Abstract
Brain tissues from Alzheimer’s Disease (AD) patients show increased levels of oxidative DNA damage and 7,8-dihydro-8-oxoguanine (8-oxoG) accumulation. In humans, the base excision repair protein 8-oxoguanine-DNA glycosylase (OGG1) is the major enzyme that recognizes and excises the mutagenic DNA base lesion 8-oxoG. Recently, two polymorphisms of OGG1, A53T and A288V, have been identified in brain tissues of AD patients, but little is known about how these polymorphisms may contribute to AD. We characterized the A53T and A288V polymorphic variants and detected a significant reduction in the catalytic activity for both proteins in vitro and in cells. Additionally, the A53T polymorphism has decreased substrate binding, while the A288V polymorphism has reduced AP lyase activity. Both variants have decreased binding to known OGG1 binding partners PARP-1 and XRCC1. We found that OGG1−/− cells expressing A53T and A288V OGG1 were significantly more sensitive to DNA damage and had significantly decreased survival. Our results provide both biochemical and cellular evidence that A53T and A288V polymorphic proteins have deficiencies in catalytic and protein binding activities that could be related to the increase in oxidative damage to DNA found in AD brains.
Keywords: 8-oxoguanine-DNA glycosylase (OGG1), DNA damage, DNA repair, oxidative stress, 8-oxoguanine (8-oxoG), base excision repair (BER), Alzheimer’s Disease
Introduction
Alzheimer’s Disease (AD) is the most common form of dementia in adults. Currently, AD affects more than 35 million individuals worldwide and this number is expected to double every 20 years [1]. Approximately 5% of AD cases are early-onset caused by mutations in known genes including APP, PSEN1, or PSEN2 [2–6]. The role of genetics in the remaining cases is unclear, but it is proposed that these cases of AD may arise from accrual of spontaneous mutations. One hypothesis to explain these mutations proposes they are produced as a consequence of oxidative stress [7].
Cells are in a state of oxidative stress when more reactive oxygen species (ROS) are produced than the antioxidant mechanisms in place can overcome. ROS are chemically active molecules formed in the body as a consequence of normal cellular metabolism, as well as from environmental sources, such as ionizing radiation and UV light. There are a number of reasons that the brain is particularly susceptible to oxidative stress including: 1) more exposure to oxygen (accounting for only 2% of body weight, but for 20% of oxygen consumption), thus creating more opportunities for production of oxygen radicals [8]; 2) neuronal membrane lipids are enriched for polyunsaturated fatty acids which are easily oxidizable [8, 9]; 3) neurotransmitter metabolism produces ROS [10]; 4) neurons are post-mitotic, and thus do not proceed through the cell cycle where repair of damage by oxidants would occur; 5) increased iron levels are found in brain tissues of AD patients and can result in the transfer of a valence electron which produces ROS, [11, 12]; and 6) low levels of antioxidant enzymes have been detected in brain tissues [8]. It has also been shown that oxidative DNA damage is one of the earliest detectable events in AD pathogenesis [13].
ROS can interact with DNA to induce many types of DNA damage, including the mutagenic base lesion 7,8-dihydro-8-oxoguanine (8-oxoG). It has been shown that brain tissues from aged and patients suffering from neurodegeneration (AD, Parkinson’s Disease, Huntingdon’s Disease, Amyotrophic Lateral Sclerosis) show increased levels of both nuclear and mitochondrial DNA damage, including 8-oxoG lesions [14–21]. Oxidatively modified DNA bases, such as 8-oxoG, are repaired mainly by the base excision repair (BER) pathway. The initial step in this repair pathway is the recognition and subsequent excision of the DNA base lesion by a DNA glycosylase. There are multiple glycosylases in mammalian cells which act on different DNA lesions. Recognition of 8-oxoG lesions is primarily completed by 8-oxoguanine-DNA glycosylase (OGG1). OGG1 is a bifunctional enzyme, having both DNA glycosylase and AP lyase activities. There are 21 known polymorphisms in OGG1 that are found in the general population or are disease specific variants [22, 23], 13 of which are disease associated. Of these, 10 are linked with various cancers, some tenuously, including lung, prostate, and oropharyngeal cancers [24]. In 2007, a study looking at brain tissues from late-stage AD patients identified three mutations in OGG1 that were not found in the control patients. Two of these mutations are single nucleotide polymorphisms (SNPs) resulting in the amino acid substitutions A53T and A288V [23]. The A288V polymorphism (rs3219012) is relatively rare, occurring in ~1% of general population, while the A53T polymorphism is currently only considered to be disease associated. Previous studies done by Mao et al. and Sidorenko et al. using an 8-oxoG DNA substrate found that both polymorphisms in OGG1 resulted in decreased catalytic activity although other biochemical aspects of these proteins were not investigated [23, 25].
In this paper we have examined the A53T and A288V polymorphic OGG1 proteins to further define the biochemical defects that could contribute to the accumulation of DNA damage and 8-oxoG lesions observed in AD patient brain tissues found by others. Our work shows a statistically significant decrease in catalytic activity for both polymorphic OGG1 proteins compared to WT. The decrease in catalytic activity may be explained by a decreased ability of the A53T protein to bind to DNA substrates and decreased lyase activity for the A288V protein. In addition, we found decreased binding of polymorphic OGG1 proteins to Poly(ADP-ribose) polymerase 1 (PARP-1) and X-ray cross-complementing protein 1 (XRCC1), and a decreased ability of the polymorphisms to activate PARP-1. OGG1−/− mouse embryo fibroblasts (MEFs) expressing the polymorphisms had decreased long term cell survival and were more sensitive to DNA damaging agents. Together, the results presented in this manuscript show that the A53T and A288V polymorphisms alter the protein function in a manner that may increase susceptibility to disease..
Materials and Methods
Cell Culture and Transfections
HeLa cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1X Penicillin Streptomycin (Gibco, Grand Island, NY). Wild-type mouse embryo fibroblasts (MEF) and OGG1−/− MEFs were maintained in DMEM containing 10% FBS and 1X PenStrep (Gibco) and have been described previously [26, 27].
MEF’s were transfected using XtremeGENE HP transfection reagent (Roche, Indianapolis, IN). Titration experiments were completed to find the optimal ratio of X-tremeGENE HP:µg DNA, as well as the amount of time required for complex formation before addition to the cells.
Transfections were performed according to manufacturer’s recommendations and a 3:1 ratio of X-tremeGENE HP:µg DNA was used with a 20-minute complex formation time.
Plasmid Construction
Plasmids containing the A53T and A288V polymorphic variants were generated by site directed mutagenesis of the pCMV-Tag2B-WT OGG1, pET-28a WT OGG1, and pEGFP-C1 WT OGG1 plasmids (constructions previously described [28, 29]) using the following primers (underlined bolded letters represent the changed bases): A53T For 5’-GGGAGCAAAGTCCTACACACTGGAGTGGTG-3’, A53T Rev 5’-CACCACTCCAGTGTGTAGGACTTTGCTCCC-3’, A288V For 5’-CTACCACGTCCCAGGTGAAGGGACCGAGCCC-3’ and A288V Rev 5’-GGGCTCGGTCCCTTCACCTGGGACGTGGTAG-3’. The pGEX4T1-A35T and pGEX4T1 A288V vectors were created by digestion of the pCMV-Tag2B A53T and pCMV-Tag2B A288V plasmids with SmaI and XhoI restriction enzymes, followed by ligation into the same sites in pGEX4T1 plasmid (GE Healthcare, Piscataway, NJ). All plasmids were verified by bidirectional sequencing.
Purification of Recombinant Proteins
For His-tag protein purification pET-28a WT OGG1, A53T OGG1, and A288V OGG1 plasmids were transformed into BL21 competent cells (Invitrogen, Grand Island, NY). Bacterial cultures were grown for 8 hours at 37 °C, diluted 1:200 into LB broth pre-warmed at 37°C and grown until an OD of 0.3–0.4 was reached. The proteins were then induced with 100 µM isopropyl h-D-thio-galactoside (IPTG) and grown overnight at 30°C at a speed of 100 rpm. The following day, bacteria were harvested by centrifugation, resuspended in phosphate buffered saline (PBS) with protease and phosphatase inhibitors, lysozyme (1 mg/ml), DNAse (1 mg/ml) and PMSF (10 µM), and incubated on ice for 30 minutes. The cells were lysed by sonication (8, 10-second pulses with 5 seconds off) after the addition of a final volume 1.5% sarcosine (from a 10% (w/v) solution in water). After centrifugation, the supernantant was passed through a 0.45-µm syringe filter and applied to a column containing Ni-NTA agarose beads (Qiagen, Valencia, CA) prewashed with 500 mM NaCl, 100 mM Tris-HCl (pH 8.0) and 50 mM Imidazole. The columns were rotated at 4°C for 1 hour and washed twice with ice cold PBS containing 1% Triton-X 114. Proteins were eluted with 500 mM NaCl, 100 mM Tris-Hcl (pH 8.0) and 200 mM Imidazole, and dialyzed in PBS overnight at 4°C. His-tags were removed by incubation with biotinylated thrombin (0.3 U/ml protein) (Novagen, Madison, WI), followed by two incubations with p-Aminobenzamidine-Agarose beads (Sigma) to remove the thrombin. Additional purification was performed by ion exchange chromatography using an AKTA FPLC purification system (GE Healthcare). Proteins were passed through a Q HP column (GE Healthcare) and bound to an S HP column (GE Healthcare) at 150 mM NaCl. Over a linear NaCl gradient of 150 mM−1 M, the proteins were eluted at ~350 mM NaCl. Fractions containing purified proteins were dialyzed against 20 mM Tris–HCl (pH 7.4), 300 mM NaCl, 10% glycerol, aliquoted, and stored at −80°C. Protein concentration was determined using the Bradford Assay (BioRad) and verified on a polyacrylamide gel.
