Abstract
miRNAs originate from primary transcripts (pri-miRNAs) with characteristic stem-loop structures. Accurate processing of pri-miRNAs is required for functional miRNAs. Here, using pri-miR166 family as a paradigm, we report the decisive role of pri-miRNA terminal loops in miRNA biogenesis. We found that multi-branched terminal loops in pri-miR166s substantially suppressed miR166 expression in vivo. Unlike canonical processing of pri-miRNAs, terminal-loop-branched (TLBed) pri-miRNAs can be processed by Dicer-like1 (DCL1) complexes bi-directionally: from base to loop and from loop to base, resulting in productive and abortive processing of miRNAs, respectively. In either case, DCL1 complexes canonically cut pri-miRNAs at a distance of 16-17 base pairs (bp) from a reference single-stranded loop region. DCL1 also adjusts processing sites toward an internal loop through its helicase domain. Thus, these results provide new insight into the poorly understood processing mechanism of pri-miRNAs with complicated secondary structures.
miRNA biogenesis starts with transcription of long pri-miRNAs, typically by RNA polymerase II. Pri-miRNAs are characterized by stem-loop structures consisting of a terminal loop, an upper stem, a duplex of miRNA and its complementary strand (miRNA/*), a lower stem, and flanking single-stranded (ss) basal segments (Fig.1a). In animals, pri-miRNAs are first processed by an RNase III enzyme Drosha and its partner DGCR8 protein (also known as Pasha)1. DGCR8 associates with the base of the stem–loop structure and sets the catalytic site of Drosha 11 bp away from ssRNA-double-stranded (ds) RNA junction2. The resultant products, also known as precursor miRNAs (pre-miRNAs), are further cleaved by another RNase III enzyme Dicer to release ~21bp miRNA/* duplexes2. Mature miRNAs are loaded into Argonaute (AGO)-containing RNA induced silencing complexes (RISCs) and guide AGOs to repress their complementary targets1,3-10.
Figure 1.
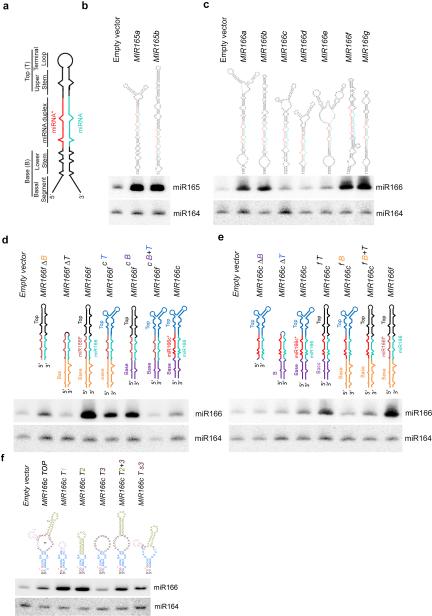
Secondary structures of pri-miRNAs affect miRNA abundance in vivo. (a) Schematic structure of a representative pri-miRNA. (b and c) sRNA blot analysis of miR165 and miR166 in T1 transgenic plants overexpressing nine MIR165 and MIR166 family members. Predicted secondary structures of pri-miR165/166s are shown on the top. miRNA and miRNA* are marked as turquoise and red, respectively. Total RNA was prepared from a pool of T1 transformants (n>200 for each construct). sRNA blots were probed using 5′ end 32P-labelled oligo probes complementary to the indicated miRNAs. miR164 serves as a loading control. (d and e) sRNA blot analysis of miR166 in the T1 stable transformants overexpressing deletion (Δ) or element-swapped mutants of pri-miR166f (d) and pri-miR166c (e). Bases (B), miRNA/*s, and Tops (T) of pri-miR166c and pri-miR166f are shown in different colors in the schematic structures. sRNA blot assays were performed as in Panels (b and c). (f) sRNA blot analysis of miR166 in the T1 stable transformants overexpressing MIR166c with various deletions in the terminal loop. Schematic structures of terminal regions of pri-miR166c deletion mutants are shown on the top. Loops # 1, 2, and 3 are shown in pink, greenish yellow and maroon, respectively. sRNA blot assays were done as in Panels (b and c). Related uncropped images can be found Supplementary 9.
In Arabidopsis, DCL1, one of four Dicer homologs, orchestrates the entire process including the pri-miRNA to pre-miRNA conversion and the pre-miRNA to miRNA/* release5,11. DCL1 has two cofactors, Serrate (SE), and a dsRNA-binding protein—Hyponastic Leaves1 (HYL1) among others12. HYL1 and SE were proposed to play a role analogous to DGCR8 by acting as a molecular ruler for miRNA processing in plants13-20. Plant pri-miRNA hairpins are heterogeneous in length and structure, with variable positioning of the miRNA/* duplex21,22. Genetic studies have demonstrated that an imperfectly paired lower stem of ~15bp below the miRNA/* duplex is a key element for the initial pri-miRNA cleavage, leading to a proposal of a “15-nucleotide (nt)” model that initial pri-miRNA processing often occurs at a distance of ~15-nt from either ssRNA-dsRNA junctions or internal unstructured regions23-26. In contrast, the terminal-loop and upper stem are largely tolerant to mutations for some pri-miRNAs24, but not for others27,28. Moreover, many plant pri-miRNAs harbor branched terminal loops (BTLs) or multiple bulges or internal loops in the lower and upper stems. How pri-miRNAs with complicated secondary structures are recognized and processed remains elusive.
miR165-166 are two related miRNAs that differ in sequence by only 1-nt, and both target the HD-ZIP III transcripts29,30 to regulate meristem development and organ polarity. They are encoded by nine loci in the Arabidopsis genome (MIR165a-b; MIR166a-g). The pri-miR165-166s contain diversified structures from linear foldbacks to complicated branched loops at variable locations in the pri-miRNAs. Here, we showed that BTLs markedly decreased miR166 abundance in the majority of cases. DCL1 complexes processed the TLBed pri-miRNAs bi-directionally: either canonically from base to loop resulting in miRNA production; or non-canonically from loop to base leading to abortive processing. DCL1 complexes cut pri-miRNAs at 16-17bp away from referent ss-dsRNA junctions for either canonical or non-canonical processing, while DCL1 could also adjust the processing sites near an internal loop through its helicase domain. Thus, these results shed light on the complicated relationship between pri-miRNA structures and miRNA biogenesis in plants.
RESULTS
BTLs of pri-miRNAs repress miRNA accumulation in vivo
We recently observed that transgenic plants harboring different pri-miR165-166s, even expressed under the same constitutive 35S promoter, exhibited diversified phenotypic abnormalities, ranging from severe pinhead, radial or curled leaves, to nearly wild-type phenotypes (Supplementary Fig.1a,b). This phenotypic severity is proportional to miR165-166 amount (Fig. 1b,c). Mfold analysis of the pri-miRNAs31 revealed two major types of secondary structures: one is a roughly linear foldback, whereas the other harbors BTLs. Interestingly, pri-miR165-166s with linear hairpins generally showed stronger morphological defects and more abundant miRNA accumulation than the ones with BTLs except for pri-miR165a and pri-miR166a (Fig.1b,c). These observations indicate that the secondary structures of pri-miRNAs play a decisive role in miRNA abundance.
Pri-miR166c and pri-miR166f exhibited the lowest and highest miR166 levels. Both pri-miRNAs contain two internal loops and one bulge in their lower stems. However, pri-miR166c has BTLs whereas pri-miR166f harbors a less structured terminal region (Fig. 1c). To delineate structural determinants that affect miRNA expression, we conducted base-top swapping experiments between pri-miR166c and pri-miR166f. Switching either the Base or Top of pri-miR166c to pri-miR166f substantially decreased miR166 accumulation and attenuated transgenic phenotype (Fig. 1d and Supplementary Fig. 1c). On the other hand, swapping of pri-miR166f Top to pri-miR166c led to enhanced miR166 accumulation and more severe phenotypes. Interestingly, this enhancement was not observed with transgenic plants expressing chimeric pri-miR166cf B that harbors the Base of pri-miR166f in the pri-miR166c context (Fig. 1e and Supplementary Fig. 1d). These results were reproducible with either extended or concise forms of pri-miRNAs in Nicotiana benthamiana (N. Bentha.) (Supplementary Fig. 1a,f,g). Thus, a combination of Base, Top, and miRNA/* in an appropriate pri-miRNA context is required for maximum miRNA accumulation.
Pri-miR166c BTLs contain two small lobe loops (labeled as #1 and #2) attached to a large terminal loop (labeled as #3) (Fig. 1f, MIR166c TOP). Deletion of loop #3 (MIR166c T1 and MIR166c T2), reduction in its size (MIR166c Ts3), and linearization of the foldback (MIR166c T2+3) substantially increased miR166 accumulation and enhanced the severity of transgenic phenotypes; whereas the large terminal loop in the Top (MIR166c T3) demonstrated a pattern similar to pri-miR166c (Fig. 1f and Supplementary Fig. 1e). Together, we concluded that the size, shape, and distance of terminal loops relative from miRNA/* duplexes can be critical for miRNA accumulation in vivo.
