Abstract
SAMHD1 is a host restriction factor for human immunodeficiency virus 1 (HIV-1) in cultured human cells. SAMHD1 mutations cause autoimmune Aicardi-Goutières syndrome and are found in cancers including chronic lymphocytic leukaemia. SAMHD1 is a triphosphohydrolase that depletes the cellular pool of deoxynucleoside triphosphates, thereby preventing reverse transcription of retroviral genomes. However, in vivo evidence for SAMHD1’s antiviral activity has been lacking. We generated Samhd1 null mice that do not develop autoimmune disease despite displaying a type I interferon signature in spleen, macrophages and fibroblasts. Samhd1−/− cells have elevated deoxynucleoside triphosphate (dNTP) levels but, surprisingly, SAMHD1 deficiency did not lead to increased infection with VSV-G-pseudotyped HIV-1 vectors. The lack of restriction is likely attributable to the fact that dNTP concentrations in SAMHD1-sufficient mouse cells are higher than the KM of HIV-1 reverse transcriptase (RT). Consistent with this notion, an HIV-1 vector mutant bearing an RT with lower affinity for dNTPs was sensitive to SAMHD1-dependent restriction in cultured cells and in mice. This shows that SAMHD1 can restrict lentiviruses in vivo and that nucleotide starvation is an evolutionarily conserved antiviral mechanism.
Keywords: Aicardi-Goutières syndrome, HIV-1, SAMHD1, type I interferon
Introduction
Human dendritic cells (DCs) and other myeloid cells in culture are largely refractory to infection with human immunodeficiency virus 1 (HIV-1) (Manel et al, 2010). The closely related virus HIV-2 encodes the protein Vpx, which can relieve restriction of HIV-1 in trans by targeting a host factor for proteasomal degradation (Goujon et al, 2007; Kaushik et al, 2009). Recent work has identified SAMHD1 as the cellular protein targeted by Vpx (Hrecka et al, 2011; Laguette et al, 2011). SAMHD1 has an N-terminal sterile alpha motif and a C-terminal HD domain that hydrolyses deoxynucleoside triphosphates (dNTPs), generating nucleosides and inorganic triphosphate (Goldstone et al, 2011; Powell et al, 2011). Experiments in vitro suggest that SAMHD1-mediated reduction of cellular dNTP concentrations may block HIV-1 reverse transcription and, consequently, restrict virus infection (Kim et al, 2012; Lahouassa et al, 2012). Consistent with that model, SAMHD1 depletion by Vpx delivery or RNA interference renders many human myeloid cells and resting T cells more permissive to HIV-1 infection (Berger et al, 2011; Hrecka et al, 2011; Laguette et al, 2011; Baldauf et al, 2012; Descours et al, 2012; Kim et al, 2012; Lahouassa et al, 2012). Moreover, monocytes and resting T cells from SAMHD1-deficient individuals more efficiently support HIV-1 replication (Berger et al, 2011; Baldauf et al, 2012; Descours et al, 2012). Collectively, these in vitro observations define SAMHD1 as a host restriction factor for HIV-1 in cultured human cells (Ayinde et al, 2012).
SAMHD1 deficiency in humans results in Aicardi-Goutières syndrome (AGS), a hereditary autoimmune encephalopathy that mimics congenital virus infection and is characterized by type I interferon (IFN) production (Crow and Livingston, 2008; Rice et al, 2009). AGS can also be caused by mutations in the TREX1, RNASEH2A-C or ADAR1 genes (Crow and Livingston, 2008; Rice et al, 2012). TREX1-deficient mice develop spontaneous and IFN-dependent multi-organ autoimmunity, particularly of the heart (Morita et al, 2004; Stetson et al, 2008; Gall et al, 2012). Interestingly, TREX1-deficient cells display cytoplasmic accumulation of DNA from endogenous retroviruses or after HIV-1 infection (Stetson et al, 2008; Yan et al, 2010). This DNA then triggers an STING-dependent innate antiviral pathway that culminates in IRF3-dependent IFN induction (Stetson et al, 2008; Yan et al, 2010; Gall et al, 2012). Disease in TREX1-deficient mice can be ameliorated by administering reverse transcriptase (RT) inhibitors (Beck-Engeser et al, 2011). As such, it appears that a failure to control endogenous and exogenous retroviruses and to degrade their nucleic acid products underlies autoimmunity caused by TREX1 deficiency. However, it is unclear whether SAMHD1 behaves similarly and whether its loss-of-function precipitates disease by permitting accumulation of retroviral nucleic acid products that chronically trigger IFN induction pathways.
To further study the molecular basis of AGS and the in vivo relevance of SAMHD1 as an antiviral host factor, we generated SAMHD1-deficient mice. Here, we show that such mice do not develop autoimmunity, even though they show evidence of spontaneous IFN production in selected tissues and cells. The levels of all four dNTPs were increased in Samhd1−/− cells, but infection with VSV-G-pseudotyped HIV-1 vectors was comparable in wild-type and Samhd1−/− DCs, macrophages and other cells, as well as in SAMHD1-deficient and control mice. These data can be explained by the fact that baseline dNTP concentrations in SAMHD1-sufficient mouse DCs were 10-fold higher than those reported for human cells and exceeded the KM of HIV-1 RT for dNTPs. Consistent with this notion, introduction of a point mutation into the viral polymerase that lowers its affinity for dNTPs revealed potent SAMHD1-dependent restriction of the resulting attenuated HIV-1 vector in cells and in vivo. Therefore, nucleotide restriction is an evolutionarily conserved mechanism for defense from retroviruses. Our data also suggest that HIV-1 has evolved a polymerase that is active at low dNTP concentrations, thereby partly circumventing some of the effect of SAMHD1 restriction.
