Abstract
Background
Signaling through the endothelin receptor B (EDNRB) is critical for the development of the enteric nervous system (ENS) and mutations in endothelin system genes cause Hirschsprung’s aganglionosis in humans. Penetrance of the disease is modulated by other genetic factors. Mutations affecting retinoic acid (RA) signaling also produce aganglionosis in mice. Thus, we hypothesized that RA and endothelin signaling pathways may interact in controlling development of the ENS.
Methods
Rat immunoselected ENS precursor cells were cultured with the EDNRB ligand endothelin-3, an EDNRB-selective antagonist (BQ-788), and/or RA for 3 or 14 days. mRNA levels of genes related to ENS development, RA- and EDNRB-signaling were measured at 3 days. Proliferating cells and cells expressing neuronal, glial, and myofibroblast markers were quantified.
Results
Culture of isolated ENS precursors for 3 days with RA decreases expression of the endothelin-3 gene and that of its activation enzyme. These changes are associated with glial proliferation, a higher percentage of glia, and a lower percentage of neurons compared to cultures without RA. These changes are independent of EDNRB signaling. Conversely, EDNRB activation in these cultures decreases expression of RA receptors β and γ mRNA and affects the expression of the RA synthetic and degradative enzymes. These gene expression changes are associated with reduced glial proliferation and a lower percentage of glia in the culture. Over 14 days in the absence of EDNRB signaling, RA induces the formation of a heterocellular plexus replete with ganglia, glia and myofibroblasts.
Conclusions
A complex endothelin-RA interaction exists that coordinately regulates the development of rat ENS precursors in vitro. These results suggest that environmental RA may modulate the expression of aganglionosis in individuals with endothelin mutations.
Introduction
During embryonic development, vagal neural crest cells must migrate caudally, proliferate, differentiate and organize into ganglionated plexuses in order to form a fully functional enteric nervous system (ENS) [1,2]. Failure to do so in a spatiotemporally regulated manner results in varying lengths of terminal aganglionosis, which manifests as the common congenital disorder Hirschsprung’s disease. Mutations in genes encoding the endothelin (EDN) and RET (rearranged during transfection) signaling pathways account for the majority of cases of Hirschsprung’s disease [3,4]. Mutations in the genes encoding the EDN receptor B (EDNRB), its ligand EDN3, and the transcription factor SOX10 cause Waardenburg-Shah syndrome (also known as Waardenburg syndrome type IV), comprised of pigment cell abnormalities and Hirschsprung’s disease. The EDNRB gene encodes a G-protein coupled receptor that is expressed on ENS precursors during development [5,6]. EDN3, is a 22 kDa peptide that is activated by the EDN converting enzyme 1 (ECE1) [7,8]. It is expressed in a spatiotemporally controlled manner by the gut mesenchyme, with expression preceding the arrival of precursor cells and continuing during their migration through the hindgut [5,9]. In vitro, EDN3 inhibits neuronal differentiation and stimulates proliferation of enteric neuronal precursors [10,11]. Rodents carrying null mutations in Ednrb exhibit colonic aganglionosis attributable to an early modest reduction in the number of enteric neural crest-derived stem cells and migration failure in the hindgut [6,10–15]. The variable length of aganglionosis in rodents carrying EDN3 or Ednrb mutations and the low penetrance of the Hirschsprung’s phenotype in humans carrying EDN3 or EDNRB mutations is partially explained by studies showing a genetic interactions between Ednrb, Ret, and Sox10 mutations [16,17]. Environmental factors that influence EDNRB signaling in ENS development have not been investigated.
Retinoic acid (RA) is a derivative of dietary vitamin A that is generated from retinaldehyde in its final synthetic step by three distinct retinaldehyde dehydrogenases (RALDH), all of which are expressed in the fetal bowel [18,19]. Retinaldehyde dehydrogenase 2 (RALDH2), is expressed in the gut mesenchyme during development, but its regulation is poorly understood [20,21]. RA forms a complex with its cognate RA receptors (RAR α, β, and γ), translocates to the nucleus, and binds to RA receptor elements encoded in the genome to affect gene transcription [22]. RA is inactivated by the cytochrome P450 26 family of enzymes [23,24]. Cyp26a1 transcripts are detected in the outer mesenchyme of the murine esophagus and stomach during development [25]. Cyp26a1 transcripts are also found in the mesenchyme and ENS of the developing small intestine, and CYP26A1 may be critical for maintaining RA gradients in the developing embryo [21,26]. RA affects multiple processes in ENS development: Excess RA induces a delay in ENS precursor migration into the intestine caudal to the cecum in intestinal explant cultures [27] – a phenomenon that mimics EDNRB signaling deficiency. However, a paucity of RA in utero, induced by maternal dietary deprivation of vitamin A in susceptible rodents, is also associated with aganglionosis [28]. A role for RA in migration is also supported by in vitro evidence that RA modulates neural crest cell polarity and cytoskeletal arrangement. In addition, RA enhances the proliferation of RET+ ENS neuronal precursors and enhances neuronal differentiation of this subset of cells in vitro [21].
EDNRB and RA signaling must be tightly regulated to produce normal ENS development [5,15,29], but their combined effects on the development of the ENS are unknown. Work in other systems suggests that RA reduces EDN signaling. RA suppresses Edn 1 expression in cultured hepatic stellate cells, a prostate cancer cell line, renal glomerular cells and endothelial cells [30–33]. To date, however, a relationship between RA and EDN3 or Ednrb gene expression has not been established. We studied the relationship of EDNRB and RA signaling on immunoselected rat p75-neurotrophin receptor expressing (p75NTR+) ENS precursors. These cells are neural crest cells that form the neurons and glia of the ENS and are commonly cultured in vitro as a model of ENS development [10,21,34–36]. We report that the combination of exogenous EDN3 and RA exerts unique effects on these ENS precursors compared to the addition of each compound alone. Specifically, we demonstrate that RA and EDN3 exert opposing effects on in vitro enteric glial development and Sox10 expression. We further suggest that RA is a suppressor of EDNRB signaling, that EDN3 disrupts RA signaling and metabolism, and that this interaction results in dramatic morphologic differences in these in vitro cultures.
Materials and Methods
Ethics Statement
This study conformed to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH Publication No. 85–23, revised 1996) and was approved by the Nationwide Children’s Hospital Institutional Animal Care and Use Committee (IACUC protocol numbers AR09-00042 and AR11-00012, Institutional Animal Assurance Number A3544-01)
ENS precursor cell isolation and enrichment
Rat ENS precursors were isolated from inbred Wistar-Kyoto (WKY-NHsd) rats using a method adapted from Sato and Heuckeroth [21]. Briefly, prenatal gut tissue from the stomach to the colon, was dissected from rat embryos on gestational day E14.5, and dissociated into a cell suspension by incubating with collagenase IV (10 mg/ml; Gibco) and 0.05% trypsin-EDTA (50%; Gibco), treating with DNase type I (250µg/ml), and passing the suspension through a nitex filter (38 micron pore) to remove debris. Cells were then labeled with an antibody to the p75NTR (1:200, clone IgG 192, Calbiochem) followed by a secondary antibody conjugated to magnetic beads (Miltenyi Biotech). Cells bound by magnetic beads were then positively magnetically selected in a MACS separation column (Miltenyi Biotech), and eluted outside of the magnetic field. Selected cells were used for all subsequent experiments. The purity of the selected cell suspension was assayed by determining the proportion of p75NTR expressing cells after elution. The cell suspension (5x105 cells/ml) was cytocentrifuged, fixed in 4% formaldehyde and immunolabelled with antibody to p75NTR (1:200, clone IgG 192, Calbiochem) followed by secondary fluorescent antibody-labeling with anti-mouse IgG Alexa Fluor 488. Selected cells consistently comprised around 10% of the total cell yield (data not shown), and 77% of these cells retained p75NTR positivity when re-labeled within two hours of isolation. In contrast, only 7% of the non-selected of cells were p75NTR+. These data contrast with previous work performed in mice and rats where the proportion of p75NTR+ cells among selected cells approached 90% [10,21], possibly reflecting interspecies and/or subtle methodologic differences. As such, we assume that our cell population, while not homogenous, still contains a large enriched fraction of ENS precursors.
Medium and culture conditions
For all experiments, we modified the medium used by Kruger et al. by omitting chick embryo extract and vitamin A [15]. The medium consists of DMEM-low glucose medium (Gibco) supplemented with Penicillin/streptomycin (1X, BioWhittaker), Normocin (50µg/ml, InvivoGen), N2 (1%, Gibco), B27 without vitamin A (2%, Invitrogen), β-mercaptoethanol (50µM, Sigma-Aldrich), Glial Cell Line-Derived Neurotrophic Factor (50ng/ml, R&D systems), and basic Fibroblast Growth Factor (20ng/ml, R&D systems). Selected cultures were also treated with RA (117nM, Sigma-Aldrich), EDN3 (100nM, Calbiochem), and/or the selective antagonist of the EDNRB, BQ-788 (5µM, Sigma-Aldrich) in the following combinations: EDN3-RA-, EDN3+RA-, EDN3-RA+, and EDN3+RA+. The concentrations of RA and EDN3 used were adopted from Kruger et al., as a basis for comparison to their work [15]. In all cultures without EDN3, the EDNRB signaling inhibitor BQ-788 was added to inhibit endogenous EDN3 signaling. An analogous RA signaling inhibitor was not added in RA-free cultures because RA abundance in dissociated cell cultures in the absence of added vitamin A or RA has previously been shown to be negligible [21]. Once added, all compounds were present for the life of the cultures, within the limits of their metabolism, as the medium was not changed for the duration of the culture. All culture dishes were pre-coated with poly-D-lysine (150µg/ml, Biomedical Technology) and laminin (20µg/ml, Invitrogen), and cells were cultured in a reduced oxygen environment in a chamber (Billups-Rothenberg) equilibrated with a gas mixture containing 1% O2/6% CO2/balance N2, generating an actual concentration inside the chamber of 3–6% oxygen [37].
RNA isolation and gene expression profiling
ENS precursors were cultured and supplemented with different combinations of EDN3 and RA as above, in 6-well culture plates (Corning Costar) for 3 or 14 days. After culture, adherent cells were washed with PBS, and RNA was isolated using the RNeasy RNA extraction kit (Qiagen) per the manufacturer’s instructions. Genomic DNA was eliminated during the column purification using RNase-free DNase (Qiagen). cDNA was generated from total RNA using the Maxima First Strand cDNA Synthesis Kit (Fermentas) and quantitative PCR was performed using Maxima Probe/ROX qPCR master mix (Fermentas) with the following conditions: 95°C for 10 min x1; 95°C for 15 sec, 60°C for 1 min for 40 cycles. Intron-spanning primers pairs with a Tm of 59-60oC generating amplicons of 63-122 base pairs were synthesized by Integrated DNA Technologies Incorporated (Table 1). Amplicon-specific fluorescent probes were purchased from Roche. Using the ΔΔCT method, raw CT values were normalized to β-actin. EDN3-RA- cultures were assigned a value of 1, and all expression data are reported relative to these values. Data are averages of 3-4 biological replicates and 2 technical replicates. –RT controls were also performed.
Table 1. Primer – probe pairs used to amplify RA and EDN related genes.
| gene | forward | reverse | Probe # |
|---|---|---|---|
| Rara | 5'-ggcatgtccaaggagtcg-3' | 5'-cgcaccttctcgatgagttc-3' | 10 |
| Rarb | 5'-agcccaccaggaaacctt-3' | 5'-gcactggaattcgtggtgta-3' | 53 |
| Rarg | 5'-ctcatcaccaaggtcagcaa-3' | 5'-atctgcgctggagttcgt-3' | 53 |
| Raldh2 | 5'-ggacgcttctgaaagaggac-3' | 5'-ccgccatttagtgattccat-3' | 120 |
| Cyp26a1 | 5'-gagagaggagagaggctggata-3' | 5'-ggctgcactggctgtagttt-3' | 49 |
| Ednrb | 5'-ctgttggcttccccttcac-3' | 5'-tgtagtccaaaaccagcaaaaa-3' | 18 |
| Edn3 | 5'-agaagcaggagactggaggtc-3' | 5'-cagctaaggctggtggactt-3' | 41 |
| Ece1 | 5'-cggaggacagcaagaacatag-3' | 5'-caggctctcctcgaatgc-3' | 80 |
| Ret | 5'-cacagccttccgtctgaaa-3' | 5'-tctgggaggcgttttcttt-3' | 76 |
| Sox10 | 5'-atgtcagatgggaacccaga-3' | 5'-gtctttggggtggttggag-3' | 21 |
Immunocytochemistry
Cells were cultured in 8-well chamber slides (Thermo Scientific) at low density (1000 cells/well) with different combinations of EDN3 and RA as above. After three or fourteen days, cells were fixed in 3.7% paraformaldehyde, permeabilized in triton x-100, and incubated with primary antibody overnight at 4oC. After washing, secondary antibody was added. Primary antibodies: rabbit anti-peripherin (1:1000, Chemicon), mouse anti-smooth muscle actin (SMA) (1:400, Sigma), mouse anti-S100β (1:500, Sigma). Secondary antibodies (Invitrogen): anti-rabbit Alexa Fluor 546, and anti-mouse Alexa Fluor 488, anti-rabbit Alexa Fluor 488. Nuclei were counterstained with prolong gold antifade with DAPI mounting medium (Invitrogen).
