Abstract
Proline dehydrogenase (Prodh) and Δ1-pyrroline-5-carboxylate dehydrogenase (P5Cdh) are two key enzymes in the cellular biogenesis of glutamate. Recombinant Prodh and P5Cdh proteins of the chestnut blight fungus Cryphonectria parasitica were investigated and showed activity in in vitro assays. Additionally, the C. parasitica Prodh and P5Cdh genes were able to complement the Saccharomyces cerevisiae put1 and put2 null mutants, respectively, to allow these proline auxotrophic yeast mutants to grow on media with proline as the sole source of nitrogen. Deletion of the Prodh gene in C. parasitica resulted in hypovirulence and a lower level of sporulation, whereas deletion of P5Cdh resulted in hypovirulence though no effect on sporulation; both Δprodh and Δp5cdh mutants were unable to grow on minimal medium with proline as the sole nitrogen source. In a wild-type strain, the intracellular level of proline and the activity of Prodh and P5Cdh increased after supplementation of exogenous proline, though the intracellular Δ1-pyrroline-5-carboxylate (P5C) content remained unchanged. Prodh and P5Cdh were both transcriptionally down-regulated in cells infected with hypovirus. The disruption of other genes with products involved in the conversion of arginine to ornithine, ornithine and glutamate to P5C, and P5C to proline in the cytosol did not appear to affect virulence; however, asexual sporulation was reduced in the Δpro1 and Δpro2 mutants. Taken together, our results showed that Prodh, P5Cdh and related mitochondrial functions are essential for virulence and that proline/glutamate pathway components may represent down-stream targets of hypovirus regulation in C. parasitica.
Introduction
Proline dehydrogenase (Prodh), also known as proline oxidase (POX) in mammals, was first isolated in 1978 from Escherichia coli and shown to have the ability to catalyze the conversion of proline to Δ1-pyrroline-5-carboxylate (P5C) [1]. A year later, the ortholog of Prodh in the yeast Saccharomyces cerevisiae, Put1-encoded proline dehydrogenase, was shown to be able to convert proline to P5C, which is then converted to glutamate by P5C dehydrogenase (P5Cdh) encoded by Put2 [2]. The Put1 gene is present as a single copy in the yeast genome [3], and its product possesses a membrane anchor that causes it to loosely associate with the inner mitochondrial membrane [4]. Intracellular L-proline was found to accumulate to a higher level in put1 mutants, conferring increased tolerance to freezing and desiccation stress [5] and inhibition to cellular apoptosis induced by H2O2 [6]. Put2 localizes to the mitochondrial matrix and was shown to enhance cell growth by improving anaerobic arginine catabolism [7]. Under aerobic conditions, put2 mutants suffer from cellular toxicity due to the accumulation of P5C, which induces an increased level of reactive oxygen species (ROS) in the cell [8]. However, the biological functions of Prodh and P5Cdh have not been previrously reported in the pathogenic fungi.
Cryphonectria parasitica is the pathogen responsible for the destructive chestnut blight that swept the once-dominant chestnut forests in North America, and interactions between C. parasitica and its host have been a major focus of modern plant pathology since its first report in 1904 [9]. Efforts in genetics [10], [11], biochemistry [12], molecular biology [13], and, recently, “omics” [14], [15] have been put forward to elucidate the regulation of C. parasitica virulence with regard to chestnut. Of particular significance is the discovery of hypoviruses and their use in the dissection of the components and regulation of fungal virulence [16]. It is now known that the trimeric G protein signaling pathway [17]–[21], the inositol triphosphate (IP3)/Ca2+/calmodulin signaling pathway [22], and the mitogen-activated protein kinase (MAPK) signaling pathway are essential for C. parasitica virulence [23]–[26]. In addition to these pathways, genes functioning in the methylation pathway [27] and in apoptosis [28] were also found to be required for virulence.
A mitochondrial dysfunction mutant [29] was shown to have a hypovirulence phenotype and gene expression patterns very similar to those caused by hypovirus infection [30], suggesting that a hypovirus may exert its effect by perturbating an important mitochondrial function. However, this mitochondrial function that is essential for C. parasitica virulence remains unknown. Additionally, the association of Prodh and P5Cdh with hypovirus has not yet been reported.
In this study, we analyzed the effects of the genes involved in the proline/glutamate pathway on virulence and other traits and on global gene expression patterns in C. parasitica using gene knockout technology. We demonstrate that Prodh and P5Cdh are essential for virulence and that Prodh, Pro1 and Pro2 are required for sporulation in C. parasitica. Evidence was also obtained showing that the accumulation of both Prodh and P5Cdh transcripts was suppressed by hypovirus infection.
Materials and Methods
Fungal strains and growth conditions
The hypovirus CHV1-EP713-infected C. parasitica strain EP713 (ATCC 52571), its isogenic virus-free parent EP155 (ATCC 38755), and a highly efficient homologous recombination strain, Δku80 [31], were maintained on PDA (Difco) plates under a constant light-dark (12 h/12 h) cycle at 25°C. For liquid culture, EP complete medium [32] was used, and the cultures were incubated at 28°C with shaking at 200 rpm. Preparation of the primary inocula for liquid cultures was performed as previously described [33]. Radial growth on the plates was assessed by measuring the diameter of the colonies [34].
Alignment and phylogenetic analysis
Prodh and P5Cdh in the species of interest were identified by searching the NCBI genomic BLAST databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the amino acid (aa) sequences of the proteins of interest were downloaded from the NCBI protein database (http://www.ncbi.nlm.nih.gov/protein/). Alignment was performed using the alignment program in the Vector NTI 11.0 software. Phylogenetic trees were constructed with amino acid sequences using Neighbor-joining, minimum-evolution, and maximum-parsimony methods in the MEGA4.0 software. The sequence alignment data were bootstrapped with 1,000 resamplings of the alignments to assess the robustness of the lineages in the trees. The trees were visualized using the software TreeView.
Prodh and P5Cdh enzymatic activity assays
Total RNA was isolated from vegetative hyphae as previously described [33], and cDNA was prepared using the First Strand cDNA Synthesis Kit from Roche Applied Science (Mannheim, Germany). The cDNA of Prodh (JGI protein ID 277618) from C. parasitica without the mitochondrial transit peptide (mTP) (aa positions 1–21) and the cDNA of P5Cdh (JGI protein ID 344979) from C. parasitica without the mTP (aa positions 1–45) were generated by site-directed mutagenesis PCR; the resulting fragments were named ProdhΔ21 and P5CdhΔ45, respectively. ProdhΔ21 and P5CdhΔ45 were subcloned into the pGEX-4T-1 vector using the EcoR I, Hind III, and Sal I restriction sites to generate recombinant expression constructs pGEX-4T-1- ProdhΔ21 and pGEX-4T-1- P5CdhΔ45, respectively. Positive clones were selected by transformation of the constructs into E. coli BL21 (DE3) pLysS and screened on LA plates containing chloramphenicol (34 μg/mL) and ampicillin (50 μg/mL). The selected single colonies were inoculated into 5 mL of LB broth containing chloramphenicol and ampicillin at appropriate concentrations and incubated overnight to generate the primary inocula. A primary inoculum of 10 mL was then used to inoculate 1 L of LB containing the necessary antibiotics and incubated at 37°C with shaking at 200 rpm until OD600 = 0.4; ProdhΔ21 expression was then induced with 0.1 mM IPTG for 6 h at 16°C and P5CdhΔ45 expression with 0.3 mM IPTG for 8 h at 18°C. The bacteria were then collected by centrifugation at 2,500×g for 10 min at 4°C. A 1-g bacterial pellet was resuspended in 10 mL of PBS buffer, and lysozyme and DNase I were added to a final concentration of 0.2 mg/mL and 20 μg/mL, respectively. The cell suspension was gently stirred for 30 min at 4°C and then centrifuged at 12, 000 × g for 10 min at 4°C to remove the cell debris. The supernatant was applied to a GST Sefinose resin column from Sangon Biotech (Shanghai, China) for affinity purification of the recombinant protein. The recombinant proteins were eluted with 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0). The quality of recombinant proteins was analyzed by SDS-PAGE. The ProdhΔ21 protein was further concentrated using an Amicon 10-kDa cutoff filter from Millipore Corporation (Massachusetts, USA), and the P5CdhΔ45 protein was concentrated using an Amicon 30-kDa cutoff filter. The enzymatic activity of 2 μM ProdhΔ21 protein was assayed in 5 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 2 mM DTT, 20 μM FAD, and 4 mM 2-aminobenzaldehyde (2-AB); the K m of ProdhΔ21 was determined at 30°C using proline as a substrate. The enzymatic activity of 2.2 μM P5CdhΔ45 protein was assayed in 5 mM HEPES (pH 7.5), 1 mM MgCl2, 0.5 mM DTT, and 0.4 mM P5C (freshly prepared and adjusted to pH 7.0 with 10 M KOH before use); the K m of P5CdhΔ45 was measured at 30°C using NAD as a substrate. The absorbance of P5C-2-AB was measured at 443 nm (ε443 = 2590 cm–1·M–1), and the absorbance of NADH was measured at 340 nm (ε340 = 6220 cm–1·M–1) using a BIO-TEK μQuant microplate reader; the data were analyzed using KC junior software.
The mycelial activity of Prodh and P5Cdh was assayed according to an established protocol used in yeast [2]. Briefly, a 7-day-old mycelium grown on a PDA plate was collected, and 0.2 g of mycelium was immediately immersed in liquid nitrogen for 10s and then quickly ground into a powder. The powder was placed into an EP tube containing 1 mL of 0.1 M HEPES buffer (pH 7.5) with 3 mM MgCl2 and kept on ice. The samples were vortexed vigorously to break the cell wall; the mixture was centrifuged at 8000×g to remove cell debris, and the supernatant was used in enzymatic assays. For the Prodh activity assay, 0.4 mL of 10% proline was added to 0.4 mL of supernatant, vortexed for 10 s, and incubated without shaking at 30°C for 30 min. The assay was developed colorimetrically by the addition of 0.1 mL of 2-AB (6 mg/mL in 20% ethanol) to the reaction mixture; the reaction proceeded for 30 min before being terminated by the addition of 0.5 mL of 10% trichloroacetic acid. The absorbance of the supernatant was measured at 443 nm (ε443 = 2590 cm–1·M–1). The P5Cdh activity was measured by determining the P5C-dependent reduction of NAD. A 4-μL aliquot of 50 mM P5C and 30 μL of 50 mM NAD were added to 0.4 mL of cell extract, mixed by vortexing vigorously for 10 s, and incubated without shaking at 30°C for 30 min. The absorbance of the supernatant was measured at 340 nm (ε340 = 6220 cm–1·M–1).