GST-tagged recombinant proteins were purified using standard procedures [30] and protein purity and concentrations were determined by Bradford Assay and verified on a polyacrylamide gel.
Purification of Nuclear Extracts
Nuclear extracts were purified from OGG1−/− MEFs transfected with WT or polymorphic GFP-tagged OGG1 containing constructs using a protocol modified from Schreiber et al [31]. Four confluent 10cm dishes of cells were washed twice with cold PBS, scraped from the dishes in 2 ml volume, and centrifuged for 3 minutes at 500xg. The pellet was resuspended in 1 ml Buffer A (10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA (pH 8), 1 mM DTT, and HALT protease inhibitors (Pierce)), then pelleted. The final pellet was resuspended in 1 ml of Buffer A and incubated on ice for 15 minutes. After the addition of 100 µl 10% NP-40, cells were vortexed for 10 seconds and then centrifuged at max speed for 30 seconds. The supernatant was removed and the nuclear pellet was resuspended in 40 µl Buffer B (10 mM HEPES, 400 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA (pH 8), and HALT protease inhibitors (Pierce)). The nuclear pellet was rotated at 4°C for 20 min, centrifuged at 4°C for 5 minutes, then aliquoted and stored at −80°C. 10 µl of each nuclear supernatant was run on a polyacrylamide gel and probed with anti-GFP antibodies (Millipore) to determine the amount of GFP-tagged protein in each sample.
Substrate Labeling and DNA Incision Assays
An HPLC purified 30-mer oligonucleotide
(GAAGAGAGAAAGAGAXAAGGAAAGAGAGAA) containing an 8-oxoG base at position X and a complementary oligonucleotide containing a C opposite X were obtained from Midland Certified Reagent Company (Midland, TX). The 5′- 32P-labeling of the duplex oligonucleotide was performed using 20 µM single stranded oligonucleotides and 10 µCi of [γ- 32P] ATP as previously described [32], for a final substrate concentration of 200 nM. To measure OGG1 catalytic activity, 32 nM or 64 nM FPLC purified WT OGG1, A53T OGG1, or A288V OGG1 were incubated in 10-µl reactions containing 200 nM radiolabeled substrate in incision buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 0.15 µg/µl BSA) for 15 minutes at 37°C. All molar values indicate final concentration in the reaction. An equal volume 2X loading buffer (1X Tris borate EDTA, 90% deionized formamide, 20 mM EDTA, 0.1% bromphenol blue, 0.1% xylene cyanol) was added to each sample, samples were heated at 95°C for 5 min, and run on 15% polyacrylamide gels containing 7 M Urea. Radioactivity was measured using a Storm Phosphorimager and quantified using ImageQuant software (GE Healthcare). Reactions using purified nuclear extracts from cells transfected with WT or polymorphic GFP-tagged OGG1 were performed as described above using equal concentrations of WT or polymorphic protein as determined by analysis on a polyacrylaminde gel. Reactions using APE1 (New England Biolabs, Ipswich, MA) were completed as described above using 64 nM OGG1 proteins, with the addition of a final concentration of 45 pM APE1 in a 15-µl reaction volume.
Substrate Labeling and AP Lyase Assay
An HPLC purified 30-mer oligonucleotide containing a Uracil base
(GAAGAGAGAAAGAGAUAAGGAAAGAGAGAA) and a complementary oligonucleotide containing a C opposite U were obtained from Midland Certified Reagent Company (Midland, TX). The 5′- 32P-labeling of the duplex oligonucleotide was performed using 20 µM single stranded oligonucleotides and 10 µCi of [γ-32P] ATP as previously described [32], for a final substrate concentration of 200 nM. 80nM labeled substrate was reacted with 20 nM UDG (a generous gift from James Stivers, Department of Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine) in the presence of 5X UDG reaction buffer (New England Biolabs) for 30 minutes at 37°C to create the AP site. The AP lyase assay was completed by incubating 64 nM FPLC purified WT OGG1, A53T OGG1, or A288V OGG1 protein with 1.6 nM radiolabeled substrate in incision buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 0.15 µg/µl BSA) for 15 minutes at 37°C. All molar values indicate final concentration in the reaction. An equal volume 2X loading buffer (1X Tris borate EDTA, 90% deionized formamide, 20 mM EDTA, 0.1% bromphenol blue, 0.1% xylene cyanol) was added to each sample, and run on 15% polyacrylamide gels containing 7 M Urea. Radioactivity was measured using a Storm Phosphorimager and quantified using ImageQuant software (GE Healthcare).
Determination of dissociation constants by electrophoretic mobility shift assay (EMSA)
Duplexed 5’- 32P labeled DNA substrates for the EMSA assays were created using the 8-oxoG lesion containing oligonucleotide and complementary oligonucleotides containing an A, T, or G base opposite the 8-oxoG lesion as described in the previous section. Binding affinities were measured by incubating FPLC purified WT OGG1, A53T OGG1, or A288V OGG1 proteins of varying concentrations between 32 nM and 375 nM, with 33 nM DNA substrate in 20 mM TrisHCl (pH 7.4), 0.15 µg/µl BSA, 15% glycerol for 5 min on ice. Samples were mixed with 6X Orange G loading buffer containing 30% Ficcoll (w.v. in water) and bound complexes were separated from free substrate by electrophoresis on native 6% polyacrylamide gels for 1 hour at 4°C in cold 0.5 X TBE buffer. Radioactivity was measured using a Storm Phosphorimager and quantified using ImageQuant software (GE Healthcare). Dissociation constants were calculated by combining data from three independent experiments for each protein following the method of Schofield [33] and curve fitting the bound fraction versus protein concentration using linear regression analysis with separate linear, quadratic, and cubic terms to account for the curvilinearity. The equation used for each Kd calculation is indicated on the corresponding graph in Supp. Fig. 2A and B. Lower and upper 95% confidence limits were calculated from the best fit lines for each protein on each DNA template.
GST Precipitation Assays
Precipitation assays were performed as previously described [27]. Briefly, HeLa cells were washed with PBS, lysed in IP buffer and incubated with 10µg of the appropriate GST fusion protein for 1 hour at 4°C. The samples were washed with IP buffer, and heated to 95° C to release bound proteins from the beads. Samples were separated by SDS-PAGE and probed by immunoblotting with anti-PARP-1 monoclonal antibodies (Enzo Life Sciences, Farmingdale, NY; Clone C-2–10), anti-XRCC1 monoclonal antibodies (Neo Markers, Kalamazoo, MI) and were stained with Ponceau S to visualize loading of the GST fusion proteins.
PARP-1 Activation Assays
To determine whether the polymorphic forms of OGG1 were able to activate PARP-1, we performed assays to measure the accumulation of of Poly(ADP-ribose) (PAR), a measure of PARP-1 activity. 10 µg of GST alone, GST-WT OGG1, GST-A53T OGG1, or GST-A288V OGG1 were washed in PBS containing 0.1% TritonX-100, followed by incubation in the presence or absence of 7.5 ng PARP-1 (Trevigen, Gaithersburg, MD) for 10 minutes. PARP cocktail, containing NAD+ (Trevigen), and activated DNA (Trevigen) were added and reactions were allowed to proceed at room temperature for 30 minutes. The samples were mixed with sample buffer, boiled at 95°C for 5 minutes, and run on a 12% polyacrylamide gel. Immunoblots were probed with anti-PAR monoclonal antibodies (Trevigen) and anti-GST antibodies (Cell Signaling, Boston, MA) to visualize the fusion protein.
Thermal Shift Assays
To determine the melting temperature (Tm) of the purified OGG1 proteins, we performed a thermal shift assay using the Sypro orange thermofluor to monitor protein unfolding and determine the melting temperature for each protein [34]. The point of inflection of each resulting curve is defined as the melting temperature (Tm). Comparison of the thermally induced melting points may reveal a stabilization (increased Tm) or destabilization (decreased Tm) for the proteins tested. For this assay, 1 µg of FPLC purified protein was mixed with a final concentration of 1X or 2X Sypro Orange (diluted in water) (Molecular Probes, Invitrogen) and buffer (20 mM Tris-HCl (pH 7.4), 300 mM NaCl, 10% glycerol) in a 20 µl reaction in duplicate wells of a 96-well plate (Bio-Rad). The plates were heated in an iCycler iQ Real Time PCR Detection System (Bio-Rad) from 20 to 70°C in increments of 0.5°C every 15 seconds. The intensities obtained were plotted versus temperature and the sigmoid curves were fit by using nonlinear regression as described in [35] for 2 independent experiments of two preparations of each protein.
