from muscle mitochondria obtained 3 months post-induction (Fig. 1D).
To test whether mito-PstI cleaved mtDNA, we measured the levels of mtDNA in skeletal muscle at different time points during recovery. Although variable levels of mtDNA were detected immediately after induction, we observed significant and persistent decreases in quadriceps at 1 and 3 months after the short 5-day induction (Fig. 1E). This was a puzzling observation as not only the levels of expression of mito-PstI were low in muscle immediately after induction, and we could not detect mito-PstI in mitochondria after the induction was stopped (Fig. 1D).
Mito-PstI expression leads to the degradation of linearized mtDNA molecules, but also generates low levels of mtDNA deletions (26,30). Previously, we reported that mtDNA deletion loads were formed and maintained in neurons when crossing our mito-PstI mouse to a central nervous-specific CamKIIα-tTA line (26). We compared the skeletal muscle mtDNA deletion loads from systemic mito-PstI mice at 6 months of age with loads detected in the cortex from CamKIIα mito-PstI mice, with both models undergoing the same induction paradigm. Large-scale (∼10.6 kB) and small-scale (∼3.8 kB) deletions were detected in both mouse models, but at low levels, estimated to be <1% (Supplementary Material, Figure S1) (26). It is likely that persistent mtDNA depletion causes more significant pathological changes than the low levels of mtDNA deletions detected in the muscle.
Systemic mito-PstI mice developed locomotive dysfunctions
We noticed a visible lack of ambulatory activity, and apparent weakness in the systemic mito-PstI animals at 6 months of age, which was 3 months after the transient mito-PstI induction. To quantify this phenotype, we performed an endurance treadmill test on 6-month-old males and females. Both systemic mito-PstI males and females performed poorly in this test (Fig. 2A). Noting the perceived lack of ambulatory activity, we also placed systemic mito-PstI male mice and controls in an activity monitoring chamber for a 12 h period during the nocturnal cycle. The ambulatory movements of systemic mito-PstI males were significantly reduced compared with controls (Fig. 2B). These observations at 6 months of age confirmed that a muscle phenotype was manifested months after the transient mito-PstI expression.
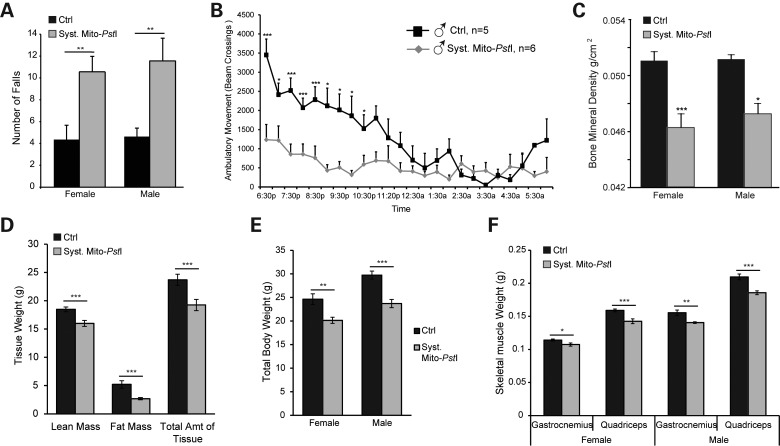
Figure 2.
Muscle wasting phenotypes characterized in systemic mito-PstI mice. (A) Treadmill performance of control and systemic mito-PstI mice at 6 months of age, measured by number of falls during a 5 min running test at 9 m/min (n = 7–12 per group). (B) Ambulatory movement of control and systemic mito-PstI male mice at 6 months of age, measured by activity cage counting beam-crossings during a 12 h period. (C) Bone mineral density measured by DEXA scan in 6-month old control and systemic mito-PstI mice (n = 7–12 per group). (D) DEXA scan quantification (grams) of lean mass, fat mass and total amount of tissue from 6-month-old control and systemic mito-PstI female mice (n = 7–12 per group). (E) Total body weight of female and male mice at 6 months of age (n = 8–12 per group). (F) Weight of gastrocnemius and quadriceps (grams) of control and systemic mito-PstI mice at 6 months of age (n = 4–10 per group). Values are presented as mean±SEM (*P< 0.05, **P< 0.01, ***P< 0.001).
Using the DEXA scan technology, we determined that there was a loss of muscle mass in our systemic mito-PstI model. This analysis showed significant declines in lean, fat, and total tissue mass of both systemic mito-PstI males and females at 6 months of age (Fig. 2D, Supplementary Material, Figure S2A). Systemic mito-PstI females also had a significant decrease in the amount of tissue in the hind limb, while male mice only showed significant decreases in fat mass (Supplementary Material, Figure S2B–S2C). Consistent with decreased skeletal muscle function, both male and female systemic mito-PstI mice had lower levels of bone mineral density in the hind limb when compared with controls (Fig. 2C). Autopsy studies at 6 months of age showed decreases in total body weight, as well as in individual skeletal muscle weights in systemic mito-PstI mice of both sexes (Fig. 2E–F). These data confirmed the weakness and atrophy of the skeletal muscle in our systemic mito-PstI mice.
Systemic mito-PstI mice presented no signs of systemic or muscle inflammation
Chronic systemic inflammation can inflict devastating degenerative effects including muscle wasting, and has been implied in many age-related diseases (31–33). To determine whether this was the cause of muscle degeneration, we measured the levels of circulating pro-inflammatory cytokines in 6-month-old control and systemic mito-PstI mice. To our surprise, we found significantly lower levels of IL-6, MCP-1, IL-12, and trends of decreased tumour necrosis factor alpha and interferonγ in the serum of mito-PstI mice (Supplementary Material, Figure S3A–S3F). We also noticed a significant decrease in the levels of IL-10, an anti-inflammatory cytokine usually upregulated by IL-6, possibly because IL-6 was not activated in systemic mito-PstI mice (Supplementary Material, Figure S3B).