Abortive processing of pri-miR166c lowers miR166 production
Pri-miR166c and pri-miR166f had comparable primary transcript levels in vivo (Supplementary Fig. 2a), indicating that the differential accumulation of miR166 in their transformants was regulated at a posttranscriptional level. We hypothesized that different secondary structures of pri-miR166c and pri-miR166f may alter their processing efficiency and/or accuracy by DCL1 complexes. To test this hypothesis, we developed an in vitro system of DCL1 reconstitution assays (Supplementary Fig. 2b-d). We coexpressed 35S-driven 2Flag-4Myc-DCL1, -6Myc-HYL1, and -SE-3HA in N. Bentha because DCL1–HYL1–SE can form an efficient miRNA processor15,32. The purified DCL1 complexes cleaved the 5′ end labeled pri-miR166f transcript at the expected position, primarily generating a 26-nt fragment, which is the 5′ flanking segment (f1, 26 nt) (Fig. 2a,b). We performed a parallel experiment with an internally labeled pri-miR166f and observed six major fragments: f1, the 3′ flanking segment (f2, 35-nt), pre-miR166f (f3, 89-nt), the upper stem and terminal loop (f4, 47nt), and miR166/* duplexes (f5 and f6, ~ 21-22-nt). Hence, the in vitro DCL1 reconstitution assays were able to recapitulate the process of miRNA biogenesis in vivo.
Figure 2.
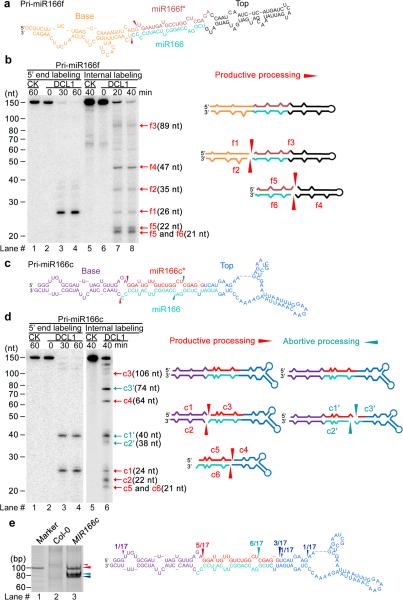
Concomitant presence of productive and abortive processing of pri-miR166c but not of pri-miR166f. (a) Predicted secondary structure of in vitro transcribed pri-miR166f. Red arrows indicate the expected initial processed sites of pri-miR166f. (b) In vitro DCL1 reconstitution assay with the 5′ end or internally labeled pri-miR166f transcripts. Immunoprecipitates were prepared through two-step affinity purification using antibodies to Flag and then to Myc from N. bentha expressing 2Flag-4Myc-DCL1, 6Myc-HYL1, and SE-3HA complexes (DCL1) and prepared from mock-infiltrated plants (CK, referring to control check). RNAs recovered from the reaction mix were fractionated on 15% denaturing gels and detected by phosphor imaging. The positions of intact substrates, cleavage products, and reaction time are marked. Schematic illustration of the intermediate and final cleavage products (f1-f6) of pri-miR166f by DCL1 is shown on right. (c) Predicted secondary structure of in vitro transcribed pri-miR166c. Red and turquoise arrows show the expected initial productive processing and the unexpected abortive processing sites of pri-miR166c, respectively. (d) In vitro DCL1 reconstitution assays with 5′ end or internally labeled pri-miR166c transcripts. The assays were conducted as in Panel (b). Schematic illustration of the intermediate and final cleavage products (c1-6 and c1′-c3′) of pri-miR166c by DCL1 is shown on right. (e) Productive- and abortive-processing fragments were detected by 5′ RACE experiments in transgenic plants expressing 35S-MIR166c in vivo. The PCR products from the 5′ RACE experiments were recovered, sequenced, and mapped on the pri-miR166c and their counts are shown on right. Related uncropped images can be found Supplementary 9.
In sharp contrast to the pri-miR166f processing, incubation of 5′ end labeled pri-miR166c with DCL1 complexes generated equal amounts of two major fragments (Fig. 2c,d). One fragment (c1, 24-nt) yielded functional pre-miRNA representing productive processing. However, the other fragment (c1′, 40-nt) suggested abnormal, abortive cleavage in pri-miR166c because the cleaved product did not contain the entire miR166 sequence. We further confirmed the presence of two processing patterns for pri-miR166c with internally labeled pri-miR166c transcripts which yielded two sets of cleavage fragments: c1-c6 fragments, equivalent to f1-f6 from pri-miR166f above, resulting from the constructive processing; and c′1-c′3 fragments, derived from abortive processing.
To determine whether abortive processing of pri-miR166c was present in planta, we performed experiments of 5′ rapid amplification of cDNA ends (5′ RACE). Although we did not recover clear intermediate processing products from pri-miR166c in wild-type plants, we identified distinct processing intermediates from 35S-MIR166c transgenic plants (Fig. 2e). These processing products indeed largely corresponded to the cleavage positions found in our in vitro analysis, indicating that abortive processing of pri-miR166c likely accounts for the lower accumulation of miR166 from pri-miR166c in vivo (Fig. 2d and Supplementary Fig. 3a-e).
Because we were unable to recover SE in our purified DCL1–HYL1 complexes, we repeated the DCL1 assays but added purified SE (Supplementary Fig. 2d-f). SE had no obvious effect on the cleavage pattern and accuracy for pri-miR166c or pri-miR166f, suggesting that this protein might not directly contribute to the processing of pri-miRNAs. In agreement with our notion, recent work demonstrated that SE functions as a scaffold for C-terminal domain phosphatase-like1 (CPL1) to dephosphorylate HYL1, therefore modulating its activity33.
DCL1 processes pri-miR166c in two opposite directions
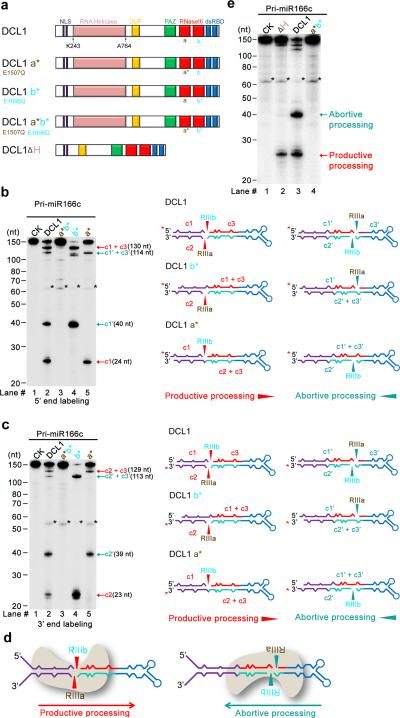
The presence of two sets of pri-miR166c processing products suggests that the DCL1-centered microprocessor recognizes two locations in pri-miR166c for cleavage. To understand how DCL1 complexes recognize pri-miR166c, we modified DCL1 to examine the relative binding orientation of DCL1 complexes to pri-miR166c. Dicers contain an RNA helicase domain, a PAZ domain, one or two dsRNA-binding domains (dsRBDs) and two RNase III (RIIIa and RIIIb) domains that cut dsRNAs concomitantly (Fig. 3a). Bioinformatics analysis revealed that residues Glu1507 and Glu1696 of DCL1 are highly conserved in RIIIa and RIIIb domains of Dicers among eukaryotic organisms34. We generated DCL1 (E1507Q) and DCL1 (E1696Q) point mutations, which inactivate the RIIIa and RIIIb domains respectively (Supplementary Fig. 4a). We reasoned that incubation of these semi-active DCL1 mutants with pri-miR166c may produce partially processed fragments, which could be used to deduce the binding orientations of DCL1 in the pri-miR166c. While DCL1a* b* (E1507Q E1696Q) totally abolished the RNase III activity (Fig. 3b,c, lanes #3), DCL1a* (E1507Q) generated c1 (24-nt) and c1′+c3′ (114-nt) fragments when the 5′ end labeled pri-miR166c was used in the assay (Fig. 3b, lane #5). In parallel, DCL1a* produced two fragments corresponding to the c2′ (39-nt) and c2+c3 (129-nt) segments when the 3′ end labeled pri-miR166c was used as a substrate (Fig. 3c, lane #5). These results indicate that DCL1 RIIIb cut the 5′ arm of the lower stem in productive processing and the 3′ arm (the site of miR166c) of pri-miR166c in abortive processing. On the other hand, DCL1b* (E1696Q) generated c1′ (40-nt) and c1+c3 (130-nt) fragments when the 5′ end labeled pri-miR166c was applied (Fig. 3b, lane #4), whereas it yielded c2 (23-nt) and c2′+c3′ (113-nt) fragments when the 3′ end labeled pri-miR166c was used (Fig. 3c, lane #4). These results denote that DCL1 RIIIa cut the 3′ arm of the lower stem in productive processing and the 5′ arm of pri-miR166c (the site of miR166c*) in abortive processing. In addition, wild-type DCL1 also generated intermediate cleavage fragments (Fig 3b,c, lane #2, c1+c3; c1′+c3′), indicating imperfectly coordinated processing at both sites. Taken together, DCL1 has two opposite binding orientations in pri-miR166c (Fig. 3d): one orientation processes pri-miR166c through its lower stem at the base, resulting in constructive production of pre-miRNAs, while the other orientation launches pri-miR166c through BTLs, leading to destruction of pre-miRNAs.
