Abstract
Differentiated sex chromosomes evolved because of suppressed recombination once sex became genetically controlled. In XX/XY and ZZ/ZW systems, the heterogametic sex became partially aneuploid after degeneration of the Y or W. Often, aneuploidy causes abnormal levels of gene expression throughout the entire genome. Dosage compensation mechanisms evolved to restore balanced expression of the genome. These mechanisms include upregulation of the heterogametic chromosome as well as repression in the homogametic sex. Remarkably, strategies for dosage compensation differ between species. In organisms where more is known about molecular mechanisms of dosage compensation, specific protein complexes containing noncoding RNAs are targeted to the X chromosome. In addition, the dosage-regulated chromosome often occupies a specific nuclear compartment. Some genes escape dosage compensation, potentially resulting in sex-specific differences in gene expression. This review focuses on dosage compensation in mammals, with comparisons to fruit flies, nematodes, and birds.
Keywords: dosage compensation, X upregulation, X inactivation, epigenetics, evolution, sex chromosomes
INTRODUCTION
Species in which sex determination is coupled to differentiated sex chromosomes exist throughout the animal kingdom. In many diploid species, males are the heterogametic sex XY, and females the homogametic sex XX. Other species, for example, birds, have the opposite pattern, i.e., males are ZZ and females ZW. Once the sex chromosomes cease recombining they acquire a different size and gene content, and a natural type of aneuploidy appears in the heterogametic sex. The X (or Z) chromosome is often large and the Y (or W) is usually small. Chromosomal aneuploidy is not well tolerated, and mechanisms triggered by abnormal gene dosage exist to maintain the stoichiometry between gene products involved in common pathways but originating from different chromosomal loci. Efficient and stable mechanisms of dosage compensation have evolved to relieve the natural aneuploidy caused by the strikingly different gene content of present-day sex chromosomes. Two major aims of sex chromosome dosage compensation have been identified: (a) to balance expression between the sex chromosomes and the rest of the genome and (b) to equalize expression between the sexes. The question of how organisms deal with the dosage imbalance due to differentiation of the sex chromosomes is the central issue we address here. Our main focus is on mammalian systems, but we refer to other organisms where comparisons are informative.
HISTORICAL PERSPECTIVE
Muller (114a) was the first to propose that lack of recombination between the sex chromosomes would lead to degeneration of the heterogametic chromosome. This concept became known as Muller’s ratchet (45). Later, Ohno (122) articulated the idea that heteromorphic sex chromosomes evolved from an ordinary pair of chromosomes that carried a sex determinant. Studies based on comparisons of closely related species within a group, for example, mosquitoes or snakes, support this concept. Persistence of remnant paralogs on the vertebrate sex chromosomes also upholds ancient homology between them. In some organisms, the Y chromosome was completely lost, for example, in the nematode Caenorhabditis elegans. In the fruit fly Drosophila melanogaster, the Y is a largely heterochromatic chromosome that acquired male advantageous genes from autosomal copies (12). In these two model organisms, sex is determined by the number of X chromosomes per diploid genome rather than by a Y-linked gene as in mammals.
Both Muller and Ohno clearly envisioned that differentiation of the sex chromosomes into entities of very different size would lead to mechanisms of dosage compensation (114a, 122). Muller initially favored the correct hypothesis, hyperexpression of the Drosophila male X, but later proposed that repression of both Xs in females would achieve equality between the sexes. This latter hypothesis, although incorrect in fruit flies, is a main regulatory mechanism in C. elegans XX hermaphrodites. Meanwhile, Lyon discovered X inactivation in mammals; indeed, she interpreted the coat color variegation observed in XX female mice with an X-linked Tabby mutation as representing random silencing of one X chromosome (98). In light of X inactivation, Ohno considered the fate of the single active X chromosome versus autosomal gene expression and suggested that there may also be a second mechanism to upregulate X-linked genes to avoid “a great peril” due to hemizygous gene expression (122). These early studies that set the stage for decades of investigation in the regulation of the sex chromosomes showed that organisms have dealt with dosage compensation in very different ways (Figure 1) (see sidebar, Main Mechanisms of Dosage Regulation). Mechanisms of dosage compensation have in common the need for regulatory systems that address the unique and fascinating challenge of specifically targeting a particular chromosome for regulation often in only one sex.
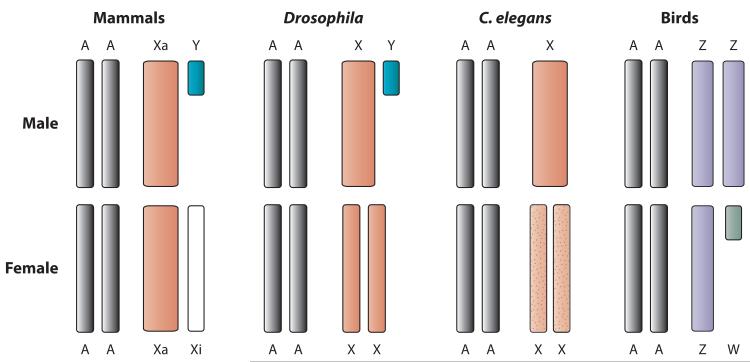
Figure 1.
Main types of sex chromosome dosage compensation. Male and female sexes are indicated. From left to right: Mammals, in which expressed genes on the active X (Xa) are upregulated in both sexes and genes on the inactive X (Xi) are silenced in females; Drosophila, in which the X is upregulated in males only; Caenorhabditis elegans, in which the X is upregulated in both sexes and downregulated in hermaphrodites (mottled); birds, in which there is apparent partial upregulation of the Z chromosome in both sexes and partial or stochastic downregulation in males. Dots represent both increased expression and repression. Abbreviation: AA, autosomes.
SEX CHROMOSOME EVOLUTION
Little is known of the initial stages of sex chromosome evolution (25). The proto-sex chromosomes may have become differentiated via genetic or epigenetic events (70). In humans, recombination between the proto-sex chromosomes was suppressed by large Y inversions, as shown by mapping paralogs with copies on the X and Y and grouping them in evolutionary strata on the basis of sequence similarities (85, 139). Among the few remaining X/Y gene pairs, some have similar functions, whereas others have acquired a divergent function on the Y. Aside from these genes, the Y has accumulated mutations and lost many genes, fulfilling Muller’s ratchet theory. Interestingly, a subset of genes are conserved on the Y of humans, chimpanzees, and rhesus monkeys, suggesting that gene loss was initially rapid but ceased 25 mya when these species diverged (66). Many of the X paralogs of this subset of conserved genes escape X inactivation with both sexes having retained two functional copies (Figure 2). Thus, the question arises of whether adverse dosage effects may have contributed to the preservation of this X/Y gene set (see below).
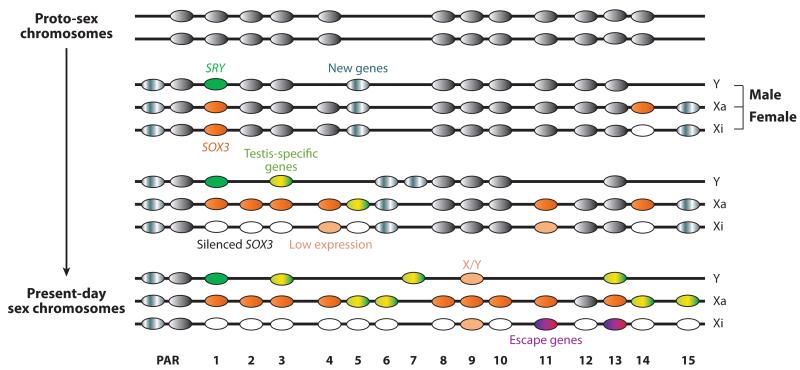
Figure 2.
Schematic of potential evolutionary pathways of mammalian sex-linked genes. From top to bottom: evolution of homologous genes originally located on the proto-sex chromosomes ( gray) to their full differentiation into Y-linked genes (Y) and genes on the active X (Xa) and inactive X (Xi). New genes have been acquired by the X and Y (mottled gray) and thus are not remnants of the proto-sex chromosomes; some may be unique to the Y (gene 7) or the X (gene 15), others may be acquired by both sex chromosomes (genes 5-6), depending on how much recombination between the sex chromosomes still occurred at that time. Pseudoautosomal region (PAR) genes are homologous on the sex chromosomes; however, note that PAR genes can also derive from additions to the sex chromosomes later in evolution. Gene 1 represents the sex determinant SRY derived from the X-linked gene SOX3, which becomes upregulated on the Xa (orange) and silenced on the Xi (white). Genes 2, 4-6, 8, 10-12, and 14-15 are examples of genes progressively lost from the Y. Genes 3 and 13 evolved from X/Y pairs by acquiring a testis function on the Y (mottled yellow/green). Their X paralog becomes silenced (gene 3) or escapes X inactivation (gene 13). Most genes on the Xa become upregulated (orange), but some do not (gene 12), and others acquire reproduction-related functions (mottled yellow/green) (genes 5, 6, 14, 15). A majority of genes (genes 1-6, 8, 10, 12, 14, 15) on the Xi become silenced (empty). A few genes escape X inactivation on the Xi (genes 9, 11, 13); some of these retain a functional Y paralog (gene 9, X/Y gene pair), resulting in equal sex expression, although expression is usually lower on the Xi and Y than on the Xa ( pale orange); others have lost the Y paralog (gene 11) or have a differentiated Y paralog expressed in testis (gene 13). Escape genes may acquire a female advantageous role (genes 11 and 13) (mottled purple/pink). Overall, both the Y and X chromosomes have acquired a number of new testis-specific (or reproduction-related) genes (mottled yellow/green).