In addition, we also performed H&E staining of muscle sections, and we did not find signs suggesting an inflammatory myopathy (e.g. necrosis, mononuclear infiltration or central nuclei) (Supplementary Material, Figure S3G). These results suggested that muscle phenotypes observed in systemic mito-PstI mice were not caused by a systemic or local inflammatory response. We also found no changes in the autophagy marker LC3II/LC3I in muscle of 6-month-old mito-PstI-induced mice (not shown).
Mild mitochondrial defects in skeletal muscle of systemic mito-PstI mice
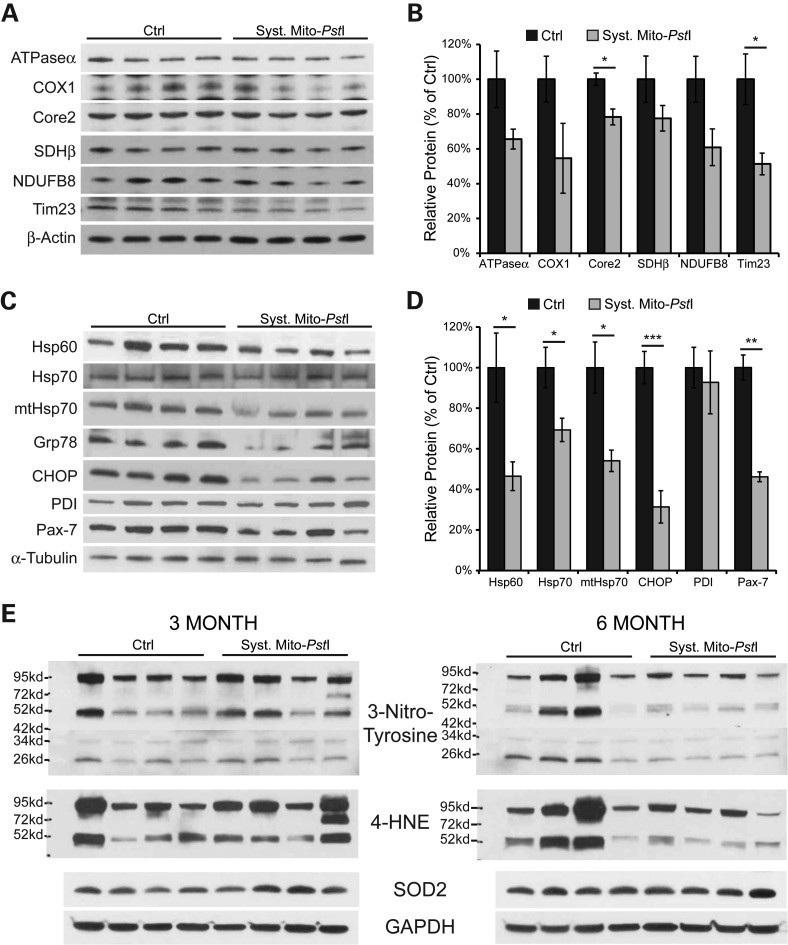
The mtDNA copy number is associated with mitochondrial function, and low mtDNA levels have been suggested to participate in the etiology of many age-related degenerative diseases (34,35). The quadriceps of the systemic mito-PstI mice had a decrease in mtDNA copy number, which could reduce OXPHOS function (Fig. 1D). We performed western blots for subunits of the OXPHOS complexes I-V, and the membrane protein Tim23. Interestingly, in muscle homogenates from 6-month-old systemic mito-PstI mice, we observed a significant decrease in complex III subunit Core2 and inner membrane transporter Tim23 (Fig. 3A and B). The other mitochondrial proteins appeared to be mildly decreased in mito-PstI mice, but the changes did not reach significance (Fig. 3A and B). This general decline in mitochondrial markers, including the previously detected decreases in mtDNA levels (Fig. 1E), suggested a reduction in mitochondrial mass in the skeletal muscle.
Figure 3.
Systemic mito-PstI mice showed a substantial decrease in chaperones and satellite cell markers, without profound mitochondrial defect or oxidative stress. (A) Western blotting analysis of mitochondrial proteins: ATPaseα, Core2, COX1, SDHβ, NDUFB8 and Cyt c in 6-month-old control and systemic mito-PstI mice. Antibody against β-actin was used as loading control (n = 4 per group). (B) Optical density (OD) quantification of proteins of interest normalized to β-actin using ImageJ software, and presented as percentage of control. (C) Western blotting analysis of chaperones Hsp60, Hsp70, mtHsp70; ER stress markers Grp78, CHOP, PDI; and satellite cell marker Pax-7 in 6-month-old control and systemic mito-PstI mice. Antibody against α-tubulin was used as loading control (n = 4 per group). (D) OD quantification of proteins of interest normalized to α-tubulin using ImageJ software, and presented as percentage of control. (E) Steady-state levels of different oxidative stress markers (3-nitrotyrosine, 4-HNE and SOD2) were analyzed by western blotting in muscle homogenates from 3- and 6-month-old control or systemic mito-PstI mice. GAPDH was used as a loading control (n = 4 per group). Values are presented as mean ± SEM (*P< 0.05, **P< 0.01, ***P< 0.001).
To further dissect a potential mitochondrial defect, we performed cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) activity staining on freshly frozen quadriceps sections. While we observed no obvious differences in the staining for SDH, there was a mild decrease in the intensity of COX staining in some of the fibers from systemic mito-PstI mice (Supplementary Material, Fig. S4A). Because histological stainings are not quantitative, we also performed enzymatic activity assays on quadriceps homogenates. We did not detect any difference in citrate synthase (CYT SYN) activity, and only a trend for a decrease of COX activity in systemic mito-PstI mice (Supplementary Material, Fig. S4B). These data showed that although there was a reduction in many mitochondrial proteins, it was mild and not observed for all mitochondrial proteins tested.
