Abstract
The activity of natural killer cells depends on the balance between activating and inhibitory signals coming from their receptors. Among these are the killer cell immunoglobulin-like receptors (KIR) that recognize specific HLA class I allotypes. Here we characterized KIR genetic diversity and their HLA ligands in the population of Curitiba, Paraná State (n=164), and compared it with other worldwide populations. The distribution of 2DL4 alleles was also analyzed. The Curitiba population did not differ significantly from European and Euro-descendant populations, but as an admixed population showed higher genetic diversity. We found 27 KIR profiles, many of them uncommon in European populations, in agreement with the elevated historically recent gene flow in the study population. The frequencies of KIR genes and their respective HLA ligands were distributed independently and none of the analyzed individuals lacked functional KIR–HLA ligand combinations. KIR gene frequencies of 33 worldwide populations were consistent with geographic and ethnic distribution, in agreement with demography being the major factor shaping the observed gene content diversity of the KIR locus.
Keywords: KIR genes, KIR diversity, Brazilian population, HLA ligands, 2DL4 allele frequencies
Natural killer (NK) cells are crucial in innate immunity and influence the outcome of antigen-specific immune responses (Biron et al. 1999). They also contribute to placental development and the outcome of pregnancy (Trowsdale and Moffett 2008). NK cell function depends on complex cellular interactions mediated by membrane-bound and soluble gene products. Of prime importance are the inhibitory and activating killer cell immunoglobulin-like receptors (KIR) that bind major histocompatibility complex class I molecules. KIR are encoded by a cluster of homologous genes located on human chromosome 19q13.4 (Wende et al. 1999; Liu et al. 2000). KIR diversity is observed as presence/absence of genes, resulting in expansion and contraction of KIR haplotypes (Wende et al. 2000; Martin et al. 2003) and further diversity is provided by allelic variation of individual KIR genes. Almost 400 KIR profiles (i.e., genotypes defined by KIR gene content) have been identified among 190 populations catalogued online (www.allelefrequencies.net—Gonzalez-Galarza et al. 2011). The presence or absence of certain KIR genes as well as some KIR–HLA receptor–ligand combinations have been shown to associate with various diseases (Khakoo and Carrington 2006; Jiao et al. 2008; Levinson et al. 2008). KIR genes may also be genetic landmarks of populations (Rayes et al. 2008) and there are suggestions that KIR gene frequencies may be influenced by host/pathogen interactions (Kulkarni et al. 2008). There is also strong evidence that HLA and KIR are co-evolving (Single et al. 2007; Norman et al. 2007)
Much interest has been shown in the analysis of KIR gene frequencies in human populations as a means to understand the evolutionary causes and functional consequences of its polymorphism. However, the data thus far has been limited to a few geographic regions around the world and few Brazilian populations are represented (Middleton et al. 2008; Rudnick et al. 2008). Regarding allelic diversity, even less is known. In this study, we aimed to characterize KIR gene content polymorphism and 2DL4 allelic diversity in an urban Brazilian population and compare this with other populations. We also tested for correlations between KIR and HLA ligand frequencies that might support the hypothesis of co-evolution of these two unlinked loci.
The population from Curitiba (the Capital city of Paraná State, Brazil, located at 25°25′ S and 49°17′ W) is of mixed, predominantly European ancestry. A detailed description can be found in Probst et al. (2000) and Braun-Prado et al. (2000). The individuals tested (n=164) were unrelated, healthy volunteers. All subjects were informed about the aims of the study and formally agreed to participate. The study has been performed according to Brazilian laws and was approved by the ethics committee of the Federal University of Paraná. A subset of individuals (n=99) was sequenced for 2DL4. The number of individuals in some analyses may vary because of quantitative and qualitative limitations of DNA. We tested for presence/absence of 15 KIR genes by PCR-SSP with two pairs of specific primers for each locus (Martin and Carrington 2008; Kulkarni et al. 2010). Unusual profiles were carefully analyzed and were retyped two to three times to validate the results. HLA-A, HLA-B, and HLA-C were previously genotyped by the PCR-SSOP method (Braun-Prado et al. 2000). The Alellefrequencies.net database (Gonzalez-Galarza et al. 2011) was used as the reference to identify KIR profiles and to obtain data for comparisons between populations. Allelic subtyping of 2DL4 was carried out by amplification of exons 3, 4, 6, and 8 using genespecific primers and the products were sequenced using the Big Dye terminator kit (Applied Biosystems). The primer sequences are available on request.
