Abstract
Elevated activity of the MAPK signaling cascade is found in the majority of human melanomas and is known to regulate proliferation, survival, and invasion. Current targeted therapies focus on decreasing the activity of this pathway; however, we do not fully understand how these therapies impact tumor biology, especially given that melanoma is a heterogeneous disease. Using a three-dimensional (3D), collagen-embedded spheroid melanoma model, we observed that MEK and BRAF inhibitors can increase the invasive potential of approximately 20% of human melanoma cell lines. The invasive cell lines displayed increased receptor tyrosine kinase (RTK) activity and activation of the Src/FAK/STAT3 signaling axis, also associated with increased cell-to-cell adhesion and cadherin engagement following MEK inhibition. Targeting various RTKs, Src, FAK, and STAT3 with small molecule inhibitors in combination with a MEK inhibitor prevented the invasive phenotype, but only STAT3 inhibition caused cell death in the 3D context. We further show that STAT3 signaling is induced in BRAF-inhibitor resistant cells. Our findings suggest that MEK and BRAF inhibitors can induce STAT3 signaling, causing potential adverse effects such as increased invasion. We also provide the rationale for the combined targeting of the MAPK pathway along with inhibitors of RTKs, SRC, or STAT3 to counteract STAT3-mediated resistance phenotypes.
Keywords: melanoma, MEK, STAT3, targeted therapy, invasion, resistance
INTRODUCTION
Melanoma arises from the transformation of melanocytes which have accumulated mutations in growth regulating genes and display increased levels of autocrine/paracrine growth factors; this eventually leads to their uncontrolled proliferation, survival, and dissemination (1). Contributing to this transformation, the mitogen-activated protein kinase (MAPK) pathway becomes constitutively activated in the majority of melanomas, most notably in tumors harboring activating RAS and RAF mutations (2). Targeting the MAPK pathway thus became a valid anti-melanoma therapeutic strategy, and current trials using RAF-selective inhibitors for patients with BRAFV600E-positive tumors are proof of the potential to treat tumors, even advanced metastatic disease (3–6). Therapeutic challenges arise however, in the form of acquired resistance in initially responsive patients, or through intrinsic resistance as shown by tumors that do not respond or even adopt more aggressive phenotypes (5, 7, 8). Mechanisms conferring resistance to RAF inhibitors are numerous but the MAPK pathway frequently becomes reactivated despite the continued presence of inhibitors designed to block its activity (9–13). In addition, other pathways and effectors appear to become engaged such as the PI3K/Akt pathway and receptor tyrosine kinases (RTKs) such as IGFR, PDGFRβ, or the HGF receptor (9, 14–16). Secondary genetic modifications were also shown to occur, such as amplifications of COT, mutations such as MEK1 (C121S), as well as RAS mutations (14, 17, 18).
Unlike RAF-selective inhibitors, MEK inhibitors downregulate MEK and ERK activity in most cells, irrespective of RAF mutation status, and can be used for a wider patient population (19, 20). However, MEK inhibitor clinical success as single agents has often been limited by toxicity and the response rate for mutant-BRAF melanoma was shown to be lower than that for RAF inhibitors (reviewed in (21)). Their use is also challenged by the fact that MEK inhibitor response does not always correlate with mutation or phospho-protein markers associated with BRAF, MEK, RAS, or PI3K activity, suggesting the need to rely on transcriptional pathway signatures or discover new predictive targets (20). The development of novel and more potent MEK inhibitors such as trametinib could allow MEK to resurface as an exciting clinical target, especially for mutant BRAF melanomas where improved survival was recently observed (22); however, their potential and drawbacks are still under investigation.
While single agents BRAF and MEK inhibitors have shown limitations, their combination has been proposed to obtain sustained MAPK pathway inhibition. However, studies already predict resistance occurring following this treatment strategy (23). Alternatively, the combination of a MAPK pathway inhibitor and that of an agent targeting a different pathway, such as PI3K, could provide more effective results (9, 24–27). Given that melanoma is a complex and heterogeneous disease comprised of genetically and biologically diverse cell types, we anticipate that not all melanomas will respond to the same extent to a given treatment strategy and alternative therapies and targets must be available to curb disease progress.
In this study, we explore the intrinsic resistance mechanisms of metastatic melanomas against MEK inhibition by interrogating a panel of genetically diverse human melanoma cell lines. We observed that MEK inhibition (by UO126 and AZD6244) increased the invasion potential of a cohort of human melanoma cell lines in a variety of models including skin reconstructs. MEK inhibition in this cohort of melanomas is associated with increased RTK activity and the activation of the STAT3 pathway. We further show that downregulation of STAT3 activity or upstream RTKs prevent the invasive phenotype. Finally, we observed that invasion and STAT3 pathway activation is a hallmark of BRAF inhibitor-resistant melanoma cell lines.
RESULTS
MEK inhibition causes increased migration/invasion of melanoma cell lines
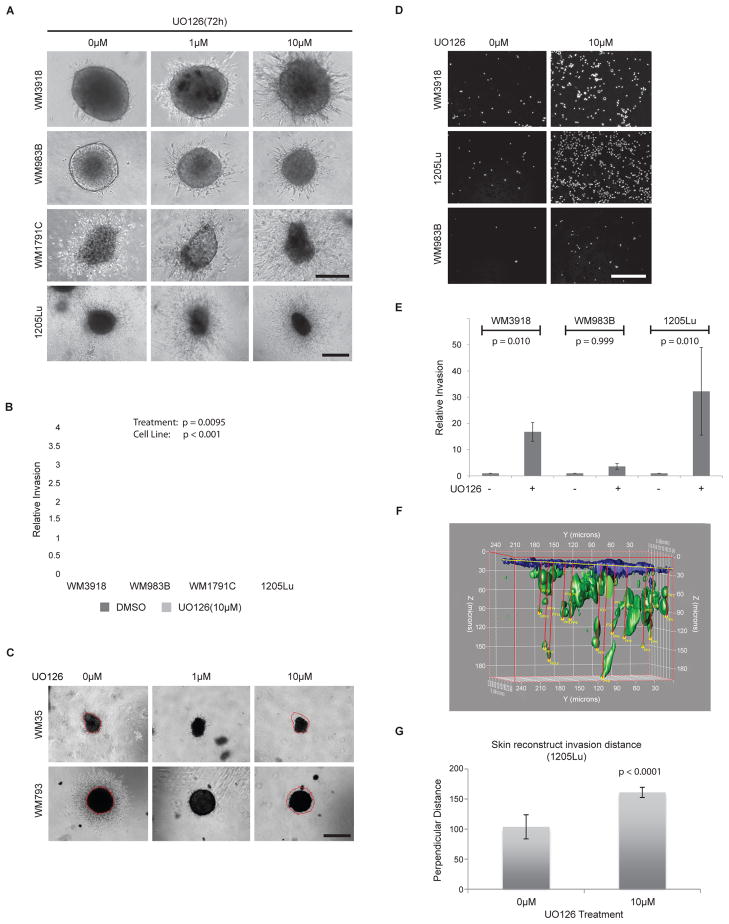
We investigated the effects of MEK inhibition in multiple melanoma cell lines, using the MEK inhibitors UO126 and AZD6244, in a collagen-embedded spheroid assay that mimics the three-dimensional (3D) skin tissue microenvironment (24). Following 72 h treatment with UO126 or AZD6244, we observed that 4/20 (20%) metastatic melanoma cell lines tested displayed significantly increased invasion (Figure 1A, 1B; Suppl. Figure 1A), while spheroid size was not greatly affected. MEK inhibition however, reduced invasion and spheroid size in the non-metastatic cell lines WM35 and WM793 (Figure 1C), in agreement with previous work indicating their sensitivity to MEK inhibitors (24). The BRAF and NRAS mutation status of each cell line did not predict the increased invasion and MEK mutations were not identified to contribute to such a phenotype (Table 1).
Figure 1. MEK inhibition causes increased migration/invasion in a cohort of melanoma cell lines.
A) 3D melanoma spheroids growing in a collagen matrix and treated with increasing doses of the MEK inhibitor UO126 for 72 h show invasive edges. Sample images are shown. Scale bar represents 200 microns for all panels except 1205Lu (600 microns). B) Quantitation of melanoma cell motility from spheroid edge in cell lines grown as spheroids as shown for (A). Values indicate the relative spheroid invasion area compared to the vehicle control-treated spheroids. The invasion observed in the treated spheroids was significantly higher than in the untreated spheroids (p=0.0095), controlling for cell line (p<0.0001). Experiments were conducted in triplicate. C) The early melanoma stage cell lines WM35 and WM793 are sensitive to UO126 and display reduced growth and invasion upon a 72 h treatment. Spheroid size differences are delineated in red, based on circumference of the untreated samples. Scale bar represents 600 microns. D) Transwell invasion assay showing DAPI staining of single cells invading through a collagen-coated semi-porous membrane in the presence of UO126 (10μM). Cells were allowed to invade for 72 h and representative images are shown. Scale bar represents 300 microns. E) Graph showing relative invasion for transwell invasion experiments as conducted in (D). Experiments were conducted in triplicate; (WM3918, p=0.010; WM983B, p=0.999; 1205Lu, p=0.010). F) Computer generated 3D model of a skin reconstruct section depicting invading 1205Lu melanoma cells (green). The model is based on 2-photon images and allows a 3D view of the invasion profile of GFP-tagged melanoma cells in a skin reconstruct, as well as the quantitation of the perpendicular distance traveled by individual 1205Lu cells from the basement membrane. Multiple skin reconstruct sections were imaged, recreated in 3D, and quantitated based on this model and results are summarized in (G). G) Histogram summarizing the average distance travelled by invasive 1205Lu cells (in microns) in skin reconstructs exposed to vehicle control or UO126 (10uM). The perpendicular distance was measured over three different sections per condition, tracking the 5 most invasive cells per section using the 3D modeling system shown in (F). This distance was significantly higher for MEK-inhibitor treated cells than for untreated cells (p<0.0001). Error bars represent standard errors.
Table 1.
Basic genetic information does not predict invasive response to MEK inhibition in 3D culture.
| Cell line | Stage | BRAF | N-RAS | Increased invasion upon MEK inhibition |
|---|---|---|---|---|
| WM3918 | MET | WT | WT | yes |
| WM983B | MET | V600E | WT | yes |
| 1205Lu | MET | V600E | WT | yes |
| 451Lu | MET | V600E | WT | in BRAF inhibitor- resistant 451Lu cells |
| WM1791C | N/A | WT | WT | yes |
| WM35 | RGP | V600E | WT | none |
| WM793 | VGP | V600E | WT | none |
| WM39 | VGP | V600E | WT | none |
| WM164 | MET | V600E | WT | none |
| WM858 | MET | V600E | WT | none |
| WM88 | MET | V600E | WT | none |
| WM1799 | N/A | V600E | WT | none |
| WM3248 | VGP | V600E | WT | none |
| WM1366 | VGP | WT | Q61L | none |
| Sbcl2 | N/A | WT | Q61K | Could not be quantified |
| C8161 | MET | WT | WT | Could not be quantified |
| WM1361A | VGP | WT | Q61R | Could not be quantified |
| WM2032 | MET | WT | Q61R | None |
| WM3211 | RGP | WT | WT | None |
| Melanocytes | WT | WT | None |
MEK mutation status was assessed in: WM3918, WM983B, 1205Lu, 451Lu, WM1791C, WM35, WM793. Only WM983B carried a MEK2 exon 2 synonymous mutation 6599, C>CT V64V.
Abbreviations: RGP, radial growth phase; VGP, vertical growth phase; MET, metastasis; WT, wild type; N/A, not available.
We further conducted a transwell migration/invasion assay through a layer of collagen and confirmed the increased migration potential of melanoma cells as single cell entities (Figures 1D, 1E). Invasion was increased at least 3 fold for all treated cell lines compared to untreated cells, and was significant for 2 out of 3 cell lines using this invasion assay; however, this was likely due to the small number of invading cells in one cell line. To ascertain that the invasive phenotype was also applicable to a more complex microenvironment, we used a human skin reconstruct model featuring the co-culture of multiple cell types including keratinocytes and fibroblasts. This skin-like model demonstrated that MEK inhibitor-treated melanoma cells invade deeper into the dermis than vehicle control-treated cells (Figures 1F, Suppl. Figure 1B, 1C). Even for metastatic cell lines that are initially invasive such as 1205Lu, significant differences were observed in UO126-treated skin reconstructs (Figure 1G). Our data indicate that the MEK inhibitor-induced invasive phenotype occurs in multiple invasion assays, in the presence of multiple cell types, and in a human skin reconstruct model.
Receptor tyrosine kinases are activated upon MEK inhibition
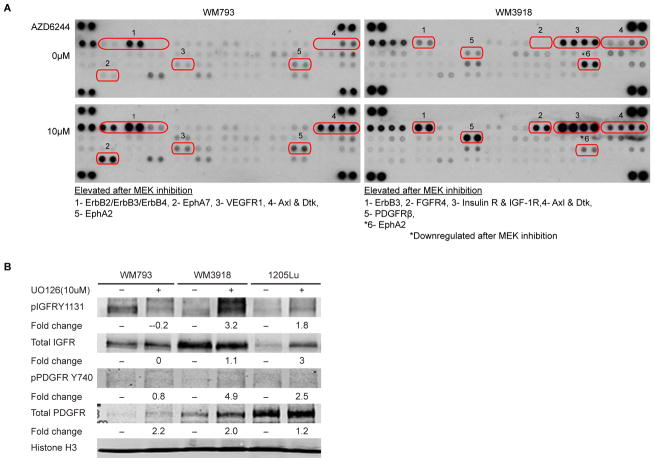
The potential of MEK inhibitors to halt invasion and spheroid growth in early stage melanomas while activating spheroid invasion in some metastatic cell lines allowed us to explore the distinct signaling mechanisms involved in melanoma intrinsic resistance to MEK inhibition. We thus conducted a phospho-antibody array comparing a cell line that is resistant and invades upon MEK inhibition (WM3918) versus a cell line that is sensitive to MEK inhibition (WM793) (28), in the presence or absence of a MEK inhibitor (AZD6244). We found that treatment with the MEK inhibitor for 72 h led to increased phosphorylation of selected RTKs in both a sensitive and resistant cell line (Figure 2A). However, there were distinct differences between the RTKs activated in the invasive cell line and the MEK-inhibitor-sensitive cell line. Phosphorylation of platelet-derived growth factor receptor beta (PDGFRβ), fibroblast growth factor receptor (FGFR), insulin receptor, and insulin-like growth factor 1 receptor (IGF-1R) were selectively increased in WM3918. Interestingly, IGFR and PDGFR have also been associated with resistance to BRAF inhibitors in melanoma (9, 25). We further confirmed the increased expression of IGFR and PDGFR following UO126 treatment in the invasive melanoma cell lines (WM3918, 1205Lu) compared to the MEK sensitive cell line WM793 (Figure 2B). The Erythropoietin-producing hepatocellular kinase A2 (EphA2) was the only phospho-RTK downregulated in the MEK-resistant and invasive cell line, while it was upregulated in the sensitive cell line.
Figure 2. MEK inhibition causes RTK activation and engages the Src/FAK/STAT3 signaling axis.
A) WM793 and WM3818 cells were treated with AZD6244 (10 μM) or DMSO vehicle control for 72h. Whole-cell lysates were incubated on RTK antibody arrays where each RTK antibody is spotted in duplicate. Differentially expressed RTKs are listed and indicated by a number. B) Western blot analysis examining phosphorylated and total levels of IGFR and PDGFR in melanoma cell lines that are non-invasive (WM793) or invasive (WM3918, 1205Lu) following MEK inhibition. Cells were treated with UO126 (10 μM) for 72h before collection of immunolysates. Fold change in band intensity between untreated and treated samples is shown. Histone H3 is the loading control. C) Western blot analysis of UO126-treated WM983B and WM3918 immunolysates extracted from 2D (top) and 3D collagen-embedded cultures (bottom). Phosphorylated levels and total levels of Src, FAK, STAT3, MEK, and ERK are shown, as well as the HSP90 loading control. D) Immunofluorescence staining of phospho-FAK576/577 and phospho-STAT3Y705 (red) in control and UO126 (10μM) treated cells for 72 h. Cell nuclei are DAPI-stained (blue). Scale bar represents 50 microns. E) Western blot analysis of immunolysates from a melanoma cell line sensitive to UO126 (WM793). Phosphorylated levels and total levels of FAK, STAT3, ERK, and MEK are shown, as well as the Hsp90 loading control.
Invasion following MEK inhibition is associated with activation of the Src/FAK/STAT3 signaling axis in 2D and 3D culture
While our previous studies indicated the importance of the PI3K/Akt pathway for melanoma survival following MAPK pathway inhibition (9, 24), we hypothesized that other pathways are also activated and could contribute to the intrinsic resistance of melanoma cells to MAPK inhibitors. For example, IGF-I/IGF-IR is able to mediate activation of the Signal transducers and activators of transcription-3 (STAT3) in vitro and in vivo via the Janus kinase (JAK) (29). Also, Src kinase activity can activate STAT3 preassembled with PDGFR receptors (30), and STAT3 binding to FGFR was shown to be activated by receptor amplification (31). The involvement of the Src/STAT3 axis in mediating growth and survival of melanoma cells was previously demonstrated (32). We therefore examined the activation of the Src, FAK, and STAT3 signaling axis in melanoma cell lines that become invasive following MEK inhibition. Figure 2C (and Suppl. Figure 2) data show that in 2D culture, MEK inhibition can lead to the phosphorylation of Src, FAK, and STAT3, while MEK and ERK phosphorylation is downregulated. Given that the invasive phenotype is difficult to observe in 2D culture and signaling in 2D versus 3D cultures may be distinct, we also confirmed STAT3 activation in cell extracts isolated from our 3D collagen-embedded spheroid cultures (Figure 2C, lower panels). We further confirmed increased FAK phosphorylation and STAT3 localization to the nucleus (indicating activation) upon MEK inhibition by immunofluorescent imaging (Figure 2D). Interestingly, western blot analysis indicates that whereas MEK inhibition upregulates Src/FAK/STAT3 activity and causes increased invasion in metastasis-derived cell lines, MEK inhibition in early stage melanoma cells (e.g. WM793) does not induce Src/FAK/STAT3 activity or invasion (Figure 2E). These data suggest a potential role for STAT3 in melanoma cells’ intrinsic resistance to MEK inhibition.
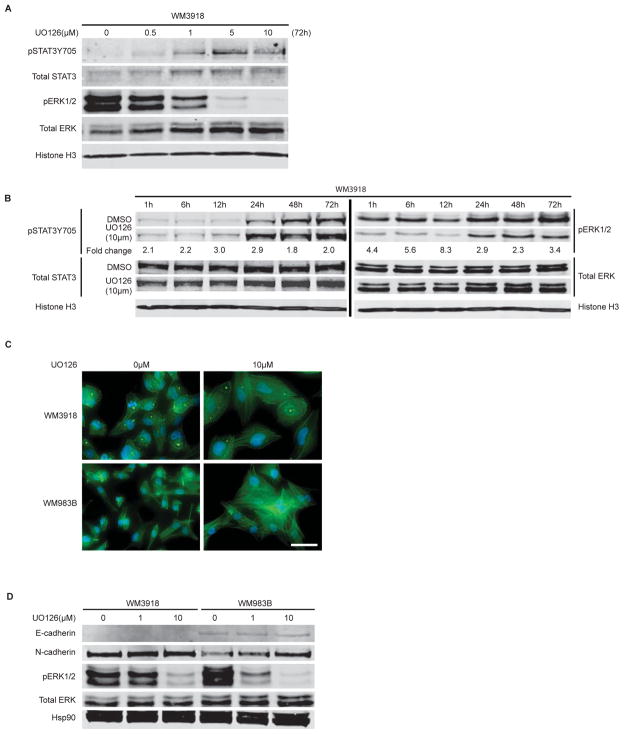
STAT3 phosphorylation increases in a MEK inhibitor dose- and time-dependent manner
There is an inverse correlation between ERK and STAT3 phosphorylation in invasive cell lines (exemplified by WM3918) as the MEK inhibitor UO126 dose increases (Figure 3A). The MEK inhibitor AZD6244 also displays this correlation (Suppl. Figure 3). We also noted an increase in STAT3 phosphorylation over time, most notably occurring at 12h post-compound addition (Figure 3B). While it was shown previously that cell-to-cell adhesion alone can mediate STAT3 phosphorylation (33), MEK inhibitor addition could potentiate this effect. To explore if increased cadherin engagement could be involved in UO126-mediated STAT3 phosphorylation, especially given the altered cell morphology and increased adhesion seen following compound addition (Figure 3C), we evaluated levels of E- and N- cadherin following UO126 addition in WM3918 and WM983B cells. We observed an increase in N-cadherin expression in a single cell line (WM983B) while E-cadherin levels remained even (Figure 3D), suggesting their possible yet incomplete involvement in potentiating STAT3 phosphorylation following MEK inhibition.
Figure 3. STAT3 activation correlates with decreased ERK activity and enhanced cadherin engagement.
A) Dose-dependent activation of STAT3 (Y705 phosphorylation) and inhibition of ERK by UO126 using western blot analysis. Cells were treated for 72h before collection of immunolysates. B) Time-dependent activation of STAT3 upon treatment with UO126 (10μM). Fold change in band intensity between untreated and treated samples is shown for phosphorylated STAT3 and ERK. C) F-actin staining using AlexaFluor488 Phalloidin staining (green) of control or UO126 (10μM) treated cells for 72 h. Cell nuclei are DAPI-stained (blue). Scale bar represents 50 microns. D) Western blot analysis of E- and N-cadherin levels, as well as phosphorylated ERK, following 72 h treatment with increasing doses of UO126.
Targeting the STAT3 pathway prevents the invasive phenotype induced by MEK inhibition
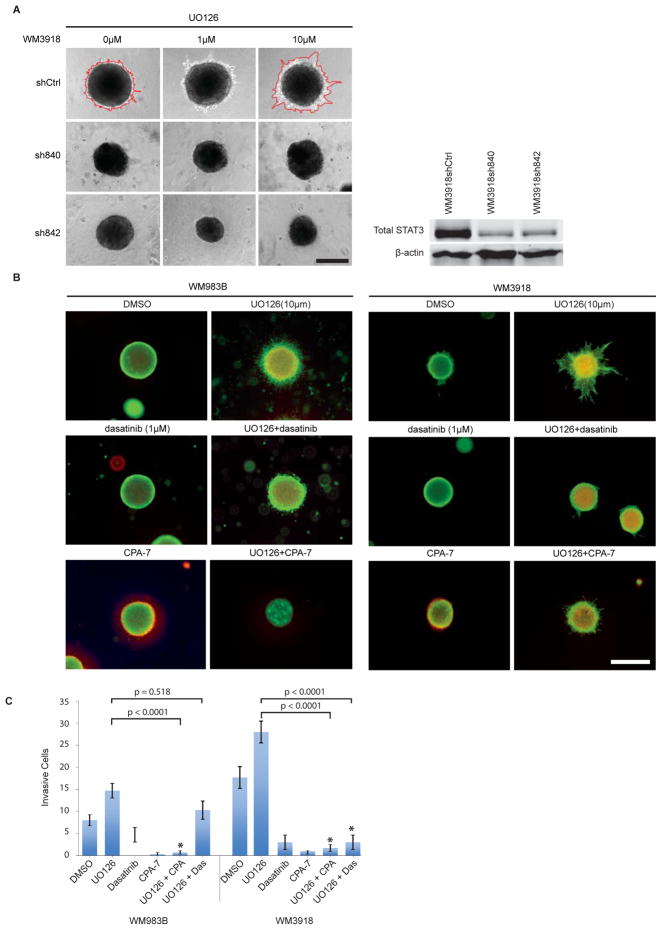
The complex signaling mechanisms found in melanoma indicate that multiple pathways are involved in mediating biological responses to a given inhibitor. To confirm the importance of the STAT3 pathway in mediating resistance to MEK inhibition, we knocked down STAT3 in our MEK-induced invasive melanoma cell lines. Two independent shRNAs were transduced via lentiviral infection and each shRNA caused a reduction in total STAT3 levels (Figure 4A). We selected invasive cell lines and their respective STAT3 knockdown counterparts to grow as spheroids in collagen or expose to a transwell invasion assay. As shown in Figure 4A, STAT3 knockdown prevented the appearance of an invasive phenotype in the presence of UO126. STAT3 knockdown in a different cell line, while less efficient, also reduced invasion as assessed using a transwell invasion assay (Suppl. Figure 4). We next examined whether we could offset the MEK inhibitor-induced invasive phenotype by combining a MEK inhibitor with compounds targeting multiple RTKs and Src family kinases (dasatinib), or STAT3 (CPA-7) (34). Using a fluorescent stain for live and dead cells, we detected increased intra-spheroid cell death and reduced invasion in the combination treatments compared to single agent MEK treatment (Figure 4B). Using a transwell invasion assay, we observed that both dasatinib and CPA-7 had anti-invasive properties and prevented MEK-inhibitor induced invasion; CPA-7 was more potent than dasatinib in halting this invasion (Figure 4C).
Figure 4. Targeting the STAT3 pathway can reverse the invasive phenotype induced by MEK inhibition.
A) STAT3 knockdown cells were grown as spheroids, embedded in collagen and allowed to invade for 72 h in the presence of UO126 or vehicle control. Spheroid invasion differences are delineated in red and representative images are shown. Scale bar represents 300 microns. Experiments were conducted in triplicate. Right panel: STAT3 knockdown efficiency following melanoma cell infection with a lentiviral vector. B) UO126 (MEK inhibitor, 10μM), Dasatinib (SRC/RTK inhibitor, 1uM), and CPA-7 (STAT3 inhibitor, 10μM), were added to collagen-embedded spheroids for 72 h as single agents or combinations as indicated. Survival and invasion were monitored using a live/dead assay. Green fluorescence indicates metabolically active (live) cells, red fluorescence indicates membrane compromised (dead) cells. Experiments were conducted in triplicate and representative images are shown. Scale bar represents 600 microns. C) Graph depicting the invasion of melanoma cells using a transwell invasion assay following treatment with UO126 (MEK inhibitor, 10μM), Dasatinib (SRC/RTK inhibitor, 1uM), and CPA-7 (STAT3 inhibitor, 10μM), or combinations thereof. The ANOVA was significant for drug treatment but effects were different for the two cell lines. Specific comparisons show significant differences between UO126 and UO126+CPA-7 treated cells for both cell lines and a significant difference between UO126 and UO126+dasatinib for WM3918 only.
STAT3 phosphorylation is increased in BRAF-resistant melanoma cells
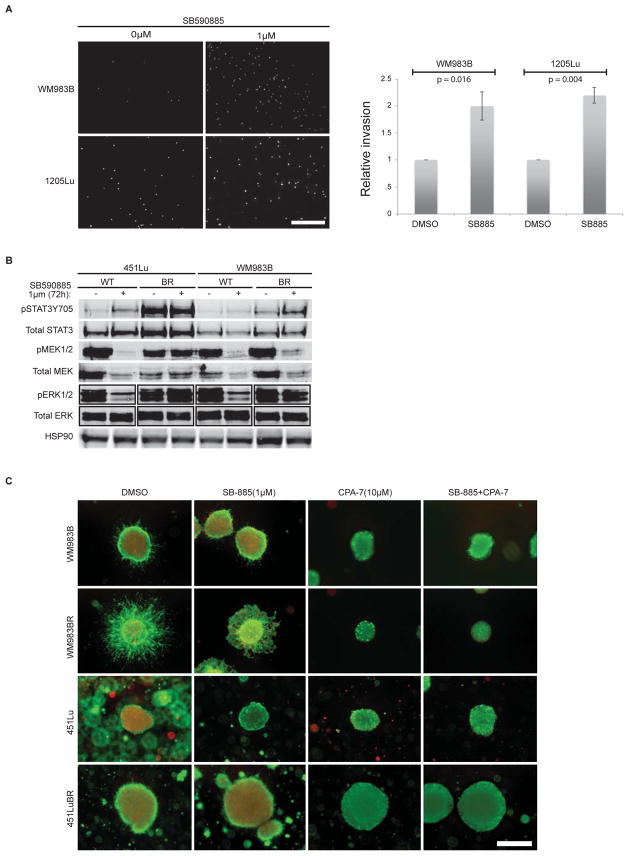
Given the current use of BRAF inhibitors for the treatment of BRAF V600E mutant melanomas, we explored whether MAPK inhibition using a BRAF inhibitor instead of a MEK inhibitor could also cause increased invasion and STAT3 phosphorylation in metastatic melanoma cell lines. The addition of the BRAF inhibitor SB590885 (SB-885) significantly increased the invasion of each two BRAFV600E mutant cell lines, but only transiently (following a 20 h treatment) (Figure 5A), before cell death was observed (data not shown). While BRAFV600E cell lines are sensitive to the addition of a BRAF inhibitor, they still upregulate the STAT3 pathway, most notably in their counterparts that were rendered BRAF inhibitor-resistant (BR) (Figure 5B) (9). BRAF inhibitor-resistant cell lines also formed more invasive spheroids (without MEK inhibition); however, they were sensitive to the addition of the STAT3 inhibitor CPA-7 (alone and in combination with a BRAF inhibitor), which caused reduced spheroid size in one cell line and abrogated the invasive phenotype (Figure 5C).
Figure 5. STAT3 phosphorylation is increased in BRAF-resistant melanoma cells.
A) Transwell invasion assay showing DAPI staining of single melanoma cells invading through a collagen-coated semi-porous membrane in the presence of SB590885 (1μM). Cells were allowed to invade for 20 h. Representative photomicrographs are shown and scale bar represents 600 microns. A histogram shows the quantitation of melanoma cell invasion from transwell assays conducted in triplicate for two cell lines. Relative number of invasive cells in treated compared to the vehicle control-treated cells are shown. Relative invasion was significantly different for both cell lines (WM938B, p=0.016 and 1205Lu, p=0.004). Error bars depict standard errors. B) Western blot analysis of immunolysates from BRAFV600E mutant melanoma cell lines treated with SB590885 or vehicle control for 72 h. WT indicates the parental cell lines; BR indicates the BRAF inhibitor-resistant cell line counterpart. Phosphorylated and total levels of STAT3, MEK, and ERK are shown, as well as the HSP90 loading control. C) SB590885 (SB-885; 1μM) and CPA-7 (10μM) were added to collagen-embedded spheroids for 72 h. Spheroids were generated from BRAFV600E mutant melanoma cell lines (parental or BRAF inhibitor-resistant (BR)). Survival and invasion were monitored using a live/dead assay and representative images are shown. Green fluorescence indicates metabolically active (live) cells; red fluorescence indicates membrane compromised (dead) cells. Independent experiments were conducted in triplicate. Scale bar represents 600 microns.
Taken together, our data suggest that in a cohort of resistant melanoma cells, BRAF or MEK inhibitors cause the upregulation of RTKs and engagement of downstream signaling pathways such as the Src/FAK/STAT3 network, which contribute to an invasive phenotype.
DISCUSSION
In this study, we explored novel compensatory mechanisms employed by melanoma cells in order to resist MAPK inhibition, with an emphasis on MEK inhibition. While most preclinical studies focused on MEK-inhibition have been conducted in the two-dimensional culture setting, which limits certain observations, here we noticed that a cohort of human melanoma cell lines grown as 3D spheroids embedded in collagen displayed an enhanced invasive phenotype. MEK-inhibitor induced invasion was validated using other invasion models and was shown for multiple cell lines. Our findings were initially generated using the MEK inhibitor UO126; however, this compound is not clinically applicable, thus we confirmed our observations with AZD6244 which showed similar results. Invasion did not correlate with the mutation status of BRAF or MEK, indicating that a more extensive knowledge of the genetic or signaling cellular background is required in order to predict which cell lines will respond adversely. Indeed, the invasion observed occurred despite the persistent inhibition of ERK phosphorylation, even following 72 h treatment with UO126 or AZD6244, suggesting the important role of other pathways to compensate and mediate this cellular response. Interestingly, others have previously shown that MAPK inhibition can induce a more aggressive phenotype or cell migration in a subgroup of cell lines when plated on a basement membrane-like matrix (8, 35). Our results are in accordance with these findings, extend the observations to more complex 3D culture models, and add MEK inhibitors to the list of small molecules that need to be carefully characterized and monitored in certain patients. Indeed, as reported for VEGF-R inhibitors, there is potential to increase the metastatic potential of tumor cells by using small molecule inhibitors in the wrong cellular background (36, 37). While for VEGF-R-mediated increased metastasis, hypoxia was suggested as a possible facilitator of invasion, our invasion observations using the 3D spheroid model with MEK inhibition did not change even following 5 days of growth in 1% O2 hypoxic conditions (data not shown). Instead, our data support the hypothesis that a re-wiring of pathways following MAPK small molecule inhibitor addition and engagement of counteracting networks cause an invasive phenotype (36).
We have previously shown the involvement of the IGFR/PI3K pathway along with reactivation of MAPK signaling in counteracting BRAF inhibition (38). Our phospho-RTK array and western blot results following MEK inhibitor addition also show the involvement of IGF-1R. While we do not discount that IGFR/PI3K signaling is involved in the intrinsic resistance of melanoma to MEK inhibition, as it is for BRAF inhibition, here we focused on identifying novel signaling networks that could contribute to resistance and invasion following MAPK pathway inhibition. IGF-R, PDGF-R, and FGF-R signaling has been previously shown to activate the STAT3 pathway (29–31); additionally, studies carried out in STAT3 null cells demonstrated that STAT3 represents an essential effector pathway of Rho GTPases in regulating multiple cellular functions including actin cytoskeleton reorganization, gene activation, and cell migration (39). Thus, we hypothesized and then confirmed that STAT3 is activated following MEK inhibitor addition (in both 2D and 3D culture).
The regulation of STAT3 activity is complex and is the point of convergence for many tyrosine kinase signals (32). In addition to growth factors and cytokine stimulation, cell-to-cell adhesion and cadherin-engagement were also shown to promote STAT3 activation and promote cell survival (33, 40). We observed that confluence in our melanoma cells increases STAT3 phosphorylation, but the signal is potentiated in the presence of a MEK inhibitor. This observation can be explained by previous findings demonstrating that cadherins and integrins associate directly with RTKs to upregulate a number of pathways and support cellular responses such as invasion, proliferation, and survival (reviewed in (41)). Thus, both the engagement of cell adhesion molecules and the contribution of multiple RTKs activity could contribute to STAT3 activation in aggressive melanoma cells. Indeed, examination of E- and N-cadherin levels following MEK inhibitor addition showed that higher N-cadherin levels were induced. This was observed in only one treated cell line, suggesting a partial and cell-dependent involvement of cadherins in STAT3 activation. Crosstalk between the MAPK and STAT3 pathways was previously described at the Ser727 site on STAT3, a phosphorylation site that has its own role and regulation of STAT3 activity (42, 43). MEK inhibition in both our sensitive and resistant cell lines showed a decrease in phospho-STAT3 Ser727 levels; therefore, the regulation of this site alone does not explain the differential response of our melanomas to MEK inhibition (data not show). While the mechanism of STAT3 activation following MEK inhibition may be complex, we sought to show its role in mediating resistance and invasion in our melanoma models. Indeed, our STAT3 knockdown studies indicate that downregulated STAT3 prevents MEK-induced invasion. Furthermore, a tool compound inhibiting STAT3 (CPA-7) also showed that targeting STAT3 prevents invasion in different assays and displays cytotoxic effects in 3D melanoma spheroids. Small molecule inhibitors of STAT3 are not yet approved for therapeutic use, but clinical trials are ongoing to identify effective drugs (for example OPB-31121, Otsuka Pharma.) However, compounds targeting upstream effectors of the STAT3 pathway such as RTKs, Src, and JAKs are available and have already been assessed in the clinic. Our study shows that targeting RTKs with dasatinib does not cause extensive spheroid cell death compared to targetingSTAT3 directly, in agreement with previous studies (44). However, higher drug concentrations may be warranted to achieve cell death when a MEK inhibitor is present since the drug target activities, namely that of the RTKs and Src, are upregulated. In accordance with our findings, recent MEK and SRC inhibitor combinations have shown anti-melanoma activity (45). Also encouragingly, inhibition of the JAK2/STAT3 signaling pathway was shown to impede the migratory and invasive potential of human glioblastoma cells and the effects were observed in both PTEN-expressing and PTEN-deficient cells (46). The combination of MEK inhibitors and effectors of the STAT3 pathway may therefore be useful in curbing melanoma progression and increase patient survival.
Finally, we extended our observations of an increasingly activated STAT3 pathway in BRAF-resistant melanoma cell lines, which are also more invasive compared to their parental counterparts. These observations indicate that SRC, JAK, and STAT3 inhibitors could also be useful for BRAF inhibitor resistant melanoma cases. In sum, our studies indicate that despite harboring high MAPK pathway activity, not all melanomas respond favorably to inhibition of the pathway and can engage an intrinsically resistant response with potential adverse effects such as induced invasion. We identify STAT3 as a pathway upregulated upon MEK inhibition in a cohort of aggressive melanoma cell lines, and in cell lines rendered resistant to BRAF inhibition. Our study also highlights the usefulness of 3D culture methods to further characterize compounds and their potential adverse effects. A future challenge will be to identify which patients will most benefit from a MEK, BRAF, and SRC/JAK/STAT3 inhibition combination. Such a treatment strategy may be useful for all patients, or could provide a novel option to patients displaying intrinsic or acquired resistance to MAPK pathway inhibitors. Additionally, given the role of STAT3 in evading the immune response, it would be interesting to explore the correlation between STAT3 activity and response to immunotherapies.
MATERIALS AND METHODS
Cell culture and reagents
Human melanoma cell lines have been previously described (47, 48). All cell lines were cultured in DMEM supplemented with 5% fetal bovine serum and grown at 37°C in 5% CO2. The consistency of cellular genotypes and cell line identities were confirmed by DNA fingerprinting using Coriell’s microsatellite kit. UO126 was purchased from Promega; AZD6244 and dasatinib were purchased from Chemietek (Indiannapolis, IN); PLX4720 was provided by Plexxikon/Roche, CPA-7 was a gift from Dr. L. Raptis (Queen’s University, Canada). All compounds were stored in DMSO as 10mM stocks and stored at −20°C. Lentiviral shRNAs (TRCN0000020842 and TRCN0000020840 for STAT3) were in the pLKO.1puro vector and were obtained from Sigma (St. Louis, MO).
Mutation Analysis
MEK: Specific exons of the MEK1 and MEK2 genes were amplified by PCR with the following primers: MEK1 exon 2 F 5′-AGCCTCCCACTTTGATTATCTGTCT-3′ and R 5′-AGTCTTCCTTCTACCCTGGTCCCC-3′; MEK1 exon 3 F 5′-AGTGTGAACCAAGTCCTCAAGGTC-3′ and R 5′-CCAGATGCCCCAGATAGTGATGT-3′; MEK1 exon 6 F 5′-CATTTCATCTCCTGACAGTTGC-3′ and R 5′-CCTCCTCCCTCACTTCTTGTC-3′; MEK2 exon 2 F 5′-CGCCCTGGATTCTGGTCATT-3′ and R R 5′-TTCCCAACCTGCCTCCTGTTTC-3′; MEK2 exon 3 F 5′-CTTTACGGCTGAGTCCTGTTT-3′ and R 5′-GTCTTCCTTCTCCCCAACAT-3′; and MEK2 exon 7 F 5′-CTCTATCCATTGCTGCGGTCAT-3′ and R 5′-GGTTTGGGGTGCTCACTGCTT-3′. PCR reactions contained 10ng DNA, 1uM each forward and reverse primers, and 0.25uM dNTPs. PCR settings are as follows: MEK1 exon 2 and exon 3 – 95° 2min; 35 cycles of 95° for 15 sec, 67.9° for 15 sec, 72° for 40 sec; 72° for 1 min, and then 4°; MEK1 exon 6 and MEK2 exon 2 – 95° for 3min; 8 cycles of 95° for 30 sec, 61.0° for 90 sec, 72° for 60sec; 35 cycles of 95° for 30 sec, 57.0° for 90sec, 72° for 60 sec; 72° for 10min, and then 4°; MEK2 exon 3 – 95° for 2min; 35 cycles of 95° for 15sec, 61.9° for 15 sec, 72° for 40 sec; 72° for 1 min and then 4°; and MEK2 exon 7 – 95° for 10 mins; 8 cycles of 95° for 30 sec, touchdown gradient from 70° to 60° (−0.5°C/cycle), 72° for 60 sec; 35 cycles 95° for 30 sec, 65° for 90sec, 72° for 60 sec; 72° for 10min, and then 4°. PCR reactions were then cleaned using ExoSAP-IT®, sequenced in the forward and reverse directions, with respective primers, using Big Dye® Terminator v1.1 Cycle Sequencing Kit, and analyzed on Applied BioSystems DNA Analyzer. Mutations were analyzed with Mutation Surveyor® DNA sequencing analysis software. BRAF: Genomic DNA from samples were analyzed for mutations in BRAF by the nucleotide extension assay using the iPlex platform (Sequenom, Inc, San Diego, CA) (49–52).
Three-dimensional spheroid assay
Melanoma spheroids were prepared (53), and stained (54) as previously described. Collagen-embedded spheroids were treated with inhibitors for 72 h before staining or imaging. Spheroids were imaged using a TE2000 Nikon inverted fluorescence microscope.
Invasion assay
Transwell invasion chambers (Becton Dickinson), 24-well format, were used according to the manufacturer’s instructions and 50μL of collagen mix (as described above) were used to coat the membrane prior to use. Briefly, cells were suspended in 100μL serum-free medium before being loaded into an upper invasion chamber. Cells were allowed to adhere for 4h with the upper chamber sitting in a well containing 750μL of freshly collected conditioned medium from each respective cell line grown for 72h. Inhibitors or the vehicle control DMSO were then added in 100μL serum-free medium for a final concentration as indicated, and the cells were allowed to migrate for 72h towards conditioned media. Prior to imaging, noninvasive cells were removed from the upper chamber and the remaining invasive cells were stained with DAPI. Fluorescent staining was examined using a Nikon TE2000 inverted microscope.
Western blots and RTK array analyses
Proteins were extracted as previously described (55) and 50μg of cell extract were resolved on a 10% polyacrylamide-SDS gel before being transferred onto a polyvinylidene membrane (Millipore, Billerica, MA). Membranes were handled and imaged as previously described (56), or they were analyzed using the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE). All primary antibodies were from Cell Signaling Technologies, Beverly, MA. The secondary antibodies IRDye 700 and IRDye 800 were from Li-Cor. To identify the relative levels of phosphorylation of RTKs, we used a human phospho-RTK array kit (ARY001; R&D Systems Minneapolis, MN), according to manufacturer instructions.
Immunofluorescence
Cells were grown on a glass coverslip and were treated with DMSO or MEK inhibitor for 72 h prior to fixation and imaging as previously described (55). Antibodies used were anti-FAK pY576/577, anti-pSTAT3Y705 (Cell Signaling Technologies), AlexaFluor488 Phalloidin, anti-rabbit AlexaFluor488 secondary antibodies (Life Technologies, Carlsbad, CA). S100 (DAKO) and isotype-matched IgG control antibodies were used to stain fixed skin reconstruct samples.
Human skin reconstructs
Human skin reconstructs were generated as described previously (57). Briefly, inserts of tissue culture trays (Organogenesis, Canton, MA) were coated with 1 ml bovine collagen I mixture (Organogenesis) and layered with 3 ml collagen I mixture containing 7.5×104 fibroblasts. After 4 to 7 days of incubation at 37°C, melanoma cells were mixed with keratinocytes and seeded on top of the dermal reconstructs at a ratio of 1:5 (melanoma cells to keratinocytes). Cells were cultured in skin reconstruct media (Suppl. Materials and Methods) then raised to the air—liquid interface on day 5, via feeding from below with medium. Two weeks later, skin reconstructs were harvested for imaging or fixed in 10% neutral buffered formalin for 4–6 hours before being processed by routine histological methods. The exposure of skin reconstructs to inhibitors involved the pre-treatment of melanoma cells (with vehicle control or 10μM of MEK inhibitor) prior to mixing with the keratinocytes, then the addition of compound (same concentration) to the feeding media in the last 72h of skin reconstruct growth.
Two-photon microscopy imaging
Deep tissue imaging was performed using a Prairie Ultima II 2-photon microscope (Prairie Technologies, Inc, Middleton, WI). The system incorporates a fixed-stage upright Olympus BX61WI microscope, a Chameleon XR femtosecond pulsed Ti:Sapphire laser (Coherent Laser Group, Santa Clara, CA) with excitation ranging from 710–980nm, and a 4 channel, filter based detection system. Unfixed tissue samples were directly imaged with a 40X PlanFl/IR water immersion lens to a standardized depth of 200 microns. Excitation at 910nm provided signal from the GFP labeled melanoma cells and also provided a Second Harmonic Signal (SHG) to visualize the upper collagen layer. Stacks of 100 slices were generated in 2 channels and 3D reconstruction and analysis was accomplished using ImagePro Plus 3D software (Media Cybernetics, Silver Spring Maryland).
Statistical analysis
The two-way analysis of variance was used to test for treatment effects on invasion controlling for cell line after taking the natural logarithm of invasion. When there was a significant interaction, comparisons were done for each cell line. T-tests using Satterwaithe’s method, when variances were unequal were used to test for cell line-specific treatment effects on distance traveled in the skin reconstruct experiments. Tukey’s procedure was used to evaluate pairwise differences when an ANOVA was significant. Error bars represent standard errors.
Supplementary Material
Acknowledgments
Financial support: Our research is supported by grants from the National Cancer Institute (P01 CA114046, PO1 CA025874, P30 CA010815, RO1 CA117881).
We thank P. Sompalli, and F. Raman for technical assistance. We thank GlaxoSmithKline for SB590885, and Dr. L. Raptis (Queen’s University, Canada) for CPA-7. We also thank R. Delgiacco of the Wistar Histology Facility. AV, JV, KLN, and MH are members of the ITMAT-University of Pennsylvania. This work was supported by NIH grants PO1 CA114046, P01 CA025874, P30 CA010815, R01 CA117881, and by the Adelson Medical Research Foundation (AMRF).
Footnotes
This is an original research article in the area of molecular oncology of human tumors.
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Haass NK, Smalley KS, Herlyn M. The role of altered cell-cell communication in melanoma progression. J Mol Histol. 2004 Mar;35(3):309–18. doi: 10.1023/b:hijo.0000032362.35354.bb. [DOI] [PubMed] [Google Scholar]
- 2.Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007 Apr 20;25(12):1606–20. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 3.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010 Sep 7; doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT, Li L, Brafford PA, van den Eijnden M, Halloran MB, Sproesser K, et al. PLX4032, a potent inhibitor of the B-Raf V600E oncogene, selectively inhibits V600E-positive melanomas. Pigment Cell Melanoma Res. 2010 Dec;23(6):820–7. doi: 10.1111/j.1755-148X.2010.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011 Jun 30;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012 Feb 23;366(8):707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010 Aug 26;363(9):80919. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010 Apr;23(2):190–00. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, Santiago-Walker AE, Letrero R, D’Andrea K, Pushparajan A, Hayden JE, Dahlman Brown K, Laquerre S, McArthur GA, Sosman JA, Nathanson KL, Herlyn M. Acquired Resistance to BRAF Inhibitors Mediated by a RAF Kinase Switch in Melanoma Can Be Overcome by Cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johannessen CMBJ, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, Garraway LA. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010 Nov 24; doi: 10.1038/nature09627. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010 Mar 18;464(7287):427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010 Jan 22;140(2):209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010 Mar 18;464(7287):431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 14.Nazarian RSH, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010 Nov 24; doi: 10.1038/nature09626. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012 Jul 4; doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012 Jul 4; doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting Therapeutic Resistance to RAF Inhibition in Melanoma by Tumor Genomic Profiling. J Clin Oncol. 2011 Mar 7; doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010 Dec 16;468(7326):968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006 Jan 19;439(7074):358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010 Mar 15;70(6):2264–73. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nissan MH, Solit DB. The “SWOT” of BRAF inhibition in melanoma: RAF inhibitors, MEK inhibitors or both? Curr Oncol Rep. 2011 Dec;13(6):479–87. doi: 10.1007/s11912-011-0198-4. [DOI] [PubMed] [Google Scholar]
- 22.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved Survival with MEK Inhibition in BRAF-Mutated Melanoma. N Engl J Med. 2012 Jun 4; doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 23.Gowrishankar K, Snoyman S, Pupo GM, Becker TM, Kefford RF, Rizos H. Acquired Resistance to BRAF Inhibition Can Confer Cross-Resistance to Combined BRAF/MEK Inhibition. J Invest Dermatol. 2012 Mar 22; doi: 10.1038/jid.2012.63. [DOI] [PubMed] [Google Scholar]
- 24.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006 May;5(5):1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 25.Shi H, Kong X, Ribas A, Lo RS. Combinatorial Treatments That Overcome PDGFR{beta}-Driven Resistance of Melanoma Cells to V600EB-RAF Inhibition. Cancer Res. 2011 Aug 1;71(15):5067–74. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010 Nov 1;70(21):8736–47. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, et al. Combinations of BRAF, MEK, and PI3K/mTOR Inhibitors Overcome Acquired Resistance to the BRAF Inhibitor GSK2118436 Dabrafenib, Mediated by NRAS or MEK Mutations. Mol Cancer Ther. 2012 Apr;11(4):909–20. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 28.Haass NK, Sproesser K, Nguyen TK, Contractor R, Medina CA, Nathanson KL, et al. The mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor AZD6244 (ARRY-142886) induces growth arrest in melanoma cells and tumor regression when combined with docetaxel. Clin Cancer Res. 2008 Jan 1;14(1):230–9. doi: 10.1158/1078-0432.CCR-07-1440. [DOI] [PubMed] [Google Scholar]
- 29.Zong CS, Chan J, Levy DE, Horvath C, Sadowski HB, Wang LH. Mechanism of STAT3 activation by insulin-like growth factor I receptor. J Biol Chem. 2000 May 19;275(20):15099–105. doi: 10.1074/jbc.M000089200. [DOI] [PubMed] [Google Scholar]
- 30.Wang YZ, Wharton W, Garcia R, Kraker A, Jove R, Pledger WJ. Activation of Stat3 preassembled with platelet-derived growth factor beta receptors requires Src kinase activity. Oncogene. 2000 Apr 20;19(17):2075–85. doi: 10.1038/sj.onc.1203548. [DOI] [PubMed] [Google Scholar]
- 31.Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res. 2010 Apr 15;70(8):3391–401. doi: 10.1158/0008-5472.CAN-09-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu G, Bowman T, Huang M, Shivers S, Reintgen D, Daud A, et al. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene. 2002 Oct 10;21(46):7001–10. doi: 10.1038/sj.onc.1205859. [DOI] [PubMed] [Google Scholar]
- 33.Vultur A, Cao J, Arulanandam R, Turkson J, Jove R, Greer P, et al. Cell-to-cell adhesion modulates Stat3 activity in normal and breast carcinoma cells. Oncogene. 2004 Apr 8;23(15):2600–16. doi: 10.1038/sj.onc.1207378. [DOI] [PubMed] [Google Scholar]
- 34.Littlefield SL, Baird MC, Anagnostopoulou A, Raptis L. Synthesis, characterization and Stat3 inhibitory properties of the prototypical platinum(IV) anticancer drug, [PtCl3(NO2)(NH3)2] (CPA-7) Inorg Chem. 2008 Apr 7;47(7):2798–804. doi: 10.1021/ic702057q. [DOI] [PubMed] [Google Scholar]
- 35.Zipser MC, Eichhoff OM, Widmer DS, Schlegel NC, Schoenewolf NL, Stuart D, et al. A proliferative melanoma cell phenotype is responsive to RAF/MEK inhibition independent of BRAF mutation status. Pigment Cell Melanoma Res. 2011 Apr;24(2):326–33. doi: 10.1111/j.1755-148X.2010.00823.x. [DOI] [PubMed] [Google Scholar]
- 36.Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009 Mar 3;15(3):232–9. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009 Mar 3;15(3):220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010 Dec 14;18(6):683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J Biol Chem. 2005 Apr 29;280(17):17275–85. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 40.Arulanandam R, Vultur A, Cao J, Carefoot E, Elliott BE, Truesdell PF, et al. Cadherin-cadherin engagement promotes cell survival via Rac1/Cdc42 and signal transducer and activator of transcription-3. Mol Cancer Res. 2009 Aug;7(8):1310–27. doi: 10.1158/1541-7786.MCR-08-0469. [DOI] [PubMed] [Google Scholar]
- 41.Christofori G. Changing neighbours, changing behaviour: cell adhesion molecule-mediated signalling during tumour progression. EMBO J. 2003 May 15;22(10):2318–23. doi: 10.1093/emboj/cdg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weng YI, Aroor AR, Shukla SD. Ethanol inhibition of angiotensin II-stimulated Tyr705 and Ser727 STAT3 phosphorylation in cultured rat hepatocytes: relevance to activation of p42/44 mitogen-activated protein kinase. Alcohol. 2008 Aug;42(5):397–406. doi: 10.1016/j.alcohol.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi M, Oka M, Iwasaki T, Fukami Y, Nishigori C. Role and Regulation of STAT3 Phosphorylation at Ser727 in Melanocytes and Melanoma Cells. J Invest Dermatol. 2012 Mar 15; doi: 10.1038/jid.2012.45. [DOI] [PubMed] [Google Scholar]
- 44.Buettner R, Mesa T, Vultur A, Lee F, Jove R. Inhibition of Src family kinases with dasatinib blocks migration and invasion of human melanoma cells. Mol Cancer Res. 2008 Nov;6(11):1766–74. doi: 10.1158/1541-7786.MCR-08-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferguson J, Arozarena I, Ehrhardt M, Wellbrock C. Combination of MEK and SRC inhibition suppresses melanoma cell growth and invasion. Oncogene. 2012 Feb 6; doi: 10.1038/onc.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senft C, Priester M, Polacin M, Schroder K, Seifert V, Kogel D, et al. Inhibition of the JAK-2/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. J Neurooncol. 2011 Feb;101(3):393–403. doi: 10.1007/s11060-010-0273-y. [DOI] [PubMed] [Google Scholar]
- 47.Satyamoorthy K, Li G, Gerrero MR, Brose MS, Volpe P, Weber BL, et al. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res. 2003 Feb 15;63(4):756–9. [PubMed] [Google Scholar]
- 48.Iliopoulos D, Ernst C, Steplewski Z, Jambrosic JA, Rodeck U, Herlyn M, et al. Inhibition of metastases of a human melanoma xenograft by monoclonal antibody to the GD2/GD3 gangliosides. J Natl Cancer Inst. 1989 Mar 15;81(6):440–4. doi: 10.1093/jnci/81.6.440. [DOI] [PubMed] [Google Scholar]
- 49.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007 Mar;39(3):347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 50.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009 Jul 7;16(1):21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ragoussis J, Elvidge GP, Kaur K, Colella S. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics research. PLoS Genet. 2006 Jul;2(7):e100. doi: 10.1371/journal.pgen.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalley KS, Herlyn M. Targeting intracellular signaling pathways as a novel strategy in melanoma therapeutics. Ann N Y Acad Sci. 2005 Nov;1059:16–25. doi: 10.1196/annals.1339.005. [DOI] [PubMed] [Google Scholar]
- 54.Smalley KS, Contractor R, Nguyen TK, Xiao M, Edwards R, Muthusamy V, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008 Jul 15;68(14):5743–52. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vultur A, Buettner R, Kowolik C, Liang W, Smith D, Boschelli F, et al. SKI-606 (bosutinib), a novel Src kinase inhibitor, suppresses migration and invasion of human breast cancer cells. Mol Cancer Ther. 2008 May;7(5):1185–94. doi: 10.1158/1535-7163.MCT-08-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010 May 14;141(4):583–94. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukunaga-Kalabis M, Martinez G, Liu ZJ, Kalabis J, Mrass P, Weninger W, et al. CCN3 controls 3D spatial localization of melanocytes in the human skin through DDR1. J Cell Biol. 2006 Nov 20;175(4):563–9. doi: 10.1083/jcb.200602132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.