Cellular Localization
To determine cellular localization, OGG1−/− MEFs were seeded in a 4-well chamber slide and transfected with pEGFP-C1, pEGFP-C1-WT OGG1, pEGFP-C1-A53T OGG1, or pEGFP-C1-A88V OGG1 plasmids as described above. 24 hours after transfection the cells were incubated with a final concentration of 1 µM Hoechst stain and 50 nM Mitotracker Red CMXRos (Invitrogen) in serum-free DMEM media for 30 minutes. The cells were then placed in PBS and live fluorescent images were taken on a Zeiss Observer D1 microscope with an AxioCam1Cc1 camera at a set exposure time.
Colony Formation Assay
For the colony formation assays, OGG1−/− MEFs were transfected with pEGFP-C1, pEGFP-C1-WT OGG1, pEGFP-C1-A53T OGG1, or pEGFP-C1-A88V OGG1 plasmids as described above, and cells were seeded 24 hours later in triplicate wells (3000/well) of a 6-well plate. The following day cells were left untreated or treated with 20 µM menadione or 50 µM H2O2. Menadione treatments were performed for 30 minutes in serum-free media, after which they were placed into DMEM without damaging agents. Cells were allowed to grow for 7 days. Cell media with or without H2O2 was changed 3 days after initial treatment. Colonies were stained with crystal violet, and only colonies with >50 cells were counted.
Results
A53T and A288V OGG1 polymorphisms have reduced catalytic activity
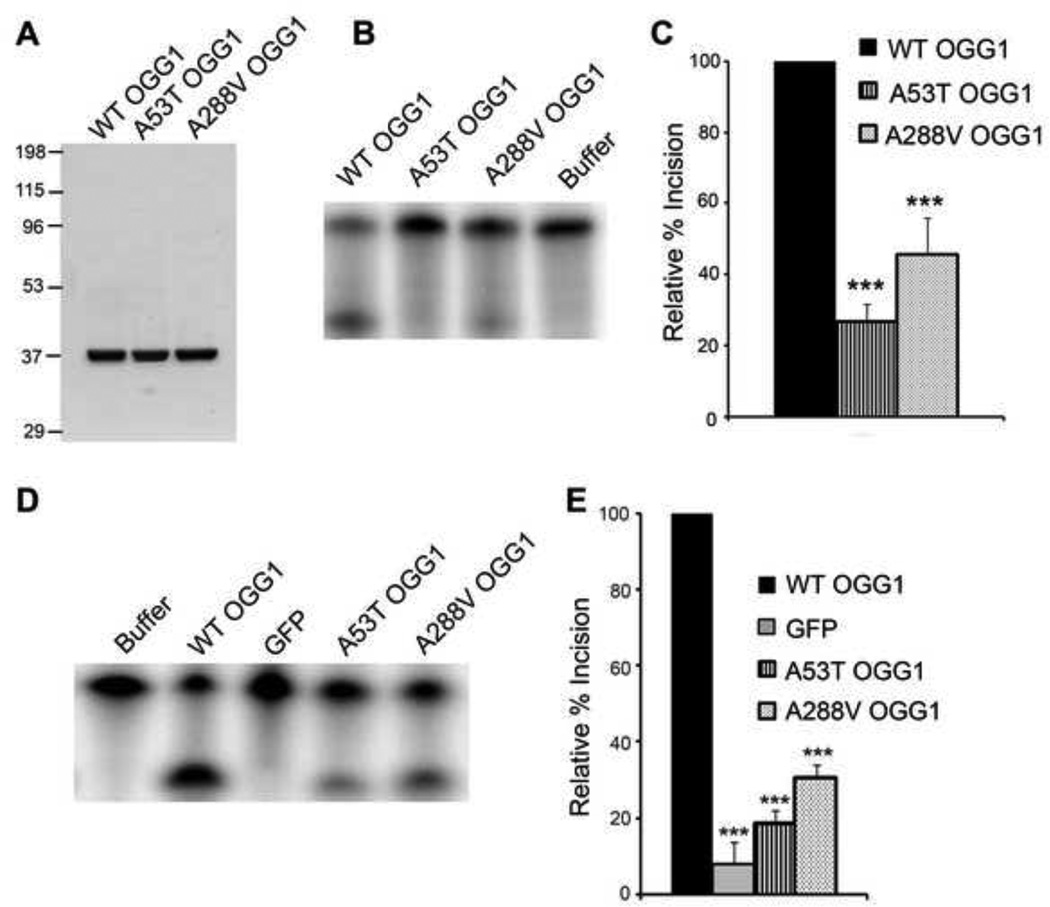
To characterize the biochemical properties of the A53T and A288V polymorphisms, we first FPLC purified WT OGG1 and proteins containing the OGG1 polymorphisms (Fig. 1A). These proteins were then used in in vitro incision assays to determine the ability of the proteins to excise a DNA substrate containing an 8-oxoG lesion. WT or polymorphic OGG1 proteins were reacted with an excess of radiolabeled DNA substrate. We observed a decrease in incision activity for both of the polymorphic proteins (Fig. 1B, C). There was a significant decrease in incision ability for both the A53T (~75% reduction) and A288V (~60% reduction) polymorphic proteins. Similarly, we tested the incision activity of nuclear extracts obtained from OGG1−/− MEFs transfected with WT or polymorphic GFP-tagged OGG1 proteins. As expected, expression of WT OGG1 significantly increased 8-oxoG incision compared to cells expressing GFP control, which would lack OGG1 expression. Similar to the experiments with purified proteins, we observed significant reductions in the incision activity of the nuclear extracts isolated from cells expressing the A53T (~80% reduction) and A288V (~70% reduction) polymorphic proteins compared to wild-type (Fig. 1D, E). These results verify that there is a defect in catalytic activity for both of the OGG1 polymorphisms.
Fig 1. Decreased incision activity of OGG1 polymorphisms.
(A) The indicated OGG1 recombinant proteins were expressed in E. coli, purified using a Ni-NTA column, followed by a thrombin cleavage step and subsequent FPLC purification. Gel Code Blue staining of the wild-type (WT), A53T, and A288V recombinant proteins (2.5µg). (B) OGG1 recombinant proteins (64 nM) were incubated with a 5′-end-labeled oligonucleotide duplex containing an 8-oxoG lesion. After a 15 minute incubation at 37°C, the samples were run on a 15% polyacrylamide gel containing 7 M urea and imaged by a phosphorimager. A representative experiment is shown. (C) The histogram represents the mean ± S.E.M. from nine independent experiments. The percent incision was calculated by taking the amount of cleaved substrate (lower band) normalized to the amount of cleaved + uncleaved substrate (lower + upper bands). The data were normalized to the incision activity of OGG1 alone (100%). (D) Nuclear extracts from OGG1−/− MEFs transfected with WT or polymorphic GFP-tagged OGG1 were reacted with a 5′-end-labeled oligonucleotide duplex containing an 8-oxoG:C mispair as in B. A representative experiment is shown. (E) The histogram represents the mean ± S.E.M. from 5 independent experiments. Percent incision was calculated as in C. ***p< 0.001 comparing WT and variant forms of OGG1 using one-way ANOVA and Tukey's post-hoc test.
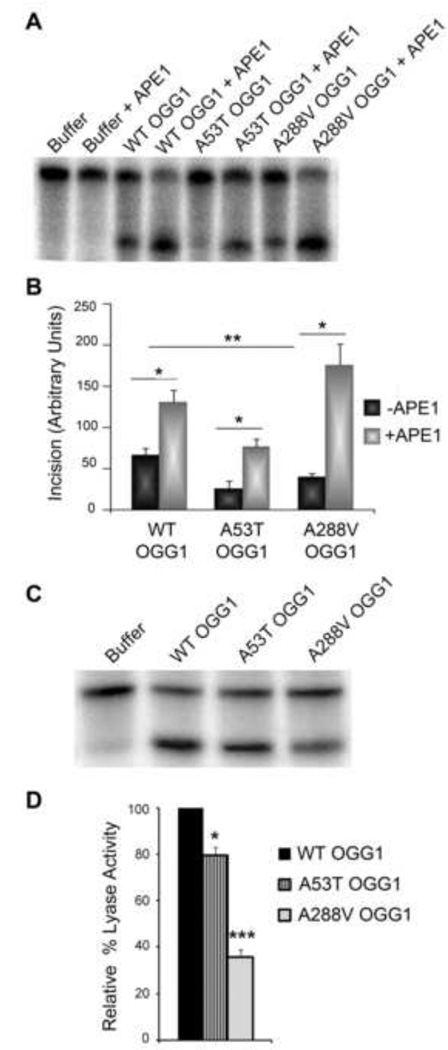
To further elucidate the effect of the OGG1 polymorphisms on catalytic activity, we wanted to determine whether APE1, a known stimulator of OGG1 activity [36–38], was able to stimulate the catalytic activity of the polymorphic proteins to the same level as WT OGG1. To determine this we performed incision assays in the presence of APE1 protein (Fig. 2A). Similar to what we have shown in Fig. 1B and 1C, a significant decrease in catalytic activity in the absence of APE1 was observed for both of the polymorphic mutants compared to WT (Fig. 2B). In each case, the addition of APE1 stimulated the catalytic activity of each enzyme tested even in the catalytically impaired polymorphic enzymes (WT 1.9-fold, A53T 2.9-fold, and A288V 4.3-fold) (Fig. 2B). Although APE1 stimulated the activity of the A53T protein, the level of incision was still significantly lower than WT stimulated by APE1 (p<0.01). APE1 stimulated the A288V polymorphic protein above APE1 stimulation of WT but this increase was not statistically significant.