Figure 3.
DCL1 complexes process pri-miR166c bi-directionally: from lower stem to loop and from loop to lower stem. (a) Schematic illustration of DCL1 domains and DCL1 mutants. DCL1 a*, b*, a* b*, and DCL1 (ΔH) refer to RNaseIII a (E1507Q), b (E1696Q) point mutation, double mutations (E1507Q E1696Q), and a helicase-domain deletion mutant, respectively. (b) In vitro cleavage assays with the 5′ end labeled pri-miR166c transcripts by DCL1 and DCL1 mutants. Immunoprecipitation, cleavage assays, and RNA processing were performed as in Figure 2b. Black asterisks show the residual non-specific cleavage frequently present in the assay system. Schematic illustration of the cleavage products is shown on right. Red asterisks on transcripts indicate the 32P-labelling positions. Red and turquoise arrows show the productive and abortive processing sites, respectively. (c) In vitro cleavage assays with the 3′ end labeled pri-miR166c transcripts by DCL1 and DCL1 mutants. The cleavage assays and schematic illustration were essentially the same as in Panel (b). Note: the 3′ end labeling added extra “Cytosine” on the 3′ end of pri-miR166c and thus, the lengths of c2 and c2′ fragments in Figure 3c are 1-nt longer than those in Figure 2d. (d) Schematic for two orientations of DCL1 in processing of pri-miR166c and resultant constructive or destructive processing patterns. (e) In vitro cleavage assays with the 5′ end labeled pri-miR166c transcripts by DCL1(ΔH). The cleavage assays and schematic illustration were the same as in Panel (b). Related uncropped images can be found Supplementary 9.
Helicase domain of DCL1 modulates its catalytic activity32. To study whether the helicase domain plays a role in pri-miR166c processing, we repeated the cleavage assays using a truncated DCL1 (Δ helicase domain). We found that the deletion of the helicase domain completely abolished DCL1 activity for abortive processing and to some extent, it compromised productive processing of pri-miR166c (Fig. 3e). Consistent with this result, ATP is required for abortive processing but only partially necessary for productive processing (Supplementary Fig. 4b). Thus, the helicase-mediated unwinding of pri-miR166c structures, which is driven by ATP hydrolysis, is required for abortive processing.
DCL1 processes pri-miRNAs at 16-17bp from a reference site
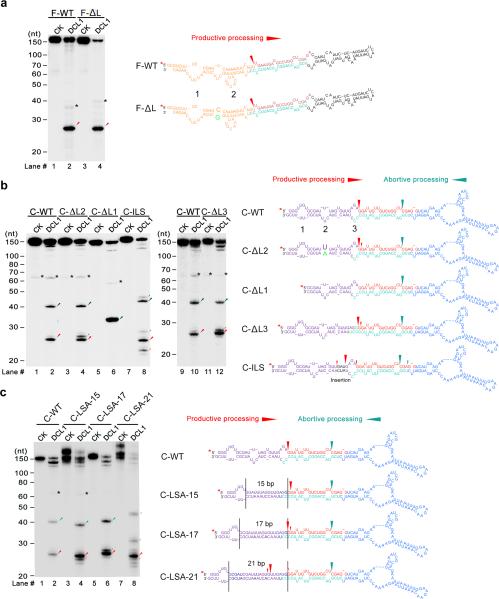
Given the bidirectional activity of DCL1 complexes, we wanted to investigate how the cleavage sites are determined. Multiple internal loops or bulges present in the lower stems of pri-miR166c and pri-miR166f essentially mimic ssRNA-dsRNA junctions and thus, may serve as the reference sites for guiding DCL1 catalytic activity. To investigate this possibility, we first annealed internal loop #2, which is ~11bp away from the miR166/* duplex, in the lower stems of pri-miR166f (Fig. 4a, lane #4, F-ΔL) and pri-miR166c (Fig. 4b, lane #4, C-ΔL2) and examined how the cleavage patterns were affected. Annealing internal loop #2 of either pri-miR166c or pri-miR166f did not alter the predominant cleavage patterns. Thus, the length of the molecular ruler in the pri-miRNA processing in plants is not ~11bp as observed in animals2.
Figure 4.
DCL1 initiates cleavage at 16-17bp from the ssRNA-dsRNA junction in pri-miRNAs. (a) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts of a pri-miR166f mutant. Schematic illustration of the cleavage products by DCL1 is shown on right. Two internal loops in the lower stems of pri-miR166f are shown as numbers 1 and 2. (b) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts of pri-miR166c deletion and insertion mutants in the lower stem. Schematic illustration of the cleavage products by DCL1 is shown on right. Three internal loops in the lower stems of pri-miR166c are shown as numbers 1, 2 and 3, respectively. (c) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts of pri-miR166c mutants with fully complementary lower stems. Schematic illustration of the cleavage products by DCL1 is shown on right. For panels (a-c), immunoprecipitation, cleavage assays and RNA processing were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are marked. Black asterisks show the residual non-specific cleavage frequently present in the assay system. Red asterisks on transcripts indicate the 32P-labelling positions. Large and small red arrows show the predominant and minor cleavage sites while turquoise arrows indicate abortive processing sites.
We next deleted internal loop #1 in pri-miR166c, which is 17-nt away from the miR166/* duplex if the 5′ arm is counted (16-nt if the 3′ arm is counted). Only one major product (~33-nt) was produced in the DCL1 reconstitution assay (Fig. 4b, lane #6, C-ΔL1). Further studies with the DCL1 RNase III a or b-compromised mutants (Supplementary Fig. 5) indicated that the 33-nt cleavage product resulted from abortive processing starting from BTLs to the miRNA/* duplex. This result indicated that the internal loop #1 was required for the canonical processing of pri-miR166c and likely served as the counting site for DCL1 processing. To further define the length of the molecular ruler of the pri-miRNA microprocessor, we created pri-miR166c mutants by either annealing internal loop #3 (Fig. 4b, lane #12, C-ΔL3) or inserting three more nucleotides between internal loops #2 and #3 in the lower stem (Fig. 4b, lanes #8. C-ILS). In either case the length of the predominant cleavage products remained the same. These results indicated that the distance between the reference and processing sites is ~16-17bp long in plants.
The presence of internal loops and bulges between the reference sites and processing sites in the pri-miRNAs might affect the measurement accuracy of the microprocessor in the pri-miRNAs. To examine this possibility, we created another set of pri-miR166c mutants harboring completely linear lower stems but of different lengths (Fig. 4c). DCL1 cleaved the pri-miRNA mutants in a way similar to the wild-type pri-miR166c (Fig. 4b,c), further validating that the processing sites in pri-miRNA were 16-17bp away from the ssRNA reference site. Together, our in vitro results support the “15nt model” observed in previous genetic analyses23-26 and more precisely pinpoint cleavage sites of DCL1 complexes at 16-17bp away from reference sites.
DCL1 adjusts processing sites close to an internal loop
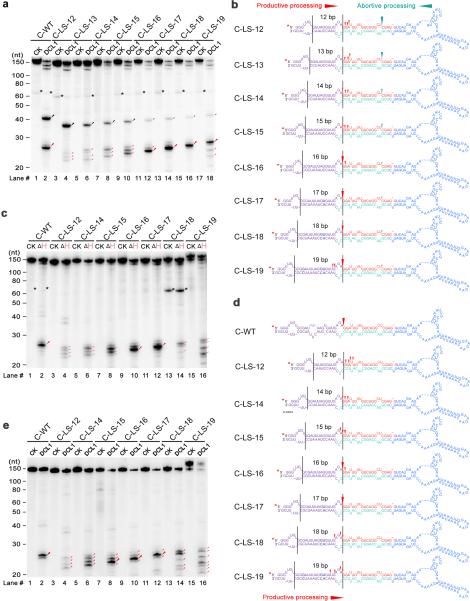
Plant pri-miRNAs are highly heterogeneous in structure and many of them lack lower stems with the optimal distances of 16-17bp length between reference ssRNA regions and miRNA/* duplexes25. We wondered if plant DCL1 complexes have some flexibility for measuring distances and/or an additional sensor system to identify processing sites that allow for the precise cleavage of pri-miRNAs with structural and length heterogeneity. To facilitate our experiments, we annealed the internal loop #2 and a small bulge between internal loops #2 and #3 in the lower stem of pri-miR166c (Fig. 5a,b, C-LS-17). Next, we generated a series of deletions or insertions based on the C-LS-17 backbone for DCL1 cleavage assays. In agreement with the optimal length of the molecular ruler identified above (Fig. 4), the cleavage efficiency of pri-miRNAs was highest when the miRNA/* duplex was 17bp away from the reference internal loop (Fig. 5a, lane #14, C-LS-17). Deviation from this optimal distance compromised the productivity and accuracy of DCL1 complexes. Specifically, if the distance was shorter than 16bp, DCL1 inaccurately cut the pri-miRNAs at multiple locations, resulting in a pool of heterogeneous fragments (Fig. 5a,b). This result is consistent with previous genetic results that show the sequence and structure of the lower stem (within ~15nt away from the miRNA/* duplex) is critical for accurate processing of pri-miRNA24-26. Interestingly, when the lower stem is shorter than the optimal length, more products resulting from abortive processing were observed while fewer products were generated from productive processing (Fig. 5a,b). The inverse correlation between abortive and productive processing on the same pri-miRNAs suggests a steric hindrance for launching DCL1 complexes from two opposite directions on the same pri-miRNA.
