Abstract
The invasive dengue vector Aedes aegypti has persisted for > 200 years in South Florida in the United States. We tested the hypotheses that Florida’s landscape creates dispersal barriers and corridors and that long-distance human-aided dispersal structures populations of Ae. aegypti. We evaluated the phylogeography of 362 individuals from Florida’s East and West Coasts with a 760-bp (418- and 342-bp fragments of ND5 and ND4, respectively) mitochondrial sequence. Populations from these two coasts were not significantly differentiated, suggesting that limited urbanization in central Florida is not a strong barrier to gene flow. Evidence for long-distance dispersal between Ft. Lauderdale and the West and Ft. Myers and the East indicates the importance of human-aided dispersal. West Coast populations showed no genetic differentiation, indicating that West Coast rivers and bays did not significantly impede gene flow. Phylogeographic analysis of haplotypes showed two distinct matrilines with no geographic patterns, suggesting multiple introductions or balancing selection.
Introduction
Aedes aegypti (yellow fever mosquito) is the major vector of arboviruses causing yellow fever and dengue.1 Recent Ae. aegypti-driven outbreaks of Chikungunya in the Indian Ocean and Italy2,3 and dengue in South America4,5 and Key West, Florida6 emphasize the continuing threat of Ae. aegypti to public health. Ae. aegypti invaded the Western Hemisphere about three centuries ago and established persistent populations in the states of Florida, Georgia, Louisiana1 (K. Caillouet, personal communication), Arizona,7 and Hawaii, despite intense eradication efforts in the 1950s and 1960s.8
Ae. aegypti is an urban container-dwelling species9 that preferentially feeds on human hosts.10 Human population density, road connectivity, transportation, and urbanization that facilitate human-aided long-distance dispersal (i.e., transport of immatures in water-filled containers like tires or transport of adults in vehicles) likely aided the expansion of Ae. aegypti populations.5,11 In Florida, Ae. aegypti is now largely limited to urban areas on the East and West Coasts and some densely populated urban areas in central Florida.12,13 Although the small-scale genetic structure of Ae. aegypti in Florida is likely to be distinct from the structure in Asia and Latin America because of differential dispersal potential associated with greater urban sprawl and the lack of opportunities to breed indoors in Florida,12,14 the larger-scale population genetic structure is expected to be similar to the structure observed elsewhere, showing panmixia because of the wide dispersal capability of the species.15 Ae. aegypti population history in Florida is complex as a result of founder effects,16 adaptation,17 multiple introductions, dispersal, bottlenecks, and expansions.18,19
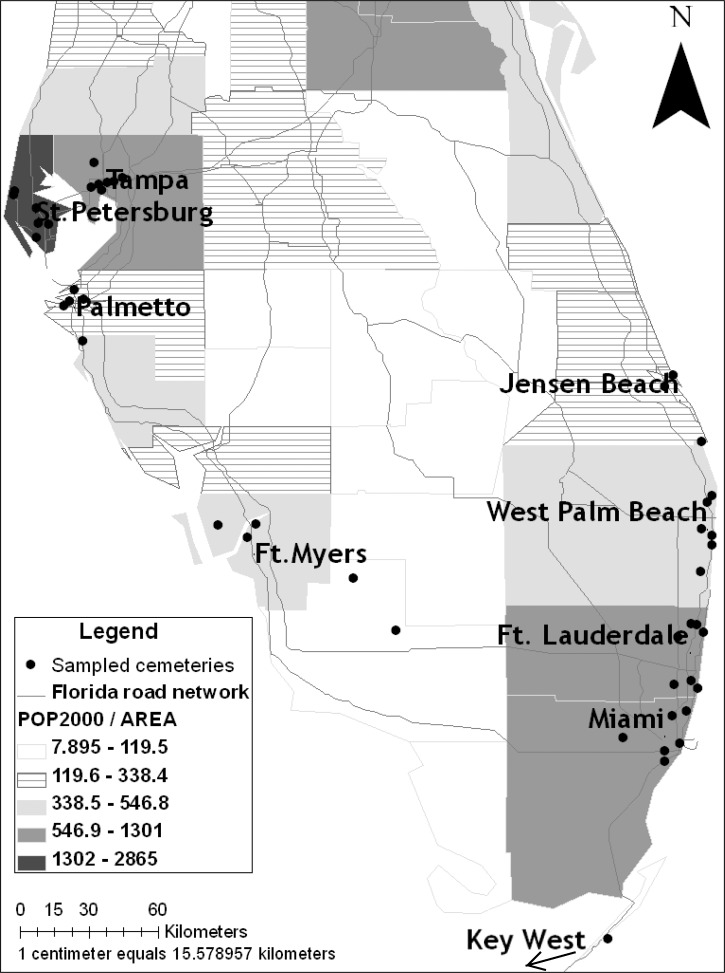
Geographic expansion furthered by dispersal is a key reason for persistence of Ae. aegypti in multiple continents.20 However, barriers to dispersal can place limits on the ultimate range and potential impact of invasive species. A detailed understanding of dispersal barriers may ultimately inform strategies to control this vector.16,21 In this study, we test the hypothesis that landscape barriers (e.g., rivers, saltwater bays, and non-urban habitat) and dispersal corridors (e.g., roads and contiguous urban habitat) for Ae. aegypti affect gene flow patterns at the scale of the Florida peninsula. For example, wide river mouths are evident on the West Coast of Florida, but they are mostly absent on the East Coast of Florida (Figure 1), and human population density is high and evenly distributed along the East Coast, especially in the south, but heterogeneous for much of the West Coast (Figure 1). In Southwestern Florida, we postulate that rural expanses are barriers to Ae. aegypti because of insufficient human hosts and oviposition containers, both of which are necessary for dispersal of Ae. aegypti. We examined phylogeographic patterns based on mitochondrial DNA (mtDNA) variation of Ae. aegypti on the East and West Coasts of South Florida, including the Florida Keys. Specifically, we sampled from urban cemeteries, where water-filled vases offer permanent or semipermanent larval habitats to sustain Ae. aegypti populations.13 We sampled containers within cemeteries, cemeteries within cities, and cities within coasts in a nested sampling pattern to examine the phylogeography and population structure of Ae. aegypti across Florida.