Decreases in the satellite cell pool but no signs of oxidative stress in skeletal muscle of mito-PstI-induced mice
We analyzed the levels of the muscle satellite cell marker Pax-7, and found a marked decrease in quadriceps homogenates of systemic mito-PstI mice at 6 months (Fig. 3C–D). Interestingly, we and others have observed that stem cells are preferentially affected by mtDNA damage, probably mediated by local ROS signaling (36)(unpublished data). We examined the extent of oxidative damage in muscle homogenates from 3-month-old and 6-month-old mice. Paradoxically, oxidative stress markers such as protein adducts of nitrosylation (N-tyrosine) and lipid peroxidation (4-hydroxynonenal, 4-HNE) were lower in the systemic mito-PstI muscle homogenates at 6 months of age (Fig. 3E). The levels of mitochondrial superoxide dismutase 2 (SOD2) were similar between control and systemic mito-PstI mice at both 3 and 6 months. We further performed immunohistochemistry of muscle sections with an antibody against 8-hydroxyguanosine/8-hydroxydeoxyguanosine (8-OHG/8-OHdG), a marker of nucleic acid oxidation, and found similar levels of oxidative stress in control and systemic mito-PstI mice immediately after DOX induction and at 6 months of age (Supplementary Material, Fig. S4C). Therefore, no signs of chronic oxidative stress were observed in mature muscle.
Decreases in molecular chaperones in skeletal muscle of mito-PstIinduced mice
Several studies have shown that endurance training upregulates the expression of genes involved in chaperoning (37). Because systemic mito-PstI mice had significantly less ambulatory movement than controls, we wondered whether there would be a reduction in the levels of chaperones. We analyzed the levels of the cytoplasmic chaperones Hsp60 and Hsp70, mitochondrial chaperone mtHsp70 (Grp75), ER stress markers Grp78, CHOP and PDI. Interestingly, in a quadriceps homogenate from 6-month-old systemic mito-PstI mice, we observed significant decreases in steady-state protein levels of all markers listed above, except for PDI (Fig. 3C–D). PDI (protein disulfide isomerase) catalyzes formation and isomerization of intra- and inter- molecular disulfide bonds post translation (38). The level of PDI remained unchanged in systemic mito-PstI mice probably due to the importance of maintaining protein modification capacity, while a whole panel of molecular chaperones was likely decreased because of reduced metabolic stress associated with declines in ambulatory movement.
Loss of active acetylcholinesterase at neuromuscular junction in systemic mito-PstI mice
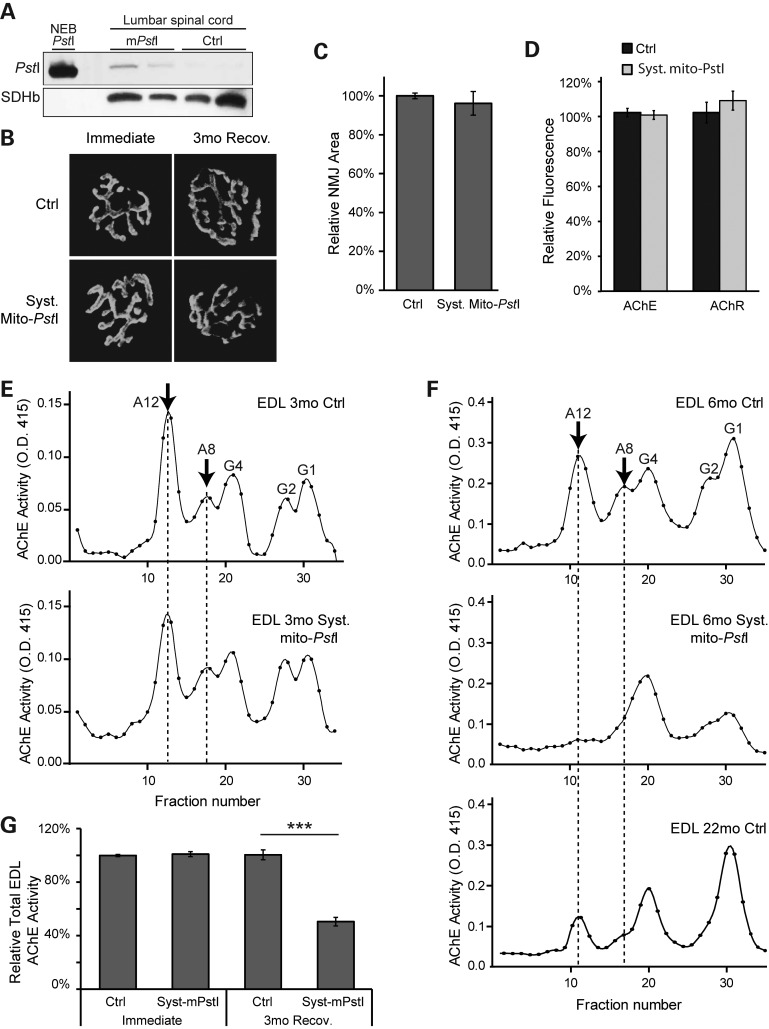
Although we observed a mild mitochondrial defect in the skeletal muscle of systemic mito-PstI mice, this is unlikely to be the triggering factor leading to the sarcopenia-like phenotype. As innervation plays a critical role in muscle contraction, we decided to study the function of motor unit in these mice. We first confirmed that PstI was also induced in the motor neurons in the spinal cord of systemic mito-PstI mice immediately after the 5-day DOX induction (Fig. 4A). We then looked at the morphology of neuromuscular junctions (NMJs) in control and systemic mito-PstI mice, immediately or 3 months after the 5-day DOX induction. We did not detect any difference in the overall morphology of the NMJs at these time points (Fig. 4B). In 6-month-old mice, we also examined the area covered by NMJs, as well as fluorescent intensity of reagents labeling AChE and the acetylcholine receptor (AChR). These values were all similar between control and systemic mito-PstI mice (Fig. 4C–D).
