Abstract
The modified DNA base 5-hydroxymethylcytosine (5hmC) is enriched in neurons where it may contribute to gene regulation and cellular identity. To determine how 5hmC influences gene expression in an in vivo neuronal population, we assessed the patterning and function of the base along the developmental lineage of the main olfactory epithelium—from multipotent stem cells through neuronal progenitors to mature olfactory sensory neurons (mOSNs). We find that 5hmC increases over gene bodies during mOSN development with substantial patterning occuring between the progenitor and mOSN stages. Although gene-body 5hmC levels correlate with gene expression in all three developmental cell types, this association is particularly pronounced within mOSNs. Overexpression of Tet3 in mOSNs markedly alters gene-body 5hmC levels and gene expression in a manner consistent with a positive role for 5hmC in transcription. Moreover, Tet3 overexpression disrupts olfactory receptor expression and the targeting of axons to the olfactory bulb, key molecular and anatomical features of the olfactory system. Our results suggest a physiologically significant role for gene-body 5hmC in transcriptional facilitation and the maintenance of cellular identity independent of its function as an intermediate to demethylation.
Keywords: epigenetics, olfaction, neurodevelopment
The functional repertoire of DNA modifications has expanded recently with the discovery of a derivative of 5-methylcytosine (5mC)—5-hydroxymethylcytosine (5hmC)—in mammalian genomes (1, 2). This modified base is enriched in embryonic stem cells (ESCs) and several adult tissues, with particular enrichment within the nervous system and bone marrow (1, 3). The modification is generated by a conserved family of enzymes, the Ten-eleven translocation family of methylcytosine dioxygenases (Tet1, Tet2, and Tet3), which show variable levels of expression across tissues (2, 4). Work in neural systems (5, 6) and ESCs (7–11) has found that 5hmC is concentrated on putative intergenic enhancers and gene bodies where levels of the base positively correlate with transcription.
Despite substantial work in ES cells describing the localization of 5hmC, its association with protein factors, and the effects of its manipulation on gene expression, it is still unclear how the modified base influences gene expression within a neuronal system. Several models describe 5hmC as an intermediate to a demethylated state (4, 9, 12–16). However, it remains possible that 5hmC influences transcription not only via the eventual lack of cytosine modification but also directly through interactions with the chromatin environment or through the recruitment or exclusion of binding proteins. Recent work suggests that 5hmC patterning persists at some loci during the development of the central nervous system (6). In addition, the recent identification of protein factors that bind 5hmC suggests that the modified base may be interpreted directly as an epigenetic mark (17–19).
It is difficult to assess the stability and function of the modified base in a mitotic population, such as ESCs where continual division may mask persistent 5hmC patterning, or in a fully developed tissue, such as the adult brain, where comparisons with previous developmental stages are challenging. To better understand how 5hmC influences developmental gene regulation, we characterized its patterning and function in an in vivo developmental system, the mouse main olfactory epithelium (MOE). The MOE continuously regenerates over the life of the animal, such that the major neurogenic cell types are simultaneously present in adulthood. Using genetic and cell-surface markers, we isolated distinct neurodevelopmental stages from the MOE and defined their 5hmC, 5mC, and transcriptional states through next-generation sequencing approaches. Our analysis focused on three cell types representing three stages of neurodevelopment. Horizontal basal cells (HBCs), located in a single layer along the basal lamina, are quiescent multipotent cells that can produce all of the cell types of the MOE in response to injury (20). Globose basal cells (GBCs), located apical to the HBC layer, are mitotically active progenitors and can produce both neuronal and nonneuronal cell types (21). Finally, mature olfactory sensory neurons (mOSNs) are terminally differentiated primary sensory neurons and compose the majority of cells within the adult MOE.
Results
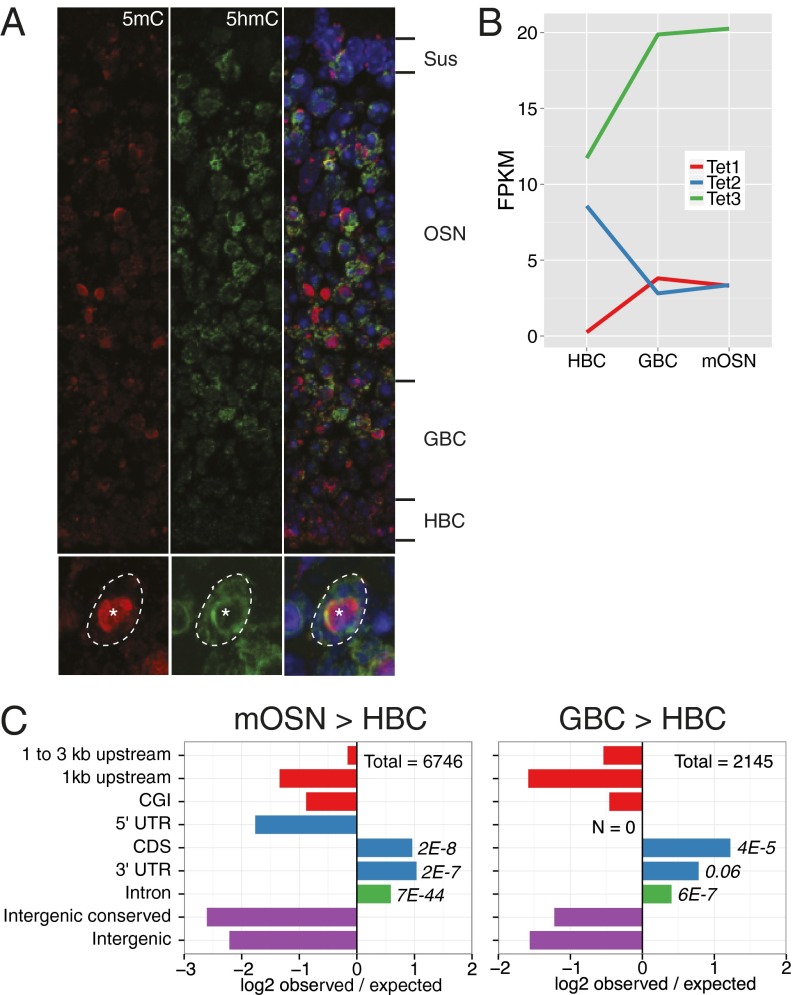
To initially assess global changes of 5mC and 5hmC abundances across MOE neurogenesis, we stained sections of the MOE from postnatal day 0 animals using antibodies that specifically recognize the two modified bases (4). 5hmC is substantially enriched in neurons and neuronal progenitors relative to basal stem cells and apical, nonneuronal sustentacular cells (Fig. 1A, Upper). In contrast, global levels of 5mC are relatively constant across the tissue. Within the nucleus (Fig. 1A, Lower), 5hmC and 5mC occupy largely complementary compartments: 5hmC is diffusely localized within the nucleus yet excluded from the central pericentromeric heterochromatin focus (22), where 5mC is enriched. This apparent increase in 5hmC levels during differentiation coincides with the up-regulation of Tet3, the most highly expressed Tet family member in the MOE (Fig. 1B).