Results
Samhd1−/− mice are healthy, although they display an IFN signature in some tissues and cells
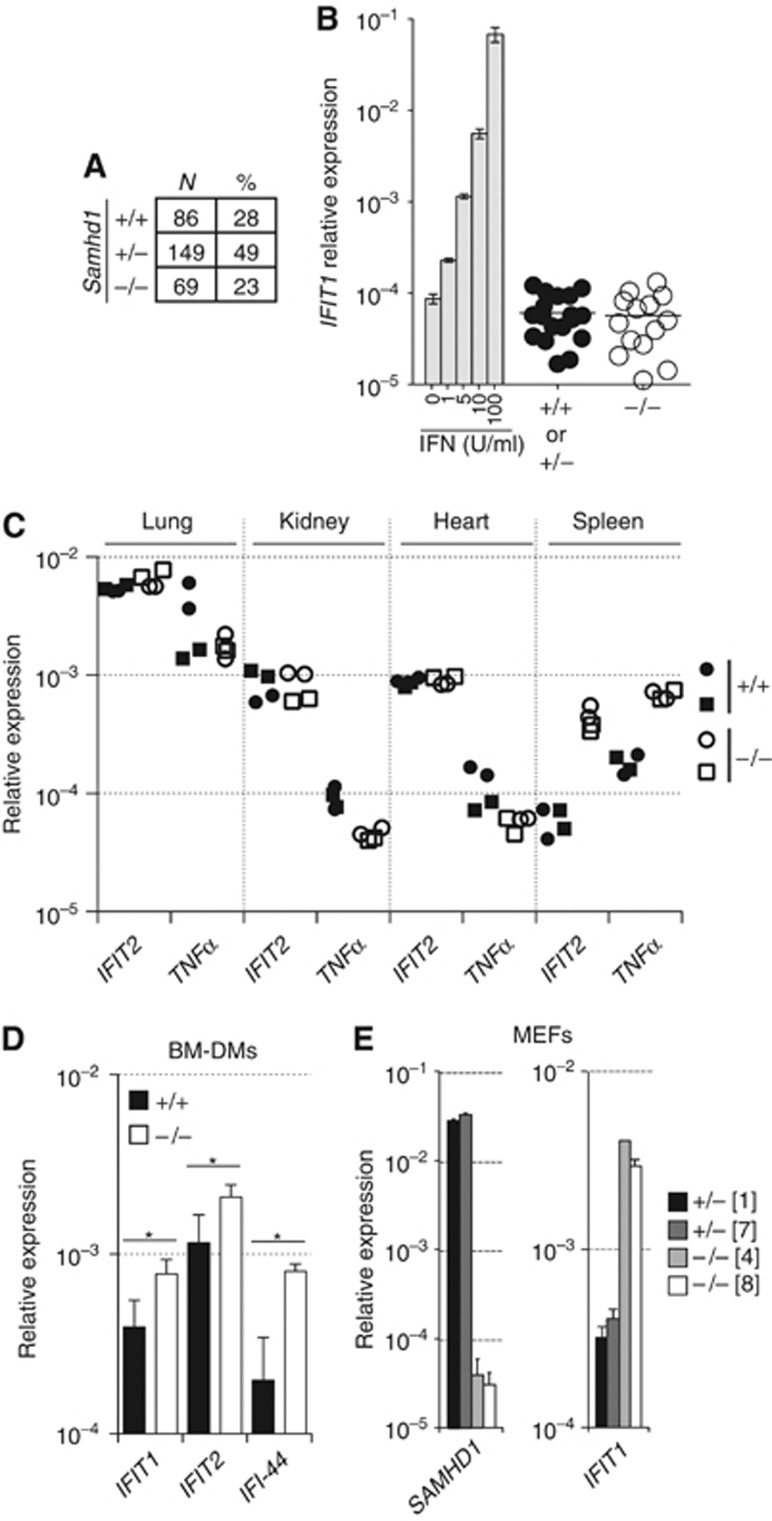
We generated SAMHD1-deficient mice in a mixed or a pure C57BL/6 background (Supplementary Figure S1A–E). Independent of background, Samhd1−/− animals were born at Mendelian ratios (Figure 1A; Supplementary Figure S1F) and developed normally. They were indistinguishable from their wild-type or heterozygous littermates, were fertile and did not show signs of disease even after ageing for over 24 months. Unlike serum samples from AGS patients (Goutieres et al, 1998), sera from SAMHD1-deficient and littermate control mice did not contain detectable IFN as measured by a sensitive bioassay based on induction of the IFN-stimulated gene (ISG), IFIT1 (Figure 1B). Transcript levels of the ISG IFIT2 or of the pro-inflammatory cytokine TNFα were also not increased in lung, kidney and heart from Samhd1−/− mice when compared to tissues from wild-type mice (Figure 1C). However, in spleen, IFIT2 and TNFα mRNA levels were upregulated seven- and four-fold, respectively (Figure 1C). Next, we analysed whether Samhd1−/− cells in culture produce IFN spontaneously. mRNA levels for ISGs were two- to four-fold increased in SAMHD1-deficient bone marrow-derived macrophages (BM-DMs) (Figure 1D). Similarly, SAMHD1-deficient primary mouse embryonic fibroblasts (MEFs) showed a 10-fold increase in IFIT1 mRNA levels when compared to control Samhd1+/− MEFs, which we found to express SAMHD1 mRNA and protein (Figure 1E; Supplementary Figure S1C). In summary, although SAMHD1-deficient mice do not display detectable amounts of circulating IFN or ISG upregulation in most tissues, an IFN signature is evident in Samhd1−/− spleens, macrophages and fibroblasts. Nevertheless, these mice are healthy and do not develop autoimmune disease.
Figure 1.
Spontaneous IFN production by cells and mice lacking SAMHD1. (A) Numbers (N) and percentages (%) of Samhd1+/+, Samhd1+/− and Samhd1−/− offspring of heterozygous Samhd1+/− breedings. Data are pooled from 5D6 (pure B6 background) and 1F8 (mixed S6/B6 background) mice. See Supplementary Figure S1F for further details. (B) Serum samples were obtained from Samhd1+/+, Samhd1+/− and Samhd1−/− littermates by tail bleeding and were analysed for the presence of IFN by bioassay. Samples were added to NIH3T3 cells and induction of the IFN-stimulated mRNA IFIT1 was assessed by RT Q-PCR. Data from samples of 5D6 and 1F8 mice were pooled and the differences between SAMHD1-sufficient and SAMHD1-deficient mice were not statistically significant. This was also the case when 5D6 and 1F8 samples were analysed separately. IFN-A/D was used as a positive control at the indicated concentrations; bars represent the mean of duplicate measurements and error bars show the range. (C) Littermate animals of the indicated Samhd1 genotypes were sacrificed at the age of 9 months and RNA was extracted from lung, kidney, heart or spleen. IFIT2 and TNFα mRNA levels were determined by RT Q-PCR. Circles and squares correspond to individual animals (duplicate measurement). (D) IFIT1, IFIT2 and IFI-44 mRNA levels in BM-DMs of the indicated Samhd1 genotypes cultured for 12 days were determined by RT Q-PCR. Bars represent the mean of four BM-DM cultures from independent mice and error bars show the standard deviation. *P<0.05 (unpaired t-test). (E) Primary MEFs of the indicated Samhd1 genotypes were obtained by crossing Samhd1+/− and Samhd1−/− animals. The expression levels of SAMHD1 (one primer in exon 2) and IFIT1 mRNAs were determined by RT Q-PCR. Bars correspond to cell lines derived from individual embryos and show the mean of triplicate measurements; error bars show the standard deviation. The number in square brackets corresponds to the cell line identification code. (B–E) Relative expression levels compared to GAPDH mRNA are shown. (C–E) Data from 5D6 mice and cells (pure B6 background) are shown.