Figure 1.
Sampled cemeteries in south Florida. Counties are differentiated based on human population density (population census 2000) compared with land area (square mile); road connectivity in south Florida is displayed. Black dots represent the cemeteries sampled within each city. < ‐‐‐ = Key West is ∼ 100 km south west.
Materials and Methods
Between June and October of 2006, we sampled cemeteries (hereafter called sites) in South Florida and collected larvae from all water-filled vases at each site where Ae. aegypti was present (Figure 1 and Supplemental Table 1). Because many of these sites harbored Ae. aegypti, Ae. albopictus, and Ae. triseriatus, we brought all larvae to the laboratory, identified them individually, and reared only Ae. aegypti to adulthood. Individual adults were placed in 95% ethanol on the day of eclosion.
DNA extraction and sequencing.
DNA was extracted from individual mosquitoes using a well-established DNAzol protocol.22 Published primers were used to amplify a 452-bp region of the ND5 mtDNA sequence (forward: 5′-TCCTTAGAATAAAATCCCGC-3′; reverse: 5′-GTTTCTGCTTTAGTTCATTCTTC-3′)23 and a 394-bp region of the ND4 mitochondrial sequence (forward: 5′-GTD YAT TTA TGA TTR CCT AA-3′; reverse: 5′-CTT CGD CTT CCW ADW CGT TC-3′).24 We chose ND4 and ND5, because previous studies on Ae. aegypti11,24–30 and other mosquitoes23,31,32 showed variability in the ND4 and ND5 sequences that yielded reliable gene genealogies suitable for inferring population structure and contributions of historical and current demographic processes. Both forward and reverse strands were sequenced using the ABI 3100 automated sequencer at the Keck Center, University of Illinois, Urbana-Champaign, IL. When singleton mutations were detected, those fragments were resequenced in both directions to exclude the probability of Taq polymerase amplification error.33 Sequences were edited using Sequencher (ver. 3.0) and aligned using ClustalW option in SeaView.34 The ND5 and ND4 gene fragments were trimmed to 418 and 342 bp, respectively, and they were then concatenated for a final 760-bp-long gene sequence (Hudson, Kreitman & Aguade test, P = 0.8); final analyses were based on the edited 760-bp fragment.
Statistical analysis.
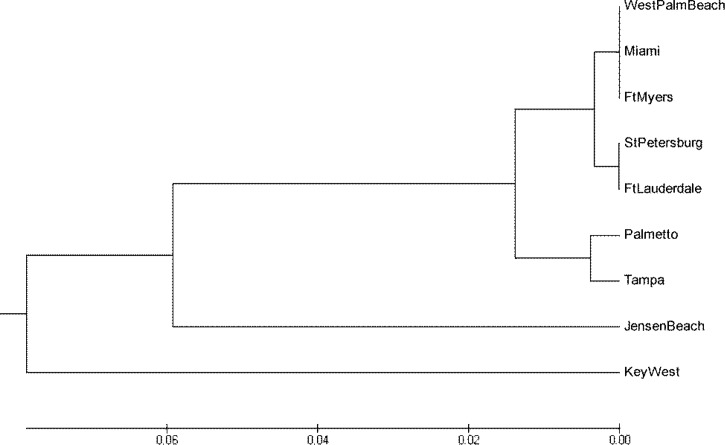
Haplotype diversity, number of haplotypes, and average number of nucleotide differences (π) for cemeteries within each city and among cities along the two coasts were calculated using DnaSP 5.0.35 Identification of haplotypes, calculation of pairwise FST, analysis of molecular variance (AMOVA), and testing hypotheses of differentiation between the East and West Coasts and between cities within each coast were conducted using ARLEQUIN 3.1.36 Effective migration rates were calculated as Nem = (1 − FST)/4FST.37 We used Tajima’s D statistic38 to assess deviations from neutrality that can indicate population history using DnaSP 5.0.35 For example, Tajima's D values below zero suggest increasing population size or purifying selection, whereas values above zero are consistent with decreasing population size and balancing selection; migration can result in a range of values.39,40 As a rule of thumb, values greater than +2 or less than −2 are likely to be biologically significant.40 An unrooted Unweighted Pair Group Method with Arithmetic mean (UPGMA) tree based on pairwise FST estimates was constructed to display genetic distances within and between East and West Coasts using MEGA 3.41
We used one-tailed Mantel tests with genetic (FST) and geographic distances as well as log-transformed genetic distance (FST/1 − FST) and log-transformed geographic distance across all sites42 to test for isolation by distance (IBD).37 We also used a Mantel test with restricted randomization,43,44 stratified the data into East and West Coast groups, and thus, tested for IBD within coasts for the entire set of nine sites. Finally, we performed a Mantel test with restricted randomization, stratifying the data into East Coast, West Coast, and Florida Keys, to determine if the population at Key West had a disproportionate effect on patterns of IBD. In all cases, we used 999,000 permutations with the program RT v. 2.143 for Mantel tests. Euclidean distances between pairs of sites were calculated using ArcGIS. We implemented the spatial analysis of molecular variance (SAMOVA)45 algorithm to identify groups of sampled populations (i.e., K groups) that are maximally differentiated from one another without any a priori assumptions about population structure. We performed 1,000 annealing processes for two to four groups (K = 2–4).