Sex chromosomes also evolved by translocation with autosomes. For example, the X chromosome is larger in eutherian mammals than in marsupials, probably because of translocation of autosomal material to its short arm. Such additions are followed by attrition on the heterogametic chromosome (56). In eutherian mammals, one or two homologous regions of meiotic pairing, the pseudoautosomal region(s) (PARs), persist. PAR genes being expressed from both sex chromosomes would presumably not be subject to dosage compensation, as verified for genes in human PAR1 (Figure 2) (73). However, certain genes located in human PAR2 are silenced both on the inactive X and on the Y (35). The gene content of the mammalian PAR(s) evolved rapidly, with swift changes in the position of the PAR boundary even between closely related species (43, 126). PAR regions that become sex specific would be expected to rapidly become dosage compensated, but this process may not always be complete (see below).
Although mammalian sex chromosomes have been evolving for a long time, other species offer a glimpse of early events in sex chromosome differentiation (7). For example, recent acquisition of a sex-determining gene in stickleback fish led to rapid divergence of their sex chromosomes and absence of dosage compensation (88, 129). Species can also switch back and forth between temperature-sensitive systems of sex determination associated with homomorphic sex chromosomes and gene-based or chromosome-based systems associated with differentiating sex chromosomes (132). Environmental sex determination does not cause a dosage imbalance but has the disadvantage of being at the mercy of changes in surroundings. Gene-based and chromosome-based systems ensure a more stable distribution of sex but eventually cause imbalance in the genome. Mank et al. (103) have argued that dosage compensation mechanisms might in fact be rare, as they are so complex and onerous that species with rapid divergence of the sex chromosomes probably would not evolve such mechanisms. However, dosage compensation of the X chromosome can quickly evolve de novo in plants, as shown in a new study that demonstrates not only that dosage compensation is not limited to the animal kingdom but also that it can evolve at the same time as Y degeneration (115).
REPRODUCTION-RELATED GENES ON THE SEX CHROMOSOMES
Evolution of the sex chromosomes is accompanied not only by the emergence of dosage compensation mechanisms but also by functional specialization of existing or newly recruited genes (155). The mammalian Y chromosome carries not only the testis-determining gene SRY (82, 146) but also a number of testis-expressed genes essential for normal male fertility (140, 147). The accumulation of genes beneficial to male-specific functions on the Y is easily understood because this chromosome is present solely in males. Such genes either diverged from their X paralog or evolved from a transposed autosomal copy (56) (Figure 2). In some species, a number of Y-linked genes expressed in testis have been duplicated manyfold (66). Why are multicopy gene families so prevalent on the Y? One answer is that strong selection amplifies a mutated Y gene that has become less active in order to restore sufficient function (56). In addition to guarding against complete degeneration, gene families provide opportunities for rapid evolution of new functions (61, 84, 140).
The Y chromosome is not alone in having amassed genes with sex- or reproduction-related functions. The accumulation of genes expressed in testis on the mammalian X is also remarkable (78, 114, 139, 158). As many as 12% of X-linked genes in human and 18% of X-linked genes in mouse represent multicopy testis-expressed genes. Many of these genes have been acquired by the X; thus, the X and Y have evolved by a combination of decay and differentiation from an ancestral autosomal pair, together with transposition and selection of a number of new genes from autosomes and from duplications (10) (Figure 2). In males, the hemizygous X is a favored location for immediate expression of male-biased recessive mutations (76, 136). Aside from protein-coding genes, the X also accumulated testis-expressed microRNAs whose function remains to be fully explored (175). Although a number of X-linked genes are expressed in testis before and after meiosis, the sex chromosomes are actually silenced during meiosis itself. This meiotic sex chromosome inactivation (MSCI) is triggered by the largely unpaired nature of the X and Y compartment, which attracts a specific epigenetic machinery involving the histone H2AX (154). Backup copies of essential X-linked genes exist on autosomes, often as retroposons, to ensure expression during MSCI (157). In contrast to mammals, both Drosophila and C. elegans actually have few male-biased X-linked genes, probably because of the early onset of sex chromosome silencing in germ cells.
Genes important in female reproduction are also enriched on the mammalian X (78). Female-biased genes could arise by dominant or recessive mutations because only one X copy is expressed in a given cell (155). The human X chromosome contains many genes expressed in the brain as well, and X-linked forms of mental retardation are 3.5 times more common than autosomal forms, even after correcting for the ease of discovering X-linked disorders (138, 148). Sexual selection may help the accumulation of intelligence genes on the X; females could choose more resourceful males, resulting in rapid selection for recessive X-linked mutations in males (173). Alternatively, beneficial alleles may have also been selected in females, for example, in relation to prolonged care of the young (70). Many of the same genes expressed in mammalian testis are also expressed in brain. The concept of a smart and sexy X chromosome has been championed by Marshall Graves (104), who also pointed out that such genes may provide an engine of speciation by influencing mating barriers. Gene expression evolution is faster on the X chromosome, potentially reflecting faster functional adaptation (20). The highly specialized nature of the sex chromosomes should be taken into account when evaluating their dosage regulation. Indeed, the mammalian X contains a relatively smaller percentage of genes expressed in somatic tissues and implicated in functional networks that involve autosomal genes (39, 131).
Remarkably, the Z chromosome of birds has diverged along a similar path as the mammalian X (Figure 1). Although these sex chromosomes arose from completely different ancestral autosomes, they both became highly enriched in reproduction-related genes not found in their respective ancestral autosomes (10). In fact, 15% of the chicken Z chromosome is occupied by an array of multicopy testis-expressed genes. Such convergent evolution must be taken into account when considering dosage compensation in these organisms (see below). Interestingly, although both the present-day X and Z chromosomes are relatively gene-poor, their respective ancestral autosomes had even fewer genes; thus, the X and Z independently acquired hundreds of genes, mostly members of testis-expressed multicopy families (10) (Figure 2). Complete sequencing of additional species in conjunction with analyses of gene expression will help refine this model.
EFFECTS OF ANEUPLOIDY
The heterogametic sex represents a case of natural aneuploidy because each sex chromosome is present in one copy, whereas the rest of the genome is diploid (15). It is therefore instructive to consider what happens in cases of autosomal aneuploidy. Autosomes are present in two copies in most somatic cells of mammals, whereas germ cells are haploid. Aneuploidy is a deviation from these states that can affect entire chromosomes after nondisjunction or small chromosomal regions after partial deletions or duplications. Constitutional or acquired aneuploidies cause birth defects and cancer. In humans, constitutional trisomies for a few whole autosomes are, although debilitating, compatible with survival, whereas monosomies are lethal; thus, haploinsufficiency causes particularly harmful developmental defects. In contrast, sex chromosome aneuploidy is comparatively well tolerated because of the paucity of essential genes on the Y and inactivation of all but one upregulated X copy per diploid genome. Nonetheless, monosomy X causes Turner syndrome in humans, and the presence of more than three X chromosomes (e.g., tetrasomy) causes severe phenotypes due both to abnormal expression of genes that escape X inactivation, including most genes located in the PAR, and to developmental effects (see below).
The current explosion in discovery of specific syndromes associated with small deletions or duplications in the human genome has not been accompanied by vigorous investigation of their effects on dosage of gene expression, which is sorely needed to facilitate interpretation of clinical findings. For some genes, increased dosage has been directly implicated in specific abnormal features, for example, SOD (superoxide dismutase) in Down syndrome (44). However, the effects of abnormal gene dosage are not clear in different tissues. A complicating factor is the wide spectrum of clinical findings even in the well-defined trisomies. When considering expression levels of individual genes linked to chromosome 21 or to the corresponding mouse chromosomes in models of Down syndrome, much of the observed inconsistency is apparently unrelated to severity of phenotypes (142). Developmental pathways are stochastically altered by aberrant gene dosage, leading to unpredictable phenotypes, as suggested by Epstein (44). Phenotypic variability further depends on genetic background, for example, additional second hits in the genome of individuals with similar deletions or duplications but different phenotypes (55).
Subsets of genes located in aneuploid chromosomal regions do not show the expected change in expression (e.g., 1.5-fold increase in trisomy or twofold decrease in monosomy), suggesting the existence of mechanisms that dampen deleterious imbalances (47, 65). In addition to feedback mechanisms that directly regulate single genes, feed-forward and general buffering mechanisms may operate by modulating transcription or translation and/or RNA or protein degradation. These mechanisms may elicit epigenetic modifications, possibly over large chromosomal domains. The X chromosome is clearly an extreme example of such corrective expression adjustment. In Drosophila, dampening of abnormal levels of autosomal gene expression provides a stronger adjustment for monosomic than trisomic genes (151), and dampening contributes to a significant portion of the twofold upregulation of the male X chromosome (176).