Figure 4.
Systemic mito-PstI mice present defects in the NMJ. (A) Western blotting using an anti-PstI antibody to detect mito-PstI expression in spinal cord homogenate from systemic mito-PstI mice after a 5-day induction at 3 months of age. NEB (New England Biolabs) PstI restriction enzyme was used as a molecular weight control. Antibody against SDHβ was used as a loading control. (B) Representative fluorescent images of the NMJ of quadriceps muscle labeled with α-bungarotoxin (BTX) in mice immediately after the 5-day DOX induction and after a 3-month recovery. (C and D) Quantification of the size of quadriceps muscle NMJ and BTX fluorescent intensity of 6-month-old control and systemic mito-PstI mice (n = 5 per group). (E and F) Representative sucrose gradient profiles of AChE activity of EDL muscle at OD 415 nm showing the different oligomeric forms of AChE expressed in 3-month (E) and 6-month (F) old control and systemic mito-PstI mice. The arrow indicates the predominant forms of AChE at the neuromuscular synapse. (G) Quantification of total AChE activity of EDL muscle in 3- and 6-month-old mice (n = 4 per group).
To determine whether the NMJs were functionally intact, we further determined the profile of AChE oligomeric forms by velocity sedimentation using extracts of the extensor digitorum longus (EDL) muscle. The AChE activity profiles were essentially the same in control and systemic mito-PstI mice immediately after DOX induction (Fig. 4E). However, although we were still able to detect the peak corresponding to the newly synthesized, monomeric (G1) and dimeric (G2) endoplasmic reticulum-resident forms of the enzyme, there was a decrease in the functional asymmetric A8 and A12 AChE forms in our 6-month-old systemic mito-PstI mice, resembling the profile of the aged 22-month-old controls (Fig. 4F). The asymmetric A8 and A12 forms consist of two or three tetramers of catalytic subunits and are the active forms of the enzyme located at NMJs. When quantified, the total AChE activity was >50% lower in 6-month-old mito-PstI mice compared with controls (Fig. 4G). Even though the NMJs in systemic mito-PstI mice appeared to be structurally intact, the diminished AChE activity would affect muscle contraction and/or coordination, resulting in the decreased locomotive performance of these mice.
DISCUSSION
As recently discussed in a viewpoint article, ‘muscle atrophy is not always sarcopenia’ (39). However, the muscle wasting observed in our model has several features resembling age-related muscle loss, including reduction in satellite cells and alterations in the NMJ. These defects in satellite cells and NMJs in our model likely led to the observed reduced locomotive activity, compounding and accelerating muscle loss. Still, not all features observed in bona fide sarcopenia are reproduced in our model (e.g. fiber type grouping), possibly because these require more time to develop. The observation that the mtDNA damage did not lead to a severely bioenergetically-impaired muscle was unexpected because previous reports showed that mature muscle accumulates mtDNA mutations over time, potentially contributing to the declines in mitochondrial function at the individual fiber level during natural aging (14,15). However, studies on sarcopenia are increasingly pointing to stem cells and NMJ dysfunctions as major players in the process (40–42). In agreement, our study suggests that systemic age-related mtDNA alterations may affect muscle mostly by impairing these processes rather than affecting mature muscle (Fig. 5). Therefore, we believe our model reflects key alterations observed during muscle aging.
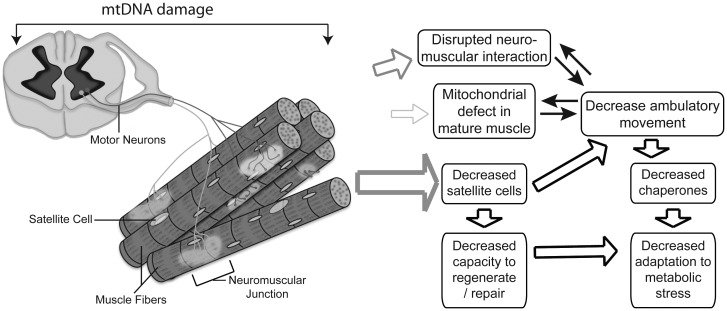
Figure 5.
Proposed mechanism by which ubiquitous mtDNA damage leads to sarcopenia. Proposed mechanism showing the consequences of low levels of systemic mtDNA damage to motor neurons, at the NMJ, muscle fibers and muscle satellite cells. The thickness of the open arrows reflects the proposed contribution to the muscle wasting phenotype. We propose that impairment of muscle satellite cell pool, due to its vulnerability to mtDNA damage, compromises skeletal muscle capacity to keep up with optimal regeneration and maintenance.
The mtDNA Mutator mouse model that expresses a proofreading deficient catalytic subunit in polymerase γ suggested that somatic mtDNA mutations were capable of causing a variety of progeroid phenotypes in mammals (43,44). Nevertheless, it is still not clear how low level damage to mtDNA elicits this phenotype. The original mitochondrial theory of aging suggests that increasing oxidative stress with age either directly causes cellular damage by oxidizing proteins and organelles, or further exacerbates mitochondrial dysfunction in a positive feedback manner (45). However, the Mutator mouse model actually showed unchanged or even decreased oxidative damage in multiple mature tissues examined (43,44), including skeletal muscle (44), suggesting other mechanisms employed by mitochondrial defects triggered the aging signaling pathways. In our model, we did not detect a severe mitochondrial dysfunction or oxidative damage in the muscle 3 months after the ubiquitous/transient expression of mito-PstI. Nevertheless, this does not rule out the possibility that ROS was transiently and locally produced, acting, rather than a strong oxidative agent, as either signaling molecules to modulate nuclear gene expression or damaging compounds to highly susceptible cells, such as progenitor cells. In fact, ROS appears to be involved in the decrease in stem cells in the Mutator mouse (36), even though mature tissues showed no evidence of oxidative stress (44).