Results
Within cemeteries, the number of individual vases that held Ae. aegypti varied between 1 and 19. A total of 68 mitochondrial haplotypes was identified from 362 sequenced Ae. aegypti individuals (Supplemental Table 2). A single haplotype (AEF6) was shared across all of nine cities sampled, whereas AEF4 was present in all of the inland cities (Supplemental Table 2). A total of 20 haplotypes was shared in at least two of the sampled cities; a majority of the haplotypes (48), however, were found only within a single city (Tampa = 11, St. Petersburg = 5, Palmetto = 4, Ft. Myers = 2, Miami = 9, Ft. Lauderdale = 8, West Palm Beach = 5, and Jensen Beach = 4). The nucleotide sequences were characterized by 35 polymorphic sites, of which 33 sites were parsimony-informative. Nucleotide substitutions were identified at 35 of 760 sites, of which 91.42% were transitions.
Haplotype and nucleotide diversities were comparable on the West Coast (0.870 and 0.01362, respectively) and the East Coast (0.901 and 0.01329, respectively) (Supplemental Table 3). There was a moderate level of genetic differentiation when considering all of the cemeteries sampled across South Florida (FST = 0.05708, Nm = 8.26). Within cities, across cemetery sites, values of FST ranged between 0 (Ft. Myers, Jensen Beach, and West Palm Beach) and 0.17430 (St. Petersburg). The hierarchical AMOVA of all sites, except Key West, did not detect any significant differentiation between the East and West Coasts (FCT = −0.00159, P > 0.05); however, the differentiations among cities within each coast (FSC = 0.02835) and within cities (FST = 0.02681; both P < 0.01) were significant (Supplemental Table 4). In a separate AMOVA comparing three groups (East Coast, West Coast, and Key West), results were similar: the coasts and Key West were not differentiated from one another (FCT = 0.0067, P > 0.2) (Table 1). An AMOVA of the West Coast sites alone confirmed extensive gene flow among cities along this coast (FCT = −0.00240, P > 0.05) and to a lesser extent, cemeteries within cities (FSC = 0.05128, P > 0.05) but significant differentiation within cemeteries (FST = 0.04901, P < 0.0430). In contrast, a similar analysis for the East Coast sites alone detected significant differentiation only among cities (FCT = 0.05577, P = 0.0146) (Supplemental Table 4).
Table 1.
Partitioning of variance components and FST of partial ND4 and ND5 mitochondrial haplotypes identified from cities in south Florida, including Key West
| Source of variation | df | Sum of squares | Variance components | Percent variation | Ф Statistics |
|---|---|---|---|---|---|
| East Coast, West Coast, and Key West | |||||
| Among groups | 2 | 25.243 | 0.03443 | 0.67 | 0.00670 |
| Among cities within groups | 6 | 67.042 | 0.14769 | 2.88 | 0.02895* |
| Within cities | 354 | 1,753.599 | 4.95367 | 96.45 | 0.03546† |
| Total | 362 | 1,845.884 | 5.13580 | ||
P < 0.05.
P < 0.001.
df = degrees of freedom.
Pairwise comparison of genetic differentiation between cities indicated significant differences between Jensen Beach and both Ft. Lauderdale and Key West after Bonferroni correction (P < 0.0013) (Supplemental Table 5). The FST-based UPGMA tree (Figure 2) highlights likely long-distance dispersal between Ft. Lauderdale and the West as well as Ft. Myers and the East, a close connection between Tampa and Palmetto separate from other sites, and the genetic distinctness of the Key West and Jensen Beach populations. The haplotype network identified AEF6 (the haplotype shared across all sampled cities) as most ancestral (Supplemental Figure 1 and Supplemental Table 2). Analyses of mismatch frequencies of pairwise sequence differences and segregating sites display bimodal distributions (Supplemental Figures 2 and 3). Although this result is consistent with two distinct clades, these clades are less obvious in the network, which had no clear spatial patterns in haplotype distribution. Tajima’s D tests for deviations from neutrality were significant for all cities in the West Coast but none of the East Coast cities (Supplemental Table 6). Although not always significant, Tajima’s D values were greater than two for many sites (Supplemental Table 6), which can be indicative of a high degree of dispersal.