The carrier frequency (F) of each KIR gene (presence/absence) was obtained by direct counting. KIR gene frequencies were estimated using Bernstein’s formula fG= 1–√(1–F), where F corresponds to the frequency of carriers. Frequencies of the two pairs of “allelic genes”, 2DL2–2DL3 and 3DL1–3DS1, were estimated by two approaches: (1) like the other KIR genes as just described, considering them as independent from each other, and (2) considering that they segregate as alleles of one locus with three alleles, two of which are codominant (the “genes”) and the third one being recessive (absence of the gene). Haplotype A and B frequencies were also calculated by Bernstein’s formula. The linkage disequilibrium (LD) parameter D corresponds to the difference between the estimated two-loci haplotype frequencies and the haplotype frequencies expected at equilibrium and was calculated as described by Mattiuz et al. (1970). D’, the LD coefficient standardized by the maximum value it can take given the allele frequencies (Lewontin 1964), was estimated with the Arlequin software package version 3.16 (Excoffier et al. 2005).
Thirty-two previously described worldwide populations were compared with the study population by the exact test of population differentiation included in the Arlequin software package v. 3.16. The non-informative pseudogenes and framework genes were excluded from the analysis. Genetic distances were estimated by Nei’s method (Nei 1972) using PHYLIP—version 3.6 (Felsenstein 2004). A dendrogram was drawn by the neighbor-joining method (Saitou and Nei 1987) and visualized with the TreeView software (Page 1996). The expected frequency of co-occurrence of KIR and their HLA class I ligands was estimated and compared to the observed frequency using the chi-square test. The receptor/ligand pairs tested were KIR3DL1/S1+HLA Bw4, KIR 2DL2/3+HLA-C group 1, and KIR2DL1+HLA-C group 2. In addition, we counted in each individual the number of functional KIR–HLA combinations to estimate their average number and to determine the proportion of individuals in the population lacking functional receptor/ligand combinations.
The carrier frequencies of the KIR genes in the study population varied from 31.9% (2DS5) to 100% (2DL4, 3DL2, and 3DL3). With the exception of 2DS4 (93.6%), no other activating KIR gene reached a carrier frequency of 60%. Overall, the frequencies of the inhibitory KIR genes were higher and more homogeneous among worldwide populations than for the activating genes (Table 1) and KIR gene frequencies in the study population are in agreement with the predominantly European ancestry of this population. Among all the KIR genes, only 2DL2 and 2DL3 differed significantly in frequencies between the Curitiba population and other populations with European ancestry (Table 1). The frequency of 2DL2 differed significantly between Curitiba and France and 2DL3 differed between Curitiba and some European and Euro-descendant populations, including the population from Belo Horizonte. The reason for this heterogeneity is not clear but it may reflect the ethnic admixture of some of these populations. It may also be related to the partially overlapping functions of these genes. 2DL2 and 2DL3 segregate as alleles of one locus and both bind HLA-C group 1 molecules although ligand binding is stronger for 2DL2 (Moesta et al. 2008). Thus, 2DL2 and 2DL3 may be exposed to less severe functional constraints in comparison to other inhibitory KIR genes. Relaxation of selection may also permit greater fluctuation of frequencies as a consequence of demographic factors. The two-locus LD pattern was similar to other population studies (see Supplementary Figure 1).
Table 1.
KIR gene frequencies in the population of Curitiba and in worldwide populations
| n | F (%) | Curitiba 153 fG |