Proliferation assay
The click-it EDU proliferation assay (Invitrogen) was performed proliferation assay was performed according to the manufacturer’s instructions. Cells were cultured for 66 hours, followed by EDU (10µM) incorporation for 6 hours. After incorporation, cells were washed, fixed and labeled with Alexa Fluor 488 or 594 click-it reagent. Immunolabelling for different cell lineages and DAPI counterstaining were then performed as described in the immunocytochemistry section.
Microscopy
Cells were visualized with a Zeiss AxioScope A1 epifluorescent microscope with a Zeiss Axiocam HRc digital camera and images were recorded using Axiovision software. An AMG EVOS “FL” Epifluorescence microscope was used for some experiments. All images were processed and analyzed using NIH ImageJ software, and ImageJ was used for manual cell counting.
Statistical analysis
All quantitative experiments were repeated 3-9 times. To evaluate the individual contributions of exogenous EDN3 and RA, as well as any interaction effects between the two compounds, a two-way ANOVA (SigmaPlot) was used. The All Pairwise Multiple Comparison Procedure (Tukey Test) was used to determine the individual contributions of RA (comparing RA- vs. RA+) and EDN3 (comparing EDN3- vs. EDN3+) to changes observed in culture. Note that each comparison can be made in the presence or absence of the other compound. For instance, comparing EDN3-RA- to EDN3+RA- provides information about an EDN3 effect, as does comparing EDN3-RA+ to EDN3+RA+, but the latter is also in the presence of RA. The definition of an interaction is thus: The direction and/or magnitude of an observed change in response to one compound are affected by the presence of the other compound. A p value <0.05 is deemed significant in all experiments. Graphs were generated using Graphpad Prism 6.
Results
RA and EDN3 have opposing effects on S100β+ cell prevalence
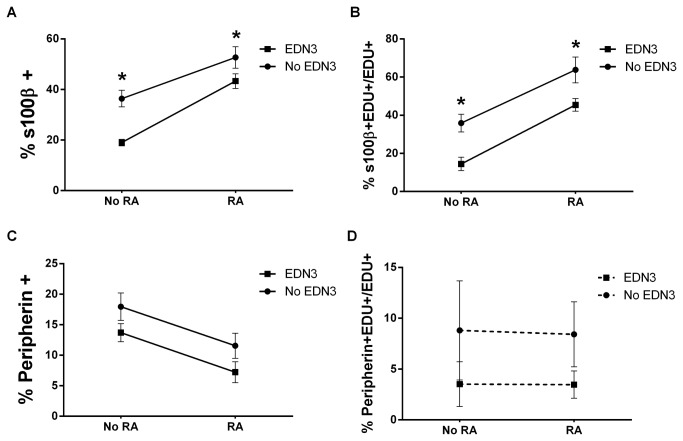
To determine how RA and EDN3 together and separately influence the in vitro cellular composition of the ENS, p75NTR+ immunoselected cells were isolated and exposed in parallel to four separate culture environments: 1) EDN3-RA- was devoid of exogenous RA, vitamin A, and EDN3 and the EDN signaling inhibitor BQ-788 was added to further inhibit any endogenous EDNRB signaling; 2) EDN3+RA-, where only EDN3 was added, permits EDNRB activation only; 3) EDN3-RA+, where RA was added with BQ-788, permitting RA activation only; and 4) EDN3+RA+, where EDN3 and RA were added and duel activation of EDNRB and RA is presumed to occur. These combinations permitted us to compare the effects of RA-signaling in the presence and absence of EDN3 and, conversely, the effects of EDN3 signaling in the presence and absence of RA. After 3 days of culture, the proportions of peripherin, SMA, and S100β immunoreactive cells were determined immunocytochemically (Figure 1 and Table 2). We found that the proportion of S100β+ cells, bearing a morphological resemblance to glia, was greatly enriched by RA. Comparing EDN3-RA- to EDN3-RA+, the proportion of S100β+ cells increased from 36.4% to 52.6% of the total cells in culture, a 45% increase (p<0.001). Similarly, the proportion more than doubled from 19.0 to 43.3% (p<0.001) when comparing EDN3+RA- to EDN3+RA+. Thus, regardless of whether EDN3 is present, RA is associated with a statistically significant increased percentage of S100β+ cells. In contrast, a comparison of EDN3-RA- to EDN3+RA- reveals a net decrease in S100β+ cell prevalence from 36.4% to 19.0%, a 47% decrease (p<0.001). Likewise, comparing EDN3-RA+ to EDN3+RA+ reveals a net decrease in S100β+ cell prevalence from 52.6% to 43.3%, an 18% decrease (p=0.043). Collectively, these data demonstrate that RA and EDN3 treatment have opposite effects on the composition of 3-day ENS precursor cultures, with RA enriching S100β+ cells and EDN3 depleting them.
Figure 1. RA increases S100β+ and decreases peripherin+ cell prevalence while EDN3 decreases S100β+ cell prevalence.
p75NTR+ cells were grown for 72 hours in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. The proportion (A and C) and proliferating fraction (B and D) of peripherin- and S100β-immunoreactive cells were quantified. Solid lines denote a statistically significant difference (p<0.05) between “No RA” and “RA”, and broken lines indicate a p value >0.05. Asterisk denotes a statistically significant difference (p<0.05) between “No EDN3” and “EDN3”. (A) The proportion of S100β+ cells was enriched by RA and decreased in response to EDN3 (B) RA increased, and EDN3 decreased the fraction of proliferating S100β+ cells. (C) the proportion of peripherin+ cells decreased in response to RA treatment, but EDN3 did not have an effect. (D) Neither RA nor EDN3 had an effect on proliferation of peripherin+ cells.
Table 2. Changes in cell composition in response to RA and EDN3.
| Comparison of mean proportion of immunolabelled cells (p value) |
|||
|---|---|---|---|
| %peripherin+ | %SMA+ | %S100β+ | |
| RA- vs. RA+ (EDN3-) | 17.9 vs. 11.5 (p=0.023) | 32.4 vs. 23.4 | 36.4 vs. 52.7 (p=0.001) |
| RA- vs. RA+ (EDN3+) | 13.7 vs. 7.2 (p=0.022) | 27.2 vs. 25.3 | 19.0 vs. 43.3 (p<0.001) |
| EDN3- vs. EDN3+ (RA-) | 17.9 vs. 13.7 | 32.4 vs. 27.2 | 36.4 vs. 19.0 (p<0.001) |
| EDN3- vs. EDN3+ (RA+) | 11.5 vs. 7.2 | 23.4 vs. 25.3 | 52.7 vs. 43.3 (p<0.043) |
| Interaction present? | No | No | No |
This table summarizes the effects of RA and EDN3 on the cell composition of ENS precursor cultures after 3 days. Immunoselected cells were cultured in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. The proportion of peripherin, SMA, and S100β immunoreactive cells as a percentage of all cells counted in each condition were compared. A two way ANOVA was employed to identify the contribution of each compound to changes in the proportion of immunolabelled cells. Significant changes are in bold and p values are noted. Data is the summary of nine biological replicates, and ten random microscope fields were counted and summed per condition. Abbreviations: SMA = α-smooth muscle actin; EDN3 = endothelin-3; RA = Retinoic acid;
RA decreases the prevalence of peripherin+ cells
Concomitant with the RA-associated increase in S100β+ cells in culture, there was a decrease in the proportion of peripherin+ cells, signifying differentiating neurons (Figure 1 and Table 2). With the addition of RA, the proportion of peripherin+ cells decreased from 18 to 11.5% (EDN3-RA- vs. EDN3-RA+), a 35% decrease (p=0.023), and from 13.7 to 7.2% (EDN3+RA- vs. EDN3+RA+), a 47% decrease (p=0.022). EDN3 did not have a statistically significant effect on the percentage of peripherin+ cells and no combinations of EDN3 and RA affected the prevalence of SMA+ cells.
RA increases, and EDN3 decreases, the proliferation of S100β+ cells
To understand the mechanism underlying the compositional differences in response to EDN3 and RA exposure, we performed an EDU incorporation assay to study proliferative differences (Figure 1 and Table 3). In the presence of RA, there was an increase in the fraction of proliferating (% EDU+/DAPI+) cells overall, without respect to lineage. EDN3 also promoted proliferation, but its effect was not significant in the presence of RA. Upon examination of specific lineages, RA promoted the proliferation of S100β+ cells: the proportion of EDU+S100β+ out of all EDU+ cells nearly doubled from 35.9 to 63.8% in response to RA (p<0.001) and tripled when EDN3 was also present (p<0.001). Conversely, the proportion of proliferating S100β+ cells decreased in response to EDN3 from 35.9 to 14.4% (p=0.006) in the absence of RA and from 63.8 to 45.4% (p=0.015) in the presence of RA. There were no significant changes in the proportion of proliferating peripherin+ and SMA+ cells in any culture condition. Collectively, these data demonstrate that RA stimulates the proliferation of S100β+ cells in vitro, and that EDN3 has an opposing effect on S100β+ proliferation thereby reducing the number of S100β+ cells in the culture.
Table 3. Lineage-specific and overall changes in proliferation in response to RA and EDN3.
| Comparison of mean proportion of proliferating cells (p value) |
||||
|---|---|---|---|---|
| %EDU+/DAPI+ | %EDU+peripherin+/EDU+ | %EDU+SMA+/EDU+ | %EDU+S100β+/EDU+ | |
| RA- vs. RA+ (EDN3-) | 25.8 vs. 45.1 (p<0.001) | 8.8 vs. 8.4 | 28.5 vs. 18.3 | 35.9 vs. 63.8 (p<0.001) |
| RA- vs. RA+ (EDN3+) | 33.0 vs. 48.8 (p<0.001) | 3.5 vs. 3.5 | 19.2 vs. 8.4 | 14.4 vs. 45.4 (p<0.001) |
| EDN3- vs. EDN3+ (RA-) | 25.8 vs. 33.0 (p=0.014) | 8.8 vs. 3.5 | 28.5 vs. 19.2 | 35.9 vs. 14.4 (p=0.006) |
| EDN3- vs. EDN3+ (RA+) | 45.1 vs. 48.8 | 8.4 vs. 3.5 | 18.3 vs. 8.4 | 63.8 vs. 45.4 (p=0.015) |
| Interaction present? | No | No | No | No |
This table summarizes the effects of RA and EDN3 on the proliferation of ENS precursor cultures after 3 days. Immunoselected cells were cultured in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. The overall proportion of proliferating cells (EDU+/DAPI+), and the proportion of proliferating cells of each lineage as a percentage of all proliferating cells (EDU+) were quantified and compared between the groups. A two way ANOVA was employed to identify the contribution of each compound to changes in the proportion of proliferating cells. Significant changes (p<0.05) are in bold. Abbreviations: SMA =α-smooth muscle actin; DAPI = 4',6-diamidino-2-phenylindole; EDU = 5-ethynyl-2´-deoxyuridine; EDN3 = endothelin-3; RA = Retinoic acid;
RA and EDN3 interact to affect Sox10 expression
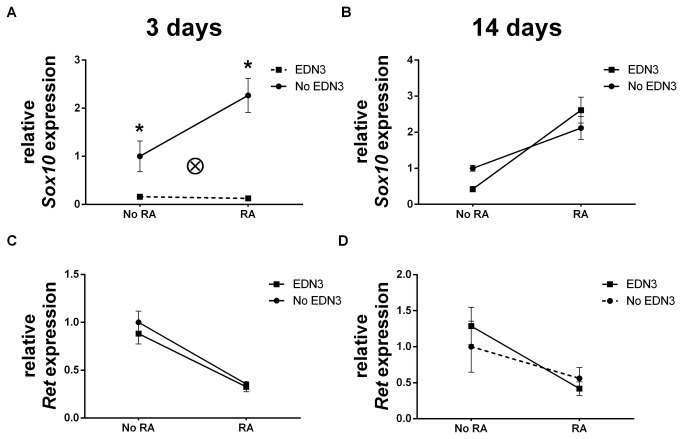
SOX10 expression is associated with ENS precursor maintenance and glial differentiation [38–41]. EDN3 maintains SOX10 levels in vivo and SOX10 regulates EDN signaling by acting directly on the Ednrb promoter [42,43]. We therefore wanted to assess the relationship of EDN and Sox10 expression in the relatively controlled environment of our defined culture system and determine whether RA impacts this relationship (Figure 2 and Table 4). RA and EDN3 interacted in the regulation of Sox10: In the absence of EDN3, RA doubled Sox10 levels at 3 days (p=0.003). However, the concurrent addition of EDN3 abolished this phenomenon. After 14 days, RA was still associated with a 2- to 6-fold increase in Sox10 abundance, but this association was no longer affected by EDN3 Sox10 levels sharply declined by 84% (p=0.029) to 94% (p<0.001) in response to EDN3, depending on whether RA was also present. These data reveal a bi-directional interaction between EDN3 and RA in the modulation of Sox10, with RA favoring, and EDN3 antagonizing Sox10 expression.