Figure 5.
DCL1 complexes adjust cleavage sites toward the edge of internal loops through the helicase domain. (a) In vitro DCL1 reconstitution assays with ATP. (b) Schematic illustration of the cleavage products from the panel (a). (c) In vitro DCL1 reconstitution assays with DCL1 (ΔH) and ATP. (d) Schematic illustration of the cleavage products from the panel (c). (e) In vitro DCL1 reconstitution assays without ATP. Schematic illustration of the cleavage products from the panel (e) was similar to (d) and thus omitted here. In panels (a-e), the substrates were the 5′ end labeled transcripts of pri-miR166c mutants with varying distances between internal loops and the reference sites. Immunoprecipitation, cleavage assays and RNA processing were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Black asterisks show non-specific cleavage products frequently present in the assay system. Large and small red arrows show the predominant and minor cleavage sites while turquoise arrows indicate abortive processing sites. Black lines mark the locations of reference sites and internal loops. Red asterisks on the transcripts indicate 32P-labelling positions. Note: predominant cleavages at the edges of internal loops in C-LS-18 and C-LS-19 disappeared in (c) and (e) compared to those in (a).
On the other hand, when the distance is 1-2bp longer than the optimal 17bp, the predominant cleavage sites remained at the same location–at the edge of an internal loop, while minor cleavage products resulting from inaccurate cleavage were observed, (Fig. 5a, lanes #16 and 18; Fig. 5b). This result suggested that DCL1 complexes could sense the loop and adjust their catalytic sites toward the edge of the loop for cleavage. Intriguingly, this cleavage pattern is reminiscent of a recent observation with animal Dicers that can sense loop regions to determine pre-miRNA processing35. Consistent to this notion, a number of miRNA/* duplexes reside nearby an internal loop ~18-19bp away from reference sites of pri-miRNAs in a genome-wide analysis28. Hence, DCL1 complexes can tolerate slight deviations from the optimal 17bp distance between the reference site and the miRNA/* duplex, if the processing position is close to an internal loop.
To further study how DCL1 complexes recognize the internal loop for processing, we examined a possible role of DCL1 helicase domain. Similar to DCL1, DCL1 (ΔH) predominantly cleaved pri-miRNAs at the sites of 16-17bp away from the ssRNA reference sites, whether or not the processing sites were at the edge of the internal loops (Fig. 5c,d and Supplementary Fig. 4c). These results further validated that the canonical ruler of the microprocessor is 16-17bp in length. In contrast, DCL1 (ΔH) generated multiple cleavage products when the internal loops deviated 1-2bp from the optimal processing sites, suggesting that pri-miRNAs were inaccurately processed. This result was also line with a previous study that deletion of the helicase domain leads to inaccuracy of miRNA biogenesis32. The deletion of the entire helicase domain potentially changes the DCL1 overall structure, resulting in the compromised processing accuracy. To test this possibility, we repeated the cleavage assays with wild-type DCL1 in the absence of ATP and GTP. In this scenario, the cleavage patterns were essentially identical to the ones with DCL1 (ΔH) (Fig. 5e and Supplementary Fig. 4d). Together, these results indicated that the helicase domain inspects an internal loop for the processing authenticity, a situation reminiscent of fly Dicer1 that recognizes ss terminal-loop structures of pre-miRNAs through its helicase domain36.
Structure determinants for pri-miRNA abortive processing
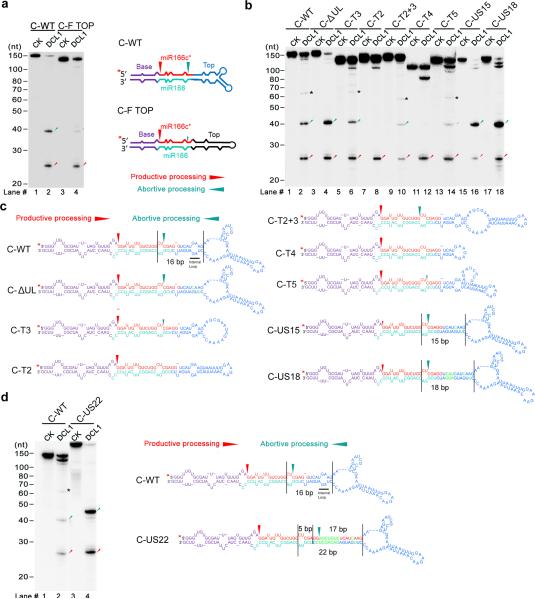
Swapping the upper stem and terminal region of pri-miR166c with the counterparts of the pri-miR166f abolished abortive processing (Fig. 6a), suggesting that the large terminal loop might be disguised as an ssRNA-dsRNA reference site, leading to the non-canonical recruitment of DCL1 complexes. To further pinpoint the critical structures required for the abortive processing, we created a series of mutations in the upper stem and the terminal loops of pri-miR166c (Fig. 6 b,c). Decrease in the size of the terminal loop either completely blocked or substantially decreased abortive processing (Fig. 6b, lane #8; C-T2; lane #12; C-T4; lane #14, C-T5; Supplementary Fig. 5b). Additionally, a terminal loop with less complicated top structures also alleviated abortive processing to a certain extent (Fig. 6b, lane #10, C-T2+3). On the other hand, the presence of ~15-18bp-long linearized upper stem close to a large terminal loop favored abortive processing over productive processing (Fig. 6b, lanes #16 and 18). These in vitro results are consistent with miR166 accumulation in the transgenic plants expressing these constructs (Fig. 1f). Together, a large terminal loop in pri-miRNAs readily triggers non-canonical launching for a microprocessor, resulting in abortive processing.
Figure 6.
Secondary structural elements required for abortive processing in pri-miR166c. (a) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts of chimeric pri-miR166c containing the top region of pri-miR166f. Schematic illustration of the cleavage products by DCL1 is shown on right. (b) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts of pri-miR166c mutants with various deletions, insertions and base-pairings in the upper stem and terminal loop. (c) Schematic illustration of the cleavage products by DCL1 from panel (b). (d) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts of a pri-miR166c mutant with a full complementary upper stem. Immunoprecipitation, cleavage assays, and RNA processing were performed as described Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Black asterisks show non-specific cleavage products frequently present in the assay system. Red asterisks on the transcripts indicate 32P-labelling positions. Black lines show the distance between the indicated positions. Red arrows show the expected productive cleavage sites while turquoise arrows indicate abortive processing sites.
Similar to the scenario observed in productive processing, all abortive processing sites tended to occur close to internal loops (Fig. 5a and Fig. 6c). To precisely validate the length of the molecular ruler used in the abortive processing, we first annealed a small internal loop (Fig. 6b,c, lane #4, C-ΔUL) that is 11bp away from abortive processing sites. The abortive processing pattern remained unchanged. Furthermore, we created a pri-miR166c mutant by increasing the length of the upper stem between the internal loop and terminal loop (Fig. 6d, C-US22). DCL1 reconstitution assays with pri-miR166c-US22 revealed that the abortive processing site was exactly 17bp away from the large terminal loop. This result indicated that the molecular ruler for abortive processing was identical to the one used in productive processing (Figs. 4 and 5).
Many other pri-miR165-166s in Arabidopsis also harbor complicated terminal structures. To examine whether abortive processing was applied to these pri-miRNAs, we tested pri-miR166d in DCL1 reconstitution assays. As expected, pri-miR166d, similar to pri-miR166c, also displayed abortive processing patterns (Supplementary Fig. 6). These results further indicate that abortive processing of pri-miRNAs accounted for the low accumulation of miR166 in the transgenic lines (Fig. 1). Because pri-miR165a and pri-miR166a have branched terminal loops but yield higher in vivo levels of miR165-166 expression, we wanted to learn how DCL1 complexes process these pri-miRNAs. In vitro assays showed that pri-miR165a, in contrast to pri-miR166c, was predominantly cleaved in a productive manner (Supplementary Fig. 6). The preference for productive processing over abortive processing of pri-miR165a is likely due to its relatively smaller terminal loop size and the shorter distance between the terminal loop and the miRNA/* duplex. More intriguingly, pri-miR166a could never be cut by DCL1 in our reconstitution system (Zhu et al., unpublished), suggesting that there might be an alternative pathway bypassing the canonical DCL1-mediated processing in vivo.
