Abstract
Background
Inner ear development involves signaling from surrounding tissues, including the adjacent hindbrain, periotic mesenchyme and notochord. These signals include SHH, FGFs, BMPs and WNTs from the hindbrain and SHH from the notochord. Zic genes, which are expressed in the dorsal neural tube and act during neural development, have been implicated as effectors of these pathways. This report examines whether Zic genes’ involvement in inner ear development is a tenable hypothesis based on their expression patterns.
Results
In the developing inner ear of both the chick and mouse, all of the Zic genes were expressed in the dorsal neural tube and variably in the periotic mesenchyme, but expression of the Zic genes in the otic epithelium was not found. The onset of expression differed among the Zic genes; within any given region surrounding the otic epithelium, multiple Zic genes were expressed in the same place at the same time.
Conclusions
Zic gene expression in the region of the developing inner ear is similar between mouse and chick. Zic expression domains overlap with sites of WNT and SHH signaling during otocyst patterning, suggesting a role for Zic genes in modulating signaling from these pathways.
Keywords: Zic gene expression, inner ear, embryogenesis, mouse, chick
Introduction
The Zic (zinc finger of the cerebellum) genes comprise a family of transcription factors found in both vertebrates and invertebrates (reviewed in Ali et al., 2012) that are related to the Drosophila Zic homologue, odd-paired (opa) (Nusslein-Volhard and Wieschaus, 1980; Benedyk et al., 1994). Among vertebrate species, humans and mice have five Zic genes (Zic1-5); other species have more (zebrafish, 8 Zic genes; frog, 6 Zic genes) or fewer (chicken, 4 Zic genes) Zic genes. Structurally, ZIC proteins are characterized by a zinc finger region that contains five tandem C2H2 zinc finger domains (Aruga, 2004; Merzdorf, 2007; Ali et al., 2012). The amino acid sequence of zinc fingers 2 though 5 is almost identical among Zic1-5 for a given species, while the sequence outside of this region can be highly variable (Aruga, 2004; Grinberg and Millen, 2005).
The Zic genes are involved in a multitude of developmental processes. Zic gene function has predominantly been studied during neural development, where they have roles in controlling neural patterning, in generation of neural crest, in cerebellar development, and in the formation of the neural tube (Aruga, 2004; Merzdorf, 2007). However, Zic genes also function in somite myogenesis (Pan et al., 2011), left-right asymmetry (Herman and El-Hodiri, 2002), and retinal development. Although there have been a number of studies focused on Zic gene function, the exact role these genes play in vivo has yet to be elucidated. In vitro, ZIC2 inhibits WNT/β-catenin signaling through a direct interaction with TCF4 (Pourebrahim et al., 2011) and ZIC proteins interact with GLI transcription factors to either suppress or enhance GLI-mediated transactivation (Koyabu et al., 2001; Mizugishi et al., 2001; Pan et al., 2011). In Xenopus, Zic genes both activate and inhibit WNT/β-catenin signaling (Merzdorf and Sive, 2006; Pourebrahim et al., 2011; Fujimi et al., 2012), and in zebrafish, Zic genes regulate expression of shh and nodal and modulate Hedgehog-mediated gene expression (Maurus and Harris, 2009; Sanek et al., 2009).
Given the importance of Zic gene expression in the neural tube, as well as the ability of Zic genes to modulate WNT and SHH signaling, key pathways known to be involved in dorso-ventral patterning of the inner ear (Riccomagno et al., 2002; Riccomagno et al., 2005), surprisingly little attention has been paid to Zic gene expression and function in and around the developing inner ear. One early study identified Zic2 as one of the first genes up-regulated in the regenerating sensory epithelium of the chicken inner ear after noise exposure (Gong et al., 1996). Conflicting data exist regarding the location of Zic gene expression during inner ear development (Warner et al., 2003; McMahon and Merzdorf, 2010). Although some Zic expression patterns have been described in the mouse (reviewed in Merzdorf, 2007; Ali et al., 2012), to date Zic expression in the developing mouse inner ear has not been examined. Given the discrepancies in the Zic gene expression patterns described in the earlier studies, we have undertaken a more extensive comparative spatiotemporal analysis of Zic mRNA expression in the otic regions of both the chick and mouse during the early stages of inner ear development.
Results
Evolutionary Conservation of the Zic Genes Across Species
Phylogenetic analysis of ZIC proteins from mouse (ZIC1-5), human (ZIC1-5), chicken (ZIC1-4), zebrafish (Zic1/Opl, Zic1-like, Zic2a and b, and Zic3-6), frog (Zic1, Zic2a and 2b, and Zic3-5), and fly (Odd-paired; OPA) indicate significant conservation among species (Fig. 1A). ZIC proteins from across species clustered together into five groups, with each representing one of the 5 Zic genes. In addition to these 5 clusters, Zic6 and Zic1-like from zebrafish grouped together with the ancestral protein from the fly, odd-paired, into a 6th cluster. The ZIC1, ZIC2, and ZIC3 clusters group together, the ZIC5 and odd-paired clusters group together, and the ZIC4 proteins form two groups (mouse/human, and chick/frog/fish) that cluster together away from all of the other Zics. Within each of the five major clusters (ZIC1-5), the mouse and human genes are closely related to one another, and more distantly related to those from chicken, zebrafish, and frog.
Figure 1. Comparison of Zic genes across species.
(A) Phylogenetic tree showing the relationships among the Zic genes from human, mouse, chick, zebrafish, and frog, as well as the ancestral gene odd-paired (opa) from Drosophila. The complete amino acid sequences were used for the construction of the tree. (B) Percent identity and similarity of the amino acid sequences among the mouse, chick, and human ZIC proteins. The chick ZIC2, ZIC3, and ZIC4 protein sequences have not been finalized, so comparisons to these protein sequences result in much lower percent identity and similarity. Abbreviations: Dm, Drosophila melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Xl, Xenopus laevis
Identity and similarity comparisons among the complete amino acid sequences of the mouse, human, and chicken ZIC proteins indicates that the mouse and human ZIC1, ZIC2, and ZIC3 proteins are almost identical, but the ZIC4 and ZIC5 proteins differ slightly (Fig. 1B). Chicken Zic1 is very similar to both mouse and human ZIC1 proteins, but chicken Zic2-Zic4 differ greatly from mouse and human ZIC2-4, with Zic4 being the most different. Zic1-3 are the most similar between chicken and mouse and human, while Zic4 is not as conserved. ZIC1-3 contain a Zic-opa conserved (ZOC) domain, resulting in a higher level of homology among these proteins, while ZIC4 and ZIC5 do not. The ZOC domain is required for activation of target gene transcription in vitro, as well as for binding I-mfa, a myogenic repressor protein (Mizugishi et al., 2004). Further, the intron-exon boundaries are conserved between ZIC1-3, but not between ZIC1-3 and either ZIC4 or ZIC5 (Grinberg and Millen, 2005). This suggests that the Zic4 and Zic5 genes are the most divergent and may have evolved novel functions. ZIC1 is almost identical in mouse, human, and chicken and therefore may have the same function in all three organisms. ZIC2 and ZIC3 are almost identical between mouse and human, but differ when compared to chick Zic2 and Zic3, suggesting that they may have different functions in birds and mammals.