Complementation of yeast mutants
The S. cerevisiae strains 24099 and 21000 (His–, Leu–, Ura–) and their parent strain BY4743 were purchased from Invitrogen Corporation (California, USA). The sequences of Put1, Put2 and Sdh1-mTP (1–156bp of the coding sequence) from yeast were amplified using genomic DNA, and Prodh and P5Cdh were amplified using C. parasitica mRNA with the primers listed in Table S1. The full-length Put1 and Put2 sequences were directly ligated into the pYES2 vector. Mitochondrial transit peptides in Prodh and P5Cdh were identified with the MitoPro II software version 1.0, and the DNA sequences for these signal peptides were replaced with the Sdh1-mTP sequence and fused to the 5′-end of ProdhΔ21 or P5CdhΔ45 by fusion PCR [35]. The final PCR products were cloned into pUC19 for sequence verification before cloning into the pYES2 vector to yield the appropriate complementation constructs, which were transformed into yeast by the LiAc/PEG method [36]. The transformants were selected on a synthetic medium with 20 g/L glucose supplemented with a mixture of amino acids without uracil. For evaluation of growth on organic nitrogen sources, the yeast strains were cultured in 1 mL of YPD medium over night with shaking; the overnight cultures were centrifuged and the resulting pellets were washed once with 1 mL of sterile water and centrifuged again. The pellets were resuspended in 10 μL of sterile water and used to streak on synthetic medium plates with 20 g/L galactose and the specified nitrogen sources.
Deletion of Prodh, P5Cdh, and other genes involved in the proline/glutamate pathway
In addition to Prodh (JGI protein ID 277618) and P5Cdh (JGI protein ID 344979), there are seven other genes involved in the proline/glutamate pathway in C. parasitica: Pro1 (JGI protein ID 335454), Pro2 (JGI protein ID 343220), Pro3 (JGI protein ID 74323), Car1 (JGI protein ID 99673), Car2 (JGI protein ID 86840), Put3 (JGI protein ID 331838), and Put4 (JGI protein ID 330367). The primer pairs used in the construction of gene disruption mutants for these genes are listed in Table S1. A gene replacement strategy was employed to generate the null mutants. Replacement cassettes were constructed using a double-joint PCR [35] in which the hygromycin B resistance gene hph was fused to the 5′- and 3′- flanking fragments of the target genes in a molar ratio of 3:1:1. The PCR reaction cycle consisted of 94°C for 2 min, followed by 15 cycles of 94°C for 30 s, 58°C for 2 min and 72°C for 4 min, with a final extension of 5 min at 72°C. The pUCHyg plasmid was used for the hph template DNA and the flanking regions were generated from the genomic DNA of C. parasitica by PCR. Next, a nested PCR was performed using the fused DNA as a template; the primers used for the nested PCR are listed in Table S1. Gel electrophoresis-purified RCR products were directly used to transform protoplasts of the C. parasitica strain Δku80 [31]. A typical transformation reaction contained 8 × 107 protoplasts in a volume of 100 µL.
The functional complementation of each gene-knockout mutant was performed using a wild-type gene fragment containing a 1.5-kb promoter region, the complete coding region, and a 0.9-kb terminator region, according to an established protocol [37]. Briefly, a full-length target gene was amplified with gene-specific primer sets and cloned into the pUCG418 vector to generate the complementation construct. The construct was then used to transform a mutant null for the gene of interest. The protoplast preparation and transformation were performed as previously described [38], and selection was performed with 30 μg/mL hygromycin or 25 μg/mL neomycin in the regeneration medium. To ensure a true positive, three rounds of screening on selection plates were performed, and the selected transformants were subjected to single-spore purification.
Genomic DNA was extracted from vegetative hyphae as described [39], and a Southern blot analysis was performed with the DIG High Prime DNA Labeling and Detection Starter Kit II from Roche Applied Science (Mannheim, Germany).
Determination of the intracellular concentration of proline and P5C
The intracellular levels of proline and P5C in mycelia were assayed as previously described for yeast [2]. Briefly, vigorously growing C. parasitica was cultured on an agar plate at 25°C for 7 days. The mycelium was collected and dried, and 0.1 g was homogenized in 5 mL of 3% aqueous sulfosalicylic acid and transferred to a boiling water bath for 10 min to extract the intracellular amino acids. The homogenate was filtered through Whatman filter paper. For the determination of the proline content, 2 mL of filtrate was reacted with 2 mL of glacial acetic acid and 2.5 mL of acid-ninhydrin (2.5 g of ninhydrin dissolved in 6 mL of glacial acetic acid and 4 mL of 6 M phosphoric acid) in a Corning tube for 1 h at 100°C. The reaction was stopped by incubation on ice, and the mixture was extracted with 5 mL of toluene. The toluene phase was separated, and the concentration of proline was measured at OD520. For the determination of the P5C content, 1 mL of filtrate was added to 0.1 mL of trichloroacetic acid. A volumn of 0.5 mL of 2-AB at 6 mg/ml in 20% ethanol was then added to the mixture, and the reaction was allowed to proceed for 1 h. After centrifugation at 10,000×g for 10 min, the absorbance of the clear supernatant was measured at 443 nm.
Sporulation
The strains tested were cultured on PDA plates at 25°C for 14 days under laboratory bench-top conditions with a day/night cycle of 12 h/12 h. The conidial spores were collected as a suspension solution in 0.2% Tween 20 and counted under a light microscope with the aid of a hemocytometer [33].
Virulence assay
Dormant stems of Chinese chestnut (Castanea mollissima) were used in virulence assays of the fungal strains with six replicates for each fungal strain. The inoculated stems were kept at room temperature in a plastic bag to maintain moisture, and cankers were measured at 4 weeks after inoculation [37].
Results
Identification of genes encoding proline dehydrogenase and P5C dehydrogenase in C. parasitica
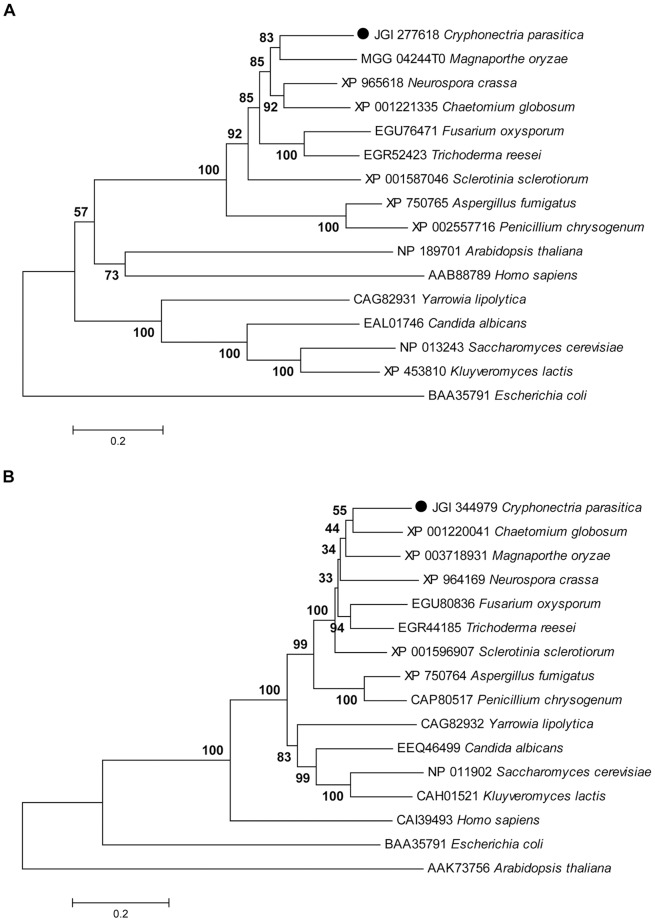
Using the complete amino acid sequence of the Put1 proline dehydrogenase (EC 1.5.99.8) enzyme in S. cerevisiae, the ortholog in the C. parasitica EP155 genome (http://genome.jgi-psf.org/Crypa2/Crypa2.home.html) was identified and designated Prodh. Prodh (JGI protein ID 277618) was predicted to encode a 438-aa protein with a predicted molecular mass of 47.6 kDa and a pI of 8.59. A BLASTp search of Prodh against the non-redundant protein sequences (nr) database revealed that it has a conserved ProDH domain (60–434 aa). Prodh in C. parasitica shares 65% identity with the putative Prodh of Magnaporthe oryzae and 28% identity with Put1 of S. cerevisiae (Figure S1A). The phylogenetic tree constructed with Prodh from microbial, plant, and animal sequences showed that the Prodh sequences from ascomycete fungi cluster as a clade, Prodh from plants and humans as a second clade, and Prodh from yeasts a third clade, with E. coli Prodh being the most distal clade (Figure 1A). In terms of aa sequence identity, the Prodh proteins are highly diverse among the clades (less than 30% in identity); even in the same clade for ascomycetes, the highest identity is only 65%.
Figure 1. Phylogenetic trees of Prodh and P5Cdh across kingdoms.
The phylogenetic trees were generated from amino acid sequences using the Neighbor-joining, minimum-evolution, and maximum-parsimony methods in the MEGA4.0 software. The sequence alignment data were bootstrapped with 1,000 resamplings. The scale bar indicates 0.02 nucleotide substitutions per position. The amino acid sequences of the Prodh and P5Cdh orthologs were downloaded from the NCBI protein database, and accession number for each protein is placed before the name of the protein. A, The tree for Prodhs; B, the tree for P5Cdhs.
The Put2 ortholog, named P5Cdh in the C. parasitica genome was identified using the Put2 aa sequence to blast the genome of C. parasitica. P5Cdh (JGI protein ID 344979) was deduced to encode a 594-aa protein with a predicted molecular mass of 64.2 kDa and a pI of 8.65. A BLASTp search of P5Cdh against the nr database revealed that it contains a conserved ALDH_F4-17_P5CDH domain (57–580 aa). P5Cdh shares 66% identity with the putative P5Cdh of M. oryzae and 51% identity with Put2 of S. cerevisiae (Figure S1 B). The phylogenetic tree constructed with the P5Cdh proteins from microbial, plant, and animal sequences showed a clear lineage, with P5Cdhs from ascomycete fungi clustering as a clade, those from yeasts as a second clade, those from humans as a third clade, those from bacterium as a fourth clade, and those from plants being the most distal clade. Within the ascomycetes, P5Cdhs from Penicillium and Aspergillus form a sub-clade (Figure 1B). In terms of aa sequence identity, P5Cdhs are less diverse within clades than Prodhs.