Figure 2. Culture in the presence of RA increases Sox10, but EDN3 suppresses this effect.
p75NTR+ cells were grown for 72 hours in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative Sox10 (A–B) and Ret (C–D) mRNA levels were measured by quantitative RT-PCR after 3 (A, C) and 14 (B, D) days. Expression is normalized to β-actin and the -RA/-EDN3 condition was standardized to a value of 1. Solid lines denote a statistically significant difference (p<0.05) between “No RA” and “RA”, and broken lines indicate a p value >0.05. Asterisk denotes a statistically significant difference (p<0.05) between “No EDN3” and “EDN3”. (A) In the absence of EDN3, RA increased Sox10 levels; however, the concurrent addition of EDN3 abolished this phenomenon. There was a statistically significant interaction (ⓧ; p=0.019) between EDN3 and RA in their effect on Sox10 levels at 3 days. This interaction is specific to Sox10 expression and was not observed with Ret. (B) RA treatment was associated with increased Sox10 levels but EDN3 had no effect at 14 days. (C) RA decreased Ret levels at 3 days regardless of whether EDN3 was present, but EDN3 did not affect Ret gene expression. (D) Decreased Ret levels in response to RA persisted at 14 days, but only in the presence of EDN3
Table 4. Changes in ENS development-related mRNA levels in response to RA and EDN3.
| Comparison of relative mean mRNA levels |
||||
|---|---|---|---|---|
| (p value) |
|
|||
| Target gene: |
Ret
|
Sox10
|
||
| 3d | 14d | 3d | 14d | |
| RA- vs. RA+ (EDN3-) | 1.0 vs. 0.356 | 1.0 vs. 0.560 | 1.0 vs. 2.267 | 1.0 vs. 2.116 |
| (p<0.001) | (p=0.003) | (p=0.012) | ||
| RA- vs. RA+ (EDN3+) | 0.881 vs. 0.327 | 1.286 vs. 0.419 | 0.160 vs. 0.127 | 0.423 vs. 2.613 |
| (p<0.001) | (p=0.032) | (p<0.001) | ||
| EDN3- vs. EDN3+ (RA-) | 1.0 vs. 0.881 | 1.0 vs. 1.286 | 1.0 vs. 0.160 | 1.0 vs. 0.423 |
| (p=0.029) | ||||
| EDN3- vs. EDN3+ (RA+) | 0.356 vs. 0.327 | 0.560 vs. 0.419 | 2.267 vs. 0.127 | 2.116 vs. 2.613 |
| (p<0.001) | ||||
| Interaction present? | No | No | Yes (p=0.019) | No |
This table summarizes the effects of RA and EDN3 on the gene expression in ENS precursor cultures after 3 and 14 days. Immunoselected cells were cultured in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative mRNA levels were measured by quantitative RT-PCR. Levels are normalized to β-actin and the - RA/- EDN3 condition was standardized to a value of 1. Relative mRNA levels were compared between the groups. A two way ANOVA was employed to identify the contribution of each compound to changes in the proportion of proliferating cells. Significant changes (p<0.05) are in bold. Abbreviations: 3d = 3-day culture; 14d = 14-day culture; EDN3 = endothelin-3; RA = Retinoic acid;
RA exerts a stable suppressive effect on Ret expression
Since Ret deficiency is associated with ENS precursor defects and intestinal aganglionosis [44] we explored the possibility that EDN3 and/or RA also contribute to the transcriptional regulation of Ret in these cells. We cultured p75NTR+ cells for 3 and 14 days with different combinations of EDN3 and RA, as above, and mRNA levels were quantified. Although the addition of EDN3 did not significantly affect Ret gene expression, RA was associated with a 62% net decrease in Ret levels at 3 days (p<0.001) and 67% at 14 days (p=0.032) (Figure 2 and Table 4). With these data, RA may be added to the factors that regulate the transcription of Ret in ENS precursors, although further investigation is necessary to determine whether this occurs by direct action on the Ret gene or whether this reflects changes in the cell composition of the culture.
RA is associated with a net decrease in EDN-related gene expression
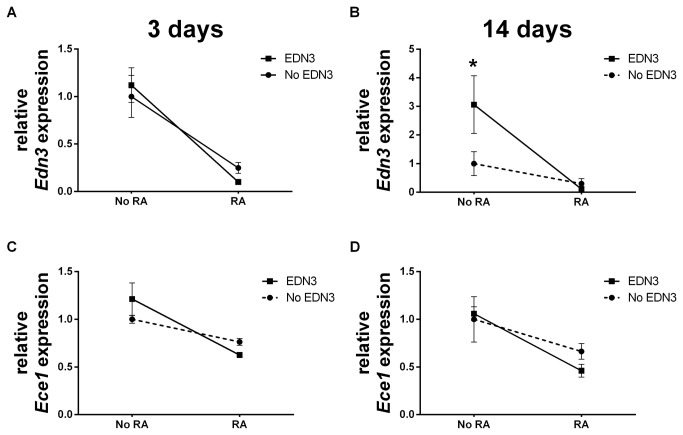
In light of the opposing actions of EDN3 and RA on lineage predominance and Sox10 expression in our in vitro system, we sought to determine how, if at all, RA and EDN3 affect each other at the gene expression level (Figure 3 and Table 5). We observed that the addition of RA was associated with a net decrease in the level of EDN3 by 75% (p=0.004) in the absence of exogenous EDN3, and by 91% (p<0.001) in the presence of exogenous EDN3 (p<0.001). Similarly, adding RA was associated with a decrease in the relative abundance of Ece1 by 48% (p<0.001), albeit only in the presence of EDN3 Neither RA nor EDN3 were associated with changes in the expression of the Ednrb gene at 3 days. Collectively, these data demonstrate that culture of ENS precursors in the presence of RA leads to a decrease in abundance of EDN3 and Ece1 mRNA.
Figure 3. RA decreases EDN3 and Ece1 mRNA levels, depending on whether exogenous EDN3 is present.
p75NTR+ cells were grown for 72 hours in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative EDN3 (A-B) and Ece1 (C-D) mRNA levels were measured by quantitative RT-PCR after 3 (A, C) and 14 (B, D) days. Levels are normalized to β-actin and the -RA/-EDN3 condition was standardized to a value of 1. Solid lines denote a statistically significant difference (p<0.05) between “No RA” and “RA”, and broken lines indicate a p value >0.05. Asterisk denotes a statistically significant difference (p<0.05) between “No EDN3” and “EDN3”. (A) At 3 days, RA was associated with a net decrease in the level of EDN3 in the absence and presence of exogenous EDN3 (C) Similarly, RA was associated with a decrease in the relative abundance of Ece1, albeit only in the presence of EDN3 After 14 days, RA still decreased the relative abundance of EDN3 (B) and Ece1 (D) mRNA.
Table 5. Changes in EDN-related mRNA levels in response to RA and EDN3.
| Comparison of relative mean mRNA levels |
||||||
|---|---|---|---|---|---|---|
| (p value) |
|
|
||||
| Target gene: |
EDN3
|
Ece1
|
Ednrb
|
|||
| 3d | 14d | 3d | 14d | 3d | 14d | |
| RA- vs. RA+ (EDN3-) | 1.0 vs. 0.248 | 1.0 vs. 0.299 | 1.0 vs. 0.763 | 1.0 vs. 0.663 | 1.0 vs. 1.512 | 1.0 vs. 15.201 |
| (p=0.004) | ||||||
| RA- vs. RA+ (EDN3+) | 1.12 vs. 0.099 | 3.062 vs. 0.110 | 1.213 vs. 0.625 | 1.059 vs. 0.461 | 1.512 vs. 1.246 | 2.8 vs. 22.873 |
| (p<0.001) | (p=0.006) | (p<0.001) | (p=0.014) | (p=0.017) | ||
| EDN3- vs. EDN3+ (RA-) | 1.0 vs. 1.120 | 1.0 vs. 3.062 | 1.0 vs. 1.213 | 1.0 vs. 1.059 | 1.0 vs. 1.512 | 1.0 vs. 2.8 |
| (p=0.030) | ||||||
| EDN3- vs. EDN3+ (RA+) | 0.248 vs. 0.099 | 0.299 vs. 0.110 | 0.763 vs. 0.625 | 0.663 vs. 0.461 | 1.553 vs. 1.246 | 15.201 vs. 22.873 |
| Interaction present? | No | No | No | No | No | No |
This table summarizes the effects of RA and EDN3 on the gene expression in ENS precursor cultures after 3 and 14 days. Immunoselected cells were cultured in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative mRNA levels were measured by quantitative RT-PCR. Levels are normalized to β-actin and the - RA/- EDN3 condition was standardized to a value of 1. Relative mRNA levels were compared between the groups. A two way ANOVA was employed to identify the contribution of each compound to changes in the proportion of proliferating cells. Significant changes (p<0.05) are in bold. Abbreviations: 3d = 3-day culture; 14d = 14-day culture; EDN3 = endothelin-3; RA = Retinoic acid;
EDN3 treatment leads to a net reduction in RA receptor gene expression
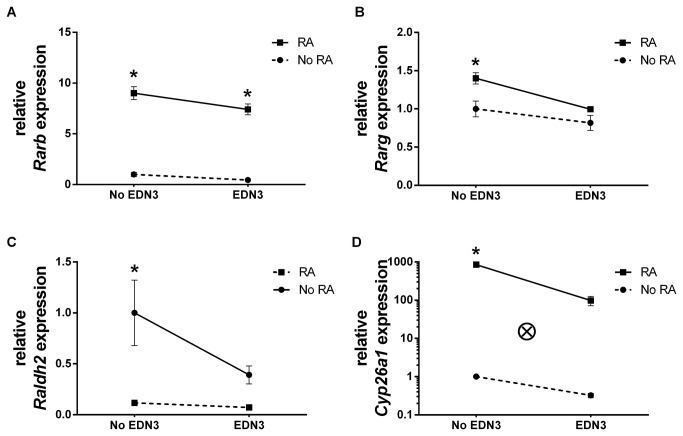
Since RA was associated with decreased abundance of EDN3 and Ece1 gene expression, we next examined whether the converse was true: Is EDN3 associated with changes in the expression of RA signaling related genes in 3-day cultures? EDN3 was associated with a net decrease in Rarb and Rarg RNA transcripts (Figure 4 and Table 6): in the presence of RA, EDN3 decreased Rarb and Rarg by 18% (p=0.023) and 28% (p=0.005), respectively. In the absence of RA, EDN3 did not contribute to RA receptor expression. Consistent with previous studies, adding RA was associated with increased levels of Rarb (p<0.001) and Rarg (p=0.005) RNA transcripts [45–54]. No significant effects on Rara were observed with either compound. These experiments suggest that EDN3 and RA competitively alter the abundance of Rarb and Rarg.