Figure 2.
FST-based UPGMA tree showing the relationship between sampled cities in south Florida.
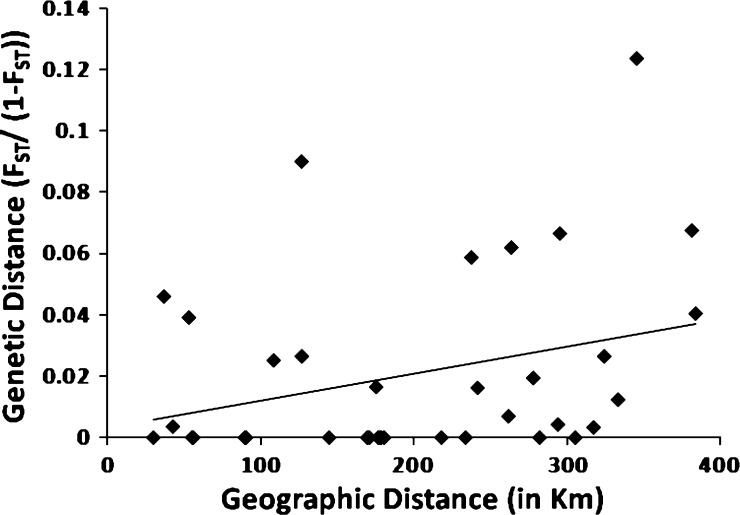
Unrestricted analysis of all nine sites yielded a significant positive correlation between genetic and geographic distances (one-tailed Mantel test, RM2 = 0.093, P = 0.0399) (Figure 3). Correlation of log–log distances approached significance (one-tailed Mantel test, RM2 = 0.077, P = 0.0635).42 Because our analyses also showed differentiation of East versus West Coast populations and the Key West population, this apparently significant correlation in unrestricted analysis may be misleading. Our restricted randomization Mantel test, stratifying populations into East and West Coasts, yielded a marginally non-significant relationship between geographic and genetic distances (one-tailed Mantel test, RM2 = 0.103, P = 0.0510). In contrast, a restricted randomization Mantel test, with randomizations stratified into East Coast, West Coast, and Florida Keys, yielded a clearly non-significant relationship between genetic and geographic distances (one-tailed Mantel test, RM2 = 0.019, P = 0.2017). Thus, it seems that most of the apparent relationship of genetic and geographic distances derives from populations on the two coasts (and thus, geographically far apart) being genetically distinct and the population at Key West being both distinct and geographically distant from most others.
Figure 3.
Correlation of genetic [FST/(1 − FST)] distance of A. aegypti partial ND4 and ND5 mtDNA sequences and geographic distance across all sampled cities in South Florida [negative values of FST/(1 − FST) = 0; P = 0.2017].
SAMOVA on the mainland cities alone (excluding Key West) identified two optimal groups (Jensen Beach versus all others) based on the most significant FCT index (Supplemental Table 7). SAMOVA including Key West showed four optimal groups (Table 2), suggesting that there were at least three putative barriers to dispersal or distinct population histories of Ae. aegypti in this geographic region. These groupings are consistent with the FST-based UPGMA tree (Figure 2), which also shows Key West as the most distinct population, with Jensen Beach next and Palmetto as divergent from all remaining populations except Tampa.
Table 2.
Fixation indices corresponding to the groups of populations inferred by SAMOVA analysis for Ae. aegypti populations in south Florida (with Key West) tested for partial ND4 and ND5 mitochondrial sequences
| No. of groups | Group composition | FSC | FST | FCT |
|---|---|---|---|---|
| 2 | 1. Key West; 2. all other populations | 0.0233* | 0.0974* | 0.07592 |
| 3 | 1. Key West; 2. Jensen Beach; 3. all other populations | 0.0145* | 0.0884* | 0.07492* |
| 4 | 1. Key West; 2. Jensen Beach; 3. Palmetto; 4. all other populations | −0.0003* | 0.04486 | 0.04515† |
P < 0.05.
P < 0.001.
Discussion
Long-distance human-aided dispersal.