The ubiquitous/transient mitochondrial defect could affect the NMJ either directly, by impairing metabolic reactions and possibly AChE assembly or perhaps more likely, as a response to the decreased regenerative potential due to a reduction in the satellite cell pool. Abnormalities in the NMJ have been found in MyoD(−/−) mice with disrupted lineage progression of satellite cells (46). The NMJ goes through a series of structural and functional changes during the course of aging (47,48). It was suggested that these changes were due to the degeneration and regeneration of muscle fiber segments at the synapse, progressively yet permanently (49). More recently, a new approach, caloric restriction, was examined for its effect on age-related changes of NMJs. Both exercise and caloric restriction were able to attenuate age-related changes in mouse NMJs; however, they achieved this goal by different mechanisms: exercise decreased the fragmentation and partially reversed structural changes of NMJs, while caloric restriction preserved motor neurons and attenuated increases in muscle fiber turnover (50). In addition, a recent study found that aging muscle shows a progressive reduction in markers of mitochondrial function and the NMJ (40).
A compelling observation in this study is that transient mtDNA damage reduced the level of early myogenic Pax-7 positive progenitor cells. Satellite cells appear to be more sensitive to mtDNA defects as was recently shown for other stem cell types such as hematopoietic progenitors in the Mutator mouse (36). We also found that thymic progenitors were the most sensitive and susceptible to mtDNA damage in our systemic mito-PstI mouse model (unpublished data). Damage to mtDNA in progenitor phases can also lead to aberrant differentiation into mature cell types (51) (unpublished data). These findings highlight the importance of mtDNA integrity for stem and progenitor cell types, suggesting that satellite cells could be particularly sensitive to mtDNA damage and contribute to aging processes by impairing regeneration of post-mitotic skeletal muscle.
In summary, we presented a mouse model with ubiquitous/transient mtDNA damage leading to a muscle wasting phenotype resembling some age-related features. While mtDNA depletion in skeletal muscle can affect energy production, we propose that the primary cause of the sarcopenia-like phenotype is related to alterations in the satellite cell pool. Although the factors contributing to sarcopenia are complex and feedback-prone, the main finding of this study is that systemic but mild damage to mtDNA, an event previously associated with normal aging, is likely to cause wasting not by directly affecting mature muscle, but rather by affecting more sensitive satellite cell pools.
MATERIALS AND METHODS
Animals
The generation of mito-PstI transgenic mice was previously described (26). Mito-PstI male and female animals were pure C57BL/6J backcrossed at least 10 generations, and mito-PstI animals were crossed with Rosa26-rtTA animal of the same genetic background (Jackson Laboratories) (27). Mito-PstI mice were also crossed with CamKIIα-tTA mouse lines (Jackson laboratories) as previously described (26). These experimental mice were male on pure C57BL/6J background.
All mice-related procedures were performed according to a protocol approved by the University of Miami Institutional Animal Care and Use Committee. When mito-PstI was induced at 3 months of age, we supplied both control and systemic mito-PstI animals with a 10 g/kg DOX diet (Bioserv©) for 5 days and afterwards replaced with a standard rodent diet. All experiments and comparisons described were performed on age- and gender-matched animals.
Analysis of the NMJ
AChE enzyme activity and oligomeric forms were assessed as described (52). Briefly, hind limb muscles were homogenized in 10 volumes of extraction buffer consisting of 20 mm sodium borate buffer, pH 9.0, 1 M NaCl, 1.0% Triton X-100, 10 mM EDTA, and protease inhibitors. Aliquots were analyzed by velocity sedimentation on 5–20% sucrose gradients, and the fractions were assayed for AChE activity using the colorimetric Ellman assay (53). For semi-quantitative analysis of the NMJs, mouse thigh muscles were labeled with Alexa-555-Btx and Alexa-488 Fasciculin-2, and digital images were taken at two wavelengths appropriate to each fluorophore using a Princeton Instruments Micromax camera mounted on a Leica DMR-A microscope. Images were captured using Slidebook 4.0 software and measured using a calibrated optical micrometer. For analysis of AChE and AChR, the images were captured using identical exposure parameters and the fluorescence intensity at each wavelength expressed as relative units. Confocal images were captured using LSM710 confocal microscope (Zeiss).
Treadmill test
Endurance was evaluated using a six lane treadmill with motivation grid designed for rodents (Columbus Instruments). Animals were given one training day to adapt to the equipment and motivation grid. On the test day, mice were required to run at a speed of 9 m/min for 3 min with the number of falls onto the motivation grid recorded.
Activity monitoring
Spontaneous self-initiated movement was recorded using an activity cage setup (Columbus Instruments) designed for mice. Animals were separately housed in a novel cage environment 30min prior to their dark cycle and monitored for a 20 or 24 h period undisturbed. The number of infrared beam breaks that occurred inside of the cage was counted to determine ambulatory activity.
Western blotting
Protein extracts were prepared from the skeletal muscles we analyzed that were homogenized with a hand-held homogenizer (Omni) in phosphate-buffered saline (PBS) containing a 1× protease inhibitor cocktail (Roche). Samples were then snap-frozen in liquid nitrogen and stored in −80°C until used. Upon use, sodium dodecyl sulfate (SDS) was added to the homogenate at the final concentration of 4%. Homogenates were then centrifuged at 15 000g, and the supernatant was collected for analysis. Protein extracts from cells were isolated from trypsinized cells washed once in PBS 1× protease inhibitor cocktail (Roche). Cells were then lysed with RIPA buffer (62.5 mm Tris; 150 mm NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA pH 7.4) and sonicated. Lysed cells were centrifuged at 15 000g and the supernatant was collected for analysis.