Bi-directional processing extends beyond pri-miR166 family
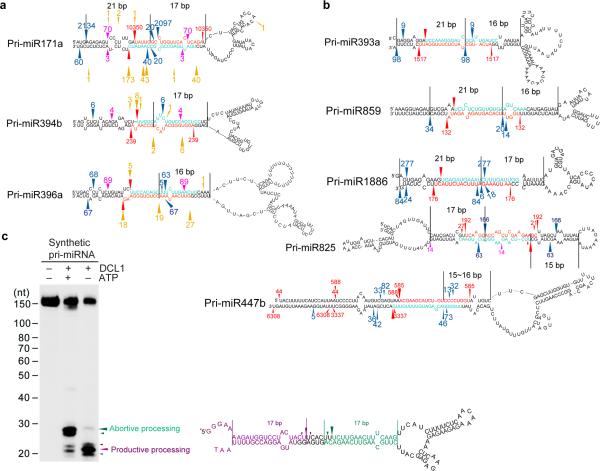
We found 77 out of 232 Arabidopsis hairpins from miRBase had BTLs. To examine if bi-directional processing of pri-miRNAs occurs beyond pri-miR166 families, we first explored eight previously described degradome datasets37-40 for evidence of abortive processing. Degradome sequencing determines the RNAs with 5′-monophosphates, which include remnants of DCL1-catalyzed pri-miRNA processing27 among other types of processed RNAs. We defined abortive processing as degradome-inferred remnants ending between 15 and 17-nt from the loop-proximal end of the miRNA-containing helix. Of the 155 hairpins with normal, unbranched loops, six had evidence of abortive processing (3.9%). At a higher proportion, seven of the 77 (9.1%), of the hairpins with BTLs had such evidence (Fig. 7a and Supplementary Fig. 7a).
Figure 7.
Bi-directional processing of pri-miRNAs (BTLs) extends beyond pri-miR166 family in Arabidopsis. (a) Examples recovered from analyses of degradome and sRNA libraries. (b) Examples recovered only from analysis of sRNA libraries. For panels (a and b), miRNA and mRNA* sequences are marked in turquoise and red. Red arrows ( ) and numbers show productive processing sites and counts of miRNA* reads. Blue arrows (
) and numbers show productive processing sites and counts of miRNA* reads. Blue arrows ( ) and numbers show positions and counts of sRNAs resulting from apparent abortive processing events. Pink arrows (
) and numbers show positions and counts of sRNAs resulting from apparent abortive processing events. Pink arrows ( ) and numbers show positions and counts of sRNAs resulting from yet unappreciated processing events. Gold arrows (
) and numbers show positions and counts of sRNAs resulting from yet unappreciated processing events. Gold arrows ( ) and numbers show 5′ positions and counts of degradome reads22,41-43. The distance from abortive processing sites to terminal loops or between processing sites is shown between two black lines. Note: reads of miRNAs are not shown because they are not unique for individual pri-miRNA paralogs. Minor forms of miRNA/*s that are progressively away from predominant miRNA/* loci are not shown and can be referred to in Supplemental Tables 2 and 3. (c) In vitro DCL1 reconstitution assays with a synthetic TLBed pri-miRNA. Schematic illustration of the cleavage products by DCL1 is shown on right. Protein immunoprecipitation, cleavage assays and RNA processing were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Red asterisks on transcripts indicate the 32P-labelling positions. Turquoise and purple arrows indicate abortive and productive processing sites, respectively.
) and numbers show 5′ positions and counts of degradome reads22,41-43. The distance from abortive processing sites to terminal loops or between processing sites is shown between two black lines. Note: reads of miRNAs are not shown because they are not unique for individual pri-miRNA paralogs. Minor forms of miRNA/*s that are progressively away from predominant miRNA/* loci are not shown and can be referred to in Supplemental Tables 2 and 3. (c) In vitro DCL1 reconstitution assays with a synthetic TLBed pri-miRNA. Schematic illustration of the cleavage products by DCL1 is shown on right. Protein immunoprecipitation, cleavage assays and RNA processing were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Red asterisks on transcripts indicate the 32P-labelling positions. Turquoise and purple arrows indicate abortive and productive processing sites, respectively.
Products derived from abortive processing of pri-miRNAs, exemplified by pri-miR166c, are likely unstable and as such many may escape detection by degradome sequencing. We argue that, if intermediate processing fragments from abortive processing are nearly self-complementary, progressive cleavage by DCL1 complexes might generate some small RNA (sRNA) remnants other than miRNA/* duplexes. We next mined high-throughput sRNA sequencing datasets41 to identify any sRNA byproducts derived from potential abortive processing. We indeed observed many sRNAs indicative of bi-directional processing (Fig. 7b and Supplementary Fig. 7b). Some of these pri-miRNAs were identical to the ones recovered from the degradome-inferred approach above (Fig. 7a). In one instructive case, pri-miR825 harbors branched loops in its lower stem and a normal loop in the upper stem. Interestingly, the directions of productive and abortive processing are also switched for pri-miR825. Similar studies on rice also recovered several pri-miRNAs including pri-miR166c suggesting that presence of bi-directional processing of the same pri-miRNAs extends beyond Arabidopsis (Supplementary Fig. 7c).
To further test if there is a general predisposition towards bi-directional processing for TLBed pri-miRNAs, we engineered an artificial miniature TLBed pri-miRNA (Fig. 7c). In vitro DCL1 reconstitution assay with this synthetic pri-miRNA indeed revealed the concurrent presence of both productive and abortive processing. This result further suggested that bi-directional processing of pri-miRNAs with large BTLs might be a common scenario in plants.
Terminal loops influences stability of pre-miRNAs
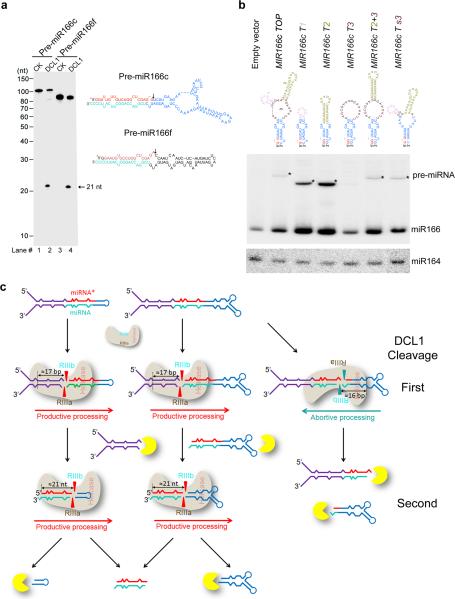
The presence of abortive processing of pri-miR166c was not solely responsible for the lowest miR166 accumulation in vivo because half of pri-miR166c was still processed through productive processing (Fig. 2c). To better understand whether the TLBed pre-miRNAs were processed in a way similar to their pri-miRNAs, we repeated the in vitro DCL1 reconstitution assays with 5′ end labeled pre-miR166c and pre-miR166f. We only observed one cleavage fragment (21-nt) in processing of both pre-miR166c and f (Fig. 8a), indicating that DCL1 complexes accurately processed the pre-miRNAs, and a large terminal loop did not trigger abortive processing for pre-miR166c.
Figure 8.
Terminal loop affects steady-state abundance but not processing pattern of pre-miRNAs. (a) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts with pre-miR166c and pre-miR166f. Protein immunoprecipitation, cleavage assays, and processing of RNA products were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Schematic illustration of the cleavage products by DCL1 is shown on right. Red asterisks on the transcripts indicate 32P-labelling positions. Black arrows show expected processing sites. (b) RNA blot analysis of pre-miR166 in the stable transformants expressing 35S-MIR166c mutants. RNA blot was probed using 32P-labelled oligo probes complementary to junction regions of miR166 and upper stem in pre-miR166c. Black triangles indicate pre-miRNAs. miR164 was probed as a loading control. (c) A model for bi-directional processing of pri-miRNA by DCL1. BTLs regulate miRNA biogenesis by triggering abortive processing of pri-miRNAs and destabilizing pre-miRNAs. Exo- or endo-ribonucleases are shown in yellow.
Finally, we compared the steady-state abundance of pre-miRNAs which contain different terminal structures. RNA blot analysis of total RNAs prepared from stable transgenic plants or the N. bentha transient system showed that pre-miRNAs with small terminal loops (MIR166c T1 and T2 were easier to detect than the ones with large terminal loops (MIR166c TOP and MIR166c T3) (Fig. 8b and Supplementary Fig. 8). In addition, a large internal loop in the upper stem or multi-branched terminal structures also affected pre-miRNA stability (Fig. 8b and Supplementary Fig. 8, MIR166c T2+3 and MIR166c Ts3). These results indicate that the secondary structures of terminal regions in pre-miRNAs regulated their stability, further controlling homeostasis of miRNA accumulation in vivo.