Figure 4. EDN3 is associated with decreased levels of RA receptor mRNA and RA metabolic enzyme mRNA.
p75NTR+ cells were grown for 72 hours in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative Rarb (A), Rarg (B), Raldh2 (C), and Cyp26a1 (D) mRNA levels were measured by quantitative RT-PCR. Levels are normalized to β-actin and the -RA/-EDN3 condition was standardized to a value of 1. Solid lines denote a statistically significant difference (p<0.05) between “No EDN3” and “EDN3”, and broken lines indicate a p value >0.05. Asterisk denotes a statistically significant difference (p<0.05) between “No RA” and “RA”. EDN3 was associated with a net decrease in Rarb (A) and Rarg (B) mRNA transcripts in the presence of exogenous RA. In the absence of exogenous RA, EDN3 did not have an effect on RA receptor levels. (C) EDN3 was also associated with a decrease in Raldh2 abundance, but not when RA was present. (D) EDN3 also reduced Cyp26a1 mRNA levels, but only in the presence of RA. Consistent with autoregulation, RA treatment was responsible for a reduction in Raldh2 levels (C) and an increase in Cyp26a1 (D), in the absence of exogenous EDN3 However, associations of RA with Raldh2 and Cyp26a1 levels were lost when EDN3 was simultaneously present. There was a statistically significant interaction (ⓧ; p<0.001) between EDN3 and RA in their effect on Cyp26a1 levels at 3 days.
Table 6. Changes in RA receptor-related mRNA levels in response to RA and EDN3.
| Comparison of relative mean mRNA levels |
||||||
|---|---|---|---|---|---|---|
| (p value) |
||||||
| Target gene: |
Rara
|
Rarb
|
Rarg
|
|||
| 3d | 14d | 3d | 14d | 3d | 14d | |
| RA- vs. RA+ (EDN3-) | 1.0 vs. 0.953 | 1.0 vs. 1.124 | 1.0 vs. 9.005 | 1.0 vs. 183.758 | 1.0 vs. 1.4 | 1.0 vs. 0.854 |
| (p<0.001) | (p=0.002) | (p=0.005) | ||||
| RA- vs. RA+ (EDN3+) | 0.846 vs. 1.028 | 1.315 vs. 0.751 | 0.438 vs. 7.406 | 1.589 vs. 222.325 | 0.817 vs. 0.995 | 1.320 vs. 1.031 |
| (p=0.001) | (p<0.001) | |||||
| EDN3- vs. EDN3+ (RA-) | 1.0 vs. 0.846 | 1.0 vs. 1.315 | 1.0 vs. 0.438 | 1.0 vs. 1.589 | 1.0 vs. 0.817 | 1.0 vs. 1.32 |
| EDN3- vs. EDN3+ (RA+) | 0.953 vs. 1.028 | 1.124 vs. 0.751 | 9.005 vs. 7.406 | 183.758 vs. 222.325 | 1.4 vs. 0.995 | 0.854 vs. 1.031 |
| (p=0.023) | (p=0.005) | |||||
| Interaction present? | No | No | No | No | No | No |
This table summarizes the effects of RA and EDN3 on the gene expression in ENS precursor cultures after 3 and 14 days. Immunoselected cells were cultured in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative mRNA levels were measured by quantitative RT-PCR. Levels are normalized to β-actin and the - RA/- EDN3 condition was standardized to a value of 1. Relative mRNA levels were compared between the groups. A two way ANOVA was employed to identify the contribution of each compound to changes in the proportion of proliferating cells. Significant changes (p<0.05) are in bold. Abbreviations: 3d = 3-day culture; 14d = 14-day culture; EDN3 = endothelin-3; RA = Retinoic acid;
EDN3 and RA regulate RA metabolic enzyme gene expression in a complex fashion
RA homeostasis is regulated, in part, by the RA synthesizing enzyme RALDH2, and the RA degrading enzyme CYP26A1 [18]. Since EDN3 was shown to reduce the abundance of RA receptor mRNA, we next explored the possibility that EDN3 alters the availability of RA by affecting Raldh2 and Cyp26a1 expression in vitro (Figure 4 and Table 7). The responses of these genes to EDN3 and RA were complex. EDN3 was associated with a 61% (p=0.024) decrease in Raldh2 abundance, but this decrease was abolished when RA was present. EDN3 also significantly reduced Cyp26a1 mRNA levels by 88% when RA was added concomitantly (p<0.001). RA treatment was responsible for a reduction by 88% in Raldh2 levels (p<0.003) and a >800-fold increase in Cyp26a1 (p<0.001), in the absence of exogenous EDN3 However, a statistically significant association of RA with Raldh2 and Cyp26a1 levels were not observed when EDN3 was simultaneously present, suggesting that EDN3 somehow disrupts this process. Together, these findings suggest that EDN3 is capable of impacting RA levels via an effect on the gene expression of RA metabolic machinery.
Table 7. Changes in RA metabolism enzyme-related mRNA levels in response to RA and EDN3.
| Comparison of relative mean mRNA levels |
||||
|---|---|---|---|---|
| (p value) |
||||
| Target gene: |
Raldh2
|
Cyp26a1
|
||
| 3d | 14d | 3d | 14d | |
| RA- vs. RA+ (EDN3-) | 1.0 vs. 0.117 | 1.0 vs. 4.748 | 1.0 vs. 846.015 | 1.0 vs. 1.682 |
| (p=0.003) | (p=0.007) | (p<0.001) | ||
| RA- vs. RA+ (EDN3+) | 0.391 vs. 0.072 | 0.619 vs. 0.257 | 0.325 vs. 98.403 | 0.139 vs. 6.461 |
| (p<0.001) | ||||
| EDN3- vs. EDN3+ (RA-) | 1.0 vs. 0.391 | 1.0 vs. 0.619 | 1.0 vs. 0.325 | 1.0 vs. 0.139 |
| (p=0.024) | ||||
| EDN3- vs. EDN3+ (RA+) | 0.117 vs. 0.072 | 4.748 vs. 0.257 | 846.015 vs. 98.403 | 1.682 vs. 6.461 |
| (p=0.003) | (p<0.001) | (p=0.004) | ||
| Interaction present? | No | Yes (p=0.023) | Yes (p<0.001) | Yes (p=0.01) |
This table summarizes the effects of RA and EDN3 on the gene expression in ENS precursor cultures after 3 and 14 days. Immunoselected cells were cultured in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Relative mRNA levels were measured by quantitative RT-PCR. Levels are normalized to β-actin and the - RA/- EDN3 condition was standardized to a value of 1. Relative mRNA levels were compared between the groups. A two way ANOVA was employed to identify the contribution of each compound to changes in the proportion of proliferating cells. Significant changes (p<0.05) are in bold. Abbreviations: 3d = 3-day culture; 14d = 14-day culture; EDN3 = endothelin-3; RA = Retinoic acid;
The relationship of EDN and RA signaling and metabolism changes as ENS precursors differentiate
Our next objective was to determine the changes in gene expression in later cultures and correlate these with earlier expression changes and phenotypic changes. We evaluated the expression of EDN- and RA-related genes after 14 days (Tables 4-7). EDN3 and Ece1 abundance continued to be lower in cultures treated with RA, with decreases of 96% (p=0.006) and 56% (p=0.014), respectively, when exogenous EDN3 was concurrently present (Figure 3). In contrast to observations in our short term cultures where RA was not associated with Ednrb expression, we observed an 8-fold increase (p=0.017) in Ednrb transcript levels in response to RA treatment (Table 5). The relationship between EDN3 and Rar expression that was seen at 3d was not observed at 14 days (Figure 4). Nevertheless, RA, EDN3, and the combination of RA and EDN3 continued to have distinct significant effects on Raldh2 and Cyp26a1 levels indicating an interaction between the compounds in the modulation of gene expression: EDN3 was associated with a net 95% reduction (p=0.003) in Raldh2 levels and net 3.8-fold increase in Cyp26a1 when RA was also present. RA augmented Raldh2 levels almost 5-fold at 14 days, but this relationship was abolished when EDN3 was concomitantly present. RA was associated with a 45-fold increase in Cyp26a1 expression (p<0.001), but only in the presence of EDN3 Collectively, these data reflect the existence of a bi-directional interaction between EDN3 and RA in the modulation of RA-related gene expression. We hypothesize that this interaction occurs in vivo and impacts the local levels of bioavailable RA.
RA induces the formation of an organized heterocellular plexus
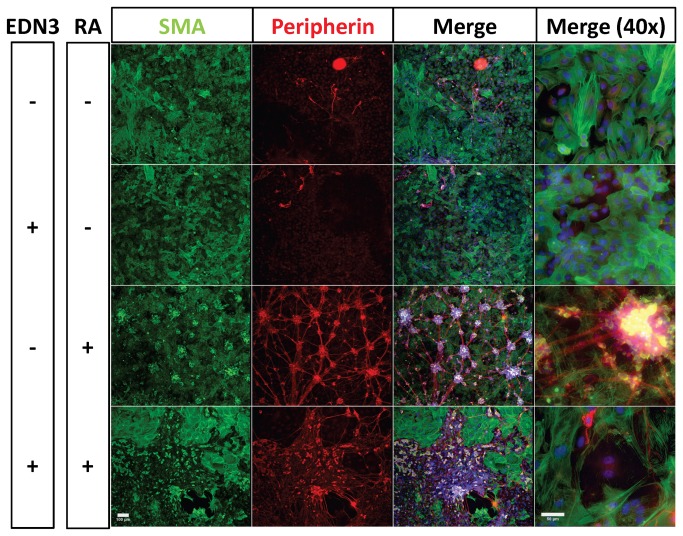
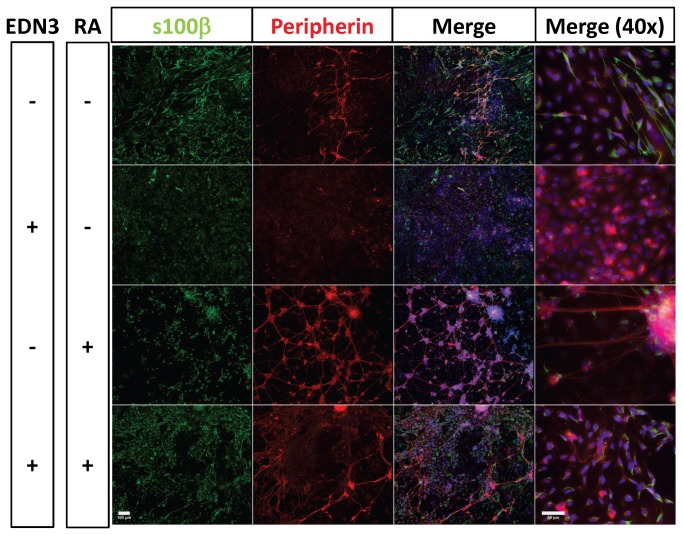
To understand how the changes in S100β+ cell prevalence induced by RA and EDN3 relate to terminal culture morphology, we studied ENS precursor cultures after 14 days (Figures 5 and 6). High cell densities, as well as the 3-dimensional structure of these cultures precluded accurate quantitative comparisons of the different lineages. Nevertheless, distinct features of each condition were evident. Cells cultured in the absence of EDN3 and RA (EDN3-RA-) were predominantly myofibroblastic in nature, although they did develop sparse neurons, and more numerous and more brightly staining glia. EDN3+RA- cultures formed an almost-uniform amorphous sheet of SMA+ cells possessing myofibroblast morphology. EDN3-RA+ cultures were distinct in that neurons formed a plexus pattern, punctuated by large heterocellular ganglia, abundant peripherin+ neurites and cell bodies, abundant S100β+ cells, and non-myofibroblast-like SMA+ cells. Cells morphologically resembling myofibroblasts were also abundantly present, but resembled a feeder monolayer beneath the plexus. EDN3+RA+ treatment resulted in the emergence of many peripherin+ cells with neuronal morphology and long complex neurites, but these neurons were disorganized, and did not form a discernible plexus. Furthermore, myofibroblasts, while excluded from neuronal regions, were still present in large number. Other SMA+ cells were also observed among the neurons, but lacked myofibroblast morphology and the lineage of these cells is unclear. Weakly staining S100β+ cells with glial cell morphology were also found amidst the neurons. The glia in these cultures were more fusiform and were mainly associated with neurons. Overall, RA presence supported the formation of neurons and glia but activation of EDNRB in these same cultures resulted in a significant difference in the arrangement of the neuronal elements.