Fig. 2. Decreased AP lyase activity and differential stimulation of polymorphic OGG1s by APE1.
(A) OGG1 recombinant proteins (64nM) were incubated at 37°C for 15 minutes in the presence (+) or absence (−) of 45pM APE1, along with a 5′-end-labeled oligonucleotide duplex containing an 8-oxoG lesion. The cleavage products were analyzed on a 15% polyacrylamide gel containing 7M Urea and imaged on a phosphorimager. A representative experiment is shown. (B) The histogram represents the mean ± S.E.M. from five independent experiments. The incision value was calculated by taking the amount of cleaved substrate (lower band) normalized to the amount of cleaved + uncleaved substrate (lower + upper bands). *p< 0.05, **p<0.01 for the indicated comparisons using one-way ANOVA and Tukey's post-hoc test. (C) To examine AP lyase activity, OGG1 recombinant proteins were incubated with a 5′-end-labeled DNA substrate containing an AP site. The cleavage products were analyzed on a 15% polyacrylamide gel containing 7M Urea and imaged on a phosphorimager. A representative experiment is shown. (D) The histogram represents the mean ± S.E.M. from four independent experiments. The percent lyase activity was calculated as in B. *p<0.05, ***p< 0.001 comparing WT and OGG1 variants using one-way ANOVA and Tukey's post-hoc test.
Since the OGG1 polymorphic proteins had reduced catalytic activity yet were able to be stimulated by APE1, we examined whether either of these proteins had defects in AP lyase activity. To test this, we used a DNA substrate containing a uracil base to create a radiolabeled double stranded substrate that after addition of UDG contains an AP site. The AP site containing substrate was reacted with 64 nM of each OGG1 protein to determine the ability of the proteins to cleave the AP site. We found that both of the polymorphic proteins have a defect in AP lyase activity (Fig. 2C, D). However, the defect is much more pronounced in the A288V polymorphism (~60% reduction in AP lyase activity). Interestingly, we found a similar decrease in both the incision and AP lyase activity for the A288V protein. These results suggest that the defective AP lyase activity may account for the decreased catalytic activity of the A288V polymorphism.
A53T OGG1 polymorphism exhibits decreased binding to 8-oxoguanine substrates
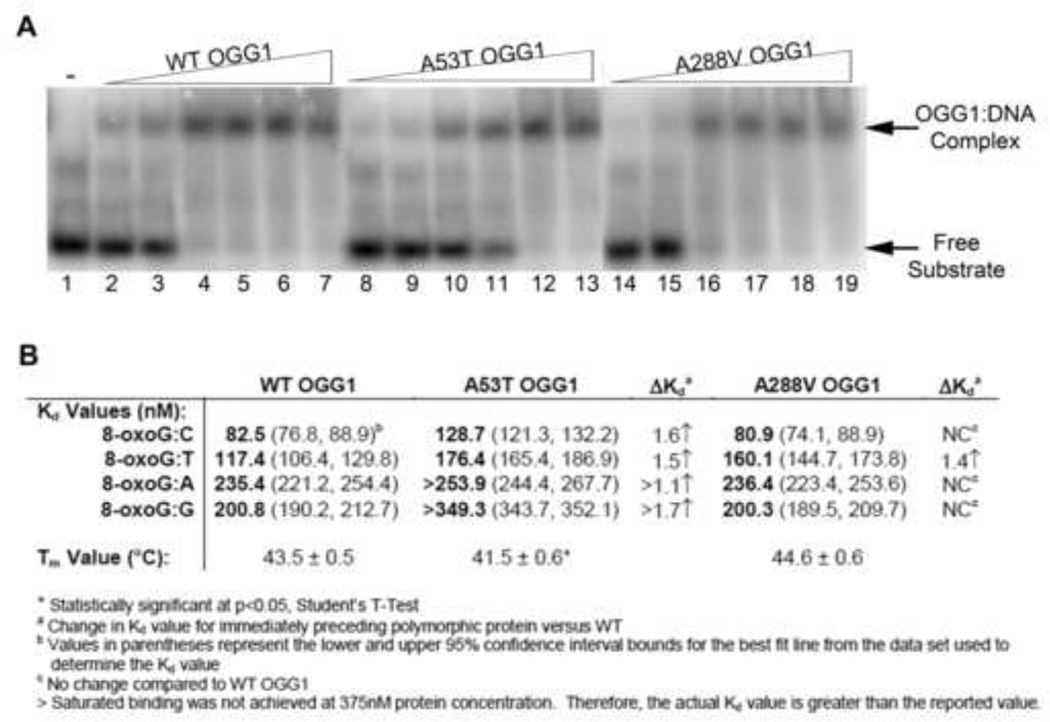
To assess whether the decrease in catalytic activity we observed for the A53T and A288V OGG1 proteins was due to a defect in protein binding to the DNA substrate, we performed electrophoretic mobility shift assays (EMSA) using radiolabeled substrates. Increasing concentrations of WT or polymorphic OGG1 proteins were incubated with a DNA substrate that contained an 8-oxoG lesion paired with one of the four DNA bases to allow for formation of the protein:DNA complex. The samples were immediately run on a native polyacrylamide gel to visualize the complex formation. For the 8-oxoG:C substrate, we observed similar binding of WT and A288V OGG1 proteins to the DNA substrate (representative gel of 8-oxoG:C template shown in Fig. 3A; other templates in Supp Fig. 1). However, for the A53T protein we found that increased protein concentrations were required for efficient formation of the protein:DNA complex, indicating a decreased binding affinity toward this DNA substrate. Similarly, the A53T protein showed diminished binding affinity for the 8-oxoG:T, G, and A substrates (Fig. 3B, Suppl. Fig. 1–2). The dissociation constants (Kd) were determined for each protein with each DNA template (Fig. 3B). The graphs of the data and best fit lines used to determine Kd are shown in Supp. Fig. 2A,B. For WT OGG1, we found that binding to the 8-oxoG:C and 8-oxoG:T DNA templates resulted in similar Kd values, indicating that WT OGG1 binds well to these templates. For the 8-oxoG:G and 8-oxoG:A DNA substrates we observed a large increase in Kd values, indicating that these are non-preferred DNA substrates for OGG1. Although the A288V protein exhibited similar Kd values to WT on the 8-oxoG:C, G, and A templates the A288V protein did show a statistically increased Kd value for the 8-oxoG:T template. Similarly, a statistically significant increase in Kd was found for the A53T polymorphic protein on all templates compared to WT, indicating a defect in DNA binding for this protein (Fig. 3B, Suppl. Fig. 1–2). It is interesting to note, that the binding curves for the A53T polymorphic protein appear very sigmoidal in nature (with the exception of the 8-oxoG:C template, which appears similar to WT OGG1), which differs from the WT and A288V proteins. This binding pattern would indicate cooperative binding of the DNA substrate to the protein active site for the A53T protein.
Fig. 3. Binding of WT and polymorphic OGG1 proteins to 8-oxoG:C substrate.
(A) Increasing concentrations of recombinant OGG1 proteins were incubated with 33.3 nM 5′-end-labeled oligonucleotide duplex containing an 8-oxoG lesion paired with A, T, C, or G for 5 minutes on ice. The bound complexes were separated from free probe on a 6% native polyacrylamide gel and imaged on a phosphorimager. A representative experiment for the 8-oxoG:C substrate is shown. The presence of the higher band indicates the formation of the protein:substrate complex. Lane 1: Buffer alone. Lanes 2, 8, 14: 32 nM protein. Lanes 3, 9, 15: 64 nM protein. Lanes 4, 10, 16: 125 nM protein. Lanes 5, 11, 17: 190 nM protein. Lanes 6, 12, 18: 250 nM protein. Lanes 7, 13, 19: 375 nM protein. (B) Dissociation constants calculated from EMSAs and melting temperatures from thermal stability assays for wild-type and polymorphic OGG1 proteins (see Suppl. Fig. 1–3 for graphs used for calculations).