Enzymatic activity of recombinant Prodh and P5Cdh
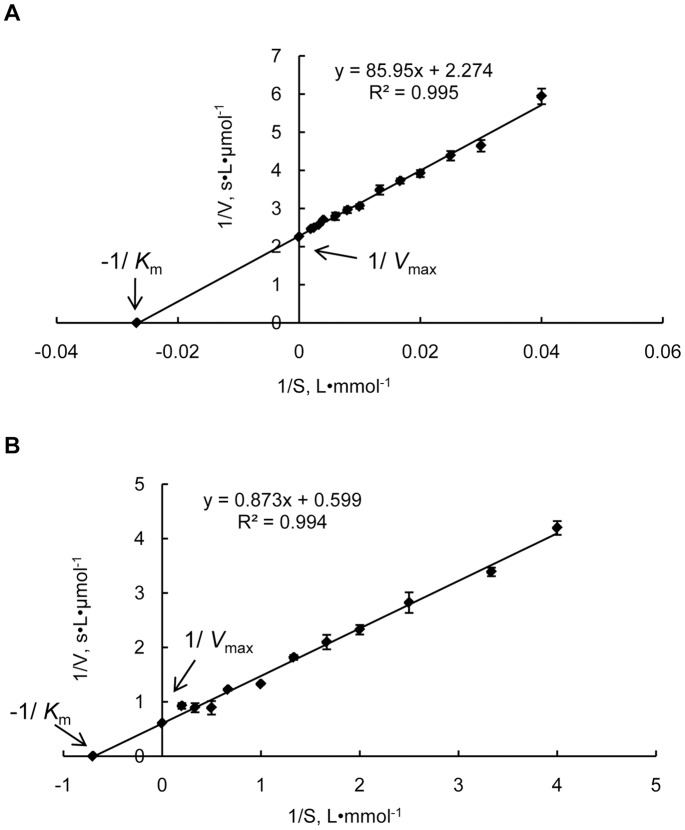
cDNAs from JGI protein ID 277618 (hereafter named Prodh) and from JGI protein ID 344979 (thereafter named as P5Cdh) without their mitochondrial targeting sequences were expressed in E. coli to generate recombinant proteins GST-ProdhΔ21 and GST-P5CdhΔ45, both with an N-terminal GST tag to facilitate purification. The purified GST-ProdhΔ21 and GST-P5CdhΔ45 proteins were soluble and appeared as clear single bands by SDS-PAGE, with an apparent molecular mass of approximately 72 kDa and 92 kDa, respectively, which matches well with their calculated molecular masses plus the 26-kDa GST-tag. In assays using L-proline as a substrate, the GST-ProdhΔ21 protein displayed typical Michaelis-Menten kinetics, with a maximum enzymatic speed of 0.044±0.010 μmol·s–1, a K m of 38.15±3.523 mM, and a k cat of 0.022±0.005 s–1 at 30°C (Figure 2A).
Figure 2. Lineweaver–Burk double reciprocal plots for the purified recombinant proteins GST-ProdhΔ21 and GST-P5CdhΔ45.
Steady-state kinetic parameters V max and K m for the purified recombinant proteins GST-ProdhΔ21 and GST-P5CdhΔ45 were determined by nonlinear regression to fit the data of the Michaelis–Menten equation. The data represent the mean of three independent experiments. A, Prodh activity was assayed based on the dehydrogenation of proline. The reaction velocity was determined at 30°C using 0–500 mM proline as the substrate. The P5C-2-AB concentration was measured at 443 nm; B, P5Cdh activity was assayed based on the dehydrogenation of P5C. The reaction velocity was determined at 30°C using 0–5 mM NAD as the substrate and the electron acceptor. The NADH concentration was measured at 330nm.
In assays using the P5C as a substrate and NAD as the terminal electron acceptor, GST-P5CdhΔ45 displayed typical Michaelis-Menten kinetics, with a maximum enzymatic speed of 1.69±0.111 μmol·s–1, a K m of 1.48±0.145 mM, and a k cat of 0.076±0.005 s–1 at 30°C (Figure 2B).
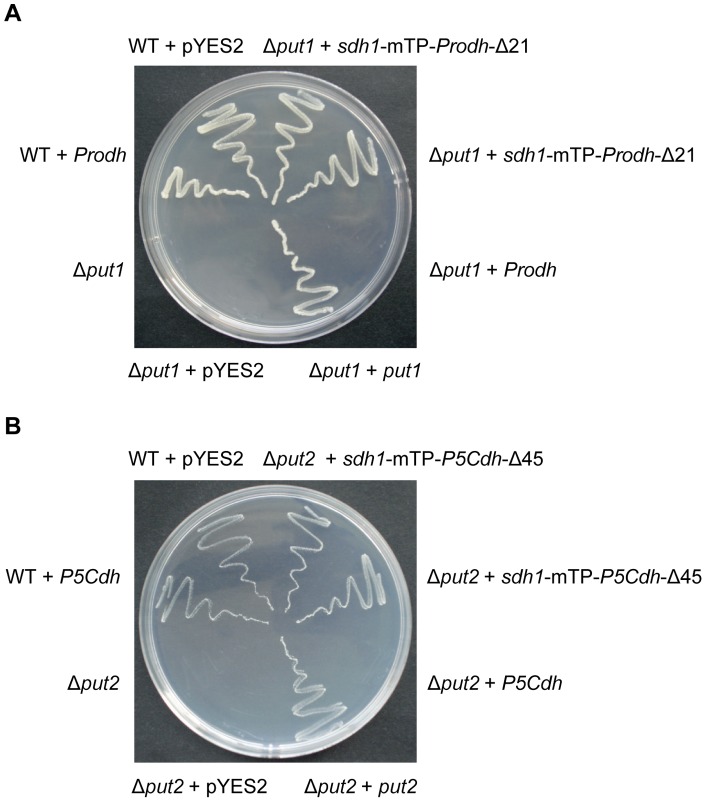
Prodh and P5Cdh can biologically complement yeast put1 and put2 mutants
The put1 and put2 (equivalent to Prodh and P5Cdh, respectively) null mutants of S. cerevisiae cannot grow on a medium with proline as the sole nitrogen source [2]. To test whether the full-length Prodh gene could complement the Put1 mutant, we expressed Prodh in the yeast put1 yeast mutant 24099. Transformation of the Δput1 mutant with pYES2-Prodh, which harbored the full-length cDNA of Prodh, did not enable the cells to metabolize proline. However, after replacing the Prodh mTP (aa 1–21) with that of the flavoprotein subunit of yeast succinate dehydrogenase (Sdh1, aa 1–52), the Prodh fusion was able to restore the proline metabolism to the Δput1 strain (Figure 3A). Similarly, expression of P5Cdh with an mTP from Sdh1 enabled the Δput2 strain to grow on the medium with proline as the sole nitrogen source (Figure 3B).
Figure 3. Heterologous complementation of yeast Δput1 and Δput2 mutants by Prodh and P5Cdh from C. parasitica.
The yeast wild-type (WT) and mutant strains transformed with or without complementation constructs were streaked onto minimal medium (MM) with 2% galactose as the carbon source and 10 mM urea or proline as the sole nitrogen source. The WT strain grew well on all media whereas the mutants survived only on 10 mM urea. The mutants were able to grow on MM supplemented with 10 mM proline when the native Put1 or Put2 of the yeast or C. parasitica Prodh or P5Cdh fused to the yeast Sdh1 mTP was re-introduced into the corresponding mutant. A, Complementation of the yeast Δput1 mutant with Prodh; B, complementation of the yeast Δput2 mutant with P5Cdh. The photographs were taken on the 7th day.