f A | Czech 125 F (%) |
BHorizonte 90 |
Argentina 102 |
France 108 |
Ireland 200 |
Palestine 105 |
India 145 |
USAfricans 58 |
Senegal 118 |
Camoros 54 |
Singapore 47 |
Hong Kong 100 |
Japan 132 |
China 106 |
S. Korea 154 |
Amazon 40 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2DL1 | 97.4 | 0.84 | – | 95.0 | 96.7 | 96.1 | 97.0 | 96.0 | 83.0*** | 99.3 | 98.3 | 100.0 | 100.0 | 100.0 | 99.0 | 100.0 | 99.0 | 99.4 | 93.0 |
| 2DL2 | 59.4 | 0.36 | 0.36 | 59.0 | 52.2 | 62.5 | 50.0*** | 51.0 | 62.0 | 62.8 | 56.9 | 55.0 | 41.0* | 28.2*** | 28.0*** | 11.4*** | 17.3*** | 14.3*** | 65.0 |
| 2DL3 | 84.6 | 0.61 | 0.55 | 86.0 | 94.4* | 86.5 | 91.0*** | 93.0* | 85.0 | 81.9 | 82.0 | 90.0 | 93.0 | 100.0** | 98.0*** | 100.0*** | 99.0*** | 99.4*** | 80.0 |
| 2DL4 | 100.0 | 1.00 | – | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 2DL5 | 51.9 | 0.31 | – | 52.0 | 58.9 | 55.7 | 47.0 | 44.0 | 63.0 | 71.0*** | 53.4 | 52.0 | 62.7 | 39.1 | 45.0 | 35.6** | 68.7* | 38.3** | 85.0*** |
| 2DP1 | 94.7 | 0.77 | – | 94.0 | 96.7 | 96.1 | 97.0 | 97.0 | NT | 99.3 | 98.3 | 100.0 | 100.0 | 100.0 | 99.0 | 100.0 | 99.0 | NT | NT |
| 2DS1 | 42.1 | 0.24 | – | 43.0 | 37.8 | 45.4 | 36.0 | 34.0 | 44.0 | 62.8*** | 22.4 | 13.0*** | 17.0*** | 28.3 | 40.0 | 33.3 | 34.3 | 37.7 | 88.0*** |
| 2DS2 | 59.2 | 0.36 | – | 57.0 | 53.3 | 54.5 | 51.0 | 52.0 | 64.0 | 62.8 | 46.6 | 42.0** | 30.0*** | 28.3*** | 28.0*** | 11.4*** | 17.3*** | 16.9*** | 58.0 |
| 2DS3 | 32.5 | 0.18 | – | 36.0 | 38.9 | 28.6 | 31.0 | 28.0 | 37.0 | 53.8*** | 27.6 | 24.0 | 20.0 | 17.4* | 25.0 | 13.6*** | 12.5*** | 16.2*** | 10.0** |
| 2DS4 | 93.6 | 0.75 | – | 92.0 | 95.6 | 95.0 | 96.0 | 95.0 | 88.0 | 86.1*** | 100.0 | 100.0** | 96.0 | 97.8 | 94.0 | 99.2 | 94.2 | 94.2 | 78.0** |
| 2DS5 | 31.9 | 0.17 | – | 26.0 | 32.2 | 35.4 | 27.0 | 26.0 | 27.0 | 51.0*** | 37.9 | 30.0 | 37.0 | 21.7 | 26.0 | 22.0 | 23.1 | 26.6 | 90.0*** |
| 3DL1 | 92.7 | 0.73 | 0.74 | 94.0 | 95.6 | 95.0 | 96.0 | 95.0 | 88.0 | 87.4 | 100.0* | 99.0* | 98.0 | 97.8 | 94.0 | 99.2** | 94.2 | 94.2 | 65.0*** |
| 3DS1 | 37.3 | 0.21 | 0.26 | 38.0 | 41.1 | 42.0 | 44.0 | 35.0 | 39.0 | 62.2*** | 13.8*** | 4.0*** | 15.0** | 30.4 | 39.0 | 31.9 | 32.7 | 36.4 | 70.0*** |
| 3DL2 | 100.0 | 1.00 | – | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 3DL3 | 100.0 | 1.00 | – | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
The asterisks indicate p values for the populations that differed significantly from Curitiba: 0.01≤p<0.05;
0.001≤p<0.01, and
p<0.001. References are listed in Table 3. The populations are named as in the dendrogram
n number of individuals in the sample, F carrier frequency, fG gene frequency obtained by Bernstein’s method, fA the gene frequency, considering the allelic status of 2DL2 and 2DL3, and of 3DL1, 3DS1, NT not tested
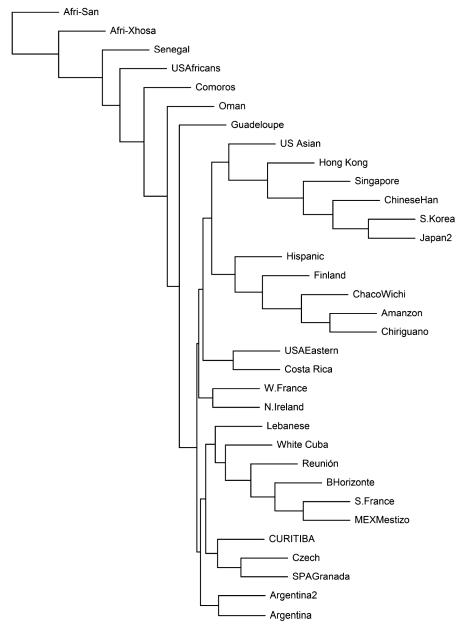
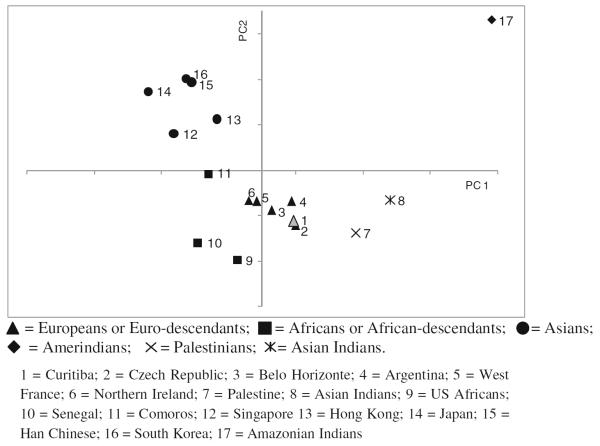
We characterized genetic distances based on KIR gene carrier frequencies of 33 populations, and the dendrogram generated is consistent with geography and ancestry (Fig. 1). In general, the European populations tended to cluster together including the Curitiba population, but Finland was somewhat separated from other Europeans. The most divergent are the Africans and they did not cluster in a single clade. It should however be stressed that internal nodes of the dendrogram do not represent a common ancestral population and that grouping of populations may result not only from common ancestry but also from gene flow (admixture), natural selection, or demographic factors resulting in random genetic drift, bottleneck, and founder effects. The principal component analysis (Fig. 2) also revealed that common ancestry is the major factor resulting in similarity of KIR gene frequencies, followed by gene flow and possibly random demographic factors.