Nucleic acids and viruses induce normal IFN responses in Samhd1−/− cells and mice
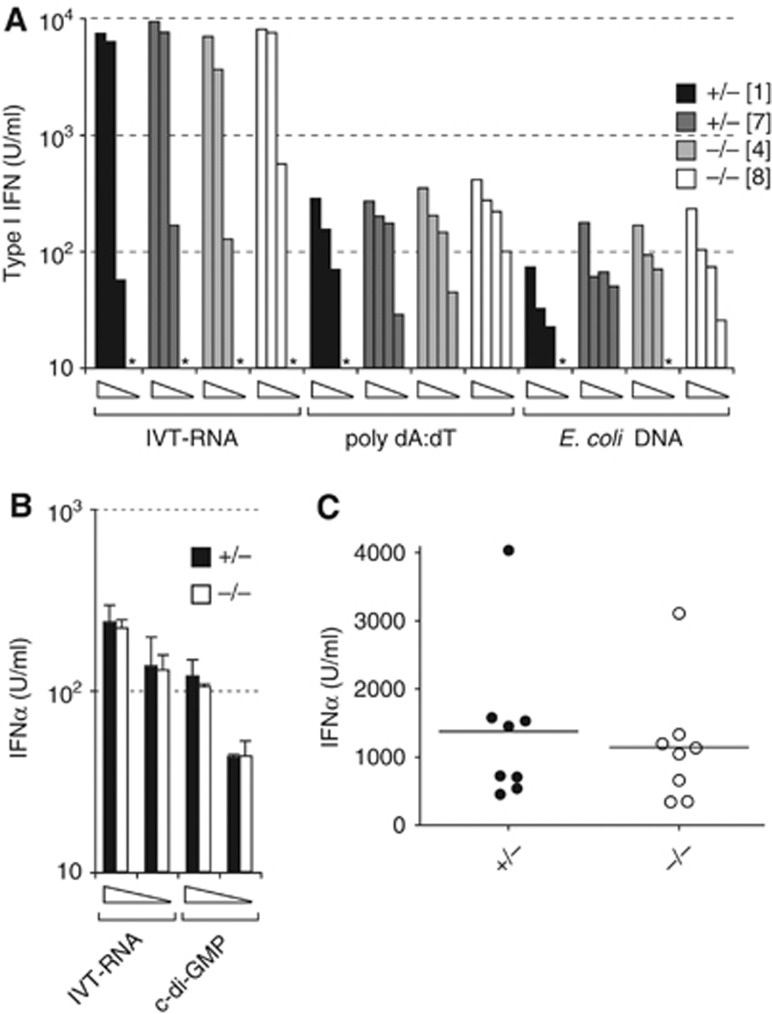
Next, we induced IFN by treating SAMHD1-deficient cells and mice with nucleic acids or by infecting them with viruses. IFN induction in Samhd1−/− and control MEFs was similar in response to transfection of in vitro-transcribed RNA, poly dA:dT or E. coli DNA (Figure 2A). Similarly, SAMHD1-sufficient and SAMHD1-deficient BM-DMs produced comparable amounts of IFNα when stimulated with in vitro-transcribed RNA or cyclic diguanylate monophosphate, an STING agonist (Figure 2B). The induction of IFNβ and IFIT1 transcripts and of mRNAs for the pro-inflammatory factors IL-6, TNFα and IL-1β was similar in SAMHD1-depleted and control BM-derived DCs (BM-DCs) in response to transfected E. coli DNA (Supplementary Figure S2). When assessed in vivo, Samhd1−/− mice produced normal levels of IFNα after encephalomyocarditis virus (EMCV) infection when compared to controls (Figure 2C). These data suggest that SAMHD1 is either redundant or not involved in the pathways that trigger IFN in response to nucleic acids or virus infection.
Figure 2.
Cells and mice lacking SAMHD1 produce normal levels of IFN in response to nucleic acid or viral challenge. (A) Primary MEFs (Figure 1E) were transfected with 2000, 400, 80 or 16 ng of in vitro-transcribed RNA (IVT-RNA), poly dA:dT or E. coli DNA. After overnight incubation, IFN in culture supernatants was quantified by LL171 bioassay. *None detectable. (B) BM-DMs from 5D6 mice of the indicated Samhd1 genotypes were transfected with 1000 or 100 ng of IVT-RNA or cyclic diguanylate monophosphate (c-di-GMP). Six hours later, IFNα in culture supernatants was analysed by ELISA. Error bars show the standard deviation of triplicate measurements. (C) Mice of the indicated genotypes were infected with EMCV by intraperitoneal injection. Serum IFNα was determined by ELISA 24 h later. Data from samples of 5D6 and 1F8 mice were pooled and the differences between Samhd+/− and Samhd1−/− mice were not statistically significant. This was also the case when 5D6 and 1F8 samples were analysed separately.