Fig. 1.
Developmental 5hmC patterning in the MOE. (A, Upper) MOE coronal section from a P0 mouse stained for 5hmC (green), 5mC (red), and DNA (DAPI, blue). Major developmental layers are indicated. (Lower) Outlined OSN nucleus; asterisk indicates pericentromeric condensation. (B) Tet family member expression levels within HBCs, GBCs, and mOSNs. (C) Intersections of differential 5hmC peaks in mOSN vs. HBC (Left) and GBC vs. HBC (Right) with genomic features. Differential peaks were identified in each biological replicate individually and then intersected for a consensus set. Values are log2 ratios of observed versus expected number of peaks within a given region. Significance values are one-sided Fisher-test false discovery rates. CGI, CpG island.
To develop genomic maps of 5hmC across MOE neurogenesis, we sorted Olfactory marker protein (OMP)-expressing mOSNs, Neurogenin1-expressing GBCs, and Intercellular Adhesion Molecule 1-expressing HBCs from the MOEs of 8-wk-old mice using genetic and cell-surface markers (Fig. S1A) (23). We then purified genomic DNA from these populations and performed DNA immunoprecipitation (DIP) coupled to next-generation sequencing using anti-5hmC antibodies [hydroxymethylated DNA immunoprecipitation (hmeDIP-seq); see Dataset S1 for metadata]. Two biological replicates of each hmeDIP-seq library were generated. Unless otherwise noted, subsequent analyses were performed using averages of the replicates. We also performed methylated DIP (meDIP)-seq in these populations to follow the relative changes of 5hmC/5mC ratios during differentiation. The ability of these antibodies to discriminate among cytosine-, 5mC-, and 5hmC-containing templates was verified by DIP-quantitative PCR (qPCR) (Fig. S1B). To correlate epigenetic signatures with the transcriptional state, we used expression analyses performed in these cell lines by mRNA-seq. (23).
Genome-wide correlations of the DNA modification datasets give an initial impression of the dynamics and stability of the modifications across mOSN development. In agreement with the developmental relationships between the stages, 5hmC levels are more highly correlated between mOSN/GBC (Pearson R = 0.78) and GBC/HBC (R = 0.75) stages than those in mOSN/HBC (R = 0.70). In addition, differential peak analysis of 5hmC indicates that most de novo patterning occurs within mOSNs (mOSN/HBC peak number = 6,746; GBC/HBC = 2,145) and is strongly associated with exons and introns (Fig. 1C).
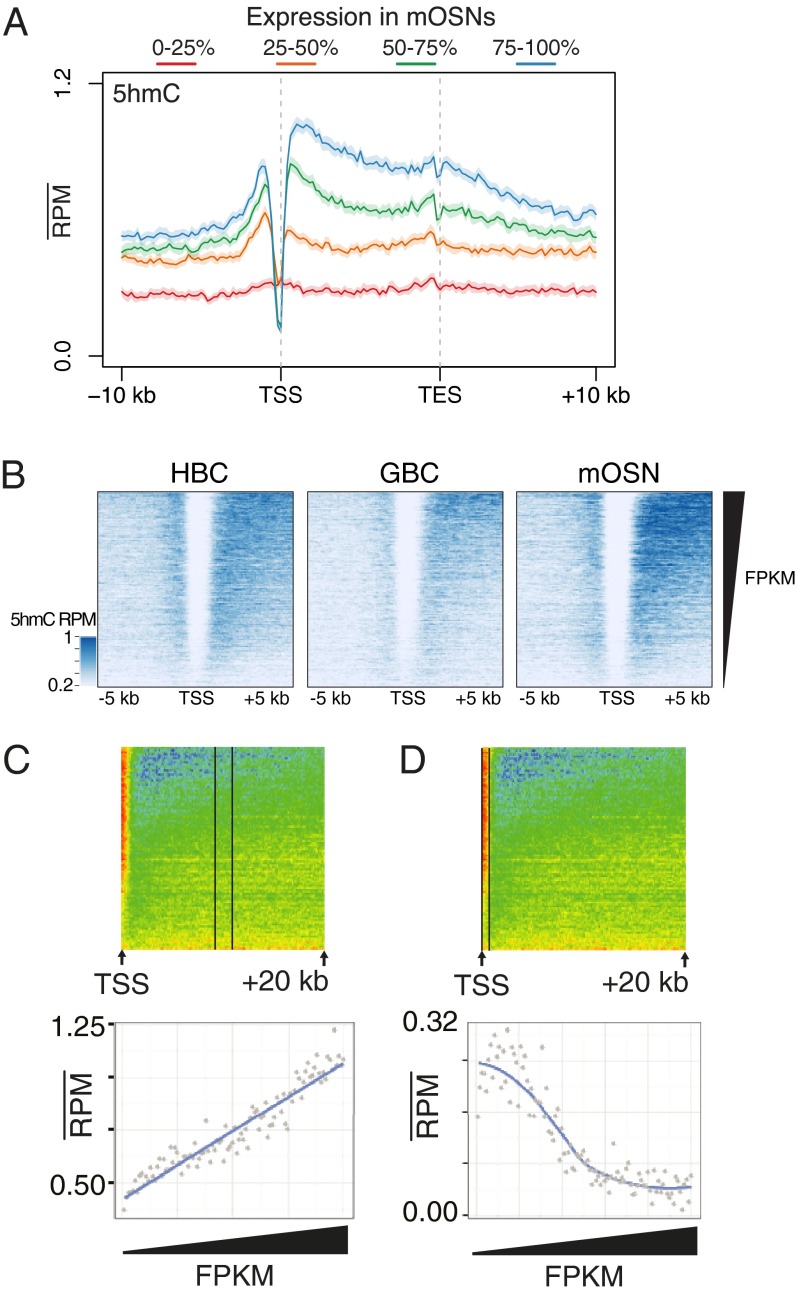
