Abstract
The Resistance to Dieldrin gene, Rdl, encodes a GABA-gated chloride channel subunit that is targeted by cyclodiene and phenylpyrazole insecticides. The gene was first characterized in Drosophila melanogaster by genetic mapping of resistance to the cyclodiene dieldrin. The 4,000-fold resistance observed was due to a single amino acid replacement, Ala301 to Ser. The equivalent change was subsequently identified in Rdl orthologs of a large range of resistant insect species. Here, we report identification of a duplication at the Rdl locus in D. melanogaster. The 113-kb duplication contains one WT copy of Rdl and a second copy with two point mutations: an Ala301 to Ser resistance mutation and Met360 to Ile replacement. Individuals with this duplication exhibit intermediate dieldrin resistance compared with single copy Ser301 homozygotes, reduced temperature sensitivity, and altered RNA editing associated with the resistant allele. Ectopic recombination between Roo transposable elements is involved in generating this genomic rearrangement. The duplication phenotypes were confirmed by construction of a transgenic, artificial duplication integrating the 55.7-kb Rdl locus with a Ser301 change into an Ala301 background. Gene duplications can contribute significantly to the evolution of insecticide resistance, most commonly by increasing the amount of gene product produced. Here however, duplication of the Rdl target site creates permanent heterozygosity, providing unique potential for adaptive mutations to accrue in one copy, without abolishing the endogenous role of an essential gene.
The single point mutation in the Resistance to dieldrin (Rdl) gene represents one of the most significant cases of target site resistance to an insecticide yet observed. Cyclodiene resistance was reported in 62% of insecticide resistant species in the 1980s, following widespread use of cyclodiene insecticides, including dieldrin, which started in the 1950s (1). The nature of the genetic target, Rdl, was discovered after dieldrin was discontinued because of the widespread evolution of resistance in many species. Rdl was first discovered in Drosophila melanogaster using a positional cloning approach. High homology to human GABA receptors confirmed it was the first insect ligand-gated chloride channel subunit identified (2–4). A point mutation in the chloride channel pore-lining domain, replacing alanine 301 with serine, was present in all resistant D. melanogaster strains (5). This mutation provided 4,000-fold resistance when homozygous and lower levels of resistance in heterozygotes (2, 6). The homologous mutation was subsequently found in a large number of cyclodiene-resistant species from many insect orders (7–9), as well as a glycine replacement at the homologous site in some resistant strains of Drosophila simulans and other species (5, 10, 11).
Characterization of deficiency lines and inversions in D. melanogaster showed that Rdl is an essential gene (3). Thus, the Ala301 to Ser or Gly mutation in Rdl exhibits unique properties, providing high levels of dieldrin resistance without abolishing the role of the RDL receptor (12). Electrophysiological studies showed that the 301 replacement affects cyclodiene sensitivity by two mechanisms: inhibiting direct binding and allosterically modifying the Rdl receptor to disrupt the antagonist-favored conformation (13). These differences in channel properties have little effect on overall fitness in D. melanogaster other than temperature sensitivity. Resistant adults exposed to temperature stress (38 °C) showed delayed recovery indicated by temporarily impaired flight (14). Laboratory studies of D. melanogaster and D. simulans showed no decline in Ser or Gly301 allele frequencies in population cages after 1 y in the absence of cyclodiene insecticide selection (15). More recently, the Ala301 to Ser change was shown to decrease sleep latency (16). Because fitness testing was conducted in the laboratory, the true extent of fitness costs may have been underestimated. Conversely, in the blowfly Lucilia cuprina, field studies have shown that in the absence of dieldrin, the Rdl resistance allele is at a dramatic selective disadvantage (17). Resistant individuals were more severely selected against during overwintering than other points throughout the year (18).
RDL is highly conserved in insects, and the universal nature of the Ala to Ser/Gly resistance mutation exemplifies this conservation. However, an influx of genomic information from insect species has shown that some lineages have multiple Rdl loci, with three copies present in Lepidopteran genomes and two present in the aphid Acyrthosiphon pisum (19–21). Before sequencing the A. pisum genome, two Rdl copies were reported in the peach potato aphids Myzus persicae and M. nicotinae (10). The presence of two copies in all three species suggests it is an ancient duplication present in the ancestral aphid lineage.
Gene amplification has previously been implicated as a major evolutionary avenue to attaining insecticide resistance, primarily in the form of increased gene expression of detoxification enzymes (22). Organophosphorus chemical resistance in the mosquito, Culex pipiens, is mediated by esterase overproduction due to regulatory changes and 250-fold increased copy number of esterase B3 (23). In D. melanogaster, an allelic series at the cytochrome P450 Cyp6g1 locus involves a gene duplication and a variety of transposable element (TE) insertions. The most derived alleles are correlated with increased enzyme production and multiinsecticide resistance, including dichlorodiphenyltrichloroethane (DDT) (24).
Here we report the occurrence of a recent duplication spanning the genomic region of Rdl in D. melanogaster. Because Rdl is an essential gene (3) and a major insecticide target site for cyclodiene and phenylpyrazole insecticides, copy number variation allows for evolutionary flexibility, where adaptive mutations may accumulate in one copy, whereas WT function is maintained in the other. The duplication creates a situation of permanent heterozygosity, providing advantages in the presence and absence of insecticide.
Results
Identification of the Rdl Gene Duplication.
We examined the available genome sequence for the Rdl gene in 168 D. melanogaster lines [The Drosophila Genomic Reference Panel (DGRP) (25)]. The DGRP was established in 2003 from an outbred population collected at a farmers market in Raleigh, NC. Isofemale lines were inbred for 20 generations (25). Among the inbred lines, there were 10 nonsynonymous polymorphisms in Rdl (Table S1; Fig. S1). The Rdl resistance mutation, Ser301, was present in four lines (Ral-317, Ral-318, Ral-378, and Ral-491); however, in the first three, the mutation was heterozygous with Ala. These three lines also contained a second heterozygous, nonsynonymous mutation, Met360 to Ile. Met360 is one of four RNA-editing locations in Rdl. Editing to Val occurs in 10% of adult head RdlBD transcripts, where RdlBD is the most common splice isoform of four alternately spliced transcripts (26).