Infection of SAMHD1-deficient cells and mice with retroviruses and retroviral vectors
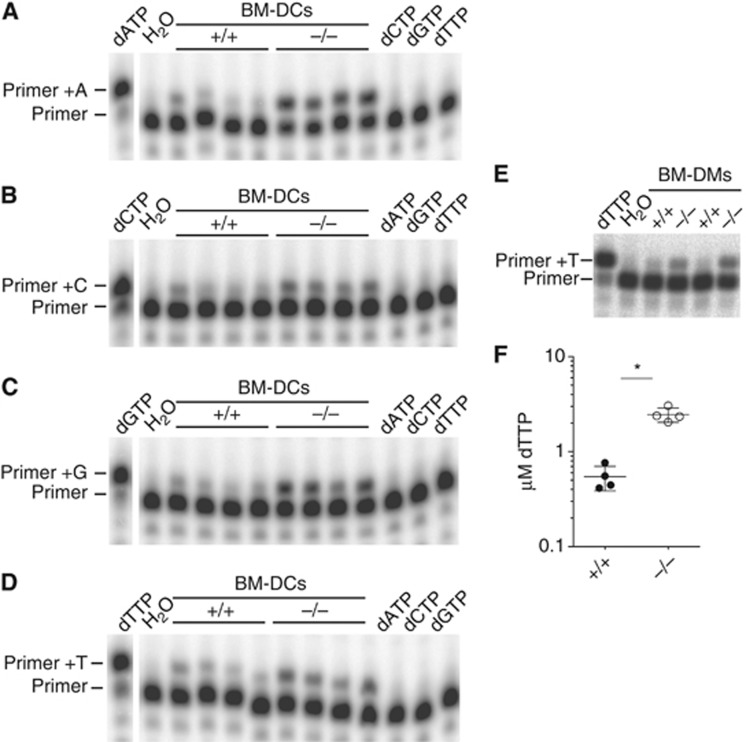
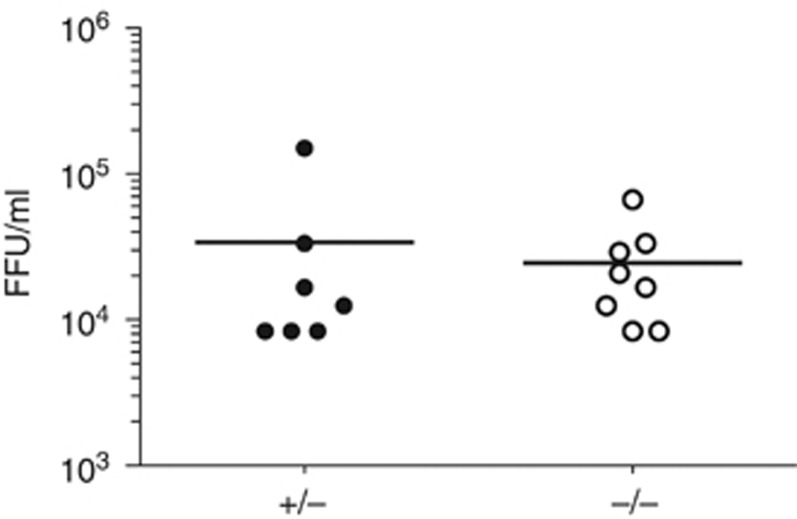
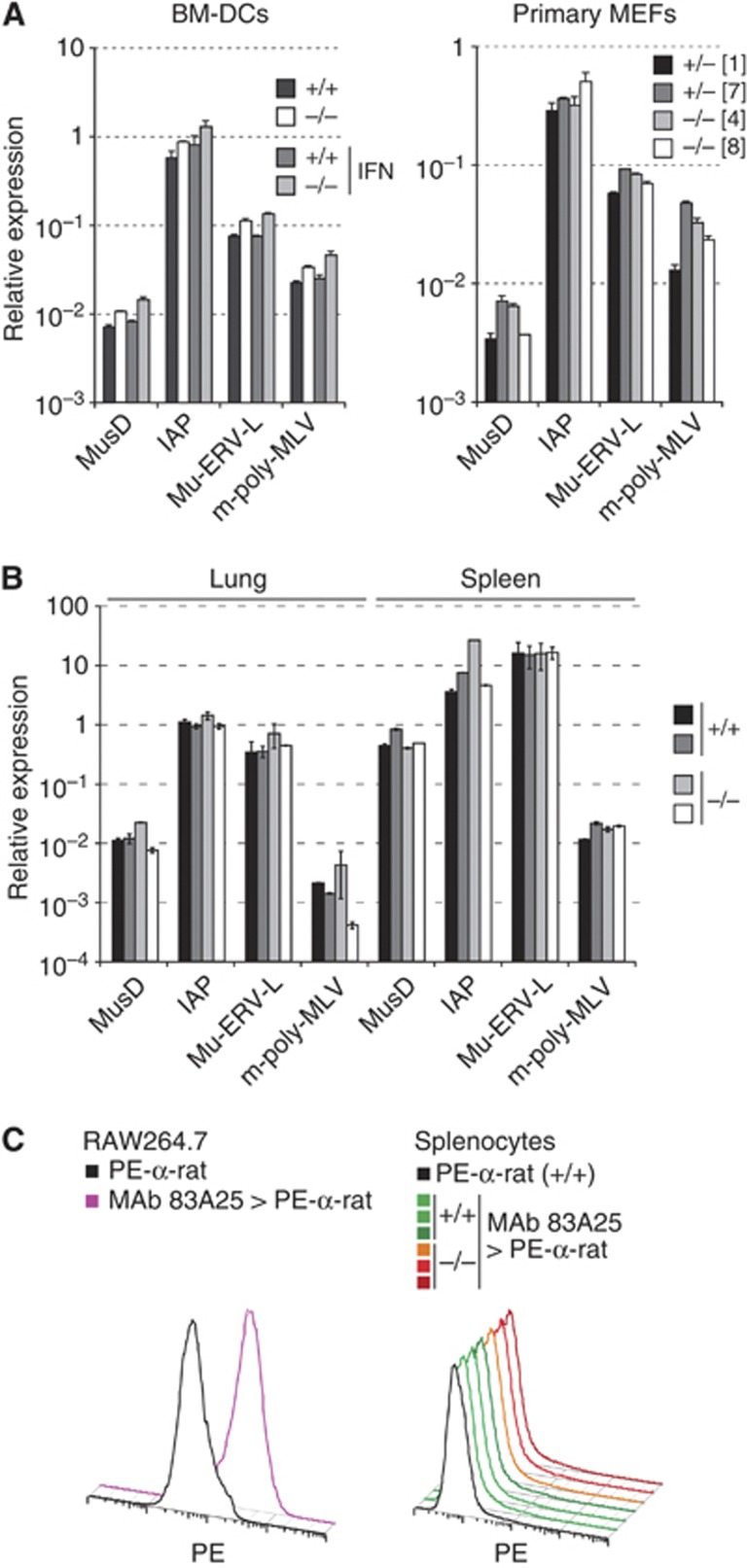
Human SAMHD1 restricts HIV-1 in vitro by degrading dNTPs (Goldstone et al, 2011; Hrecka et al, 2011; Laguette et al, 2011; Powell et al, 2011; Baldauf et al, 2012; Kim et al, 2012; Lahouassa et al, 2012). Using a primer extension assay (Diamond et al, 2004), we found increased concentrations of all four dNTPs in Samhd1−/− BM-DCs (Figure 3A–D). Similarly, dTTP levels were elevated in SAMHD1-deficient BM-DMs (Figure 3E). We independently confirmed these observations using a different fluorescence-based assay (Wilson et al, 2011) (Figure 3F and data not shown). These data substantiate earlier biochemical findings with human SAMHD1 (Goldstone et al, 2011; Powell et al, 2011) and show that dNTP hydrolysis is an evolutionarily conserved function of the protein. To test whether SAMHD1 and dNTP hydrolysis impacts infection with murine retroviruses, we infected neonatal mice with the gamma-retrovirus Moloney murine leukaemia virus (Mo-MLV) and determined viral titers in the serum 2 weeks after infection. We were unable to detect differences in viral load between Samhd1−/− animals and heterozygous littermates (Figure 4). However, we cannot exclude that SAMHD1 controls viral titers at earlier or later time points. SAMHD1 deficiency also had no impact on RNA levels of the MusD, IAP, Mu-ERV-L and m-poly-MLV retroelements in BM-DCs and MEFs, or in lung and spleen tissue (Figure 5A and B). Similarly, we did not detect increased staining of Samhd1−/− spleen cells with an antibody reacting with the envelope glycoproteins of many classes of endogenous murine leukaemia viruses (Figure 5C). Thus, SAMHD1 appears to be dispensable or redundant for in vivo control of replication of exogenous Mo-MLV and for restricting RNA or protein expression from the endogenous retroelements we tested. Nevertheless, it remains possible that SAMHD1 controls other retroelements and/or prevents cDNA synthesis.
Figure 3.