Proteins were quantified using a Dc Protein Assay kit with a Lowry HS method (BioRad). Proteins were loaded on either a 12% SDS-acrylamide gel or 4–20% SDS-polyacrylamide gradient gel (BioRad) depending on the predicted molecular weight. The gel was transferred to polyvinylidene fluoride membrane (BioRad).
Membranes were blocked in 1:1 of PBS with Odyssey blocking solution (LI-COR Biosciences) for 1 hour at room temperature. Primary antibodies used were anti-PstI (54), rodent OXPHOS cocktail (MitoSciences), Tim23 (BD Biosciences), Hsp60 (Cell Signaling), Hsp70 (Cell Signaling), mtHsp70 (Cell Signaling), Grp78 (Cell Signaling), CHOP (Cell Signaling), PDI (Cell Signaling), Pax-7 (Abcam), α-tubulin (Sigma), and actin (Sigma). Primary antibody was incubated overnight at 4°C. HRP conjugated anti- mouse or rabbit secondary antibodies (Cell Signaling) were used at 1:3000–1:5000 dilutions. Secondary antibodies were incubated for 1 h at room temperature. Blots were visualized with Pico Supersignal West Chemiluminescent substrates (ThermoScientific). Films were developed with a Konica Minolta developer. Optical density was quantified using Image J software.
Crude mitochondrial isolation
Mitochondria were isolated from freshly isolated skeletal muscle as previously described (55). Final mitochondrial pellets were resuspended into ice-cold mitochondria incubation buffer containing 130 mm KCl, 2 mm KH2PO4, 2 mm MgCl2, 10 mm HEPES and 1 mm EDTA (pH 7.2). Protein concentration was determined by the Bradford protein assay (BioRad).
Real-time PCR
DNA was extracted from quadriceps using a phenol: chloroform method. The following primer pairs were used for the quantification of mtDNA copy number compared with total DNA: ND1 and β-actin (internal genomic DNA control). Primers and sequences are as follows: ND1 3281F, CAGCCTGACCCATAGCCATA; ND1 3364B, ATTCTCCTTCTGTCAGGTCGAA. Primers for genomic DNA are as follows: β-actin F, GCGCAAGTACTCTGTGTGGA; β-actin B, CATCGTACTCCTGCTTGCTG.
To detect mtDNA deletion events, total DNA was used in a qPCR to detect potential recombination events that involve mtDNA deletions around the PstI restriction endonuclease sites. Primers used and sequences are as follows: small deletion (∼3.8 kb): 8378F, TTCAACCAATGGCATTAGCA; 12281B, GATGTCTCCGATGCGGTTAT; for large deletion: 5549F, AATTGGAGGCTTTGGAAACT; 15501B, CTAGCTTATATGCTTGGGGAAA (∼10.6 kb) (26).
A maxima SYBR Green/ROX qPCR master mix (Fermentas) was used to perform a real-time PCR with the CFX96 Realtime PCR system (Bio-Rad) under the suggested PCR conditions from the manufacturer. The relative quantity was corrected for relative PCR amplification efficiency using BioRad CFX Manager Software. A comparative Ct method was used to determine the relative abundance of mtDNA compared with control samples (56).
Spectrophotometer assays
OXPHOS assays were previously described (57). Briefly, homogenates from skeletal muscles were prepared using a tissue homogenizer (Omni) in PBS plus protease inhibitor cocktail (Roche) on ice. cytochrome c 2 mm reduced with sodium dithionite was added to homogenates in a buffer (10 mm KH2PO4, 1 mg/ml BSA, 120 mm lauryl maltoside). Samples were read at 550 nm with the slope reading taken for 2 min at 37°C. Potassium cyanide 240 μm was used to inhibit the reaction to ensure that the slope was specific to COX. Readings were normalized to protein concentration determined by Bradford methodology.
Homogenates for citrate synthase were added to a buffer [50 mm Tris–HCl pH 7.5, 20 mm acetyl CoA, 10 mm 5,5′-dithiobis (2-nitrobenzoic acid) 0.2% triton X-100] and performed at 412 nm with 50 mm oxaloacetate to start the reaction. Readings were obtained for 5min at 30°C, and normalized to protein concentration determined by Bradford methodology.
In situ cytochrome c and succinate dehydrogenase activity assay
Freshly isolated quadriceps were embedded in OCT compound (Sakura) and immediately frozen in isopentane-cooled liquid nitrogen. Each muscle sample was cut into 10 µm transverse sections and stained for SDH and COX activities as described previously (58).
Inflammatory cytokines quantification
Blood was taken from the left ventricle of deeply anesthetized mice before euthanization. Blood was allowed to clot on ice, and serum was isolated at 1000g in a bench top centrifuge (Eppendorff 5424) for 15 min at 4°C. For complete platelet removal, the serum was re-spun at 10 000g for 10 min at 4°C. Serum was used in BD cytometric bead array mouse inflammation cytokine kit according to the manufacturer's instructions (BD Biosciences). Samples were analyzed on a BD LSR Fortessa cell analyzer (BD Biosciences).
DEXA scan
DEXA scans were performed using a Lunar PIXImus DEXA scan according to manufacturer's instructions. Default software was used to quantify the measurements.
Statistics
A two-tailed, unpaired Student's t-test was used to determine the statistical significance between the different groups. Data are expressed as mean ± SEM, and the numbers of observations/animals used in each experimental series are included in the figure legends. *P< 0.05, **P< 0.01, ***P< 0.005.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by the National Institutes of Health Grants AG036871, NS079965 and EY010804 (C.T.M.) and NS057994 (R.L.R.). The Muscular Dystrophy Association (C.T.M.) and the AHA 11Pre7610007 (A.M.P.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Wayne E. Balkan for the use and direction of the DEXA scan. We also thank the Flow Cytometry Core at the UM Sylvester Comprehensive Cancer Center for assistance and the Lois Pope LIFE Center's light microscopy core for the use of their microscopes.