Genomic heterozygosity within an inbred line may indicate copy number variation (27). We confirmed Ala301/Ser301 heterozygosity by HaeII restriction digest of PCR products from Ral-318 and Ral-378 individuals (Materials and Methods; Fig. S2). Residual heterozygosity still occurs in these inbred lines (25), so to distinguish this from our prediction that the Rdl gene was duplicated, we followed the inheritance of the variants. If they were alleles, alternate segregation would be observed, whereas if it was a duplication, they would cosegregate. Ral-318 and Ral-378 were crossed to an Rdl WT line, w1118. F1 offspring were tested for genotype at the 301 site using the HaeII restriction digestion assay. One hundred percent were heterozygous for the Ala301/Ser301 polymorphism (n = 32; Fig. S2), indicating cosegregation of alleles and supporting the duplication hypothesis.
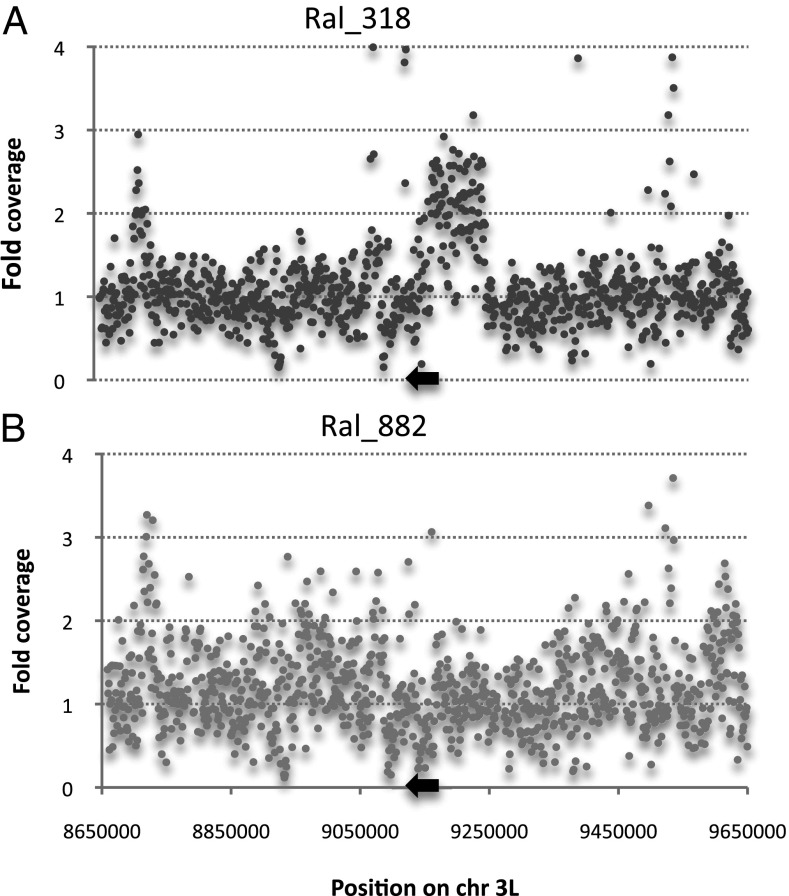
The assembled genome sequence Ral-318 and Ral-378 showed regions of heterozygosity spanning ∼110 kb surrounding Rdl. Genome coverage showed a greater number of sequence reads across the putative duplication region compared with adjacent regions of the 3L chromosome arm (Fig. 1). Alignment of reads in the region revealed the putative duplication arrangement. The first breakpoint occurred at the site of a Roo TE long terminal repeat (LTR) in intron 7 of nervous wreck (nwk), the gene directly 5′ of Rdl. The second breakpoint occurred 113 kb downstream, the site of a second Roo TE LTR in intron 1 of the glutamate receptor IB (Glu-RIB) locus. The rearrangement results in two tandemly arranged copies of Rdl and surrounding genes within the 113-kb duplication (Fig. 2). Five genes were fully duplicated across the 1-kb region, as well as partial duplication of the 3′ portion of nwk and the 5′ portion of Glu-RIB (Fig. 2). Including Rdl Ala301/Ser and Met360/Ile, 15 heterozygous nonsynonymous differences were identified between the duplicated regions (Table S2).
Fig. 1.
Genome sequence coverage of 1 Mb of Chr3L spanning the Rdl locus in DGRP sequenced lines. (A) Duplication line Ral-318 and (B) single copy line Ral-882. Values are normalized against the average read depth for each line. The black arrow indicates the approximate position of Rdl. Fold coverage is greater than 1 across the putative duplication region for Ral-318 compared with the single copy line 882. Similar results were observed for Ral-378.
Fig. 2.
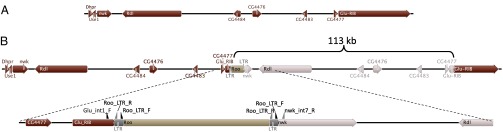
Topology of the duplication. (A) Genomic structure surrounding the Rdl locus in reference single-copy strains. (B) Duplication structure in Ral-317, -318, and -378. The 5′ duplication breakpoint occurs within intron 7 of nwk, the gene directly upstream of Rdl; 113 kb of sequence is duplicated, ending in intron 1 of Glu-RIB. Five genes are completely duplicated (Table S2), and the 3′ portion of nwk and 5′ portion of Glu-RIB are partially duplicated. Between the partial Glu-RIB and nwk segments remains a Roo TE, as indicated by LTRs and long PCR products (Fig. S3). The region surrounding the Roo TE is shown in greater detail, indicating position of primer binding sites (Table S3).
Six primers were designed to regions of nwk, Glu-RIB, and the Roo LTR (Table S3). Combinations therein and sequencing of positive products confirmed the predicted duplication topology. The presence of an internal Roo element within the duplication structure was confirmed with long PCR spanning the Glu-RIB intron 1 to nwk intron 7 (Fig. S3; Fig. 2). A feature of Roo TEs is insertion site duplication, where 5 bp of genomic DNA is duplicated at either side of the point where the TE inserts (28). When ectopic recombination occurs between two different TE insertions, the flanking sequences should differ on each side of the TE. We examined the genomic sequence in Ral-317, -318, and -378 at both Roo insertion sites. We found the nwk-Roo had a different 5-bp insertion site duplication to the Glu-RIB-Roo (AATCT and ACCTG, respectively).