5hmC levels within the gene body [from transcription start site (TSS) to transcription end site] positively correlate with transcription in mOSNs (Pearson R = 0.21, P < 2.2E-16) and to a lesser extent in GBCs and HBCs (R = 0.16, P < 2.2E-16; R = 0.14, P < 2.2E-16, respectively) (Fig. 2 A and B and Fig. S2A). As in other systems (6, 24), we observe a dichotomy between the correlation of 5hmC levels and transcription that depends on genic location: although gene-body 5hmC levels linearly increase with transcription across the full range of the transcriptional output (Fig. 2C), the TSSs of active genes are depleted of the modified base (Fig. 2D). It is possible that the differential patterning of 5hmC on the TSS and gene body produces differential effects on transcription. To test this model, we performed a cell-free transcription assay using a linear template with a CMV promoter or transcribed region containing unmodified cytosines, 5mC, or 5hmC (Fig. S2B). We find that the presence of either 5mC or 5hmC in the promoter region has a deleterious effect on transcription yet has little consequence when restricted to the transcribed region, suggesting that 5hmC within the gene body may require interactions with the chromatin environment to exert an effect.
Fig. 2.
Relationships between 5hmC levels and gene transcription. (A) Aligned gene profiles of mOSN 5hmC levels split by mRNA fragments per kilobase per millions of reads (FPKM) quartiles in mOSNs. Error is 95% bootstrap confidence interval. (B) HBC, GBC, and mOSN 5hmC flanking TSSs sorted by FPKM in each cell type. (C and D) Mean mOSN 5hmC values within regions extending (C) 9–11 kb within the gene body and (D) from the TSS to 500 bp downstream. The x axis is the index of increasing FPKMs. In C, linear regression adjusted R-squared = 0.86.
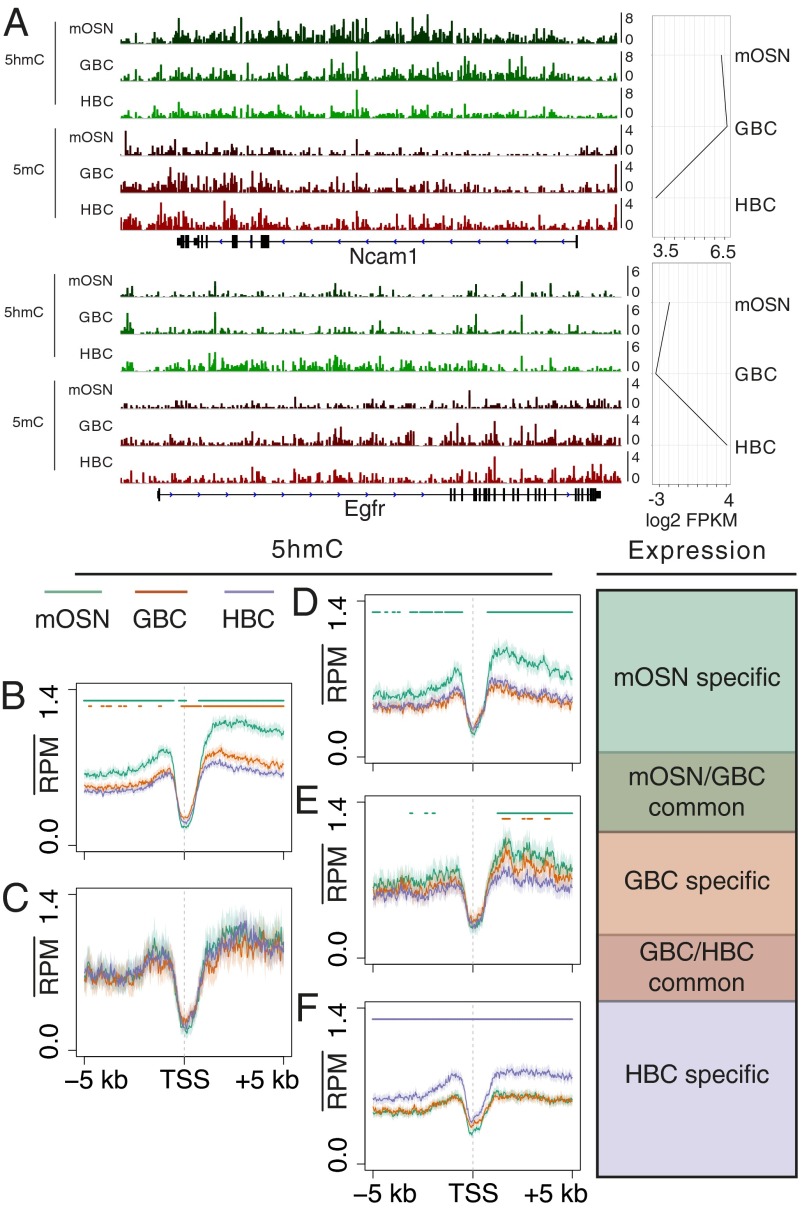
The significant increases of 5hmC seen over gene elements as OSN differentiation progresses suggest that 5hmC patterning may contribute to the definition of the neuronal progenitor and neuronal stages. To further examine the developmental patterning of 5hmC, we characterized 5hmC levels associated with developmentally regulated genes. Inspection of the modification patterning across two representative genes, Ncam1 and Egfr, which are differentially expressed between HBCs and mOSNs reveals a dynamic interplay between 5hmC and 5mC at different developmental stages (Fig. 3A). Substantial enrichment of 5hmC is seen over the Ncam1 gene body only within the cell types in which it is actively transcribed. Accordingly, 5mC is depleted from the 5′ portion of the gene upon its transcription in mOSNs. A similar although weaker enrichment of 5hmC is seen across the Egfr locus only in HBCs, whereas 5mC gene-body levels remain unchanged. 5hmC and 5mC DIPs using independent sorted populations and qPCR analysis of five differentially expressed genes (Ncam1, Ebf1, Neurog1, Egfri, and Notch2) support this pattern (Fig. S3 A and B).
