Abstract
Mitofusin 2 (Mfn2) is a key protein in mitochondrial fusion and it participates in the bridging of mitochondria to the endoplasmic reticulum (ER). Recent data indicate that Mfn2 ablation leads to ER stress. Here we report on the mechanisms by which Mfn2 modulates cellular responses to ER stress. Induction of ER stress in Mfn2-deficient cells caused massive ER expansion and excessive activation of all three Unfolded Protein Response (UPR) branches (PERK, XBP-1, and ATF6). In spite of an enhanced UPR, these cells showed reduced activation of apoptosis and autophagy during ER stress. Silencing of PERK increased the apoptosis of Mfn2-ablated cells in response to ER stress. XBP-1 loss-of-function ameliorated autophagic activity of these cells upon ER stress. Mfn2 physically interacts with PERK, and Mfn2-ablated cells showed sustained activation of this protein kinase under basal conditions. Unexpectedly, PERK silencing in these cells reduced ROS production, normalized mitochondrial calcium, and improved mitochondrial morphology. In summary, our data indicate that Mfn2 is an upstream modulator of PERK. Furthermore, Mfn2 loss-of-function reveals that PERK is a key regulator of mitochondrial morphology and function.
Keywords: cell death and autophagy, cell metabolism, mitochondria
Introduction
Endoplasmic reticulum (ER) stress is triggered in response to a variety of stimuli that lead to a number of alterations of the ER, such as dysregulated redox homoeostasis, abnormal protein folding, and reduced calcium levels. Pathologies such as neurodegeneration, diabetes, cancer, and cardiovascular disease are characterized by the presence of ER stress (Hotamisligil, 2010). At least three proteins located in the ER membrane (PERK, IRE-1α, and ATF6) respond to ER stress stimuli by triggering the Unfolded Protein Response (UPR). These pathways activate several transcription factors involved in cell survival during the early stages of stress and in apoptotic cell death during acute or chronic stress. The PERK branch participates in the reduction of protein synthesis and activation of autophagy, apoptosis, and chaperone expression (Harding et al, 1999; Lu et al, 2004b). PERK-stimulated CHOP favors apoptosis (Harding et al, 2000; Rouschop et al, 2010), and, conversely, it has also been reported that CHOP protects neuronal cells after ischaemia reperfusion (Halterman et al, 2010). PERK activation decreases protein synthesis through eIF2α phosphorylation (Lu et al, 2004a), and, in addition, sustained PERK activation induces GADD34 expression, which relieves protein synthesis inhibition through eIF2α dephosphorylation (Novoa et al, 2001).
The IRE-1α branch activates the expression of XBP-1 transcription factor, which is involved in the synthesis of chaperones and other ER proteins. The IRE-1α pathway is also required for autophagy activation (Ogata et al, 2006). Interestingly, XBP-1 deficiency promotes autophagy in neuronal cells (Hetz et al, 2009). This observation points to a complex role of the IRE-1α pathway in autophagic regulation.
The transcription factor ATF6 is also stimulated during ER stress. In addition, calcium signals participate in this activation (Xu et al, 2004). This transcription factor induces XBP-1 and chaperones and participates in cell survival. Thus, reduced apoptosis occurs in response to ischaemia reperfusion in hearts overexpressing ATF6 (Doroudgar et al, 2009). Tumour cell survival also requires ATF6 through the activation of the mTOR pathway (Schewe and Aguirre-Ghiso, 2008; Doroudgar et al, 2009). ATF6 also mediates the synthesis of proteins and lipids required for ER biogenesis and participates in the synthesis of proteins required to activate ERAD, a mechanism for the degradation of ER proteins (Yamamoto et al, 2004; Maiuolo et al, 2011). Recent data demonstrate that PERK facilitates the stimulation of the ATF6 pathway (Teske et al, 2011). These studies indicate that UPR branches are exquisitely regulated and that several proteins are involved in the cross talk between branches.
Mitofusin 2 (Mfn2) is a GTPase protein localized in the outer mitochondrial membrane and in mitochondrial-associated membranes. In addition to controlling ER morphology, this protein participates in mitochondrial fusion and regulates the transfer of calcium from the ER to mitochondria (Santel and Fuller, 2001; Legros et al, 2002; Bach et al, 2003; Ishihara et al, 2004; de Brito and Scorrano, 2008). Moreover, Mfn2 deficiency upregulates markers of the UPR in liver, skeletal muscle, and cultured cells (Ngoh et al, 2012; Sebastian et al, 2012), and normalization of ER stress by treatment with tauroursodeoxycholic acid ameliorates deficient insulin signalling in liver-specific Mfn2-knockout mice (Sebastian et al, 2012). Here we demonstrate that Mfn2 is a novel PERK regulator and that it is required for the normal activation of apoptosis and autophagy during the ER stress response. Moreover, Mfn2 deficiency causes mitochondrial dysfunction through sustained activation of PERK.
Results
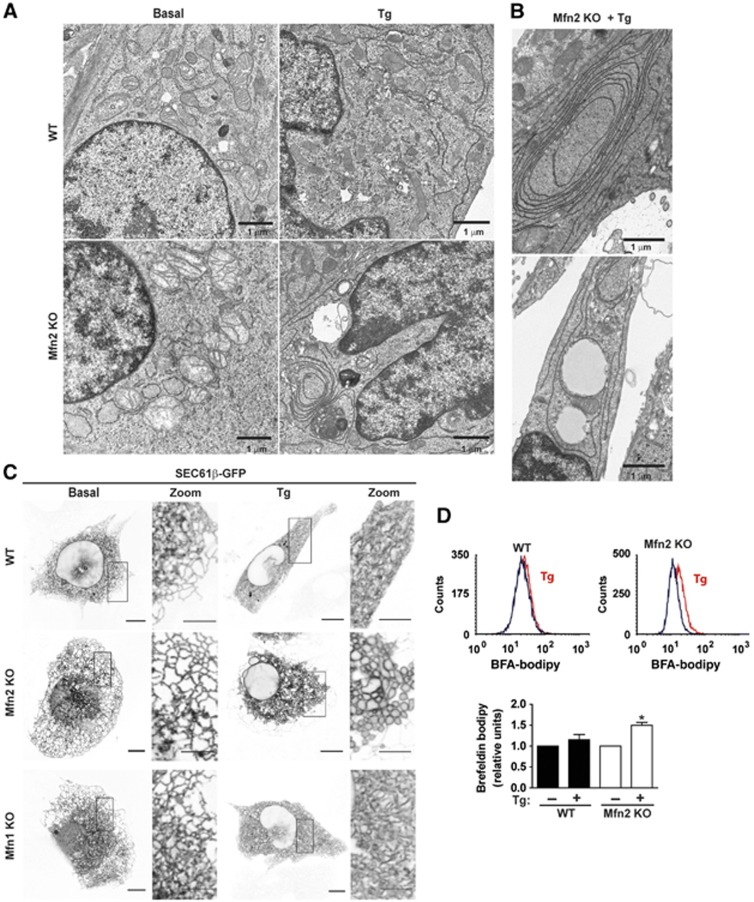
Mfn2 knockout (KO) cells show massive ER expansion in response to ER stress
On the basis of the observations linking Mfn2 to ER biology, we examined the response of Mfn2-deficient cells to agents that cause ER stress. To this end, mouse embryo fibroblasts (MEFs) were incubated with 1 μM thapsigargin (Tg) for 12 h in order to induce ER stress. Under these conditions, Mfn2 KO MEFs underwent dramatic ER expansion, characterized by multilamellar structures, compared to wild-type (WT) MEFs (Figure 1A and B). To provide further evidence of the effects of ER stress on ER morphology, cells were transfected with a plasmid encoding an RFP protein that is localized in the ER luminal compartment (ER-RFP), and thereafter treated with Tg and visualized by epifluorescence microscopy. Under basal conditions, Mfn2 ablation caused ER fragmentation, which was detectable with the luminal ER marker (Supplementary Figure S1A). This finding is in keeping with prior data (de Brito and Scorrano, 2008). Supplementary Figure S1A also shows increased ER luminal expansion during ER stress (induced by Tg, tunicamycin, or brefeldin A) in Mfn2 KO cells compared to WT cells. Movie 1 (Supplementary Data) shows the formation of the expanded ER in Mfn2 KO cells upon the induction of ER stress. Labelling of the ER membrane by transfection of Sec61β-GFP also showed a dramatic ER expansion in Mfn2 KO cells in response to Tg (Figure 1C). However, labelling of ER membranes with Sec61β-GFP did not show alterations in ER morphology in these cells under basal conditions (Figure 1C), which is in keeping with prior data in COS cells (Friedman et al, 2011). No alterations in ER morphology were detected in Mfn1 KO cells in response to Tg (Figure 1C). Reexpression of Mfn2 in Mfn2 KO cells rescued normal mitochondrial morphology and prevented ER expansion in response to ER stress (Supplementary Figure S1B–E).
Figure 1.
Mfn2 ablation promotes abnormal ER expansion during ER stress conditions. (A) WT or Mfn2 KO MEFs (Mfn2 KO cells) were treated with 1 μM Tg for 12 h and then processed for EM visualization of the ER morphology. Scale bar: 1 μm. (B) EM images of Tg-treated Mfn2 KO cells show accumulation of ER membrane stacking. Scale bar: 1 μm. (C) WT, Mfn2 KO or Mfn1 KO cells were transfected with the Sec61β-GFP plasmid and treated with 1 μM Tg for 24 h. Confocal microscopy images show ER vacuolization in Mfn2 KO cells treated with Tg. Scale bar: 10 μm. Insets show × 10 zoomed images. Scale bar: 5 μm. (D) WT and Mfn2 KO cells were treated with Tg 1 μM for 3 h and then incubated with brefeldin A-bodipy to stain ER and Golgi. Representative flow-cytometry histograms (upper panel). Mean fluorescence intensity was used to quantify ER expansion (n=5) (lower panel). Data are mean±s.e.m. *P<0.05 versus WT group.