The primers designed to detect the nwk-Roo TE also amplified a product from the RdlR MD-RR line, first isolated in 1990 (29), suggesting this line carried a Roo TE LTR at this site. Sequencing this product showed the same 5-bp target site duplication adjacent to the Roo LTR (AATCT) as Ral-317, -318, and -378. Previous characterization of RdlR MD-RR showed that it contained both Ser301 and Ile360 replacements (6), indicating a common haplotype to the Ral lines. In contrast, the nwk-Roo product was not amplified in another single-copy Ser301 line from the DGRP, Ral-491, nor did this line contain the Met360 to Ile replacement. Long PCR was performed on RdlR-MDRR to assess whether a duplication was also present, although we were unable to amplify the nwk-Roo-GluRIB product (Fig. S3). Sequencing of introns and cDNA from multiple regions of Rdl revealed no heterozygous SNPs, confirming it was homozygous for Ser301 and lacked extensive heterozygosity as seen in the duplication lines.
Both Copies of Rdl Are Expressed.
cDNA was produced from RNA extracted from heads of 1-d-old adults from Ral-318 and -378. The Rdl exon 7 and 8 region, spanning the 301 and 360 sites, was amplified and cloned. Forty-five individual clones were sequenced to identify whether both copies of Rdl were expressed and if the two derived nonsynonomous variants were found in the same copy. This tissue and life stage were chosen to determine whether Met360 to Val RNA editing still occurred correctly in Ile360 mutants (26).
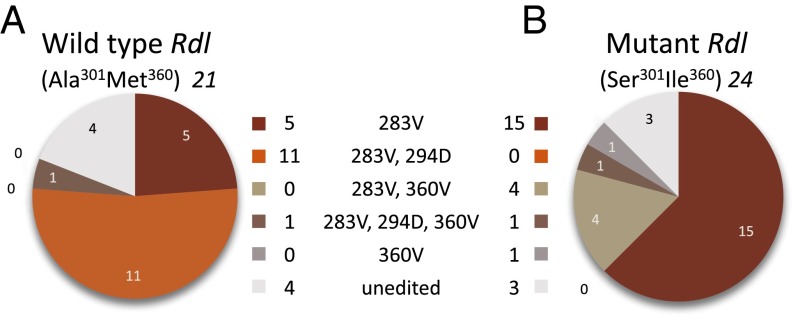
The clones fell into two categories: one characterized by the presence of the Ser301 and Ile360 mutations in the same transcript, and the other category was WT, indicating that both copies of duplicated Rdl are expressed. The frequency of sequences containing the Ser301Ile360 mutations was 24 of 45 (53%), indicating approximately the same expression level as the WT copy (21 of 45, 47%; χ2 = 0.2; P = 0.655; Table S4). The expressed clones showed variation in RNA editing between the two copies (Fig. 3; Table S5). Editing of codon 360 to Val occurred in Ser301Ile360 mutants in 6 of 24 mutant clones as opposed to 1 of 21 WT clones, although this difference was not significant (Fisher exact test, P = 0.10). There was a significant loss of Asn294 to Asp editing in the Ser301Ile360 mutant, with 12 clones edited at the 294 codon in WT, but only 1 edited in mutant clones (Fisher exact test, P = 0.00013).
Fig. 3.
RNA editing frequency. Frequency of RNA editing at three of the four known editing sites (Ile283 to Val, Asn294 to Asp, Met360 to Val) in Rdl transcripts within the amplified exon 7–8 region, spanning codons 301 and 360. (A) Editing in WT Ala301Met360 sequenced clones compared with (B) mutant Ser301Ile360 clones. Frequencies are shown in Tables S4 and S5.
Resistance to Dieldrin.
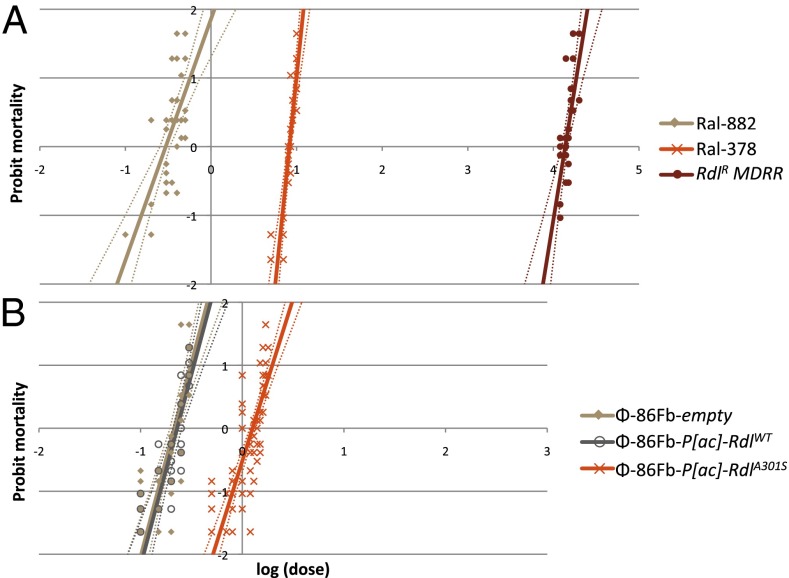
Dieldrin toxicity assays were conducted on adults from three naturally derived lines: RdlR MD-RR (single copy; Ser301); Ral-378 (duplication; Ala301Met360/Ser301Ile360); and Ral-882, (single copy; WT Rdl). Dosage–mortality curves were generated (Fig. 4A). Compared with WT, RdlR showed >45,000-fold resistance to dieldrin. Ral-378, containing both Rdl genotypes, showed significant resistance, with 27-fold survival over WT (Table S6).
Fig. 4.
Dosage mortality response curves for dieldrin. (A) Natural populations: Ral- 882 (WT, Ala301Met360); Ral-378 (duplication, Ala301Met360/Ser301Ile360); and RdlR MD-RR (single copy, Ser301Ile360). (B) Transgenic lines: Φ-86Fb-empty control, Φ-86Fb-P[ac]-RdlWT (WT Rdl Ala301 insertion) and Φ-86Fb-P[ac]-RdlA301S (mutant Rdl Ser301 insertion). Fold changes are shown in Table S5.