Fig. 3.
Dynamic patterning of 5hmC at developmentally regulated genes. (A) 5hmC and 5mC tracks for two representative genes, Ncam1 and Egfr, with different expression paths across mOSN, GBC, and HBC cell types. Scale is reads per million. (B–F) 5hmC TSS profiles in mOSN, GBC, and HBC stages grouped into five developmental expression sets: (B) mOSN/GBC common (n = 2,577), (C) GBC/HBC common (n = 281), (D) mOSN-specific (n = 893), (E) GBC-specific (n = 557), and (F) HBC-specific (n = 1,566). Bars indicate a significant difference (Materials and Methods) between the profile of the correspondingly colored track and the HBC profile (B–D), the GBC profile (E), or the mOSN profile (F).
The positive correlation between 5hmC gene-body levels and transcription suggests that 5hmC patterning may simply reflect the transcriptional status of a gene within a given cell type. To determine if 5hmC levels faithfully track with differential expression across the three cell types, we generated five sets of differentially expressed genes—mOSN/GBC-common, GBC/HBC-common, mOSN-specific, GBC-specific, and HBC-specific (Dataset S2)—and computed the 5hmC profiles flanking the TSSs of each set (Fig. 3 B–F). mOSN/GBC common genes exhibit an elevation of gene-body 5hmC in the GBC and mOSN stages (Fig. 3B) with higher levels in the mOSN stage. Similarly, mOSN- (Fig. 3D) and HBC-specific (Fig. 3F) genes exhibit elevated gene-body 5hmC within the mOSN and HBC populations, respectively, again supporting the general association between 5hmC levels and gene expression. However, within the bodies of GBC-specific (Fig. 3E) genes, the timing of 5hmC elevation is more complicated. These genes exhibit slight increases in 5hmC levels within the GBC stage that then persist into the mOSN stage. Thus, transcriptionally down-regulated genes may retain elevated levels of 5hmC within mOSNs, indicating that, although there is a close association between gene-body 5hmC levels and transcription, the presence of the modified base is likely not sufficient for gene activation.
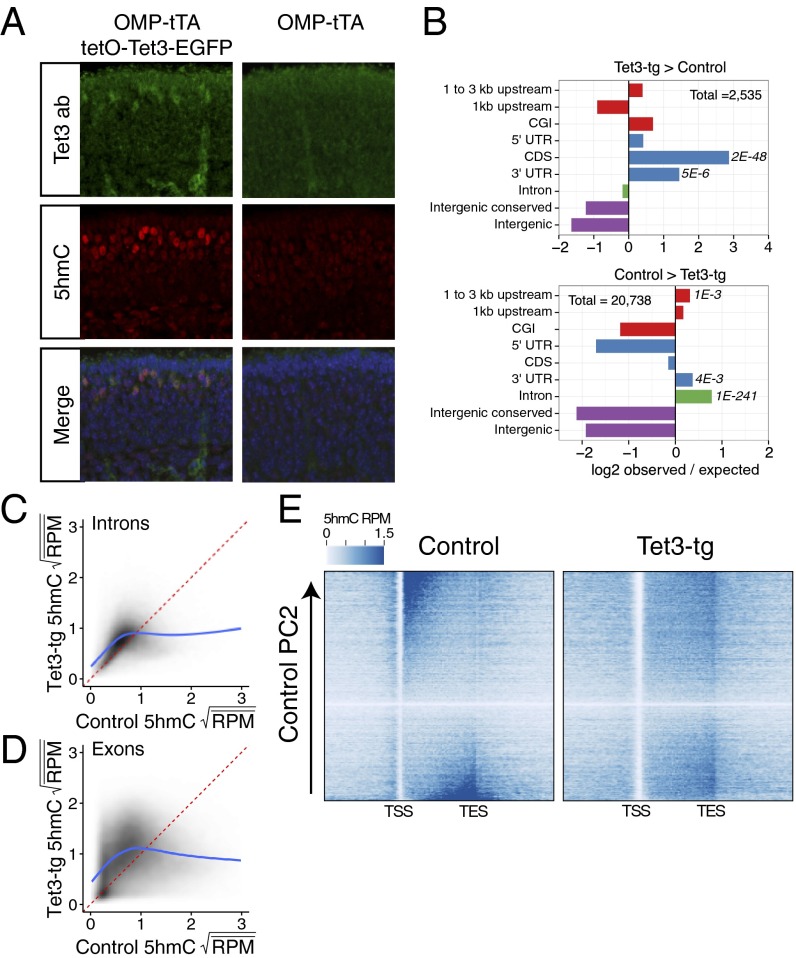
The dynamic patterning of gene-body 5hmC across the mOSN developmental lineage suggests that the modified base plays a key role in defining the identity of differentiated cell types. To test this model we used a genetic strategy to alter the 5hmC landscape of mOSNs. It is likely that the modification state of each locus reflects the steady state of methylation, hydroxymethylation, and demethylation reactions. Therefore, an increase of Tet3 activity in mOSNs may bias loci with low 5hmC levels toward increased hydroxymethylation and loci with high 5hmC levels toward demodification. With this model in mind, we generated a tetO-Tet3-ires-EGFP transgenic mouse, which we crossed into a background containing OMP-ires-tTA to drive expression in mOSNs. This genetic strategy allowed specific expression of transgenic Tet3 (Tet3-tg) and the bicistronic EGFP reporter in approximately half of the mOSNs (49 ± 12% as determined by FACS). By immunofluorescence, Tet3 protein level is increased in Tet3-tg expressing MOEs and colocalizes with increased levels of 5hmC (Fig. 4A).