Fig. 1.
Nei’s genetic distances of 33 worldwide populations visualized in a neighbor-joining dendrogram. Population references are given in Table 3
Fig. 2.
Principal components analysis of 17 worldwide populations, including the population of Curitiba
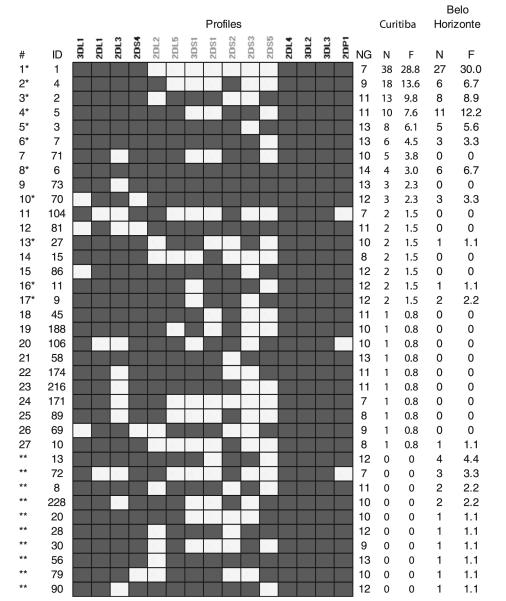
The profiles observed in the study population are listed in Fig. 3. For comparison, the profiles found in Brazilians from Belo Horizonte (Middleton et al. 2008) are also shown. We identified 27 profiles, eight of which accounted for 77.3% of all observed profiles. The profile representing homozygosity for haplotype A (profile 1 in Fig. 3) was the most frequent (F=28.8%), as reported for other European populations. The frequency of A and B haplotypes were estimated to be 0.54 and 0.46, respectively. The less frequent profiles that are uncommon in Europeans or Euro-descendants reflect the heterogeneous ancestry of this population. Apart from Europeans (Portuguese, Italian, Polish, German, Ukrainian, and others), Africans, as well as Amerindians who had lived in South America before the colonization, contributed to the present gene pool of our study population (Probst et al. 2000). Previous results from our group showed that the ancestry of this population is approximately 87% European, 8% African, and 5% Amerindian (Braun-Prado et al. 2000; Probst et al. 2000 and unpublished results). Therefore, the occurrence of profiles characteristic of non-Europeans and a higher diversity than that seen in other European populations is not unexpected. KIR haplotypes could be more informative as ancestry markers, but the recessive absence of KIR genes makes it difficult to accurately infer haplotypes in samples of unrelated individuals and segregation analysis would require analysis of a large sample of extended families. The KIR complex also has the potential to be a good ancestry marker after more studies describing its allele diversity become available.
Fig. 3.
KIR profiles observed in the population from Curitiba and Belo Horizonte. Profiles found in Curitiba (present study) and Belo Horizonte, another urban Brazilian population (Middleton et al. 2008). * Profiles found in both populations. ** Profiles not found in Curitiba. ID is the identification number in Allefrequencies.net (Gonzalez-Galarza et al. 2011); NG is the number of genes per profile (excluding pseudogenes), N number of individuals with a given profile, F relative frequency (%). Genes shown in gray are found exclusively on B haplotypes. Filled boxes indicate presence of a KIR gene and open boxes indicate absence of the gene
We next compared 2DL4 allele frequencies between Curitiba and Belo Horizonte, another urban Brazilian population (Table 2, top) and found that there were significant differences in allele frequencies (See Table 3 for references of Belo Horizonte and other populations used for comparisons). However, this difference can be explained by the low typing resolution used in the Belo Horizonte study, where only exons 3 and 4 that encode the extracellular domains were sequenced (Williams et al. 2004; Middleton et al. 2008). The allele pairs 00202 and 00801, and 00201 and 00802, differ only in an adenine deletion in exon 6, which encodes the transmembrane domain. Differences in exons 6 and 8 also discriminate between 00501 and 011, and between 00102 and 00103, respectively. When the ambiguous allele pairs were grouped (Table 2, bottom), their frequencies did not differ significantly (p=0.67). This is noteworthy because of the differential