To isolate the contribution of the Rdl Ser301 mutation to resistance from within the 113-kb genomic duplication, a transgenic model was generated using the P[acman]-attB vector. The transgenic construct incorporated 55.7 kb of Rdl genomic DNA, but excluded all other genes present in the duplication. Both WT Ala301 and mutant Ser301 lines were generated in a controlled genetic background, differing by only a single base pair mutation. Three lines were tested for dieldrin resistance: Φ-86Fb-empty [attP 86Fb line (30)]; Φ-86Fb-P[ac]-RdlWT (Rdl Ala301 insertion); and Φ-86Fb-P[ac]-RdlA301S (Rdl Ser301 insertion). Thus, the insert lines replicated the Rdl duplication, containing two copies of Rdl: one transgenic and one endogenous copy.
Φ-86Fb-empty and Φ-86Fb-P[ac]-RdlWT lines showed no difference in survival (Fig. 4B; Table S6), indicating an extra copy of WT Rdl did not modify resistance in the transgenic Φ-86Fb-P[ac]-RdlWT line. However, Φ-86Fb-P[ac]-RdlA301S had increased survival, with sixfold resistance to dieldrin (Fig. 4B; Table S6). The moderate resistance observed highlighted the semidominant nature of Rdl resistance (2), with combined effects of transgenic Ser301 and endogenous WT Ala301.
Recovery from Heat Shock.
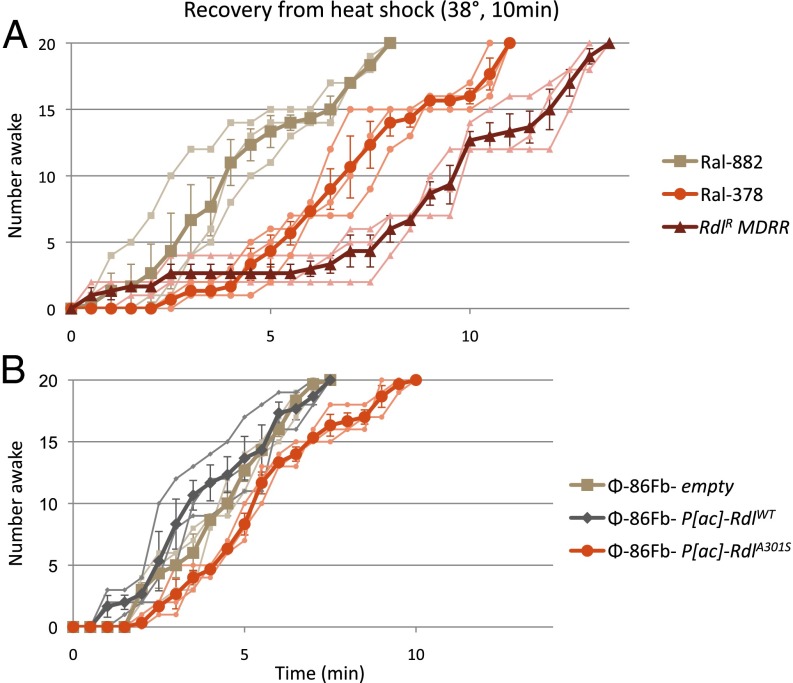
To examine temperature sensitivity associated with the Rdl Ser301 mutation (14), heat shock recovery tests were conducted in lines derived from natural populations and transgenic lines examined previously for dieldrin resistance. Adult females were exposed to 38 °C for 10 min, and recovery time was observed. Among the naturally derived lines, genotype had a significant affect on recovery time [F(2,6) = 41.861, P < 0.0001]. Pairwise comparison showed a significant difference between WT Ral-882 and Ral-378 (Ala301Met360/Ser301Ile360) (P = 0.004) and Ral-882 and RdlR MD-RR (Ser301) (P < 0.0001), with the Ral-378 and RdlR comparison yielding P = 0.051 (Bonferroni correction). RdlR was the slowest to recover from heat shock, with a median recovery time of 10 min. Ral-378 showed an intermediate recovery of 7 min, and WT Ral-882 showed the fastest recovery time (4 min; Fig. 5A). Genotype also significantly affected recovery time in the transgenic lines [F(2,6) = 19.767, P = 0.002]. Φ-86Fb-empty and Φ-86Fb-P[ac]-RdlWT were not significantly different from each other (P = 0.585); however, Φ-86Fb-P[ac]-RdlA301S had a significantly increased temperature recovery time compared with both controls (P = 0.011 and P = 0.003, respectively; Fig. 5B), with a median recovery time of 5.5 min compared with 4.5 (Φ-86Fb-empty) and 3.5 min (Φ-86Fb-P[ac]-RdlWT).
Fig. 5.
Recovery from heat shock. (A) Ral-882 (WT); Ral-378 (duplication, Ala301Met360/Ser301Ile360); and RdlR MD-RR (single copy, Ser301). Three individual replicates are graphed with the average (±SEM) superimposed for each line. (B) Transgenic lines: Φ-86Fb-empty, Φ-86Fb-P[ac]-RdlWT, and Φ-86Fb-P[ac]-RdlA301S.
Discussion
Investigating the genome sequence of 168 naturally derived inbred lines of D. melanogaster revealed a 113-kb tandem duplication encompassing the major insecticide target site, Rdl. Two nonsynonymous polymorphisms were present between the duplicated copies: at the insecticide resistance site, Ala301 to Ser, and an RNA-edited site, Met360 to Ile. Both mutations were found in the same copy, expressed at equivalent levels to the WT copy. The duplication and associated polymorphisms have implications in posttranscriptional RNA editing, dieldrin resistance, and heat shock recovery.
When resistance is dominant, heterozygotes often present intermediate phenotypes in both fitness and resistance to insecticide. This offset may allow resistant alleles to persist in populations in the absence of insecticide (31). Duplications enable the maintenance of permanent heterozygosity and have previously been shown to modify fitness in insecticide resistance context. In Culex pipiens, a point mutation in the acetylcholine esterase-1 (Ace-1) locus is associated with high levels of resistance to organophosphates; however, this mutation reduces Ace-1 activity by 60%, incurring a significant fitness cost (22). Combining a resistant and susceptible allele by gene duplication offsets part of the fitness cost (32). A fitness offset may also be the case for Rdl in the duplicated lines, which show expression of both the resistant Ser301Ile360 copy and the Ala301Met360 WT copy at equal levels and subsequently display intermediate levels of resistance (Fig. 4) and heat shock recovery (Fig. 5). Given that D. melanogaster is not generally considered to be a pest, it is likely to be incidentally exposed to insecticides at a lower frequency and concentrations that would be the case for pest species. The intermediate resistance associated with the duplication may be protective against such exposure.