Figure 5. Effects of RA and EDN3 on the terminal culture morphology of ENS precursors.
p75NTR immunoselected cells were grown for 14 days in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Culture morphology was evaluated by immunofluorescence microscopy. Peripherin expression (red) is consistent with neuronal morphology and myofibroblasts are indicated by expression of α-SMA (green). The third and fourth columns include DAPI staining for nuclei. The first three columns were photographed at 10x (Scale bar = 100 µm) and the fourth column was photographed at 40x (Scale bar = 50 µm). In the absence of EDN3 and RA (EDN3-RA-), SMA+ myofibroblasts predominated, but sparse neurons were also seen. EDN3+RA- treated cultures formed a homogeneous sheet of SMA+ myofibroblasts. In EDN3-RA+ cultures, neurons formed a plexus punctuated by large multicellular ganglia, abundant peripherin+ neurites and cell bodies, and non-myofibroblast-like SMA+ cells. SMA+ myofibroblasts were also present beneath the plexus. With EDN3+RA+ treatment, many peripherin+ neurons and long complex neurites were seen, without forming a plexus. Myofibroblasts, while excluded from neuronal regions, were still present in large number.
Figure 6. Effects of RA and EDN3 on the terminal culture morphology of ENS precursors.
p75NTR immunoselected cells were grown for 14 days in the presence of RA, EDN3, RA with EDN3, or neither compound. When EDN3 was absent, the EDN signaling inhibitor BQ-788 was added to inhibit endogenous EDNRB signaling. Culture morphology was evaluated by immunofluorescence microscopy. Peripherin expression (red) is consistent with neuronal morphology and glia are indicated by expression of S100β (green). The third and fourth columns include DAPI staining for nuclei. The first three columns were photographed at 10x (Scale bar = 100 µm) and the fourth column was photographed at 40x (Scale bar = 50 µm). Without EDN3 and RA (EDN3-RA-) sparse neurons developed, but S100β+ glia were more numerous and more brightly staining. EDN3+RA- cultures did not contain glia or neurons. EDN3-RA+ cultures formed discrete heterocellular ganglia with abundant peripherin+ cell bodies and S100β+ glia, linked by thick peripherin+ neurites in a plexus pattern. EDN3+RA+ treatment also contained many peripherin+ neurons and long complex neurites, but these neurons were disorganized, and did not form a plexus. Weakly staining S100β+ cells with glial cell morphology were also found amidst the neurons. The glia in these cultures were more fusiform and were mainly associated with neurons.
Discussion
Both RA and EDNRB signaling play critical roles in the formation of the ENS. Evidence from disparate sources [5,20] suggests that the two morphogens spatiotemporally overlap during ENS development. Furthermore, RA has been shown to modulate EDN signaling in other cell types [30–33], although the reverse has not been demonstrated. The fact that both are implicated in the pathogenesis of aganglionosis prompted us to investigate their combined effects and to determine whether there exists a relationship between the two pathways in ENS precursor cultures. Our findings suggest that 1) RA supports enteric glial proliferation resulting in an increased abundance of enteric glia and decreased neurons in culture, 2) EDNRB signaling depletes enteric glia by suppressing their proliferation but promotes proliferation overall, 3) RA suppresses Ret gene expression and this is associated with a decreased proportion of neurons in culture, 4) RA signaling is associated with a decrease in the abundance of EDN3 mRNA, 5) EDN3 alters the abundance of mRNA encoding RA receptors and metabolic enzymes, and 6) RA and EDNRB signaling interact to affect the morphology of mature cultures of ENS precursors in vitro.
Enteric neurons develop from neural crest-derived precursors that persist in the adult gut and share many properties with enteric glia, including the expression of SOX10, and p75NTR [55,56]. Multipotent ENS precursors and mature glia also express EDNRB [13,57–61]. The distinction between neurogenic precursors and enteric glia has been blurred by the striking finding that enteric glia are capable of undergoing neurogenesis [55,56,62]. Our data demonstrate that RA stimulates proliferation of S100β+ cells at 3 days, resulting in increased abundance of these cells. Whether these cells represent mature glia or a specialized stem cell population is unclear. Interestingly, Sox10, but not Ednrb, expression is also increased after 3 days in the presence of RA. Increased Sox10 expression is consistent with a study showing a dose-dependent positive effect of RA on chimeric SOX10-GFP protein expression in murine embryonic stem cells, resulting in the acquisition of markers of neural crest progenitors [63]. If we interpret the lack of a concomitant increase in the EDNRB+ population to indicate stable numbers of ENS progenitors, our results suggest that the increase in S100β and Sox10 expression observed in the presence of RA reflects an increase in the number of mature glia. Culture in the presence of the EDNRB ligand dramatically suppresses Sox10 expression, even in the presence of RA, revealing a significant statistical interaction in the relationship of RA and EDN3 with Sox10 expression. The underlying biological mechanism for this association is unknown and will require further study. Nevertheless, effects on Sox10 do not appear to be solely a reflection of a decreased percentage of S100β+ glia in the culture because Sox10 expression is reduced even in EDN3+RA+ cultures, where S100β+ cells are increased. Previous studies demonstrate that EDNRB activation of ENS progenitors induces proliferation of a SOX10+ cell population [5]. One explanation for our findings is that over the course of the three day culture, in which secondary and tertiary effects may occur, EDN3-RA+ culture promotes the development of SOX10+S100β+ cells while EDN3+RA+ culture promotes the development of SOX10-S100β+ cells. These two populations may represent distinct subpopulations of enteric glia. Joseph et al. observed that 26% of S100β+ gut cells expressed the ENS stem cell marker integrin alpha2 and were capable of forming multilineage neurospheres in culture [55]. They hypothesized that enteric glia may be heterogeneous in their ability to form neurons. While it is established the multipotent ENS precursors express EDNRB, EDNRB expression in subpopulations of cells along the neural and glial differentiation pathways has not been studied making conclusions regarding the lack of change of EDNRB expression in response to RA difficult to interpret. RA does not affect the total number of S100β+RET+ mouse p75NTR-immunoselected ENS precursors after 7 days in culture [21]. It is likely that the S100β+ cells we studied are a different population. RA treatment was also associated with a statistically significant decrease in the abundance of peripherin+ cells bearing neuronal morphology. This is surprising because RA increases neuronal differentiation in other neuronal lineages [21,64–67]. As such, we speculate that alterations in the proportion of neurons are a reflection of relatively increased glial abundance rather than a direct effect of RA on neurogenesis. Since the proportion of each lineage was measured separately, we could not discern the relative ratios of each lineage, so appraisal of the effects of RA on neurogenesis in this system requires further study.
Contrasting our observations with those of Sato and Heuckeroth [21], our RA-induced decrease in neuronal abundance may reflect subtle but important differences in our culture method. Specifically, there were differences in culture medium components (neurobasal vs. DMEM), RA concentration (10-6 vs. 10-7M), and oxygen concentration during culture incubation (21% vs. ~5%). As the half-life of RA and its metabolites is affected by these parameters, it is reasonable to assume that conditions that most closely match physiologic conditions in utero better reflect the underlying biology. The limitations of reductionist in vitro systems highlight the necessity for further in vivo work to shed light on how retinoid deficiency or excess might affect the developing ENS.
In contrast to RA, we found that EDN3 decreases S100β+ cell proliferation and decreases their abundance in 3-day cultures. Our data contrast with another study showing that EDN3 promotes the expansion of glia from multipotent precursors. However, the cells assayed were derived from quail trunk neural crest at an earlier developmental stage, and a distinct glial marker (Schwann Cell Myelin Protein) was employed [68]. Furthermore, enteric glia express functional EDNRB receptors and elaborate EDNs, and rats in which functional EDNRB receptors are absent feature accelerated expression of S100 (a Schwann cell marker) in developing nerves [61,69]. This aligns with our observation that EDN3 acts as negative regulator of S100β-expressing cells. Despite effects on glial abundance, EDN3 did not affect proliferation or abundance of neurons or myofibroblasts in culture.
In addition to lineage-specific proliferative changes, we also observed an increase in the overall proportion of proliferating cells in response to RA and in response to EDN3 While the increase in response to RA treatment may be attributable to proliferation amongst the glial lineage discussed above, we did not identify any lineage-specific increases in response to EDN3 in our experiments. As such, we may infer that the global increase in proliferation in response to EDN3 also occurs subtly in one or more specific lineages, but below the threshold of statistical significance, or occurs in a population not detected by us, i.e. peripherin-SMA-S100β- cells. The nature of this population is unclear, but based on previous work we can speculate that these cells may be immature neural crest-derived cells, as EDN3 has been shown to stimulate proliferation of enteric neuronal precursors under similar culture conditions [5,10,11].
The RET receptor is expressed on proliferating neurogenic and gliogenic precursors and several proteins, including SOX10, are known to regulate its transcription [70,71]. These precursors continue to express RET as they differentiate into neurons but lose RET expression if they differentiate into glia [35,72–74]. EDN and RET signaling act synergistically to permit normal development of the ENS [17,75] and interaction probably occurs at the level of common downstream intracellular signals [5]. The effect of RA on Ret expression is less clear and likely depends on the model system being studied. RA is known to be a potent activator of Ret in neuroblastoma cell lines [76–78], but down-regulates Ret in models of cardiac development [79]. Niederreither et al. observed that deletion of Raldh2 is associated with a decrease in Ret-expressing neuroblasts, suggesting that RA is necessary to maintain Ret expression or Ret-expressing cells [29]. In a murine cell population resembling ours, RA was shown to stimulate the proliferation of a subpopulation of RET+ neuroblasts [21]. We observed a decrease in Ret mRNA levels in RA-treated ENS precursor cultures. These data are consistent with the gliogenic effect and the decline in peripherin+ cells in our 3-day cultures in response to RA. Curiously, an RA-induced decrease in Ret mRNA was also noted in 14-day cultures coinciding with an RA-induced increase in neuronal abundance. This contrasts with an RA-associated increase in the abundance of RET+TUJ+ neurons in a similar model [21]. Our data therefore suggest that our culture conditions (which differ from those used by Sato and Heuckeroth – see above) are more conducive to the development of RET-negative neurons. Alternatively, Ret gene expression is likely not proportional to RET surface protein expression.
As glia and Sox10 appear to be responsive to both RA and EDN3 signaling, we hypothesized that the two signals could reciprocally control each other. Since RA modulates EDN signaling in other contexts, we sought this phenomenon in ENS precursor cultures [30–33]. RA treatment decreased EDN3 and the EDN3 activating enzyme, Ece1 mRNA levels in our ENS precursor cultures. The mechanism by which RA modulates EDN3 and Ece1 expression is unclear. Direct transcriptional repression is unlikely because the EDN3 and Ece1 promoters are not purported to contain canonical RA response elements (RAREs). Nevertheless, it is possible that RA regulates expression via non-canonical RAREs, directly via RA receptor independent mechanisms, or indirectly via other intermediates. However, given the increase in s100β+ cells in response to RA, a more plausible explanation is that the glia we studied scarcely express EDN3/Ece1 and their expansion is therefore associated with a net decrease in EDN3/Ece1. While EDN3 transcript levels remained low in the differentiated 14 day cultures, Ednrb transcripts are significantly increased by culture in the presence of RA. The increase in Ednrb transcript levels in 14 day cultures may be the result of long-term RA suppression of EDN3 resulting in a compensatory increase in Ednrb expression or may be related to altered phenotypic differentiation. Alternatively, the increased abundance of Ednrb reflects the expansion of enteric glia and neurons in RA-treated cultures, both of which are known to express this receptor [43,61]. Further study of the expression of ECE1, EDRNB, EDN3 and RA-related proteins will be necessary to address these issues.