Conflicts of Interest Statement: The authors declare no conflict of interest.
REFERENCES
- 1.Sehl M.E., Yates F.E. Kinetics of human aging: I. Rates of senescence between ages 30 and 70 years in healthy people. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2001;56:B198–208. doi: 10.1093/gerona/56.5.b198. [DOI] [PubMed] [Google Scholar]
- 2.Jackson A.S., Janssen I., Sui X., Church T.S., Blair S.N. Longitudinal changes in body composition associated with healthy ageing: men, aged 20–96 years. Br. J. Nutr. 2012;107:1085–1091. doi: 10.1017/S0007114511003886. [DOI] [PubMed] [Google Scholar]
- 3.Marcell T.J. Sarcopenia: causes, consequences, and preventions. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2003;58:M911–6. doi: 10.1093/gerona/58.10.m911. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson B.E., Irving D. The numbers of limb motor neurons in the human lumbosacral cord throughout life. J. Neurol. Sci. 1977;34:213–219. doi: 10.1016/0022-510x(77)90069-7. [DOI] [PubMed] [Google Scholar]
- 5.Visser M., Pahor M., Taaffe D.R., Goodpaster B.H., Simonsick E.M., Newman A.B., Nevitt M., Harris T.B. Relationship of interleukin-6 and tumor necrosis factor-alpha with muscle mass and muscle strength in elderly men and women: the Health ABC Study. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2002;57:M326–32. doi: 10.1093/gerona/57.5.m326. [DOI] [PubMed] [Google Scholar]
- 6.Shefer G., Van de Mark D.P., Richardson J.B., Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev. Biol. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryall J.G., Schertzer J.D., Lynch G.S. Cellular and molecular mechanisms underlying age-related skeletal muscle wasting and weakness. Biogerontology. 2008;9:213–228. doi: 10.1007/s10522-008-9131-0. [DOI] [PubMed] [Google Scholar]
- 8.Moyes C.D., Hood D.A. Origins and consequences of mitochondrial variation in vertebrate muscle. Annu. Rev. Physiol. 2003;65:177–201. doi: 10.1146/annurev.physiol.65.092101.142705. [DOI] [PubMed] [Google Scholar]
- 9.Short K.R., Bigelow M.L., Kahl J., Singh R., Coenen-Schimke J., Raghavakaimal S., Nair K.S. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl Acad. Sci. USA. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayo T., Allison D.B., Weindruch R., Prolla T.A. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc. Natl. Acad. Sci. USA. 2001;98:5093–5098. doi: 10.1073/pnas.081061898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn J.M., Sonu R., Vogel H., Crane E., Mazan-Mamczarz K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 13.Cortopassi G.A., Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bua E., Johnson J., Herbst A., Delong B., McKenzie D., Salamat S., Aiken J.M. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbst A., Pak J.W., McKenzie D., Bua E., Bassiouni M., Aiken J.M. Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2007;62:235–245. doi: 10.1093/gerona/62.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detmer S.A., Vande Velde C., Cleveland D.W., Chan D.C. Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot-Marie-Tooth type 2A. Hum. Mol. Genet. 2008;17:367–375. doi: 10.1093/hmg/ddm314. [DOI] [PubMed] [Google Scholar]
- 17.Kong J., Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J. Neurosci. 1998;18:3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 19.Buford T.W., Cooke M.B., Manini T.M., Leeuwenburgh C., Willoughby D.S. Effects of age and sedentary lifestyle on skeletal muscle NF-kappaB signaling in men. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2010;65:532–537. doi: 10.1093/gerona/glp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safdar A., Hamadeh M.J., Kaczor J.J., Raha S., Debeer J., Tarnopolsky M.A. Aberrant mitochondrial homeostasis in the skeletal muscle of sedentary older adults. PloS One. 2010;5:e10778. doi: 10.1371/journal.pone.0010778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirkinezos I.G., Hernandez D., Bradley W.G., Moraes C.T. Regular exercise is beneficial to a mouse model of amyotrophic lateral sclerosis. Ann. Neurol. 2003;53:804–807. doi: 10.1002/ana.10597. [DOI] [PubMed] [Google Scholar]
- 22.Wenz T., Diaz F., Hernandez D., Moraes C.T. Endurance exercise is protective for mice with mitochondrial myopathy. J. Appl. Physiol. 2009;106:1712–1719. doi: 10.1152/japplphysiol.91571.2008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Safdar A., Bourgeois J.M., Ogborn D.I., Little J.P., Hettinga B.P., Akhtar M., Thompson J.E., Melov S., Mocellin N.J., Kujoth G.C., et al. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc. Natl Acad. Sci. USA. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Rosa E.F., Silva A.C., Ihara S.S., Mora O.A., Aboulafia J., Nouailhetas V.L. Habitual exercise program protects murine intestinal, skeletal, and cardiac muscles against aging. J. Appl. Physiol. 2005;99:1569–1575. doi: 10.1152/japplphysiol.00417.2005. [DOI] [PubMed] [Google Scholar]
- 25.Wenz T., Rossi S.G., Rotundo R.L., Spiegelman B.M., Moraes C.T. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc. Natl Acad. Sci. USA. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Fukui H., Moraes C.T. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S.E., Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickrell A.M., Fukui H., Wang X., Pinto M., Moraes C.T. The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. J Neurosci. 2011;31:9895–9904. doi: 10.1523/JNEUROSCI.6223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava S., Moraes C.T. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacman S.R., Williams S.L., Moraes C.T. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37:4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degens H. The role of systemic inflammation in age-related muscle weakness and wasting. Scand. J. Med. Sci. Sports. 2010;20:28–38. doi: 10.1111/j.1600-0838.2009.01018.x. [DOI] [PubMed] [Google Scholar]