DISCUSSION
A canonical pathway for pri-miRNA processing is that RNase III proteins and their partners define their initial cut at a position away from the ssRNA-dsRNA junction in the lower stems to release pre-miRNAs2. A non-canonical pathway for miRNA biogenesis also exists for some plant pri-miRNAs that harbor a long linear stem-loop structure (pri-miR159a and pri-miR319a)--DCL1 initiates sequential cuts close to the terminal loop rather than at the Base until the release of miRNA/* duplexes28. Here, we propose a new action mode for pri-miRNA processing–DCL1 protein processes the exact same pri-miRNAs in two directions: either from the lower stem to the terminal loop or vice versa (Fig. 8c). This scenario occurs as a result of the structural heterogeneity of plant pri-miRNAs: Many pri-miRNAs including pri-miR166c contain large terminal loops, which may “disguise” an ssRNA-dsRNA junction to recruit DCL1 complexes for cleavage. This outside-in mechanism has two outcomes: if DCL1 goes from base to loop, it produces mature miRNAs; otherwise, if it goes from the opposite direction, it will destroy them.
Given that pri-miRNAs could be processed forward or backward in plants, how do DCL1 complexes process pri-miRNAs with large terminal loops? Our results have shown that the binding orientation of DCL1 complexes in pri-miRNAs is a key factor in determining whether productive or abortive processing events occur. Although how exactly DCL1 complexes associate with pri-miRNAs or vice versa is unclear in plants, some structural elements in pri-miRNA may contribute to the preference of DCL1 association (Fig. 8c). First, large terminal loops and long upper stems promote non-canonical processing (Fig. 6a-c). Second, the presence of an ssRNA region in the lower stem is required for canonical DCL1 function because the deletion of this segment completely suppressed productive processing (Fig. 4b). Third, the presence of an internal loop in the lower stem likely suppresses productive processing and facilitates abortive processing (Fig. 5a). Similar results have also been reported in animal systems2, suggesting that in both plants and animals, internal loop structures in the stem influence the ratio of the productive and abortive processes2. It should be noted that, in our study, Mfold analysis 41 did suggest the presence of large internal loops in the lower stem of pri-miR166a and pri-miR166b. These pri-miRNAs were efficiently processed in vivo but not in vitro. It is tempting to propose that the secondary structures of these pri-miRNAs might be different from predicted. Alternatively, these pri-miRNAs may have unappreciated processing mechanisms that bypass the canonical pri-miRNA processing.
We here reported numerous TLBed pri-miRNAs in Arabidopsis and additional cases in rice indicative of bi-directional processing. Such a processing way may be also present but not easily detected in many other TLBed pri-miRNAs, exemplified by pri-miR166c, due to extremely low expression of pri-miRNAs or instability of abortive processing products. On the other hand, we have no reason to exclude presence of bi-directional processing in the pri-miRNAs with linear-hairpin structures but with large internal or terminal loops as observed in in vitro assays (Fig. 6) and in planta (Supplementary Fig. 7a).
It has been contemplated that DCL1 complexes initially processes pri-mRNAs at a distance of ~15nt from either an ssRNA-dsRNA junction or an internal unstructured region23-26. Here we have provided direct biochemical evidence to boost this model and more precisely pinpoint the cleavage sites. We have also demonstrated that the canonical ruler system is applicable to both productive and abortive processing (Fig. 8c). The reference sites for microprocessor recognition can be a large internal or terminal loop and is not necessarily counted from the ssRNA segments at the end of pri-miRNAs. Intriguingly, genome-wide analysis shows that half of pri-miRNAs, which are most likely subjected to canonical miRNA processing, harbor an unpaired region or loop 17-nt away from the loop-distal DCL1 cleavage sites in the lower stem25. That said, the microprocessor in plants also has a certain degree of plasticity and allows productive processing from structures slightly different from the optimal 16-17bp, especially in scenarios where the processing sites are near an internal loop. We propose that DCL1 may utilize a non-canonical ruler system to sense the loop region and adjust the cleavage sites in pri-miRNAs, thus permitting more fitness to generate uniform miRNAs (Fig. 5). The sensing of the internal loop is apparently through the helicase domain of DCL1 (Fig. 5). However, DCL1 complexes do not favor distances shorter than 16bp from the ssRNA-dsRNA reference site to the potential processing position. Our biochemical results are consistent with the previous computational discovery that Arabidopsis pri-miRNAs have evolved to rarely contain unpaired regions or loops 12-14nt away from the cleavage sites25. Taken together, our results provide new insight into our understanding of the complicated processing mechanism of pri-miRNAs in plants and beyond.
ONLINE METHODS
Vector construction
The majority of constructs were made using a Gateway system (Invitrogen)42,43. The primers for the construction of all vectors are listed in Supplementary Table 1.
MIRNA genes, including concise and extended forms, were cloned from Arabidopsis genomic DNA. These MIRNA genes were introduced into pENTR/D vectors, confirmed by sequencing and finally transferred into the pBA-DC destination vectors through the LR Clonase (Invitrogen). Chimeric primary miRNAs with extended form were constructed as follows:
For pENTR-pri-MIR166f c B+T (extended form), we replaced Base and Top sequences of pri-MIR166f with the counterparts of pri-miR166c with miR166f duplex intact. Briefly, pENTR-pri-MIR166c extended form was used as a template for two sequential mutagenesis PCRs using two sets of primers: c to f For and c to f Rev for PCR1 and primers c* to f*For and c* to f* Rev for PCR2, Then the mutated pENTR-pri-MIR166c (extended form) was further used as a template for overlapping PCR using two sets of primers (f*cTL For and M13 Rev; and M13 For and f*cLS Rev). The resultant PCR fragments were mixed, amplified using miR166c For and miR166c Rev primers and further cloned into pENTR/D to generate pENTR-pri-MIR166f c B+T (extended form).
For pENTR-pri-MIR166c f B+T (extended form), we changed the Base and TOP sequence of pri-MIR166c with the counterparts of pri-MIR166f using a strategy similar to that for pENTR-pri-MIR166f c B+T (extended form) above. The primers for sequential mutagenesis PCRs were f to c For and f to c Rev; f* to c* For and f* to c* Rev, respectively. The primers for overlapping PCRs were c*fTL For /M13 Rev, M13 For / c*fLS Rev, and miR166f For/miR166 Rev.
For pENTR/D-pri-MIR166f c T (extended form), we changed the top region of MIR166f with the one from MIR166c. We used pENTR-pri-MIR166f (extended form) as a template for three rounds of extension PCRs. The three groups of primers were cTL-f For 1/M13 Rev, cTL-f For 2/M13 Rev, and cTL-f For 3/M13 Rev, respectively. The resultant PCR fragment was mixed with another PCR fragment generated with M13 For and cTL-f* Rev using pENTR-pri-MIR166f (extended form) as template for overlapping PCR. Finally, the overlapping PCR product was amplified using miR166f For and miR166f Rev and cloned into pENTR/D to generate pENTR-pri-MIR166f c T (extended form).
For pENTR/D-pri-MIR166c f T (extended form), we changed MIR166c TOP with the one from MIR166f using a strategy similar to the one for chimeric vector pENTR-pri-MIR166f c T (extended form) above. The two sets of primers for extension PCRs were fTL-c For 1/M13 Rev and fTL-c For 2/M13 Rev. The primers or the overlapping PCRs were M13 For / fTL-c* Rev and miR166c For and miR166c Rev. The final PCR product was cloned into pENTR/D to generate pENTR-MIR166c f T (extended form).
For pENTR/D-pri-MIR166f c B (extended form), we changed MIR166f Base with the Base of MIR166c using a strategy similar with the chimeric plasmids above. The two sets of primers for the extension PCRs were fTL-c For 1/M13 Rev and fTL-c For 2/M13 Rev. The primers for the overlapping PCRs were M13 For /fTL-c* Rev and miR166c For / miR166c Rev. The resultant PCR product was cloned into a pENTR/D vector. This pENTR vector was used as a template for an additional round mutagenesis PCR using the primers of cLS GT to ag For and cLs GT to ag Rev to create pENTR-pri-MIR166f c B (extended form).
For pENTR/D-pri-MIR166c f B (extended form), we replaced MIR166c Base to MIR166f Base in MIR166c backbone. The extension PCRs were conducted with three sets of primers (cTL-f1 For / M13 rev; cTL-f For 2/M13 Rev and cTL-f For 3/ M13 Rev. Next, the overlapping PCR was performed using the primers of M13 For /cTL-c* Rev, and miR166f For/miR166f Rev. The resultant PCR product was cloned into pENTR/D. This pENTR vector was used as a template for an additional round mutagenesis PCR using the primers of fLS a to gt For and fLS a to gt Rev to create pENTR-pri-MIR166c f B (extended form).