The enhanced ER expansion in Mfn2 KO cells upon Tg treatment was further documented by labelling ER–Golgi with brefeldin A-bodipy (Hetz et al, 2006) (Figure 1D).
Deficiency of Mfn1 or Mfn2 was also induced in MEFs by lentiviral expression of shRNAs, which caused mitochondrial fragmentation (Supplementary Figure S2A and B). Labelling of the ER membrane by transfection of Sec61β-GFP also showed marked ER expansion in Mfn2-silenced MEF cells treated with Tg (Supplementary Figure S2C). Mfn1 deficiency did not cause ER expansion upon Tg treatment (Supplementary Figure S2C).
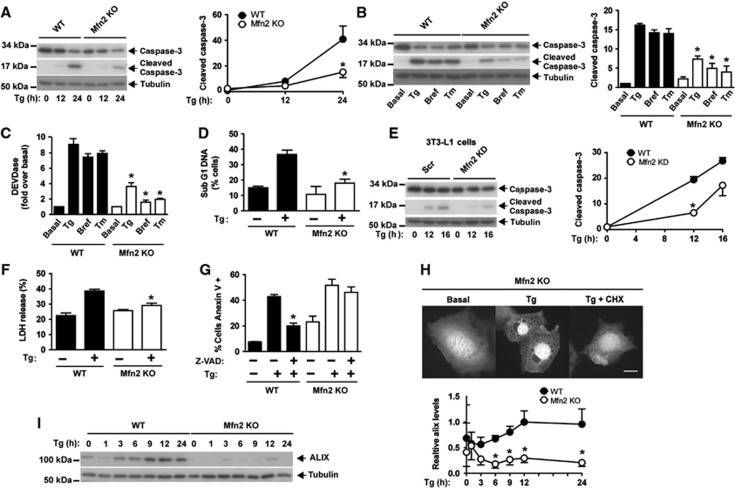
Mfn2 KO cells show defective apoptosis in response to ER stress
On the basis of the initial observations indicating that Mfn2 ablation causes abnormal ER expansion in response to ER stress, next we examined whether Mfn2 loss-of-function also alters the biological responses driven by this stress. In this regard, apoptosis was induced through activation of caspase 3 (Rao et al, 2002) in response to ER stress. Mfn2 loss-of-function significantly enhanced caspase activity in MEFs and in C2C12 cells under basal conditions (Supplementary Figure S3A) (see Supplementary Figures S2 and S4 for details on Mfn2 deficiency induced in these cells). WT MEFs showed caspase 3 activation and increased caspase activity in response to tunicamycin, brefeldin A, or Tg (Figure 2A–C). Under these conditions, Mfn2 ablation greatly reduced caspase 3 processing and caspase activity (Figure 2A–C), and reexpression of Mfn2 normalized the activation of this enzyme (Supplementary Figure S3B). Reduced apoptosis was also detected by cytometry assays (subG1 DNA fragmentation) in Mfn2 KO cells (Figure 2D). Mfn1 KO cells showed reduced caspase 3 processing and caspase activity upon Tg, tunicamycin, or brefeldin A treatment, although the values observed were higher than in Mfn2 KO cells (Supplementary Figure S3C and D). Silencing of Mfn2 in 3T3-L1 or in C2C12 cells also reduced caspase 3 processing in response to ER stress (Figure 2E and Supplementary Figure S3F). In contrast, agents that induce apoptosis independently of ER stress, such as staurosporine or TNFalpha/cycloheximide (CHX), activated caspase 3 processing to a greater extent in Mfn2 KO cells compared to WT cells (Supplementary Figure S3E).
Figure 2.
Mfn2 ablation prevents caspase activation during ER stress and promotes paraptosis-like cell death. (A) WT and Mfn2 KO cells were treated with 1 μM Tg for 12 or 24 h. Total and cleaved caspase 3 levels were detected by western blot. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. (B, C) WT and Mfn2 KO cells were treated with 0.5 μg/ml tunicamycin (Tm), 100 ng/ml brefeldin A (Bref), or 1 μM Tg for 24 h. Total and cleaved caspase 3 levels were detected by western blot (B) and caspase activity (C) by measurement of DEVD-AFC substrate processing. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. (D) Flow-cytometry analysis of the Sub G1 DNA fragmentation in methanol-fixed WT and Mfn2 KO cells after incubation with or without 1 μM Tg for 24 h. Data are given as mean±s.e.m. (n=3). *P<0.05 versus WT group. (E) Scr (stably expressing scrambled shRNA) and Mfn2 knockdown (KD) (stably expressing shRNA directed against Mfn2) 3T3-L1 fibroblasts were incubated in the presence or absence of 1 μM Tg for 24 h. Total and cleaved caspase 3 were detected by western blot. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. (F) WT and Mfn2 KO cells were treated with 1 μM Tg for 24 h. Lactate dehydrogenase (LDH) release was analyzed by flow cytometry to assess necrotic cell death. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. (G) WT and Mfn2 KO cells were incubated for 24 h with or without 1 μM Tg in the presence or absence of z-VAD-fmk and stained for annexin V/PI. Data are mean±s.e.m. (n=4). *P<0.05 versus WT+Tg group. (H) Mfn2 KO cells transfected with the pEGFP plasmid were incubated with 1 μM Tg alone or in combination with 2 μM CHX for 24 h (3 h of pre-incubation with CHX). Florescence microscopy images show that CHX prevents cytoplasmic vacuolization. Scale bar: 10 μm. (I) WT (black circles) and Mfn2 KO cells (white circles) were incubated with 1 μM Tg for varying times, and ALIX protein was detected by western blot. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group.
Source data for this figure is available on the online supplementary information page.
Next, we analyzed whether Mfn2 KO cells die as a result of another form of cell death upon ER stress. Lactate dehydrogenase release (an index of cell necrosis) increased in control cells during ER stress, but this did not occur in Mfn2-ablated cells (Figure 2F). We explored whether Mfn2 KO cells undergo a paraptotic-like death, which is characterized by swelling of the mitochondria and of the ER, reduced ALIX expression (Sperandio et al, 2000; Yoon et al, 2010), and involves externalization of phosphatidylserine (PS) independently of caspases (Wang et al, 2004). In keeping with this, Mfn2 KO cells became annexin V-positive in response to Tg, and PS externalization was not inhibited with the pan-caspase inhibitor z-VAD (Figure 2G). In addition, in response to ER stress, Mfn2 KO cells underwent vacuolization, which was rescued by CHX (Figure 2H), and showed lower ALIX protein expression (Figure 2I). Propidium iodide (PI) permeability assays coupled to annexin V labelling in control and Mfn2 KO cells showed increased PI/annexin V labelling in response to Tg. This labelling was inhibited by addition of CHX in Mfn2 KO cells but not in WT cells (Supplementary Figure 3G). In summary, our data show that Mfn2 loss-of-function impairs the normal activation of apoptosis during ER stress, and instead paraptosis-like cell death is triggered. This observation suggests that Mfn2 plays a key role in the maintenance of a normal response to ER stress.
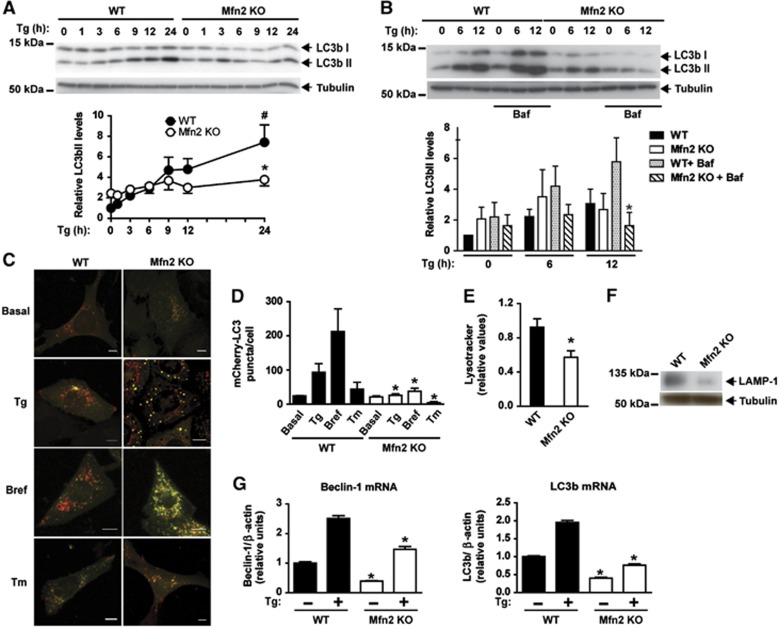
Mfn2 KO cells show defective autophagy in response to ER stress
ER stress also activates autophagy (Ogata et al, 2006), and the accumulation of multilamellar ER structures detected in Mfn2−/− cells (Figure 1) pointed to alterations in the clearance process. To analyze autophagy in cells subjected to ER stress, control and Mfn2 KO cells were treated with Tg for a range of times, and autophagy was assayed by immunodetection of LC3b-I and LC3b-II. ER stress caused an increased abundance of LC3b-II in control cells, which is coherent with the activation of autophagosome formation (Figure 3A). Under basal conditions, Mfn2 KO cells showed a slightly increased LC3b-II abundance (Figure 3A). ER stress-induced LC3b-II was inhibited in Mfn2 KO cells (Figure 3A), thereby suggesting a defect in autophagy progression. Incubation with the lysosomal inhibitor bafilomycin A1 increased LC3b-II levels in WT cells but not in Mfn2 KO ones (Figure 3B). Mfn2 reexpression normalized LC3b-II levels in Mfn2 KO cells treated with Tg (Supplementary Figure S5A). These observations indicate that autophagosomal formation is impaired during ER stress upon ablation of Mfn2 in MEFs.