Ae. aegypti dispersal depends on two main factors: the availability and movement of containers and human transport.7,46–48 If Ae. aegypti dispersal within Florida occurs only by flying adults, then numerous, uniformly distributed aquatic container habitats would seem to be necessary to act as stepping stones for gene flow and long-distance dispersal over time. However, unlike many developing countries where Ae. aegypti populations have been studied, peridomestic aquatic container habitats, such as domestic water storage containers, seem to be relatively rare in the United States.14 Consequently, we postulated that relatively low human population density and associated scarcity of peridomestic habitat in Central South Florida as well as the West Coast rivers and water bodies would impede dispersal of Ae. aegypti (Figure 1) between the coasts. We find, however, no significant differentiation between the East and West Coast populations, suggesting that practically no large-scale barriers to gene flow exist. This result explains the presence of considerable human-aided long-distance dispersal of Ae. aegypti in South Florida (Supplemental Table 4). Such human-aided dispersal may explain both the clustering of Ft. Myers’ populations with the populations on the East Coast and the clustering of Ft. Lauderdale’s populations with St. Petersburg’s population on the West Coast (Figure 2). The absence of a significant pattern of isolation by distance also suggests a prominent role for human-aided dispersal, a likely mechanism disrupting any population structure based on dispersal by adult flight.
Tajima’s D values of Ae. aegypti populations in Miami and the West Coast are unusually high and positive, and they yield an overall significant positive value across all sampled cities. Although population decrease and balancing selection can result in significant positive Tajima’s D values, values more than two are rare. Simulation models indicate that extremely high D values (i.e., > 2) are six to seven times more likely with an average migration rate of 0.1/generation.40 Migration rather than local bottlenecking or selection on mtDNA seems the most parsimonious explanation for the high D values for West Coast cities and Miami, especially because the UPGMA tree (Figure 2) and the pairwise effective number of migrants also suggest high immigration into those cities, with the exception of Ft. Lauderdale. It is beyond the scope of our study to test alternative hypotheses of bottlenecks versus dispersal to explain these large Tajima’s D values (R. Nielsen, personal communication).
The Key West population exhibited a negative, albeit non-significant Tajima’s D, suggesting either population expansion or purifying selection. Key West is known for its extensive mosquito control program (http://keysmosquito.org/), and it is a popular tourist destination, which may contribute to human-aided immigration of Ae. aegypti into the island. Hence, repeated bottleneck–recolonization events may be common in Key West, and they may contribute to the differentiation of this population evident in SAMOVA (Table 2).
Cryptic barriers to gene flow.
In contrast to the West Coast, where gene flow was extensive (Table 2 and Supplemental Tables 4 and 5), the differentiation among East Coast cities was unexpectedly significant given the almost contiguous road network and urban areas that should provide corridors for natural dispersal as well as human-aided dispersal. Although effective vector control in East Coast cities could have created patterns of asynchronous local bottlenecking and recolonization, we found no evidence supporting such population fluctuations, suggesting that there may be cryptic barriers to gene flow (e.g., patchy distribution of peridomestic habitat) in the urban landscape. Study of Ae. aegypti populations across the east and west sides of Uriah Butler Highway in Trinidad, West Indies, using mtDNA and microsatellite DNA showed a similar pattern, where cryptic barriers in the urban landscape limited dispersal.49 Another detailed, fine-scale analysis of local landscapes within and between East Coast cities using nuclear markers would be necessary to identify such barriers.
Invasion history.
As an invasive species in South Florida, we might expect that Ae. aegypti would show reduced genetic diversity,16,19 but the evidence is mixed; also, comparable data are limited. For example, we identified 68 haplotypes from 362 individuals (0.886 haplotype diversity [Hd]) for the combined ND4 and ND5 mtDNA fragments, and we identified 12 haplotypes (Hd = 0.713) for ND5 and 42 haplotypes (Hd = 0.817) for ND4 when analyzing the fragments separately. By comparison, native mosquitoes have greater genetic diversity. For example, Ae. vexans31 displayed 34 ND5 haplotypes among 54 individuals (0.953 Hd), and Culex tarsalis32 displayed 64 ND4 haplotypes among 170 individuals (0.887 Hd). Genetic diversity of Ae. aegypti in Southern Florida is consistent with a complex history of long-distance dispersal and extinction–recolonization since its invasion.50–53
Comparison of Ae. aegypti population structure with other geographic areas.
Our sequence and phylogenetic analyses of the ND4 and ND5 mtDNA fragments documented two matrilineal clades that had no geographic structure, similar to Ae. aegypti studied in several other geographic areas.11,15,24–30,54 Discovery of two ND4 mtDNA clusters of Ae. aegypti in Northeastern Mexico provided the first evidence of introductions of two independent mitochondrial lineages or a newly introgressed matriline.24,47 Other ND4 mtDNA studies came to similar conclusions for Ae. aegypti populations in Thailand,25 Peru,26 Venezuela,27 Brazil,28,54 and several sites in South America and Africa29,30 and for Ae. japonicus, a recent invader into the United States.55 An alternative explanation could be that two clades exist in the source population (African continent),30 which could have been introduced into Florida through a single invasion event. To identify the source populations for such introductions into the Americas, comparisons29 of two clusters of Ae. aegypti ND4 haplotypes from the Americas with haplotypes from three localities each in Africa and Asia/Polynesia indicated that one cluster was more similar to Asia/Polynesia (Singapore, Cambodia, and Tahiti) and Africa (Uganda and Guinea), whereas the other cluster was more closely related to Senegal in Africa. Although most of the above studies have used the ND4 partial sequence, our analysis of both ND4 and ND5 partial sequences again showed a similar pattern, supporting the multiple introduction hypothesis. Although nuclear copies of mtDNA may explain clustering of haplotypes into two distinct clades,24–30,54,56,57 we confirmed the absence of nuclear copies in our Ae. aegypti mtDNA sequences.