contribution of Europeans to these two populations: while the European ancestry component of Belo Horizonte is essentially Iberian, in Curitiba, apart from the Portuguese, other Mediterranean and central and eastern Europeans account for a high proportion of the immigrants (Probst et al. 2000). Further, the French (Buhler et al. 2009) and the USA populations (Gedil et al. 2005) typed at the same high resolution as in our study, show 2DL4 allele frequencies similar to those seen in Curitiba (p>0.05; data not shown). Thus, the similarity between Curitiba and Belo Horizonte provides additional evidence of low differentiation across Europeans for 2DL4. Based on the adenine deletion in exon 6, nucleotide position 811, two groups of 2DL4 alleles have been defined, referred to as 9A (the deleted) and 10A (Witt et al. 2000). The two groups differ markedly with respect to 2DL4 expression and function. Deletion alleles cannot be expressed as membrane-bound receptors due to excision of the transmembrane domain as a consequence of the frame shift caused by the adenine deletion. (Goodridge et al. 2003; Goodridge et al, 2007). In our study population, 57.6% of the individuals have a deleted 9A allele in at least one chromosome and 18.2% are homozygous for 9A and therefore lack cell surface 2DL4 receptors. It has been suggested that these two allele groups are maintained at intermediate frequency by balancing natural selection (Witt et al. 2000; Goodridge et al. 2003). This hypothesis is supported by the high frequency of both the deleted and the undeleted allele groups. 2DS4 also exhibits deletion and nondeletion allelic variants. A group of 2DS4 alleles have a deletion of 22 bp in exon 5, encompassing codons 131 to 137 and the first nucleotide of codon 138 within the D2 extracellular domain (Maxwell et al. 2002; Hsu et al. 2002). This results in a frame shift yielding a truncated protein with no transmembrane or cytoplasmic domains and is therefore not expressed on the cell surface. The truncated 2DS4 carrier frequency in Curitiba was 75%. In France this allele group is also very common (carrier frequency 84%), while in Senegal the frequency is lower (carrier frequency 48%) (Denis et al. 2005). 2DS4 is the only classical activating receptor found on the A haplotype, and interestingly we estimated that in Curitiba 33% of individuals homozygous for haplotype A (corresponding to 13.6% of the detected profiles) are homozygous for the deletion, and therefore lack a functional classical activating KIR.
Table 2.
KIR2DL4 allele frequencies in Curitiba and in Belo Horizonte
| Allele | Curitiba (n=99) |
Belo Horizonte (n=90) | |
|---|---|---|---|
| f A | F (%) | F (%) | |
| 2DL4*00102/5 | 0.28 | 44.4 | 50.0 |
| 2DL4*00501 | 0.21 | 35.4 | 53.3 |
| 2DL4*00802 | 0.15 | 28.3 | 0.0 |
| 2DL4*00801 | 0.13 | 23.2 | 0.0 |
| 2DL4*011 | 0.11 | 15.2 | 0.0 |
| 2DL4*00103 | 0.06 | 10.1 | 0.0 |
| 2DL4*006 | 0.04 | 5.1 | 9.0 |
| 2DL4*010 | 0.01 | 2.0 | 0.0 |
| 2DL4*00104 | 0.01 | 1.0 | 0.0 |
| 2DL4*00202 | 0.00 | 0.0 | 27.9 |
| 2DL4*00201 | 0.00 | 0.0 | 28.7 |
| Allele groupsa | |||
| 2DL4*00102/5+00103 | 0.34 | 54.5 | 50.0 |
| 2DL4*00501+011 | 0.32 | 50.5 | 53.3 |
| 2DL4*00802+00201 | 0.15 | 28.3 | 28.7 |
| 2DL4*00801+00202 | 0.13 | 23.2 | 27.9 |
Data for Belo Horizonte from Middleton et al. (2008)
n number of individuals, fA allele frequency, F(%) carrier frequency
Alleles were grouped considering the ambiguities of the low resolution typing employed for the analysis of Belo Horizonte’s population (Middleton et al. 2008)
Table 3.