- 32.Schaap L.A., Pluijm S.M., Deeg D.J., Visser M. Inflammatory markers and loss of muscle mass (sarcopenia) and strength. Am. J. Med. 2006;119:526 e9–17. doi: 10.1016/j.amjmed.2005.10.049. [DOI] [PubMed] [Google Scholar]
- 33.Chung H.Y., Kim H.J., Kim K.W., Choi J.S., Yu B.P. Molecular inflammation hypothesis of aging based on the anti-aging mechanism of calorie restriction. Microsc. Res. Tech. 2002;59:264–272. doi: 10.1002/jemt.10203. [DOI] [PubMed] [Google Scholar]
- 34.Durham S.E., Bonilla E., Samuels D.C., DiMauro S., Chinnery P.F. Mitochondrial DNA copy number threshold in mtDNA depletion myopathy. Neurology. 2005;65:453–455. doi: 10.1212/01.wnl.0000171861.30277.88. [DOI] [PubMed] [Google Scholar]
- 35.Coskun P.E., Wyrembak J., Derbereva O., Melkonian G., Doran E., Lott I.T., Head E., Cotman C.W., Wallace D.C. Systemic mitochondrial dysfunction and the etiology of Alzheimer's disease and down syndrome dementia. J. Alzheimers Dis. 2010;20(Suppl 2):S293–310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahlqvist K.J., Hamalainen R.H., Yatsuga S., Uutela M., Terzioglu M., Gotz A., Forsstrom S., Salven P., Angers-Loustau A., Kopra O.H., et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Yoshioka M., Tanaka H., Shono N., Snyder E.E., Shindo M., St-Amand J. Serial analysis of gene expression in the skeletal muscle of endurance athletes compared to sedentary men. FASEB J. 2003;17:1812–1819. doi: 10.1096/fj.02-1200com. [DOI] [PubMed] [Google Scholar]
- 38.Ellgaard L., Ruddock L.W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hepple R.T. Muscle atrophy is not always sarcopenia. J. Appl. Physiol. 2012;113:677–679. doi: 10.1152/japplphysiol.00304.2012. [DOI] [PubMed] [Google Scholar]
- 40.Ibebunjo C., Chick J.M., Kendall T., Eash J.K., Li C., Zhang Y., Vickers C., Wu Z., Clarke B.A., Shi J., et al. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol. Cell. Biol. 2013;33:194–212. doi: 10.1128/MCB.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chai R.J., Vukovic J., Dunlop S., Grounds M.D., Shavlakadze T. Striking denervation of neuromuscular junctions without lumbar motor neuron loss in geriatric mouse muscle. PloS One. 2011;6:e28090. doi: 10.1371/journal.pone.0028090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drey M., Sieber C.C., Bauer J.M., Uter W., Dahinden P., Fariello R.G., Vrijbloed J.W. C-terminal agrin fragment as a potential marker for sarcopenia caused by degeneration of the neuromuscular junction. Exp. Gerontol. 2013;48:76–80. doi: 10.1016/j.exger.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 43.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly Y.M., Gidlof S., Oldfors A., Wibom R., et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 44.Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 45.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 46.Macharia R., Otto A., Valasek P., Patel K. Neuromuscular junction morphology, fiber-type proportions, and satellite-cell proliferation rates are altered in MyoD(−/−) mice. Muscle Nerve. 2010;42:38–52. doi: 10.1002/mus.21637. [DOI] [PubMed] [Google Scholar]
- 47.Shigemoto K., Kubo S., Mori S., Yamada S., Akiyoshi T., Miyazaki T. Muscle weakness and neuromuscular junctions in aging and disease. Geriatr. Gerontol. Int. 2010;10(Suppl 1):S137–47. doi: 10.1111/j.1447-0594.2010.00608.x. [DOI] [PubMed] [Google Scholar]
- 48.Courtney J., Steinbach J.H. Age changes in neuromuscular junction morphology and acetylcholine receptor distribution on rat skeletal muscle fibres. J. Physiol. 1981;320:435–447. doi: 10.1113/jphysiol.1981.sp013960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Lee Y., Thompson W.J. Changes in aging mouse neuromuscular junctions are explained by degeneration and regeneration of muscle fiber segments at the synapse. J. Neurosci. 2011;31:14910–14919. doi: 10.1523/JNEUROSCI.3590-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valdez G., Tapia J.C., Kang H., Clemenson G.D., Jr., Gage F.H., Lichtman J.W., Sanes J.R. Attenuation of age-related changes in mouse neuromuscular synapses by caloric restriction and exercise. Proc. Natl Acad. Sci. USA. 2010;107:14863–14868. doi: 10.1073/pnas.1002220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W., Esbensen Y., Kunke D., Suganthan R., Rachek L., Bjoras M., Eide L. Mitochondrial DNA damage level determines neural stem cell differentiation fate. J. Neurosci. 2011;31:9746–9751. doi: 10.1523/JNEUROSCI.0852-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossi S.G., Rotundo R.L. Localization of non-extractable acetylcholinesterase to the vertebrate neuromuscular-junction. J. Biol. Chem. 1993;268:19152–19159. [PubMed] [Google Scholar]
- 53.Ellman G.L., Courtney K.D., Andres V., Featherstone R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961;7:88–90. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava S., Moraes C.T. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum. Mol. Genet. 2001;10:3093–3099. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- 55.Diaz F., Thomas C.K., Garcia S., Hernandez D., Moraes C.T. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 2005;14:2737–2748. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods. 2002;26:307–316. doi: 10.1016/S1046-2023(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 58.Sciacco M., Bonilla E. Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods Enzymol. 1996;264:509–521. doi: 10.1016/s0076-6879(96)64045-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.