In summary, we have some evidence for two separate introductions of Ae. aegypti into Southern Florida, which has been postulated by other authors, and find no evidence for a significant barrier to dispersal between the East and West Coasts of Florida. We interpret our results as evidence for a prominent role of human-aided long-distance dispersal and incidence of panmixia between these coasts, which has been illustrated in the case of Ae. aegypti in the Americas using a Bayesian coalescent framework-based gene flow network model analysis.15 Significant long-distance and human-aided dispersal in Florida may have implications for vector control efforts directed at this species. Such intercity dispersal is likely to interfere with local vector eradication attempts by contributing to recolonization if local eradication is achieved. For transgenic approaches to vector control or eradication (e.g., sterile male release and release of insects with dominant lethal genes), long-distance dispersal and population connectivity can have important effects on surrounding non-target populations, but the effects depend on the control method used.21 Intercity dispersal argues for a statewide or regional approach to attempts at vector eradication or control. The evidence presented here for absence of barriers to dispersal and the importance of human-aided dispersal may be important considerations for efforts to combat recent increases in dengue activity in Florida.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to L. P. Lounibos, G. F. O’Meara, and W. J. Tabachnick for their input and laboratory space and B. Kesavaraju, P. Leisnham, and D. Bustamante for help with collecting.
Footnotes
Financial support: This study was funded by Phi Sigma and Graduate Student Association Grants from Illinois State University (to K.D.), National Institutes of Health Grant AI R15-068692-01 (to S.A.J. and S.S.L.), and a grant from Illinois State University (to S.A.J.).
Authors’ addresses: Kavitha Damal, Steven A. Juliano, and Sabine S. Loew, School of Biological Sciences, Illinois State University, Normal, IL, E-mails: Kavitha.damal@hsc.utah.edu, sajulian@ilstu.edu, and ssloew@ilstu.edu. Ebony G. Murrell, Department of Entomology, University of Wisconsin, Madison, WI, E-mail: murrell2@wisc.edu. Jan E. Conn, Wadsworth Center, NYS Department of Health, Albany, NY, E-mail: jconn@wadsworth.org.
References
- 1.Tabachnick WJ. Evolutionary genetics and arthropod-borne disease: the yellow fever mosquito. Am Entomol. 1991;37:14–26. [Google Scholar]
- 2.Josseran L, Paquet C, Zehgnoun A, Caillere N, Le Tertre A, Solet JL, Ledrans M. Chikungunya disease outbreak, Reunion Island. Emerg Infect Dis. 2006;12:1994–1995. doi: 10.3201/eid1212.060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. Chikv Study Group Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 4.Pinheiro FP, Corber SJ. Global situation of dengue and dengue haemorrhagic fever, and its emergence in the Americas. World Health Stat Q. 1997;50:161–169. [PubMed] [Google Scholar]
- 5.Gubler DJ, Reiter P, Ebi KL, Yap W, Nasci R, Patz JA. Climate variability and change in the United States: potential impacts on vector- and rodent-borne diseases. Environ Health Perspect. 2001;109((Suppl 2)):223–233. doi: 10.1289/ehp.109-1240669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson M. Dengue Virus Returns to Florida After More Than 50 Years, UF Researchers Say. 2009. news.ufl.edu/2009/11/23/dengue/ Available at. Accessed December 20, 2011.