References for the populations used in the estimation of genetic distances and principal components analysis
| Name as in the dendrogram | Population name and sample size | Reference |
|---|---|---|
| Amazon | Amazon Amerindians 40 | Ewerton et al. 2007 |
| Argentina | Argentina 102 | Unpublished data * |
| Argentina 2 | Argentina Buenos Aires 365 | Flores et al. 2007 |
| ChacoWichi | Argentina Chaco Wichis 82 | Flores et al. 2007 |
| Chiriguano | Argentina Chiriguanos 54 | Flores et al. 2007 |
| Czech | Czech Population | Pavlova et al. 2008 |
| BHorizonte | Brazil Belo Horizonte 90 | Middleton et al. 2008 |
| ChineseHan | China Zhejiang Han 104 | Jiang et al. 2005 |
| Comoros | Comoros 54 | Frassati et al. 2006 |
| Costa Rica | Costa Rica Guanacaste 117 | Carrington et al. 2005 |
| White Cuba | Cuban White 70 | Middleton et al. 2008 |
| Curitiba | Euro-descendants 164 | Present study |
| Finland | Finland Helsinki 101 | Denis et al. 2005 |
| S. France | France Southeast pop2 38 | Frassati et al. 2006 |
| W. France | France West 108 | Denis et al. 2005 |
| Guadeloupe | Guadeloupe 118 | Denis et al. 2005 |
| Hong Kong | Hong Kong Chinese 100 | Middleton et al. 2008 |
| N. Ireland | Ireland Northern pop2 154 | Middleton et al. 2008 |
| Japan2 | Japanese 132 | Yawata et al. 2006 |
| Lebanese | Lebanon 120 | Mahfouz et al. 2006 |
| MEX Mestizo | Mexico Veracruz Mestizos 51 | Contreras et al. 2007 |
| Oman | Oman 99 | Middleton et al. 2008 |
| Palestine | Palestine Jordan 105 | Norman et al. 2001 |
| Reunion | Reunion 101 | Denis et al. 2005 |
| Senegal | Senegal 90 | Denis et al. 2005 |
| Singapore | Singapore Chinese 47 | Middleton et al. 2008 |
| Afri-San | South African San 91 | Middleton et al. 2008 |
| Afri-Xhosa | South African Xhosa 50 | Middleton et al. 2008 |
| S. Korea | South Korea 154 | Whang et al. 2005 |
| SPAGranada | Spain Granada 100 WS | Middleton et al. 2007 |
| US Africans | US California African Americans 58 | Du et al. 2007 |
| US Asian | US California Asian Americans 150 | Du et al. 2007 |
| USAEastern | Euro-descendants from USA 213 | Carrington et al. 2005 |
| Hispanic | US California Hispanics 128 | Du et al. 2007 |
| India | India 145 | Kulkarni et al. 2008 |
Population data available on Allelefrequencies.net database (Gonzalez-Galarza et al. 2011)
Because KIR and HLA are both highly polymorphic and because these two gene clusters segregate independently, an individual may have receptor and no ligand or vice versa. Previous studies have also highlighted the relevance of KIR with their HLA ligands in disease pathogenesis and resistance to viral infections, thus we evaluated the KIR–HLA class I ligand combinations in our population. With respect to 2DL1, 2DL2, 2DL3, and 3DL1, the present population sample exhibited an average of 2.9 functional receptor–ligand pairs per individual and each individual carried at least two functional receptor–ligand pairs. The inhibitory 2DL1 binds HLA-C group 2 allotypes with lysine at position 80, and 2DL2/3 receptors bind HLA-C group 1 allotypes, with asparagine at the same position. 3DL1 recognizes HLA-A and HLA-B allotypes with the Bw4 motif. The Bw4 positive HLA-A molecules (A*23, A*24, A*25, and A*32) have isoleucine at position 80 (Bw4-80I) that is also present in a subset of the HLA-B Bw4 molecules. HLA-B Bw4-80I molecules exhibit stronger interactions with their cognate KIR receptors than Bw4-80T (threonine) bearing HLA-B molecules (Cella et al. 1994; Carr et al. 2005). In the present study, we observed a similar frequency of Bw4-80I from both the HLA-A and HLA-B loci (0.20 and 0.24, respectively). Of the individuals lacking HLA-B Bw4, 47% had a Bw4 HLA-A allele. The high frequency of HLA-A molecules with the Bw4 epitope suggest that these HLA-A and HLA-B allotypes may be equally important for NK cell function.
The frequencies of the HLA ligands are shown in Supplementary Table 1. All ligands and their respective receptors were distributed independently (p=0.44, data not shown). This might indicate that natural selection acting on specific receptor/ligand combinations is not a major factor determining their population frequencies. Alternatively, the KIR–HLA frequencies might be at equilibrium in this population. However, this result should be interpreted with caution, because for a statistically significant association to be observed in such a population sample, the selection pressure would need to be unusually strong.
The heterogeneous colonization and the continued migrations during the five centuries of Brazil’s history as well as the high frequency of interethnic unions, has resulted in an admixed population whose ancestry differs among geographic regions. This is the first study describing KIR2DL4 allele frequencies in a Southern Brazilian population of predominately European background and only a few South American populations have thus far been analyzed with respect to KIR gene content variation and KIR gene allelic frequencies. The European origins of the population of Curitiba are more heterogeneous than that of most other Brazilian populations. Nevertheless, its KIR gene content and 2DL4 allelic diversity are generally similar to those of most European populations. More effort should be dedicated to analyze other admixed and unique isolated populations of various ancestries existing in Brazil and all over the world in order to provide more informative data for evolutionary and functional analyses of KIR polymorphism.
Supplementary Material
Acknowledgments
We thank the staff of the Laboratory of Human Molecular Genetics at the Department of Genetics of the Federal University of Paraná for assistance and support. Special thanks to Liana Alves de Oliveira for her expertise and valuable comments. We also thank Fuh-Mei Duh and Colm O’huigin for helpful advice and Maria Dias da Silva for reading this manuscript. This project received financial support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), PRONEX, Institutos do Milênio, Fundação Araucária, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00251-011-0565-1) contains supplementary material, which is available to authorized users.