Fig. 4.
Tet3 overexpression alters 5hmC patterning. (A) Immunofluoresence of Tet3 and 5hmC on coronal MOE sections of P0 mice either expressing tetO-Tet3-EGFP in mOSNs (Left) or not (Right). (B) Differential 5hmC peak analysis and genomic feature intersections. Values are log2 ratios of observed versus expected number of peaks within a given region. (Upper) Regions enriched in Tet3-tg versus control mOSNs. (Lower) Regions enriched in control versus Tet3-tg mOSNs. Significance values are one-sided Fisher-test false discovery rates. CGI, CpG island. (C and D) Comparison of square-root mean 5hmC reads per million (RPM) in control and Tet3 overexpressing mOSNs within introns (C) and exons (D). Fitted lines are predicted values from a generalized additive model: R command: gam(y ∼ s(x, bs = ′cs′)). (E) Heatmaps of genic 5hmC in control (Left) and Tet3-tg (Right) mOSNs. Genes are ordered by control 5hmC gene-body levels according to principal component 2 (Materials and Methods).
To assess the consequences of Tet3 overexpression in these neurons, we FAC-sorted two biological replicates of control (OMP-ires-GFP) and Tet3-transgenic (Tet3-tg) mOSNs and analyzed the patterning of 5hmC by hmeDIP-seq and expression levels by rRNA-depleted RNA-seq. Differential 5hmC peak analysis indicates significant alterations of 5hmC levels within regions 1–3 kb upstream of TSSs, coding DNA sequences (CDSs), 3′ UTRs, and introns (Fig. 4B). Interestingly, these regions exhibit both increases and decreases of 5hmC depending on control levels of 5hmC (Dataset S3). Both exons (Fig. 4C) and introns (Fig. 4D) with moderate levels of 5hmC in control mOSNs (middle 10%) exhibit a slight gain of the mark in Tet3-tg (Tet3-tg/control 5hmC ratio ± SD exons = 1.38 ± 0.57, Wilcoxon P < 2.2E-16; introns = 1.23 ± 0.42, Wilcoxon P < 2.2E-16), whereas those with high levels (top 10%) exhibit a significant reduction (exons = 0.49 ± 0.23, Wilcoxon P < 2.2E-16; introns = 0.56 ± 0.25, Wilcoxon P < 2.2E-16). These opposing effects are consistent with the ability of Tet3 to oxidize 5hmC as well as 5mC; genes with high levels of 5hmC in control mOSNs may recruit sufficient levels of Tet3 to induce further oxidation, whereas those with moderate levels achieve only partial oxidation from 5mC to 5hmC. Moreover, gene-body 5hmC levels are shifted toward 3′ ends (Fig. 4D), perhaps reflecting differential recruitment of Tet3 by 5′-enriched transcriptional elongation complexes. Regions that lose 5hmC do not exhibit a complementary increase of apparent fC (Fig. S4A), suggesting that the loss of 5hmC does not result in the enrichment of additionally oxidized forms, but in a return to an unmodified state. In contrast to the bidirectional changes to 5hmC levels in gene bodies, Tet3-tg 5hmC levels at promoters and TSSs are generally reduced (Fig. S4B) and do not exhibit a complex dependency on control 5hmC levels (Fig. S4C).
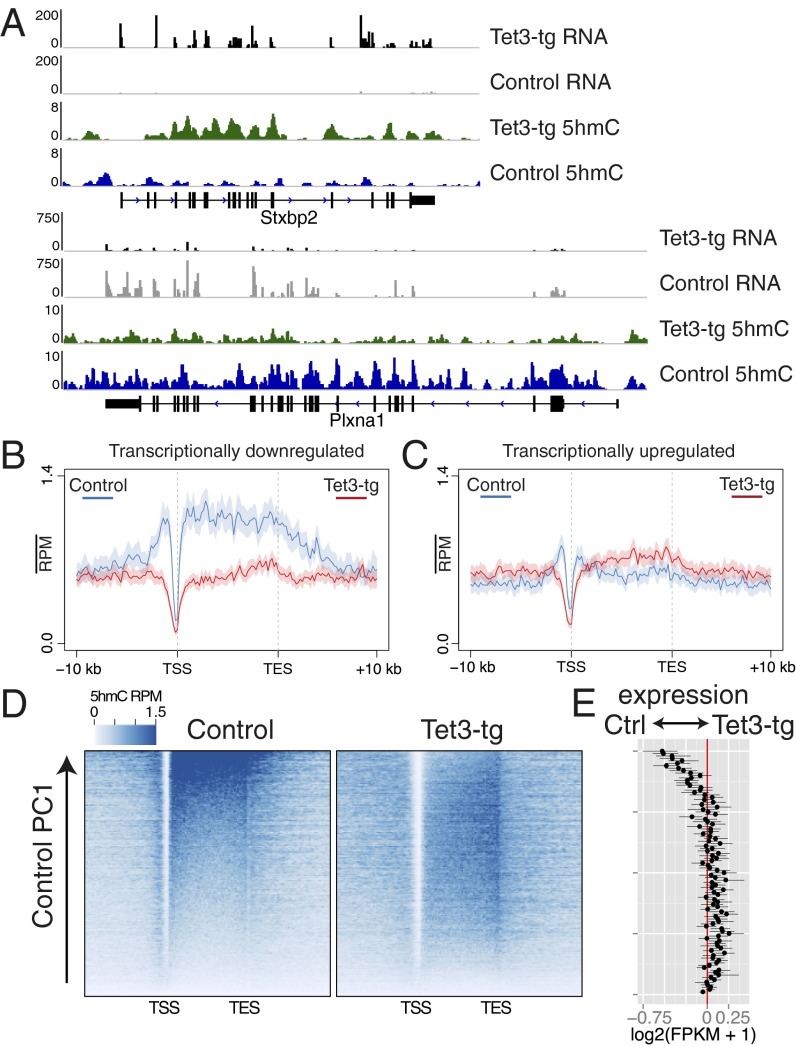