- 7.Merrill SA, Ramberg FB, Hagedorn HH. Phylogeography and population structure of Aedes aegypti in Arizona. Am J Trop Med Hyg. 2005;72:304–310. [PubMed] [Google Scholar]
- 8.Soper FL. The 1964 status of Aedes aegypti eradication and yellow fever in the Americas. Am J Trop Med Hyg. 1965;14:887–891. doi: 10.4269/ajtmh.1965.14.887. [DOI] [PubMed] [Google Scholar]
- 9.Moncayo AC, Fernandez Z, Ortiz D, Diallo M, Sall A, Hartman S, Davis CT, Coffey L, Mathiot CC, Tesh RB, Weaver SC. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg Infect Dis. 2004;10:1790–1796. doi: 10.3201/eid1010.030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrington LC, Edman JD, Scott TW. Why do female Aedes aegypti (Diptera: Culicidae) feed preferentially and frequently on human blood? J Med Entomol. 2001;38:411–422. doi: 10.1603/0022-2585-38.3.411. [DOI] [PubMed] [Google Scholar]
- 11.Duenas JC, Llinas GA, Panzetia-Dutari GM, Gardenal CN. Two different routes of colonization of Aedes aegypti in Argentina from neighboring countries. J Med Entomol. 2009;46:1344–1354. doi: 10.1603/033.046.0613. [DOI] [PubMed] [Google Scholar]
- 12.Gill J, Stark LM, Clark GG. Dengue surveillance in Florida, 1997–98. Emerg Infect Dis. 2000;6:30–35. doi: 10.3201/eid0601.000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Meara GF, Evans LF, Gettman AD, Cuda JP. Spread of Aedes albopictus and decline of Aedes aegypti (Diptera: Culicidae) in Florida. J Med Entomol. 1995;32:554–562. doi: 10.1093/jmedent/32.4.554. [DOI] [PubMed] [Google Scholar]
- 14.Reiter P, Lathrop S, Bunning M, Biggerstaff B, Singer D, Tiwari T, Baber L, Amador M, Thirion J, Hayes J, Seca C, Mendez J, Ramirez B, Robinson J, Rawlings J, Vorndam V, Waterman S, Gubler D, Clark G, Hayes E. Texas lifestyle limits transmission of dengue virus. Emerg Infect Dis. 2003;9:86–89. doi: 10.3201/eid0901.020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goncalves da Silva A, Cunha IC, Santos WS, Luz SL, Ribolla PE, Abad-Franch F. Gene flow networks among American Aedes aegypti populations. Evol Appl. 2012;5:664–676. doi: 10.1111/j.1752-4571.2012.00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O'Neil P, Parker IM, Thompson JN, Weller SG. The population biology of invasive species. Annu Rev Ecol Syst. 2001;32:305–332. [Google Scholar]
- 17.Lee CE. Evolutionary genetics of invasive species. Trends Ecol Evol. 2002;17:386–391. [Google Scholar]
- 18.Kolar CS, Lodge DM. Progress in invasion biology: predicting invaders. Trends Ecol Evol. 2001;16:199–204. doi: 10.1016/s0169-5347(01)02101-2. [DOI] [PubMed] [Google Scholar]
- 19.Kolbe JJ, Glor RE, Rodriguez Schettino L, Lara AC, Larson A, Losos JB. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–181. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- 20.Price TD, Sol D. Introduction: genetics of colonizing species. Am Nat. 2008;172((Suppl 1)):S1–S3. doi: 10.1086/588639. [DOI] [PubMed] [Google Scholar]
- 21.Yakob L, Alphey L, Bonsall MB. Aedes aegypti control: the concomitant role of competition, space and transgenic technologies. J Appl Ecol. 2008;45:1258–1265. [Google Scholar]
- 22.Huber K, Mousson L, Rodhain F, Failloux AB. Isolation and variability of polymorphic microsatellite loci in Aedes aegypti, the vector of dengue viruses. Mol Ecol Notes. 2001;1:219–222. [Google Scholar]
- 23.Birungi J, Munstermann LE. Genetic structure of Aedes albopictus (Diptera: Culicidae) populations based on mitochondrial ND5 sequences: evidence for an independent invasion into Brazil and United States. Ann Entomol Soc Am. 2002;95:125–132. [Google Scholar]
- 24.Gorrochotegui-Escalante N, Munoz ML, Fernandez-Salas I, Beaty BJ, Black WC. Genetic isolation by distance among Aedes aegypti populations along the northeastern coast of Mexico. Am J Trop Med Hyg. 2000;62:200–209. doi: 10.4269/ajtmh.2000.62.200. [DOI] [PubMed] [Google Scholar]
- 25.Bosio CF, Harrington LC, Jones JW, Sithiprasasna R, Norris DE, Scott TW. Genetic structure of Aedes aegypti populations in Thailand using mitochondrial DNA. Am J Trop Med Hyg. 2005;72:434–442. [PubMed] [Google Scholar]
- 26.Costa-da-Silva AL, Capurro ML, Bracco JE. Genetic lineages in the yellow fever mosquito Aedes (Stegomyia) aegypti (Diptera: Culicidae) from Peru. Mem Inst Oswaldo Cruz. 2005;100:539–544. doi: 10.1590/s0074-02762005000600007. [DOI] [PubMed] [Google Scholar]
- 27.Herrera F, Urdaneta L, Rivero J, Zoghbi N, Ruiz J, Carrasquel G, Martínez JA, Pernalete M, Villegas P, Montoya A. Population genetic structure of the dengue mosquito Aedes aegypti in Venezuela. Mem Inst Oswaldo Cruz. 2006;101:625–633. doi: 10.1590/s0074-02762006000600008. [DOI] [PubMed] [Google Scholar]
- 28.Paduan KDS, Ribolla PEM. Mitochondrial DNA polymorphism and heteroplasmy in populations of Aedes aegypti in Brazil. J Med Entomol. 2008;45:59–67. doi: 10.1603/0022-2585(2008)45[59:mdpahi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 29.Bracco JE, Capurro ML, Lourenço-de-Oliveira R, Sallum MAM. Genetic variability of Aedes aegypti in the Americas using a mitochondrial gene: evidence of multiple introductions. Mem Inst Oswaldo Cruz. 2007;102:573–580. doi: 10.1590/s0074-02762007005000062. [DOI] [PubMed] [Google Scholar]