Comparison of otic development in chick and mouse
The early stages of inner ear development are similar in both the chick and in the mouse (summarized in Fig. 2), and require signaling from different tissues to specify and pattern the otic placode (underlying mesoderm), otocyst (mesenchyme, neural tube, notochord), and inner ear (mesenchyme, neural tube, notochord). Initially, cells in the dorsal ectoderm adjacent to rhombomeres 5 and 6 of the hindbrain thicken, forming the otic placode (HH stage 10 in chick (Hamburger and Hamilton, 1992), embryonic day 8 to 8.5 (E8-E8.5) in mouse). Subsequently, the placode invaginates to form the otic cup, and neuroblasts begin to delaminate from the ventral region of the otic cup and migrate away to form the cochleovestibular ganglion (CVG)/statoacoustic ganglion (SAG) (HH stage 13 in chick, E8.75-E9 in mouse). The otic cup closes completely and pinches off from the overlying ectoderm, forming the otic vesicle/otocyst (HH stage 17 in chick, E9.5 in mouse). Over the next several days the otocyst undergoes complex morphological changes, transforming into a complex structure comprised of three semicircular canals, the utricle, the saccule, and the cochlea (mouse)/basilar papilla (chick).
Figure 2. Inner ear development in chicken and mouse.
Schematic depicting inner ear development in chicken from stage 13 to stage 32 (top row) and in mouse from E9.5 to E13.5 (lower row). Gray shaded regions correspond to the neural tube and the notochord. Images are based upon transverse sections through the middle of the otocyst, but are not to scale. Abbreviations: sag, statoacoustic ganglion; cvg, cochleovestibular ganglion; d, dorsal; m, medial.
Expression of the Zic genes during otic development in chick and mouse
We used in situ hybridization to examine the expression of the Zic genes (Zic1-4 in chick, Zic1-5 in mouse; refer to Fig. 3 for the location of the riboprobes within each gene) in the hindbrain and in the adjacent region of the developing inner ear in the chick (HH stage 13 to HH stage 32) and in the mouse (E9.5 to E13.5). Pax2 expression in this region, which has previously been reported for both chick (Hutson et al., 1999; Hidalgo-Sanchez et al., 2000; Sanchez-Calderon et al., 2002; Li et al., 2004; Sanchez-Calderon et al., 2005) and mouse (Nornes et al., 1990; Puschel et al., 1992; Rinkwitz-Brandt et al., 1995; Rinkwitz-Brandt et al., 1996; Lawoko-Kerali et al., 2002; Burton et al., 2004), is included in the same series of in situ hybridizations as an internal control for otic epithelial gene expression.
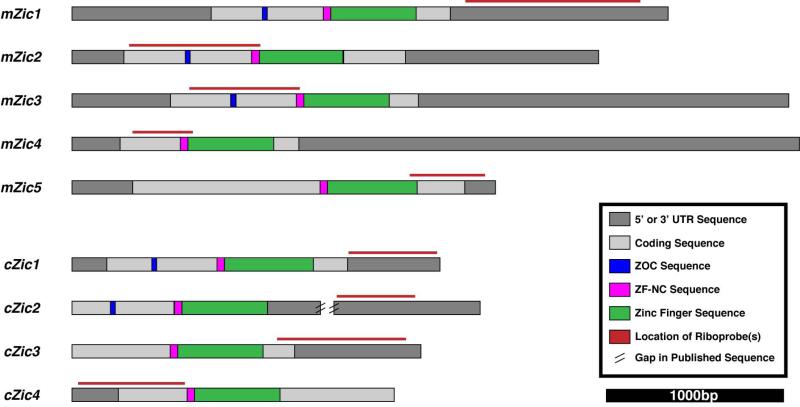
Figure 3. Location of riboprobes within the mRNA sequence of the Zic genes.
Diagrams of the mRNA sequence for each Zic gene illustrate the location of the coding sequence (light gray boxes), UTRs (dark gray boxes), and functional domains (blue, magenta, and green boxes) within each of the Zic genes. Probes for each of the Zic genes in mouse (Zic1-5) and chick (Zic1-4) were designed to target the less-conserved 5’ and 3’ UTR regions of each gene (red lines above the mRNA diagram show the location of the riboprobes). The published sequences for chick Zic2-4 are still being annotated and updated, so the most 5’ and 3’ untranslated regions of the mRNA have not been defined. Abbreviations: UTR, untranslated region; ZOC, Zic-opa conserved domain; ZF-NC, zinc-finger nucleocapsid domain.
Expression of Zic genes in the developing chick inner ear
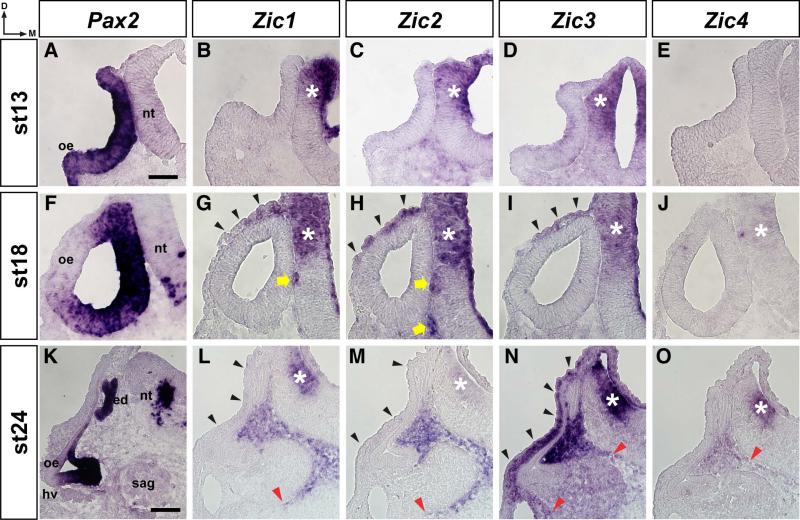
At HH stage 13, Pax2 was expressed throughout the epithelium of the otic cup but not in the adjacent neuroepithelium of the developing neural tube (Fig. 4A). By HH stage 18, the otic cup has closed to form the otocyst (Fig. 2, top row) and Pax2 expression was restricted to the medial and ventral walls of the otic epithelium (Fig. 4F). At this stage, Pax2 expression was also detected in the ventral neural tube. Between HH stages 18 and 24, the spherical otocyst elongates to form the early inner ear with identifiable dorsal (endolymphatic duct and sac) and ventral (basilar papilla) structures (top row, Fig. 2). Pax2 expression at HH stage 24 (Fig. 4K) was further restricted to the endolymphatic duct and prosensory regions of the otic epithelium, and was maintained in the ventral neural tube. In contrast to Pax2, expression of the Zic genes was not detected in the otic epithelium at HH stages 13, 18, or 24 (Fig. 4). Expression of Zic1-3 was detected in the dorsal neural tube at HH stages 13, 18, and 24, but Zic4 expression was not detected in the dorsal neural tube until HH stages 18 (weak) and 24 (asterisks in Fig. 4B, 4C, 4D, 4G, 4H, 4I, 4J, 4L, 4M, 4N, 4O). Following otocyst closure at HH stage 18, Zic1-3 expression was observed laterally around the outside of the otocyst (black arrowheads, Fig. 4G, 4H, 4I), and Zic1 and Zic2 expression was detected medially between the otic epithelium and the neural tube (yellow arrows, Fig. 4G, 4H). By HH stage 24, expression of Zic1-4 was observed medially between the developing inner ear and the neural tube, with the ventral extent of expression varying among the Zic genes (Fig. 4L, 4M, 4N, 4O; the red arrowheads indicates ventral extent of Zic-expressing cells). Weak Zic1 and Zic2 expression, as well as strong Zic3 expression, was observed lateral to the developing inner ear (black arrowheads, Fig. 4L, 4M, 4N).