Figure 3.
Mfn2 loss-of-function decreases autophagy in response to ER stress. (A, upper panel) WT (black circles) or Mfn2 KO cells (white circles) were treated with 1 μM Tg for a range of times. (A, lower) Densitometric quantification. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group; #P<0.05 versus WT non-Tg-treated group. (B, upper panel) WT or Mfn2 KO cells were treated with Tg for 0, 6 or 12 h, in the presence or absence of Bafilomycin (Baf, 100 nM). LC3b-I and LC3b-II expression was measured by western blot. (B, lower panel) Densitometric quantification of LC3b-II levels (relative to tubulin). Data are mean±s.e.m. (n=3). *P<0.05 versus WT+Baf+Tg group. (C) WT or Mfn2 KO cells stably expressing mCherry-GFP-LC3b were treated with 1 μM Tg, 100 ng/ml Brefeldin (Bref), or 0.5 μg/ml tunicamycin (Tm) for 24 h and examined by confocal microscopy. Scale bar: 10 μm. (D) Quantification of mCherry+/GFP- (mCherry-LC3) red puncta per cell (a measure of acidic autophagosomes) is shown. Data are mean±s.e.m. (n=3; 100 cells were analyzed per experiment and group). *P<0.05 versus WT group. (E) Acidic compartments of WT or Mfn2 KO cells were stained with Lysotracker Green and analyzed by flow cytometry. Data are given as mean±s.e.m. (n=3). *P<0.05 versus WT group. (F) LAMP1 expression was immunodetected in WT and Mfn2 KO cells in basal conditions. (G) Expression of Beclin-1 or LC3b transcripts in WT or Mfn2 KO cells treated with or without 1 μM Tg for 24 h. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group.
Source data for this figure is available on the online supplementary information page.
To obtain further insight into the autophagic steps that may be altered in Mfn2-deficient cells, we expressed mCherry-EGFP-LC3 protein in these cells. ER stress markedly enhanced the abundance of acidic autophagolysosomes—detected as red punctuates in mCherry-EGFP-LC3B-expressing cells—which is consistent with increased autophagic activity (Figure 3C and D). Under these conditions, Mfn2 ablation reduced the abundance of acidic autophagolysosomes during ER stress induced by Tg, brefeldin A, or tunicamycin (Figure 3C and D). mCherry-EGFP-LC3B remained as yellow punctuate structures in Mfn2 KO cells, thereby indicating its presence in autophagosomes (Figure 3C and D). In keeping with these data, reduced Lysotracker staining and LAMP1 expression were detected in these cells (Figure 3E and F). The expression of the autophagic genes LC3b or beclin-1 was also dysregulated in Mfn2 KO cells upon ER stress (Figure 3G).
In contrast to the data obtained in Mfn2 KO cells, ablation of Mfn1 did not alter ER stress-induced autophagy (Supplementary Figure S5B–D), and Lysotracker staining was enhanced compared to WT cells (Supplementary Figure S5E). Autophagy was also analyzed in 3T3-L1 fibroblasts and in C2C12 myoblasts upon Mfn2 silencing (Supplementary Figure S4). Reduced autophagic activity, assessed by LC3b-II abundance or the degradation of long-lived proteins, was detected in Mfn2-deficient cells in response to ER stress (Supplementary Figure S5F–H). In parallel, a reduced degradation rate of long-lived proteins was detected in Mfn2-deficient 3T3-L1 cells in response to tunicamycin (Supplementary Figure S5H).
In all, our data show that Mfn2 loss-of-function impairs the normal progression of autophagy during ER stress. This loss-of-function is characterized by reduced autophagosome formation, reduced lysosome abundance, and reduced autophagolysosome formation.
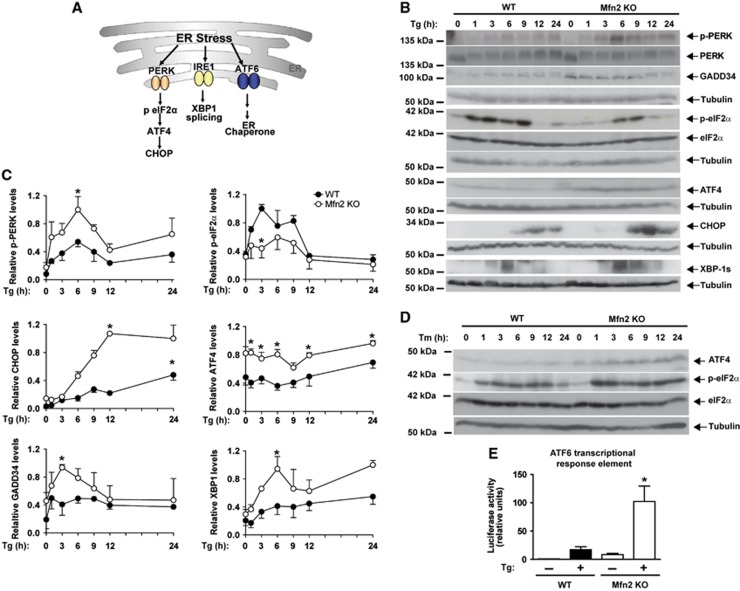
Mfn2 deficiency dysregulates PERK, IRE-1, and ATF6 UPR branches in response to ER stress
To understand how Mfn2 ablation leads to a disrupted biological response to ER stress, next we monitored the UPR response (Figure 4A). Mfn2 KO cells showed enhanced PERK phosphorylation as well as induced expression of downstream signalling proteins, ATF4, GADD34, and CHOP (Figure 4B and C). These observations indicate enhanced PERK branch signalling during ER stress in Mfn2 KO cells. Under these conditions, the activation of eIF2α was reduced in these cells during ER stress (Figure 4B and C). Tunicamycin also caused overactivation of the PERK pathway in Mfn2 KO cells, which was characterized by enhanced phosphorylation of eIF2α, and greater ATF4 expression (Figure 4D).
Figure 4.
Mfn2 modulates the UPR response. (A) Scheme on UPR branches. (B) Immunodetection of p-PERK,PERK, GADD34, p-eIF2α, eIF2α, ATF4, CHOP, and XBP-1s in WT and Mfn2 KO cells treated with 1 μM Tg for the times indicated. (C) Densitometric quantifications. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. (D) Immunodetection of p-eIF2α, eIF2α, and ATF4 in WT and Mfn2 KO cells treated with 0.5 μg/ml Tunicamycin (Tm) for the times indicated. (E) Transcriptional activity driven by ATF6. WT and Mfn2 KO cells were co-transfected with the 5xATF6-GL3 and TK-Renilla plasmids and treated with 1 μM Tg for 24 h after transfection. Data are mean±s.e.m. (n=4). *P<0.05 versus WT group.
Source data for this figure is available on the online supplementary information page.
To determine the activity of the IRE-1α branch, we monitored protein variant XBP-1s, generated through alternative transcript processing upon Tg treatment. Mfn2 ablation caused a sustained activation of XBP-1s in response to ER stress (Figure 4B and C). Furthermore, transcriptional activity driven by ATF6 was greater in Mfn2 KO cells upon Tg treatment (Figure 4E). Analysis of UPR target genes indicated that 38 genes were induced in WT MEFs by Tg (Supplementary Table S1). Almost all these genes were additionally upregulated in Mfn2 KO cells compared to WT cells, and differences were statistically significant for 22 genes (Supplementary Table S1). In all, our data indicate that Mfn2 ablation enhances the activity of the PERK/CHOP, XBP-1, and ATF6 UPR branches in response to ER stress.
PERK deficiency improves apoptosis, and XBP-1 loss-of-function enhances autophagy during ER stress in Mfn2 KO cells
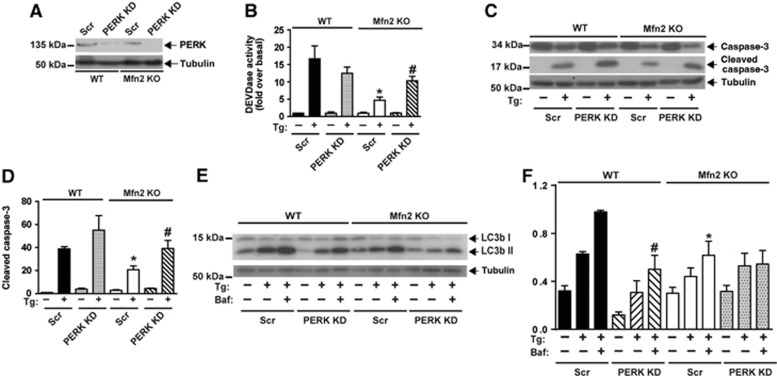
Next, we studied whether the enhanced activity of the UPR branches was responsible for the impaired responses to ER stress in Mfn2 KO cells. To this end, WT and Mfn2 KO cells were subjected to gene silencing by means of lentiviral vectors expressing shRNA directed against PERK, XBP-1, or ATF6 (Figures 5 and 6 and Supplementary Figure S6). PERK loss-of-function (Figure 5A) substantially enhanced caspase activity and caspase 3 cleavage in Mfn2 KO cells upon treatment with Tg (Figure 5B–D). In contrast, PERK silencing in WT cells did not alter either of these parameters upon treatment with this drug (Figure 5B–D). Knockdown of PERK did not enhance LC3b-II levels in response to Tg treatment in Mfn2 KO cells (Figure 5E and F).