SAMHD1 degrades dNTPs. (A) dATP levels in cell extracts from one million BM-DCs of the indicated Samhd1 genotypes were assessed using a primer extension assay that incorporates a single dATP nucleotide. dATP was used as a positive control and water, dCTP, dGTP and dTTP as negative controls. (B) As (A) for dCTP. (C) As (A) for cGTP. (D) As (A) for dTTP. (E) dTTP levels in cell extracts from BM-DMs were determined as in A. (F) Cell extracts as in A were assessed for dTTP with a fluorescence-based assay. On the basis of a dTTP standard and the cell volume of BM-DCs (2244 μm3, see Supplementary Materials and methods), dTTP concentrations were determined. Horizontal lines indicate the average of four independent samples and error bars depict the standard deviation. (A–F) Cells were on the B6 background (5D6). *P<0.05 (unpaired t-test).
Figure 4.
Infection of SAMHD1-deficient mice with Mo-MLV. Neonatal mice from Samhd1+/− × Samhd1−/− matings were infected with Mo-MLV by intraperitoneal injection. Fourteen days later, animals were sacrificed. MLV serum titers are shown as focus forming units (FFU) per ml. DNA extracted from tail samples was used for genotyping. Data from two experiments with pure B6 background mice (5D6) were pooled. The differences between the two groups are not statistically significant.
Figure 5.
Loss of SAMHD1 does not result in increased RNA and envelope glycoprotein expression of selected endogenous retroelements. (A) RNA from BM-DC cultures (left) or primary MEFs (right) was analysed by RT Q-PCR for the expression of retroelement transcripts. Some cells were treated overnight with 1000 U/ml IFN-A/D as indicated (left). Each bar corresponds to one DC culture or MEF cell line from an individual embryo; numbers in square brackets refer to the cell line identifier (right). Average relative expression levels compared to GAPDH mRNA from two measurements are shown; error bars represent the range. (B) Littermate animals of the indicated Samhd1 genotypes were sacrificed at the age of 9 months and RNA was extracted from lung and spleen. Retroelement transcripts were analysed as in A. Each bar corresponds to samples from an individual animal. (C) Cultured RAW264.7 cells and freshly isolated splenocytes of the indicated Samhd1 genotypes were stained with monoclonal antibody 83A25 and secondary PE-conjugated α-rat antibody and then analysed by flow cytometry. As a control, an aliquot of cells was stained with PE-α-rat alone. 83A25 reacts with the envelope glycoproteins of endogenous murine leukaemia viruses expressed in RAW264.7 cells. (A–C) Cells and mice were on the B6 background (5D6).
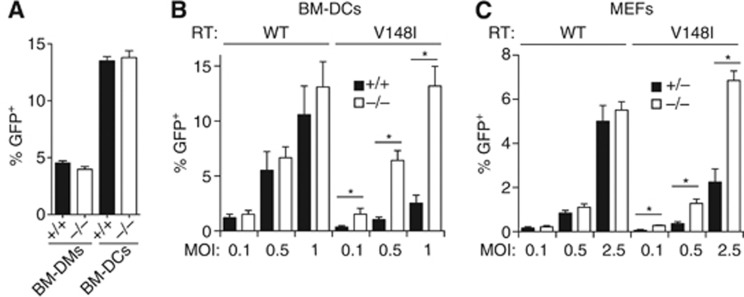
We extended the analysis to infection with single-round, VSV-G-pseudotyped HIV-1 vectors encoding GFP (referred to as HIV-1-GFP below). These minimal, replication-defective lentivirus vectors transduce quiescent cells, which have lower dNTP concentrations than the cycling cells targeted by Mo-MLV (Baldauf et al, 2012; Lahouassa et al, 2012), and have internal promoters driving the GFP reporter. We infected BM-DCs or BM-DMs in vitro after their differentiation with GM-CSF or M-CSF, respectively, at a time when the cells are no longer cycling. Surprisingly, for both myeloid cell types, the efficiency of transduction with HIV-1-GFP was independent of SAMHD1 (Figure 6A). Similarly, transduction with a vector encoding the HIV-1 accessory proteins Vif, Vpr, Vpu and Nef was unchanged in Samhd1−/− BM-DCs (Figure 6B). IFN was not induced to detectable levels upon HIV-1-GFP infection in either wild-type or SAMHD1-deficient BM-DCs (Supplementary Figure S3). Next, we infected freshly isolated non-dividing splenocytes with HIV-1- and MLV-based vectors and identified DCs in the mixture by CD11c staining. Similar numbers of CD11c+ cells were transduced by both vectors independently of Samhd1 genotype (Supplementary Figure S4A). We also infected fresh bone marrow and then cultured the transduced cells with GM-CSF to promote DC differentiation. Transduction efficiencies were identical between SAMHD1-deficient and control DC precursors (Supplementary Figure S4B) and were similarly comparable in SAMHD1-sufficient and SAMHD1-deficient immortalized MEFs (Supplementary Figure S4C). Collectively, these data suggest that SAMHD1 is largely ineffective at restricting reverse transcription of HIV-1 in mouse cells.
Figure 6.
SAMHD1 inhibits pseudotyped HIV-1 in vitro. (A) BM-DMs and BM-DCs of the indicated Samhd1 genotypes were infected with VSV-G-pseudotyped HIV-1-GFP (pCSGW/p8.91, MOI=1). Virus stocks were titrated in 293T cells and multiplicities of infection (MOIs) were calculated using 293T infectious units. Twenty-four hours after infection, the fraction of GFP-expressing cells was determined by flow cytometry. Averages from two different bone marrow donors are shown and error bars represent the range. (B) BM-DCs of the indicated Samhd1 genotypes were infected with VSV-G-pseudotyped HIV-1-GFP (pRRLsin.eGFP/pCMVΔ8.2) and with a vector that has a single amino-acid substitution in the RT (pRRLsin.eGFP/pCMVΔ8.2 V148I). Twenty-four hours after infection, DCs were identified by CD11c staining and percentages of GFP-expressing cells were determined by flow cytometry as further detailed in Supplementary Figure S5. Averages from three DC cultures per group from independent mice are shown and error bars represent the standard deviation. (C) Primary MEFs of the indicated Samhd1 genotypes were infected with the viruses from B. Twenty-four hours after infection, GFP expression was analysed by flow cytometry. Averages from three infections are shown and error bars represent the standard deviation. (A–C) Cells were on the B6 background (5D6). *P<0.05 (unpaired t-test). The differences between the samples infected with wild-type RT HIV-1-GFP were not statistically significant.