In aneuploid genomes, gene expression is altered not just in the aneuploid region but also globally, suggesting enhanced gene expression instability unseen in euploid genomes, which may partly explain overlapping features of different chromosomal disorders (47, 65). As pointed out by Birchler (15), analyses of gene expression in aneuploid samples should be interpreted cautiously because normalization of the data may obscure expression changes throughout the genome. Absolute measurements of gene expression would help sort this out, but these are difficult and require co-isolation of DNA and RNA from the same sample. Furthermore, there may be a great deal of cell-to-cell variability that is lost when analyzing tissue samples or cell cultures. Some cell types are remarkably tolerant to aneuploidy, for example, human oocytes, which display a staggering level of chromosomal imbalance (up to 20% of oocytes have a chromosomal abnormality) (48). Interestingly, both X chromosomes are expressed in these cells. Conversely, some tissues may not tolerate dosage imbalance; for example, gene expression in the brain may be particularly dosage sensitive, hence favoring stable compensation (96, 117).
The structure and gene content of the sex chromosomes may reflect the sensitivity of some genes to haploinsufficiency. Notably, the paucity of orthologs of haploinsufficiency genes previously identified in yeast on the mammalian and C. elegans X chromosomes suggests constraints on evolution of their gene content (36). Among genes especially sensitive to haploinsufficiency are those encoding transcription factors whose abnormal levels affect the entire genome, often in multiple tissues. A subset of genes may be spared deleterious effects of deletion due to redundancy (143). In principle, highly abundant proteins would also be less dosage sensitive. This is relevant to the regulation of the Drosophila X chromosome, in which medium-expressed genes appear to be more sensitive to disruption in dosage than highly expressed genes (60, 80). In humans, a subset of X-linked genes expected to be dosage-sensitive on the basis of the inclusion of their products in large protein complexes are efficiently dosage compensated (131). It will be interesting to further characterize genes sensitive to dosage disruptions on the mammalian X chromosome. As pointed out above, dosage-sensitive genes may be retained longer as X/Y gene pairs on the sex chromosomes if they do not tolerate the loss of the Y copy.
NATURAL VARIATIONS IN GENE DOSAGE AND GENE EXPRESSION
Is natural aneuploidy caused by variation in copy number tolerated or even beneficial? Inherited or de novo benign copy number variants (CNVs) are abundant in normal individuals and can be mosaic, thus representing somatic events (54). CNVs range in size from 1-50 bp to several Mb and probably cover approximately 5% of any human genome, possibly explaining as much as 17% of the variation in gene expression (33). CNVs can influence expression of genes within the variant and in adjacent regions, suggesting long-distance effects (64). CNVs are also frequent between species, highlighting their role in evolution with gains as well as losses playing an important role in generating new traits (27, 124). Genes involved in adaptive evolution, for example, immune defense, metabolism, and sensory and brain function, often have variable dosage (38, 130). For genes whose CNV is not immediately favorable, dosage compensation mechanisms based on feedback or buffering may help dampen adverse effects. However, specific modules of genes implicated in the same pathway probably remain single copy because changes in their expression are not tolerated, underscoring the maintenance of stoichiometry (17). As stated above, the mammalian sex chromosomes favor reproduction-related gene families with variable copy number between species and individuals. The rest of the X, however, is highly conserved and tightly controlled (122). This is because complex mechanisms of dosage compensation are targeted to the X, where newly added regions have to be incorporated in X-specific regulatory systems.
An added level of natural variation in gene expression is caused by allele-specific expression. Autosomal genes are usually expressed from both alleles but can occasionally display skewed or even monoallelic expression, which could contribute to evolution and have profound effects on dosage (81, 121). Imprinted genes are normally expressed either from the maternal or paternal allele, and severe phenotypes result from aberrant expression (9). Imprinted gene clusters have features redolent of the inactive X: Long noncoding RNAs and epigenetic changes are often involved in their regulation, suggesting common regulatory and evolutionary pathways (120). Monoallelic expression also characterizes the immunoglobulin, T-cell receptor, interleukin, olfactory, and vomeronasal genes to insure expression of a single allele in a given cell (18, 101). In aggregate, approximately 15% of human autosomal genes appear to be expressed from one allele at random, but their role is often poorly defined (53).
The X chromosome represents an extreme case of monoallelic expression, which is potentially beneficial for tight expression control of tissue-specific genes, such as testis-expressed genes (70). However, monoallelic expression can also be associated with increased expression noise. Dosage compensation mechanisms to enhance expression of the X may help reduce natural fluctuations within a cell. This noise represents random production and decay of low-copy transcripts and/or protein products, or possibly unequal distribution of products at cell division (67, 134). Although enhanced expression noise may be favorable to fast adaptive responses to stimuli, reduced noise is clearly beneficial for gene products that should remain relatively constant (19). For example, high expression of transcription factors may be particularly important to ensure promoters have a greater chance of being turned on. Elegant experiments in C. elegans have shown that the impact of random fluctuations in gene expression is buffered by normal biallelic expression or by expression of a closely related gene (22). For the single active X chromosome, increased expression may be critical to stabilize expression.
MOLECULAR MECHANISMS OF SEX CHROMOSOME REGULATION
Eutherian Mammals: X Upregulation in Both Sexes and X Inactivation in Females
The evidence for X upregulation in mammals was initially based on analysis of a single gene, Clcn4-2, in different mouse species with either an X-linked or autosomal copy (1). Subsequently, global analyses of gene expression in multiple species and tissues demonstrated upregulation of expressed X-linked versus autosomal genes (39, 58, 116). RNA-seq analyses initially appeared to contradict results obtained by expression arrays (165), but new analyses have confirmed upregulation of expressed X-linked genes in mammals (39, 77, 93). A major confounding factor is the inclusion of genes with extremely low expression, which artificially lowers median X:autosome expression ratios (165). It turns out that genes with no or low expression in somatic tissues are more abundant on the X than autosomes and represent testis-specific genes (39). Thus, the reproduction-related features of genes on the X must be taken into account when examining their dosage regulation in somatic tissues (Figure 3). Interestingly, the subset of genes subject to efficient X upregulation and X inactivation in humans are included in large protein complexes that would be particularly sensitive to disruptions in dosage (131). Two recent studies have compared expression of a subset of conserved ancestral genes on the mammalian X chromosome to expression of their orthologs in chicken where the genes are autosomal (75, 94). Cross-species comparisons of RNA-seq data to measure gene expression levels should be interpreted with caution, given the significant differences in the depths of genome annotation between species. On the basis of these analyses, the authors concluded that ancestral X-linked genes were not upregulated during evolution from autosomal to X-linked status. One study acknowledges that some X-linked genes are upregulated and also proposes that balanced expression between X-linked and autosomal genes may be partly mediated by repression of autosomal genes implicated in common pathways (75). This is similar to the reverse dosage model in Drosophila (see below) (14).
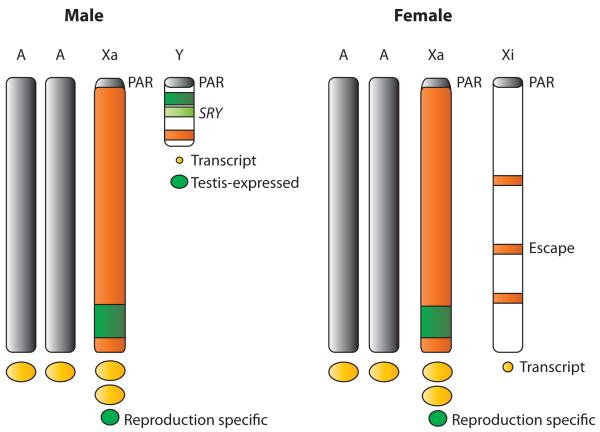
Figure 3.
Schematic of the mammalian sex chromosomes in males and females. Expressed genes on the active X (Xa) produce a higher level of transcripts compared with genes on the autosomes (AA). The pseudoautosomal region (PAR) is not upregulated. The Y and X chromosomes contain many testis-expressed (or reproduction-specific) genes ( green). The dots under the chromosomes represent the amount of gene product. Genes that escape X inactivation on the inactive X (Xi) and their few paralogs on the Y produce a small number of transcripts (small yellow dots).
Although much is known about the onset of X inactivation in early embryogenesis, the timing of X upregulation is unknown. Our original analyses show a balanced X:autosome expression ratio in haploid germ cells, suggesting a lack of X upregulation in these cells (116). The progressive increase in X-linked gene expression observed in differentiating male and female embryonic stem (ES) cells is the best evidence so far that upregulation gets established during early embryogenesis (92, 93). Single-cell RNA-seq (68) will help determine the precise timing of X upregulation during embryo development and in different tissues.