Figure 5.
PERK deficiency improves apoptosis during ER stress in Mfn2-ablated cells. (A) Immunodetection of PERK in WT and Mfn2 KO cells stably expressing a scrambled shRNA (Scr) or a shRNA directed against PERK (PERK KD). (B) Caspase activity was detected by measurement of DEVD-AFC substrate processing in WT or Mfn2 KO cells subjected to PERK silencing and treated with 1 μM Tg for 24 h. Data are mean±s.e.m. (n=3). *P<0.05 versus WT; #P<0.05 versus Scr group. (C, D) Total and cleaved caspase 3 levels were detected by western blot. Densitometric quantification is shown in D. Data are mean±s.e.m. (n=3). *P<0.05 versus WT; #P<0.05 versus Scr group. (E, F) Immunodetection of LC3b-I and -II in WT and Mfn2 KO cells subjected to PERK silencing and treated as indicated with Tg or Baf (1 μM Tg; 100 nM Baf) for 6 h. Densitometric quantification is shown in F. Data are mean±s.e.m. (n=3). *P<0.05 versus WT; #P<0.05 versus Scr group.
Source data for this figure is available on the online supplementary information page.
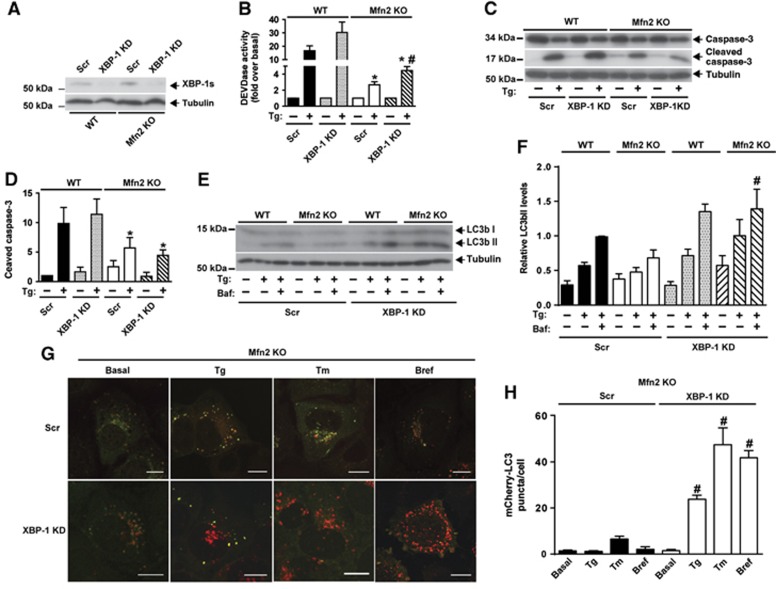
Figure 6.
XBP-1 loss-of-function enhances autophagy during ER stress in Mfn2-ablated cells. (A) Immunodetection of XBP-1s in WT and Mfn2 KO cells stably expressing a scrambled shRNA (Scr) or a shRNA directed to XBP-1 (XBP-1 KD), and treated with Tg for 6 h. (B) Caspase activity was detected by measurement of DEVD-AFC substrate processing. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. #P<0.05 versus Scr group. (C, D) Total and cleaved caspase 3 levels were detected by western blot. WT and Mfn2 KO cells subjected or not to XBP-1 silencing (Scr, and KD) and treated with 1 μM Tg for 24 h. Data are mean±s.e.m. (n=3). *P<0.05 versus WT group. (E, F) Immunodetection of LC3b-I and -II in Scr and XBP-1 KD WT or Mfn2 KO cells treated as indicated (1 μM Tg; 100 nM Baf) for 6 h. Densitometric quantification is shown in F. Data are mean±s.e.m. (n=3). #P<0.05 versus Scr group. (G) Mfn2 KO cells stably expressing mCherry-GFP-LC3b and subjected or not to XBP-1 silencing (Scr or KD) were treated with 1 μM Tg, 100 ng/ml Bref, or 0.5 μg/ml Tm for 24 h and examined by confocal microscopy. Scale bar: 10 μm. (H) Quantification of mCherry+/GFP- (mCherry-LC3) red puncta per cell (a measure of acidic autophagosomes) is shown. Data are mean±s.e.m. (n=3; 100 cells were analyzed per experiment and group). #P<0.05 versus Scr group.
Source data for this figure is available on the online supplementary information page.
XBP-1 loss-of-function (Figure 6A) moderately increased caspase activity but did not enhance caspase 3 cleavage in response to 6 h of Tg treatment (Figure 6B–D). In contrast, XBP-1 knockdown markedly enhanced LC3b-II abundance in Mfn2 KO cells upon treatment with this drug (Figure 6E and F). XBP-1 did not alter any of the parameters studied in WT cells (Figure 6B–F). In keeping with these observations, XBP-1 loss-of-function increased the number of acidic autophagolysosomes, detected as red punctuates in mCherry-EGFP-LC3B-expressing Mfn2 KO cells subjected to treatment to Tg, tunicamycin, or brefeldin A (Figure 6G and H).
ATF6 loss-of-function (Supplementary Figure S6A) accelerated ER vacuolization (Supplementary Figure S6B) in Mfn2 KO cells subjected to ER stress. Under these conditions, ATF6-silenced cells showed no changes in LC3b-II levels (Supplementary Figure S6C) and underwent higher cell death (Supplementary Figure S6D), and showed unaltered caspase activity and caspase 3 cleavage upon Tg treatment (Supplementary Figure S6E and F). Knockdown of ATF6 did not alter either of these two parameters in response to Tg treatment in WT cells (Supplementary Figure S6E and F).
Mfn2 deficiency causes a sustained activation of PERK, which mediates defective mitochondrial morphology and function under basal conditions
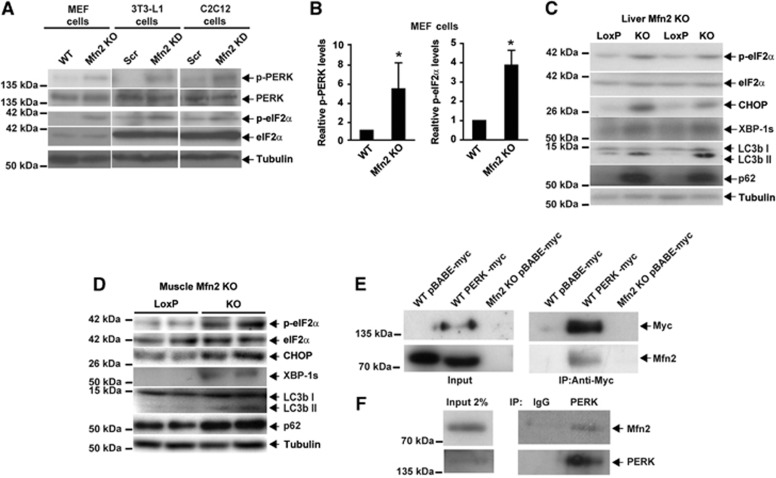
In studies oriented to monitor the activity of UPR branches upon ER stress, data pointed to PERK stimulation under basal conditions (Figure 4B). On the basis of these data, next we studied the effect of Mfn2 loss-of-function on the basal activation state of the PERK pathway in MEFs, and in 3T3-L1 and C2C12 cells. Mfn2 loss-of-function caused sustained PERK stimulation, characterized by enhanced PERK and eIF2α phosphorylation in all three cell types (Figure 7A and B). Chronic activation of the PERK pathway was also detected in Mfn2 KO livers and skeletal muscle and was characterized by enhanced eIF2α phosphorylation and CHOP expression (Figure 7C and D). Enhanced XBP-1 abundance was also found in the liver and muscle of Mfn2 KO mice (Figure 7C and D). Furthermore, the abundance of LC3-II and p62, markers of autophagy, were enhanced in the tissues of these mice (Figure 7C and D), thereby suggesting impaired autophagy.
Figure 7.
Mfn2 regulates PERK activity. (A) Immunodetection of p-PERK and p-eIF2α in Mfn2 KO MEFs, Mfn2 knockdown 3T3-L1 fibroblasts, and Mfn2 knockdown C2C12 myoblasts. (B) Densitometric quantification of p-PERK and p-eIF2α in MEFs. Data are mean±s.e.m. (n=3). *P<0.05 versus WT cells. (C, D) Immunodetection of p-eIF2α, eIF2α, CHOP, XBP-1s, LC3b, and p62 in Mfn2-deficient liver (C) or skeletal muscle (D) from tissue-specific KO mice. (E) Co-immunoprecipitation of Mfn2 and PERK in WT or Mfn2 KO cells stably expressing PERK-myc or an empty vector (pBABE-myc; negative control). (F) Co-immunoprecipitation of endogenous Mfn2 and PERK in WT MEF lysates. PERK was immunoprecipitated with a C-terminal antibody and co-immunoprecipitation of Mfn2 was detected by western blot.