Gene duplications associated with insecticide resistance have been predominantly involved in increased gene expression of metabolic enzymes resulting in enhanced insecticide detoxification (24, 33, 34). However, beyond enhanced expression, gene duplication in a target site provides a unique opportunity for the redundant copy of the duplicate pair to accumulate mutations. Some mutations may be adaptive in certain environmental scenarios that are otherwise detrimental to the original function of the gene (33). Gene duplications are one of the major adaptive forces in eukaryote evolution, but the persistence of duplicate copies is not favored unless each copy acquires a specific role. After duplication occurs, the majority of duplicate pairs result in one gene undergoing rapid deleterious mutations leading to pseudogenization (35). However, a duplicate pair is preserved if beneficial mutations accrue. Either one member acquires mutations that provide a new function (neofunctionalization) or the two copies share the original function of the progenitor gene by subfunctionalizing (35, 36).
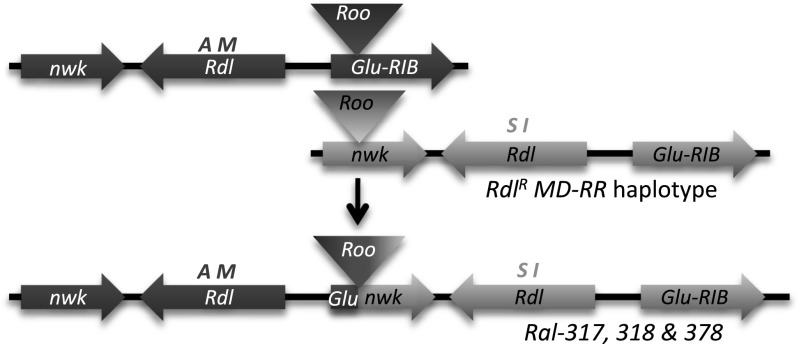
Duplications may arise via a number of mechanisms including unequal crossing over or replication slippage (32). For the Rdl duplicated lines, our data suggest ectopic recombination in neighboring Roo TEs initiated the genome rearrangement. TEs and genome rearrangements have previously been implicated in adaptation to environmental pressures such as insecticide resistance (24, 37, 38). TEs contribute substantially to adaptive evolution and have the capacity to generate deletions, duplications, and regulatory changes with wide-ranging phenotypic effects that cannot be achieved by point mutations (39). Roo elements are the most common TE in the D. melanogaster genome (40), and are frequent initiators of chromosomal rearrangements such as duplications and deletions (41). PCR analysis indicated a Roo LTR was present in intron 7 of nwk in the original RdlR MD-RR line. The RdlR haplotype may therefore be implicated as a precursor for the duplication. Recombination between the RdlR nwk-Roo haplotype and a downstream Glu-RIB-Roo haplotype would result in the generation of the 113-kb tandem arrangement we observe in Ral-317, -318, and -378 (Fig. 6). Observation of different insert site duplication sequences flanking the Roo LTRs supports our theory of ectopic recombination between two different Roo TEs in generating this duplication structure. Our diagnostic PCRs detect upstream nwk-Roo elements at a low frequency in populations of D. melanogaster from Australian populations, although no nonduplicated populations have yet been identified with the corresponding downstream Glu-RIB-Roo (SI Text).
Fig. 6.
Schematic showing a putative origin of the duplication in the Ral lines, based on ectopic recombination of a heterozygous RdlR MD-RR/WT line, containing the nwk intron 7 Roo TE and the Glu-RIB intron 1 Roo TE in trans.
Permanent Heterozygosity, Resistance, and Temperature Sensitivity.
A functional GABA receptor consists of five subunits. Pentamers formed in homozygous Ser301 mutants contain only resistant subunits; however, in duplication lines, Ala301 and Ser301 alleles from the two Rdl copies would result in heteromeric RDL receptors containing a mixture of resistant and susceptible subunits. The same receptor composition also occurs in Φ-86Fb-P[ac]-RdlA301S, containing transgenic Ser301 and endogenous Ala301 Rdl, and is functionally equivalent to single copy Ala301/Ser heterozygotes, with heteromeric receptors and semidominant, intermediate resistance levels (2).
The role of Ser301 in resistance is well established (5, 6). However, to distinguish it as the causal mutation for the phenotypes assayed, transgenic lines containing a 55.7-kb Rdl-only duplication were generated. The transgenic construct eliminated other genes and mutations contained in the 113-kb natural duplication, and isolated the Ser301 mutation in the absence of other nonsynonymous replacements including Ile360, present in duplication lines and the original RdlR MD-RR line (6). A distinct resistance and temperature sensitive phenotype emerged, even when eliminating background factors in the natural populations, which may consist of many generations of coadapted modifications. The 14 other nonsynonymous changes in the 113-kb duplication, or indeed other genes elsewhere in the genome, may have an additive effect on the two phenotypes assayed here in Ral-378. Additionally, reduced RNA editing efficiency at codon 294, and increased editing at codon 360 (Fig. 3; Table S5) may affect RDL receptor properties and phenotypes alongside the Ser301 and Ile360 mutations. However, the contribution of Ser301 is verified by the transgenic experiments, where the Rdl insertion lines differ by a single base pair and illustrate the significance of this point mutation in genetically identical lines.
Rdl Copy Number Variation in Insects.
Previous studies reported two copies of Rdl in the peach-potato aphid, Myzus persicae: one copy with serine at the equivalent 301 site and the other containing alanine. Cyclodiene resistance was attributed to a glycine change in the alanine copy (10). The recent release of the pea aphid (A. pisum) genome also revealed two copies of Rdl, with Ala in one copy and Ser in the other (21). In Lepidoptera, three copies of Rdl are present, and in Bombyx mori these copies share 75–91% similarity at the protein level (19). However, one copy has Ala, one has Ser, and the third has a glutamine at the equivalent 301 site (Fig. S4). This site is therefore polymorphic in species with multiple copies of Rdl, suggesting a reduction in functional constraint within an otherwise highly conserved domain. It may explain to some extent why the 301 site is amenable to resistance mutations in single copy species.
In Drosophila, this is the first example of copy number variation at the Rdl locus. Identification of the present D. melanogaster duplication was facilitated by the availability of high-depth genome sequences of a large number of inbred lines expected to be homozygous. We did not detect the duplication in a survey of Australian populations (SI Text) or in the genome sequences available from 139 African D. melanogaster lines (27), suggesting a rare event led to the arrangement discovered in the DGRP. It is possible that other duplications exist at the Rdl locus, which were not detectable by our specific diagnostic PCRs.