Figure 4. Zic expression in the otic region of early stage chick embryos.
In situ hybridization on 12μm transverse sections through the otocyst of stage 13 (A-E), stage 18 (F-J), and stage 24 (K-O) chick embryos using probes for Pax2 (A, F, K), Zic1 (B, G, L), Zic2 (C, H, M), Zic3 (D, I, N), and Zic4 (E, J, O). Note that Pax2 is expressed in sensory regions of the otic epithelium, while Zic gene expression is restricted to the surrounding mesenchyme. Abbreviations: oe, otic epithelium; ed, endolymphatic duct; nt, neural tube; sag, statoacoustic ganglion; hv, head vein; d, dorsal; m, medial. Asterisks identify Zic expression in the neural tube (B, C, D, G, H, I, J, L, M, N, O). Black arrowheads indicate Zic-expressing cells between the outer epithelium and the otic epithelium (G, H, I, L, M, N). Yellow arrow indicates Zic-expressing cells between neural tube and otic epithelium (G, H). Red arrowheads indicate the ventral-most extent of Zic expression in the mesenchyme (L, M, N, O). Scale bar in A, 50μm (applies to A-J); scale bar in K, 100μm (applies to K-O).
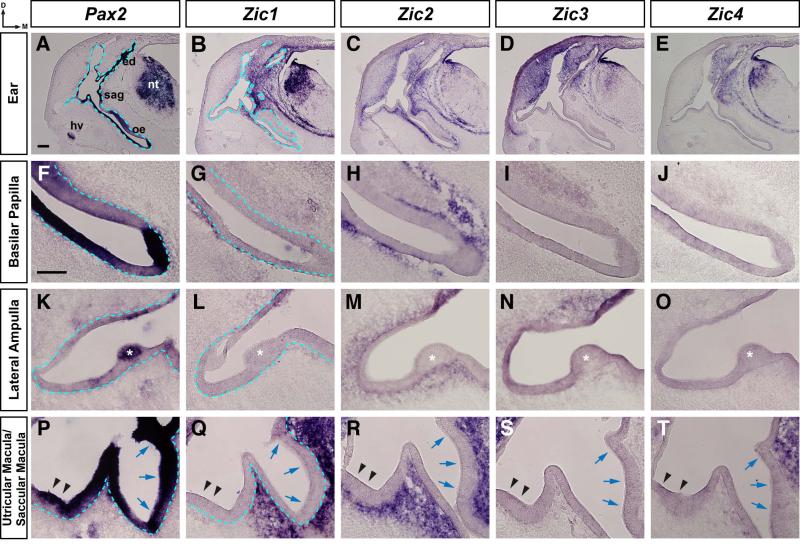
Between HH stages 24 and 32, the developing inner ear has undergone further morphological changes, resulting in an inner ear with clearly defined structures with identifiable prosensory domains (Fig. 2, top row; Fig. 5). Pax2 was expressed in the endolymphatic duct and the neuroepithelium of the neural tube (Fig. 5A; blue dashed line marks border between otic epithelium and the surrounding mesenchyme) and in the emerging sensory patches within the otic epithelium of the basilar papilla (Fig. 5F), the crista of the lateral semicircular canal (asterisk, Fig. 5K), the utricular macula (arrowheads, Fig. 5P), and the saccular macula (blue arrows, Fig. 5P). Zic1-4 expression was not detected in the otic epithelium at HH stage 32 (Fig. 5). However, Zic1-4 expression was detected in the dorsal neural tube and adjacent to the otic epithelium (Fig. 5B, 5C, 5D, 5E; a blue dashed line in Fig. 5B marks the border between the otic epithelium and the surrounding mesenchyme). Zic2 was the only Zic gene expressed surrounding the entire otic epithelium, including the developing basilar papilla (Fig. 5H), lateral semicircular canal (Fig. 5M), and the utricular and saccular maculae (Fig. 5R). Expression of the other Zic genes was restricted to the mesenchyme adjacent to specific regions of the otic epithelium. Zic1 expression was limited to the dorsomedial region of the mesenchyme between the otic epithelium and neural tube, but was also detected adjacent to a portion of the ventral basilar papilla (Fig. 5G), next to the dorsolateral wall of the ear (Fig. 5L), and surrounding the saccular macula but not the utricular macula (Fig. 5Q). Zic3 expression was restricted to cells surrounding the dorsal region of the developing inner ear, including cells adjacent to the dorsal wall of the lateral semicircular canal (Fig. 5N), but was not found in cells adjacent to the basilar papilla (Fig. 5I) or the utricular and saccular maculae (Fig. 5S). Zic4 had the most restricted expression pattern, expressed in the mesenchyme in the dorsomedial region between the otic epithelium and neural tube, including in cells adjacent to the dorsomedial portion of the saccular macula (Fig. 5T), but not in cells adjacent to the basilar papilla (Fig. 5J) or in cells adjacent to the lateral semicircular canal (Fig. 5O).
Figure 5. Zic expression in the otic region of stage 32 chick embryos.
In situ hybridization on 25μm transverse sections through the otocyst of stage 32 chick embryos using probes for Pax2 (A, F, K, P), Zic1 (B, G, L, Q), Zic2 (C, H, M, R), Zic3 (D, I, N, S), and Zic4 (E, J, O, T). Expression at low magnification in the ear (A-E), and at higher magnification in the basilar papilla (F-J), Lateral ampulla (K-O) and utricular/saccular maculae (P-T). Note that Pax2 is expressed in sensory regions of the otic epithelium, while Zic gene expression is restricted to the surrounding mesenchyme. In panels 5L-5N, the otic epithelium at the upper right corner is darkened as a result of tissue folding. Abbreviations: oe, otic epithelium; ed, endolymphatic duct; nt, neural tube; gl, ganglion; hv, head vein; d, dorsal; m, medial. Asterisks identify the lateral ampulla (K, L, M, N, O). Black arrowheads indicate the utricular macula and blue arrows denote the saccular macula (P, Q, R, S, T). Blue dashed line defines border between the otic epithelium and the mesenchyme (first two columns). Scale bar in A, 200μm (applies to A-E); scale bar in F, 100μm (applies to F-T).