Source data for this figure is available on the online supplementary information page.
The negative relationship between Mfn2 expression and PERK activity points to a physical interaction. To test this hypothesis, lysates from cells stably expressing a myc-tagged version of PERK were immunoprecipitated with anti-myc antibodies, and the immunoprecipitates were subjected to western blot assays. Immunoprecipitation assays revealed physical interaction of PERK and endogenous Mfn2 (Figure 7E). Assays were also performed in MEFs that expressed endogenous Mfn2 and PERK upon immunoprecipitation with anti-PERK antibodies. Indeed, we detected a physical interaction between endogenous PERK and Mfn2 (Figure 7F). Our data indicate that the absence of Mfn2 enhances PERK phosphorylation, which is coherent with a model in which Mfn2 negatively controls PERK.
Under basal conditions, Mfn2 KO MEFs lacked mitochondrial filaments and instead showed mitochondrial swelling, as visualized by Mitotracker Red staining (which labels the mitochondrial matrix) (Supplementary Figure S7A), or after transfection of cells with BAK-GFP (which labels the outer mitochondrial membrane) (Supplementary Figure S7B). Mfn2 ablation also caused major alterations in mitochondrial function, such as enhanced ROS production (Supplementary Figure S7C) and reduced respiration (Supplementary Figure S7D). Mitochondrial calcium overload was measured with the Rhod-2 probe, which was specifically detected in mitochondria under our experimental conditions (Supplementary Figure S7E). Mfn2 ablation caused enhanced mitochondrial Ca2+ overload (Supplementary Figure S7E). All these alterations in mitochondrial function are coherent with previous reports (Bach et al, 2003; Chen et al, 2005; de Brito and Scorrano, 2008). Mitochondrial alterations in morphology, ROS production, and mitochondrial respiration were reversed by reexpression of Mfn2 in Mfn2 KO cells (Supplementary Figures S1C, S7F and G).
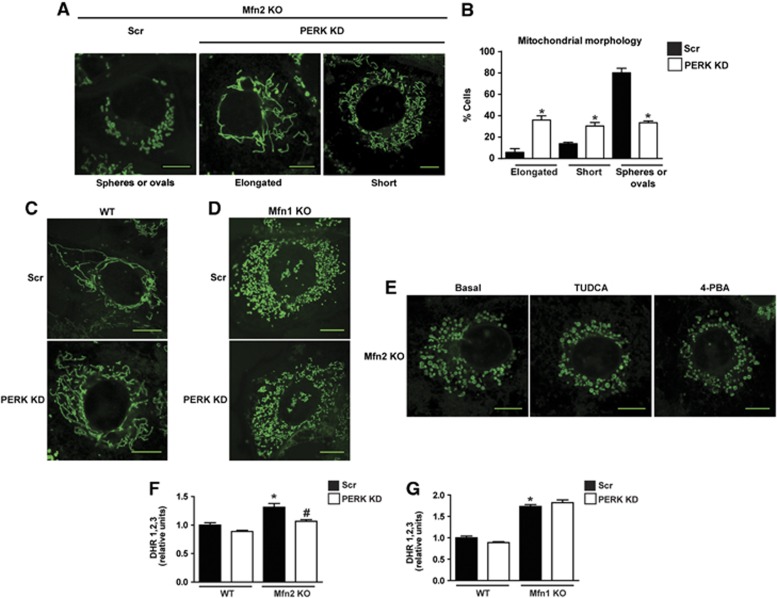
The mitochondrial alterations secondary to Mfn2 ablation have been attributed to the role of Mfn2 as a mitochondrial fusion protein. Alternatively, we reasoned that Mfn2 deficiency may affect mitochondrial function as a result of PERK activation. To test this hypothesis, Mfn2 KO cells were subjected to PERK silencing. Surprisingly, PERK silencing ameliorated the defective mitochondrial morphology, thus swollen mitochondria were much less abundant, and instead mitochondria formed short and even long and thinner filaments (Figure 8A and B). This effect of PERK silencing was not detected in WT or Mfn1 KO cells (Figure 8C and D). Moreover, the addition of the chemical chaperones TUDCA or 4-phenyl butyric acid to Mfn2 KO cells did not alter the morphology of their mitochondria (Figure 8E). Silencing of XBP-1 or ATF6 did not modify mitochondrial morphology in WT or Mfn2 KO cells (Supplementary Figure 8A). In addition, the effects of PERK silencing on normalizing mitochondrial morphology were not associated with changes in the expression of mitochondrial fusion or fission proteins in Mfn2 KO cells (Supplementary Figure 8B and C).
Figure 8.
PERK knockdown rescues mitochondrial morphology and excessive ROS production in Mfn2 KO cells. (A) Representative confocal images of mitochondrial morphology in scrambled (Scr) and PERK KD Mfn2 KO cells stained with MitoTracker Green. (B) Mitochondrial morphology quantification. Data are mean±s.e.m. (n=3; 100 cells were analyzed per experiment and group). *P<0.05 versus Scr group. (C, D) Representative confocal images of mitochondrial morphology in scrambled (Scr) and PERK KD WT cells (C) or scrambled (Scr) and PERK KD Mfn1 KO cells (D) stained with MitoTracker Green. (E) Representative confocal images of mitochondrial morphology in Mfn2 KO cells treated with 1 μM TUDCA or 10 mM 4-phenyl butyric acid for 6 h and stained with MitoTracker Green. (F) Flow-cytometry quantification of ROS levels in Scr and PERK KD WT and Mfn2 KO cells (G) or Scr and PERK KD WT and Mfn1 cells using DHR1,2,3. Data are mean±s.e.m. (n=4). *P<0.05 versus WT group, #P<0.05 versus Scr group. Scale bars: 10 μm.
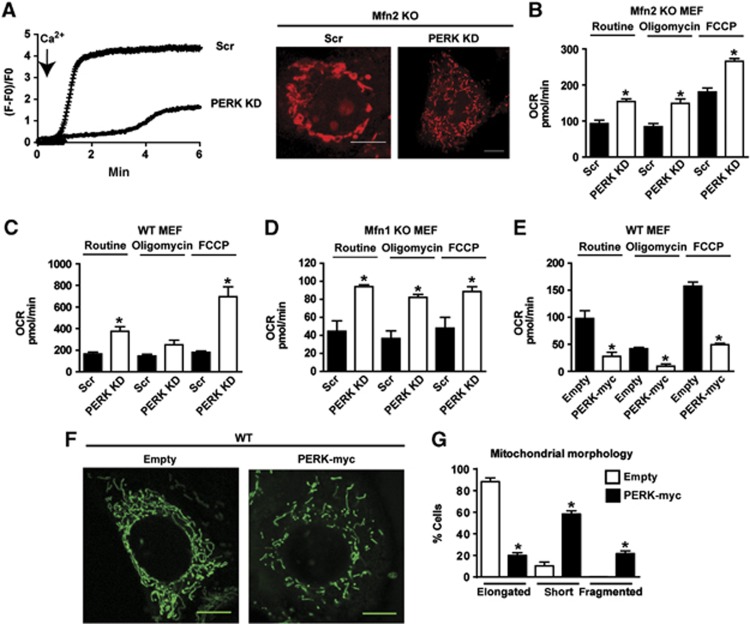
PERK silencing also reduced ROS production (Figure 8F) and mitochondrial Ca2+ overload in Mfn2 KO cells (Figure 9A). In contrast, PERK loss-of-function did not alter ROS in WT or Mfn1 KO cells (Figure 8F and G). In addition, PERK silencing enhanced routine and maximal uncoupled respiration in Mfn2 KO cells (Figure 9B). The stimulatory effects of PERK silencing on cell respiration were also detected in WT and Mfn1 KO cells (Figure 9C and D). In keeping with these data, PERK overexpression modified mitochondrial function and morphology in WT cells. Thus, mitochondrial respiration was markedly repressed in response to PERK overexpression (Figure 9E), and mitochondria were fragmented (60% of the cells overexpressing PERK showed fragmented or round mitochondria) (Figure 9F and G).
Figure 9.
PERK knockdown normalizes mitochondrial Ca2+ overload and mitochondrial respiration. (A) Mitochondrial calcium overload in Scr and PERK KD Mfn2 KO cells. Cells were loaded with Rhod-2 and then treated with 2.5 mM CaCl2 (left). Calcium uptake was monitored by confocal microscopy. Representative confocal images of mitochondrial morphology in WT and Mfn2 KO cells stained with Rhod-2 and incubated with 2.5 mM CaCl2 for 5 min (right). Scale bars: 10 μm. (B–E) Mitochondrial oxygen consumption (OCR) was measured in Scr and PERK KD Mfn2 KO cells (B), Scr and PERK KD WT cells (C), Scr and PERK KD Mfn1 KO cells (D), and WT cells stably expressing PERK-myc (E). Data are mean of three independent experiments. The following parameters were measured: oxygen consumption under routine conditions (DMEM with 5.5 mM glucose), maximal respiratory capacity reached after uncoupling with FCCP, and respiratory leak, measured after inhibition of ATP synthase with oligomycin. Data are mean±s.e.m. *P<0.05 versus Scr group or versus the empty plasmid group. (F, G) Representative confocal images of mitochondrial morphology in WT cells stably expressing PERK-myc stained with MitoTracker Green. Scale bars: 10 μm. Quantitative analysis of mitochondrial morphology is shown in G. Data are mean±s.e.m. (n=3). *P<0.05 versus the empty plasmid group.