Molecular differences between the active X chromosome and autosomes are starting to emerge. We reported the first evidence of higher RNA polymerase II (PolII) occupancy at the 5′ end of expressed X-linked versus autosomal mouse genes, which is consistent with increased transcription (39). Another study confirmed enrichment for PolII as well as for specific histone modifications associated with active chromatin, including H3K4me3 and H3K36me3 on active X alleles (170). In contrast, the inactive X compartment excludes PolII, and histone modifications change to those associated with repressed chromatin (26, 62). It is possible that the X-inactivation machinery specifically counteracts modifications associated with X upregulation, such as histone acetylation, which decorates the two active X chromosomes in undifferentiated female ES cells (119). More than one mechanism of upregulation could operate on individual mammalian X-linked genes. Even in Drosophila, not all X upregulation is mediated by the MSL (male-specific lethal) complex, and some X-linked genes are apparently not upregulated (87). In addition, dosage compensation may be selected for a subset of genes with other genes simply being affected by the same mechanism. As discussed above an important function of X upregulation may be to reduce expression noise from single expressed alleles (172). It is noteworthy that transcripts from mammalian X-linked genes are less susceptible to decay compared with autosomal genes, suggesting increased RNA stability that could contribute up to 30% upregulation (171). Single-cell transcriptome analyses (68) and in situ studies will facilitate comparisons between X and autosomes in terms of fluctuations in cell- and tissue-specific expression. The high X-linked gene expression we observed in the brain (117) should foster stable and robust gene expression in an organ that may not tolerate sudden changes in gene expression.
Much more is known about mechanisms of X inactivation (62, 113, 128). X inactivation is triggered by the noncoding RNA Xist in early development when pluripotency factors are downregulated. Specific activators influence the probability of initiating silencing, for example, Rnf-12, whose dosage is critical (74). In mice, Xist coats the X and recruits the Polycomb complexes to install repressive chromatin modifications, including H3K27me3 and H2AK119ub. Xist is regulated by multiple factors, including its antisense transcript Tsix and other noncoding RNAs (Jpx and Ftx) located near Xist (137). Eventually, the inactive X becomes late replicating, CpG islands are methylated, and SMCHD1 and macroH2A are recruited to secure silencing (52). In somatic cells, Xist RNA is not required for maintenance of silencing, which is extremely stable because of the multiple layers of epigenetic regulation. Reactivation of individual X-linked genes is rarely observed in aging (160). Interestingly, X inactivation can be reversed in induced pluripotent stem cells following the activation of stem-cell factors (79).
X reactivation due to loss of Xist occurs in primordial female germ cells (5, 63, 153). An important question is whether these cells have X hyperexpression. Mouse ES cells derived from the inner cell mass of female blastocysts have two active X chromosomes, resulting in a high X:autosome expression ratio of approximately 1.4 but not 2, possibly due to dampening and/or partial upregulation. Tellingly, undifferentiated ES cells with two active X chromosomes are difficult to maintain in culture and often lose or silence one X (177). Furthermore, mouse embryos engineered to retain two fully active X chromosomes have a severe phenotype reminiscent of tetrasomy (111). These studies suggest that the presence of two active X chromosomes can be tolerated at specific developmental stages but cannot be sustained throughout development. Tight dosage control in mice is associated with early paternal X inactivation at the 2-4-cell stage, which led to the idea that imprinted X inactivation was the primitive state. However, random X inactivation may be the norm, as shown in human and rabbit (123). In these species, the onset of X inactivation is delayed compared with mouse, and Xist can be biallelically expressed. In these species, cells apparently tolerate the presence of one, two, or zero active Xs with no tight X dosage regulation up to the blastocyst stage.
Marsupial and Monotreme Dosage Compensation
Both X upregulation and X inactivation are well documented in marsupials (56, 75). Rather than being random, marsupial X inactivation is imprinted toward the paternal X chromosome, similar to the situation in extraembryonic mouse tissues. Molecular mechanisms of silencing are not completely worked out, but there are significant differences with eutherian mammals. The noncoding RNA Xist is absent in marsupials (42), which employ a different noncoding RNA Rsx to trigger silencing (57). There are some similarities between marsupials and eutherians, for example, the inactive X becomes enriched in histone H3K27me3 (156), but DNA methylation is not clearly involved in silencing. In marsupial cultures, cells with both monoallelic and biallelic X expression are observed by RNA-FISH, suggesting that partial paternal expression is stochastic (4). X inactivation is apparently more stable in tissues (100). Contrary to the prevailing view that paternal X inactivation is a remnant of MSCI, the marsupial X, like the eutherian X, becomes de novo inactivated in early embryos (100). Monotremes have multiple sex chromosomes that are related to the chicken Z rather than the human X, which complicates the analysis of their regulation. Male-to-female expression ratios vary between one and two for different loci, and again partial inactivation is explained by stochastic frequencies of monoallelic and biallelic expression (37).
Drosophila melanogaster: X Upregulation in Males
Because so much is known about dosage compensation in Drosophila, it is useful to review its major features, which may help in understanding mammalian systems. We refer the reader to extensive reviews of the molecular mechanisms that upregulate expression from the male X chromosome (31, 51, 152). Sex in fruit flies is triggered by the X:autosome ratio and upregulation of the male X is tied to the sex determination pathway (30) (Figure 1). In male somatic cells, a protein complex containing noncoding RNAs called the MSL complex is targeted to the X. An important component of the complex, MOF, specifically acetylates histone H4 at lysine 16, which opens the chromatin and increases gene expression. Upregulation of the male X in somatic cells is due to an increase in transcription elongation (86), but transcription initiation, pause release, and RNA stability also play a role (31). Notably, PolII is enriched at the promoters of expressed X-linked versus autosomal genes in male Drosophila (32), similar to that observed at the 5′ end of ex-pressed mouse X-linked genes (39). The MSL complex is not sufficient to achieve a precise doubling of gene expression, and other components are also involved, including some repressive factors (51, 176). Thus, X upregulation in fruit flies is probably due to a combination of buffering, feedback, and feed-forward mechanisms. Surprisingly, male germ cells apparently employ a different mechanism of X upregulation that does not involve the MSL complex (39, 58). Although the prevailing model suggests mediation of compensation mainly by the MSL complex, an alternative model involves an inverse dosage effect that affects the whole genome (14). In this model, sequestration of MOF on the male X prevents upregulation of autosomal expression due to a lower dose of the X chromosome in males but allows upregulation of the X, leading to a balanced genome (13). This model can also account for dosage compensation in triple X metafemales, which the MSL model does not address. These vigorously debated models would benefit from measurements of absolute gene expression levels of the X and the autosomes (13), which are seldom performed.
Caenorhabditis elegans: X Upregulation in Both Sexes and X Repression in Hermaphrodites
C. elegans is another model organism in which dosage compensation mechanisms have been extensively studied. Sex is determined by the X to autosome ratio, males being X0 and hermaphrodites XX (109) (Figure 1). X upregulation has been documented in somatic cells, but little is known about the molecular mechanisms of enhanced gene expression (39, 58). Silencing of the sex chromosomes in C. elegans germ cells confounded a recent study that claimed the absence of X upregulation in the worm (165). However, analyses of mutant worms with no germ cells yielded a balanced median X:autosome expression ratio of one (39). In XX hermaphrodites, both X chromosomes are repressed to avoid hyperexpression (Figure 1). The composition and mechanism of action of the repressive dosage compensation complex (DCC) in hermaphrodites have been extensively reviewed (109). In brief, the DCC assembles from ten proteins, including five members of the condensin protein family, one of which, DPY27, becomes specifically recruited to the X.
Bird Dosage Compensation
In contrast to mammals, birds have a ZZ/ZW sex determination system, females being the heterogametic sex (Figure 1). Incomplete dosage compensation is evident based on Z-linked gene expression, with male to female ratios ranging from 1.2 to 1.4 in chicken, finch, and crow. However, when comparing Z versus autosomal expression, females have Z:A expression ratios higher than 0.5 (0.6-0.8), indicating partial dosage compensation (Figure 1) (75, 107, 164). Compensated genes in one bird species tend to be the same in other species, suggesting that there is a subset of dosage-sensitive genes. Partial dosage compensation in birds may result from piecemeal upregulation of Z-linked genes especially in females, perhaps using buffering mechanisms. In addition, stochastic Z repression is observed in male cells (96). Interestingly, a subset of Z genes in chicken, but not in finch, associate with a noncoding RNA MHM (male hypermethylated) and recruit H4K16 acetylation for upregulation in females (69, 107). Thus, H4K16ac may be involved in upregulation not only in Drosophila but also in birds. However, Mank & Ellegren (102) pointed out that this dosage-compensated region may simply represent female-biased genes. One can speculate that adjustment of expression from the Z chromosome reflects its gene content. It is possible that the Z carries many genes whose variable expression between the sexes is neutral or actually advantageous in evolving sex-specific traits, such as plumage, testis function, or sex determination itself, as shown for the testis determinant Dmrt1 that has higher expression in males. As with the X, the Z chromosome is somewhat gene-poor but does contain a large number of amplified gene families expressed in testis (10) (see above). The convergent evolution of the X and Z is puzzling because in birds the heterogametic sex is female. However, strong selection for highly expressed testis genes favors the absence of dosage compensation and a double dose of Z in males, whereas similar selective forces favors upregulation of the active X in mammals.