Expression of Zic genes in the developing mouse inner ear
At E9.5, Pax2 was expressed in ventromedial wall of the otocyst but not in the adjacent neuroepithelium of the developing neural tube of the mouse (Fig. 6A). By E10.5, the otocyst has begun to elongate (Fig. 2, bottom row) and Pax2 expression remained in the lateral and ventromedial wall of the otocyst (Fig. 6G). Between E10.5 and E11.5, the otocyst undergoes further morphological changes, elongating along its dorso-ventral axis and becoming compacted along its medio-lateral axis, forming the endolymphatic duct at the dorsal end and the start of the cochlear duct at the ventral end (Fig. 2, bottom row). At E11.5, Pax2 was expressed in the neural tube, in the endolymphatic duct, and in the developing sensory patches of the otic epithelium, but not in the mesenchyme surrounding the developing inner ear (Fig. 6M; a blue dashed line marks the border between the otic epithelium and the surrounding mesenchyme). In contrast to Pax2, expression of Zic1-5 was not detected in the otic epithelium at E9.5, E10.5, or E11.5 (Fig. 6). Zic1-5 expression was seen in the dorsal neural tube at E9.5, E10.5, and E11.5 (asterisks in Fig. 6B-6F, 6H-6L, 6N-6R). Zic1- and Zic2-expressing cells were found in the mesenchyme between the ventral neural tube and the otic epithelium and adjacent to the dorsolateral side of the otocyst at E9.5 and E10.5, although the Zic2-expressing cells were not seen as far ventrally as were the Zic1-expressing cells (yellow arrow and black arrowheads, Fig. 6B, 6C, 6H, 6I). Zic3- and Zic4-expressing cells were also found adjacent to the dorsolateral side of the otocyst at E10.5 (black arrowheads, Fig. 6J, 6K). By E11.5, Zic1 expression was seen in cells located medially between the otic epithelium and the neural tube (Fig. 6N; a blue dashed line marks the border between otic epithelium and the surrounding mesenchyme), while Zic2- and Zic5-expressing cells surrounded the entire otic epithelium, although Zic5 was expressed more strongly adjacent to the dorsal region of the otic epithelium (Fig. 6O, 6R). Zic3 and Zic4 expression was restricted to the mesenchyme surrounding the dorsal half of the otic epithelium, with Zic3 expression extending further ventrally than Zic4 on the lateral side of the otic epithelium and Zic4 expression extending further ventrally than Zic3 on the medial side of the otic epithelium (Fig. 6P, 6Q; red arrowheads denote the ventral extent of expression).
Figure 6. Zic expression in the otic region of mouse embryos between E9.5 and E11.5.
In situ hybridization on 12μm transverse sections through the otocyst of E9.5 (A-F), E10.5 (G-L), and E11.5 (M-R) mouse embryos using probes for Pax2 (A, G, M), Zic1 (B, H, N), Zic2 (C, I, O), Zic3 (D, J, P), Zic4 (E, K, Q), and Zic5 (F, L, R). Note that Pax2 is expressed in the otic epithelium, while Zic gene expression is restricted to the surrounding mesenchyme. Abbreviations: otc, otocyst; oe, otic epithelium; nt, neural tube; ed, endolymphatic duct; gl, ganglion; hv, head vein; d, dorsal; m, medial. Asterisks identify Zic expression in the neural tube (B-F, H-L, N-R). Black arrowheads indicate Zic-expressing cells between the surface ectoderm and the otic epithelium (B, C, H, I, J, K). Yellow arrows indicate Zic-expressing cells between neural tube and otic epithelium (B, C, H, I). Red arrowheads indicate the ventral-most extent of Zic expression in the mesenchyme (P, Q). Blue dashed line defines border between the otic epithelium and the mesenchyme (M, N). Scale bar in A, 50μm (applies to A-L); scale bar in M, 200μm (applies to M-R).
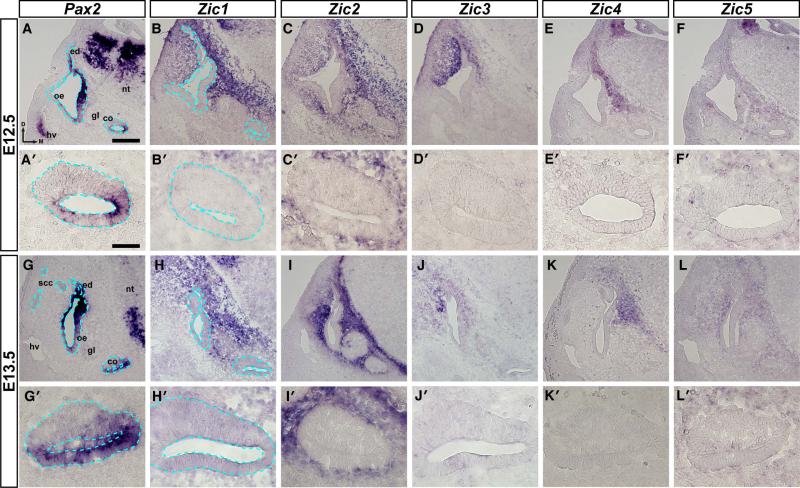
At E12.5 and E13.5, Pax2 was expressed in the neural tube, endolymphatic duct, and the developing sensory patches of the otic epithelium (Fig. 7A, 7G; a blue dashed line marks the border between the otic epithelium and the surrounding mesenchyme), including the developing cochlear duct (Fig. 7A’, 7G’), but was not found in the mesenchyme surrounding the developing inner ear. Zic1-5 were expressed in the dorsal neural tube (data not shown), but in contrast to Pax2, they were not expressed in the otic epithelium (Fig. 7B-F, 7B’-F’, 7H-L, 7H’-L’). Zic1 was expressed in cells in the mesenchyme adjacent to all portions of the otic epithelium except for the ventral portions of the ear and the CVG (Fig. 7B, 7B’, 7H, 7H’; a blue dashed line marks the border between the otic epithelium and the surrounding mesenchyme). Zic2 and Zic5 expression was seen in cells in the mesenchyme that surrounded the entire otic epithelium, including the cochlea, although the expression of Zic5 was much weaker (Fig. 7C, 7C’, 7F, 7F’, 7I, 7I’, 7L, 7L’). Expression of Zic3 was restricted to cells surrounding the dorsal half of the developing inner ear and was strongest in cells adjacent to the dorsolateral region of the otic epithelium (Fig. 7D, 7D’, 7J, 7J’). Zic4 expression was the most restricted of the Zic genes at these stages, with Zic4-expressing cells only detected medially in the mesenchyme between the neural tube and otic epithelium (Fig.7E, 7E’, 7K, 7K’).
Figure 7. Zic expression in the otic region of E12.5 and E13.5 mouse embryos.