Since the cessation of cyclodiene insecticide use, the frequency of the Rdl Ser301 resistance allele varies from undetectable levels in houseflies (42) to relatively high levels in cockroaches and fleas (9, 43). The high stability of these compounds may result in exposure to persistent residues in the environment, maintaining selection for the resistance allele in natural populations. Additionally, increasing household and field use of phenylpyrazole insecticides, which also target the insect RDL GABA receptor, may select for resistance alleles (9, 44). In the early 1990s, the Ser301 mutation frequency in populations of D. melanogaster in the United States was estimated at 1% (45). This value is similar to the current estimate of 2.4% in the DGRP lines extracted from a population North Carolina and 3% in our survey of Australian populations (25) (SI Text).
The duplication identified here creates adaptive potential for accumulation of resistance mutations in one copy that may be detrimental to the endogenous role of Rdl. Although the Ala301 to Ser/Gly replacement does not result in lethality and in many species has a negligible fitness effect (15), other replacements may result in enhanced resistance but have a greater impact on fitness. Ser301 provides low resistance to the phenylpyrazole fipronil that does not impact the use of this insecticide in the field (9, 43, 46, 47). However, fipronil-resistant strains isolated from two species of planthopper have been shown to contain an Ala to Asn, rather than Ser or Gly, mutation at the equivalent 301 site, found only in the heterozygous state (48, 49). The lack of homozygous Asn individuals was proposed to be a result of lethality, supported by the reduced GABA median effective concentration (EC50) observed in electrophysiological data (49). Mutations resulting in lethality can only be viable in a heterozygous state, or more permanently, in a duplication of the type described in this study. Continuing use of phenylpyrazole insecticides may result in increased prevalence of Asn replacements and enhanced resistance in other relevant species. Duplications generating permanent heterozygosity would facilitate the maintenance and spread of such mutations.
Conclusion
Duplications are a major source of selectable genetic variation and provide evolutionary flexibility for adaptation to new functional niches. Here we characterize a recent duplication in a major insecticide target site, Rdl, in D. melanogaster. The duplication is associated with insecticide resistance and RNA editing mutations in one copy and one WT copy. The resulting expression of both WT and mutant transcripts results in intermediate resistance to dieldrin while reducing a temperature-sensitive fitness cost, creating a functional, permanent, and heritable heterozygosity. This discovery highlights considerations for continued use of insecticides targeting Rdl receptors in insects. A number of pest species exhibit Rdl copy number variation and therefore have flexibility to accumulate resistance mutations while retaining sufficient WT function. In species such as D. melanogaster, that are ancestrally single copy for Rdl, evidence of novel copy number variation at this target site provides a platform for future adaptation to environmental pressures, such as the ongoing use of insecticides.
Materials and Methods
Drosophila melanogaster Lines.
RdlR MD-RR (Bloomington: 1675) was originally isolated in Maryland in 1990, with 4,000-fold dieldrin resistance, and was used in the initial characterization of the Rdl Ser301 mutation (2, 6, 29). The sequence of Rdl from 168 DGRP genomes (25) was extracted using reference genome coordinates, and polymorphisms were annotated manually. Sequence coverage and read depth of the Rdl genomic region was analyzed using MAQ (http://maq.sorceforge.net/maq-man.shtml). Two DGRP lines were used in phenotypic analysis of dieldrin resistance and heat shock recovery: one randomly selected Rdl WT line, Ral-882, and one duplication line, Ral-378 (Bloomington: 28255, 28187). An additional DGRP duplication line, Ral-318 (Bloomington: 28168), was used alongside Ral-378 to genetically verify the presence of a duplication by F1 analysis with w1118 (Bloomington: 3605), a control line containing WT Rdl, used in reciprocal crosses with the putative duplication lines (Fig. S2). Transgenic integration of the Rdl locus was performed using the attB-P[acman]-ApR plasmid into chromosome III of the Φ-86Fb-attP line (Bloomington: 24749). The Φ-86Fb-empty line was used as a control for the two generated insertion lines: Φ-86Fb-P[ac]-RdlWT (WT Rdl insertion) and Φ-86Fb-P[ac]-RdlA301S (Ser301 mutant Rdl insertion).
Generation of P[acman]-Rdl Lines.
Recombineering was carried out according to the methods used in Venken et al. (50). A 55.7-kb fragment surrounding the Rdl locus was obtained by the cloning of left and right arms (Table S3) into the attB-P[acman] vector and homologous recombination with BAC-1E12 (RP98 library, Drosophila Genomic Resources Centre). The Ala301 to Ser replacement was generated using the galK counter selection method (51). Both WT and A301S constructs were integrated into Φ-86Fb-attP, and transgenics were identified by eye color and PCR confirmation. Expression of the Ser301 transgene was detected by HaeII restriction digest of exon7 from cDNA generated from the Φ-86Fb-P[ac]-RdlA301S line.
Generation of RNA, cDNA, Cloning, and Sequencing.
RNA was extracted from 1-d-old adult heads from lines Ral-378 and Ral-318 using TRIzol reagent, and cDNA was synthesized with SuperScript III Reverse Transcriptase [oligo(dT)20; Invitrogen], following the manufacturer’s instructions. PCR primers were designed to amplify exon 7 and 8, spanning the two polymorphisms at codons 301 and 360 (Rdl_Ex7_F/Ex8_R; Table S3). Products were cloned into pGEM T-easy (Promega) before sequencing (Macrogen).
Diagnostic HaeII Restriction Digest.
The Ala301 to Ser polymorphism occurs in exon 7 of Rdl as a result of a T to C base pair substitution, removing a HaeII restriction site (5); 268 bp of exon 7 was amplified (Rdl_Ex7_F/R (Table S3), and HaeII digest was used to differentiate between Ser301 (205 and 63 bp), Ala301 (148, 57, and 63 bp), or heterozygous lines (205, 148, 63, and 57 bp; Fig. S2).
Insecticide Screens.