COUNTING Xs: POLYSOMY AND POLYPLOIDY
How are X chromosomes counted? During normal mouse development, the two X chromosomes interact with each other, which is a process thought to represent counting and choice (105). This kissing or contact-dependent mechanism may be a special feature of mouse X inactivation in which Tsix plays an important role. Other mammalian species, such as human and rabbit, may depend on a largely stochastic mechanism of counting and choice followed by cell selection (112, 123). In diploid cells with any number of X chromosomes, a single X remains active, whereas all other copies are silenced, thus insuring correct dosage. Nonetheless, X aneuploidy causes dosage imbalance due to genes that escape from X inactivation (see below) and to effects prior to X inactivation in early embryos. Additional effects after X reactivation in germ cells may cause infertility (63). For example, in XXY germ cells from Klinefelter individuals, X reactivation occurs and dosage of X-linked genes normally repressed by MSCI may be too high. In this case, only germ cells that have lost one X can proceed through meiosis. In germ cells from X0 Turner individuals, the unpaired X is silenced by MSCI, hampering ovarian development. In contrast, XXX females are usually fertile because two of the Xs would pair and remain active and the unpaired one would be silenced (63). Interestingly, flies with three X chromosomes can survive, indicating that repression of the X must take place (15).
Polyploidy can also be informative and aid our understanding of X dosage regulation. Polyploidy is tolerated in many organisms despite massive changes in dosage throughout the genome. For example, plants and amphibians can have very large polyploid genomes. Polyploidy generated by whole-genome duplication theoretically causes no chromosome imbalance (59) and can even be advantageous, perhaps by increasing variation in gene expression due to rapid genetic and epigenetic changes (125). In mammals, polyploidy is tolerated only in certain somatic tissues, for example, liver and giant trophoblastic cells, potentially to increase gene expression and help tissue regeneration (89). Cancer cells are often polyploid, a way to generate genomic diversity with subsequent frequent loss or rearrangements.
Congenital polyploidy causes severe birth defects. Human triploid and tetraploid fetuses are spontaneously aborted. The question of the regulation of the X in the presence of three sets of autosomes has long fascinated researchers (50, 71). How many active and inactive X chromosomes would these cells have? In Drosophila, triploidy is associated with tripling of gene dosage on the single X (16). Human or mouse triploid embryos with a single active X chromosome (XYY) are very rare, probably because of massive X-to-autosome imbalance. We examined cells from triploid human fetuses to address the question of whether X or autosomal expression may become adjusted (40). Absolute autosomal expression levels per gene copy are similar in triploid versus diploid cells, indicating no apparent global effect on autosomes. In triploid cells with two active X chromosomes, a basic doubling of X-linked gene expression takes place without further adjustment, but in cells with a single active X, gene expression is adjusted upward presumably by an epigenetic mechanism. These findings are reminiscent of the greater expression tuning of genes in monosomic versus trisomic chromosomal regions (176). Expression of a subset (~7%) of X-linked genes is apparently proportional to the number of autosomal sets in human triploid cells, suggesting that these genes are efficiently dosage compensated (40).
TARGETING AND ORGANIZING THE X: MOTIFS AND NUCLEAR POSITION
An important question in dosage compensation is how to target the regulatory machinery to the X alone while sparing the autosomes. This process is best understood in flies and worms. In Drosophila, approximately 150-300 high-affinity MSL recognition sites exist on the X, from which the MSL complex spreads to low-affinity sites (51). These GA dinucleotide-rich entry sites are not exclusive to the X, being enriched only twofold. Thus, selective X targeting of the MSL complex is not completely explained. One attractive hypothesis is that the X occupies a specific territory in the nucleus, facilitating spreading of the MSL complex (31). This nucleation theory is akin to the core component of the inactive X in mammals (see below). Nuclear pore elements interact with the MSL complex, suggesting a specific X location favorable to enhanced transcription (108). In C. elegans, X-specific motifs recruit the repressive DCC in hermaphrodites via 200 rex (recruitment elements on the X) sites that contain a specific 12-bp MEX motif from which the DCC spreads to dox (dependent on X) sites often located at highly active promoters (34, 106). Some genes escape repression even if they bind the DCC, suggesting that additional factors, perhaps related to chromosome configuration, are important (109). In both flies and worms, the dosage compensation machinery is capable of altering autosomal expression if targeted there.
Are there motifs on the mammalian X for either silencing or upregulation? The remarkable conservation of the X as a linkage group, probably due to unique dosage regulation mechanisms, suggests the existence of X-specific motifs. Comparisons between homologous regions on the X and autosomes help find X-specific sequences. Clcn4-2, represents a rare opportunity to do this, as it is X-linked in most mammals and autosomal in one branch of Mus (127, 141). When located on the X, Clcn4-2 is subject to both X upregulation and X inactivation, thus providing a useful model. At the autosomal locus, the size of Clcn4-2 is dramatically reduced by deletions of large portions of its introns. AT-rich sequence motifs deleted on the autosome but preserved on the X are abundant throughout the entire X (118). Whether these motifs are implicated in either X upregulation and/or X inactivation remains to be investigated. Interestingly, proteins that bind AT motifs such as SATB1 are implicated in X inactivation (2). Additional motifs potentially facilitating X inactivation have been identified in humans by comparing genes subject to X inactivation with escape genes (159). The mapping of Xist entry sites using novel methods to define global RNA-chromatin interactions (ChIRP-seq) is eagerly awaited to further define X inactivation-related motifs akin to the previously postulated way stations (29, 49, 145). An important factor may be YY1, which facilitates Xist binding to the chromatin (72). LINE (long interspersed repeat elements) repeats abundant on the X have also been proposed as way stations for the propagation of silencing (99). However, some Oryzomys species lack active LINEs and still have X inactivation, suggesting that other elements may be implicated in these rodents (23). In mouse, LINE elements nucleate the condensed inactive X compartment, thus organizing the Barr body (8) in which genes surround the periphery of the core, with escape genes being the most peripheral (26, 28, 150). The inactive X often visits the nucleolus, probably to help maintain its heterochromatic structure (174).
As with X inactivation, X upregulation could involve sequence motifs that attract an epigenetic machinery to the whole X chromosome. However, although X inactivation can spread to autosomal portions of X-autosome translocations (144), there is no evidence so far of spreading of X upregulation. Alternatively, 5′ end promoter mutations that enhance transcription and/or 3′ end mutations that enhance stability of message may have evolved gene-by-gene when the Y paralog degenerated. It will be interesting to determine whether the 5′ end or the 3′ end sequences of X-linked genes differ from those of autosomal genes, which could attract specific epigenetic modifications to the active X. The active X is often located at the nuclear periphery, which may facilitate upregulation by proximity to nuclear pores, as shown in Drosophila (108). Elegant 4C (circular chromosome conformation capture) studies have shown striking differences in conformation between the active and inactive Xs in mammals, but no specific differences were noted between active X and autosomes (150).
INCOMPLETE DOSAGE COMPENSATION AND SEX DIFFERENCES
Escape From X Inactivation
Exceptional genes remain expressed from the inactive X in mammals (Figure 2 and 3). Surveys show that approximately 15% and 3%-6% of genes escape X inactivation in human and mouse, respectively (24, 150, 168). Domains of escape in human and other species contain multiple adjacent genes, whereas in rodents each domain contains one or two genes (11). Interestingly, genes that produce a long noncoding RNA have been discovered in domains of escape (97, 135), suggesting a similar regulation to imprinting domains. This is an attractive hypothesis, given that long noncoding RNAs are known to have long-range effects that influence chromatin structure. Escape genes are enriched in PolII and in active chromatin marks on the inactive X (83, 168). Barrier elements such as CTCF may help insulate escape domains but their role is not completely understood (46, 91).
Are escape genes subject to X upregulation on the active X? PAR genes are apparently not upregulated, which is consistent with near-equal sex dosage (73). A priori, if escape genes outside the PAR represent early stages of sex chromosome evolution, such genes would not be expected to be upregulated. However, we find no significant differences in overall expression levels of escape genes compared with inactivated genes in males, suggesting that genes outside the PAR are already upregulated on the active X. Although many escape genes still have a Y copy, this paralog may have acquired a male-specific function, justifying upregulation of the X copy (41). The higher density of genes that escape X inactivation in regions recently diverged from the Y suggests that X inactivation may follow Y degeneration (56). However, once X inactivation became controlled by a center from which epigenetic changes could efficiently spread, as shown in female individuals with X:autosome translocations, it is conceivable that forward silencing of the inactive allele forced upregulation of the active allele and allowed degeneration or differentiation of the corresponding Y-linked paralog in males. Expression from the Y and from the inactive X is usually much lower than from the active X, consistent with upregulation of the allele on the active X only (11), implying that the machinery to upregulate the active X may be excluded from the inactive X.
Some genes have variable levels of escape between cells, tissues, and individuals, suggesting a role in phenotypic variability (21). Escape genes in humans, mice, and elephants show by single-cell RNA-FISH a pattern of stochastic transcription from one or both alleles (3). However, by allele-specific reverse transcription polymerase chain reaction analyses in single cells, the escape gene Kdm5c shows consistent biallelic expression in cells from adult mouse tissues (95). Interestingly, stochastic silencing of Kdm5c occurs in embryos, followed by reactivation, possibly due to lack of DNA methylation, to secure silencing (46, 95). Thus, the complexity of mechanisms of dosage compensation, including escape from X inactivation, provides opportunities for evolution of variability in gene expression possibly beneficial to females.