SAMHD1 can restrict an RT mutant HIV-1 in vitro and in vivo
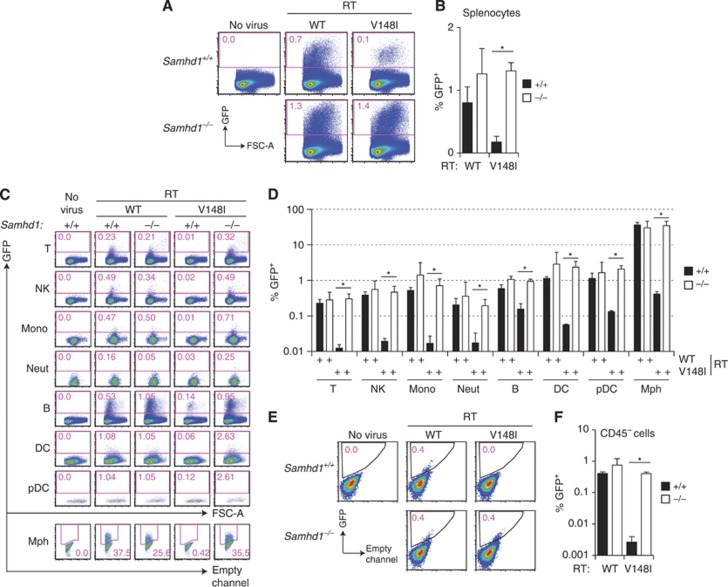
One possible explanation for the above results is that baseline dNTP concentrations in mouse cells are high enough to support HIV-1 reverse transcription, even in the presence of SAMHD1. Consistent with this idea, we found that wild-type BM-DCs contained around 0.5 μM dTTP (Figure 3F), while the KM of HIV-1 RT for dTTP is 0.07 μM (Diamond et al, 2004). We therefore tested a modified HIV-1-GFP encoding a mutant RT with lower affinity for dNTPs (V148I) (Diamond et al, 2003; Lahouassa et al, 2012). Infection of differentiated BM-DCs with HIV-1-GFPV148I resulted in about five-fold increased transduction in Samhd1−/− cells compared to SAMHD1-sufficient control cells (Figure 6B; Supplementary Figure S5). Similarly, HIV-1-GFPV148I transduction rates were three- to five-fold higher in Samhd1−/− primary MEFs (Figure 6C). To test whether the same is true in vivo, we infected SAMHD1-deficient and wild-type animals with the different VSV-G-pseudotyped HIV-1-GFP vectors by intravenous injection. We analysed transduction of splenocytes 5–6 days after infection by flow cytometry. In mice infected with wild-type RT HIV-1-GFP, transduction efficiency was equivalent between control and SAMHD1-deficient animals and resulted in around 1% of all cells in the spleen expressing GFP (Figure 7A and B; Supplementary Figure S6). Similarly, when we infected mice with HIV-1-GFP derived from a different construct that does not encode the accessory proteins, transduction efficiencies of total splenocytes were not significantly different between Samhd1+/+ and Samhd1−/− mice (Supplementary Figure S7). We then identified individual cell populations in the spleen by cell surface marker staining, including lymphoid cells (B cells (B220+, CD11c−), T cells (CD3+, NK1.1−), NK cells (NK1.1+, CD3−)) and myeloid cells (DCs (CD11c+, MHCII+), plasmacytoid DCs (CD11c+, B220+), macrophages (F4/80+, autofluorescence+), neutrophils (CD11bhigh, Gr1high) and inflammatory monocytes (CD11bint, Gr1int)) (gating strategies in Supplementary Figure S6). A percentage of cells between 0.2% (neutrophils) and 36% (macrophages) of these populations were transduced in both wild-type and Samhd1−/− animals with no differences observable between the genotypes (Figure 7C and D; Supplementary Figure S7). In contrast, in vivo infection with HIV-1-GFPV148I resulted in an eight-fold increase in transduction of Samhd1−/− splenocytes compared to control mice (Figure 7A and B). Moreover, transduction of all myeloid and lymphoid cell populations that we analysed, as well as of non-haematopoietic cells (CD45−), was much greater in SAMHD1-deficient mice (Figure 7C–F). These data show that SAMHD1 can restrict infection by some lentiviruses in a plethora of cell types in vivo. Given that the V148I mutation lowers binding of the viral RT to dNTPs and that SAMHD1 deficiency results in increased intracellular pools of dNTPs (Figure 3), our data show that mouse SAMHD1 mediates lentiviral restriction in vivo and in vitro by controlling dNTP availability.
Figure 7.
SAMHD1 restricts pseudotyped HIV-1 in mice. C57BL/6 wild type and SAMHD1-deficient (5D6) animals were injected intravenously with HIV-1-GFP vectors (pRRLsin.eGFP/pCMVΔ8.2 or pCMVΔ8.2 V148I, 5 × 107 293T infectious units). Six days later, transduction of splenocytes was analysed by flow cytometry. (A) GFP expression in total splenocytes was analysed after pre-gating on DAPI and autofluorescence-negative cells as shown in Supplementary Figure S6. Numbers represent percentages of GFP+ cells. Representative examples are shown. (B) Averages from A from three mice per group. (C) Leukocyte subsets were identified as shown in Supplementary Figure S6, and GFP expression was analysed in these populations. Numbers represent percentages of GFP+ cells. Representative examples are shown. T, T cells; NK, NK cells; Mono, inflammatory monocytes; Neut, neutrophils; B, B cells; DC, dendritic cells; pDC, plasmacytoid dendritic cells; Mph, macrophages. (D) Averages from three mice per group analysed as in C. (E) Transduction of non-haematopoietic splenocytes was analysed by gating on CD45- and DAPI-negative cells. Representative FACS plots of CD45− DAPI− cells are shown. Numbers represent percentages of GFP+ cells. (F) Averages from E three mice per group. (B, D, F) Error bars represent the standard deviation. *P<0.05 (unpaired t-test). The differences between the samples from mice infected with wild-type RT HIV-1-GFP were not statistically significant.
Discussion
The recent identification of SAMHD1 as an AGS locus and HIV-1 restriction factor in human myeloid cells has led to marked interest in this hitherto obscure protein (Ayinde et al, 2012; Yan and Chen, 2012). Here, we generated SAMHD1-deficient mice and found evidence for spontaneous IFN production in some cell types (e.g., BM-DMs and MEFs) and tissues (e.g., spleen). However, this did not lead to the development of patent autoimmune disease. We speculate that exogenous insults including microbial infections and other pro-inflammatory events may contribute to AGS development in humans, and that the absence of these triggers in mice housed in clean animal facilities prevents overt phenotypes in Samhd1−/− mice. For example, both environmental factors and the intestinal microbiota contribute to the control of endogenous retroviruses (Young et al, 2012). It will be interesting to determine whether experimental modulation of retroelements results in disease in Samhd1−/− mice.
Recent large-scale sequencing projects identified SAMHD1 mutations in several types of cancer, including in chronic lymphocytic leukaemia (CLL), a B-cell malignancy with disease onset in older individuals (Schuh et al, 2012; Landau et al, 2013), and in lung and colon carcinomas (Imielinski et al, 2012; Liu et al, 2012; Muzny et al, 2012). The availability of SAMHD1-deficient mice will allow dissection of the contribution of this protein in vivo to the development of cancer. This is likely to occur in conjunction with additional mutations (Schuh et al, 2012), a notion that is supported by our observation that Samhd1−/− mice are healthy.