Discussion
Mfn2 is a mitochondrial fusion protein with a lower GTPase activity than that of Mfn1 (Ishihara et al, 2004; Neuspiel et al, 2005). On the basis of the mitochondrial fusion activity of Mfn2, the alterations in mitochondrial morphology and function detected upon silencing or ablation of this gene have been attributed to reduced mitochondrial fusion (Bach et al, 2003; Chen et al, 2003; Pich et al, 2005). Here we provide evidence that an ER-related mechanism is responsible for the development of swollen mitochondria, enhanced Ca2+ overload, increased ROS production, and reduced mitochondrial respiration in Mfn2-deficient cells. This view is based on a number of observations, namely, (a) Mfn2 KO or Mfn2-silenced cells showed sustained PERK activation under basal conditions; (b) PERK silencing ameliorated the mitochondrial network and respiration and reduced ROS production and mitochondrial Ca2+ in Mfn2 KO cells; (c) PERK overexpression caused mitochondrial fragmentation and reduced mitochondrial respiration in cells; and (d) there is a physical interaction between Mfn2 and PERK.
Our data support the notion that Mfn2 lies upstream of PERK and that under basal conditions Mfn2 maintains PERK inactive. We detected sustained activation of PERK in skeletal muscle, liver, and pre-adipose cells under conditions of Mfn2 loss-of-function; therefore, the activation of PERK may be independent of the cellular context. We also propose that the interaction between Mfn2 and PERK occurs in mitochondrial–ER contact sites. In favour of this view, both Mfn2 and PERK are localized in this compartment (de Brito and Scorrano, 2008; Verfaillie et al, 2012).
Our study is also the first to demonstrate that PERK controls mitochondrial morphology and function, as well as oxidative stress in cells. In this respect, we propose that the alterations detected in tissues upon Mfn2 ablation, such as enhanced ROS production, defective mitochondrial respiration, and deficient insulin signalling and glucose handling (Sebastian et al, 2012), are, at least in part, the result of enhanced PERK activity.
Here we report that Mfn2 loss-of-function causes the dysregulation of UPR branches under basal or ER stress conditions. Under the former, we detected enhanced PERK and eIF2α phosphorylation in Mfn2 KO cells or tissues. In addition, Mfn2-deficient cells exposed to ER stress (by incubation with agents such as Tg or tunicamycin) showed increased activation of the three major UPR branches, that is, PERK, IRE-1α, and ATF6, and enhanced expression of UPR genes. In connection with the enhanced ER stress-induced PERK pathway, which was characterized by greater expression of the proteins ATF4, GADD34, and CHOP, tunicamycin also caused enhanced phosphorylation of eIF2α, whereas Tg reduced eIF2α phosphorylation. This reduction may be explained through the complex effects of Tg on calcium homoeostasis (Inesi et al, 1998; Michelangeli and East, 2011), and an interaction between these effects and Mfn2 loss-of-function during ER stress. Our data agree with reports by Zhao et al (2012), who found increased phosphorylation of eIF2α and enhanced expression of UPR target genes in Mfn2 KO cardiac cells. In contrast, Ngoh et al (2012) analyzed the impact of ER stress on the PERK pathway in MEFs transduced with adenoviruses for 24 or 48 h before exposure to ER stress. They found that at short times (0.5 h) the phosphorylation of eIF2α or the expression of GADD34 or p58IPK were similarly induced in control and Mfn2-deficient cells in response to ER stress. However, at longer exposure (18 h) to ER stress, Mfn2-deficient cells showed reduced induction. These discrepancies could be attributable to differences between primary cultures, immortalized MEFs, or the use of adenoviral transduction.
ER stress activates autophagy, which is a key factor driving survival under unfavourable conditions (Ogata et al, 2006; Yorimitsu et al, 2006; Kouroku et al, 2007). It also induces autophagic genes via PERK/eIF2α/ATF4. Thus, ATF4 silencing, expression of a non-phosphorylatable form of eIF2α, or expression of a dominant-negative PERK inhibits ER stress-induced autophagy (Kouroku et al, 2007; Rzymski et al, 2010). There is also evidence that the IRE-1-JNK pathway is required for autophagy activation after ER stress. Thus, IRE-1α-deficient cells or cells treated with a JNK inhibitor show inhibited ER stress-induced autophagy (Ogata et al, 2006). In contrast, XBP-1 deficiency causes increased autophagy (Hetz et al, 2009). Mfn2 deficiency has been reported to impair basal autophagy in MEFs and cardiac myocytes (Hailey et al, 2010; Zhao et al, 2012). Our data indicate that Mfn2 ablation causes a deficient ER stress-induced autophagy. Mfn2-ablated cells showed a number of autophagic defects upon ER stress, namely: (a) deficient expression of autophagic genes such as beclin-1 or LC3b; (b) reduced autophagosomal proliferation; (c) reduced lysosomal abundance; and (d) decreased autophagolysosome formation. A defective response to ER stress was observed in MEFs, as well as in 3T3-L1 pre-adipocytes and C2C12 myoblasts, thereby supporting the view that these effects are independent of the cell context. We also found that the enhanced IRE-1α/XBP-1 activity upon ER stress was responsible for the reduced ER stress-induced autophagy of Mfn2-deficient cells, and silencing of XBP-1 (but not of PERK) rescued normal LC3-II formation and improved the formation of acidic autophagosomes in Mfn2-ablated cells subjected to ER stress. These effects were not detected in WT cells, thus suggesting the specificity of the Mfn2-deficient state. Neither were the effects detected under basal conditions of Mfn2 KO cells. These observations suggest that the function of XBP-1 was limited to ER stress-induced autophagy and not to basal autophagy. These data also support the notion that several mechanisms repress basal and ER stress-induced autophagy in Mfn2 KO cells.
We also report that Mfn2 KO cells are unable to stimulate apoptosis or to activate caspase 3 in response to ER stress, and instead trigger a paraptotic-like cell death pathway. Our data contrast those reported by Ngoh et al (2012), which indicated that ER stress exacerbated ER stress-induced apoptosis in Mfn2-deficient MEFs. Ngoh et al (2012) used 10-fold lower concentrations of Tg than those used in our study, and as a result they found that caspase activation was increased only 2-fold (in contrast to the 10-fold increase in our study). It is possible that the conditions used by Ngoh et al (2012) did not fully induce apoptosis in WT cells, as the UPR response was sufficient to deal with and resolve mild ER stress. However, under greater ER stress, cells showed a full apoptotic response.
Our data also indicate that overactivation of the PERK pathway is responsible for defective induction of apoptosis in response to ER stress. PERK loss-of-function rescued apoptosis during ER stress in the context of Mfn2 ablation. However, PERK silencing did not alter apoptosis in WT cells, and in turn apoptosis was not modified by silencing of XBP-1 or ATF6 in Mfn2 KO cells. The lack of capacity of Mfn2 KO cells to induce apoptosis during ER stress is surprising given that CHOP, one of the key regulators of apoptosis during ER stress (Tabas and Ron, 2011), is induced under these conditions. CHOP promotes BIM expression and suppresses the pro-survival protein Bcl2, which leads to the permeabilization of the mitochondrial outer membrane (McCullough et al, 2001; Puthalakath et al, 2007). Moreover, CHOP promotes mitochondrial calcium overload through the ERO1α-IP3R-CAMKII pathway (Seimon et al, 2006; Timmins et al, 2009; Li et al, 2010). However, this protein has also been associated with protection against cell death (Halterman et al, 2010; Rouschop et al, 2010).
Mfn2 overexpression has been reported to induce apoptosis in tumoural cells and to trigger ROS-mediated apoptosis through caspase activation (Guo et al, 2007; Shen et al, 2007; Wu et al, 2008). Moreover, the overexpression of this protein promotes spontaneous apoptosis and suppresses tumour growth in vivo (Rehman et al, 2012). In contrast, several reports have shown that Mfn1 or Mfn2 loss-of-function increases sensitivity to apoptotic stimuli and that these proteins also interact with Bcl2 family members (Sugioka et al, 2004; Karbowski et al, 2006; Brooks et al, 2007; Hoppins et al, 2011; Leboucher et al, 2012). The increased sensitivity to apoptosis caused by loss-of-function is similar in both Mfn1 and Mfn2 ablation and is associated with mitochondrial fragmentation (Sugioka et al, 2004). Interestingly, a recent report demonstrates that Mfn2 and Bax/Bak are required for Ca2+-induced mPTP opening. This novel role of Mfn2 and proteins of the Bcl2 family in necrosis indicates that mitochondrial dynamics coordinate the cell death pathway in a stimulus-dependent manner (Whelan et al, 2012).
Mitochondrial–ER coupling regulates metabolism, calcium signalling, and apoptosis (Simmen et al, 2005; de Brito and Scorrano, 2008; Bravo et al, 2011; Sebastian et al, 2012). Several reports indicate that depletion of the proteins involved in mitochondrial–ER contact sites, such as PACS-2, SERCA1T, and PML, blocks the apoptotic program (Simmen et al, 2005; Chami et al, 2008; Giorgi et al, 2010). Induction of the NogoB protein reduces ER–mitochondria coupling and inhibits apoptosis (Sutendra et al, 2011). Thus, ER–mitochondria coupling mediated by Mfn2 is crucial for metabolic homoeostasis and the regulation of cell death. On the basis of these observations, we propose that, under ER stress, Mfn2 is a key protein that determines cell fate via its role in ER–mitochondria coupling.