Phenotypic characterization of Prodh and P5Cdh deletion mutants
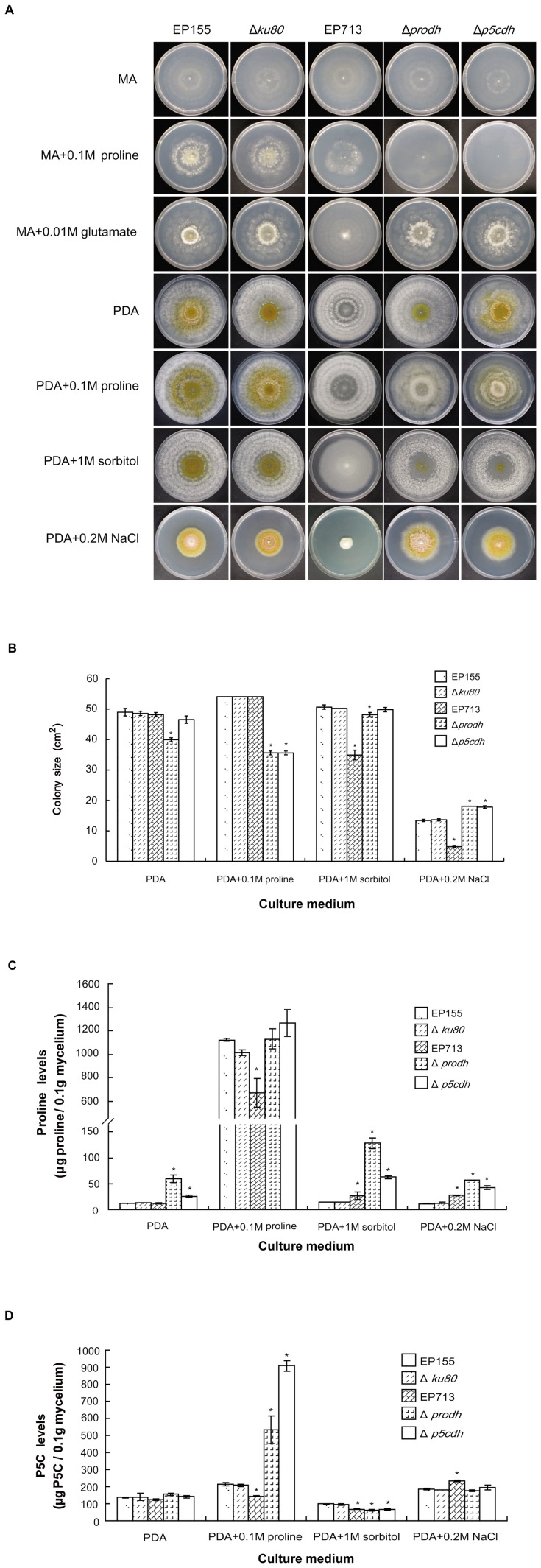
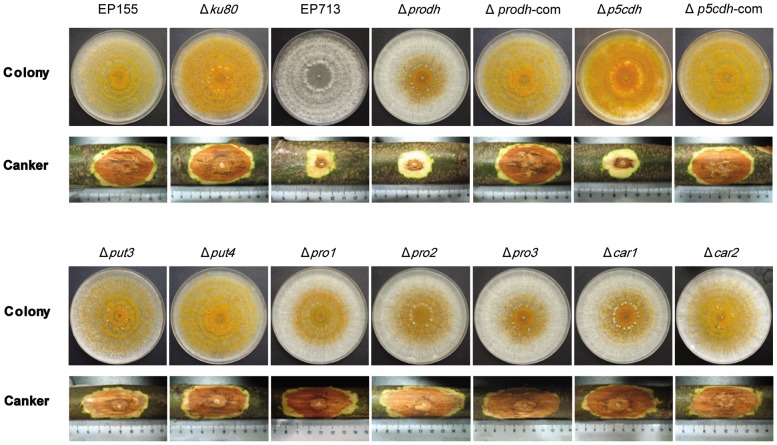
To explore the effects of Prodh and P5Cdh deletions on the phenotypic and physiological traits of C. parasitica, prodh- and p5cdh-null mutants were constructed using gene replacement and confirmed by Southern analyses (Figure S2). A total of 19 Δprodh and 9 Δp5cdh transformants were obtained. Complemented Δprodh and Δp5cdh strains were generated using the wild-type alleles of the Prodh and P5Cdh genes. The Δprodh mutants were all alike and displayed less orange pigmentation than the wild-type EP155 strain. The Δp5cdh mutants were similar to EP155 but with a slightly intensified orange pigmentation. The complemented Prodh mutant (Δprodh-com) and complemented P5Cdh mutant (Δp5cdh-com) were indistinguishable from the wild-type strain on PDA (Figure 4A).
Figure 4. Growth in stressed conditions and intracellular proline and P5C of Δprodh and Δp5cdh mutants.
A, Growth on minimal agar (MA) and MA supplemented with proline and glutamate at 25°C for 14 days; growth on PDA and PDA supplemented with proline, sorbitol or NaCl at 25°C for 7 days. B, Colony size of the strains grown on PDA and PDA supplemented with proline, sorbitol or NaCl at 25°C for 7 days. C, Intracellular proline content of the strains cultured on PDA and PDA supplemented with proline, sorbitol or NaCl at 25°C for 7 days. Mycelia were collected from the plates, and the intracellular proline level was assayed according to an established protocol [2]. D, Intracellular P5C content of the strains cultured on PDA and PDA supplemented with proline, sorbitol or NaCl at 25°C for 7 days. Mycelia were collected from the plates, and the intracellular P5C level was measured according to an established protocol [2]. The error bars represent standard deviations. The statistical significance was determined using Student's t-test (*p<0.05). The assays were repeated 3 times.
Proline is toxic to Δprodh and Δp5cdh mutants
The deletion of Prodh or P5Cdh rusults in the loss of the ability to utilize proline as the sole nitrogen source [5]. To test whether Prodh or P5Cdh perform the same function in the C. parasitica, the minimal agar (MA) with glucose as the sole carbon source and without any nitrogen source was used as the base medium with different nitrogen salt supplementation. On MA, all strains grew as a thin layer, with little mycelium mass after 14 days at 25°C (Figure 4A). The growth of the wild-type EP155 strain, the knockout strain Δku80, and the hypovirus-infected EP713 strain was improved on MA plates when the plates were supplemented with 0.1 M proline; in contrast, the Δprodh and Δp5cdh mutants were not able to grow on MA supplemented with 0.1 M proline. However, when MA was supplemented with glutamate, the Δprodh and Δp5cdh mutants grew as well as the wild-type cells, demonstrating that the poor growth of the Δprodh and Δp5cdh mutants was due to their inability to utilize proline and convert it to glutamate (Figure 4A). All tested strains grew well on PDA, but their response to the addition of extra proline varied: EP155, Δku80, and EP713 grew better and faster when supplemented with 0.1 M proline than on PDA, but the Δprodh and Δp5cdh mutants grew worse than on PDA (Figure 4B), demonstrating that a higher level of proline was toxic to the Δprodh and Δp5cdh mutants.
Osmotic response and salt sensitivity of prodh and p5cdh mutants
When cultured on solid medium with 1 M sorbitol to create osmotic stress, EP155, Δku80, Δprodh, and Δp5cdh grew at similar rates, with orange pigmentation and conidial spores at the center of the colonies, whereas the Δprodh and Δp5cdh colonies were lush and cotton-like. EP713 showed clear signs of stress under this condition, with a thin and flat colony and a reduced growth rate. Under 0.2 M NaCl stress, all the strains were severely inhibited in growth, with EP713 being the most sensitive, whereas the Δprodh and Δp5cdh mutants out-grew the parental Δku80 and EP155 strains (Figure 4A & B).
Relationship between Prodh and P5Cdh and intracellular accumulation of proline and P5C
As shown in Figure 4C, the proline content in the Δprodh mutant was 3 times higher (59 g/0.1 g mycelium) and 1.5 times higher in Δp5cdh mutant (26 g/0.1 g mycelium) than in EP155, Δku80, and EP713 (12 g/0.1 g mycelium) when cultured on PDA. Using proline production on PDA as a reference, the supplementation of 0.1 M proline increased the intracellular proline to a very high level (660–1200 g/0.1 g mycelium) for all strains, suggesting that proline could be efficiently taken up by the cells. Although 1 M sorbitol did not affect the proline levels in EP155 or Δku80, the proline levels in EP713, Δprodh, and Δp5cdh significantly elevated (2.0-fold). Supplementation of 0.2 M NaCl did not significantly alter the proline levels relative to the levels produced on PDA for any of the strains, with the exception of EP713 in which the intracellular proline level increased by approximately 2-fold.
The intracellular P5C levels were similar (130 g/0.1 g mycelium) for all the strains on PDA. Using PDA as a reference, P5C in the Δprodh strain increased from 156 to 533 g/0.1 g mycelium and accumulated to an even higher level in the Δp5cdh strain, from 142 to 907 g/0.1 g mycelium, when PDA was supplemented with 0.1 M proline (Figure 4D).
The addition of 1 M sorbitol slightly decreased P5C accumulation in EP155 and Δku80 but significantly decreased it (from 130 to 60 g/0.1 g mycelium) in EP713, Δprodh, and Δp5cdh. The addition of 0.2 M NaCl elevated the intracellular P5C levels for all the strains by approximately 60–70% (Figure 4D).
Prodh and P5Cdh are responsible for intracellular Prodh and P5Cdh activities
In an assay for proline dehydrogenase activity using cell extracts, the Δprodh mutant exhibited a very low level of proline dehydrogenase activity, approximately 1/10 of the activity observed in the parent strain Δku80 under various culture conditions, confirming that Prodh is the major enzyme responsible for proline dehydrogenation in the cells (Table 1). When grown on PDA, the deletion of P5Cdh resulted in significantly lower Prodh activity (40% of Δku80), suggesting that P5Cdh has a feedback influence on Prodh (Table 1). It was also noted that the Prodh activity in EP713 was significantly lower than in EP155 (16% of EP155). The Prodh activity was induced by 3-8-fold for all strains (except the Δprodh mutant) when proline was added to the medium, with the Prodh activity in Δp5cdh increasing the most (8-fold). Although the enzymatic activity in EP713 increased with the supplementation of external proline, the activity only accounted for 25% of that in EP155. The addition of 1.0 M sorbitol or 0.2 M NaCl to the medium significantly suppressed the Prodh activity in all strains.
Table 1. Intracellular Prodh activity of C. parasitica strains in various culture mediaa.
| PDA | PDA+0.1M proline | PDA+1M sorbitol | PDA+0.2M NaCl | |
| EP155 | 0.425±0.059 | 1.218±0.072 | 0.257±0.018 | 0.238±0.068 |
| Δku80 | 0.249±0.053 | 0.824±0.134 | 0.239±0.012 | 0.281±0.063 |
| EP713 | 0.069±0.006 | 0.313±0.044 | 0.238±0.005 | 0.135±0.023 |
| Δprodh | 0.024±0 | 0.028±0.006 | 0.027±0.016 | 0.018±0.004 |
| Δp5cdh | 0.099±0.024 | 0.862±0.069 | 0.078±0.008 | 0.114±0.009 |
Prodh activity was expressed as 1 μM of P5C formed per min per 100 mg of mycelium. The data are from three replicates.
Similar assays were performed for the P5C dehydrogenase activity, which was very low in the Δp5cdh mutant compared to the parental strain Δku80 under all the tested conditions, confirming that P5Cdh is the major enzyme for P5C dehydrogenation in C. parasitica. As shown in Table 2, deletion of Prodh did not significantly affect the accumulation of P5C in the cells, nor did a hypovirus infection (EP713). The addition of exogenous 0.1 M proline to PDA significantly induced P5C dehydrogenase activity in all the strains except the Δp5cdh mutant. The supplementation of 1 M sorbitol or 0.2 M NaCl did not substantially influence the P5C levels in any of the strains (Table 2).
Table 2. Intracellular P5Cdh activity of C. parasitica strains in various culture mediaa.
| PDA | PDA+0.1M proline | PDA+1M sorbitol | PDA+0.2M NaCl | |
| EP155 | 1.993±0.074 | 3.304±0.091 | 1.826±0.154 | 1.992±0.249 |
| Δku80 | 1.831±0.14 | 4.396±0.269 | 1.379±0.051 | 1.783±0.326 |
| EP713 | 1.594±0.016 | 3.421±0.131 | 1.817±0.135 | 1.889±0.153 |
| Δprodh | 1.16±0.067 | 3.847±0.081 | 1.402±0.124 | 1.843±0.205 |
| Δp5cdh | 0.138±0.019 | 0.194±0.027 | 0.135±0.014 | 0.178±0.004 |
P5Cdh activity was expressed as 1 μM of NADH formed per min per 100 mg of mycelium. The data are from three replicates.