Cultured human resting T cells and several types of myeloid cells are largely refractory to in vitro HIV-1 infection and this inhibition can be relieved by SAMHD1 depletion using Vpx delivery or RNA interference knockdowns (Hrecka et al, 2011; Laguette et al, 2011; Baldauf et al, 2012; Descours et al, 2012). Surprisingly, HIV-1-GFP vectors were not sensitive to restriction by mouse SAMHD1. These experiments were conducted with two HIV-1-GFP vectors, which encode or lack the accessory proteins Vif, Vpr, Vpu and Nef, respectively, establishing that our results are not attributable to antagonism of mouse SAMHD1 by viral factors. Instead, we found that the dTTP concentration in mouse BM-DCs (0.5 μM) (Figure 3F) is about one order of magnitude greater than the one determined for human monocyte-derived macrophages (MDMs) (around 0.05 μM) (Diamond et al, 2004; Kennedy et al, 2010; Lahouassa et al, 2012). The KM of HIV-1 RT for dTTP has been estimated to be 0.07 μM (Diamond et al, 2004). Therefore, in contrast to human MDMs, dNTP pools are not limiting in mouse BM-DCs, likely explaining the lack of SAMHD1-dependent restriction. Why mouse BM-DCs contain more dNTPs than human MDMs remains an open question for future work. Human and mouse SAMHD1 proteins have around 70% sequence identity (Supplementary Figure S8). When the mouse protein was overexpressed in human U937 cells, it was equally efficient at reducing dNTP levels compared to human SAMHD1 (Lahouassa et al, 2012). However, it is possible that species differences in metabolic control and/or differences in the regulation of SAMHD1 function impact on actual dNTP pools. Two recent reports demonstrated that the antiviral activity of human SAMHD1 is negatively regulated in cycling cells by phosphorylation of threonine 592, while this modification is absent in non-cycling cells (Cribier et al, 2013; White et al, 2013). In the mouse, alternative splicing generates two SAMHD1 isoforms, both of which are deleted in our knockout animals (Supplementary Figure S1D). These isoforms differ at the C-terminus, and the phosphorylation motif is present only in isoform 1 (Supplementary Figure S8). It is therefore conceivable that human and mouse SAMHD1 proteins are differentially regulated, and this comparison will be an interesting topic for future studies.
Using a sensitive bioassay, we did not find evidence for induction of IFN in both SAMHD1-deficient and SAMHD1-sufficient cells during infection with the replication-defective lentiviral vectors used here (Supplementary Figure S3). Similarly, human monocyte-derived DCs depleted of SAMHD1 by Vpx delivery failed to upregulate the ISG CD86 upon infection with such minimal lentiviral vectors (Manel et al, 2010). However, infection of human cells lacking SAMHD1 with replication-competent vectors or HIV-1 results in IFN and cytokine production (Manel et al, 2010; Berger et al, 2011; Puigdomenech et al, 2013). The availability of SAMHD1-deficient mice will allow further dissection of the pathways that induce IFN in response to lentiviral infection.
Importantly, we found markedly increased sensitivity of SAMHD1-deficient mice and cells to transduction with an attenuated virus with decreased binding of RT to dNTPs. This mutant polymerase reaches maximal activity in a primer extension assay between 1 and 2.5 μM dNTP (Diamond et al, 2004). This nicely fits with our dNTP quantification: Samhd1−/− BM-DCs contain around 2.5 μM dTTP (Figure 3F) and, as such, are able to support replication of the RT mutant virus, which is restricted in wild-type cells containing only 0.5 μM dTTP.
Taken together, our data show for the first time that SAMHD1 can restrict viruses in vivo in a living animal. Of note, the importance of dNTP hydrolysis to the antiviral function of SAMHD1 has been questioned recently (White et al, 2013) and both nuclease activity of the protein and nucleic acid binding have been reported (Goncalves et al, 2012; Beloglazova et al, 2013; Tungler et al, 2013). We do not exclude the possibility that SAMHD1 may counteract infection in multiple ways; however, our comparison of a wild-type RT with a mutant RT with decreased binding affinity to dNTPs shows that depletion in dNTP pools is key to the antiviral function of SAMHD1 and, indeed, that ‘dNTP starvation’ is an evolutionarily conserved host defense strategy.
Our results also suggest that HIV-1 may have been under SAMHD1 pressure to evolve an RT with high affinity for dNTPs and that many virus laboratory strains isolated from patients may therefore show a low sensitivity to SAMHD1-mediated dNTP depletion in the mouse cells used here both in vitro and in vivo. Consistent with the notion of selection, when we analysed 2336 HIV-1 B-clade sequences deposited in the LANL database, we found that 1.33% had sequence variations in RT at the 148 position. Furthermore, a V148C mutation has been identified in the RT of a simian immunodeficiency virus infecting African green monkeys and this polymerase has similar biochemical properties compared to the V148I enzyme (Skasko et al, 2009). It is interesting to speculate that the V148I mutation may therefore resemble a primordial HIV-1 before SAMHD1-mediated innate immune selection in human patients.
HIV-1 and related lentiviruses replicate in species encoding SAMHD1 and SAMHD1 and pathogenic viruses likely impose selective pressure on each other (Laguette et al, 2012; Lim et al, 2012; Zhang et al, 2012). Notably, Vpx activity is species specific, and mouse SAMHD1and zebrafish SAMHD1 escape Vpx-mediated degradation (Supplementary Figure S9; Ahn et al, 2012; Lahouassa et al, 2012). Our data suggest that HIV-1’s strategy for partially overcoming restriction by SAMHD1 relates to its ability to replicate when nucleotide levels are low rather than encoding a Vpx-like protein that causes SAMHD1 degradation, as is the case for HIV-2. Consistent with this notion, in a primer extension assay, HIV-1 RT was found to be more efficient at low dNTP concentrations than HIV-2’s polymerase (Boyer et al, 2012). As such, HIV-1 partially escapes SAMHD1 restriction and in many instances is able to infect a small proportion of human myeloid cells (Duvall et al, 2007; Hrecka et al, 2011; Laguette et al, 2011; Lahouassa et al, 2012). How this alternate evasion mechanism impacts on HIV-1 transmission and pathogenesis remains unclear, but it likely governs HIV-1 infection of DCs. This is important because DC infection with HIV-1 in vitro, in the presence of Vpx, leads to maturation and IFN production (Manel et al, 2010). In vivo, DC infection would likely influence the nature and potency of immune responses directed against HIV-1, possibly leading to a more successful outcome for the host. Thus, the relationship between SAMHD1 and lentiviruses dictates the cell tropism of the virus and could have profound influence on the nature of the antiviral immune response. The study of these relationships will help us to understand the differences between pathogenic and non-pathogenic lentiviral infections and will likely inform vaccine design.