In situ hybridization on 12μm transverse sections through the otocyst of E12.5 (A-F, A’-F’) and E13.5 (G-L, G’-L’) mouse embryos using probes for Pax2 (A, A’, G, G’), Zic1 (B, B’, H, H’), Zic2 (C, C’, I, I’), Zic3 (D, D’, J, J’), Zic4 (E, E’, K, K’), and Zic5 (F, F’, L, L’). Note that Pax2 is expressed in the otic epithelium, while Zic gene expression is restricted to the surrounding mesenchyme. Abbreviations: oe, otic epithelium; ed, endolymphatic duct; co, cochlea; scc, semicircular canals; nt, neural tube; gl, ganglion; hv, head vein; d, dorsal; m, medial. Blue dashed line defines border between the otic epithelium and the mesenchyme (A, A’, B, B’, G, G’, H, H’). Scale bar in A, 200μm (applies to A-L); scale bar in A’, 50μm (applies to A’-L’).
Identifying Zic-expressing cells
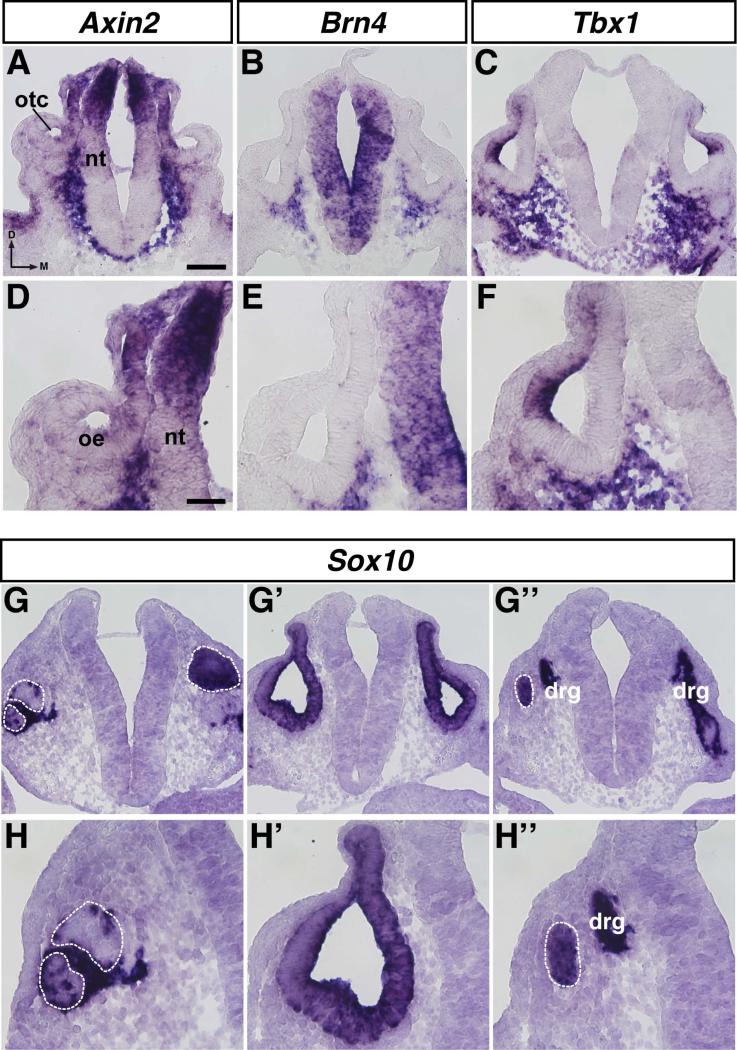
We examined the expression of Axin2, Brn4, Tbx1, and Sox10 in sections at similar levels through the otocyst at E10.5 to attempt to characterize the Zic-expressing cells in the mesenchyme outside of the neuroepithelium and otic epithelium. Axin2, which identifies cells with active Wnt signaling (Jho et al., 2002), was expressed in the dorsal neural tube, the dorsomedial otic epithelium, and in cells adjacent to the neural tube along its entire dorso-ventral axis (Fig. 8A, 8D). A subset of Axin2-expressing cells adjacent to the dorsal neural tube also expressed Zic1-4 (cf. Figs. 6H, 6I, 6J, 6K and Figs. 8A, 8D). In the mesenchyme between the ventral neural tube and the otic epithelium, Axin2 expression overlapped with the expression of Zic1 and Zic2 (cf. Figs. 6H, 6I and Figs. 8A, 8D). Brn4, a marker of condensing mesenchyme (Phippard et al., 1998; Riccomagno et al., 2002), was expressed throughout the neural tube and in cells adjacent to the ventrolateral and ventromedial otic epithelium, but was not expressed in a band of cells immediately adjacent to the neural tube (Fig. 8B, 8E). This expression overlapped with that of Zic1 and Zic2 in the mesenchyme between the ventral neural tube and the otic epithelium; however, the region of Brn4+ cells only partially overlapped the region of Zic+ cells (cf. Figs. 6H, 6I and Figs. 8B, 8E). Expression of Tbx1, a marker of the periotic mesenchyme (Riccomagno et al., 2002; Vitelli et al., 2003; Raft et al., 2004), was also detected in the lateral wall of the otocyst, in most of the cells below the ventral otic epithelium, and between the ventral neural tube and the otic epithelium (Fig. 8C, 8F). Most of the Brn4-expressing cells also expressed Tbx1, which was expected given that Tbx1 marks the periotic mesenchyme, and Brn4 is expressed in condensing cells of the periotic mesenchyme (cf. Figs. 8B, 8E and Figs. 8C, 8F). The mesenchymal expression of Zic1 and Zic2 between the ventral neural tube and the otic epithelium overlapped with Tbx1 expression (cf. Figs. 6H, 6I and Figs. 8C, 8F).
Figure 8. Characterization of Zic-expressing cells in the periotic mesenchyme of E10.5 mouse embryos.
In situ hybridization on 12μm transverse sections through the otocyst of E10.5 mouse embryos using probes for Axin2 (A, D), Brn4 (B, E), Tbx1 (C, F), and Sox10 (G-G’’, HH’’). Abbreviations: oe, otic epithelium; otc, otocyst; nt, neural tube; drg, dorsal root ganglion; d, dorsal; m, medial. White dashed line indicates anterior (G, H) or posterior (G’’, H’’) edge of otocyst. Scale bar in A, 100μm (applies to A-C, G-G’’); scale bar in D, 50μm (applies to D-F, HH’’).
Expression of Sox10, which identifies neural crest cells and is also expressed by cells in the otic epithelium (Watanabe et al., 2000; Breuskin et al., 2009; Bronner, 2012), was examined at three different axial levels through the otocyst: anterior to the otocyst (Fig. 8G, 8H), through the otocyst (Fig. 8G’, 8H’), and posterior to the otocyst (Fig. 8G’’, 8H’’). Anterior to the otocyst, Sox10 expression was detected in the wall of the otocyst but not in the mesenchyme (Fig. 8G, 8H; white dashed lines outline the otocyst). Similar expression was observed through the otocyst (Fig. 8G’, 8H’). Posterior to the otocyst, Sox10 expression was again found in the wall of the otocyst (white dashed line) as well as in neurons of the neural crest-derived dorsal root ganglion adjacent to the neural tube (drg; Fig. 8G’’, 8H’’).