Two example gene loci, Stxbp2 and Plxna1, demonstrate both the increase and the decrease, respectively, of gene-body 5hmC levels in Tet3-tg mOSNs (Fig. 5A). Interestingly, the expression levels of these genes track with Tet3-induced 5hmC changes. An analysis of differentially expressed genes (Bayes factor ≥ 20) (Materials and Methods and Dataset S4) indicates that transcriptionally down-regulated genes (Fig. 5B, n = 1621) exhibit a significant depletion of gene-body 5hmC (Wilcoxon P = 5.7E-18), whereas up-regulated genes (Fig. 5C, n = 938) exhibit a modest, but insignificant, increase of gene-body 5hmC. We examined the general association between gene-body 5hmC and expression by ordering genes according to their gene-body 5hmC levels in either control or Tet3-tg mOSNs using principal component analysis (Fig. 5 D and E). Strikingly, the fold change of control to Tet3-tg mOSN gene expression reflects alterations to gene-body 5hmC: genes that lose gene-body 5hmC are substantially down-regulated whereas those that gain 5hmC are slightly up-regulated. Notably, these transcriptional effects positively correlate with 5hmC levels at the gene body (Pearson R = 0.12, P < 2.2E16) and not at the TSS (±500 bp of TSS, R = −0.02, P = 0.01).
Fig. 5.
5hmC alterations modulate gene expression. (A) rRNA-depleted RNA (intersected with exons) and 5hmC levels in Tet3-tg and control mOSNs for two representative genes. The y axis is RPM. (B and C) 5hmC profiles over genes transcriptionally down-regulated (B) or up-regulated (C) in Tet3-tg relative to control mOSNs. (D) Heatmaps of gene-body 5hmC in control and Tet3-tg mOSNs ordered by control 5hmC gene-body levels according to principal component 1 (Materials and Methods). (E) Difference between Tet3-tg and control log2(FPKM + 1) values of genes along the ordering used in D. Values shown are the average of these genes grouped into 100 bins. Error bars are 95% bootstrap confidence intervals.
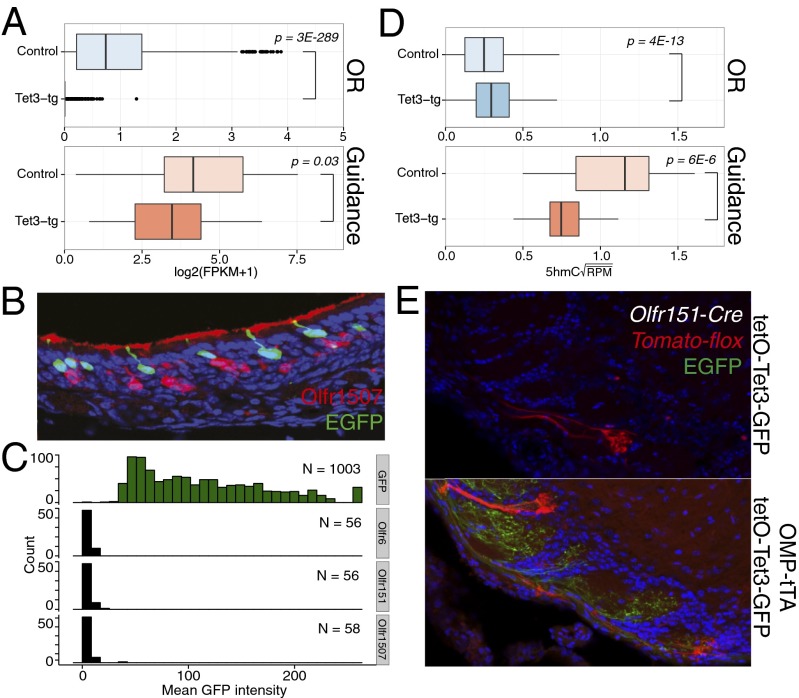
The most highly expressed mOSN-specific genes, such as olfactory receptors (ORs) and the guidance molecules involved in the wiring of olfactory neurons, are transcriptionally down-regulated in Tet3-tg mOSNs (1,071 of 1,082 expressed ORs and 21 of 25 expressed guidance molecules) (Fig. 6A and Dataset S4). By immunofluoresence, we find no overlap between OR-expressing mOSNs and Tet3-tg mOSNs (Fig. 6 B and C). Consistent with the general effects described above, 5hmC is significantly depleted within gene bodies encoding guidance molecules in Tet3-tg mOSNs (Fig. 6D). However, the same relationship between OR down-regulation and 5hmC levels cannot be established for ORs likely due to technical reasons; each OR is expressed in only 0.1% of the cells in the MOE, and activating epigenetic marks cannot be detected on these genes in mixed mOSN populations (23). Moreover, OR genes have a high AT content (25), which reduces the number of modifiable cytosines. As a result, the levels of 5hmC in OR genes are extremely low in this mixed mOSN population, possibly below the sensitivity of hmeDIP.
Fig. 6.
Tet3 overexpression disrupts mOSN axonal targeting. (A) Boxplots of rRNA-depleted RNA log2(FPKM + 1) values of olfactory receptors (ORs) and axon guidance molecules (defined in Materials and Methods) in control and Tet3-tg mOSNs. P values from Wilcoxon two-sided rank-sum test. (B) Immunofluorescent staining of Olfr1507 demonstrating mutually exclusive expression with Tet3-tg. (C) Quantification of GFP intensities within GFP-, Olfr6-, Olfr151-, and Olfr1507-positive OSNs in OMP-tTA/tetO-Tet3-GFP MOEs. (D) Square-root mean 5hmC RPM of ORs and axon guidance molecules with a log2(FPKM + 1) ≥ 2 in either genotype. P values from Wilcoxon two-sided rank-sum test. (E) Analysis of OSN axon targeting to the olfactory bulb in Olfr151-Cre/Tomato-floxed animals under control Tet3 expression (Upper) or Tet3 overexpression in mOSNs (Lower).