In all, this report uncovers a missing molecular link in the UPR. The observation that Mfn2 controls the UPR upon ER stress and that it is an upstream regulator of PERK reveals a previously unsuspected role for a protein most recognized for its key role in mitochondrial fusion. The Mfn2–PERK interaction also uncovers a new mechanism for the regulation of PERK. In keeping with the initial observations by de Brito and Scorrano (2008), our data support a major role of Mfn2 in mitochondrial–ER contact sites. Under basal conditions, Mfn2 suppresses PERK activation through direct interaction, and loss of interaction in Mfn2-eficient cells affects ROS production, mitochondrial morphology, respiration, and mitochondrial Ca2+ overload. Furthermore, Mfn2-deficient cells show an exaggerated activation of the UPR pathways, PERK, IRE-1α, and ATF6, and an enhanced response of PERK and XBP-1 is responsible for the deficient activation of apoptosis and autophagy, respectively. Our data show that Mfn2 plays a unique role in orchestrating mitochondrial metabolism and the UPR. These observations allow us to propose that Mfn2 senses the cellular metabolic state and coordinates the ER stress response.
Materials and methods
Reagents
Antibodies
Plasmids
Cells and cell culture
SV-40-transformed WT, Mfn1 KO, and Mfn2 KO MEFs were a gift from D.C. Chan (Division of Biology, California Institute of Technology, UA). Mfn KO cells, HEK293T, 3T3-L1, and C2C12 cell lines were from ATCC. MEFs, HEK293T, and C2C12 cells were grown in DMEM (Invitrogen) with 10% FBS and 100 U/ml of penicillin/streptomycin (Invitrogen), whereas 3T3L-1 cells were cultured in DMEM (GIBCO, Invitrogen 12800), 10% FBS, and 1.5 g/ml sodium bicarbonate at 37°C in a humidified atmosphere of 5% CO2/95% O2. Cells were starved of FBS for 3 h before the treatment with ER stress-inducing agents.
Animal care and generation of animal models
Western blotting assay
Lentivirus production and cell infection
Lentiviral vectors were packed using pMDLg/pRRE, pRSV-Rev, and pMD2.G plasmids. HEK293T cells were transfected with pLKO.1-puro plasmid or pLenti-GIII-CMV-hMFN2-HA and a third-generation packing system for 24 h at 37°C and incubated for additional 24 h at 33°C to facilitate lentiviral production. After 48 h of lentiviral particle production, MEFs, 3T3-L1, or C2C12 cells were infected with filtered lentiviral medium (derived from HEK293T cultures) supplemented with 2 μg/ml polybrene. Cells were then selected by incubation with 2.5 mg/ml puromycin in the complete medium.
Ecotropic retroviral packing
HEK293T cells were co-transfected with pCL-Eco and pBABE-puro mCherry-EGFP-LC3B, PERK-myc, or empty vector. After 48 h of retroviral particle production, MEFs, at 50% of confluence, were infected with retroviral medium (derived from HEK293T cultures) supplemented with 2 μg/ml polybrene (Sigma-Aldrich). Puromycin (2.5 μg/ml) was used to select stably transfected cells.
Confocal microscopy
For live imaging studies, cells were plated on 22-mm glass coverslips and transfected with 0.5 μg Sec61β-GFP, or BAK-GFP for 24 h using Metafectin (Bio-Rad). Cells were placed in a chamber under culture conditions (DMEM at 37°C and 5% CO2), and live cells were visualized using a Leica SP2 Confocal Microscope. Time-lapse microscopy analysis for ER expansion was performed in cells expressing ER-RFP. Cells were maintained in a chamber at 37°C equilibrated with 5% CO2, and images were recorded every 5 min using a Nikon TE200 Inverted Microscope. Images were then processed with ImageJ software (NIH).
mCherry-GFP-LC3b confocal microscopy
WT and Mfn2 KO MEFs, as well as WT and Mfn2 KO MEFs stably transduced with lentiviruses encoding scrambled or XBP-1 shRNA, were studied. These cells were also transduced with a lentivirus expressing mCherry-GFP-LC3. mCherry/GFP-expressing cells were selected by flow cytometry, plated in 22-mm glass coverslips, and treated with ER stress inducers for 24 h. After treatment, the cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline. All the cell images were obtained using SP2 Leica microscope. Autophagosomes and acidic autophagosomes were measured by confocal counting of the cells as mCherry+/GFP+ (yellow) puncta and mCherry+/GFP− (red) puncta, respectively. At least 50–100 cells/sample were counted in triplicate samples per condition and per experiment.
Labelling of mitochondrial compartment and morphological analysis
Cells were loaded with 100 nM Mitotracker Green or Mitotracker Red CMXRos for the last 20 min of incubation in culture medium. They were then washed with culture medium, and live cells were analyzed in an inverted Leica SP2 Confocal Microscope. Images were collected using a 488-nm excitation laser and 505–530-nm emission spectrum for Mitotracker Green, or excited at a wavelength of 561 nm and emission detected in red fluorescence for Mitotracker Red CMXRos. They were then processed with ImageJ software (NIH). Cells were classified on the basis of their mitochondrial morphology: oval/spheres (only observed in Mfn2 KO cells), short (<6 μm), or long (>6 μm) mitochondrial filaments. At least 100 cells/sample were counted in triplicate samples/condition/experiment.
Transmission electron microscopy
Cell death assays
Autophagy flux analysis and protein degradation assay
RNA extraction and Real-Time PCR
Luciferase reporter assay
MEFs were transfected for 24 h with 0.5 μg of 5XATF6-GL3 encoding a luciferase reporter construct for ATF6 activity and 0.5 μg TK Renilla by using Metafectin (Bio-Rad), following the manufacturer’s instructions. Transfected cells were then incubated with 1 μM Tg for an additional 24 h. Luciferase activity was determined with the Dual-GLO Luciferase Assay System (Promega) in a luminometer Lumat LB 9507 (Berthold Technologies).
Oxygen consumption measurements in MEFs
MEFs (WT, Mfn1 KO, and Mfn2 KO) were plated in SeaHorse Bioscience XF24 plates. After 48 h, a Seahorse Bioscience XF24 extracellular flux analyzer was used to measure oxygen consumption. The instrument was calibrated the day before the experiment, following the manufacturer’s instructions. On the day of the experiment, the injection ports on the sensor cartridge were loaded with 1.25 mM oligomycin (complex V inhibitor) to distinguish the percentage of oxygen consumption devoted to ATP synthesis and the percentage of oxygen consumption required to overcome the natural proton leak across the inner mitochondrial membrane: 1 mM FCCP was used to calculate the ‘spare’ respiratory capacity of cells, which is defined as the quantitative difference between maximal uncontrolled OCR and the initial basal OCR; and 0.1 mM rotenone (complex I inhibitor) and 0.1 mM antimycin A (complex III inhibitor) were used to calculate the remaining respiration caused by oxidative side reactions. During the sensor calibration, MEFs were kept in a 37°C incubator without CO2 in 700 μl of respiration buffer (DMEM, 5 mM glucose, 2 mM glutamate, 31.6 mM NaCl and Phenol Red). Plates were immediately placed into the calibrated Seahorse XF24 flux analyzer for mitochondrial bioenergetic analysis.
Measurement of ROS
Cells were loaded with freshly prepared 10 μM dihydrorhodamine 123 (λexcitation=543 nm; λemission=560 nm) for 30 min and then subjected to flow cytometry in a FACS Scan (Becton Dickinson, San Jose, CA).
Measurement of mitochondrial calcium
Cells were plated on coverslips and preloaded with 6 μM Rhod-2/0, 02% Pluronic F-127 in Krebs buffer without Ca2+ (145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM HEPES-Na, and 5.6 mM glucose, pH 7.4), and then incubated for 3 h at 4°C. They were washed two times with Krebs buffer. Mitochondrial Ca2+ uptake was monitored at 23°C in an inverted Leica SP2 confocal microscope. Cells were excited at 561 nm, and emission was detected in red fluorescence. Basal fluorescence was monitored for 1 min, and cells were then treated with 2.5 mM CaCl2. Fluorescence was analyzed using Image J software. The increase in mitochondrial calcium uptake was expressed as F-F0/F0 where F is fluorescence intensity and F0 is basal fluorescence intensity.
Immunoprecipitation
Cells stably expressing retroviral PERK-myc were homogenized with IP buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM Na4P2O7, 20 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and Roche protease inhibitor mixture) and centrifuged at 10 000 g for 30 min at 4°C. One milligram of protein lysate was incubated overnight at 4°C with an anti-c-Myc Agarose Affinity Gel antibody and then washed five times with IP buffer. To immunoprecipitate endogenous PERK, 1 mg of protein lysate was pre-cleared with IgG–agarose beads (Santa Cruz Biotechnology) for 2 h, and then the lysate was incubated overnight at 4°C with ImmunoCruzTM IP/WB Optima B beads (Santa Cruz Biotechnology), pre-incubated with an anti-PERK C-terminal antibody (Sigma-Aldrich) and then washed five times with IP buffer. Immunoblots were performed with specific antibodies against Mfn2 and PERK and detected with ImmunoCuzTM IP/WB secondary antibody (Santa Cruz Biotechnology).
Expression of results and statistical methods
Data are presented either as mean±s.e.m. of a number of independent experiments (ranging from 3 to 15). Data were subjected to analysis of variance, and comparisons between groups were performed using a protected Tukey’s t-test. A value of P<0.05 was chosen as the limit of statistical significance.