Sex-Specific Differences and Disease Susceptibility
Sex-specific differences are usually ascribed to hormonal effects. The mouse four-core genotype system pioneered by Burgoyne and Arnold (6) has helped sort out effects of hormones from those of the sex chromosomes on phenotypes by producing XX males and XY females. This has revealed sex differences that depend on the number of X chromosomes as well as on the presence or absence of the Y. Interestingly, the sex chromosome makeup also influences autosomal expression independently of the phenotypic sex (162). Thus, the number of X chromosomes has a profound effect on the entire genome in this mouse model. This is in contrast to human triploid cells with one or two active X chromosomes in which no apparent autosomal effects were detected (40).
Higher female than male expression is often observed for genes that escape X inactivation, although apparent sex differences in transcription levels can be compensated at the level of translation (166, 167). In some instances, for example, RPS4X and RPS4Y, partial expression from the inactive X and from the Y equalizes sex expression (161). Johnston et al. (73) argued that, except for a few genes, dosage compensation between males and females is virtually complete in human lymphoblastoid cell lines. However, tissue-specific differences may exist (169). It is remarkable that a subset of escape genes is conserved in multiple mammalian species; for example, KDM5C/KDM5D and KDM6A/UTY have remained on the sex chromosomes for more than 145 million years. Such genes may simply not have had time to acquire dosage compensation or they could be dosage sensitive. Alternatively, we favor the possibility that escape from X inactivation may be beneficial to females (11, 21). Specific examples remain to be fully investigated. One intriguing possibility is a role in X inactivation itself. X-linked genes rarely have lower expression in females; however, this is the case for PAR genes because of spreading of silencing on the inactive X (73). Paternally imprinted X-linked genes represent another special category with lower expression in females because of random X inactivation; one example is the Xlr cluster in mouse, which influences sex-specific behavior (133, 163). Additional sex differences result from biallelic X expression during specific stages of female embryogenesis and from specific sex chromosome expression in germ cells. Finally, the inactive X could exert heterochromatin sink effects and attract repressive factors that would enhance expression of the rest of the genome in females, similar to effects of the largely heterochromatic Y chromosome in Drosophila (6).
Dosage compensation mechanisms have a profound influence on the manifestation of sex chromosome disorders. Added or missing copies of the X cause abnormal phenotypes because of increased or decreased expression of escape genes and abnormal effects prior to X inactivation during development (11, 63). Structural anomalies of the human X also cause abnormal phenotypes dependent on the sex of the patient. For example, duplications of the X are deleterious in males because of hyperexpression but not in females in whom the duplicated X is usually inactivated (90). Small-ring X chromosomes that lack XIST can cause severe phenotypes in females because of the absence of silencing, whereas large rings that retain XIST do not (90). Males are more susceptible to X-linked disorders than females, who are often protected either because random X inactivation ensures a sufficient number of normal cells and gene products or because skewing of X inactivation favors normal cells. Skewing, which can be cell or tissue specific has been well documented in females for many human diseases caused by constitutional X-linked mutations (21, 110). In diseases in which mutations are acquired, such as cancer, oncogenes subject to X inactivation would have increased expression in females only if the active X is mutated or the inactive allele reactivated epigenetically. Furthermore, tumor suppressor genes normally subject to X inactivation may need only one hit to exert their deleterious effects because of the clonal nature of tumors (149). X-autosome translocations represent another cause of expression changes in cancer cells because of position effects.
CONCLUDING REMARKS
Once sex chromosomes became differentiated, dosage compensation mechanisms evolved to avoid the resulting deleterious effects of genome imbalance. To balance expression throughout the genome, organisms have evolved fascinating means of enhancing expression and of silencing the sex chromosomes. Special features of dosage compensation mechanisms are emerging to explain how they operate at a chromosomal scale. Targeting of regulatory mechanisms to the sex chromosomes is well understood for only some organisms, and it is already clear that the machinery varies widely between species even though there may be some common features. Mammalian X inactivation employs both noncoding RNAs and epigenetic modifications to achieve multiple layers of control for silencing. In contrast, little is known about the mechanism of upregulation of expressed genes on the active X. Functional hemizygosity of X-linked genes has greatly influenced the gene content and regulation of the sex chromosomes. Although much about the evolution of sex determination and dosage compensation is now understood in its outline, further studies in additional species will add to our understanding of the forces that shape the sex chromosomes and influence their regulation.
MAIN MECHANISMS OF DOSAGE REGULATION.
Three types of gene expression control have been identified in aneuploids:
General dampening of abnormally increased or decreased gene expression due to aneuploidy.
Feedback control of expression of individual genes.
Feed-forward mechanisms specific to the sex chromosomes.
SUMMARY POINTS.
Sex chromosome differentiation occurs because of suppressed recombination in species with genetic sex determination. Different mechanisms of dosage compensation have evolved to deal with the resulting dosage imbalance. In mammals, upregulation of expressed genes on the active X chromosome balances expression with the autosomes, whereas X inactivation silences one X in females. In Drosophila, the X chromosome is upregulated in males only, and in C. elegans upregulation of the X takes place in both sexes together with repression of the Xs in hermaphrodites. Inverse dosage mechanisms that regulate the autosomes may also contribute to dosage compensation.
Mammalian and avian sex chromosomes are enriched in genes important for sexual reproduction, such as genes expressed in testis.
Autosomal aneuploidy is deleterious, and monoallelic expression is associated with increased stochastic expression noise, suggesting that adjustment of X expression by upregulation is important.
Dosage compensation may use a combination of mechanisms, including a general buffering caused by aneuploidy, feedback mechanisms to control individual genes, and feed-forward mechanisms to control entire chromosomes. Different mechanisms may control specific subsets of genes.
Dosage compensation is not efficient in some organisms, such as birds and fishes; this leads to sex differences in gene expression levels that may play a role in phenotypic sex attributes.
Molecular mechanisms of dosage compensation specific to the sex chromosomes involve specific complexes of proteins and noncoding RNAs as well as specific features of chromosomal structure and location. Targeting of the X chromosome involves enrichment in specific motifs on the X.
A number of genes escape dosage compensation. In particular, genes that escape X inactivation in mammals are associated with sex-specific differences. Such genes may be important in female-specific functions.
FUTURE ISSUES.
An important evolutionary question regards when dosage compensation mechanisms arise. Does upregulation of a specific X-linked gene happen as soon as its Y-linked paralog is lost, degraded, or differentiated? In mammals and C. elegans with two forms of dosage regulation, are both X upregulation and X repression timed to coincide with Y loss or degeneration? What is the impact on autosomal gene expression?
The molecular mechanisms of dosage compensation are not completely understood in mammals, especially those responsible for X upregulation. Is upregulation mediated by a protein complex targeted to the active X, or does it result from a gene-by-gene enhancement of expression during evolution? Furthermore, what are the effects of disrupting dosage compensation mechanisms on disease?
Analyses of gene expression in single cells will help identify events associated with the transition from haploidy to diploidy and vice-versa in terms of X expression.
How sex-specific differences in gene expression affect phenotypes is not well understood and will require examining dosage compensation in different tissues and developmental stages. The role of escape from X inactivation in female-specific functions and in diseases prevalent in females should be further investigated.
ACKNOWLEDGMENTS
I would like to thank the members of my laboratory, in particular, X. Deng, D.K. Nguyen, and J.B. Berletch for their contributions and discussions of the subject of this review. I thank X. Deng (University of Washington, Seattle), J.A. Marshall Graves (La Trobe University, Melbourne, Australia), and R. Monnat (University of Washington, Seattle) for their critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (GM079537, GM046883, MH083949).