Contributor Information
D. G. Augusto, Laboratório de Genética Molecular Humana, Departamento de Genética, Universidade Federal do Paraná, Caixa Postal 19071, Curitiba 81531-980, Brazil
L. Zehnder-Alves, Laboratório de Genética Molecular Humana, Departamento de Genética, Universidade Federal do Paraná, Caixa Postal 19071, Curitiba 81531-980, Brazil
M. R. Pincerati, Departamento de Genética e Biologia Evolutiva, Universidade de São Paulo, Rua do Matão 277, São Paulo 05508-090, Brazil
M. P. Martin, Cancer and Inflammation Program, Laboratory of Experimental Immunology, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, USA
M. Carrington, Cancer and Inflammation Program, Laboratory of Experimental Immunology, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, USA; Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology and Harvard University, Boston, MA 02114, USA
Maria Luiza Petzl-Erler, Laboratório de Genética Molecular Humana, Departamento de Genética, Universidade Federal do Paraná, Caixa Postal 19071, Curitiba 81531-980, Brazil.
References
- Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- Braun-Prado K, Vieira Mion AL, Farah Pereira N, Culpi L, Petzl-Erler ML. HLA class I polymorphism, as characterised by PCR-SSOP, in a Brazilian exogamic population. Tissue Antigens. 2000;56:417–427. doi: 10.1034/j.1399-0039.2000.560504.x. [DOI] [PubMed] [Google Scholar]
- Buhler S, Di Cristofaro J, Frassati C, Basire A, Galicher V, Chiaroni J, Picard C. High levels of molecular polymorphism at the KIR2DL4 locus in French and Congolese populations: impact for anthropology and clinical studies. Hum Immunol. 2009;70(11):953–959. doi: 10.1016/j.humimm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175(8):5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- Carrington M, Wang S, Martin MP, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180(4):1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras G, Aláez C, Murguía A, García D, Flores H, Gorodezky C. Distribution of the killer cell immunoglobulin-like receptors in Mexican Mestizos. Tissue Antigens. 2007;69:125–129. doi: 10.1111/j.1399-0039.2006.76212.x. [DOI] [PubMed] [Google Scholar]
- Denis L, Sivula J, Gourraud PA, et al. Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens. 2005;66:267–276. doi: 10.1111/j.1399-0039.2005.00473.x. [DOI] [PubMed] [Google Scholar]
- Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007;59:1–15. doi: 10.1007/s00251-006-0168-4. [DOI] [PubMed] [Google Scholar]
- Ewerton PD, Leite Mde M, Magalhães M, Sena L, Melo dos Santos EJ. Amazonian Amerindians exhibit high variability of KIR profiles. Immunogenetics. 2007;59:625–630. doi: 10.1007/s00251-007-0229-3. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval LG, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington; Seattle: 2004. Distributed by the author. [Google Scholar]
- Flores AC, Marcos CY, Paladino N, et al. KIR genes polymorphism in Argentinean Caucasoid and Amerindian populations. Tissue Antigens. 2007;69:568–576. doi: 10.1111/j.1399-0039.2007.00824.x. [DOI] [PubMed] [Google Scholar]
- Frassati C, Touinssi M, Picard C, et al. Distribution of killer-cell immunoglobulin-like receptor (KIR) in Comoros and Southeast France. Tissue Antigens. 2006;67:356–367. doi: 10.1111/j.1399-0039.2006.00592.x. [DOI] [PubMed] [Google Scholar]
- Gedil MA, Steiner NK, Hurley CK. Genomic characterization of KIR2DL4 infamilies and unrelated individuals reveals extensive diversity in exon and intron sequences including a common frameshift variation occurring in several alleles. Tissue Antigens. 2005;65:402–418. doi: 10.1111/j.1399-0039.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acid Research. 2011;39:D913–D919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge JP, Witt CS, Christiansen FT, Warren HS. KIR2DL4 (CD158d) genotype influences expression and function in NK cells. J Immunol. 2003;171:1768–1774. doi: 10.4049/jimmunol.171.4.1768. [DOI] [PubMed] [Google Scholar]
- Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, Hurley CK, Christiansen FT, Witt CS. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework aplotypes, each with multiple subsets. J Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. 2005;65:556–563. doi: 10.1111/j.1399-0039.2005.00412.x. [DOI] [PubMed] [Google Scholar]
- Jiao YL, Ma CY, Wang LC, Cui B, Zhang J, You L, Chen ZJ, Li JF, Zhao YR. Polymorphisms of KIRs gene and HLA-C alleles in patients with ankylosing spondylitis: possible association with susceptibility to the disease. J Clin Immunol. 2008;28:343–349. doi: 10.1007/s10875-008-9183-6. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Single RM, Martin MP, Rajalingam R, Badwe R, Joshi N, Carrington M. Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics. 2008;60:121–129. doi: 10.1007/s00251-008-0279-1. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. KIR genotyping by multiplex PCR-SSP. Methods Mol Biol. 2010;612:365–375. doi: 10.1007/978-1-60761-362-6_25. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson RD, Du Z, Luo L, Holland GN, Rao NA, Reed EF, Rajalingam R. KIR and HLA gene combinations in Vogt-Koyanagi-Harada disease. Hum Immunol. 2008;69:349–353. doi: 10.1016/j.humimm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP. Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4. Immunogenetics. 2000;51:659–669. doi: 10.1007/s002510000183. [DOI] [PubMed] [Google Scholar]