- 30.Moore M, Sylla M, Goss L, Burugu MW, Sang R, Kamau LW, Kenya EU, Bosio C, de Lourdes Munoz M, Sharakova M. Dual African origins of global Aedes aegypti sl populations revealed by mitochondrial DNA. PLoS Negl Trop Dis. 2013;7:e2175. doi: 10.1371/journal.pntd.0002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szalanski AL, Owens CB, Lewter JA, Broce AB. Genetic structure of Aedes vexans (Diptera: Culicidae) populations from central United States based on mitochondrial ND5 sequences. Ann Entomol Soc Am. 2006;99:157–163. [Google Scholar]
- 32.Venkatesan M, Westbrook CJ, Hauer MC, Rasgon JL. Evidence for a population expansion in the West Nile virus vector Culex tarsalis. Mol Biol Evol. 2007;24:1208–1218. doi: 10.1093/molbev/msm040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simard F, Licht M, Besansky NJ, Lehmann T. Polymorphism at the defensin gene in the Anopheles gambiae complex: testing different selection hypotheses. Infect Genet Evol. 2007;7:285–292. doi: 10.1016/j.meegid.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 35.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 36.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 37.Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 38.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity (Edinb) 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 42.Legendre P, Fortin M-J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol Ecol Resour. 2010;10:831–844. doi: 10.1111/j.1755-0998.2010.02866.x. [DOI] [PubMed] [Google Scholar]
- 43.Manly BFJ. RT, A Program for Randomization Testing. Dunedin, New Zealand: University of Otago, Center for Applications of Statistics and Mathematics; 1997. [Google Scholar]
- 44.Fortin M-J, Payett S. How to test the significance of the relation between spatially autocorrelated data at the landscape scale: a case study using fire and forest maps. Ecoscience. 2002;9:213–218. [Google Scholar]
- 45.Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- 46.Huber K, Loan LL, Chantha N, Failloux AB. Human transportation influences Aedes aegypti gene flow in Southeast Asia. Acta Trop. 2004;90:23–29. doi: 10.1016/j.actatropica.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Gorrochotegui-Escalante N, Gomez-Machorro C, Lozano-Fuentes S, Fernandez-Salas I, Munoz Md L, Farfan-Ale JA, Garcia-Rejon J, Beaty BJ, Black WC., IV Breeding structure of Aedes aegypti populations in Mexico varies by region. Am J Trop Med Hyg. 2002;66:213–222. doi: 10.4269/ajtmh.2002.66.213. [DOI] [PubMed] [Google Scholar]
- 48.Paupy C, Chantha N, Reynes JM, Failloux AB. Factors influencing the population structure of Aedes aegypti from the main cities in Cambodia. Heredity (Edinb) 2005;95:144–147. doi: 10.1038/sj.hdy.6800698. [DOI] [PubMed] [Google Scholar]
- 49.Hemme RR, Thomas CL, Chadee DD, Severson DW. Influence of urban landscapes on population dynamics in a short-distance migrant mosquito: evidence for the dengue vector Aedes aegypti. PLoS Negl Trop Dis. 2010;4:e634. doi: 10.1371/journal.pntd.0000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haag CR, Riek M, Hottinger JW, Pajunen VI, Ebert D. Genetic diversity and genetic differentiation in Daphnia metapopulations with subpopulations of known age. Genetics. 2005;170:1809–1820. doi: 10.1534/genetics.104.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Premoli AC, Chischilly S, Mitton JB. Levels of genetic variation captured by four descendant populations of Pinyon pine (Pinus edulis Engelm.) Biodivers Conserv. 1994;3:331–340. [Google Scholar]
- 52.Clegg SM, Degnan SM, Kikkawa J, Moritz C, Estoup A, Owens IP. Genetic consequences of sequential founder events by an island-colonizing bird. Proc Natl Acad Sci USA. 2002;99:8127–8132. doi: 10.1073/pnas.102583399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdelkrim J, Pascal M, Samadi S. Island colonization and founder effects: the invasion of the Guadeloupe islands by ship rats (Rattus rattus) Mol Ecol. 2005;14:2923–2931. doi: 10.1111/j.1365-294X.2005.02604.x. [DOI] [PubMed] [Google Scholar]
- 54.Lima RS, Jr, Scarpassa VM. Evidence of two lineages of the dengue vector Aedes aegypti in the Brazilian Amazon, based on mitochondrial DNA ND4 gene sequences. Genet Mol Biol. 2009;32:414–422. doi: 10.1590/S1415-47572009005000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonseca DM, Widdel AK, Hutchinson M, Spichiger SE, Kramer LD. Fine-scale spatial and temporal population genetics of Aedes japonicus, a new US mosquito, reveal multiple introductions. Mol Ecol. 2010;19:1559–1572. doi: 10.1111/j.1365-294X.2010.04576.x. [DOI] [PubMed] [Google Scholar]
- 56.Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, Chang MS, Walton C. Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: implications for past and future population genetic studies. BMC Genet. 2009;10:11. doi: 10.1186/1471-2156-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Black WC, Bernhardt S. Abundant nuclear copies of mitochondrial origin (NUMTs) in the Aedes aegypti genome. Insect Mol Biol. 2009;18:705–713. doi: 10.1111/j.1365-2583.2009.00925.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.