For all concise forms of the pENTR-pri-MIR166c and pri-miR166f vectors, the short PCR products were generated using the wild-type and chimeric mutants of pENTR-pri-MIR166c and pri-miR166f (extended form) as templates and the primers of miR166c 150 For / miR166c 150 3′ Rev and miR166f 148 5′ For / miR166f 148 3′ Rev. The resultant fragments were cloned into pENTR/D to create the concise wild-type and mutant forms of pENTR-pri-MIR166c and f.
pENTR-pri-MIR166c ΔL2, -MIR166c ILS, -MIR166f ΔL, -MIR166c ΔUL, -MIR166c IUS and -MIR166c US-15 were created through mutagenesis PCRs using pENTR-pri-MIR166c (concise form) as a backbone and the corresponding primers listed in Supplementary Table 1. pENTR-pri-MIR166c US-18 and -MIR166c US-22 were created through mutagenesis PCRs using pENTR-pri-MIR166c US-15 as a backbone and the corresponding primers listed in Supplementary Table 1.
LR reactions yield ~ 40-nt single-stranded segments flanking at either 5′ end or 3′ end of lower stems of pri-miRNAs and might alter folding patterns of pri-miRNAs. To avoid this potential issue, we created a series of concise pri-miRNAs using classic restriction digestion and ligation. Briefly, PCR products were amplified using pENTR-pri-MIR166c (concise form) and pENTR-pri-MIR166f (concise form) as templates and corresponding primers listed in Supplementary Table 1. The purified PCR fragments were cut with XbaI and SacI. The resultant fragments were ligated into XbaI/SacI-treated pBA002 to produce pBA002-pri-MIR166c, pBA002-pri-MIR166f , pBA002-pri-MIR166c ΔB and pBA002-pri-MIR166f ΔB.
For pBA002-pri-MIR166c ΔT, we annealed two long oligos, C88-1 and C88-2. PCR was performed using annealed oligos as template and primers (miR166c XbaI For and miR166c SacI Rev) and amplified each other. The resultant PCR products were cut with SacI and XbaI and ligated to the same enzymes-treated pBA002.
pBA002-pri-MIR166f ΔT, -MIR166c T2+3, 2-MIR166c T3, -MIR166c T1+2, -MIR166c-T2, -MIR166c-T1, -MIR166c-T4, -MIR166c-T5 and -MIR166c-Ts3 were created using a strategy similar to pBA002-pri-MIR166c ΔT above with corresponding primers listed in Supplementary Table 1.
For pBA002-pri-MIR166c ΔL3, pENTR-pri-MIR166c (concise form) was used as a template for two sequential mutagenesis PCRs using corresponding primers listed in Supplementary Table 1. The resultant mutated pENTR-MIR166c was further used as a template for PCR reaction with two primers of miR166c XbaI For and miR166c SacI Rev. The final PCR product was cut with XbaI/SacI and subcloned into pBA002.
For pENTR-pri-MIR166c LSA-15, pBA002-pri-MIR166c ΔL3 was used as a template for one mutagenesis PCR using corresponding primers listed in Supplementary Table 1. The resultant fragments were cloned into pENTR/D vectors.
pBA-6Myc-HYL1 were generated by the LR reaction between pDONRzeo-HYL1 and pBA-6myc-DC. SE was amplified from Arabidopsis cDNA with two primers listed in Supplementary Table 1 and cloned into pENTR vector to generate pENTR-SE. The SE gene was further transferred into pBA-DC-3HA or pBA-Flag-4Myc-DC to yield pBA-SE-3HA or pBA-Flag-4Myc-SE through LR reaction.
Regarding pBA-2Flag-4Myc-DCL1, DCL1 was first amplified from pTA7002-Flag-DCL118 and sub-cloned into pBA002 to create pBA002-2Flag-4Myc-DCL1. All pBA002-2Flag-4Myc-DCL1 mutants including DCL1 (E1507Q), DCL1 (E1696Q), DCL1 (E1507Q E1696Q) and DCL1 ΔH were generated by mutagenesis PCR and overlapping PCR using the primers listed in Supplementary Table 1
Plant material and growth conditions
Plant growing conditions were previously described44. Wild type Arabidopsis thaliana plants (Col-0 and Ler) were transformed with the recombination binary vectors containing concise forms of pri-miRNAs and empty vector by the floral dip transformation method1. Seeds from infiltrated plants were selected on standard MS medium containing the appropriate selective agents: 10 mg/L glufosinate ammonium (Sigma) and 100 mg/L carbenicillin (Sigma).
RNA blot analyses
Total RNA was extracted using Trizol reagent from Agrobacterium-transfected N. benthamiana leaves or Arabidopsis tissues, including 7 (or 10) -day-old seedlings, depending on the desired assays. Northern blots were performed as described2,3,29. Each lane contained 10 μg of total RNA. Blots were hybridized with 32P-radiolabeled oligonucleotide probes complementary to the miR165 or miR166. U6 served as loading controls in transient assay. miR164a served as loading controls in blots from transgenic plants. RNA blots were detected after exposure to a phosphor plate and quantified using the Quantity One Version 4.6.9 according to the manufacturer's instructions (Bio-Rad).
Material preparation for transient and in vitro processing experiments
For transient experiments, 4-week-old N. benthamiana leaves were infiltrated with A. tumefaciens ABI harboring variety of binary plasmids. The OD600 of A. tumefaciens cultures were normally diluted to 0.4 in most experiments. For DCL1 complexes (WT or mutants) in vitro processing experiment, we infiltrated tobacco with three ABI harboring DCL1 (or DCL1 mutants), HYL1 and SE together. The OD600 of the pBA002-2Flag-4Myc-DCL1 (or DCL1 mutants) culture was 0.8. The OD600s of pBA-6Myc-HYL1, pBA-SE-3HA and pBA-Flag-4Myc-SE were diluted to 0.1. The transfected leaves were collected two days after agroinfiltration.
In vitro transcription of RNA
RNA substrates were transcribed under the T7 promoter in vitro using PCR generated templates. The templates and primers used for PCRs and the synthetic pri-miRNA are listed in Supplementary Table 1. The in vitro transcription of RNAs was carried out in one 20μl reaction incubated for 3h or overnight at 37°C: 1μl of DNA template (100ng), 4 μl of 5X transcription buffer (400mM HEPES pH 7.5, 10mM spermidine, 200mM DTT, 125mM MgCl2, 20mM each NTP), 1 μl of RNase inhibitor (Ambion), 2 μl of T7 RNA polymerase and 12 μl of water. DNase-treated RNA was fractionated by 6% polyacrylamide-8M urea gel (denaturing gel), eluted overnight from gel slices in RNA elution buffer (0.3M NaAc, pH5.5 and 2% SDS) using Thermomixer R (Effendorf) at 42°C and 1200rpm, ethanol precipitated and stored in DEPC water.
Labeling of RNA
For internal labeling, [α-32P]-UTP (PerkinElmer) was included in the NTP mixture (20mM CTP, 20mM ATP, 20mM GTP and 4mM UTP) for the in vitro transcription described above. For 5′ end labeling, the nonradioactive purified Pri- or Pre-miRNAs (5 pmol) were dephosphorylated by Alkaline Phosphatase (New England Biolabs) and labeled with [γ-32P] ATP (PerkinElmer, ) by T4 Polynucleotide kinase (New England Biolabs) according to manufacturer's protocol. For 3′ end labeling, the nonradioactive purified Pri-miRNA was ligated with [α-32P] pCp (PerkinElmer) using T4 RNA Ligase 1 (New England Biolabs) according to manufacturer's instructions. All the labeled RNAs were fractionated by 6% denaturing gel, eluted from gel slices in RNA elution buffer, ethanol precipitated with glycoblue (Ambion), and stored in buffer (100mM KCl, 30mM Tris-HCl, pH 7.5). Labeled RNAs were folded by heating to 95°C for 2 min, slowly cooled down to room temperature and normalized to ~2×103c.p.m./μl.