Glossary
- Sex chromosomes
specialized chromosomes that determine the sex of an individual; e.g., XY is male and XX is female
- Heterogametic sex
sex in which the sex chromosomes differ; e.g., XY or ZW
- Homogametic sex
sex in which the sex chromosomes are the same; e.g., XX or ZZ
- Aneuploidy
abnormal number of chromosomes or chromosomal segments
- Stoichiometry
balance of amount of products within a network
- Heteromorphic sex chromosomes
chromosomes with different size and gene content
- Paralogs
genes with a similar sequence but located on different chromosomes
- Hemizygous
genetic material present in a single copy
- Autosomes
chromosomes that are not sex chromosomes
- Pseudoautosomal region (PAR)
region of homology and pairing on the sex chromosomes
- Homomorphic sex chromosomes
chromosomes with similar size and gene content
- Haploinsufficiency
insufficiency due to the presence of a single copy instead of two copies of a gene in a diploid cell
- Monosomy
one instead of two chromosomes in a diploid organism
- Trisomy
three instead of two chromosomes in a diploid organism
- Orthologs
genes with a similar sequence and/or function between species
- Biallelic expression
expression of two copies (alleles) of a gene
- Polyploidy
increased number of whole sets of chromosomes; triploidy is three sets, tetraploidy is four sets
- X-autosome translocation
translocation between the X chromosome and an autosome
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adler DA, Rugarli EI, Lingenfelter PA, Tsuchiya K, Poslinski D, et al. Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus. Proc. Natl. Acad. Sci. USA. 1997;94:9244–48. doi: 10.1073/pnas.94.17.9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, et al. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev. Cell. 2009;16:507–16. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al Nadaf S, Deakin JE, Gilbert C, Robinson TJ, Graves JA, Waters PD. A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma. 2011;121:71–78. doi: 10.1007/s00412-011-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Nadaf S, Waters PD, Koina E, Deakin JE, Jordan KS, Graves JA. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol. 2010;11:R122. doi: 10.1186/gb-2010-11-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andina RJ. A study of X chromosome regulation during oogenesis in the mouse. Exp. Cell Res. 1978;111:211–18. doi: 10.1016/0014-4827(78)90251-3. [DOI] [PubMed] [Google Scholar]
- 6.Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2011;28:55–61. doi: 10.1016/j.tig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, et al. Are all sex chromosomes created equal? Trends Genet. 2011;27:350–57. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 9.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011;3:a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–16. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum. Genet. 2011;130:237–45. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernardo Carvalho A, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009;25:270–77. doi: 10.1016/j.tig.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhadra MP, Bhadra U, Kundu J, Birchler JA. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics. 2005;169:2061–74. doi: 10.1534/genetics.104.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birchler J, Sun L, Fernandez H, Donohue R, Xie W, Sanyal A. Re-evaluation of the function of the male specific lethal complex in Drosophila. J. Genet. Genomics. 2011;38:327–32. doi: 10.1016/j.jgg.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Birchler JA. Reflections on studies of gene expression in aneuploids. Biochem. J. 2010;426:119–23. doi: 10.1042/BJ20091617. [DOI] [PubMed] [Google Scholar]
- 16.Birchler JA, Fernandez HR, Kavi HH. Commonalities in compensation. BioEssays. 2006;28:565–68. doi: 10.1002/bies.20408. [DOI] [PubMed] [Google Scholar]
- 17.Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bix M, Locksley RM. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–54. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- 19.Bollenbach T, Kishony R. Quiet gene circuit more fragile than its noisy peer. Cell. 2009;139:460–61. doi: 10.1016/j.cell.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–48. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 21.Brown CJ, Greally JM. A stain upon the silence: genes escaping X inactivation. Trends Genet. 2003;19:432–38. doi: 10.1016/S0168-9525(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 22.Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–53. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 23.Cantrell MA, Carstens BC, Wichman HA. X chromosome inactivation and Xist evolution in a rodent lacking LINE-1 activity. PLoS One. 2009;4:e6252. doi: 10.1371/journal.pone.0006252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 25.Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–33. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 26.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–37. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Z, Ventura M, She X, Khaitovich P, Graves T, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 28.Chow JC, Heard E. Nuclear organization and dosage compensation. Cold Spring Harb. Perspect. Biol. 2010;2(11):a000604. doi: 10.1101/cshperspect.a000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cline TW, Meyer BJ. Vive la difference: males versus females in flies versus worms. Annu. Rev. Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 31.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 2012;13:123–34. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 32.Conrad T, Cavalli FM, Vaquerizas JM, Luscombe NM, Akhtar A. Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters. Science. 2012;337:742–46. doi: 10.1126/science.1221428. [DOI] [PubMed] [Google Scholar]
- 33.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 2007;39:S22–29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 34.Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science. 2004;303:1182–85. doi: 10.1126/science.1092938. [DOI] [PubMed] [Google Scholar]
- 35.D’Esposito M, Ciccodicola A, Gianfrancesco F, Esposito T, Flagiello L, et al. A synaptobrevin-like gene in the Xq28 pseudoautosomal region undergoes X inactivation. Nat. Genet. 1996;13:227–29. doi: 10.1038/ng0696-227. [DOI] [PubMed] [Google Scholar]
- 36.de Clare M, Pir P, Oliver SG. Haploinsufficiency and the sex chromosomes from yeasts to humans. BMC Biol. 2011;9:15. doi: 10.1186/1741-7007-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deakin JE, Hore TA, Koina E, Marshall Graves JA. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 2008;4:e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demuth JP, Hahn MW. The life and death of gene families. BioEssays. 2009;31:29–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- 39.Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 2011;43:1179–85. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng X, Nguyen DK, Hansen RS, Van Dyke DL, Gartler SM, Disteche CM. Dosage regulation of the active X chromosome in human triploid cells. PLoS Genet. 2009;5:e1000751. doi: 10.1371/journal.pgen.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet. Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- 42.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–55. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 43.Ellis N, Goodfellow PN. The mammalian pseudoautosomal region. Trends Genet. 1989;5:406–10. doi: 10.1016/0168-9525(89)90199-6. [DOI] [PubMed] [Google Scholar]
- 44.Epstein CJ. Principles, Mechanisms, and Models. Cambridge Univ. Press; Cambridge, UK: 1986. The Consequences of Chromosome Imbalance. [Google Scholar]
- 45.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–56. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 47.FitzPatrick DR. Transcriptional consequences of autosomal trisomy: primary gene dosage with complex downstream effects. Trends Genet. 2005;21:249–53. doi: 10.1016/j.tig.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Fragouli E, Wells D, Delhanty JD. Chromosome abnormalities in the human oocyte. Cytogenet. Genome Res. 2011;133:107–18. doi: 10.1159/000323801. [DOI] [PubMed] [Google Scholar]
- 49.Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 1983;17:155–90. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- 50.Gartler SM, Varadarajan KR, Luo P, Norwood TH, Canfield TK, Hansen RS. Abnormal X: autosome ratio, but normal X chromosome inactivation in human triploid cultures. BMC Genet. 2006;7:41. doi: 10.1186/1471-2156-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, et al. Smchd1-dependent and - independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell. 2012;23:265–79. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–40. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 54.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 2011;45:203–26. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, et al. A recurrent 16p12. 1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–14. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 57.Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487:254–58. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, et al. Global analysis of X-chromosome dosage compensation. J. Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haldane JBS. The cost of natural selection. J. Genet. 1957;55:511–24. [Google Scholar]
- 60.Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–94. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–67. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–67. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 63.Heard E, Turner J. Function of the sex chromosomes in mammalian fertility. Cold Spring Harb. Perspect. Biol. 2011;3:a002675. doi: 10.1101/cshperspect.a002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum. Mol. Genet. 2009;18:R1–8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 65.Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, et al. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–29. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 66.Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2012;463:536–39. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat. Genet. 2010;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–67. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 2010;20:512–18. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jablonka E. The evolution of the peculiarities of mammalian sex chromosomes: an epigenetic view. BioEssays. 2004;26:1327–32. doi: 10.1002/bies.20140. [DOI] [PubMed] [Google Scholar]
- 71.Jacobs PA, Szulman AE, Funkhouser J, Matsuura JS, Wilson CC. Human triploidy: relationship between parental origin of the additional haploid complement and development of partial hydatidiform mole. Ann. Hum. Genet. 1982;46:223–31. doi: 10.1111/j.1469-1809.1982.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 72.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–33. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, et al. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 75.Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–54. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 77.Kharchenko PV, Xi R, Park PJ. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat. Genet. 2011;43:1167–69. doi: 10.1038/ng.991. [DOI] [PubMed] [Google Scholar]
- 78.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–46. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 79.Kim DH, Jeon Y, Anguera MC, Lee JT. X-chromosome epigenetic reprogramming in pluripotent stem cells via noncoding genes. Semin. Cell Dev. Biol. 2011;22:336–42. doi: 10.1016/j.semcdb.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–28. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 81.Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–16. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 82.Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–52. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 83.Kucera KS, Reddy TE, Pauli F, Gertz J, Logan JE, et al. Allele-specific distribution of RNA polymerase II on female X chromosomes. Hum. Mol. Genet. 2011;20:3964–73. doi: 10.1093/hmg/ddr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuroiwa A, Handa S, Nishiyama C, Chiba E, Yamada F, et al. Additional copies of CBX2 in the genomes of males of mammals lacking SRY, the Amami spiny rat (Tokudaia osimensis) and the Tokunoshima spiny rat (Tokudaia tokunoshimensis) Chromosome Res. 2011;19:635–44. doi: 10.1007/s10577-011-9223-6. [DOI] [PubMed] [Google Scholar]
- 85.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:877–79. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 86.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–18. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Larsson J, Meller VH. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 2006;14:417–31. doi: 10.1007/s10577-006-1064-3. [DOI] [PubMed] [Google Scholar]