Taken together, these results indicate that the Zic+ cells surrounding the otocyst are not neural crest cells because Sox10 expression was not seen in the same regions in which Zic+ cells were located (Fig. 8G, 8H, 8G’, 8H’, 8G’’, 8H’’). The ventral-most Zic+ cells are most likely periotic mesenchyme, a subset of which is condensing cartilage (that will later form the otic capsule), because they expressed either Tbx1 or Tbx1 and Brn4 (Fig. 8B, 8C, 8E, 8F). The Zic+ region in the ventral mesenchyme and adjacent to the dorsal neural tube overlapped with the Axin2+ region (Fig. 8A, 8D), suggesting that WNT signaling was active in these cells.
Discussion
Distinct spatiotemporal expression of Zic genes during inner ear development in the chick and mouse
This study presents a comprehensive analysis of Zic gene expression in the developing chick and mouse inner ear. Zic genes from both mouse (Zic1-5) and chick (Zic1-4) were found to be expressed in similar patterns during inner ear development—in the dorsal neural tube and throughout the mesenchyme adjacent to the developing inner ear, but not in the otic epithelium. Each Zic gene had a unique pattern of expression at each developmental stage examined, but localized overlapping expression of multiple Zic genes often occurred. The timing of expression also differed, as the expression of Zic1-5 in mouse and Zic1-3 in chick was detected at the earliest time points analyzed in this study (E9.5 in mouse, HH stage 13 in chick), but weak expression of chick Zic4 was not found until HH stage 18, with stronger expression seen by HH stage 24. At E9.5, expression of Zic1 and Zic2 in mouse was seen in both the dorsal neural tube and in the mesenchyme surrounding the otic epithelium, while mesenchymal expression of Zic3-5 in mouse and Zic1-3 in chick was found only after the onset of expression in the neuroepithelium. The differences in spatiotemporal expression of the Zic genes suggests that each Zic gene may be playing a different role during inner ear development. Zic2, which is expressed in the mesenchyme surrounding the entire otic epithelium in both mouse and chick, may be important for the overall growth and shaping of the entire developing inner ear. The other Zic genes, which are expressed in different regions of the periotic mesenchyme, may be responsible for growth and morphological changes associated with specific inner ear structures.
Identity and possible roles of Zic-positive cells in critical tissues during inner ear development
Neural Crest
Our analyses identified Zic-positive cells outside of the neural tube, adjacent to the otic epithelium. We considered the possibility that these cells were migrating cranial neural crest. However, several lines of evidence argue against this. Previous work in the chick demonstrated that Zic1-positive cells in the trunk outside of the neural tube were not labeled with HNK-1, a marker of migratory neural crest cells (Sun Rhodes and Merzdorf, 2006). Additionally, the timing, location, and neural crest marker expression of these cells does not correlate with that of migrating cranial neural crest cells. In the mouse, cranial neural crest cells form and begin migrating at the 5 somite stage (Chan and Tam, 1988) and finish migrating by the 14 somite stage (Serbedzija et al., 1992), while in the chick, neural crest cells migrate from HH stage 9+ to HH stage 11 (Tosney, 1982). The Zic+ cells we detected in the mesenchyme were seen in much older embryos—22-24 somite stage (E9.5) and older in the mouse, and HH stage 18 and older in the chick. Second, DiI labeling studies in chick embryos and lineage tracing studies in mouse embryos indicate that neural crest cells from rhombomere 5 migrate rostrally and caudally around the otocyst and arrive in the second and third branchial arches, while neural crest cells from rhombomere 6 migrate caudally around the otocyst before arriving in the third branchial arch (Sechrist et al., 1993; Trainor et al., 2002). The sections in which Zic+ cells were detected in the mesenchyme were taken midway through the otocyst in a region where neural crest cells do not migrate. Finally, we used Sox10 expression as a marker for migrating neural crest cells in the mouse (Bronner, 2012). Sox10 expression was not detected outside of the otic epithelium in sections through the medial portion of the otocyst at E10.5, but was found in sections at the most anterior or posterior levels of the otocyst. Zic1-4+ cells at E10.5 were detected outside of the neural tube in sections through the medial portions of the otocyst, suggesting that these Zic-expressing cells were not of neural crest origin.
Periotic Mesenchyme
We examined the expression of Brn4 and Tbx1, markers of condensing mesenchyme and periotic mesenchyme, in the regions where we found Zic-expressing cells. The region of Tbx1 expression overlapped with the regions of Zic1 and Zic2 expression, but the region of Brn4 expression only overlapped slightly with the regions of Zic1 and Zic2 expression. This suggests that the majority of Zic1+ and Zic2+ cells in the ventral mesenchyme are Tbx1+ periotic mesenchyme cells, and a few of these cells are Brn4+ cells of the condensing mesenchyme. Expression of Tbx1 and Brn4 is regulated by SHH signaling (Riccomagno et al., 2002), and Zic genes regulate SHH signaling in vitro (Koyabu et al., 2001; Mizugishi et al., 2001; Pan et al., 2011), so the expression of Zic genes in the same regions where Tbx1 and Brn4 are expressed could mean that the Zic genes are involved in the SHH-dependent expression of Tbx1 and Brn4. Alternatively, the Tbx1 and Brn4 expression could be independent of Zic expression, suggesting a different role for the Zic genes. Zic genes, like Tbx1 and Brn4, could be required for proper epithelial-mesenchymal signaling. Loss of either Brn4 (Brn4−/−) or Brn4 and a single allele of Tbx1 (Brn4−/−;Tbx1+/−) in the periotic mesenchyme in mice results in improper coiling of the cochlea (Braunstein et al., 2008). Loss of one or multiple Zic genes in the periotic mesenchyme could result in similar defects in inner ear structures, with the specific structures affected depending on which Zic gene is lost.
We also examined Axin2 expression in the mesenchyme. The dorsal-most expression of Axin2 overlapped with the expression of Zic1-4, while the ventral expression overlapped with the expression of Zic1 and Zic2. Axin2 expression induced by WNT signaling acts as part of a negative feedback loop to limit the extent and duration of WNT signaling (Jho et al., 2002). WNT signaling from the dorsal hindbrain and SHH signaling from the notochord and floor plate pattern the otocyst along its dorsal-ventral axis and are critical for the development of the dorsal (WNT) and ventral (SHH) structures of the inner ear (Riccomagno et al., 2002; Riccomagno et al., 2005). In vitro, ZIC2 binding to TCF4 inhibits WNT signaling (Pourebrahim et al., 2011), so the combination of Axin2 expression and Zic expression may selectively modulate WNT signals from the dorsal hindbrain to the otocyst as well as restricting ventral WNT signals closer to the source of SHH. Additional experiments are needed to determine this, as well as co-labeling experiments to determine whether the Zic+ mesenchymal cells are Axin2+/Tbx1+/Brn4+/Sox10− as would be expected.