Immunoprecipitation and in vitro DCL1 reconstitution assay
Immunoprecipitation (IP) was performed as described29 with some modifications. Tobacco samples were ground in liquid nitrogen and protein/sRNA complexes were extracted using five volumes of IP buffer (40 mM Tris-HCl pH7.5, 300 mM KCl, 5 mM MgCl2, 5 mM DTT, 0.2mM EDTA pH 8.0, 0.2% Triton X-100, 1mM PMSF, 2% Glycerol, 0.3% (v/v) proteinase inhibitor cocktail (Sigma) and 1tablet/25ml IP buffer of EDTA-free protease inhibitor cocktail (Roche). After removal of insoluble materials by centrifugation twice at 16,000g for 15min at 4°C, extracts were incubated with anti-Flag M2 Magnetic beads (Sigma) for 2 h at 4°C. The beads were washed 3 times with the IP buffer, and tagged DCL1 complexes were eluted twice by incubation with the IP buffer containing 100 μg/ml 3× FLAG peptide (Sigma) for 30 minutes at 4°C. The Eluted fraction was incubated with anti-Myc agarose beads (sigma) for 1.5 h. The beads were washed 3 times with IP buffer and then 3 times with washing buffer (20mM Tris-HCl pH 7.5, 1mM DTT, 4mM MgCl2 and 100mM KCl). Briefly, in vitro DCL1 cleavage assays were performed in a total volume of 30 μl in 20 mM Tris-HCl pH 7.5, 53mM KCl, 4mM MgCl2, 1mM DTT, 7.5 mM ATP, 1mM GTP, 1u/μl RNase Inhibitor (Ambion), including 1μl of labeled RNA (around 2×103c.p.m.) and 15 μl of the immunopurified proteins in washing buffer. The reaction mixture was incubated at 37 °C for 10–90 min. For pre-miRNA, the reaction was 10-15min. After extracted with phenol/chloroform and ethanol precipitated with glycoblue (Ambion), the processing products were fractionated by 15% denaturing gels. RNA marker is Decade marker (Ambion), which was 5′ end labeled according to manufacturer's protocol with [γ-32P] ATP (PerkinElmer). The denaturing gel was fixed in B buffer (40% ethanol, 10% acetate acid, 5% glycerol) and dried with Gel Dryer (Bio-Rad) under gradient cycle at 80°C for 5h. The processed RNA products were detected after exposing overnight to a phosphor plate and quantified with the Quantity One Version 4.6.9 according to the manufacturer's instructions (Bio-Rad). Original images of key blots and autoradiographs used in this study can be found in Supplementary Figure 9.
Western blot analysis
Assays of western blots were performed as described29. The antibodies for immunoprecipitation were anti-Flag M2 Magnetic beads (Sigma, M8823) and anti-Myc-Ararose (Sigma, A7470). Antibodies for western blot analysis were anti-Flag (Sigma, F3165), anti-Myc (Sigma, C39560), and anti-HA (Santa Cruz, sc-7392).
RNA Secondary Structure
The secondary structures were predicted from Mfold web server (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
Semi-quantitative PCR
Total RNAs were prepared from control plants or different transformants and treated with DNase before being subjected to cDNA synthesis using Superscript III reverse transcriptase (Invitrogen) primed by oligo dT. The primary miRNA genes were amplified with the primers listed in Supplementary Table 1. The EF1a gene was included as an internal control29.
5′ RNA ligase-mediated Rapid Amplification of cDNA Ends (5′ -RLM-RACE)
RNA was isolated from wild-type Col-0 and pooled seedlings of T1 transgenic plants expressing 35S-MIR166c. One hundred pmol of 5′ adapter (rGrUrUrCrArGrArGrUrUr CrUrArCrArGrUrCrCrGrArCrGrArUrC) was directly ligated with 5 μg of total RNA. After ligation, first-strand cDNAs were synthesized using Superscript III Reverse Transcriptase (Invitrogen, CA) and pri-miR166c specific primer (5′ RACE Outer Rev). The cDNA was treated with RNaseH to remove the RNA strand and amplified for two-round using two sets of primers (5′ RACE Outer For/5′ RACE Outer Rev; 5′ RACE Inner For/5′ RACE Inner Rev). The distinct PCR products were cloned into pENTR vector (Invitrogen, CA) and sequenced using either M13 For or M13 Rev primers.
Computational analysis of degradome libraries and sRNA databases
Sequences of all Arabidopsis miRBase hairpins (miRBase 19) were retrieved from genomic coordinates with extended 50-nt on either side. Degradome datasets37-40 were downloaded from GEO. Degradome reads were mapped to the extended pri-miRNA hairpins. The 5′ positions of the degradome reads represent uncapped 5′ ends of in vivo RNA fragments. They were tallied on the sense strand relative to hairpin and plotted alongside RNA secondary structures and mature miRNA positions.
Sequences and read numbers for sRNAs in Arabidopsis (168 datasets) were downloaded from NCBI GEO under the accession numbers listed in Supplemental Table S#241. Similarly, 45 sRNA deep sequencing datasets in rice were retrieved from GEO and listed in Supplemental Table S#3. The sRNAs were mapped to miRNA precursor sequences which were downloaded from the miRBase database Release 16 by in-house C++ program. We defined four criteria for potential bi-directional processing of pri-miRNAs: 1) sRNAs are processed from 15-17-nt away from branched or large terminal loops; 2) there are no clear unstructured regions at 15-17-nt away from the sRNA remnants in the opposite direction (Supplementary Fig. 7); 3) Because miRNA/*s are not precisely processed in many cases and have some minor forms centered around the predominant miRNA/*s, we defined the sRNAs that are at least 3-nt away from an adjacent minor form of miRNA/* are derived from a processing different from productive processing (Supplementary Tables 2 and 3); 4), sRNAs are primarily 21-nt long; and present in at least three individual libraries.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Koiwa for pDONRzeo-HYL1, D. Shippen, G. Kapler, F. Qiao, P.W. Li, and M. Klein for stimulating discussions and critical review of the manuscript. We also thank Fuqu Hu, Hengyi Xu and Zhonghui Zhang, Changjun Huang, and Cheng Lu for technical assistance. The work was supported by grants from NSF (MCB-0951120) and NSF CAREER (MCB-1253369), NIH (R21AI097570) and Welch foundation (A-1777) to X.Z. A.L. was supported by NSF-REU (MCB-1232817). Y.Z. was supported by Chinese Scholarship Council.
Footnotes
AUTHOR CONTRIBUTIONS
H.Z., C.C.G., and X.Z designed experiments. Y.Z. carried out genetic research, H.Z. and C. C.G. performed biochemical studies. M.J. A. conducted degradome analysis. Y. Z. and X. W. worked on sRNA dataset analysis. Other coauthors participated in experiments or provided materials and intellectual input for the work. X.Z. and H.Z. wrote the manuscript.
REFERENCES
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu JD, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 5.Voinnet O. Origin, Biogenesis and Activity of Plant MicroRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nature Review Genetics. 2011;12:20–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazzini A, Lee M, Giraldez A. Ribosome Profiling Shows That miR-430 Reduces Translation Before Causing mRNA Decay in Zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djuranovic S, Nahvi A, Green R. miRNA-Mediated Gene Silencing by Translational Repression Followed by mRNA Deadenylation and Decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, et al. MicroRNAs Inhibit the Translation of Target mRNAs on the Endoplasmic Reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X. Plant microRNAs at a glance. Semin Cell Dev Biol. 2010;21 doi: 10.1016/j.semcdb.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Ren G, Xie M, Dou Y, Zhang S, Chi Zhang C, Yu B. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machida S, Chen H, Yuan A. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Research. 2011;39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang SW, Chen HY, Yang J, Machida S, Chua N,H, Yuan YA. Structure of Arabidopsis HYPONASTIC LEAVES1 and Its Molecular Implications for miRNA Processing. Structure. 2010;18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Z, Han M, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigg S, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 17.Han M, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNAbinding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobbes D, Rallapalli G, Schmidt D, Martin C, Clarke J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for MicroRNA accumulation and plant development, but not posttranscriptional transgene silencing. Current Biology. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch J, et al. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006;140:1192–1204. doi: 10.1104/pp.105.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie ZX, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiology. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA. 2009;107:466–471. doi: 10.1073/pnas.0913203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of MicroRNA Processing Determinants by Random Mutagenesis of Arabidopsis MIR172a Precursor. Current Biology. 2010;20:49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 25.Song L, Axtell M, Fedoroff N. RNA Secondary Structural Determinants of miRNA Precursor Processing in Arabidopsis. Current Biology. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 26.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Current Biology. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 27.Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–2121. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop to base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009;28:3646–3656. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Zhang X. Argonautes compete for miR165/166 to regulate shoot apical meristem development. Current Opinion in Plant Biology. 2012 doi: 10.1016/j.pbi.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, Axtell MJ, Fedoroff NV. The Helicase and RNaseIIIa Domains of Arabidopsis Dicer-Like1 Modulate Catalytic Parameters during MicroRNA Biogenesis. Plant Physiology. 2012;159 doi: 10.1104/pp.112.193508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manavella P, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D. Fast-Forward Genetics Identifies Plant CPL Phosphatases as Regulators of miRNA Processing Factor HYL1. Cell. 2012;151:859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg D, Kotaro N, K., Dinshaw J, Patel D, Bartel D. The Inside-Out Mechanism of Dicers from Budding Yeasts. Cell. 2011;146:262–276. doi: 10.1016/j.cell.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis P, Kay M. The Loop Position of shRNAs and Pre-miRNAs Is Critical for the Accuracy of Dicer Processing In Vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre miRNA structure by Drosophila Dicer-1. Nature Struc. & Mol. Biol. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- 37.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Current Biology. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nature Biotechnology. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 39.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Developmental Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Ma Z, Coruh C, Axtell MJ. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell. 2010;22:1090–1103. doi: 10.1105/tpc.110.073882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Henriques R, Lin S.-s., Niu Q, Chua N-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral-dip method. Nature Protocol. 2006;1:1–6. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 43.Zhang XR, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes & Development. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang XR, Garreton V, Chua NH. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes & Development. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.