Our defined culture system induced ectopic EDN3 in ENS precursors after 3 days in culture, which differs from the in vivo observation that EDN3 is expressed by the developing gut mesenchyme, not by ENS precursors [5,9,10]. Although our cultures are enriched for immunoselected p75NTR+ ENS precursors, and the media preferentially supports the growth of these cells, the cultures contain a minority of p75NTR- cells which may be gut mesenchymal cells that express EDN3 However, it is unlikely that the addition of RA to the culture decreases EDN3 expression solely by reducing the percentage of mesenchymal cells in the culture. If this were the case, we would expect an increase in many or all of the other markers of ENS precursors or their derivatives. Instead, RA treatment results in a reduction in Ret expression and no change in Ednrb or Sox10 expression (except in the absence of EDNRB activation) in 3 day cultures. Further, we did not find an increase in the percentage of cells not labeling with peripherin or S100β (Table 2). Though we think it is unlikely, we cannot eliminate the possibility that RA down-regulates EDN3 expression in the mesenchymal p75NTR- cell population. If true, this explanation for the expression of EDN3 in our cultures and its down-regulation by RA supports an interaction of RA and EDNRB signaling in the expression of Hirschsprung’s disease, albeit in a non-neural crest autonomous fashion. Alternatively, vagal crest-derived cells are capable of changing their phenotype in culture, and the expression of Edn3 in vitro may reflect the influence of the culture environment [80]. For example, our data suggest that if in vivo microenvironmental levels of RA are high, EDN3 expression by ENS precursors may be undetectable.
Until now, an influence of EDN3 on RA signaling has not been reported. We now demonstrate that EDN3 treatment reduces Rarb and Rarg levels in ENS precursor cultures. There are two ways that this may be achieved. First, EDN3 may reduce the abundance of ENS precursors that are known to express these receptors [21]. Alternatively, EDN3 treatment may affect transcript levels of these genes. The effects of EDN3 on Raldh2 and Cyp26a1 were more difficult to interpret: In the presence of RA, EDN3 decreased Cyp26a1 levels at 3 days and increased levels at 14 days. EDN3 decreased Raldh2 at 3 days and 14 days, but these phenomena were in association with the absence and presence of RA, respectively. We surmise that the diverse effects of EDN3 on the expression of RA metabolic machinery at different time points and conditions reflect a broader regulatory role for EDN3 in fine-tuning local levels of RA by interfering with RA feedback autoregulation. For instance, the EDN3 induced decrease in Cyp26a1 expression may be a response to the decrease in RA signaling via the RA receptors. Indeed, previous work by others has suggested a link between RA receptor activation and Cyp26a1 expression [81,82]. Moreover, after 14 days of culture, EDN3 may act as a brake on RA production by suppressing Raldh2. An alternative interpretation is that the emergence of diverse cell populations in extended cell culture is itself a cause for altered mRNA levels. Nevertheless, some observed gene expression changes occurring early in the culture period persisted after 14 days, despite the dramatic morphologic changes that occurred. Lastly, it should be recalled that the metabolism of RA is complex and many of its synthetic intermediates and degradation products are themselves bioactive [19,83,84]. The culture medium we used was lacking Vitamin A, an important RA precursor, thus many RA metabolites were likely also lacking. Although we have taken a reductionist approach to study the effects of RA and EDN3, it is our expectation that these studies will be complemented with in vivo and in situ models.
We observed four phenotypes in long-term cultures of ENS precursors that were dependent on the balance of EDN3 and RA: In the absence of RA, EDNRB activation led to exclusive myofibroblast differentiation and EDNRB signaling blockade allowed the emergence of glia in addition to myofibroblasts. RA supports the development of myofibroblasts, neurons and glia but with a strikingly different terminal morphology depending upon the presence or absence of EDNRB signaling. RA in the presence of EDNRB blockade supported the development heterocellular ganglia and a dense interconnected plexus. These findings may have implications regarding the molecular mechanisms that dictate the formation of the ENS in health and disease. It should be noted that long-term changes in cell phenotype, culture morphology, and gene expression were presumed to be attributable to the presence of RA and/or EDN3 However, since the study was limited to a single treatment of RA and/or EDN3 at the beginning of the culture period and the stability of EDN3 and RA in these long-term cultures is unknown, it is also possible that the observed changes are related to changes in the bioavailability of EDN3 and RA over time or that EDN3 and RA initiate specific transcriptional programs that persist in culture regardless of drug stability.
Consistent with previous reports [15,85], we found that EDN3 promoted the differentiation of ENS precursors into myofibroblasts as evidenced by their morphology and by smooth muscle actin immunoreactivity. Interestingly, the addition of RA supported the formation of neuronal and glial cells, in addition to myofibroblasts. Our data suggest that RA and EDN3 compete to promote specific lineages, with RA favoring neuronal/glial and EDN3 favoring myofibroblast formation in culture, while favoring the maintenance of an undifferentiated cell in vivo. This dichotomous differentiation pattern seen in vitro can arise by two distinct mechanisms: The first possibility is that RA and EDN3 each independently induce different lineages and the pattern that arises is dose-dependent. Alternatively, reciprocal inhibition of the opposing pathway may occur, signifying an interaction between the two signaling pathways. In order to discriminate between these two possibilities, we evaluated the morphological characteristics of cultures lacking EDN3 and RA, by simultaneously blocking EDNRB signaling and by not adding RA. Here, although the pattern was primarily myofibroblastic in nature, scant neurons were present and mature glial were represented more prominently. These data suggest that the default pathway of the ENS precursors in this culture system is permissive for myofibroblasts and glia, that EDN3 inhibits glial and neuronal differentiation, and that RA actively promotes neuronal (at 14 days) and glial differentiation.
Taken together, our long-term culture data support the existence of a reciprocal relationship between RA and EDN3 (Figure 7). This relationship may underlie some of the opposing effects that RA and EDN3 exert on ENS precursors. For instance, activation of EDNRB signaling during the development of the ENS has been associated with the inhibition of neuronal differentiation, a decrease in self-renewal and impeded migration of ENS precursors in vitro [11,15]. Conversely, RA favors neurogenesis, migration and ENS precursor proliferation [21,28]. The potential implications of a bidirectional interaction are noteworthy: If RA is capable of lowering the overall pool of EDN3 in the developing gut, then changes in local RA concentrations may be responsible for the tightly-regulated spatiotemporal expression pattern of EDN3 It is also tempting to speculate that since EDN3 modulates Cyp26a1 and Raldh2 expression in vitro, and the proteins encoded by these genes are important in establishing RA gradients in vivo, EDN3 may play a role in the in vivo maintenance of an RA gradient in the gut [26,86,87]. Furthermore, based on our data, it is possible that RA, by effectively eliminating cell stimulation by EDN3, induces intrinsic changes in cells that permit them to differentiate, migrate, and proliferate.
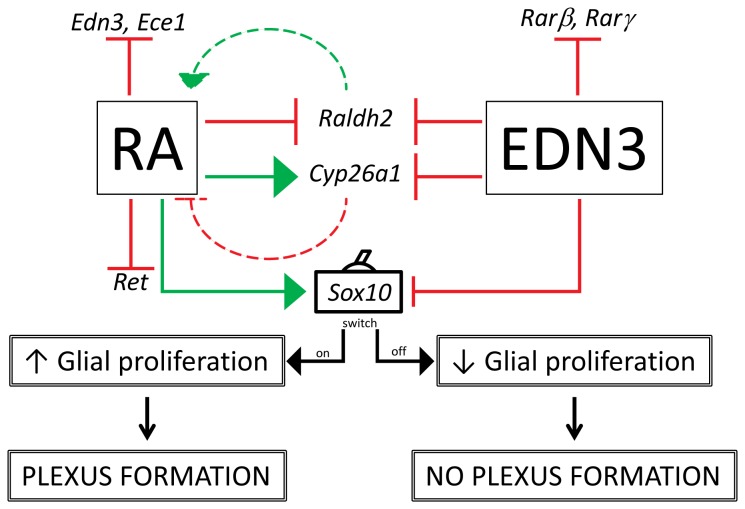
Figure 7. A hypothetical model depicting the bidirectional relationship of RA and EDN3 signaling in ENS precursors.
The above pictogram, based on our data, summarizes and integrates the observed RNA expression profiles and the culture morphological data. Solid green arrows indicate increases in mRNA levels and solid red arrows with flat arrowheads denote decreases in mRNA levels. Although RA and EDN3 levels were not quantified in our culture model, dashed green and red arrows denote putative RA synthesis and degradation, respectively. We hypothesize that RA inhibits EDN signaling in ENS precursors by modulating the gene expression of EDN3 and Ece1, and EDN3 regulates RA signaling by inhibiting RA receptor expression, thereby decreasing RA responsiveness, and fine-tuning RA metabolic enzyme expression, putatively altering RA availability. We propose that RA perpetuates glia by decreasing Ret and enhancing Sox10 expression, and that EDN3 prevents the proliferation of glia by decreasing Sox10. The persistence of glia is conducive to the formation of a heterocellular ganglionated plexus. This model suggests that control of Sox10 marks a key developmental decision point - a switch - that sustains multipotent progenitors in culture and ultimately depends on the balance of RA and EDN3
The formation of ganglia is deemed to be important for the establishment of functional circuits. RA, in concert with EDN signaling blockade, induced the formation of a distinct plexus. Notably, while the addition of EDN3 did not inhibit differentiation of the precursors, it did inhibit the formation of this plexus, suggesting that EDN3 inhibits the RA-induced clustering of neuron cell bodies and glia into ganglia. The formation of a plexus requires differentiation of precursors into glial and neuronal components, neurite and axonal elongation, 3-dimensional spatial arrangements of its constituents, the establishment of synaptic connectivity and neuronal subtype specification [88–92]. Very little is known about the molecular mechanisms controlling each of these processes. Furthermore, there are few models for studying enteric plexuses in vitro. We have developed another model of plexus formation that, by virtue of it being an in vitro culture from undifferentiated cells, is amenable to the study of many aspects of plexus development, structure and function. ENS precursors that are permitted to develop undisturbed in an hypoxic environment in the presence of RA and the EDNRB selective antagonist BQ-788 spontaneously developed into a plexus containing neurons, glia, and supportive tissue, whose 3-dimensional arrangement is very reminiscent of an authentic gastrointestinal neural plexus.
Conclusion
A complex EDN-RA interaction exists that coordinately regulates the development of rat ENS precursors in vitro. These results suggest that environmental RA may modulate the expression of aganglionosis in individuals with endothelin mutations.
Acknowledgments
We are grateful to Ms. Kerry McTigue for her assistance with the preparation of the images for publication. We also are thankful to Ms. Naoko Murakami for her help with maintaining the rat colony and Mary Cismowski for her assistance with the qPCR experiments and manuscript preparation. We also thank Scott Barton for his assistance with using the statistical software. Particular thanks go to Pam Lucchesi for her valuable feedback and advice.
Funding Statement
The authors have no funding or support to report.