Neural Tube
In both chick and mouse, all Zic genes (chick Zic1-4, mouse Zic1-5) were expressed in the dorsal neural tube. The inner ear lies adjacent to the neural tube, and alterations in specification and patterning of the neural tube are known to affect inner ear development. Signaling from the hindbrain, especially rhombomeres 5 and 6, is critical for proper patterning and development of the inner ear, as well as development of the cochleovestibular ganglion that innervates the ear (Bok et al., 2005; Kil et al., 2005; Bok et al., 2007; Choo, 2007; Vazquez-Echeverria et al., 2008; Liang et al., 2010). Neural tube closure defects, including exencephaly, in or near the hindbrain, are seen in Zic mutant mice examined to date, including Zic2kd/kd (Nagai et al., 2000), Zic3Bn (Klootwijk et al., 2000) and Zic5−/− (Inoue et al., 2004) mutant mice. In Zic2Ku/Ku mutants, rhombomeres 3 and 5 are smaller than those in wild type embryos, and ectopic follistatin expression is detected in these rhombomeres (Elms et al., 2003). In addition to effects on hindbrain development, mutations in Zic genes affect signaling from the hindbrain to the inner ear. Expression of Wnt3a, a signaling molecule from the hindbrain important for inner ear development (Riccomagno et al., 2005), is delayed in the dorsal hindbrain of both Zic2kd/kd (Nagai et al., 2000) and Zic5−/− (Inoue et al., 2004) mutant mice. Zic genes expressed in the neural tube could effect inner ear development in multiple ways, both directly and indirectly. ZIC proteins could directly interact with WNT or BMP pathway components in the neural tube to modulate WNT or BMP signaling to the developing inner ear, or ZIC proteins could promote/inhibit the expression of other factors in the dorsal neural tube. Indirectly, loss of specific Zic genes in the neural tube leads to neural tube defects (such as exencephaly), which would alter the position of the neural tube relative to the inner ear. This change in position of the neural tube relative to the developing inner ear would alter the position of specific regions of the otic epithelium relative to sources of BMP, WNT, and SHH signaling, potentially leading to malformations of the inner ear.
Investigating the function of Zic genes during inner ear development
To date, the involvement of Zic genes in inner ear development has only been examined at the level of gene expression (this study; Warner et al., 2003). Further functional studies are needed to investigate the role of the Zic genes during inner ear development. Mice with mutations in each of the Zic genes have been generated (Ali et al., 2012), but no ear phenotypes have been reported to date. One reason for this could be that multiple severe defects in the postnatal animals mask any ear phenotypes. Both Zic1 and Zic5 homozygous mutants display abnormal gait and posture characteristic of defects in the vestibular region of the inner ear, but these abnormalities were attributed to cerebellar defects (Zic1−/−) or hydrocephalus (Zic5−/−) (Aruga et al., 1998; Inoue et al., 2004). However, the majority of Zic5 mutants did not have any noticeable changes in brain morphology, so the cause of the abnormal gait and posture was never fully investigated (Inoue et al., 2004). Another reason an inner ear phenotype has not been characterized is that significant numbers of mutants die, either during embryonic stages (all Zic2Ku/Ku, ~24% of Zic3Bn) or shortly after birth (50% of Zic1−/−, all Zic2kd/kd, some Zic5−/−) (Aruga et al., 1998; Klootwijk et al., 2000; Nagai et al., 2000; Elms et al., 2003; Inoue et al., 2004), making it impossible to detect signs of inner ear defects that affect the mature function of this sensory organ, such as changes in gait or posture, abnormal behaviors such as circling, and changes in the acoustic startle response or auditory brainstem responses (ABR) (Saul et al., 2008). Studies using these mice, including recording of ABRs in any animals, including heterozygotes, that live to postnatal stages, as we have done for other mouse mutations (Bank et al., 2012) could provide insights into how individual Zic genes function during inner ear development. Further, the generation of compound Zic mutants would be useful to address questions of redundancy among the Zic genes during inner ear development, as we have shown that the Zic genes are expressed in overlapping regions of the periotic mesenchyme in both chick and mouse.
Attempts to reconcile the disparate results of earlier Zic gene expression studies
Because the results found by McMahon and Merzdorf (McMahon and Merzdorf, 2010) were found to be distinctly different from those of our earlier report (Warner et al., 2003), particularly for the expression of Zic1, we repeated and expanded the expression study of the Zic genes in the developing chick inner ear. A fourth Zic gene, Zic4, has since been identified in the chicken (McMahon and Merzdorf, 2010) and its expression pattern was also examined. In addition, we wanted to compare the expression of the Zic genes in the otic region of the chicken to the expression pattern of the Zic genes in the otic region of the mouse, which had not previously been reported. To our surprise, the expression pattern of Zic1 in the developing chick inner ear we report here is not consistent with our earlier study; however, the expression patterns of Zic2 and Zic3 reported here are consistent with our earlier study (Warner et al., 2003). Our findings for the expression patterns of Zic1-4 in the neural tube of the developing chick in the present study are consistent with an earlier study examining the expression of Zic1-4 in the developing chick embryo up to HH stage 18 (McMahon and Merzdorf, 2010). However, we found expression of both Zic1 and Zic2 in the periotic mesenchyme at HH stage 18, whereas McMahon and Merzdorf only saw Zic2 expression in the periotic mesenchyme (McMahon and Merzdorf, 2010). An explanation for this is that at HH stage 18, we detected periotic mesenchyme expression in sections taken through the ear, which allowed us to examine expression patterns in locations that would not have been discernable in the whole mounts used in the other study (McMahon and Merzdorf, 2010). This leaves us to question the differences in Zic1 expression in the developing chick inner ear that were seen in this study as contrasted with our previous study (Warner et al., 2003). All probes (this study and the previous study; Warner et al., 2003) were sequenced to confirm their identity. In this study, the probes were designed to target sequences outside of the zinc finger region where there is low homology between the Zic genes (Fig. 3). Both the chick Zic2 and Zic3 probes used in this study target similar regions as the Zic2 and Zic3 probes from our previous study (Warner et al., 2003). The Zic2 probes each target a different part of the 3’ untranslated region (UTR) of the mRNA. Similarly, the Zic3 probes target an overlapping region of the 3’ UTR, with the probe used in this study also extending both into a portion of the 3’ coding region of the mRNA and further into the 3’ UTR. The Zic1 probe used in our earlier study targeted the zinc finger region of Zic1, and this likely underscores the differences with the earlier study. The probe used in this study targets the 3'UTR of Zic1 and excludes this zinc finger region.
We believe this more detailed and thorough study now resolves the differences between the two earlier studies, confirming the Merzdorf lab's findings of the lack of Zic1 expression in the developing chick embryonic inner ear, confirming the expression patterns we found in the developing chick inner ear for Zic2 and Zic3 and extending the study of the chick Zic gene expression in the region of the developing otic region and the neural tube to characterize the expression pattern of Zic4. We have further extended the study of otic/periotic/neural tube expression of the Zic genes to the mouse, as a prelude to examining such expression in Zic mutant embryos and animals.