The A53T OGG1 polymorphism exhibits decreased thermal stability
To gain a better understanding about the stability of the polymorphic proteins, we performed a thermal unfolding assay to determine the melting temperatures (Tm) of the WT, A53T, and A288V OGG1 proteins. To do this we used the environmentally sensitive dye, Sypro Orange, to monitor OGG1 protein unfolding across a range of temperatures. As the protein unfolds, Sypro Orange will bind to the newly exposed hydrophobic regions of the protein and become unquenched. By monitoring the excitation of the Sypro Orange signal using a qPCR system, we found the Tm for WT OGG1 to be 43.5°C + 0.5, which is similar to previous data [39]. A similar Tm was found for the A288V polymorphic protein (Fig. 3B; melting curves in Supp. Fig. 3). In contrast, the A53T polymorphic protein had a decreased Tm value that was statistically significant compared to WT (Fig. 3B). This data indicates that the A53T OGG1 protein becomes unfolded at a lower temperature than the WT and A288V proteins, and thus may be less stable.
OGG1 polymorphisms alter binding to known protein binding partners
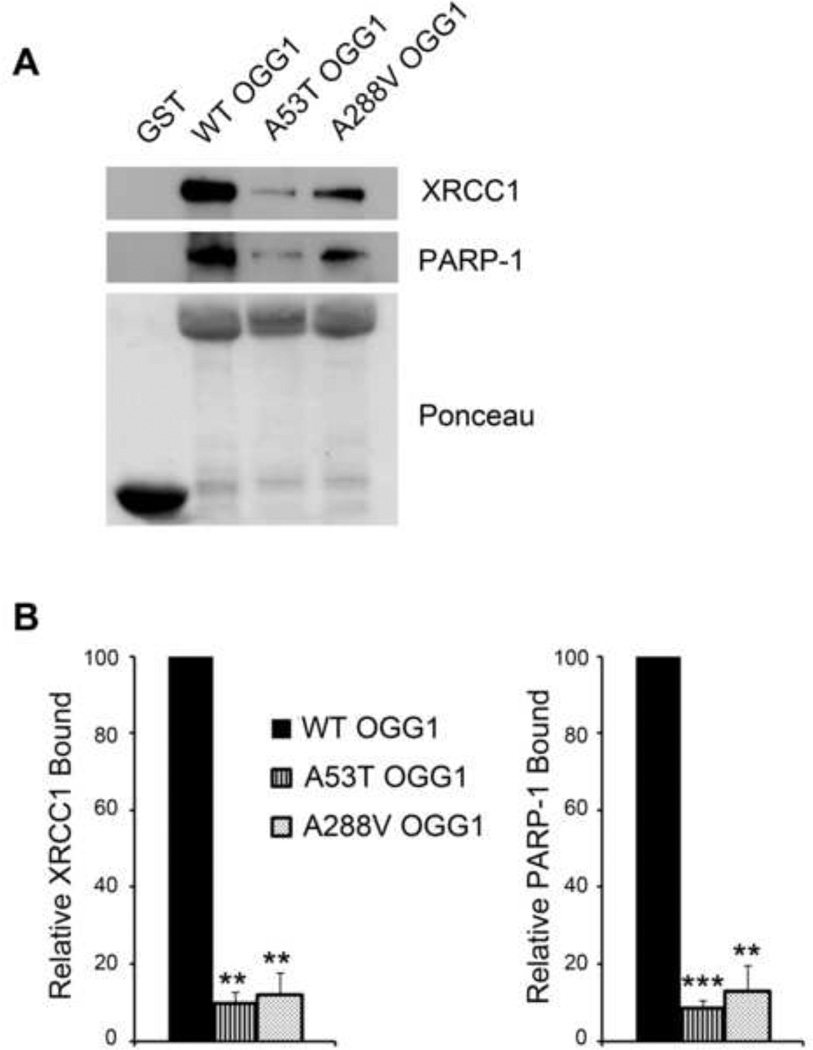
It has been previously shown that the BER repair proteins PARP-1 and XRCC1 interact directly with OGG1 [27, 40]. Given these results, and the fact that the binding between OGG1 and XRCC1 is essential to the early stages of BER, we chose to look at the binding of the polymorphic proteins to both PARP-1 and XRCC1. We purified GST-tagged WT or polymorphic proteins, verified purity by Gel Code Blue Staining (data not shown) and used these GST-tagged proteins to precipitate proteins from HeLa cell lysates. The bound proteins were run on polyacrylamide gels and immunoblotted with antibodies against PARP-1 and XRCC1 to examine the degree of protein-protein binding. We observed significantly decreased binding of the polymorphic proteins to both PARP-1 and XRCC1 (Fig. 4A), indicating that the polymorphisms result in a defect in protein-protein interactions with these BER components. We quantified the binding of each OGG1 protein to either XRCC1 or PARP-1 and found a significant decrease in protein binding for both the A53T and A288V proteins (Fig. 4B).
Fig. 4. Decreased binding of the OGG1 polymorphisms to PARP-1 and XRCC1.
(A) HeLa cell lysates were incubated with GST-tagged WT or polymorphic recombinant proteins (10 µg) and the precipitations were probed with anti-XRCC1 and anti-PARP1 antibodies. Ponceau staining was used to visualize loading of the fusion proteins. (B) The relative amount of XRCC1 or PARP-1 binding was quantified from immunoblots and normalized to the amount of GSTfusion protein. This value was then normalized to WT OGG1. The histograms represent the normalized mean ± S.E.M. from three independent experiments. **p<0.01, ***p< 0.001 comparing WT and variant forms of OGG1 using Student’s t-test.
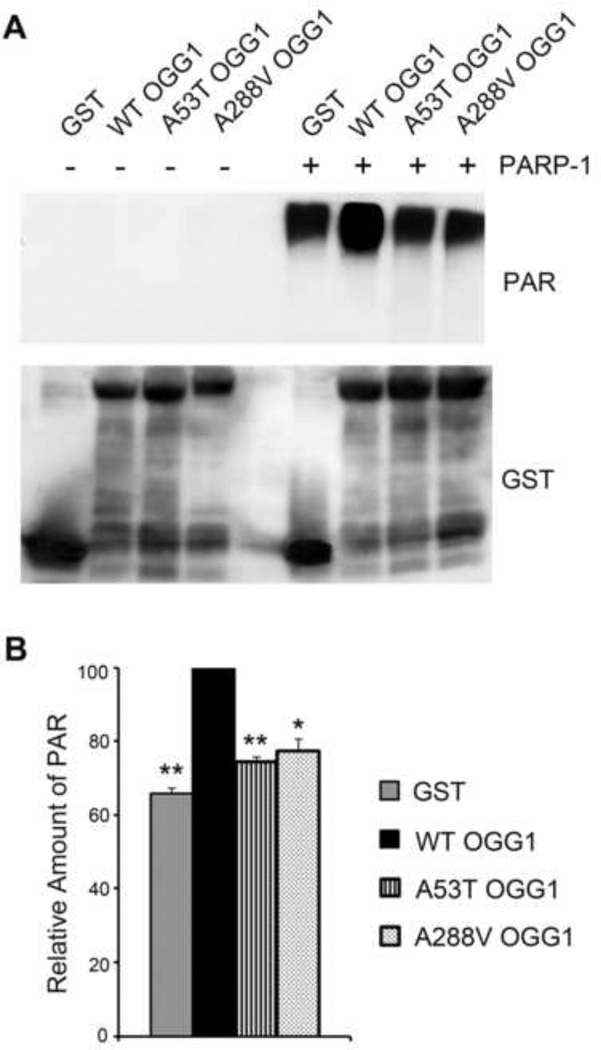
Since we found defective binding of the polymorphic proteins to PARP-1, and we have previously shown that both the S326C and R229Q OGG1 polymorphisms were less able to activate PARP-1 than WT [27], we wanted to determine whether the AD related OGG1 polymorphisms effect the ability of these proteins to activate PARP-1. We performed in vitro ribosylation reactions where GST-WT or polymorphic OGG1 proteins were incubated with PARP-1. Activated DNA and the required co-factor NAD+ were then added and the reaction was further incubated for 30 minutes. The samples were mixed with sample buffer, run on a 10% polyacrylamide gel and probed with anti- poly(ADP-ribose) (PAR) antibodies to examine the levels of PAR accumulation, a measure of PARP-1 activity. As expected, we observed no accumulation of PAR in the absence of PARP-1 (Fig. 5A). However, the level of poly(ADP-ribosyl)ation increased in the presence of PARP-1. With addition of GST-WT OGG1 we observed a modest enhancement of PARP-1 activity over control (GST alone) (Fig. 5A). In contrast, the A53T and A288V proteins did not activate PARP-1 above background levels (GST alone). A statistically significant decrease in the levels of PARP-1 automodification was observed for both the A53T and A288V polymorphic proteins (Fig. 5B), indicating that they not only have decreased binding to PARP-1, but both polymorphisms also have a reduced ability to activate PARP-1 compared to WT.
Fig. 5. Decreased activation of PARP-1 by OGG1 polymorphisms.