References
- 1. Heanue TA, Pachnis V (2007) Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies. Nat Rev Neurosci 8: 466-479. doi:10.1038/nrn2137. PubMed: 17514199. [DOI] [PubMed] [Google Scholar]
- 2. Sasselli V, Pachnis V, Burns AJ (2012) The enteric nervous system. Dev Biol, 366: 64–73. PubMed: 22290331. [DOI] [PubMed] [Google Scholar]
- 3. Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G et al. (2008) Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet 45: 1-14. doi:10.1136/jmg.2007.055129. PubMed: 17965226. [DOI] [PubMed] [Google Scholar]
- 4. Kenny SE, Tam PKH, Garcia-Barcelo M (2010) Hirschsprung’s disease. Semin Pediatr Surg 19: 194-200. doi:10.1053/j.sempedsurg.2010.03.004. PubMed: 20610192. [DOI] [PubMed] [Google Scholar]
- 5. Barlow A, de Graaff E, Pachnis V (2003) Enteric nervous system progenitors are coordinately controlled by the G protein-coupled receptor EDNRB and the receptor tyrosine kinase RET. Neuron 40: 905-916. doi:10.1016/S0896-6273(03)00730-X. PubMed: 14659090. [DOI] [PubMed] [Google Scholar]
- 6. Lee HO, Levorse JM, Shin MK (2003) The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol 259: 162-175. doi:10.1016/S0012-1606(03)00160-X. PubMed: 12812796. [DOI] [PubMed] [Google Scholar]
- 7. Inoue A, Yanagisawa M, Kimura S, Kasuya Y, Miyauchi T et al. (1989) The human endothelin family: three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc Natl Acad Sci U S A 86: 2863-2867. doi:10.1073/pnas.86.8.2863. PubMed: 2649896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC et al. (1998) Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development 125: 825-836. PubMed: 9449665. [DOI] [PubMed] [Google Scholar]
- 9. Leibl MA, Ota T, Woodward MN, Kenny SE, Lloyd DA et al. (1999) Expression of endothelin 3 by mesenchymal cells of embryonic mouse caecum. Gut 44: 246-252. doi:10.1136/gut.44.2.246. PubMed: 9895385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu JJ, Chen JX, Rothman TP, Gershon MD (1999) Inhibition of in vitro enteric neuronal development by endothelin-3: mediation by endothelin B receptors. Development 126: 1161-1173. PubMed: 10021336. [DOI] [PubMed] [Google Scholar]
- 11. Hearn CJ, Murphy M, Newgreen D (1998) GDNF and ET-3 differentially modulate the numbers of avian enteric neural crest cells and enteric neurons in vitro. Dev Biol 197: 93-105. doi:10.1006/dbio.1998.8876. PubMed: 9578621. [DOI] [PubMed] [Google Scholar]
- 12. Gariepy CE, Cass DT, Yanagisawa M (1996) Null mutation of endothelin receptor type B gene in spotting lethal rats causes aganglionic megacolon and white coat color. Proc Natl Acad Sci U S A 93: 867-872. doi:10.1073/pnas.93.2.867. PubMed: 8570650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gariepy CE, Williams SC, Richardson JA, Hammer RE, Yanagisawa M (1998) Transgenic expression of the endothelin-B receptor prevents congenital intestinal aganglionosis in a rat model of Hirschsprung disease. J Clin Invest 102: 1092-1101. doi:10.1172/JCI3702. PubMed: 9739043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Druckenbrod NR, Epstein ML (2007) Behavior of enteric neural crest-derived cells varies with respect to the migratory wavefront. Dev Dyn 236: 84-92. doi:10.1002/dvdy.20974. PubMed: 17039523. [DOI] [PubMed] [Google Scholar]
- 15. Kruger GM, Mosher JT, Tsai YH, Yeager KJ, Iwashita T et al. (2003) Temporally distinct requirements for endothelin receptor B in the generation and migration of gut neural crest stem cells. Neuron 40: 917-929. doi:10.1016/S0896-6273(03)00727-X. PubMed: 14659091. [DOI] [PubMed] [Google Scholar]
- 16. Cantrell VA, Owens SE, Chandler RL, Airey DC, Bradley KM et al. (2004) Interactions between Sox10 and EdnrB modulate penetrance and severity of aganglionosis in the Sox10Dom mouse model of Hirschsprung disease. Hum Mol Genet 13: 2289-2301. doi:10.1093/hmg/ddh243. PubMed: 15294878. [DOI] [PubMed] [Google Scholar]
- 17. Carrasquillo MM, McCallion AS, Puffenberger EG, Kashuk CS, Nouri N et al. (2002) Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet 32: 237-244. doi:10.1038/ng998. PubMed: 12355085. [DOI] [PubMed] [Google Scholar]
- 18. Blomhoff R, Blomhoff HK (2006) Overview of retinoid metabolism and function. J Neurobiol 66: 606-630. doi:10.1002/neu.20242. PubMed: 16688755. [DOI] [PubMed] [Google Scholar]
- 19. Napoli JL (2012) Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta Mol Cell Biol Lipids 1821: 152-167. doi:10.1016/j.bbalip.2011.05.004. PubMed: 21621639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niederreither K, McCaffery P, Dräger UC, Chambon P, Dollé P (1997) Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev 62: 67-78. doi:10.1016/S0925-4773(96)00653-3. PubMed: 9106168. [DOI] [PubMed] [Google Scholar]
- 21. Sato Y, Heuckeroth RO (2008) Retinoic acid regulates murine enteric nervous system precursor proliferation, enhances neuronal precursor differentiation, and reduces neurite growth in vitro. Dev Biol 320: 185-198. doi:10.1016/j.ydbio.2008.05.524. PubMed: 18561907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhinn M, Dollé P (2012) Retinoic acid signalling during development. Development 139: 843-858. doi:10.1242/dev.065938. PubMed: 22318625. [DOI] [PubMed] [Google Scholar]
- 23. White JA, Guo YD, Baetz K, Beckett-Jones B, Bonasoro J et al. (1996) Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J Biol Chem 271: 29922-29927. doi:10.1074/jbc.271.47.29922. PubMed: 8939936. [DOI] [PubMed] [Google Scholar]
- 24. Thatcher JE, Isoherranen N (2009) The role of CYP26 enzymes in retinoic acid clearance. Expert Opin Drug Metab Toxicol 5: 875-886. doi:10.1517/17425250903032681. PubMed: 19519282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abu-Abed S, MacLean G, Fraulob V, Chambon P, Petkovich M et al. (2002) Differential expression of the retinoic acid-metabolizing enzymes CYP26A1 and CYP26B1 during murine organogenesis. Mech Dev 110: 173-177. doi:10.1016/S0925-4773(01)00572-X. PubMed: 11744378. [DOI] [PubMed] [Google Scholar]
- 26. White RJ, Nie Q, Lander AD, Schilling TF (2007) Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLOS Biol 5: e304. doi:10.1371/journal.pbio.0050304. PubMed: 18031199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pitera JE, Smith VV, Woolf AS, Milla PJ (2001) Embryonic gut anomalies in a mouse model of retinoic Acid-induced caudal regression syndrome: delayed gut looping, rudimentary cecum, and anorectal anomalies. Am J Pathol 159: 2321-2329. doi:10.1016/S0002-9440(10)63082-9. PubMed: 11733381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu M, Sato Y, Lyons-Warren A, Zhang B, Kane MA et al. (2010) Vitamin A facilitates enteric nervous system precursor migration by reducing Pten accumulation. Development 137: 631-640. doi:10.1242/dev.040550. PubMed: 20110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niederreither K, Vermot J, Le Roux I, Schuhbaur B, Chambon P et al. (2003) The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development 130: 2525-2534. doi:10.1242/dev.00463. PubMed: 12702665. [DOI] [PubMed] [Google Scholar]
- 30. Chi X, Anselmi K, Watkins S, Gandhi CR (2003) Prevention of cultured rat stellate cell transformation and endothelin-B receptor upregulation by retinoic acid. Br J Pharmacol 139: 765-774. doi:10.1038/sj.bjp.0705303. PubMed: 12813000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsu JY, Pfahl M (1998) ET-1 expression and growth inhibition of prostate cancer cells: a retinoid target with novel specificity. Cancer Res 58: 4817-4822. PubMed: 9809984. [PubMed] [Google Scholar]
- 32. Lehrke I, Schaier M, Schade K, Morath C, Waldherr R et al. (2002) Retinoid receptor-specific agonists alleviate experimental glomerulonephritis. Am J Physiol Renal Physiol 282: F741-F751. PubMed: 11880336. [DOI] [PubMed] [Google Scholar]
- 33. Yokota J, Kawana M, Hidai C, Aoka Y, Ichikawa K et al. (2001) Retinoic acid suppresses endothelin-1 gene expression at the transcription level in endothelial cells. Atherosclerosis 159: 491-496. doi:10.1016/S0021-9150(01)00530-5. PubMed: 11730831. [DOI] [PubMed] [Google Scholar]
- 34. Chalazonitis A, D’Autréaux F, Pham TD, Kessler JA, Gershon MD (2011) Bone morphogenetic proteins regulate enteric gliogenesis by modulating ErbB3 signaling. Dev Biol 350: 64-79. doi:10.1016/j.ydbio.2010.11.017. PubMed: 21094638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chalazonitis A, Rothman TP, Chen J, Gershon MD (1998) Age-dependent differences in the effects of GDNF and NT-3 on the development of neurons and glia from neural crest-derived precursors immunoselected from the fetal rat gut: expression of GFRalpha-1 in vitro and in vivo. Dev Biol 204: 385-406. doi:10.1006/dbio.1998.9090. PubMed: 9882478. [DOI] [PubMed] [Google Scholar]
- 36. Morrison SJ, White PM, Zock C, Anderson DJ (1999) Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell 96: 737-749. doi:10.1016/S0092-8674(00)80583-8. PubMed: 10089888. [DOI] [PubMed] [Google Scholar]
- 37. Morrison SJ, Csete M, Groves AK, Melega W, Wold B et al. (2000) Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J Neurosci 20: 7370-7376. PubMed: 11007895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bondurand N, Natarajan D, Barlow A, Thapar N, Pachnis V (2006) Maintenance of mammalian enteric nervous system progenitors by SOX10 and endothelin 3 signalling. Development 133: 2075-2086. doi:10.1242/dev.02375. PubMed: 16624853. [DOI] [PubMed] [Google Scholar]
- 39. Mollaaghababa R, Pavan WJ (2003) The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene 22: 3024-3034. doi:10.1038/sj.onc.1206442. PubMed: 12789277. [DOI] [PubMed] [Google Scholar]
- 40. Paratore C, Eichenberger C, Suter U, Sommer L (2002) Sox10 haploinsufficiency affects maintenance of progenitor cells in a mouse model of Hirschsprung disease. Hum Mol Genet 11: 3075-3085. doi:10.1093/hmg/11.24.3075. PubMed: 12417529. [DOI] [PubMed] [Google Scholar]
- 41. Paratore C, Goerich DE, Suter U, Wegner M, Sommer L (2001) Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development 128: 3949-3961. PubMed: 11641219. [DOI] [PubMed] [Google Scholar]
- 42. Stanchina L, Baral V, Robert F, Pingault V, Lemort N et al. (2006) Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev Biol 295: 232-249. doi:10.1016/j.ydbio.2006.03.031. PubMed: 16650841. [DOI] [PubMed] [Google Scholar]
- 43. Zhu L, Lee HO, Jordan CS, Cantrell VA, Southard-Smith EM et al. (2004) Spatiotemporal regulation of endothelin receptor-B by SOX10 in neural crest-derived enteric neuron precursors. Nat Genet 36: 732-737. doi:10.1038/ng1371. PubMed: 15170213. [DOI] [PubMed] [Google Scholar]
- 44. Lantieri F, Griseri P, Ceccherini I (2006) Molecular mechanisms of RET-induced Hirschsprung pathogenesis. Ann Med 38: 11-19. doi:10.1080/07853890500442758. PubMed: 16448984. [DOI] [PubMed] [Google Scholar]
- 45. Chomienne C, Balitrand N, Ballerini P, Castaigne S, de Thé H et al. (1991) All-trans retinoic acid modulates the retinoic acid receptor-alpha in promyelocytic cells. J Clin Invest 88: 2150-2154. doi:10.1172/JCI115547. PubMed: 1661301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de The H, Marchio A, Tiollais P, Dejean A (1989) Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J 8: 429-433. PubMed: 2542014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Thé H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A (1990) Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature 343: 177-180. doi:10.1038/343177a0. PubMed: 2153268. [DOI] [PubMed] [Google Scholar]
- 48. Grummer MA, Salih ZN, Zachman RD (2000) Effect of retinoic acid and ethanol on retinoic acid receptor beta and glial fibrillary acidic protein mRNA expression in human astrocytoma cells. Neurosci Lett 294: 73-76. doi:10.1016/S0304-3940(00)01538-X. PubMed: 11058790. [DOI] [PubMed] [Google Scholar]