Experimental Procedures
Mouse Embryos
Wild type Balb/c mice were used to set up timed matings. Noon on the day on which a vaginal plug was detected was designated as E0.5. Pregnant females were sacrificed by cervical dislocation. All experiments using mice were approved by the Animal Use and Care committee of the University of Michigan and conform to all guidelines of the Unit for Laboratory Animal Medicine and those of the National Institutes of Health. The uterine horns containing the embryos were dissected out and placed in PBS with 10% FBS. Embryos were then dissected out of the surrounding maternal tissues and Reichert's membrane was removed. Embryos were fixed in 4% paraformaldehyde overnight, washed in 1X PBS (3 × 5 minutes), and then transferred to 30% sucrose in 1X PBS and rocked at 4°C overnight.
Chick Embryos
Fertilized White Leghorn chicken eggs were obtained from the Michigan State University Poultry Farm (East Lansing, MI), and placed in a humidified incubator at 37°C. Eggs were windowed and the embryos were dissected and placed into PBS with 10% FBS. The vitelline membrane was removed and embryos were dissected out of the amnion. Embryos were fixed in 4% paraformaldehyde (PFA) overnight, washed in 1X PBS (3 × 5 minutes), and then transferred to 30% sucrose in 1X PBS and rocked at 4°C overnight.
Embedding and Cryosectioning
Embryos were transferred through three progressive changes of OCT (TissueTek) to remove excess sucrose. The embryos were then transferred to embedding molds, covered with OCT, and oriented such that the anterior portion of the ear pointed down. The molds were then frozen on dry ice and stored at −80°C until sectioning. 12μm or 25μm transverse sections through the ear were cut using a Microm HM500M cryostat and collected on SuperFrost Plus slides (Fisher). Sections were air-dried on the slides at room temperature for at least 30 minutes, and then stored at −80°C.
In Situ Hybridization
In situ hybridization using digoxigenin-labeled antisense probes was performed on sections using a protocol adapted from Wilkinson and Nieto (Wilkinson and Nieto, 1993). The prehybridization, hybridization, and post-hybridization steps were performed essentially as described, except MBST (100mM Maleic Acid, 150mM NaCl, 0.1% Tween20) was used for the post-hybridization washes and the blocking solution was MBST containing 10% heat-inactivated sheep serum and 2% Blocking Reagent (Roche). For the color reaction, BM Purple (Roche) was used instead of NBT/BCIP. Following the color reaction, slides were washed three times in PBST, pH 4.5 (PBS with 0.1% Tween20), fixed (4% PFA/0.2% gluteraldehyde), washed three times in PBS, dehydrated in 70% ethanol, and dried on a slide warmer set at 60°C. Sections were then mounted under a coverslip with Glycergel Mounting Media (Dako). A minimum of 4 embryos was analyzed for each probe. Antisense probes were generated by digesting plasmids containing the cDNA sequences of either the mouse or chick genes and then transcribing with the appropriate RNA polymerase. Probes for chick Zic3 and chick Zic4 were prepared by PCR as previously described (McMahon and Merzdorf, 2010). Both the mouse and chick Zic probes were designed to target sequences outside of the zinc finger region where there is lower homology between the Zic genes (Fig. 3). Images were acquired with an Olympus BX51 microscope equipped with an Olympus camera.
Phylogenetic Tree and Sequence Analysis
The complete amino acid sequence of the Zic genes from human, mouse, chick, zebrafish, and frog were aligned with the amino acid sequence of the ancestral gene odd-paired (opa) from Drosophila using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). The alignment was then converted into a phylogenetic tree using Tree Vector (http://supfam.cs.bris.ac.uk/TreeVector). Pairwise comparisons of the amino acid sequences of human, mouse, and chick Zic genes was performed using EMBOSS Stretcher (http://www.ebi.ac.uk/Tools/psa/emboss_stretcher).
Bullet Points.
Zic genes are expressed in the dorsal neural tube and mesenchyme surrounding the developing inner ear in both mouse and chick
Zic genes are not expressed in the otic epithelium of either mouse or chick
Differential spatiotemporal expression of Zic genes (Zic1-5 in mouse, Zic1-4 in chick) is seen during inner ear development
Acknowledgements
The authors would like to thank members of the Barald lab, Dr. Ben Allen and Dr. Margaret Lomax of the University of Michigan and Dr. Ruth Arkell of ANU, Canberra, AUS for very helpful comments on the manuscript and advice on the experiments. We would also like to thank Dr. Andrew Copp (University College London) for the mouse Zic1, Zic2, and Zic4 probes, Dr. Ruth Arkell (Australian National University) for the mouse Pax2, Sox10, Zic3 and Zic5 probes, Dr. Deborah Gumucio (University of Michigan) for the mouse Axin2 probe, Dr. Douglas Epstein (University of Pennsylvania) for the mouse Brn4 and Tbx1 probes, Dr. Christa Merzdorf (Montana State University) for the chick Zic1 and Zic2 probes, and Dr. Andrea Streit (King's College London) for the chick Pax2 probe. This work was supported by NIH/NINDCD, 2 RO1 DC04184 and ARRA supplement (to KFB), the Cellular and Molecular Biology (T32GM007315), Regenerative Sciences (5T90DK070071-05), and Hearing, Balance, and Chemical Senses (5T32DC000011-32) training grants (APC) and the Undergraduate Research Opportunity Program (UROP) at the University of Michigan (IH).
References
- Ali RG, Bellchambers HM, Arkell RM. Zinc fingers of the cerebellum (Zic): transcription factors and co-factors. Int J Biochem Cell Biol. 2012;44:2065–2068. doi: 10.1016/j.biocel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Aruga J, Minowa O, Yaginuma H, Kuno J, Nagai T, Noda T, Mikoshiba K. Mouse Zic1 is involved in cerebellar development. J Neurosci. 1998;18:284–293. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank LM, Bianchi LM, Ebisu F, Lerman-Sinkoff D, Smiley EC, Shen YC, Ramamurthy P, Thompson DL, Roth TM, Beck CR, Flynn M, Teller RS, Feng L, Llewellyn GN, Holmes B, Sharples C, Coutinho-Budd J, Linn SA, Chervenak AP, Dolan DF, Benson J, Kanicki A, Martin CA, Altschuler R, Koch AE, Jewett EM, Germiller JA, Barald KF. Macrophage migration inhibitory factor acts as a neurotrophin in the developing inner ear. Development. 2012;139:4666–4674. doi: 10.1242/dev.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8:105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- Bok J, Bronner-Fraser M, Wu DK. Role of the hindbrain in dorsoventral but not anteroposterior axial specification of the inner ear. Development. 2005;132:2115–2124. doi: 10.1242/dev.01796. [DOI] [PubMed] [Google Scholar]
- Bok J, Brunet LJ, Howard O, Burton Q, Wu DK. Role of hindbrain in inner ear morphogenesis: analysis of Noggin knockout mice. Dev Biol. 2007;311:69–78. doi: 10.1016/j.ydbio.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein EM, Crenshaw EB, 3rd, Morrow BE, