Abstract
Nuclease Sensor Trio: Fluorogenic DNA sensors were developed for distinct classes of nucleases: 3′-exonuclease, 5′-exonucleases, and endonuclease. The highly selective sensors, built from very small modified DNA oligomers containing the nonnatural fluorescent base pyrene, and employing thymine as a quencher, were found to function in a variety of complex biological media.
Keywords: oligonucleotide, exonuclease, endonuclease, pyrene, quenching, sensor
Nucleases are a broad class of enzymes that hydrolyze the nucleic acid backbone for multiple biological functions including DNA replication, DNA repair, recombination, and recycling. In biological research, nucleases are widely used for processing and tailoring DNAs and RNAs, and for probing their structure as well. Many sequence-specific restriction nucleases as well as sequence-general nucleases are now widely available, and nucleases have become one of the most widely employed classes of enzymes in modern biology and biotechnology. They have been utilized in the polymerase chain reaction, in molecular cloning, gene mapping and nucleic acid analysis.[1] Importantly, phosphodiesterases are synthesized by bacteria and are used as markers of bacterial contamination; for example, assaying nuclease levels has been a standard method for measuring contamination by Staphylococcus.[2]
Nuclease activities can also be detrimental to research and to human health. The enzymes can be unwanted contaminants that may degrade the experimenter’s DNA or RNA samples. Moreover, the presence of phosphodiesterases in biological fluids and in cells presents a challenge to researchers who study the use of DNAs and RNAs as potential therapeutic agents. In humans, variable activities of cellular phosphodiesterases have been correlated with disease; for example, disruption of phosphodiesterase activities have been implicated in asthma,[3] depression,[4] schizophrenia[5] and stroke[6]. Phosphodiesterases are clinically important therapeutic targets, and a number of drugs are approved for use against these enzymes.[7,8]
The ability to rapidly measure specific nuclease activities is important in the development of these and future clinical approaches to phosphodiesterase-linked disease. Detection of nuclease activity has been traditionally performed with radioactive labeling,[9] gel electrophoresis,[10] high performance liquid chromatography[11] and enzyme-linked immunosorbent assays[12]. The recent development of nuclease sensors that yield optical signals have partly resolved some limitations of the traditional methods, which are generally time-consuming and laborious. Examples of optical nuclease sensor types include intercalating dyes,[13–15] cationic dyes,[16] conjugated polymers,[17] metal nanoparticles,[18–25] carbon nanotubes[26,27] and molecular beacons[28,29]. However, these approaches in most cases report on phosphodiester cleavage with little or no regard to location and type of bond, which can be problematic because of the overlapping types of activity that exist in the many known nuclease enzymes. One of the broadest classifications of nucleases is that of their preferred location of cleavage in the DNA or RNA strand: namely, from the 5′-end (5′-exonucleases), from the 3′-end (3′-exonucleases), and from internal linkages (endonucleases). Significantly, despite extensive studies of nucleases during the recent decades, a selective fluorogenic method to distinguish these distinct classes of nuclease activities has not yet been developed. One sensor of 3′-exonuclease activity was recently developed,[30] but sensors of endonuclease and 5′-exonuclease activity are still desired. Here, we report the development and properties of highly selective fluorogenic nuclease sensors to detect and distinguish single-strand nucleases of these three distinct classes.
Our approach to this chemosensing problem involved the use of short modified DNA strands containing designed fluorescent nucleobases.[31–34] The use of the DNA backbone renders such compounds trivial to synthesize using automated nucleic acid synthesizer. In addition, they are highly soluble in water, and easily delivered into live cells. Furthermore, the bases are efficiently stacked on one other, and as a result the fluorescent properties of such oligomers are often dramatically sequence-dependent.[35,36] Previously we have studied the remarkable quenching effect of neighboring bases on the emission of fluorescent non-natural bases.[37,38] For example, thymine (T) was found to be an efficient quencher of an adjacent fluorogenic pyrene α-deoxyriboside (Y) by a photoinduced charge transfer mechanism.[39] Other bases were less strongly quenching of this fluorophore, while adenine (A) had no effect on its emission. Recently, we reported a novel fluorogenic sensor for DNA base excision repair that takes advantage of the fact that uracil (like T) also effectively quenches pyrene.[34]
With the aim of selective reporting of distinct nuclease activities, we envisioned a series of programmed sequences of single-stranded modified nucleic acids (Scheme 1), taking advantage of the unnatural structure of fluorescent base Y and its quenching properties. We began by selecting as our initial targets three well-characterized bacterial nucleases, including one of each class (3′ and 5′ exonuclease, and endonuclease; see below). We then prepared a number of candidate fluorogenic substrates 3–10 nt in length (see Supporting Information (SI) Table S1), incorporating Y and T residues in varying numbers and positions. Preliminary enzyme substrates studies of the fifteen candidates (Figures S1-S3) led us ultimately to four best-performing probes (1–4, Scheme 1) that were studied in greater detail.
Scheme 1.
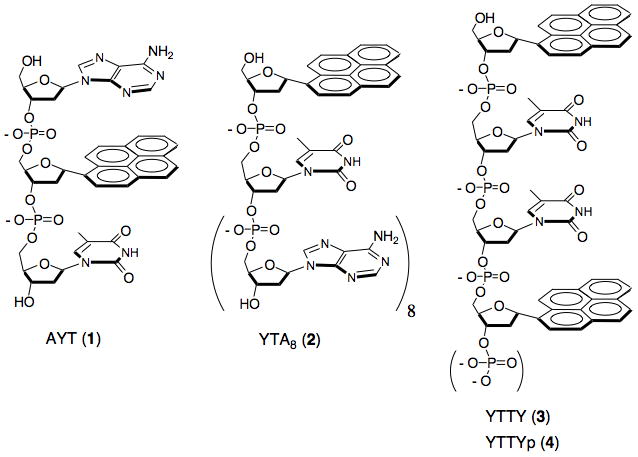
Class-selective fluorogenic nuclease sensors built from modified DNAs, using thymine residues as quenchers for fluorescent pyrene monomers (Y). Backbone cleavage by nucleases separates thymine from pyrene, resulting in a signal.
First, the short modified oligomer 5′-AYT (1) was designed as a 3′-exonuclease sensor, and as a representative enzyme target of this type we chose the bacterial enzyme Exo T, also known as RNase T,[40,41] which cleaves DNA or RNA from the 3′-end. The fluorescence of Y in AYT (1) is strongly quenched by the neighboring T (see below). In the presence of Exo T, however, the quencher T is expected to be cleaved from the end (if the neighboring unnatural pyrene does not block this activity) to give AY which would emit bright fluorescence. While thymine quenches pyrene strongly, adenine has little or no effect on its fluorescence quantum yield.[38]
The second chemosensor, 5′-YTAAAAAAAA (YTA8, 2) was as a substrate for a single-stranded DNA specific 5′-exonuclease, RecJf.[42] The length of the A8 tail at the 3′-end of 2 was designed to fulfill a substrate requirement of RecJf nuclease, which functions poorly with shorter DNA fragments.[42] In our design, RecJf nuclease is expected to cleave the 5′ terminal Y from the sensor YTA8 (2) as the initial cleavage event, releasing Y and separating it from the neighboring quenching thymine nucleobase. However, prior to our preliminary experiments it was not known whether the enzyme would be able to recognize the unnatural α-pyrene nucleotide as a substrate.
Lastly, YTTY (3) was our initial design as a sensor for a single-strand endonuclease, nuclease S1, a well-known enzyme from Staphylococcus. Similar to the other probes, the pyrene in probe 3 is quenched by nearby thymines; in this third case, any of three bond cleavages might enhance fluorescence. S1 nuclease can cleave very short oligomers of DNA or RNA, including dimers, but it was not known whether it would also cleave DNA near the large and α-anomeric pyrene residues.
During the design of the probes, we considered structure and sequence motifs that might make them selective only for one class of nuclease over the others. In our preliminary studies, we found that the dimer composed with fluorophore-quencher pair YT remained dark in the presence of Exo T or S1 nucleases (data not shown), which suggested that YT is not a good substrate for Exo T and S1. Thus, YTA8 is expected to be non-fluorogenic in the presence of Exo T and S1 even though some of its bonds can be hydrolyzed by Exo T and S1 to some extent. In addition, AYT (1) and YTTY (3) are too short to be substrates for RecJf enzyme, rendering them nonresponsive to this 5′-nuclease.[42] To make the endonuclease sensor YTTY (3) resistant to 3′-exonuclease Exo T (Figure S6), we added a phosphate group at the 3′-terminus to yield the final design of this probe (YTTYp, 4) since a terminal phosphate is known to inhibit this 3′-nuclease.[43] It still remained in question whether AYT (1) would be fluorogenic in the presence of the endonuclease S1.
To test the fluorogenic nuclease sensors, the short probe oligomers were prepared according to standard DNA synthesis protocols. The synthesized probes were purified by HPLC and characterized by MALDI-TOF-mass spectrometry and by their absorption and fluorescence spectra (Table S2; Figures S4, S5). We also measured the quantum yields, which were all very low in comparison to that of pyrene nucleotide, which is a bright fluorophore (Φfl > 0.3).[38] Although thymines quenched the pyrene residues efficiently in all three probes, the quantum yield of the quenched probe YTTYp (4, QY = 0.020) was unexpectedly high compared to AYT (1, QY = 0.0088) and YTA8 (2, QY = 0.0055). This is likely due to the intramolecular complexation of pyrenes in 4 in the excited state, as evidenced by the small residual excimer fluorescence emission of this probe (Figure S5). Excimer-forming pyrene dimers are not as strongly quenched by thymine as pyrene monomers are.[38]
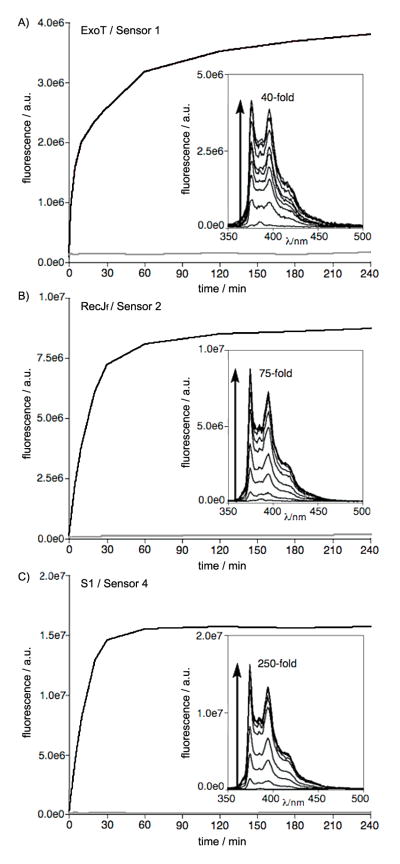
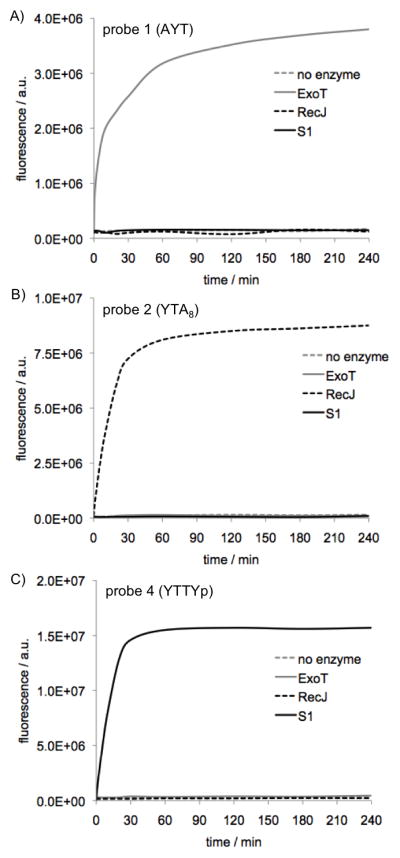
The nuclease chemosensors were tested at 100 nM with enzymes in vitro, and cleavage reactions were monitored by fluorescence increase over time (Figure 1). We were pleased to find that all three classes of nuclease sensors AYT (1), YTA8 (2) and YTTYp (4) were good substrates for their target nucleases, exhibiting rapid responses and robust fluorecence in spite of the presence of the large, hydrophobic non-natural base. All nuclease sensors showed pronounced light-up signals ranging from 40-fold (probe 1 with Exo T) to 250- fold (4 with S1) increase in fluorescence over a period of 60–180 min. Control experiments in reaction buffer without nuclease enzyme confirmed that none of the sensors yielded any signals from nonspecific degradation.
Figure 1.
In vitro responses of nuclease sensors 1, 2, and 4 with a 3′-exonuclease (A), a 5′-exonuclease (B), and an endonuclease (C), respectively (red lines). Blue lines correspond to control experiments lacking enzyme. Insets show emission spectra over the time courses. Excitation 342 nm, emission 375 nm, [nuclease sensor] = 100 nM, 25 °C. [Exo T] = 25 U/mL, [RecJf] = 900 U/mL, [S1] = 100 U/mL. See SI for details.
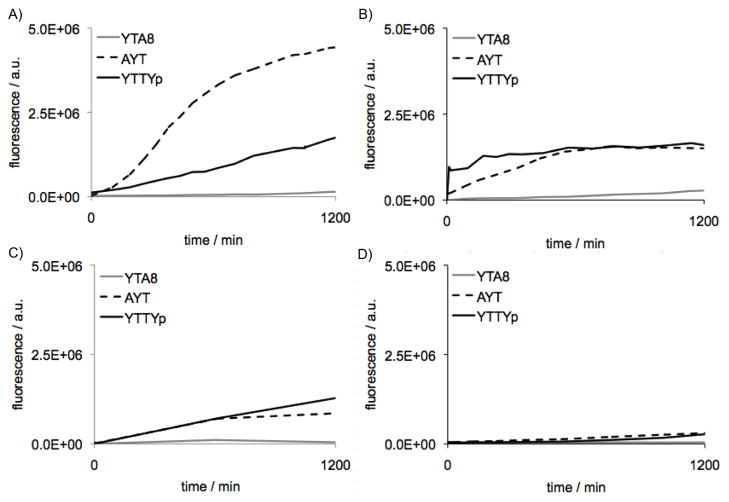
Next we examined the selectivity of the designed nuclease sensors for the three classes of nuclease activity represented in our target enzymes. As shown in Figure 2, all nuclease sensors AYT (1), YTA8 (2) and YTTYp (4) were found to be highly selective for the assigned nucleases. For example, AYT (1) remained dark in the presence of RecJf and S1, likely due to its short length and positioning of pyrene. We also noted the remarkable preference of S1 endonuclease for the tetranucleotide (YTTYp, 4) over a trinucleotide (AYT, 1). (S1 endonuclease is known to produce mono-, di- and tri-deoxynucleotides[44]). Modification of nucleotides likely affects the substrate preference of S1 endonuclease. It was reported to be possible to identify trinucleotide-nucleic acid alkylating agent adducts from enzymatically digested DNA with S1 endonuclease.[45] Exo T and S1 were also inefficient at cleaving the phosphodiesterase bond between Y and T of YTA8 (2); although they may cleave some A monomers from the sensor, this would not yield a fluorogenic signal. Finally, as designed, the 3′ terminal phosphate of YTTYp (4) successfully suppressed 3′ exonuclease activity of Exo T, as evidenced by the fact that YTTY (3) produced a robust increase in fluorescence in the presence of both Exo T and S1 enzymes (Figure S6) while 4 was highly selective for only S1 (Figure 2C).
Figure 2.
Selectivity of nuclease sensors among distinct classes of nucleases. (A) sensor 1 (AYT); (B) sensor 2 (YTA8); (C) sensor 4 (YTTYp). Conditions: 25 °C, excitation 342 nm, emission 375 nm for Exo T and S1, 374 nm for RecJf. [nuclease sensor] = 100 nM. [Exo T] = 25 U/mL, [RecJf] = 900 U/mL, [S1] = 100 U/mL.
Encouraged by these results, we proceeded to perform further tests of the fluorogenic probes in complex biological media that contain varied types of nuclease activities. Sensors 1, 2, and 4 were incubated separately in solutions of human saliva, serum, urine, and sweat (Figure 3A-D and Figure S7). Overall, sweat showed the lowest level of total nuclease activity; serum and urine exhibited several-fold higher activity, and saliva displayed the highest activity of the four. Interestingly, the fluids exhibited differences in the general classes of activity present. 5′-exonuclease-selective sensor 2 reacted slowly in all fluids tested, suggesting that this type of nuclease activity is low in all cases. Sweat and urine showed signals of similar magnitude over time for 3′-exonuclease (1) and endonuclease (4) activity. Serum yielded similar responses, but with a more rapid initial response from the endonuclease sensor. In contrast, saliva yielded a greatly different signal,[46] with a rapid and robust response from the 3′-nuclease sensor 1, followed by a slower but similarly robust signal from the endonuclease sensor 4. These three fluorogenic sensors, which were developed to be selective among three specific bacterial nucleases, are not expected to maintain this high specificity among all human nucleases, since many specialized nucleases will have distinct substrate preferences. Nevertheless, these results showed that varied nuclease activities in complex biological media can be analyzed and differentiated by use of three simple structural chemosensor variants.
Figure 3.
Reactivity of nuclease sensors 1, 2, and 4 in biological fluids. (A) 10% saliva; (B) 10% serum; (C) 10% urine; (D) 8.3% sweat. Conditions: 37 °C, excitation 342 nm, emission 375 nm. [nuclease sensor] = 100 nM
The above experiments probed nuclease activities in extracellular fluids. Next we asked whether they could be applied to nuclease activities that exist within human cells, and whether different cellular compartments contain measurably different classes of activity. To test this, we incubated the sensors 1, 2, and 4 separately with the cytoplasmic and nuclear fractions of HeLa cell lysate (Figure 4). We are not aware of previous studies of relative classes of nucleases in the nucleus, but other studies have suggested that exonucleases are predominant in the cytoplasm.[47] Sensors 1, 2 (after an initial lag), and 4 gave similar signals in the cytosolic fraction, indicating a mix of exo- and endo- nuclease activities in similar amounts. Interestingly, the nuclear fraction yielded a significantly different response: sensor 4 gave a greater signal than the other two sensors, indicating higher endonuclease activity in the nucleus relative to exonuclease activities. Taken together, the results establish that the chemosensors can function in complex biological media, and are able to indicate clear differences in the types and relative levels of nuclease activities present.
Figure 4.
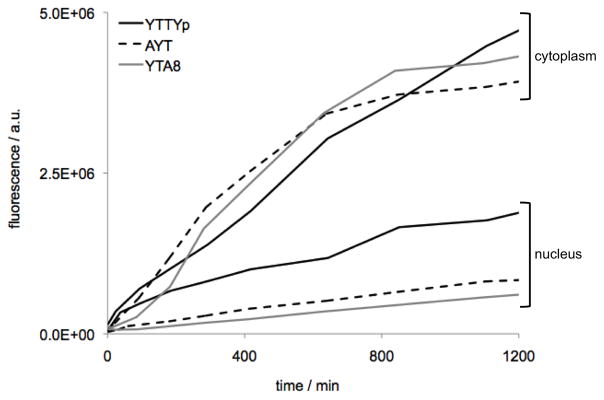
Reactivity of nuclease sensors 1, 2, and 4 in cytoplasmic and nuclear fractions of HeLa cell lysate. [nuclease sensor] = 5 uM. [cell lysate] = 1.7×1011 lysed cells/mL) at 37 °C, with excitation 342 nm, emission 375 nm.
In summary, we have developed three highly selective fluorogenic nuclease sensors built from very small modified DNA oligomers containing the nonnatural fluorescent base pyrene. Compared to most other methods for measuring nuclease activity, these nuclease sensors have advantages in 1) ease of use, requiring only simple addition to a sample; 2) small size and simplicity, which facilitates synthesis; 3) low sensor loading, thus requiring only very small amounts of the synthetic compounds; 4) a robust increase in fluorescence, exhibiting high signal over background; 5) the ability to selectively assess different classes of nuclease activities; and 6) utility in biological and cellular samples. Future work will be directed to development of sensors with varied wavelengths of signals, and to applications of the sensors in new biological contexts.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institutes of Health (GM067201 and GM072705), by a fellowship from the National Research Foundation of the Korean Ministry of Education, Science and Technology (NRF-2010-357-E00049) to J.-W.J, and by a National Science Foundation predoctoral fellowship to S.K.E.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. Vol. 2. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 2.Emswiler-Rose BS, Johnston RW, Harris ME, Lee WH. Appl Environ Microbiol. 1980;40:13–18. doi: 10.1128/aem.40.1.13-18.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen G, Jin SLC, Umetsu DT, Conti M. Proc Natl Acad Sci U S A. 2000;97:6751–6756. doi: 10.1073/pnas.97.12.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang HT, Huang Y, Jin SLC, Frith SA, Suvarna N, Conti M, O’Donnell JM. Neuropsychopharmacology. 2002;27:587–595. doi: 10.1016/S0893-133X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 5.Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS. Sci Signaling. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 6.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM. Nat Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 7.Ballard SA, Gingell CJ, Tang K, Turner LA, Price ME, Naylor AM. J Urol. 1998;159:2164–2171. doi: 10.1016/S0022-5347(01)63299-3. [DOI] [PubMed] [Google Scholar]
- 8.Endoh M, Yamashita S, Taira N. J Pharmacol Exp Ther. 1982;221:775–783. [PubMed] [Google Scholar]
- 9.Maniatis T. In: Molecular Cloning: a Laboratory Manual. Sambrook J, Fritsch EF, Maniatis T, editors. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- 10.Geiduschek E, Daniels A. Anal Biochem. 1965;11:133. doi: 10.1016/0003-2697(65)90052-7. [DOI] [PubMed] [Google Scholar]
- 11.Fliess A, Wolfes H, Rosenthal A, Schwellnus K, Blöcker H, Frank R, Pingoud A. Nucleic Acids Res. 1986;14:3463–3474. doi: 10.1093/nar/14.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeltsch A, Fritz A, Alves J, Wolfes H, Pingoud A. Anal Biochem. 1993;213:234–240. doi: 10.1006/abio.1993.1415. [DOI] [PubMed] [Google Scholar]
- 13.Tolun G, Myers RS. Nucleic Acids Res. 2003;31:e111–e111. doi: 10.1093/nar/gng111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pu F, Hu D, Ren J, Wang S, Qu X. Langmuir. 2009;26:4540–4545. doi: 10.1021/la904173j. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Pu F, Ren J, Qu X. Chem Commun (Camb) 2011;47:8133–8135. doi: 10.1039/c1cc12216a. [DOI] [PubMed] [Google Scholar]
- 16.Wang M, Zhang D, Zhang G, Tang Y, Wang S, Zhu D. Anal Chem. 2008;80:6443–6448. doi: 10.1021/ac801020v. [DOI] [PubMed] [Google Scholar]
- 17.Feng F, Tang Y, He F, Yu M, Duan X, Wang S, Li Y, Zhu D. Adv Mater. 2007;19:3490–3495. [Google Scholar]
- 18.Gill R, Willner I, Shweky I, Banin U. J Phys Chem B. 2005;109:23715–23719. doi: 10.1021/jp054874p. [DOI] [PubMed] [Google Scholar]
- 19.Ray PC, Fortner A, Darbha GK. J Phys Chem B. 2006;110:20745–20748. doi: 10.1021/jp065121l. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Han MS, Mirkin CA. Angew Chem. 2007;119:3538–3540. [Google Scholar]
- 21.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Anal Chem. 2008;80:8431–8437. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W, Lam JCF, Chiuman W, Brook MA, Li Y. Small. 2008;4:810–816. doi: 10.1002/smll.200700757. [DOI] [PubMed] [Google Scholar]
- 23.Song G, Chen C, Ren J, Qu X. ACS Nano. 2009;3:1183–1189. doi: 10.1021/nn800768z. [DOI] [PubMed] [Google Scholar]
- 24.Shen Q, Nie Z, Guo M, Zhong CJ, Lin B, Li W, Yao S. Chem Commun (Camb) 2009;28:929–931. doi: 10.1039/b818081d. [DOI] [PubMed] [Google Scholar]
- 25.Yu C, Chan KHY, Wong KMC, Yam VWW. Chem Commun (Camb) 2009;25:3756–3758. doi: 10.1039/b903080h. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, Hu P, Zhao H, Li Y, Huang C. Anal Chim Acta. 2011;706:171. doi: 10.1016/j.aca.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 27.Zhao C, Qu K, Song Y, Ren J, Qu X. Adv Funct Mater. 2011;21:583–590. [Google Scholar]
- 28.Li JJ, Geyer R, Tan W. Nucleic Acids Res. 2000;28:e52–e52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biggins JB, Prudent JR, Marshall DJ, Ruppen M, Thorson JS. Proc Natl Acad Sci U S A. 2000;97:13537. doi: 10.1073/pnas.240460997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su X, Zhu X, Zhang C, Xiao X, Zhao M. Anal Chem. 2012;84:5059–5065. doi: 10.1021/ac300745f. [DOI] [PubMed] [Google Scholar]
- 31.Dai N, Teo YN, Kool ET. Chem Commun. 2010;46:1221–1223. doi: 10.1039/b926338a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai N, Kool ET. Chem Soc Rev. 2011;40:5756–5770. doi: 10.1039/c0cs00162g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai N, Guo J, Teo YN, Kool ET. Angew Chem. 2011;123:5211–5215. doi: 10.1002/anie.201007805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono T, Wang S, Koo CK, Engstrom L, David SS, Kool ET. Angew Chem Int Ed. 2012;51:1689–1692. doi: 10.1002/anie.201108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson JN, Gao J, Kool ET. Tetrahedron. 2007;63:3427–3433. doi: 10.1016/j.tet.2006.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teo YN, Wilson JN, Kool ET. Chem Eur J. 2009;15:11551–11558. doi: 10.1002/chem.200901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson JN, Teo YN, Kool ET. J Am Chem Soc. 2007;129:15426. doi: 10.1021/ja075968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson JN, Cho Y, Tan S, Cuppoletti A, Kool ET. ChemBioChem. 2008;9:279–285. doi: 10.1002/cbic.200700381. [DOI] [PubMed] [Google Scholar]
- 39.Telser J, Cruickshank KA, Morrison LE, Netzel TL, Chan CK. J Am Chem Soc. 1989;111:7226–7232. [Google Scholar]
- 40.Viswanathan M, Dower KW, Lovett ST. J Biol Chem. 1998;273:35126–35131. doi: 10.1074/jbc.273.52.35126. [DOI] [PubMed] [Google Scholar]
- 41.Zuo Y, Deutscher MP. Nucleic Acids Res. 1999;27:4077–4082. doi: 10.1093/nar/27.20.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han ES, Cooper DL, Persky NS, Sutera VA, Jr, Whitaker RD, Montello ML, Lovett ST. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deutscher MP, Marlor CW, Zaniewski R. Proc Natl Acad Sci U S A. 1984;81:4290. doi: 10.1073/pnas.81.14.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ando T. Biochim Biophys Acta. 1966;114:158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- 45.Mohamed D, Linscheid M. Anal Bioanal Chem. 2008;392:805–817. doi: 10.1007/s00216-008-2236-0. [DOI] [PubMed] [Google Scholar]
- 46.Yaegaki K, Sakata T, Ogura R, Kameyama T, Sujaku C. J Dental Res. 1982;61:1222–1224. doi: 10.1177/00220345820610110101. [DOI] [PubMed] [Google Scholar]
- 47.Dagle JM, Weeks DL, Walder JA. Antisense Res Dev. 1991;1:11–20. doi: 10.1089/ard.1991.1.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.