Requirement of proline/glutamate pathway components for virulence and sporulation
To evaluate the contribution of the components of the proline/glutamate pathway to the virulence and conidial spore production of C. parasitica, genes encoding the components of the proline/glutamate pathway were individually knocked out, and the mutants were assayed for virulence by inoculation on the dormant chestnut stems. As shown in Figure 5, all the mutants of the proline/glutamate pathway were able to grow on PDA, demonstrating that none of these genes are essential for saprophytic growth. Although the Δcar1, Δcar2, Δpro1, Δpro2, Δpro3, Δput3, and Δput4 mutants caused normal cankers on the chestnut stems, the deletion of Prodh or P5Cdh significantly impaired the ability of the mutants to grow on chestnut stems to the same level as the hypovirus-infected strain EP713 (Figure 5, Table 3). When the level of sporulation was determined, it was found that the deletion of Pro1, Pro2, or Prodh suppressed sporulation by 1 to 2 orders of magnitude compared to the parental strain, suggesting that the regulation of virulence and sporulation is not coupled (Table 3).
Figure 5. Colony morphology and virulence assay of mutant strains.
The strains were cultured on PDA at 25°C and photographs were taken on the 14th day. Virulence assays were performed on dormant stems of Chinese chestnut (Castanea mollissima). The inoculated stems were kept at room temperature in a plastic bag to maintain moisture, and the cankers were measured at 4 weeks post-inoculation and photographed. The assays were performed with six replicates per fungal strain.
Table 3. Characterization of mutant strain sporulation and virulence.
| Strain | Conidial spores/mLa | Canker area (cm2)b |
| EP155 | 2.08×107±3.38×106 | 9.85±0.47 |
| Δku80 | 1.79×107±1.63×106 | 8.97±1.00 |
| EP713 | 0 | 1.27±0.52* |
| Δprodh | 1.76×106±1.65×105* | 1.78±0.85* |
| Δp5cdh | 2.51×107±1.03×106 | 1.58±0.85* |
| Δput3 | 2.27×107±2.23×106 | 9.95±0.74 |
| Δput4 | 2.36×107±3.13×106 | 10.57±0.90 |
| Δpro1 | 1.16×106±4.17×105* | 9.25±0.84 |
| Δpro2 | 1.01×105±5.65×104* | 8.99±0.51 |
| Δpro3 | 1.33×107±1.44×106 | 9.55±0.89 |
| Δcar1 | 1.67×107±3.13×106 | 9.07±1.02 |
| Δcar2 | 1.76×107±3.13×106 | 8.55±0.95 |
Conidiation was measured after culturing on PDA plates at 25°C for 14 days. The mean and standard deviation were calculated from three replicates. *indicates a statistically significant difference (p<0.05).
The virulence assays were performed on dormant chestnut stems. The inoculated stems were kept at 25°C for 30 days. The mean and standard deviation were calculated from three replicates. *indicates a statistically significant differences (p<0.05).
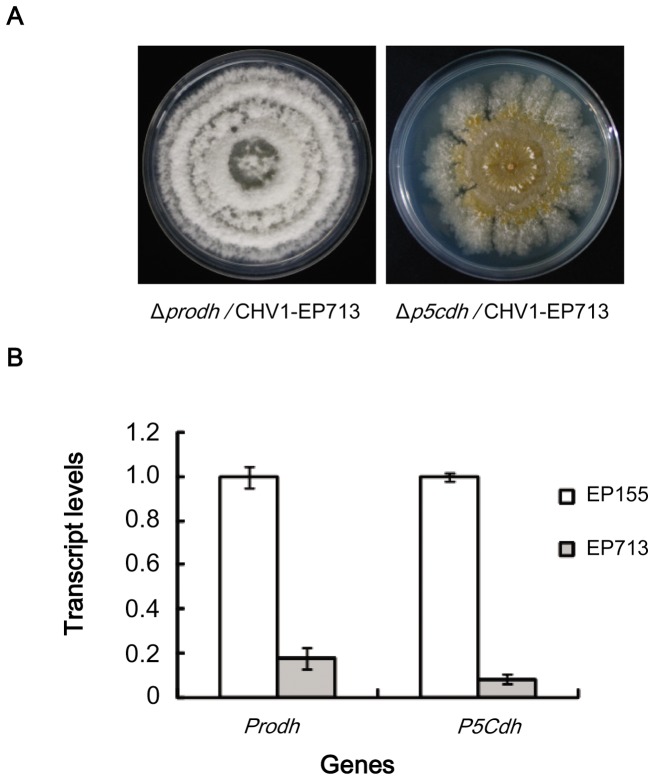
Hypovirus suppresses the expression of Prodh and P5Cdh
The paired inoculation of the hypovirus-infected strain EP713 with either the Δprodh or Δp5cdh mutant resulted in anastomosis and the conversion of the mutant strains to the hypovirus infection phenotype, as characterized by the loss of orange pigmentation and suppressed sporulation. The virus-containing colony of Δprodh/CHV1-EP713 was visually similar to EP713 but with fluffier and thicker aerial hyphae. In contrast, the virus-containing colony of Δp5cdh/CHV1-EP713 grew slowly, with irregular edges and intensified orange pigmentation (Figure 6A). Although the mutants infected with virus appeared to have different phenotypes, the viral dsRNA accumulation in each mutant was comparable with that observed in EP713 (data not shown), suggesting that the deletion of either Prodh or P5Cdh did not have an impact on hypovirus replication or maintenance. However, an examinaion of transcript levels revealed that the accumulation of Prodh and P5Cdh transcripts was down-regulated by 5- and 10-fold, respectively, in the infected strain compared to the wild-type (Figure 6B).
Figure 6. Impact of hypovirus infection on the morphology of Δprodh and Δp5cdh and on the accumulation of Prodh and P5Cdh transcripts in the wild-type strain.
A, Morphology of hypovirus-infected Δprodh and Δp5cdh mutants. The strains were cultured on PDA for 7 days; hypovirus was introduced into Δprodh and Δp5cdh from the hypovirus-infected strain EP713 by anastomosis. Paired inoculation of the Δprodh or Δp5cdh mutant with EP713 resulted in anastomosis and the conversion of the strains to the hypovirus infection phenotype. B, Transcript accumulation levels in hypovirus-free EP155 and hypovirus-infected EP713. Fungal strains were cultured on PDA at 25°C for 7 days, and mycelia were collected for mRNA isolation. The transcript levels were determined by RT-PCR using the Prodh-specific primer pair prodh-Qf/prodh-Qr and P5Cdh-specific primer pair p5cdh-Qf/p5cdh-Qr. The transcript level in EP713 is expressed as a percentage of the transcript level in EP155. The values were calculated from three independent experiments. The error bars represent standard deviations.
Discussion
Evolutionary relationship between Prodh and P5Cdh across kingdoms
Prodh is characterized by a ProDH domain (Figure S1A) [40], and enzymatic assays verified the Prodh activity of JGI 277618 in C. parasitica (Figure 2A). A similar approach lead to the identification of P5Cdh (JGI 344979) in C. parasitica having P5Cdh activity (Figure S1B; Figure 2B). Though not many Prodh enzymes or genes have been characterized in fungi, the deduced Prodh amino acid sequences available in GenBank and other databases allow for the comparison of Prodh among microbial, plant, and animal sequences. As illustrated in Figure 1, Prodh from filamentous ascomycetes cluster into a clade, whereas those from yeasts, plants, and animals form their own clades, with Prodh from the bacterium E. coli forming a distal clade. Within filamentous ascomycetes, Prodh from C. parasitica has 65% identity with Prodh from M. oryzae; however, the Prodh identity is only 28% between C. parasitica and S. cerevisiae and 23.6% between S. cerevisiae and Arabidopsis thaliana [41]. The high divergence among Prodhs suggests that the evolution of this enzyme began a long time ago. Because Prodh functions in mitochondria, it is likely that this enzyme originated from an ancient bacterium [42].
In contrast to Prodh, P5Cdhs from the species of different kingdoms are relatively more conserved, with identities of 50% or more at the amino acid level (Figure S1B). Our phylogenetic tree shows that there is no clear-cut lineage for this enzyme in species of filamentous ascomycetes, whereas we found clearly defined clades among filamentous ascomycetes, yeasts, humans, bacteria, and plants, indicating that this enzyme may have diverged in a somewhat different way from Prodh (Figure 1B). Prodh and P5Cdh are present in both saprophyte (Neurospora crassa) and pathogenic fungi (C. parasitica, M. oryzae and Fusarium oxysporum), suggesting that these enzymes perform at least some shared basic cellular functions.
Conserved biological functions of Prodh and P5Cdh
In yeast, intracellular proline confers stress tolerance to freezing, desiccation, oxidation stress, and ethanol stress [5], [43], [44]. However, few studies using other fungi have been reported with regard to Prodh and P5Cdh. Unlike the limited studies in fungi, proline dehydrogenase has been extensively studied in plants. In A. thaliana, two ProDH genes have been identified and functionally characterized. ProDH1 is a dehydration-responsive gene and is up-regulated after rehydration, accompanied by a decrease of intracellular proline [41]. ProDH1 appears to be the dominant isoform under most conditions and in most tissues, whereas ProDH2 is specifically up-regulated during salt stress [45]. Proline functions to protect plants from drought and salinity stress [46], and ProDH is one of the key enzymes that regulates proline accumulation in vivo [47]. However, proline is toxic to cells by playing a negative role in the repression of normal morphogenesis in Arabidopsis [48]. The similar negative effect of excessive proline could also arise from P5C, the catalytic product of proline by Prodh; excessive proline and P5C are toxic to cells because they induce ROS accumulation and programmed cell death [8]. There are also two ProDH genes in tobacco, NtPDH1 and NtPDH2, which respond to and regulate proline metabolism during drought stress and subsequent recovery [49]. NtPDH1 is less sensitive to dehydration or rehydration, whereas NtPDH2 responds rapidly to both conditions and is down-regulated under drought. Proline toxicity has been observed upon mutation of the Arabidopsis ortholog of P5Cdh [50]. The external application of proline caused the accumulation of P5C and programmed cell death, and proline and P5C/glutamate semialdehyde have been suspected to serve as a link between stress responses and cell death [51].
In humans, proline dehydrogenase, named POX, is induced by p53 and can regulate cell survival and mediate programmed cell death by causing G2 cell cycle arrest to reduce tumor formation and increase the production of α-ketoglutarate to impair HIF-1α signaling [52]. POX is up-regulated by oxidized low-density lipoproteins through peroxisome proliferator-activated receptor gamma and plays a key role in the regulation of protective autophagy in cancer cells [53]. It has been reported that mutation of the P5Cdh ortholog ALDH4A1 in humans causes the genetic disease type II hyperprolinemia, which is characterized by elevating levels of P5C, resulting in mental retardation and convulsions [54].
Despite the diverse biological functions of Prodh and P5Cdh in various species, the fact that Prodh and P5Cdh from a plant were able to function in yeast [41], [45], [50] suggests some conserved basic functions of these enzymes across species in different kingdoms. Although they have low sequence identity, Prodh and P5Cdh from C. parasitica were able to complement the put1 and put2 mutants of S. cerevisiae (Figure 3), demonstrating these two proteins have the same in vivo functions. The fact that both the Δprodh and Δp5cdh mutants were able to grow as well as EP155 and Δku80 on the minimal agar with 0.01 M glutamate as the sole nitrogen source confirms that the inability to convert proline to glutamate is the cause of the alteration in the phenotypes exhibited by the Δprodh and Δp5cdh mutants. Similar to the yeast, the deletion of either Prodh or P5Cdh increases resistance to salt stress with an increase in proline (Figure 4).
An interesting observation from this and previous reports is that, for proper function, the heterologous Prodh or P5Cdh proteins must be fused with the mTP from yeast to direct the correct localization of the enzymes to the mitochondrial membrane (Figure 3) [41], [45], [50]. Thus, mTP might serve as a species-specific tag for Prodh and P5Cdh from different species.
Implication of the proline/glutamate pathway and mitochondrial function in the regulation of virulence and sporulation
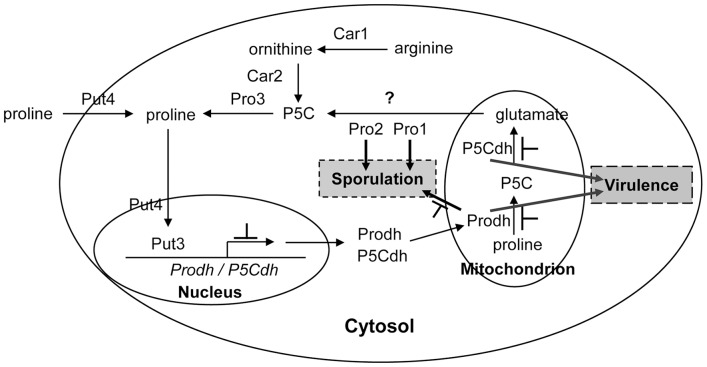
Proline catabolism in the cell involves both cytosol and mitochondria. The conversion of glutamate to proline by P5CS and P5CR and the conversion of arginine to proline by Car1 and Car2 occur in the cytosol, whereas the conversion of proline to glutamate by Prodh and P5Cdh occurs in the mitochondria (Figure 7) [55]–[59]. The deletion of genes encoding P5CS, P5CR, Car1, and Car2 did not affect the virulence (Figure 5), though deletion of Prodh and P5Cdh resulted in hypovirulence (Figure 5), demonstrating that the prodh- and p5cdh-associated mitochondrial functions are required in the regulation of virulence. This is the first report demonstrating that proline catabolism and glutamate biogenesis are indispensible for virulence in a pathogenic fungus.
Figure 7. A model of hypovirus regulation of the proline/glutamate pathway.
Proline can either be taken up into the cells from the environment by Put4 [59], or generated in the cell by the conversion of P5C by Pro3. Proline in the cytosol induces the expression of Put3 [56], which activates the transcription of Prodh and P5Cdh. Prodh and P5Cdh translocate into the mitochondria where to catalyze glutamate biogenesis; glutamate can be converted to proline via P5C when it is transported to the cytosol. The conversion of glutamate to P5C can be catalyzed by Pro1 and Pro2 [58], and P5C can then be converted to proline by P5C reductase Pro3 [57]. If the intracellular glutamate levels are insufficient for proline synthesis, proline biogenesis can be initiated from arginine, as catalyzed by Car1 (arginase), to yield ornithine; ornithine is then converted to P5C by Car2 [55]. In this network, P5C and glutamate appear to be vitally important for both virulence and sporulation. By suppressing Prodh and P5Cdh expression, the hypovirus blocks the biogenesis of P5C and glutamate in mitochondria, resulting in hypovirulence and suppressed sporulation. Although Pro1 and Pro2 do not appear to be regulated by the hypovirus at the transcriptional level, the possibility that they might be regulated at the protein level can not be ruled out, as hypovirus-encoded proteins have been detected in mitochondria [65].
Although both Prodh and P5Cdh are required for virulence, only Prodh is required for sporulation (Table 3), demonstrating that virulence and sporulation are two different processes and that Prodh is a multifunctional protein. A closer inspection of the components of the proline/glutamate pathway also revealed that Pro1 and Pro2 are required for proper sporulation (Table 3). As the gene products of Pro1 and Pro2 function to convert glutamate to P5C, it is suspected that a proper level of P5C is required for the formation of conidial spores in C. parasitica.
A mutation in the mitochondrial DNA (mtDNA) of the C. parasitica strain EP155/mit2 was previously shown to cause hypovirulence, with the production of very few asexual spores and an elevated alternative oxidase activity [29]. The fact that proline is toxic to Δprodh and Δp5cdh mutants and that P5C accumulated in the cell after proline supplementation (Figure 4) correlated to the elevated oxidase activity found in the EP155/mit2 strain, as P5C would lead to ROS accumulation, which is generated by oxidase [51], [60]. In this regard, the evidence reported in this work suggests that the mitochondrial dysfunction for hypovirulence and hypovirulence-associated traits (i.e. suppressed sporulation) could be due to the malfunction of a physiological process, i.e., proline/glutamate pathway impairment (Figure 7).
It has been reported that when the rice blast fungus M. oryzae was cultured on a medium in the absence of preferential nitrogen sources, a number of nitrogen metabolism genes, such as NPR2 and MPG1, were up-regulated. Furthermore, mutation in one or more of these genes resulted in the failure of the fungus to grow under nitrogen starvation conditions and in the loss of pathogenicity [61]. Prodh in M. oryzae was reported to be up-regulated 8.2- and 3.9-fold after 12 h and 48 h, respectively, during nitrogen starvation, suggesting its involvement in nitrogen utilization [62]. Thus, the inability of the Δprodh and Δp5cdh mutants to incite virulence cankers on dormant chestnut stems could be partly attributed to their inability to synthesize or acquire sufficient glutamate in planta.
Mechanisms of hypovirus perturbation of mitochondrial function
Although mutations in the C. parasitica Prodh and P5Cdh genes did not appear to have an impact on the accumulation of hypovirus dsRNA, viral infection did cause profound phenotypic changes to the mutants: Δprodh/CHV1-EP713 had similar colony morphology as EP713, but Δp5cdh/CHV1-EP713 showed a distinct phenotype (Figure 6A). The variation in the response to viral infection suggests a possible virus/host interaction at the levels of these gene products. The fact that a hypovirus infection profoundly down-regulates the transcription of Prodh and P5Cdh (Figure 6B) implies that there would be a shortage of Prodh and P5Cdh in the mitochondria of the host cell. Indeed, Prodh activity in EP713 was significantly lower than in the virus-free isogenic strain EP155 (Table 1). It is anticipated that insufficient Prodh and P5Cdh would jeopardize the energy supply of the cell and hamper the normal biological process of the host, including virulence and sporulation.
In contrast to its low transcript accumulation, P5Cdh activity in the cell extract assays, was the same in EP713 as in the virus-free strain (Table 2). This discrepancy is possibly due to the method used for assaying P5Cdh activity, i.e. measuring the conversion rate of NAD into NADH. We noted that the NADH dehydrogenase, capable of reducing NAD to NADH, was up-regulated 6-fold in the hypovirus-infected C. parasitica starin EP713 (our unpublished data).
Although the suppression of Prodh and P5Cdh expression may be a mechanism by which a hypovirus perturbs the mitochondrial function, this does not rule out the possibility of a direct effect of viral protein on the mitochondria. ORF A of the hypovirus CHV1-EP713 has been shown to suppress sporulation and orange pigmentation when it was transformed into a virus-free fungus strain [63]. It was later found that p29 of the polyprotein encoded by ORF A is responsible for suppressed sporulation and orange pigment production [64]. Moreover, p29 was very recently detected in mitochondria [65]. Thus, it is speculated that p29 may adversely regulate mitochondrial function by interacting with key components of the mitochondrion, of which Prodh, P5Cdh, Pro1, and Pro2 could be good candidates.
Perturbation of the trimeric G-protein signaling pathway [16]–[20] and MAPK signaling pathway [16], [23]–[26] by the hypovirus largely contributes to the multi-faceted alteration in C. parasitica phenotypes. Indeed, these pathways cover a large range of cell processes, from sensing environmental cues to the execution of cellular functions. In contrast, only a few downstream targets that execute an exact biochemical function have been reported [27], [37], [66], [67]. In this regard, the unveiling of viral perturbation of the proline/glutamate pathway in this work adds new knowledge to this colletion. Although the deletion of Prodh did not appear to perturb the trimeric G-protein and MAPK signaling pathways at the transcriptional level (data not shown), the transcript levels of Oah1, a gene encoding the hydrolase Oah1 that hydrolyzes oxaloacetate to produce oxalic acid, were significantly down-regulated in the Δprodh mutant and in the hypovirus-infected strain EP713 (Figure S3). It has been reported that Oah1 is a virulence factor in both humans and plant pathogenic fungi [68]–[71]. Its transcriptional down-regulation in the Δprodh mutant and in EP713 suggests that Oah1 expression was positively regulated by Prodh and suppressed by hypovirus infection. Because the suppression of Oah1 expression was much more severe in the hypovirus-infected cells than in the Prodh mutant, it is reasonable to conclude that in addition to regulating Prodh, hypoviruses may also regulate Oah1 via a different mechanism.