(A) PARP-1 poly(ADP-ribosyl)ation reactions were performed by incubating the GST-tagged WT or polymorphic recombinant proteins with (+) PARP-1 for 10 minutes prior to the addition of the PARP-1 cofactors NAD+ and activated DNA. Reactions without PARP-1 (−) were a negative control. Reactions were incubated further for 30 minutes, and analyzed on a 12% polyacrylamide gel and immunoblotted with anti-PAR antibodies and anti-GST antibodies as a protein loading control. The polymorphic OGG1 proteins are unable to activate PARP1 above the background level shown in the GST lane. (B) The relative amount of PAR was quantified from immunoblots and normalized to the amount of GST protein. This value was then normalized to WT OGG1. The histogram represents the mean ± S.E.M. from three independent experiments. *p<0.05, **p<0.01, comparing WT and variant forms of OGG1 using Student’s t-test.
OGG1 polymorphisms sensitize cells to DNA damaging agents and decrease cell survival
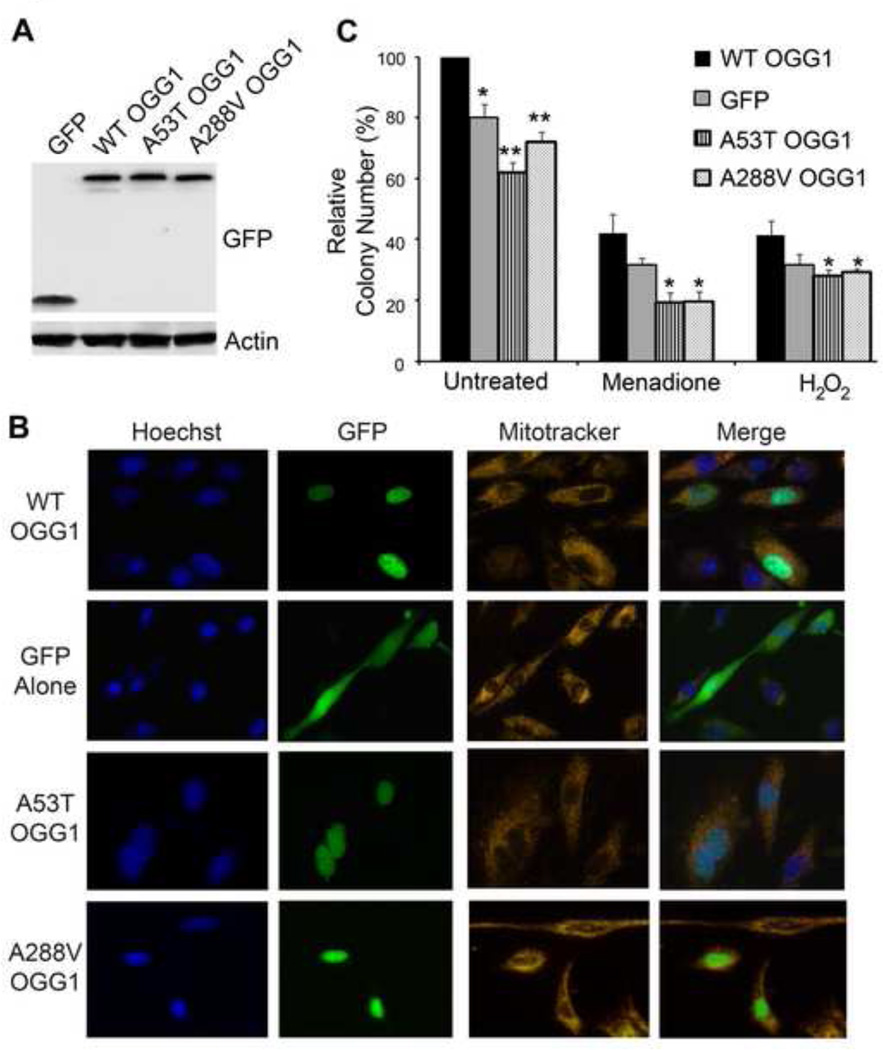
Given that the A53T and A288V polymorphisms have reduced catalytic activity, altered substrate binding in vitro, and decreased binding to known protein binding partners, we wanted to determine the consequence of the OGG1 polymorphisms in cells. OGG1−/− MEFs were transfected with plasmids containing GFP tagged WT or polymorphic OGG1 proteins. GFP-tagged constructs were used in this experiment in order to visually monitor the transfection efficiency and GFP protein expression for the duration of the experiments to ensure that the OGG1 proteins were being expressed and at similar levels. Indeed, immunoblotting MEF cell lysates expressing the GFP-tagged constructs confirmed equal expression of these proteins (Fig. 6A).
Fig. 6. Cells expressing the OGG1 variants are sensitive to DNA damaging agents and have decrease colony formation.
(A) OGG1−/− MEFs were transfected with the indicated GFP-fusion proteins and 24 hrs later cells were lysed, run on a 10% polyacrylamide gel and immunoblotted with anti-GFP antibodies to show expression of the constructs. Actin was used as a protein loading control. (B) OGG1−/− MEFs were transfected with the indicated GFP-fusion proteins. 24 hrs later, cells were incubated with Mitotracker and Hoechst stains and imaged live to determine the cellular localization of the GFP-tagged WT and polymorphic OGG1 proteins. (C) OGG1−/− MEFs were transfected with GFP-tagged constructs containing WT or OGG1 polymorphisms and equal numbers of cells (3000/well) were seeded in a 6 well plate. After 24 hours, cells were treated with the indicated DNA damaging agents and the number of colonies formed for each condition was determined 6 days later. Decreased number of colonies was observed for cells expressing the polymorphic forms of OGG1 in both the absence and presence of DNA damage. The histogram shows the normalized averages ± S.E.M. from four independent experiments. *p<0.05, **p<0.01, comparing WT and variant forms of OGG1 using Student’s T-Test.
To examine whether the OGG1 polymorphisms resulted in proper protein folding and localization to the nucleus, we looked at the cellular localization of transiently transfected WT and polymorphic OGG1 proteins in OGG1−/− MEFs. By using Hoechst as a nuclear marker and Mitotracker as a mitochondrial marker, we found that GFP-tagged WT, A35T and A288V proteins all localized predominantly to the nucleus, while as expected cells transfected with GFP alone showed diffuse cytoplasmic GFP fluorescence (Fig. 6B). These results indicate the A53T and A299V proteins localize similarly to WT when transfected in cells.
To determine the effect of the polymorphisms on long term cell survival, GFP-tagged OGG1 proteins were transfected into OGG1−/− MEFs and were either untreated or treated with 50 µM H2O2 or 20 µM menadione and colonies were counted 7 days later. Interestingly, we found that in the absence of DNA damage there was a significant decrease in colony formation for both the A53T and A288V polymorphisms compared to WT (Fig 6B). This decrease in colony formation for both the A53T and A288V proteins was greater than what was observed for the GFP alone, which has a complete absence of OGG1 protein. The A53T polymorphism resulted in a statistically significant decrease in colony formation when compared to GFP alone. Similarly, when cells expressing polymorphic OGG1 proteins were treated with either of the DNA damaging agents, a significant decrease in colony formation was observed compared to WT (both menadione and H2O2) and GFP (only menadione) (Fig. 6B). The decreased colony formation in these experiments indicates that MEFs expressing the OGG1 polymorphic proteins have reduced cell viability under both unstressed (no treatment) and stressed (DNA damaged) conditions.
Discussion
Oxidative damage accumulates in brain tissues of AD patients (reviewed in [41–43]), which may contribute to an individual’s disease susceptibility or to disease progression. When left unrepaired increased accumulation of DNA damage, specifically 8-oxoG lesions, in brain tissues of AD patients can result in mutation events that may influence or result in progression of the disease. In this study, we have characterized the biological significance of A53T and A288V, two polymorphisms in the human DNA repair protein OGG1, which have been identified in AD patients. Our data suggest that the expression of these proteins may contribute to the accumulation of oxidative damage in AD.
In this study, we have shown that the presence of either of these two OGG1 polymorphisms results in defects in catalytic activity (inability to remove the DNA lesions), decreased binding to PARP-1 and XRCC1 (inability to interact with members of the BER pathway which remove oxidatively damaged bases), and a decreased ability to activate PARP-1 (inability of PARP-1 to bind to sites of damage and to poly(ADP-ribosy)late proteins). It has been previously shown that both AD associated polymorphisms show defective incision ability [23, 25]. In agreement with these findings, we found that the A53T and A288V polymorphisms have decreased incision ability and have further explored the basis for these catalytic defects. Our data indicated that the A53T protein has dramatic defects in DNA binding ability in the presence of all substrates containing various bases opposite an 8-oxoG lesion, which may explain the decreased incision ability. The A288V protein, however, displayed decreased binding to only the 8-oxoG:T DNA substrate, indicating that defects in DNA binding do not fully explain the striking differences in incision ability compared to WT. Previous data suggested that there were possible defects in DNA binding for both the A53T and A288V proteins; however, Km values were calculated based on incision activity and only from 2 different substrates [23]. Here, we further investigated the DNA binding capacity of the polymorphisms using all 4 DNA substrates and by calculating the Kd from DNA:protein complexes using EMSA. Defects in DNA binding have been shown for other OGG1 polymorphisms including S326C, H270A/L/R, Q315A and F319A [28, 44].
The BER pathway is intricately designed with many different participating proteins interacting both directly and indirectly to facilitate the steps of the repair process. A defect in any of these interactions would result in the inability to complete removal and replacement of a DNA base lesion, possibly resulting in a DNA mutation event. One well established step of BER occurs when APE1 binds directly to OGG1 and stimulates its activity [36–38, 45]. We found that APE1 significantly stimulated the incision ability of WT OGG1. APE1 was also able to stimulate the activity of the A53T and A288V variants, suggesting that these proteins are not catalytically dead. Although the catalytic activity of the A53T protein was stimulated by APE1, the level of incision was still significantly decreased compared to the WT OGG1 level, indicating that this polymorphism has a defect in incision ability even in the presence of APE1. In contrast to A53T, the A288V polymorphism was sufficiently activated by APE1.
OGG1 is a bifunctional enzyme with both glycosylase and AP lyase activities. Similar to the AP lyase activity of OGG1, APE1 knicks the phosphodiester bond of an AP site creating a ssDNA break. We observed that APE1 stimulated the catalytic activity of the polymorphic proteins, suggesting a possible defect in the AP lyase activity of the proteins. Indeed, we found that both the A53T and A288V polymorphisms had deficiencies in AP lyase activity, but the A288V had a more dramatic defect. Reduced AP lyase activity may explain the decreased catalytic activity and the fact that these proteins are still able to be stimulated by APE1. Furthermore, these results help to explain mechanistically why the A288V has reduced catalytic activity.
We also examined whether these OGG1 polymorphisms could bind to PARP-1 and XRCC1, two other proteins involved in BER. PARP-1 binds to sites of SSBs and is involved in repair by either physically interacting with the damage site or through the poly(ADP-ribosyl)ation of proteins [46, 47], while XRCC1 forms a platform for binding of BER proteins, and specifically stimulates the formation of the OGG1 Schiff-base DNA intermediate [40, 48]. Our findings that the A53T and A288V polymorphic OGG1 proteins have decreased binding to PARP-1 and XRCC1 and are less able to activate PARP-1 suggest that expression of these polymorphisms may result in altered protein-protein and/or protein-DNA interactions that could affect the efficiency of BER. This is consistent with the previous study showing a decrease in overall BER capacity in neurons from AD patients [49].
To determine the effect that these OGG1 polymorphisms may play in cells, we examined the incision activity of nuclear extracts of cells transfected with WT or polymorphic OGG1 proteins. We found that both the A53T and A288V OGG1 proteins are expressed at levels similar to WT OGG1, but resulted in decreased incision of an 8-oxoG containing substrate, similar to the in vitro results. This indicates that even when the polymorphic OGG1 proteins have the additional components of the BER system present, they are still unable to excise an 8-oxoG lesion to the same extent as WT OGG1. One possibility to explain the decreased incision activity observed with the nuclear extracts would be improper cellular localization. The GFP-tagged OGG1 protein we used in our experiments contains a nuclear localization signal, and thus should be detected as a nuclear protein. We performed localization studies in the OGG1−/− MEFs and found that both the A53T and A288V OGG1 proteins properly localize to the nucleus in cells. Therefore, the decreased incision activity in the nuclear extracts is not due to mislocalization of the polymorphic proteins.
We did find that the biochemical defects we observed with both OGG1 polymorphisms affected cellular homeostasis. By expressing the polymorphic forms of OGG1 in OGG1 deficient MEFs, we were able to examine the long term cellular survival in the absence and presence of DNA damage. Interestingly, expression of the OGG1 variants reduced cell survival compared to WT and also compared to control cells lacking OGG1 expression. Similarly, we found that both polymorphisms sensitized the cells to DNA damaging agents compared to both WT OGG1 and OGG1 null backgrounds. These findings are interesting considering that these proteins can still be stimulated by APE1 in vivo and thus may retain some catalytic activity. This low level of catalytic activity, however, may not be sufficient to repair DNA damage and promote cell survival. Decreased cell survival could also be due to defects in other factors, such as a disruption in OGG1 binding to other DNA repair protein binding partners.
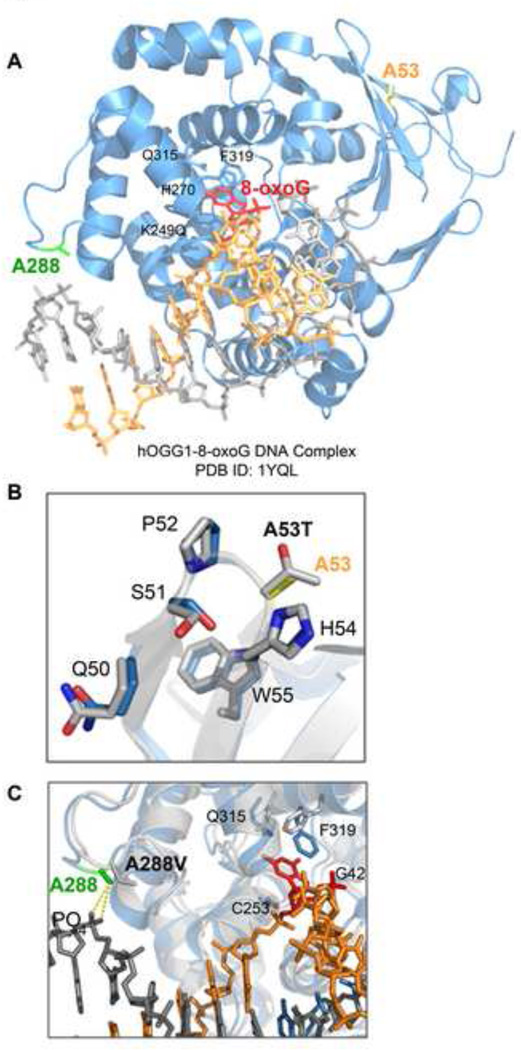
We used the three-dimensional structure of OGG1 (PDB ID:1YQL) in order to visualize how these mutations might affect protein function by analyzing the local conformation around the mutation sites in the OGG1-DNA substrate complex (Fig. 7A). The alanine at position 53 is a neutral hydrophobic amino acid that is positioned in a linker between two beta sheets that contribute to the 8-oxoG binding pocket [50] (Fig. 7B). Substitution of this alanine with threonine, a more branched polar amino acid, could change the contour of this region, disrupting the lesion binding pocket and possibly altering the flexibility of the loop region, which may explain the defects we observed in activity and DNA binding for this variant. The A288 residue resides between two alpha helices comprising the catalytic domain [50] (Fig. 7C). The substitution of alanine to valine, which is slightly larger and more branched, changes the residue from one that is a good helix former to one that is poor [51]. Based on this analysis, it is possible to speculate that the valine residue could perturb the two alpha helices of the catalytic domain, thus reducing the catalytic activity of the protein. Additionally, this residue is located in a DNA interacting region; however, our studies indicate that a substitution with a valine at this residue does not substantially affect DNA binding. This effect could be explained by the distal location of this residue to the DNA and also that the major residues responsible for the 8-oxoG recognition, such as G42, C253, Q315 and F319 (Fig. 7C), are conserved in the polymorphic proteins. It is also possible that overall protein misfolding occurs in the presence of either polymorphism, which could explain the altered binding to PARP-1 and XRCC1. However, our thermal stability assays indicate that there is not a general misfolding, but that the A53T protein is less stable. Furthermore, these proteins localize in cells similar to wild-type, further supporting the idea that these polymorphisms do not significantly affect protein folding.
Fig. 7. Location of the polymorphic residues in the three-dimensional structure of OGG1.
(A) 3D structure of human OGG1 and 7-deaza-8-azaguanine DNA complex is represented (PDB ID: 1YQK [52]). OGG1 is shown by ribbon diagrams, while DNA is shown by stick model. The amino acid residues investigated in this study are indicated in color. Additional amino acids are shown in black for reference. Protein Data Bank reference number: 1YQK [52]. The figure was prepared using PyMol software (Schrödinger). (B) View of the A53 residue within the flexible linker region between beta sheets 2 and 3 of the OGG1 structure. The yellow residue represents the WT A53 amino acid. The gray residue represents the predicted A53T polymorphism. (C) View of the A288 residue, which is in a DNA contact region. The green residue represents the WT A288 amino acid. The gray residue represents the predicted A288V polymorphism. The G42, C253, Q315, and F319 residues indicate other DNA contacting residues.
Taken together, this study shows that two AD-associated polymorphisms in OGG1, A53T and A288V, have defects in catalytic activity that may affect the ability of cells to repair the oxidative lesion 8-oxoG in DNA. Further investigation of OGG1 is needed to understand its exact role in disease and progression. Additional exploration lies in elucidating the complex defects that exist within key components of the BER pathway and how they may contribute to AD.
Supplementary Material
Highlights.