- 49. Haq R, Pfahl M, Chytil F (1991) Retinoic acid affects the expression of nuclear retinoic acid receptors in tissues of retinol-deficient rats. Proc Natl Acad Sci U S A 88: 8272-8276. doi:10.1073/pnas.88.18.8272. PubMed: 1654565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kamei Y, Kawada T, Kazuki R, Sugimoto E (1993) Retinoic acid receptor gamma 2 gene expression is up-regulated by retinoic acid in 3T3-L1 preadipocytes. Biochem J 293(3): 807-812. PubMed: 8394693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lehmann JM, Zhang XK, Pfahl M (1992) RAR gamma 2 expression is regulated through a retinoic acid response element embedded in Sp1 sites. Mol Cell Biol 12: 2976-2985. PubMed: 1320193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leroy P, Krust A, Zelent A, Mendelsohn C, Garnier JM et al. (1991) Multiple isoforms of the mouse retinoic acid receptor alpha are generated by alternative splicing and differential induction by retinoic acid. EMBO J 10: 59-69. PubMed: 1846598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leroy P, Nakshatri H, Chambon P (1991) Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc Natl Acad Sci U S A 88: 10138-10142. doi:10.1073/pnas.88.22.10138. PubMed: 1658797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sucov HM, Murakami KK, Evans RM (1990) Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc Natl Acad Sci U S A 87: 5392-5396. doi:10.1073/pnas.87.14.5392. PubMed: 2164682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Joseph NM, He S, Quintana E, Kim YG, Núñez G et al. (2011) Enteric glia are multipotent in culture but primarily form glia in the adult rodent gut. J Clin Invest 121: 3398-3411. doi:10.1172/JCI58186. PubMed: 21865643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Laranjeira C, Sandgren K, Kessaris N, Richardson W, Potocnik A et al. (2011) Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J Clin Invest 121: 3412-3424. doi:10.1172/JCI58200. PubMed: 21865647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N et al. (1994) Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell 79: 1277-1285. doi:10.1016/0092-8674(94)90018-3. PubMed: 8001160. [DOI] [PubMed] [Google Scholar]
- 58. Kapur RP, Sweetser DA, Doggett B, Siebert JR, Palmiter RD (1995) Intercellular signals downstream of endothelin receptor-B mediate colonization of the large intestine by enteric neuroblasts. Development 121: 3787-3795. PubMed: 8582288. [DOI] [PubMed] [Google Scholar]
- 59. Nataf V, Lecoin L, Eichmann A, Le Douarin NM (1996) Endothelin-B receptor is expressed by neural crest cells in the avian embryo. Proc Natl Acad Sci U S A 93: 9645-9650. doi:10.1073/pnas.93.18.9645. PubMed: 8790384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hotta R, Stamp LA, Foong JP, McConnell SN, Bergner AJ et al. (2013) Transplanted progenitors generate functional enteric neurons in the postnatal colon. J Clin Invest 123: 1182-1191. doi:10.1172/JCI65963. PubMed: 23454768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. von Boyen GB, Degenkolb N, Hartmann C, Adler G, Steinkamp M (2010) The endothelin axis influences enteric glia cell functions. Med Sci Monit 16: BR161–7: BR161- 167. PubMed: 20512083 [PubMed] [Google Scholar]
- 62. Gershon MD (2011) Behind an enteric neuron there may lie a glial cell. J Clin Invest 121: 3386-3389. doi:10.1172/JCI59573. PubMed: 21865648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kawaguchi J, Nichols J, Gierl MS, Faial T, Smith A (2010) Isolation and propagation of enteric neural crest progenitor cells from mouse embryonic stem cells and embryos. Development 137: 693-704. doi:10.1242/dev.046896. PubMed: 20147374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baldassarre G, Boccia A, Bruni P, Sandomenico C, Barone MV et al. (2000) Retinoic acid induces neuronal differentiation of embryonal carcinoma cells by reducing proteasome-dependent proteolysis of the cyclin-dependent inhibitor p27. Cell Growth Differ 11: 517-526. PubMed: 11063125. [PubMed] [Google Scholar]
- 65. Elmi M, Faigle R, Yang W, Matsumoto Y, Rosenqvist E et al. (2007) Mechanism of MASH1 induction by ASK1 and ATRA in adult neural progenitors. Mol Cell Neurosci 36: 248-259. doi:10.1016/j.mcn.2007.07.001. PubMed: 17728141. [DOI] [PubMed] [Google Scholar]
- 66. Kumar M, Bagchi B, Gupta SK, Meena AS, Gressens P et al. (2007) Neurospheres derived from human embryoid bodies treated with retinoic Acid show an increase in nestin and ngn2 expression that correlates with the proportion of tyrosine hydroxylase-positive cells. Stem Cells Dev 16: 667-681. doi:10.1089/scd.2006.0115. PubMed: 17784840. [DOI] [PubMed] [Google Scholar]
- 67. Scheibe RJ, Ginty DD, Wagner JA (1991) Retinoic acid stimulates the differentiation of PC12 cells that are deficient in cAMP-dependent protein kinase. J Cell Biol 113: 1173-1182. doi:10.1083/jcb.113.5.1173. PubMed: 1645738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lahav R, Dupin E, Lecoin L, Glavieux C, Champeval D et al. (1998) Endothelin 3 selectively promotes survival and proliferation of neural crest-derived glial and melanocytic precursors in vitro. Proc Natl Acad Sci U S A 95: 14214-14219. doi:10.1073/pnas.95.24.14214. PubMed: 9826680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brennan A, Dean CH, Zhang AL, Cass DT, Mirsky R et al. (2000) Endothelins control the timing of Schwann cell generation in vitro and in vivo. Dev Biol 227: 545-557. doi:10.1006/dbio.2000.9887. PubMed: 11071773. [DOI] [PubMed] [Google Scholar]
- 70. Bachetti T, Borghini S, Ravazzolo R, Ceccherini I (2005) An in vitro approach to test the possible role of candidate factors in the transcriptional regulation of the RET proto-oncogene. Gene Expr 12: 137-149. doi:10.3727/000000005783992106. PubMed: 16127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leon TY, Ngan ES, Poon HC, So MT, Lui VC et al. (2009) Transcriptional regulation of RET by Nkx2-1, Phox2b, Sox10, and Pax3. J Pediatr Surg 44: 1904-1912. doi:10.1016/j.jpedsurg.2008.11.055. PubMed: 19853745. [DOI] [PubMed] [Google Scholar]
- 72. Bixby S, Kruger GM, Mosher JT, Joseph NM, Morrison SJ (2002) Cell-intrinsic differences between stem cells from different regions of the peripheral nervous system regulate the generation of neural diversity. Neuron 35: 643-656. doi:10.1016/S0896-6273(02)00825-5. PubMed: 12194865. [DOI] [PubMed] [Google Scholar]
- 73. Heuckeroth RO, Lampe PA, Johnson EM, Milbrandt J (1998) Neurturin and GDNF promote proliferation and survival of enteric neuron and glial progenitors in vitro. Dev Biol 200: 116-129. doi:10.1006/dbio.1998.8955. PubMed: 9698461. [DOI] [PubMed] [Google Scholar]
- 74. Young HM, Ciampoli D, Hsuan J, Canty AJ (1999) Expression of Ret-, p75(NTR)-, Phox2a-, Phox2b-, and tyrosine hydroxylase-immunoreactivity by undifferentiated neural crest-derived cells and different classes of enteric neurons in the embryonic mouse gut. Dev Dyn 216: 137-152. doi:10.1002/(SICI)1097-0177(199910)216:2. PubMed: 10536054. [DOI] [PubMed] [Google Scholar]
- 75. McCallion AS, Stames E, Conlon RA, Chakravarti A (2003) Phenotype variation in two-locus mouse models of Hirschsprung disease: tissue-specific interaction between Ret and Ednrb. Proc Natl Acad Sci U S A 100: 1826-1831. doi:10.1073/pnas.0337540100. PubMed: 12574515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Patrone G, Puliti A, Bocciardi R, Ravazzolo R, Romeo G (1997) Sequence and characterisation of the RET proto-oncogene 5' flanking region: analysis of retinoic acid responsiveness at the transcriptional level. FEBS Lett 419: 76-82. doi:10.1016/S0014-5793(97)01435-X. PubMed: 9426223. [DOI] [PubMed] [Google Scholar]
- 77. Yamada S, Nomura T, Uebersax L, Matsumoto K, Fujita S et al. (2007) Retinoic acid induces functional c-Ret tyrosine kinase in human neuroblastoma. Neuroreport 18: 359-363. doi:10.1097/WNR.0b013e32801299b4. PubMed: 17435603. [DOI] [PubMed] [Google Scholar]
- 78. Angrisano T, Sacchetti S, Natale F, Cerrato A, Pero R et al. (2011) Chromatin and DNA methylation dynamics during retinoic acid-induced RET gene transcriptional activation in neuroblastoma cells. Nucleic Acids Res 39: 1993-2006. doi:10.1093/nar/gkq864. PubMed: 20952403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shoba T, Dheen ST, Tay SS (2002) Retinoic acid influences the expression of the neuronal regulatory genes Mash-1 and c-ret in the developing rat heart. Neurosci Lett 318: 129-132. doi:10.1016/S0304-3940(01)02491-0. PubMed: 11803116. [DOI] [PubMed] [Google Scholar]
- 80. Anderson RB, Stewart AL, Young HM (2006) Phenotypes of neural-crest-derived cells in vagal and sacral pathways. Cell Tissue Res 323: 11-25. doi:10.1007/s00441-005-0047-6. PubMed: 16133146. [DOI] [PubMed] [Google Scholar]
- 81. Hu P, Tian M, Bao J, Xing G, Gu X et al. (2008) Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn 237: 3798-3808. doi:10.1002/dvdy.21801. PubMed: 19035346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Idres N, Marill J, Chabot GG (2005) Regulation of CYP26A1 expression by selective RAR and RXR agonists in human NB4 promyelocytic leukemia cells. Biochem Pharmacol 69: 1595-1601. doi:10.1016/j.bcp.2005.02.024. PubMed: 15896339. [DOI] [PubMed] [Google Scholar]
- 83. Marill J, Idres N, Capron CC, Nguyen E, Chabot GG (2003) Retinoic acid metabolism and mechanism of action: a review. Curr Drug Metab 4: 1-10. doi:10.2174/1389200033489370. PubMed: 12570742. [DOI] [PubMed] [Google Scholar]
- 84. Ross AC, Zolfaghari R, Weisz J (2001) Vitamin A: recent advances in the biotransformation, transport, and metabolism of retinoids. Curr Opin Gastroenterol 17: 184-192. doi:10.1097/00001574-200103000-00015. PubMed: 11224677. [DOI] [PubMed] [Google Scholar]
- 85. Nagy N, Goldstein AM (2006) Endothelin-3 regulates neural crest cell proliferation and differentiation in the hindgut enteric nervous system. Dev Biol 293: 203-217. doi:10.1016/j.ydbio.2006.01.032. PubMed: 16519884. [DOI] [PubMed] [Google Scholar]
- 86. Dobbs-McAuliffe B, Zhao Q, Linney E (2004) Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev 121: 339-350. doi:10.1016/j.mod.2004.02.008. PubMed: 15110044. [DOI] [PubMed] [Google Scholar]
- 87. Villablanca EJ, Wang S, de Calisto J, Gomes DC, Kane MA et al. (2011) MyD88 and retinoic acid signaling pathways interact to modulate gastrointestinal activities of dendritic cells. Gastroenterology 141: 176-185. doi:10.1053/j.gastro.2011.04.010. PubMed: 21596042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hackett-Jones EJ, Landman KA, Newgreen DF, Zhang D (2011) On the role of differential adhesion in gangliogenesis in the enteric nervous system. J Theor Biol 287: 148-159. doi:10.1016/j.jtbi.2011.07.013. PubMed: 21816161. [DOI] [PubMed] [Google Scholar]
- 89. Faure C, Chalazonitis A, Rhéaume C, Bouchard G, Sampathkumar SG et al. (2007) Gangliogenesis in the enteric nervous system: roles of the polysialylation of the neural cell adhesion molecule and its regulation by bone morphogenetic protein-4. Dev Dyn 236: 44-59. doi:10.1002/dvdy.20943. PubMed: 16958105. [DOI] [PubMed] [Google Scholar]
- 90. Landman KA, Fernando AE, Zhang D, Newgreen DF (2011) Building stable chains with motile agents: Insights into the morphology of enteric neural crest cell migration. J Theor Biol 276: 250-268. doi:10.1016/j.jtbi.2011.01.043. PubMed: 21296089. [DOI] [PubMed] [Google Scholar]
- 91. Simpson MJ, Zhang DC, Mariani M, Landman KA, Newgreen DF (2007) Cell proliferation drives neural crest cell invasion of the intestine. Dev Biol 302: 553-568. doi:10.1016/j.ydbio.2006.10.017. PubMed: 17178116. [DOI] [PubMed] [Google Scholar]
- 92. Simpson MJ, Landman KA, Hughes BD, [!(surname)!], [!(surname)!] (2006) Looking inside an invasion wave of cells using continuum models: proliferation is the key. J Theor Biol 243: 343-360. doi:10.1016/j.jtbi.2006.06.021. PubMed: 16904698. [DOI] [PubMed] [Google Scholar]