- 88.Leder EH, Cano JM, Leinonen T, O’Hara RB, Nikinmaa M, et al. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol. Biol. Evol. 2010;27:1495–503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- 89.Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–77. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leppig KA, Disteche CM. Ring X and other structural X chromosome abnormalities: X inactivation and phenotype. Semin. Reprod. Med. 2001;19:147–57. doi: 10.1055/s-2001-15395. [DOI] [PubMed] [Google Scholar]
- 91.Li N, Carrel L. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc. Natl. Acad. Sci. USA. 2008;105:17055–60. doi: 10.1073/pnas.0807765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin H, Halsall JA, Antczak P, O’Neill LP, Falciani F, Turner BM. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno’s hypothesis. Nat. Genet. 2011;43:1169–70. doi: 10.1038/ng.992. author reply 71-72. [DOI] [PubMed] [Google Scholar]
- 94.Lin F, Xing K, Zhang J, He X. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc. Natl. Acad. Sci. USA. 2012;109:11752–57. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, et al. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 1998;18:212–13. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 96.Livernois AM, Graves JA, Waters PD. The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity. 2011;108:50–58. doi: 10.1038/hdy.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lopes AM, Arnold-Croop SE, Amorim A, Carrel L. Clustered transcripts that escape X inactivation at mouse XqD. Mamm. Genome. 2011;22:572–82. doi: 10.1007/s00335-011-9350-6. [DOI] [PubMed] [Google Scholar]
- 98.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L) Nature. 1961;190:372–73. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 99.Lyon MF. The Lyon and the LINE hypothesis. Semin. Cell Dev. Biol. 2003;14:313–18. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 100.Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr. Biol. 2009;19:1478–84. doi: 10.1016/j.cub.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 101.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–23. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 102.Mank JE, Ellegren H. Sex bias in gene expression is not the same as dosage compensation. Heredity. 2009;103:434. doi: 10.1038/hdy.2009.90. [DOI] [PubMed] [Google Scholar]
- 103.Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–44. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- 104.Marshall Graves JA. The rise and fall of SRY. Trends Genet. 2002;18:259–64. doi: 10.1016/s0168-9525(02)02666-5. [DOI] [PubMed] [Google Scholar]
- 105.Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–58. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–18. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melamed E, Arnold AP. Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol. 2007;8:R202. doi: 10.1186/gb-2007-8-9-r202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–23. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 109.Meyer BJ. Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 2010;20:179–89. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Migeon BR. Females Are Mosaic: X Inactivation and Sex Differences in Disease. Oxford Univ. Press; Oxford: 2007. [DOI] [PubMed] [Google Scholar]
- 111.Mizuno H, Okamoto I, Takagi N. Developmental abnormalities in mouse embryos tetrasomic for chromosome 11: apparent similarity to embryos functionally disomic for the x chromosome. Genes Genet. Syst. 2002;77:269–76. doi: 10.1266/ggs.77.269. [DOI] [PubMed] [Google Scholar]
- 112.Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–21. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 113.Morey C, Avner P. The demoiselle of X-inactivation: 50 years old and as trendy and mesmerising as ever. PLoS Genet. 2011;7:e1002212. doi: 10.1371/journal.pgen.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 2008;40:794–99. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114a.Muller HJ. A gene for the fourth chromosome of Drosophila. J. Exp. Zool. 1914;17:325–36. [Google Scholar]
- 115.Muyle A, Zemp N, Deschamps C, Mousset S, Widmer A, Marais GA. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 2012;10:e1001308. doi: 10.1371/journal.pbio.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 117.Nguyen DK, Disteche CM. High expression of the mammalian X chromosome in brain. Brain Res. 2006;1126:46–49. doi: 10.1016/j.brainres.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 118.Nguyen DK, Yang F, Kaul R, Alkan C, Antonellis A, et al. Clcn4-2 genomic structure differs between the X locus in Mus spretus and the autosomal locus in Mus musculus: AT motif enrichment on the X. Genome Res. 2011;21:402–9. doi: 10.1101/gr.108563.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Neill LP, Randall TE, Lavender J, Spotswood HT, Lee JT, Turner BM. X-linked genes in female embryonic stem cells carry an epigenetic mark prior to the onset of X inactivation. Hum. Mol. Genet. 2003;12:1783–90. doi: 10.1093/hmg/ddg193. [DOI] [PubMed] [Google Scholar]
- 120.O’Neill MJ. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum. Mol. Genet. 2005;14(Spec. No. 1):R113–20. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- 121.Ohlsson R, Paldi A, Graves JA. Did genomic imprinting and X chromosome inactivation arise from stochastic expression? Trends Genet. 2001;17:136–41. doi: 10.1016/s0168-9525(00)02211-3. [DOI] [PubMed] [Google Scholar]
- 122.Ohno S. Sex Chromosomes and Sex Linked Genes. Springer-Verlag; Berlin: 1967. [Google Scholar]
- 123.Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–74. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 124.Olson MV. When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–62. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 126.Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, et al. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27:358–67. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 127.Palmer S, Perry J, Ashworth A. A contravention of Ohno’s law in mice. Nat. Genet. 1995;10:472–76. doi: 10.1038/ng0895-472. [DOI] [PubMed] [Google Scholar]
- 128.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–72. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 129.Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 2004;14:1416–24. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 130.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–60. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc. Natl. Acad. Sci. USA. 2012;109:5346–51. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Quinn AE, Sarre SD, Ezaz T, Marshall Graves JA, Georges A. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol. Lett. 2011;7:443–48. doi: 10.1098/rsbl.2010.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Raefski AS, O’Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat. Genet. 2005;37:620–24. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 134.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reinius B, Shi C, Hengshuo L, Sandhu KS, Radomska KJ, et al. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rice WR. Sexually antagonistic genes: experimental evidence. Science. 1992;256:1436–39. doi: 10.1126/science.1604317. [DOI] [PubMed] [Google Scholar]
- 137.Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011;93:1935–42. doi: 10.1016/j.biochi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 138.Ropers HH. Genetics of intellectual disability. Curr. Opin. Genet. Dev. 2008;18:241–50. doi: 10.1016/j.gde.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 139.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–76. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 141.Rugarli EI, Adler DA, Borsani G, Tsuchiya K, Franco B, et al. Different chromosomal localization of the Clcn4 gene in Mus spretus and C57BL/6J mice. Nat. Genet. 1995;10:466–71. doi: 10.1038/ng0895-466. [DOI] [PubMed] [Google Scholar]
- 142.Saran NG, Pletcher MT, Natale JE, Cheng Y, Reeves RH. Global disruption of the cerebellar transcriptome in a Down syndrome mouse model. Hum. Mol. Genet. 2003;12:2013–19. doi: 10.1093/hmg/ddg217. [DOI] [PubMed] [Google Scholar]
- 143.Seidman JG, Seidman C. Transcription factor haploinsufficiency: when half a loaf is not enough. J. Clin. Invest. 2002;109:451–55. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sharp AJ, Spotswood HT, Robinson DO, Turner BM, Jacobs PA. Molecular and cytogenetic analysis of the spreading of X inactivation in X;autosome translocations. Hum. Mol. Genet. 2002;11:3145–56. doi: 10.1093/hmg/11.25.3145. [DOI] [PubMed] [Google Scholar]
- 145.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, et al. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–44. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 147.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 148.Skuse DH. X-linked genes and mental functioning. Hum. Mol. Genet. 2005;14(Spec. No. 1):R27–32. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- 149.Spatz A, Borg C, Feunteun J. X-chromosome genetics and human cancer. Nat. Rev. Cancer. 2004;4:617–29. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 150.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–83. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stenberg P, Larsson J. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma. 2011;120:213–25. doi: 10.1007/s00412-011-0319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat. Rev. Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 153.Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 155.Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 2006;7:645–53. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 156.Wakefield MJ, Keohane AM, Turner BM, Graves JAM. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc. Natl. Acad. Sci. USA. 1997;94:9665–68. doi: 10.1073/pnas.94.18.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang PJ. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol. Metab. 2004;15:79–83. doi: 10.1016/j.tem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 158.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–26. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 159.Wang Z, Willard HF, Mukherjee S, Furey TS. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput. Biol. 2006;2:e113. doi: 10.1371/journal.pcbi.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wareham KA, Lyon MF, Glenister PH, Williams ED. Age related reactivation of an X-linked gene. Nature. 1987;327:725–27. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- 161.Watanabe M, Zinn AR, Page DC, Nishimoto T. Functional equivalence of human X- and Y-encoded isoforms of ribosomal protein S4 consistent with a role in Turner syndrome. Nat. Genet. 1993;4:268–71. doi: 10.1038/ng0793-268. [DOI] [PubMed] [Google Scholar]
- 162.Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, et al. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by SRY but by sex chromosome complement as well. Dev. Cell. 2010;19:477–84. doi: 10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 163.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat. Rev. Neurosci. 2007;8:832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 164.Wolf JB, Bryk J. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics. 2011;12:91. doi: 10.1186/1471-2164-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xiong Y, Chen X, Chen Z, Wang X, Shi S, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 2010;42:1043–47. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- 166.J Xu , Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum. Mol. Genet. 2002;11:1409–19. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- 167.Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr. Patterns. 2006;6:146–55. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 168.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–22. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat. Struct. Mol. Biol. 2011;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yin S, Deng W, Zheng H, Zhang Z, Hu L, Kong X. Evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals. Biochem. Biophys. Res. Commun. 2009;383:378–82. doi: 10.1016/j.bbrc.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 172.Yin S, Wang P, Deng W, Zheng H, Hu L, et al. Dosage compensation on the active X chromosome minimizes transcriptional noise of X-linked genes in mammals. Genome Biol. 2009;10:R74. doi: 10.1186/gb-2009-10-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zechner U, Wilda M, Kehrer-Sawatzki H, Vogel W, Fundele R, Hameister H. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. 2001;17:697–701. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- 174.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 175.Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–17. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005;37:1274–79. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]