The OGG1 polymorphisms, A53T and A288V, have significantly reduced catalytic activity.
The A53T protein has a defect in DNA binding, while the A288V protein has AP lyase deficiencies.
Both polymorphisms have reduced binding to PARP-1 and XRCC1, and have reduced ability to activate PARP-1.
A53T and A288V polymorphisms reduce cell survival.
Acknowledgements
We thank Dr. Deborah Croteau (Laboratory of Molecular Gerontology, NIA) for helpful discussions and technical assistance with the EMSA assay and for critical reading of the manuscript. We thank Dr. Alan Zonderman for assistance in the statistical analysis of the EMSA data and Dr. David Wilson III for technical advice and assistance. We appreciate the helpful discussions and technical assistance from Dr. Jeff Hill, Dr. Hansen Du, and Dr. Mahdu Lal Nag and the excellent review comments provided by Dr. Sebastian Fugmann. We also thank Dr. Yie Liu for the generous gift of the wild-type and OGG1−/− primary MEFs. This research was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Abbreviations
- AD
Alzheimer’s Disease
- 8-oxoG
7,8-dihydro-8-oxoguanine
- OGG1
8-oxoguanine-DNA glycosylase
- PARP-1
Poly(ADP-ribose) polymerase 1
- XRCC1
X-ray cross-complementing protein 1
- ROS
reactive oxygen species
- BER
base excision repair
- SNP
single nucleotide polymorphism
- MEF
mouse embryo fibroblast
- PAR
poly(ADP-ribosly)ation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Alzheimer Report 2009. Alzheimer's Disease International. 2009 [Google Scholar]
- 2.Hardy J, Selkoe DJ. The Amyloid Hypothesis of Alzheimer's Disease: Progress and Problems on the Road to Therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 4.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin JF, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 5.Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, Mar L, Sorbi S, Nacmias B, Piacentini S, Amaducci L, Chumakov I, Cohen D, Lannfelt L, Fraser PE, Rommens JM, St George-Hyslop PH. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 6.Levy-Lahad E, Wasco W, Poorkaj P, Romano D, Oshima J, Pettingell W, Yu C, Jondro P, Schmidt S, Wang K, al e. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 7.William R M. Oxidative Stress Hypothesis in Alzheimer's Disease. Free Radical Biology and Medicine. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 8.Floyd RA, Carney JM. Free radical damage to protein and DNA: mechanisms involved and relevant observations on brain undergoing oxidative stress. Ann Neurol. 1992;32(Suppl):S22–S27. doi: 10.1002/ana.410320706. [DOI] [PubMed] [Google Scholar]
- 9.Butterfield DA, Castegna A, Lauderback CM, Drake J. Evidence that amyloid beta-peptide-induced lipid peroxidation and its sequelae in Alzheimer's disease brain contribute to neuronal death. Neurobiol Aging. 2002;23:655–664. doi: 10.1016/s0197-4580(01)00340-2. [DOI] [PubMed] [Google Scholar]
- 10.Coyle J, Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science. 1993;262:689–695. doi: 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- 11.Goodman L. Alzheimer's disease; a clinico-pathologic analysis of twenty-three cases with a theory on pathogenesis. J Nerv Ment Dis. 1953;118:97–130. [PubMed] [Google Scholar]
- 12.Ehmann WD, Markesbery WR, Alauddin M. Brain trace elements in Alzheimer's disease. NeuroToxicology. 1986;7:197–206. [PubMed] [Google Scholar]
- 13.Wang J, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. Journal of Neurochemistry. 2006;96:825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- 14.Mecocci P, MacGarvey U, Kaufman AE, Koontz D, Shoffner JM, Wallace DC, Beal MF. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 15.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease. Annals of Neurology. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 17.Mecocci P, Beal MF, Cecchetti R, Polidori MC, Cherubini A, Chionne F, Avellini L, Romano G, Senin U. Mitochondrial membrane fluidity and oxidative damage to mitochondrial DNA in aged and AD human brain. Mol Chem Neuropathol. 1997;31:53–64. doi: 10.1007/BF02815160. [DOI] [PubMed] [Google Scholar]
- 18.Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 19.Alam ZI, Daniel SE, Lees AJ, Marsden DC, Jenner P, Halliwell B. A generalised increase in protein carbonyls in the brain in Parkinson's but not incidental Lewy body disease. J Neurochem. 1997;69:1326–1329. doi: 10.1046/j.1471-4159.1997.69031326.x. [DOI] [PubMed] [Google Scholar]
- 20.Ferrante RJ, Browne SE, Shinobu LA, Bowling AC, Baik MJ, MacGarvey U, Kowall NW, Brown RH, Jr, Beal MF. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 21.Jacob KD, Noren Hooten N, Trzeciak AR, Evans MK. Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mechanisms of Ageing and Development. 2013;134:139–157. doi: 10.1016/j.mad.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson III DM, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2011;711:100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao G, Pan X, Zhu B-B, Zhang Y, Yuan F, Huang J, Lovell MA, Lee MP, Markesbery WR, Li G-M, Gu L. Identification and characterization of OGG1 mutations in patients with Alzheimer's disease. Nucleic Acids Research. 2007;35:2759–2766. doi: 10.1093/nar/gkm189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: A review of the functional and epidemiologic literature. Molecular Carcinogenesis. 2005;42:127–141. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 25.Sidorenko VS, Grollman AP, Jaruga P, Dizdaroglu M, Zharkov DO. Substrate specificity and excision kinetics of natural polymorphic variants and phosphomimetic mutants of human 8-oxoguanine-DNA glycosylase. FEBS J. 2009;276:5149–5162. doi: 10.1111/j.1742-4658.2009.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, Liu Y. Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres. PLoS Genet. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noren Hooten N, Kompaniez K, Barnes J, Lohani A, Evans MK. Poly(ADPribose) Polymerase 1 (PARP-1) Binds to 8-Oxoguanine-DNA Glycosylase (OGG1) Journal of Biological Chemistry. 2011;286:44679–44690. doi: 10.1074/jbc.M111.255869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Research. 2006;34:1620–1632. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill JW, Evans MK. A novel R229Q OGG1 polymorphism results in a thermolabile enzyme that sensitizes KG-1 leukemia cells to DNA damaging agents. Cancer Detection and Prevention. 2007;31:237–243. doi: 10.1016/j.cdp.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DB, Corcoran LM. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 2001. Expression and Purification of Glutathione-S-Transferase Fusion Proteins. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyaga SG, Lohani A, Evans MK. Deficient repair of 8-hydroxyguanine in the BxPC-3 pancreatic cancer cell line. Biochemical and Biophysical Research Communications. 2008;376:336–340. doi: 10.1016/j.bbrc.2008.08.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schofield MJ, Lilley DM, White MF. Dissection of the sequence specificity of the Holliday junction endonuclease CCE1. Biochemistry. 1998;37:7733–7740. doi: 10.1021/bi980399s. [DOI] [PubMed] [Google Scholar]
- 34.Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund Pr. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Analytical Biochemistry. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 35.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 36.Saitoh T, Shinmura K, Yamaguchi S, Tani M, Seki S, Murakami H, Nojima Y, Yokota J. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutation Research/DNA Repair. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 37.Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Research. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Research. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norman DPG, Chung SJ, Verdine GL. Structural and Biochemical Exploration of a Critical Amino Acid in Human 8-Oxoguanine Glycosylase. Biochemistry. 2003;42:1564–1572. doi: 10.1021/bi026823d. [DOI] [PubMed] [Google Scholar]
- 40.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the Coordination and Stimulation of Oxidative DNA Damage Repair Initiated by the DNA Glycosylase hOGG1. Journal of Biological Chemistry. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 41.Coppede F, Migliore L. DNA damage and repair in Alzheimer's disease. Curr Alzheimer Res. 2009;6:36–47. doi: 10.2174/156720509787313970. [DOI] [PubMed] [Google Scholar]
- 42.Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, Lovell M, Markesbery WR, Butterfield DA. Brain Regional Correspondence Between Alzheimer's Disease Histopathology and Biomarkers of Protein Oxidation. Journal of Neurochemistry. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 43.Halliwell B. Oxidative stress and neurodegeneration: where are we now? J Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 44.van der Kemp PA, Charbonnier J-B, Audebert M, Boiteux S. Catalytic and DNA binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket. Nucleic Acids Research. 2004;32:570–578. doi: 10.1093/nar/gkh224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidorenko VS, Nevinsky GA, Zharkov DO. Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair. 2007;6:317–328. doi: 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 47.Burkle A. DNA repair and PARP in aging. Free Radical Research. 2006;40:1295–1302. doi: 10.1080/10715760600915288. [DOI] [PubMed] [Google Scholar]
- 48.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase [beta] in cellular protection against cytotoxic DNA singlestrand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weissman L, Jo D-G, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Research. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 51.Gregoret LM, Sauer RT. Tolerance of a protein helix to multiple alanine and valine substitutions. Fold Des. 1998;3:119–126. doi: 10.1016/S1359-0278(98)00017-0. [DOI] [PubMed] [Google Scholar]
- 52.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.