Materials and methods
Generation of SAMHD1-deficient mice
The targeting construct contained a Neo cassette flanked by FRT sites and loxP sites flanking the second exon of the Samhd1 gene. We generated SAMHD1-deficient mice using mixed or pure C57BL/6 background-targeted ES cells. Two crosses with transgenic lines expressing Cre and FLP recombinases under ubiquitous promoters created heterozygous Samhd1+/− animals. These were intercrossed to generate homozygous (−/−) knockout mice.
Cells and viruses
Cell lines, BM-DCs, BM-DMs and MEFs were grown using standard protocols. VSV-G-pseudotyped retroviral vectors were produced by plasmid transfection of 293T cells. Retroviral infections were performed in the presence of polybrene, and bone marrow, spleen cells, BM-DCs and BM-DMs were transduced by spin infection.
In vivo infection models
Adult mice were infected with EMCV by intraperitoneal injection, and serum IFN levels were determined 24 h later. Mo-MLV was injected intraperitoneally into 1-day-old animals, and virus titers were analysed after 2 weeks. HIV-1-GFP was injected intravenously into adult animals, and transduction of splenocytes was analysed 5–6 days after infection.
All animal experiments were performed in accordance with the UK Animals (Scientific Procedures) Act 1986 and institutional guidelines for animal care. This work was approved by project licences granted by the UK Home Office (PPL No. 80/2309 and PPL No. 40/3583) and was also approved by the Institutional Animal Ethics Committee Review Boards at Cancer Research UK, London, and at the University of Oxford.
Supplementary Material
Acknowledgments
We thank P Borrow and K Pfafferott for the analysis of HIV-1 sequences; G Kassiotis for advice; B Chesebro for the 1912 hybridoma; Florence Margottin-Goguet for HIV-1-GFP plasmids; Leonard Evans and Stefan Bauer for the 83A25 antibody; I Rosewell, O Schulz, S Zelenay, B Schraml and P Whitney for technical help; and all members of the Immunobiology Laboratory, LRI, CRUK, for helpful discussions. This work was funded by Cancer Research UK and the UK Medical Research Council. CRS acknowledges additional financial support from Fondation Bettencourt-Schueller. JR was a recipient of an HFSP long-term postdoctoral fellowship. JM is a recipient of an EMBO long-term postdoctoral fellowship and is also supported by Marie Curie Actions (EMBOCOFUND2010, GA-2010-267146). GJT is funded by a fellowship from the Wellcome Trust and the Medical Research Council and the UCL/UCLH Comprehensive Biomedical Research Centre. YJC and DTB acknowledge the European Union's Seventh Framework Program (FP7/2007-2013) grant agreement number 241779 (NIMBL).
Author contributions: JR and CRS designed research; JR, JM, AB and RR performed research; BH, RAL, PDB, GJT, LFM, YJC and DTB contributed reagents and advice; JR, JM, AB, RR and CRS analysed data; JR and CRS wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem 287: 12550–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayinde D, Casartelli N, Schwartz O (2012) Restricting HIV the SAMHD1 way: through nucleotide starvation. Nat Rev Microbiol 10: 675–680 [DOI] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT (2012) SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18: 1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck-Engeser GB, Eilat D, Wabl M (2011) An autoimmune disease prevented by anti-retroviral drugs. Retrovirology 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem 288: 8101–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathogens 7: e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PL, Clark PK, Hughes SH (2012) HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J Virol 86: 5885–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3: 1036–1043 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Livingston JH (2008) Aicardi-Goutieres syndrome: an important Mendelian mimic of congenital infection. Dev Med Child Neurol 50: 410–416 [DOI] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schawartz O, Laguette N, Benkirane M (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B (2004) Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem 279: 51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond TL, Souroullas G, Weiss KK, Lee KY, Bambara RA, Dewhurst S, Kim B (2003) Mechanistic understanding of an altered fidelity simian immunodeficiency virus reverse transcriptase mutation, V148I, identified in a pig-tailed macaque. J Biol Chem 278: 29913–29924 [DOI] [PubMed] [Google Scholar]
- Duvall MG, Lore K, Blaak H, Ambrozak DA, Adams WC, Santos K, Geldmacher C, Mascola JR, McMichael AJ, Jaye A, Whittle HC, Rowland-Jones SL, Koup RA (2007) Dendritic cells are less susceptible to human immunodeficiency virus type 2 (HIV-2) infection than to HIV-1 infection. J Virol 81: 13486–13498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M Jr, Barber GN, Stetson DB (2012) Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480: 379–382 [DOI] [PubMed] [Google Scholar]
- Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T (2012) SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Hum Mutat 33: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutieres F, Aicardi J, Barth PG, Lebon P (1998) Aicardi-Goutieres syndrome: an update and results of interferon-alpha studies. Ann Neurol 44: 900–907 [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474: 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H et al. (2012) Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik R, Zhu X, Stranska R, Wu Y, Stevenson M (2009) A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 6: 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Gavegnano C, Nguyen L, Slater R, Lucas A, Fromentin E, Schinazi RF, Kim B (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J Biol Chem 285: 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem 287: 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M (2012) Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11: 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13: 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N et al. (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M (2012) The Ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11: 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, Klijn C, Guillory J, Bhatt D, Vartanian S, Walter K, Chan J, Holcomb T, Dijkgraaf P, Johnson S, Koeman J et al. (2012) Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 22: 2315–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR (2010) A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467: 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE (2004) Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 24: 6719–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Reid JG, Santibanez J, Shinbrot E, Trevino LR, Wu YQ, Wang M, Gunaratne P, Donehower LA, Creighton CJ, Wheeler DA et al. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem 286: 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdomenech I, Casartelli N, Porrot F, Schwartz O (2013) SAMHD1 restricts HIV-1 cell-to-cell transmission and limits immune detection in monocyte-derived dendritic cells. J Virol 87: 2846–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC et al. (2009) Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41: 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, Del Toro M et al. (2012) Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 44: 1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, Feller SM, Grocock R, Henderson S, Khrebtukova I, Kingsbury Z, Luo S, McBride D, Murray L, Menju T, Timbs A, Ross M, Taylor J, Bentley D (2012) Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 120: 4191–4196 [DOI] [PubMed] [Google Scholar]
- Skasko M, Diamond TL, Kim B (2009) Mechanistic variations among reverse transcriptases of simian immunodeficiency virus variants isolated from African green monkeys. Biochemistry 48: 5389–5395 [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungler V, Staroske W, Kind B, Dobrick M, Kretschmer S, Schmidt F, Krug C, Lorenz M, Chara O, Schwille P, Lee-Kirsch MA (2013) Single-stranded nucleic acids promote SAMHD1 complex formation. J Mol Med (Berl) 91: 759–770 [DOI] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PM, Labonte MJ, Russell J, Louie S, Ghobrial AA, Ladner RD (2011) A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res 39: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Chen ZJ (2012) Intrinsic antiviral immunity. Nat Immunol 13: 214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J (2010) The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 11: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G (2012) Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491: 774–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, de Silva S, Wang JH, Wu L (2012) Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS One 7: e37477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.