Supporting Information
Alignment of Prodh and P5Cdh from C. parasitica , M. oryzae , and S. cerevisiae. The amino acid sequences of Prodh and P5Cdh from C. parasitica, M. oryzae and S. cerevisiae were identified by searching against NCBI genomic BLAST databases and were downloaded from the NCBI protein database. Alignment of the amino acid sequences was performed using the alignment program in Vector NTI 11.0. A. The alignment of Prodh revealed that Prodh shares 65% and 28% identity with the putative protein of M. oryzae (MGG_04244T0) and Put1 of S. cerevisiae (NP_013243.1), respectively. The red bold lines indicate the conserved domains of ProDH. Identical amino acids are shaded in yellow, and blocks of similarity in green. B, The alignment of P5Cdh revealed that P5Cdh shares 66% and 51% identity with the putative protein of M. oryzae (EHA49347.1) and the Put2 of S. cerevisiae (AAB68907.1), respectively. The red bold lines indicate the conserved domains of ALDH_F4-17_P5CDH domains. Identical amino acids are shaded in yellow, and blocks of similarity in green.
(TIF)
Strategy for construction and confirmation of the knock-out mutants. A, Strategy for the construction of the Δprodh mutant. The Prodh gene structure and positions of primers used to generate the gene replacement cassette are shown at the top. An 885-bp fragment at the 5′ end and a 905-bp fragment at the 3′ end of Prodh were amplified by PCR. A hygromycin resistance gene cassette was used to replace the complete coding region and a portion of the 3′ UTR of the Prodh. B, Strategy for the construction of the Δp5cdh mutant. The P5Cdh gene structure and positions of the primers used to generate the gene replacement cassette are shown at the top. A 974-bp fragment at the 5′ end and a 1038-bp fragment at the 3′ end of P5Cdh were amplified by PCR. The hygromycin resistance gene cassette was used to replace the largest exon of P5Cdh near the 3′ end. C, Southern blot analysis of the prodh null mutant. Δprodh was developed from Δku80, which was derived from the wild-type strain EP155. Restriction digest with BglII released a 1.9 kb 3′ flanking region of Prodh from the wild-type and a 4.1 kb fragment containing the 3′ flanking region from the Δprodh. Probe 1 hybridized with the 3′ flanking region of Prodh, and probe 2 recognized the trpC promoter carried in the transformation vector cassette. D, Southern blot analysis of the Δp5cdh null mutant. Restriction digest with HindIII released a 3.5 kb 5′ region of P5Cdh from the wild-type and a 4.2 kb DNA fragment containing the 5′ region of P5Cdh from P5Cdh null mutant. Probe 3 hybridized to the 5′ flanking region of P5Cdh, and probe 2 recognized the trpC promoter carried in the transformation vector cassette.
(TIF)
Quantification of the transcript level of Oah1. The strains were cultured on PDA at 25°C for 7 days, and mycelia were collected for mRNA isolation. The Oah1 transcript accumulation levels were determined by RT-PCR using the Oah1-specific primers oah1-Qf and oah1-Qr. The transcript level in EP155 was set at 1.0, and the corresponding levels in the other strains are expressed as a percentage of the levels in EP155. The values were calculated from three independent experiments. The error bars represent standard deviations.
(TIF)
Primers used in this work.
(DOC)
Funding Statement
This work was supported in part by grants from the Natural Science Foundation of China (30130020) and Guangxi Natural Science Foundation (GNS0229001) to BC, and the Plans for Creative Training of Post-Graduates in Guangxi University (2010-01) and Innovation Project of Guangxi Graduate Education (T31057) to ZY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Scarpulla RC, Soffer RL (1978) Membrane-bound proline dehydrogenase from Escherichia coli. Solubilization, purification, and characterization. J Biol Chem 253: 5997–6001. [PubMed] [Google Scholar]
- 2. Brandriss MC, Magasanik B (1979) Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol 140: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang SS, Brandriss MC (1986) Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol Cell Biol 6: 2638–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang SS, Brandriss MC (1987) Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol 7: 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takagi H, Sakai K, Morida K, Nakamori S (2000) Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae . FEMS Microbiol Lett 184: 103–108. [DOI] [PubMed] [Google Scholar]
- 6. Chen C, Wanduragala S, Becker DF, Dickman MB (2006) Tomato QM-like protein protects Saccharomyces cerevisiae cells against oxidative stress by regulating intracellular proline levels. Appl Environ Microbiol 72: 4001–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin O, Brandriss MC, Schneider G, Bakalinsky AT (2003) Improved anaerobic use of arginine by Saccharomyces cerevisiae . Appl and Environ Microbiol 69: 1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nomura M, Takagi H (2004) Role of the yeast acetyltransferase Mpr1 in oxidative stress: regulation of oxygen reactive species caused by a toxic proline catabolism intermediate. Proc Natl Acad Sci USA 101: 12616–12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anagnostakis SL (1982) Genetic Analyses of Endothia Parasitica: Linkage Data for Four Single Genes and Three Vegetative Compatibility Types. Genetics 102: 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cortesi P, Milgroom MG (1998) Genetics of vegetative incompatibility in Cryphonectria parasitica . Appl Environ Microbiol 64: 2988–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan BQ, Li ZZ, Huang HW, Qin L (2007) Genetic diversity and population differentiation of chestnut blight fungus, Cryphonectria parasitica, in China as revealed by RAPD. Biochem Genet 45: 487–506. [DOI] [PubMed] [Google Scholar]
- 12. Dawe AL, Van Voorhies WA, Lau TA, Ulanov AV, Li Z (2009) Major impacts on the primary metabolism of the plant pathogen Cryphonectria parasitica by the virulence-attenuating virus CHV1-EP713. Microbiology-Sgm 155: 3913–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, et al. (2012) Molecular Characterization of Vegetative Incompatibility Genes That Restrict Hypovirus Transmission in the Chestnut Blight Fungus Cryphonectria parasitica . Genetics 190: 113–U573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J, Wang F, Feng Y, Mi K, Chen Q, et al. (2013) Comparative vesicle proteomics reveals selective regulation of protein expression in chestnut blight fungus by a hypovirus. J Proteomics 78: 221–230. [DOI] [PubMed] [Google Scholar]
- 15. Kim JM, Park JA, Kim DH (2012) Comparative proteomic analysis of chestnut blight fungus, Cryphonectria parasitica, under tannic acid inducing and hypovirus-regulating conditions. Can J Microbiol 58: 863–871. [DOI] [PubMed] [Google Scholar]
- 16. Nuss DL (2005) Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol 3: 632–642. [DOI] [PubMed] [Google Scholar]
- 17. Choi GH, Chen B, Nuss DL (1995) Virus-mediated or transgenic suppression of a G-protein alpha subunit and attenuation of fungal virulence. Proc Natl Acad Sci USA 92: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao S, Nuss DL (1996) Distinct roles for two G protein alpha subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci USA 93: 14122–14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kasahara S, Nuss DL (1997) Targeted disruption of a fungal G-protein beta subunit gene results in increased vegetative growth but reduced virulence. Mol Plant Microbe Interact 10: 984–993. [DOI] [PubMed] [Google Scholar]
- 20. Parsley TB, Segers GC, Nuss DL, Dawe AL (2003) Analysis of altered G-protein subunit accumulation in Cryphonectria parasitica reveals a third Galpha homologue. Curr Genet 43: 24–33. [DOI] [PubMed] [Google Scholar]
- 21. Segers GC, Regier JC, Nuss DL (2004) Evidence for a role of the regulator of G-protein signaling protein CPRGS-1 in Galpha subunit CPG-1-mediated regulation of fungal virulence, conidiation, and hydrophobin synthesis in the chestnut blight fungus Cryphonectria parasitica . Eukaryot Cell 3: 1454–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chung HJ, Kim MJ, Lim JY, Park SM, Cha BJ, et al. (2006) A gene encoding phosphatidyl inositol-specific phospholipase C from Cryphonectria parasitica modulates the lac1 expression. Fungal Genet Biol 43: 326–336. [DOI] [PubMed] [Google Scholar]
- 23. Park JA, Kim JM, Park SM, Kim DH (2012) Characterization of CpSte11, a MAPKKK gene of Cryphonectria parasitica, and initial evidence of its involvement in the pheromone response pathway. Mol Plant Pathol 13: 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rostagno L, Prodi A, Turina M (2010) Cpkk1, MAPKK of Cryphonectria parasitica, is necessary for virulence on chestnut. Phytopathology 100: 1100–1110. [DOI] [PubMed] [Google Scholar]
- 25. Choi ES, Chung HJ, Kim MJ, Park SM, Cha BJ, et al. (2005) Characterization of the ERK homologue CpMK2 from the chestnut blight fungus Cryphonectria parasitica . Microbiol-SGM 151: 1349–1358. [DOI] [PubMed] [Google Scholar]
- 26. Park SM, Choi ES, Kim MJ, Cha BJ, Yang MS, et al. (2004) Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol Microbiol 51: 1267–1277. [DOI] [PubMed] [Google Scholar]
- 27. Liao S, Li R, Shi L, Wang J, Shang J, et al. (2012) Functional analysis of an S-adenosylhomocysteine hydrolase homolog of chestnut blight fungus. FEMS Microbiol Lett 336: 64–72. [DOI] [PubMed] [Google Scholar]
- 28.Gao K, Xiong Q, Xu J, Wang K (2012) CpBir1 is required for conidiation, virulence and anti-apoptotic effects and influences hypovirus transmission in Cryphonectria parasitica. Fungal Genet Biol. [DOI] [PubMed]
- 29. Monterio-Vitorello CB, Bell JA, Fulbright DW, Bertrand HA (1995) A cytoplasmically transmissible hypovirulence phenotype associated with mitochondrial DNA mutations in the chestnut blight fungus Cryphonectria parasitica . Proc Natl Acad Sci USA 92: 5935–5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allen TD, Nuss DL (2004) Linkage between Mitochondrial Hypovirulence and Viral Hypovirulence in the Chestnut Blight Fungus Revealed by cDNA Microarray Analysis. Eukaryot Cell 3: 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lan X, Yao Z, Zhou Y, Shang J, Lin H, et al. (2008) Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr Genet 53: 59–66. [DOI] [PubMed] [Google Scholar]
- 32. Puhalla JE, Russell DW (1971) Genetics and nutritional requirements of Endothia parasitica . Phytopathology 61: 169–173. [Google Scholar]
- 33. Kim DH, Rigling D, Zhang L, Van Alfen NK (1995) A new extracellular laccase of Cryphonectria parasitica is revealed by deletion of Lac1 . Mol Plant Microbe Int 8: 259–266. [Google Scholar]
- 34. Powell WA Jr, Van Alfen NK (1987) Two nonhomologus viruses of Cryphonectria (Endothia) parasitica reduce accumulation of specific virulence-associated polypeptides. J Bacteriol 169: 5324–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, et al. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. [DOI] [PubMed] [Google Scholar]
- 36. Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360. [DOI] [PubMed] [Google Scholar]
- 37. Chen MM, Jiang M, Shang J, Lan X, Yang F, et al. (2011) CYP1, a hypovirus-regulated cyclophilin, is required for virulence in the chestnut blight fungus. Mol Plant Pathol 12: 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Churchill ACL, Ciuffetti LM, Hansen DR, Van Etten HD, Van Alfen NK (1990) Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr Genet.