- Mahfouz R, Rayes R, Mahfoud Z, Bazarbachi A, Zaatari G. Distribution of killer cell immunoglobulin-like receptors genotypes in the Lebanese population. Tissue Antigens. 2006;68:66–71. doi: 10.1111/j.1399-0039.2006.00605.x. [DOI] [PubMed] [Google Scholar]
- Martin MP, Carrington M. KIR locus polymorphisms: genotyping and disease association analysis. Methods Mol Biol. 2008;415:49–64. doi: 10.1007/978-1-59745-570-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- Mattiuz PL, Ihde D, Piazza A, Ceppellini R, Bodmer WF. New approaches to the population genetic and segregation analysis of the HL-A system. In: Terasaki P, editor. Histocompatibility testing 1970. Vol. 1971. Munksgaard; Copenhagen: 1970. pp. 193–205. [Google Scholar]
- Maxwell LD, Wallace A, Middleton D, Curran MD. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens. 2002;60:254–258. doi: 10.1034/j.1399-0039.2002.600307.x. [DOI] [PubMed] [Google Scholar]
- Middleton D, Vilchez JR, Cabrera T, Meenagh A, Williams F, Halfpenny I, Maleno I, Ruiz-Cabello F, Lopez-Nevot MA, Garrido F. Analysis of KIR gene frequencies in HLA class I characterized bladder, colorectal and laryngeal tumours. Tissue Antigens. 2007;69:220–226. doi: 10.1111/j.1399-0039.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- Middleton D, Meenagh A, Moscoso J, Arnaiz-Villena A. Killer immunoglobulin receptor gene and allele frequencies in Caucasoid, Oriental and Black populations from different continents. Tissue Antigens. 2008;71:105–113. doi: 10.1111/j.1399-0039.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;15:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. Amer Naturalist. 1972;106:283–292. [Google Scholar]
- Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52:195–205. doi: 10.1007/s002510000281. [DOI] [PubMed] [Google Scholar]
- Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- Page RD. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pavlova Y, Kolesar L, Striz I, Jabor A, Slavcev A. Distribution of KIR genes in the Czech population. Int J Immunogenet. 2008;35:57–61. doi: 10.1111/j.1744-313X.2007.00737.x. [DOI] [PubMed] [Google Scholar]
- Probst CM, Bompeixe EP, Pereira NF, de Dalalio MMO, Visentainer JE, Tsuneto LT, Petzl-Erler ML. HLA polymorphism and evaluation of European, African, and Amerindian contributionto the white and mulatto populations from Paraná, Brazil. Hum Biol. 2000;72:597–617. [PubMed] [Google Scholar]
- Rayes R, Bazarbachi A, Khazen G, Sabbagh A, Zaatari G, Mahfouz R. Natural killer cell immunoglobulin-like receptors (KIR) genotypes in two arab populations: will KIR become a genetic landmark between nations? Mol Biol Rep. 2008;35:225–229. doi: 10.1007/s11033-007-9074-6. [DOI] [PubMed] [Google Scholar]
- Rudnick CC, Franceschi DS, Marangon AV, Guelsin GA, Sell AM, Visentainer JE. Killer cell immunoglobulin-like receptor gene diversity in a Southern Brazilian population from the state of Paraná. Hum Immunol. 2008;69:872–876. doi: 10.1016/j.humimm.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-Joining mehod: a new method for reconstructing phylogenetic trees. Mol Biol and Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Moffett A. NK receptor interactions with MHC class I molecules in pregnancy. Semin Immunol. 2008;20:317–320. doi: 10.1016/j.smim.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Whang DH, Park H, Yoon JA, Park MH. Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol. 2005;66:146–154. doi: 10.1016/j.humimm.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Williams F, Meenagh A, Sleator C, Middleton D. Investigation of killer cell immunoglobulinlike receptor gene diversity: I. KIR2DL4. Hum Immunol. 2004;65:31–38. doi: 10.1016/j.humimm.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- Wende H, Volz A, Ziegler A. Extensive gene duplications and a large inversion characterize the human leukocyte receptor cluster. Immunogenetics. 2000;51:703–713. doi: 10.1007/s002510000187. [DOI] [PubMed] [Google Scholar]; Immunogenetics. 2001;52:308. Erratum in: [Google Scholar]
- Witt CS, Martin A, Christiansen FT. Detection of KIR2DL4 alleles by sequencing and SSCP reveals a common allele with a shortened cytoplasmic tail. Tissue Antigens. 2000;56:248–257. doi: 10.1034/j.1399-0039.2000.560307.x. [DOI] [PubMed] [Google Scholar]
- Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.