Dieldrin dosage–mortality analysis was carried out according to the same methods described in a DDT 24-h adult contact assay, where dieldrin powder was dissolved in acetone and coated the inside of scintillation vials (52). Five replicates of at least five doses per strain were tested to generate dose–response curves using PriProbit (ver.1.63; Fig. 4) (53). Resistance ratios and 95% CIs were estimated from dosage–mortality curves as previously described (54) (Table S6).
Temperature Sensitivity Screens.
Recovery from heat shock was conducted with minor variations to tests performed in ref. 14. Flies were reared at room temperature (22–24 °C). Three replicates of 20 5- to 8-d-old adult females were placed into glass vials equilibrated in a 38 °C water bath and heated for 10 min. Unconscious flies were tipped onto a 2.5-cm-diameter circular arena at room temperature, and flies departing from this area were scored every 30 s for 15 min (Fig. 5). Proportional data were arcsine transformed, and repeated-measures ANOVA was used to assess the significance of the effects of genotype over time. A post hoc test using the Bonferroni correction was used to compare differences between genotypes.
Supplementary Material
Acknowledgments
We thank Tom Harrop for valuable comments on the manuscript and members of the Social Insects laboratory for advice on statistics. This research was funded by an Australian Research Council grant (to P.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311341110/-/DCSupplemental.
References
- 1. Georghiou GP (1986) The magnitude of the resistance problem. Pesticide Resistance: Strategies and Tactics for Management (National Academy Press, Washington, DC)
- 2. ffrench-Constant RH, Roush RT (1991) Gene mapping and cross-resistance in cyclodiene insecticide-resistant Drosophila melanogaster. Genet Res 57(1):17–21. [DOI] [PubMed]
- 3. ffrench-Constant RH, Mortlock DP, Shaffer CD, MacIntyre RJ, Roush RT (1991) Molecular cloning and transformation of cyclodiene resistance in Drosophila: An invertebrate γ-aminobutyric acid subtype A receptor locus. Proc Natl Acad Sci USA 88(16):7209–7213. [DOI] [PMC free article] [PubMed]
- 4. ffrench-Constant RH, Rocheleau T (1992) Drosophila cyclodiene resistance gene shows conserved genomic organisation with vertebrate γ-aminobutyric acidA receptors. J Neurochem 59(4):1562–1565. [DOI] [PubMed]
- 5. ffrench-Constant RH, Steichen JC, Rocheleau TA, Aronstein K, Roush RT (1993) A single-amino acid substitution in a g-aminobutyric acid subtype A receptor locus is associated with cyclodiene insecticide resistance in Drosophila populations. Proc Natl Acad Sci USA 90(5):1957–1962. [DOI] [PMC free article] [PubMed]
- 6. ffrench-Constant R, Rocheleau TA, Steichen JC, Chalmers AE (1993) A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature 363(6428):449–451. [DOI] [PubMed]
- 7.Thompson M, Steichen JC, ffrench-Constant RH. Conservation of cyclodiene insecticide resistance-associated mutations in insects. Insect Mol Biol. 1993;2(3):149–154. doi: 10.1111/j.1365-2583.1993.tb00134.x. [DOI] [PubMed] [Google Scholar]
- 8.Andreev D, Kreitman M, Phillips TW, Beeman RW, ffrench-Constant RH. Multiple origins of cyclodiene insecticide resistance in Tribolium castaneum (Coleoptera: Tenebrionidae) J Mol Evol. 1999;48(5):615–624. doi: 10.1007/pl00006504. [DOI] [PubMed] [Google Scholar]
- 9.Bass C, Schroeder I, Turberg A, Field LM, Williamson MS. Identification of the Rdl mutation in laboratory and field strains of the cat flea, Ctenocephalides felis (Siphonaptera: Pulicidae) Pest Manag Sci. 2004;60(12):1157–1162. doi: 10.1002/ps.937. [DOI] [PubMed] [Google Scholar]
- 10.Anthony N, Unruh T, Ganser D, ffrench-Constant R. Duplication of the Rdl GABA receptor subunit gene in an insecticide-resistant aphid, Myzus persicae. Mol Gen Genet. 1998;260(2–3):165–175. doi: 10.1007/s004380050882. [DOI] [PubMed] [Google Scholar]
- 11.Du W, et al. Independent mutations in the Rdl locus confer dieldrin resistance to Anopheles gambiae and An. arabiensis. Insect Mol Biol. 2005;14(2):179–183. doi: 10.1111/j.1365-2583.2005.00544.x. [DOI] [PubMed] [Google Scholar]
- 12. ffrench-Constant RH, Pittendrigh B, Vaughan A, Anthony N (1998) Why are there so few resistance-associated mutations in insecticide target genes? Phil Trans R Soc Lond B 353(1376):1685–1693. [DOI] [PMC free article] [PubMed]
- 13.Zhang H-G, ffrench-Constant RH, Jackson MB. A unique amino acid of the Drosophila GABA receptor with influence on drug sensitivity by two mechanisms. J Physiol. 1994;479(Pt 1):65–75. doi: 10.1113/jphysiol.1994.sp020278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. ffrench-Constant RH, Steichen JC, Ode PJ (1993) Cyclodiene insecticide resistance in Drosophila melanogaster (Meigen) is associated with a temperature-sensitive phenotype. Pesticide Biochem Physiol 46(1):73–77.