- 39. Choi GH, Nuss DL (1992) Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257: 800–803. [DOI] [PubMed] [Google Scholar]
- 40. Ling M, Allen SW, Wood JM (1994) Sequence Analysis Identifies the Proline Dehydrogenase and Δ1-Pyrroline-5-carboxylate Dehydrogenase Domains of the Multifunctional Escherichia coli PutA Protein. J Mol Biol 243: 950–956. [DOI] [PubMed] [Google Scholar]
- 41. Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K (1996) A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell 8: 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kawakami R, Satomura T, Sakuraba H, Ohshima T (2012) L-proline dehydrogenases in hyperthermophilic archaea: distribution, function, structure, and application. Appl Microbiol Biotechnol 93: 83–93. [DOI] [PubMed] [Google Scholar]
- 43. Matsuura K, Takagi H (2005) Vacuolar functions are involved in stress-protective effect of intracellular proline in Saccharomyces cerevisiae . J Biosci Bioeng 100: 538–544. [DOI] [PubMed] [Google Scholar]
- 44. Kaino T, Takagi H (2008) Gene expression profiles and intracellular contents of stress protectants in Saccharomyces cerevisiae under ethanol and sorbitol stresses. Appl Microbiol Biotech 79: 273–283. [DOI] [PubMed] [Google Scholar]
- 45. Funck D, Eckard S, Muller G (2010) Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, et al. (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana . Febs Letters 461: 205–210. [DOI] [PubMed] [Google Scholar]
- 47. Peng Z, Lu Q, Verma DPS (1996) Reciprocal regulation of Delta(1)-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet 253: 334–341. [DOI] [PubMed] [Google Scholar]
- 48. Nanjo T, Fujita M, Seki M, Kato T, Tabata S, et al. (2003) Toxicity of free proline revealed in an Arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol 44: 541–548. [DOI] [PubMed] [Google Scholar]
- 49. Dobra J, Vankova R, Havlova M, Burman AJ, Libus J, et al. (2011) Tobacco leaves and roots differ in the expression of proline metabolism-related genes in the course of drought stress and subsequent recovery. J Plant Physiol 168: 1588–1597. [DOI] [PubMed] [Google Scholar]
- 50. Deuschle K, Funck D, Hellmann H, Daschner K, Binder S, et al. (2001) A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J 27: 345–356. [DOI] [PubMed] [Google Scholar]
- 51. Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, et al. (2004) The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16: 3413–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Y, Borchert GL, Donald SP, Diwan BA, Anver M, et al. (2009) Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res 69: 6414–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zabirnyk O, Liu W, Khalil S, Sharma A, Phang JM (2010) Oxidized low-density lipoproteins upregulate proline oxidase to initiate ROS-dependent autophagy. Carcinogenesis 31: 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, et al. (2012) The three-dimensional structural basis of type II hyperprolinemia. J Mol Biol 420: 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Messenguy F, Vierendeels F, Scherens B, Dubois E (2000) In Saccharomyces cerevisiae, expression of arginine catabolic genes CAR1 and CAR2 in response to exogenous nitrogen availability is mediated by the Ume6 (CargRI)-Sin3 (CargRII)-Rpd3 (CargRIII) complex. J Bacteriol 182: 3158–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. des Etages SA, Falvey DA, Reece RJ, Brandriss MC (1996) Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae . Genetics 142: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brandriss MC, Falvey DA (1992) Proline biosynthesis in Saccharomyces cerevisiae: analysis of the PRO3 gene, which encodes delta 1-pyrroline-5-carboxylate reductase. J Bacteriol 174: 5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tomenchok DM, Brandriss MC (1987) Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae . J Bacteriol 169: 5364–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jauniaux JC, Vandenbol M, Vissers S, Broman K, Grenson M (1987) Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae: Cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur J Biochem 164: 601–606. [DOI] [PubMed] [Google Scholar]
- 60. Miller G, Honig A, Stein H, Suzuki N, Mittler R, et al. (2009) Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem 284: 26482–26492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Talbot NJ, McCafferty HRK, Ma M, Moore K, Hamer JE (1997) Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol Mol Plant Pathol 50: 179–195. [Google Scholar]
- 62. Donofrio NM, Oh Y, Lundy R, Pan H, Brown DE, et al. (2006) Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea . Fungal Genet Biol 43: 605–617. [DOI] [PubMed] [Google Scholar]
- 63. Choi GH, Nuss DL (1992) A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. J EMBO 11: 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Craven MG, Pawlyk DM, Choi GH, Nuss DL (1993) Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J Virol 67: 6513–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang F, Quan R, Chen B, Wang J, Shang J, et al. (2012) Detection of hypovirus protein p29 from mitochondria of the chestnut blight fungus. Genomics and Applied Biology 31: 45–50 (in Chinese).. [Google Scholar]
- 66. Chung HJ, Kwon BR, Kim JM, Park SM, Park JK, et al. (2008) A tannic acid-inducible and hypoviral-regulated laccase3 contributes to the virulence of the chestnut blight fungus Cryphonectria parasitica . Mol Plant-Microbe Int 21: 1582–1590. [DOI] [PubMed] [Google Scholar]
- 67. Jacob-Wilk D, Moretti M, Turina M, Kazmierczak P, van Alfen NK (2012) Differential expression of the putative Kex2 processed and secreted aspartic proteinase gene family of Cryphonectria parasitica . Fungal Biology 116: 363–378. [DOI] [PubMed] [Google Scholar]
- 68. Kirkland BH, Eisa A, Keyhani AO (2005) Oxalic acid as a fungal acaracidal virulence factor. J Med Entomol 42: 346–351. [DOI] [PubMed] [Google Scholar]
- 69. Han Y, Joosten HJ, Niu W, Zhao Z, Mariano PS, et al. (2007) Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi. J Biol Chem 282: 9581–9590. [DOI] [PubMed] [Google Scholar]
- 70. Rio MC, de Oliveira BV, de Tomazella DP, Silva JA, Pereira GA (2008) Production of calcium oxalate crystals by the basidiomycete Moniliophthora perniciosa, the causal agent of witches' broom disease of Cacao. Curr Microbiol 56: 363–370. [DOI] [PubMed] [Google Scholar]
- 71. Chen C, Sun Q, Narayanan B, Nuss DL, Herzberg O (2010) Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus. J Biol Chem 285: 26685–26696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of Prodh and P5Cdh from C. parasitica , M. oryzae , and S. cerevisiae. The amino acid sequences of Prodh and P5Cdh from C. parasitica, M. oryzae and S. cerevisiae were identified by searching against NCBI genomic BLAST databases and were downloaded from the NCBI protein database. Alignment of the amino acid sequences was performed using the alignment program in Vector NTI 11.0. A. The alignment of Prodh revealed that Prodh shares 65% and 28% identity with the putative protein of M. oryzae (MGG_04244T0) and Put1 of S. cerevisiae (NP_013243.1), respectively. The red bold lines indicate the conserved domains of ProDH. Identical amino acids are shaded in yellow, and blocks of similarity in green. B, The alignment of P5Cdh revealed that P5Cdh shares 66% and 51% identity with the putative protein of M. oryzae (EHA49347.1) and the Put2 of S. cerevisiae (AAB68907.1), respectively. The red bold lines indicate the conserved domains of ALDH_F4-17_P5CDH domains. Identical amino acids are shaded in yellow, and blocks of similarity in green.
(TIF)
Strategy for construction and confirmation of the knock-out mutants. A, Strategy for the construction of the Δprodh mutant. The Prodh gene structure and positions of primers used to generate the gene replacement cassette are shown at the top. An 885-bp fragment at the 5′ end and a 905-bp fragment at the 3′ end of Prodh were amplified by PCR. A hygromycin resistance gene cassette was used to replace the complete coding region and a portion of the 3′ UTR of the Prodh. B, Strategy for the construction of the Δp5cdh mutant. The P5Cdh gene structure and positions of the primers used to generate the gene replacement cassette are shown at the top. A 974-bp fragment at the 5′ end and a 1038-bp fragment at the 3′ end of P5Cdh were amplified by PCR. The hygromycin resistance gene cassette was used to replace the largest exon of P5Cdh near the 3′ end. C, Southern blot analysis of the prodh null mutant. Δprodh was developed from Δku80, which was derived from the wild-type strain EP155. Restriction digest with BglII released a 1.9 kb 3′ flanking region of Prodh from the wild-type and a 4.1 kb fragment containing the 3′ flanking region from the Δprodh. Probe 1 hybridized with the 3′ flanking region of Prodh, and probe 2 recognized the trpC promoter carried in the transformation vector cassette. D, Southern blot analysis of the Δp5cdh null mutant. Restriction digest with HindIII released a 3.5 kb 5′ region of P5Cdh from the wild-type and a 4.2 kb DNA fragment containing the 5′ region of P5Cdh from P5Cdh null mutant. Probe 3 hybridized to the 5′ flanking region of P5Cdh, and probe 2 recognized the trpC promoter carried in the transformation vector cassette.
(TIF)
Quantification of the transcript level of Oah1. The strains were cultured on PDA at 25°C for 7 days, and mycelia were collected for mRNA isolation. The Oah1 transcript accumulation levels were determined by RT-PCR using the Oah1-specific primers oah1-Qf and oah1-Qr. The transcript level in EP155 was set at 1.0, and the corresponding levels in the other strains are expressed as a percentage of the levels in EP155. The values were calculated from three independent experiments. The error bars represent standard deviations.
(TIF)
Primers used in this work.
(DOC)