Supplementary Material
Acknowledgments
EN is the recipient of a PhD scholarship from the Ministry of Science and Innovation (FPI Program). PMC is supported by a fellowship from the ‘la Caixa/IRB International Ph.D. Programme. We thank I Castrillón, JC Monasterio, and J Seco for their technical assistance. We thank Ms T. Yates for editorial support. This study was funded by grants from the MEC (SAF2008-03803), the ‘Generalitat de Catalunya’ (grant 2009SGR915), CIBERDEM (‘Instituto de Salud Carlos III’), the FP7-European Commission (MITIN, HEALTH-F4-2008-223450), and INTERREG IV-B-SUDOE-FEDER (DIOMED, SOE1/P1/E178). AZ was the recipient of a Science Intensification Award from the University of Barcelona.
Authors contribution: JPM: conception and design of studies, data acquisition, analysis and interpretation, and writing the article; SI: design of studies, acquisition, analysis, and interpretation of data on apoptosis; JS-W, AS, CM, and PM-C: design of studies, acquisition, analysis, and interpretation of data on autophagy; EN and AD-R: design of studies, acquisition, analysis, and interpretation of data on respiration; MIH-A and DS: design of studies, acquisition, analysis, and interpretation of data on knockout mice; MP: analysis and interpretation of data; and AZ: conception and design of studies, data analysis and interpretation, and writing the article.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacin M, Vidal H, Rivera F, Brand M, Zorzano A (2003) Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem 278: 17190–17197 [DOI] [PubMed] [Google Scholar]
- Bravo R, Vicencio JM, Parra V, Troncoso R, Munoz JP, Bui M, Quiroga C, Rodriguez AE, Verdejo HE, Ferreira J, Iglewski M, Chiong M, Simmen T, Zorzano A, Hill JA, Rothermel BA, Szabadkai G, Lavandero S (2011) Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J Cell Sci 124: 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C, Wei Q, Feng L, Dong G, Tao Y, Mei L, Xie ZJ, Dong Z (2007) Bak regulates mitochondrial morphology and pathology during apoptosis by interacting with mitofusins. Proc Natl Acad Sci USA 104: 11649–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chami M, Oules B, Szabadkai G, Tacine R, Rizzuto R, Paterlini-Brechot P (2008) Role of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to mitochondria during ER stress. Mol Cell 32: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280: 26185–26192 [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610 [DOI] [PubMed] [Google Scholar]
- Doroudgar S, Thuerauf DJ, Marcinko MC, Belmont PJ, Glembotski CC (2009) Ischaemia activates the ATF6 branch of the endoplasmic reticulum stress response. J Biol Chem 284: 29735–29745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK (2011) ER tubules mark sites of mitochondrial division. Science 334: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, Bononi A, Bonora M, Duszynski J, Bernardi R, Rizzuto R, Tacchetti C, Pinton P, Pandolfi PP (2010) PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science 330: 1247–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Chen KH, Guo Y, Liao H, Tang J, Xiao RP (2007) Mitofusin 2 triggers vascular smooth muscle cell apoptosis via mitochondrial death pathway. Circ Res 101: 1113–1122 [DOI] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141: 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman MW, Gill M, DeJesus C, Ogihara M, Schor NF, Federoff HJ (2010) The endoplasmic reticulum stress response factor CHOP-10 protects against hypoxia-induced neuronal death. J Biol Chem 285: 21329–21340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312: 572–576 [DOI] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH (2009) XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev 23: 2294–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Edlich F, Cleland MM, Banerjee S, McCaffery JM, Youle RJ, Nunnari J (2011) The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol Cell 41: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140: 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inesi G, Wade R, Rogers T (1998) The sarcoplasmic reticulum Ca2+ pump: inhibition by thapsigargin and enhancement by adenovirus-mediated gene transfer. Ann N Y Acad Sci 853: 195–206 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K (2004) Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 117: 6535–6546 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ (2006) Role of Bax and Bak in mitochondrial morphogenesis. Nature 443: 658–662 [DOI] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T (2007) ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 14: 230–239 [DOI] [PubMed] [Google Scholar]
- Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM (2012) Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47(4): 547–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombes A, Frachon P, Rojo M (2002) Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell 13: 4343–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Scull C, Ozcan L, Tabas I (2010) NADPH oxidase links endoplasmic reticulum stress, oxidative stress, and PKR activation to induce apoptosis. J Cell Biol 191: 1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D (2004a) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP (2004b) Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J 23: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuolo J, Bulotta S, Verderio C, Benfante R, Borgese N (2011) Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc Natl Acad Sci USA 108: 7832–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ (2001) Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol 21: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelangeli F, East JM (2011) A diversity of SERCA Ca2+ pump inhibitors. Biochem Soc Trans 39: 789–797 [DOI] [PubMed] [Google Scholar]
- Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride H (2005) Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem 280: 25060–25070 [DOI] [PubMed] [Google Scholar]
- Ngoh GA, Papanicolaou KN, Walsh K (2012) Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem 287: 20321–20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pich S, Bach D, Briones P, Liesa M, Camps M, Testar X, Palacin M, Zorzano A (2005) The Charcot-Marie-Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet 14: 1405–1415 [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A (2007) ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Rao RV, Castro-Obregon S, Frankowski H, Schuler M, Stoka V, del Rio G, Bredesen DE, Ellerby HM (2002) Coupling endoplasmic reticulum stress to the cell death program. An Apaf-1-independent intrinsic pathway. J Biol Chem 277: 21836–21842 [DOI] [PubMed] [Google Scholar]
- Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL (2012) Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J 26: 2175–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG (2010) The unfolded protein response protects human tumour cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 120: 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29: 4424–4435 [DOI] [PubMed] [Google Scholar]
- Santel A, Fuller MT (2001) Control of mitochondrial morphology by a human mitofusin. J Cell Sci 114: 867–874 [DOI] [PubMed] [Google Scholar]
- Schewe DM, Aguirre-Ghiso JA (2008) ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumour cells in vivo. Proc Natl Acad Sci USA 105: 10519–10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian D, Hernandez-Alvarez MI, Segales J, Sorianello E, Munoz JP, Sala D, Waget A, Liesa M, Paz JC, Gopalacharyulu P, Oresic M, Pich S, Burcelin R, Palacin M, Zorzano A (2012) Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homoeostasis. Proc Natl Acad Sci USA 109: 5523–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimon TA, Obstfeld A, Moore KJ, Golenbock DT, Tabas I (2006) Combinatorial pattern recognition receptor signaling alters the balance of life and death in macrophages. Proc Natl Acad Sci USA 103: 19794–19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T, Zheng M, Cao C, Chen C, Tang J, Zhang W, Cheng H, Chen KH, Xiao RP (2007) Mitofusin-2 is a major determinant of oxidative stress-mediated heart muscle cell apoptosis. J Biol Chem 282: 23354–23361 [DOI] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G (2005) PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J 24: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio S, de Belle I, Bredesen DE (2000) An alternative, nonapoptotic form of programmed cell death. Proc Natl Acad Sci USA 97: 14376–14381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka R, Shimizu S, Tsujimoto Y (2004) Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J Biol Chem 279: 52726–52734 [DOI] [PubMed] [Google Scholar]
- Sutendra G, Dromparis P, Wright P, Bonnet S, Haromy A, Hao Z, McMurtry MS, Michalak M, Vance JE, Sessa WC, Michelakis ED (2011) The role of Nogo and the mitochondria-endoplasmic reticulum unit in pulmonary hypertension. Sci Transl Med 3: 88ra55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 13: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Wek SA, Bunpo P, Cundiff JK, McClintick JN, Anthony TG, Wek RC (2011) The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol Biol Cell 22: 4390–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins JM, Ozcan L, Seimon TA, Li G, Malagelada C, Backs J, Backs T, Bassel-Duby R, Olson EN, Anderson ME, Tabas I (2009) Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J Clin Invest 119: 2925–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie T, Rubio N, Garg AD, Bultynck G, Rizzuto R, Decuypere JP, Piette J, Linehan C, Gupta S, Samali A, Agostinis P (2012) PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ 19(11): 1880–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, Ma D (2004) An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J Cell Sci 117: 1525–1532 [DOI] [PubMed] [Google Scholar]
- Whelan RS, Konstantinidis K, Wei AC, Chen Y, Reyna DE, Jha S, Yang Y, Calvert JW, Lindsten T, Thompson CB, Crow MT, Gavathiotis E, Dorn GW 2nd, O'Rourke B, Kitsis RN (2012) Bax regulates primary necrosis through mitochondrial dynamics. Proc Natl Acad Sci USA 109: 6566–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Li Z, Zhang Y, Zhang P, Zhu X, Huang J, Ma T, Lu T, Song Q, Li Q, Guo Y, Tang J, Ma D, Chen KH, Qiu X (2008) Adenovirus-expressed human hyperplasia suppressor gene induces apoptosis in cancer cells. Mol Cancer Ther 7: 222–232 [DOI] [PubMed] [Google Scholar]
- Xu W, Liu L, Charles IG, Moncada S (2004) Nitric oxide induces coupling of mitochondrial signalling with the endoplasmic reticulum stress response. Nat Cell Biol 6: 1129–1134 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K (2004) Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem 136: 343–350 [DOI] [PubMed] [Google Scholar]
- Yoon MJ, Kim EH, Lim JH, Kwon TK, Choi KS (2010) Superoxide anion and proteasomal dysfunction contribute to curcumin-induced paraptosis of malignant breast cancer cells. Free Radic Biol Med 48: 713–726 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ (2006) Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281: 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Xiao R, Zheng M (2012) Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem 287: 23615–23625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.