The proper expression of these genes governs the correct targeting of OSN axons to discrete glomeruli in the olfactory bulb. To examine if the transcriptional misregulation caused by Tet3 overexpression disrupts this targeting, we analyzed Olfr151 glomerulus formation in the bulbs of Olfr151-Cre/floxed-Tomato mice in the presence or absence of Tet3 transgene expression (Fig. 6E). In every case examined (n = 3), Tet3 overexpression produced mistargeted axons and the formation of additional glomeruli. Consistent with the RNA-seq and immunofluorescence data, there is no detectable overlap between GFP-positive and Tomato-positive axons (Fig. 6E), indicating a lack of Olfr151 expression in Tet3-tg mOSNs. It is therefore likely that the targeting deficit seen in Tomato-positive axons is due to non-cell autonomous disruption of the glomerular map.
Discussion
Our experiments describe the alterations to the 5mC and 5hmC genomic landscapes across the neurogenic lineage of the main olfactory epithelium from multipotent stem cell to mature olfactory sensory neuron. Within the MOE, 5hmC is globally enriched in the genomes of neuronal progenitors and neurons, consistent with previous reports assaying 5hmC levels in neural tissues (3, 6). In concordance with the general positive correlation between transcription and gene-body 5hmC levels, gene-body 5hmC is associated with cell-type–specific gene expression. However, genes that are active in progenitors but silenced in neurons retain elevated levels of gene-body 5hmC in the neuronal stage, suggesting that gene-body 5hmC patterning in neurons does not always simply reflect transcriptional status.
We note different relationships between 5hmC levels and transcription depending on the location of the modification. The depletion of 5hmC at the TSSs of transcribed genes is consistent with a role for this modification in the demethylation of these sites. Although the genomic occupancy of the Tet family members has not been determined in a neuronal system, several studies have characterized the binding of Tet1 in ESCs where the protein appears to be predominantly localized at TSSs (12, 26). In addition to their ability to convert 5mC to 5hmC, the Tet family members are able to catalyze the conversion to the subsequent oxidative states of 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC), which may be acted on by DNA repair machinery to produce a fully demodified state (27, 28). Therefore, the high occupancy of Tet proteins at TSSs may drive full oxidation and demodification within these regions. In contrast, Tet protein occupancy within gene bodies and enhancers may be relatively transitory, preventing subsequent oxidation from occurring, or the interaction of the Tet proteins with regulatory factors may alter its activity. Therefore, 5hmC is likely a multifunctional modification that is used either in the demethylation pathway or as an epigenetic modification itself, depending on the genomic location of the mark. Consistent with differential role of 5hmC in TSSs versus gene bodies are the differential effects of this modification in our in vitro transcription assays, where hydroxymethylation within the promoter and TSS abolishes transcription, whereas gene-body hydroxymethylation has no measurable effects. Although these experiments cannot exclude that gene-body hydroxymethylation is just a by-product of transcriptional elongation, an attractive hypothesis proposes that 5hmC facilitates transcriptional elongation in the context of chromatin.
In this vein, the effects of Tet3 overexpression in mOSNs on 5hmC patterning and gene transcription are consistent with a direct role for 5hmC in gene regulation. The loss of gene-body 5hmC resulted in a reduction of expression, whereas the gain of 5hmC is associated with increased expression. This relationship extended on average across all genes and does not appear to be confined to defined gene classes, such as housekeeping or developmentally regulated genes. Although we are unable to exclude indirect effects of Tet3 overexpression, such a general association suggests that gene-body 5hmC patterning is a common modulator of transcriptional output, perhaps directly altering the local chromatin environment or indirectly recruiting elongation factors. Recent studies indicate that both Mbd3 (17) and MeCP2 (18) can bind 5hmC. Interestingly, as in other neuronal cell types, MeCP2 expression is highly up-regulated in mOSNs (29), and its loss leads to mOSN axons targeting deficits (30). The modulatory character of gene-body 5hmC levels suggests that the precise tuning of these levels may be necessary to maintain constant levels of expression within mature postmitotic populations such as olfactory sensory neurons. Indeed, initial OR expression and axonal targeting occurs just before the mOSN stage and the onset of Tet3 overexpression, suggesting that the effects seen here are due to the disruption of mechanisms that maintain gene expression and not those that initiate it. Through this control, gene-body 5hmC patterning may aid in establishing stable cellular identities.
Materials and Methods
Animal Care and Use.
Mice were treated in compliance with the rules and regulations of the University of California, San Francisco Institutional Animal Care and Use Committee under protocol approval no. AN084169-01.
Immunofluoresence.
Sections of postnatal C57BL/6 MOE were fixed with 3:1 methanol:acetic acid, fragmented with 1N HCl and neutralized with 100 mM Tris, pH 8. Sections were blocked and then incubated with anti-5mC (#39649) and anti-5hmC (#39791) antibodies (Active Motif) overnight at 4 °C. Tet3 immunostaining was performed on coronal MOE sections fixed with 4% (vol/vol) paraformaldehyde and using anti-Tet3 antibody (Santa Cruz, sc-139186).
Fluorescence-Activated Cell Sorting.