- 15.Aronstein K, Ode P. ffrench-Constant RH PCR based monitoring of specific Drosophila (Diptera: Drosophilidae) cyclodiene resistance alleles in the presence and absence of selection. Bull Entomol Res. 1995;85(1):5–9. [Google Scholar]
- 16.Agosto J, et al. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11(3):354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitten MJ, Dearn JM, McKenzie JA. Field studies on insecticide resistance in the Australian sheep blowfly, Lucilia cuprina. Aust J Biol Sci. 1980;33(6):725–736. [PubMed] [Google Scholar]
- 18.McKenzie JA. Selection at the dieldrin resistance locus in overwintering populations of Lucilia cuprina (Wiedemann) Aust J Zool. 1990;38(5):493–501. [Google Scholar]
- 19.Yu L-L, Cui YJ, Lang G-J, Zhang M-Y, Zhang C-X. The ionotropic γ-aminobutyric acid receptor gene family of the silkworm, Bombyx mori. Genome. 2010;53(9):688–697. doi: 10.1139/g10-056. [DOI] [PubMed] [Google Scholar]
- 20.Yuan G, Gao W, Yang Y, Wu Y. Molecular cloning, genomic structure, and genetic mapping of two Rdl-orthologous genes of GABA receptors in the diamondback moth, Plutella xylostella. Arch Insect Biochem Physiol. 2010;74(2):81–90. doi: 10.1002/arch.20361. [DOI] [PubMed] [Google Scholar]
- 21.Dale RP, et al. Identification of ion channel genes in the Acyrthosiphon pisum genome. Insect Mol Biol. 2010;19(Suppl 2):141–153. doi: 10.1111/j.1365-2583.2009.00975.x. [DOI] [PubMed] [Google Scholar]
- 22.Bass C, Field LM. Gene amplification and insecticide resistance. Pest Manag Sci. 2011;67(8):886–890. doi: 10.1002/ps.2189. [DOI] [PubMed] [Google Scholar]
- 23.Raymond M, Chevillon C, Guillemaud T, Lenormand T, Pasteur N. An overview of the evolution of overproduced esterases in the mosquito Culex pipiens. Philos Trans R Soc Lond B Biol Sci. 1998;353(1376):1707–1711. doi: 10.1098/rstb.1998.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt JM, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;6(6):e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackay TFC, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482(7384):173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones AK, et al. (2009) Splice-variant- and stage-specific RNA editing of the Drosophila GABA receptor modulates agonist potency. J Neurosci 29(13):4287–4292. [DOI] [PMC free article] [PubMed]
- 27.Pool JE, et al. Population genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012;8(12):e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linheiro RS, Bergman CM. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PLoS ONE. 2012;7(2):e30008. doi: 10.1371/journal.pone.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. ffrench-Constant RH, Roush RT, Mortlock D, Dively GP (1990) Isolation of dieldrin resistance from field populations of Drosophila melanogaster (Diptera: Drosophilidae). J Econ Entomol 83(5):1733–1737. [DOI] [PubMed]
- 30.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104(9):3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson TG. Resistance of Drosophila to toxins. Annu Rev Entomol. 2001;46:545–571. doi: 10.1146/annurev.ento.46.1.545. [DOI] [PubMed] [Google Scholar]
- 32.Labbé P, et al. Independent duplications of the acetylcholinesterase gene conferring insecticide resistance in the mosquito Culex pipiens. Mol Biol Evol. 2007;24(4):1056–1067. doi: 10.1093/molbev/msm025. [DOI] [PubMed] [Google Scholar]
- 33.Devonshire AL, Field LM. Gene amplification and insecticide resistance. Annu Rev Entomol. 1991;36:1–23. doi: 10.1146/annurev.en.36.010191.000245. [DOI] [PubMed] [Google Scholar]
- 34.Wondji CS, et al. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 2009;19(3):452–459. doi: 10.1101/gr.087916.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levasseur A, Pontarotti P. The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Biol Direct. 2011;6:11. doi: 10.1186/1745-6150-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302(5649):1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt JM, Robin C. An adaptive allelic series featuring complex gene rearrangements. PLoS Genet. 2011;7(10):e1002347. doi: 10.1371/journal.pgen.1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 39.González J, Petrov DA. The adaptive role of transposable elements in the Drosophila genome. Gene. 2009;448(2):124–133. doi: 10.1016/j.gene.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Chaux N, Wagner A. Evolutionary dynamics of the LTR retrotransposons roo and rooA inferred from twelve complete Drosophila genomes. BMC Evol Biol. 2009;9:205. doi: 10.1186/1471-2148-9-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montgomery EA, Huang S-M, Langley CH, Judd BH. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: Genome structure and evolution. Genetics. 1991;129(4):1085–1098. doi: 10.1093/genetics/129.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J-R, et al. The A302S mutation in Rdl that confers resistance to cyclodienes and limited cross-resistance to fipronil is undetectable in field populations of house flies from the USA. Pestic Biochem Physiol. 2007;88(1):66–70. [Google Scholar]
- 43.Kristensen M, Hansen KK, Jensen KM. Cross-resistance between dieldrin and fipronil in German cockroach (Dictyoptera: Blattellidae) J Econ Entomol. 2005;98(4):1305–1310. doi: 10.1603/0022-0493-98.4.1305. [DOI] [PubMed] [Google Scholar]
- 44.Cole LM, Nicholson RA, Casida JE. Action of phenylpyrazole insecticides at the GABA-gated chloride channel. Pestic Biochem Physiol. 1993;46(1):47–54. [Google Scholar]
- 45. ffrench-Constant RH (1994) The molecular and population genetics of cyclodiene insecticide resistance. Insect Biochem Molec Biol 24(4):335–345. [DOI] [PubMed]
- 46.Brunet S, Le Meter C, Murray M, Soll M, Audonnet J-C. Rdl gene polymorphism and sequence analysis and relation to in vivo fipronil susceptibility in strains of the cat flea. J Econ Entomol. 2009;102(1):366–372. doi: 10.1603/029.102.0147. [DOI] [PubMed] [Google Scholar]
- 47.Ozoe Y, et al. Fipronil-related hetercyclic compounds: Structure-activity relationships for interaction with γ-aminobutyric acid- and voltage-gated ion channels and insecticidal action. Pestic Biochem Physiol. 2000;66(2):92–104. [Google Scholar]
- 48.Nakao T, Naoi A, Kawahara N, Hirase K. Mutation of the GABA receptor associated with fipronil resistance in the whitebacked planthopper, Sogatella furcifera. Pestic Biochem Physiol. 2010;97(3):262–266. [Google Scholar]
- 49.Nakao T, et al. The A2’N mutation of the RDL gamma-aminobutyric acid receptor conferring fipronil resistance in Laodelphax striatellus (Hemiptera: Delphacidae) J Econ Entomol. 2011;104(2):646–652. doi: 10.1603/ec10391. [DOI] [PubMed] [Google Scholar]
- 50.Venken KJT, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314(5806):1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 51.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33(4):e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daborn P, Boundy S, Yen J, Pittendrigh B, ffrench-Constant R. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol Genet Genomics. 2001;266(4):556–563. doi: 10.1007/s004380100531. [DOI] [PubMed] [Google Scholar]
- 53.Sakuma M. Probit analysis of preference data. Appl Entomol Zool (Jpn) 1998;33(3):339–347. [Google Scholar]
- 54.Robertson JL, Savin NE, Preisler HK, Russell RM. Bioassays with Arthropods. 2nd Ed. Boca Raton, FL: CRC Press; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.