Mature OSNs were isolated from the MOEs of OMP-ires-GFP knock-in animals (31). GBCs were isolated from the MOEs of Ngn-GFP BAC transgenics (32). HBCs were isolated with anti-mouse CD54 (Icam1) conjugated to phycoerythrin (Biolegend). Isolation and preparation were performed as described (23). Briefly, single-cell suspensions from 2 to 10 MOEs were made using the Papain dissociation system (Worthington). MOE were dissected into Earl’s Buffered Saline Solution, minced in Papain dissociation solution (Worthington), and incubated at 37 °C for 45 min. Cells were washed once in 1:10 inhibitor solution and once in PBS. For a given assay (DIP-seq/qPCR or RNA-seq), 2E5 to 1E6 cells were used. Population purity was assayed through a limited resorting of the sorted cells. Purities of ∼90–95% were routinely achieved.
meDIP and hmeDIP-seq.
Genomic DNA was isolated, prepared for single- or paired-end Illumina sequencing using standard protocols, and immunoprecipitated as in Weber et al. (33) with minor modifications. Specifically, sorted cell gDNA was sonicated to 200–1,000 bp with a mean size of ∼400 bp and end repaired (New England Biolabs Next End Repair). Adenosine was added to 3′ ends (Klenow, 3′–5′ exo-, New England Biolabs), and 10:1 Illumina adaptor:samples were ligated onto the samples. A total of 20–50 ng of adapted genomic DNA was diluted in MB (10 mM phosphate buffer, pH 7.0, 140 mM NaCl, 0.05% Triton X-100), denatured at 95 °C for 10 min, brought to 300 µL MB, and precipitated with 0.5 μg anti-5mC or 5hmC antibodies (as used in immunofluoresence) overnight at 4 °C. Separately, 10 µL per IP of Protein A and G Dynabeads (Invitrogen) were blocked with 2 mg BSA and 2 mg yeast tRNA in 1 mL MB overnight at 4 °C. Beads and IPs were combined the next day and rotated for 3 h at 4 °C. Samples were washed four times in MB, eluted twice in 100 µL elution buffer (0.1 M NaHCO3, 1% SDS, 1 mM EDTA) by shaking at 37 °C for 15 min, phenol:chloroform:isoamyl-extracted, ethanol-precipitated, and resuspended in 50 µL TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA). Samples were then amplified with Illumina single-read or TruSeq PCR primers for 15 cycles using Phusion DNA polymerase (New England Biolabs). DIP-seq results represent averages of biological replicates unless otherwise noted.
qPCR.
DIP libraries were prepared as above and analyzed using Maxima SYBR Green (Fermentas) and the primers listed in Dataset S4. For every DIP-qPCR experiment presented here, the results represent averages from biological replicates.
RNA-seq.
Total RNA was isolated from a given cell population or tissue using TRIzol (Invitrogen) followed by TURBO DNase treatment (Ambion). mRNA sequencing libraries were prepared using either SuperScript III reverse transcription using a poly(dT) primer (Invitrogen) followed by addition of adaptors using standard techniques or FastTrack MAG Micro mRNA isolation (Invitrogen) followed by ScriptSeq (Epicentre). Control (OMP-GFP) and Tet3-tg (OMP-tTA/tetO-Tet3-EGFP) RNA-seq samples were prepared from rRNA-depleted total RNA (Invitrogen, RiboMinus) using ScriptSeq v2. Samples were amplified for 15 cycles using Phusion DNA polymerase (New England Biolabs) and Illumina single-read or ScriptSeq primers. All of the RNA-seq results represent averages of biological duplicates unless otherwise noted.
Sequencing Analysis.
A full description of the sequencing analysis can be found in SI Materials and Methods. In addition, all python and R scripts as well as R markdown files used to generate key figures can be found at https://github.com/bradleycolquitt/seqAnalysis/PNAS.
TetO-Tet3-ires-EGFP Transgenic Line.
Full-length Tet3 was PCR-amplified from MOE cDNA and cloned into the pTRE2 vector (Clontech) containing ires-EGFP. Transgenic lines were generated by pronuclear microinjection of the linearized construct into FVB founders. Transgene expression was driven by crossing into the OMP-tTA line (34).
In Vitro Transcription.
Modified promoter and transcribed regions were amplified from the HeLa Scribe positive control (Promega) using 5mC or 5hmC dNTP mixes (Zymo). Mixed modification templates were obtained by digesting both fragments with BglII, immobilizing the biotinylated promoter with streptavidin conjugated to magnetic beads (Invitrogen, M-280), ligating promoters and transcribed regions (T4 DNA Ligase, NEB), and magnetically pulling down the ligation products. In vitro transcription was performed on these products using the HeLa Scribe Kit (Promega) with α-32P-UTP. Reactions were purified and then run on a 6% acrylamide TBE-urea (89 mM Tris, 89 mM boric acid, 2 mM EDTA, 7 M urea, pH 8.3) gel.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) (DP2 OD006667, R01MH091661, Rett Syndrome Trust). B.M.C. was supported by the National Science Foundation (Graduate Research Fellowship). G.B. and W.E.A. were supported, in part, by NIH Grant 5R01MH086920. G.B. was supported by the Pew Scholars Program in the Biomedical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (GSE38604 and GSE43617).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302759110/-/DCSupplemental.
References
- 1.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruzov A, et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21(9):1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25(7):679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szulwach KE, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7(6):e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 16.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147(7):1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10(6):720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- 21.Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400(4):469–486. [PubMed] [Google Scholar]
- 22.Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151(4):724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clowney EJ, et al. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res. 2011;21(8):1249–1259. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macdonald JL, Verster A, Berndt A, Roskams AJ. MBD2 and MeCP2 regulate distinct transitions in the stage-specific differentiation of olfactory receptor neurons. Mol Cell Neurosci. 2010;44(1):55–67. doi: 10.1016/j.mcn.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Degano AL, Pasterkamp RJ, Ronnett GV. MeCP2 deficiency disrupts axonal guidance, fasciculation, and targeting by altering Semaphorin 3F function. Mol Cell Neurosci. 2009;42(3):243–254. doi: 10.1016/j.mcn.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shykind BM, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117(6):801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 33.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 34.Yu CR, et al. Spontaneous neural activity is required for the establishment and maintenance of the olfactory sensory map. Neuron. 2004;42(4):553–566. doi: 10.1016/s0896-6273(04)00224-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.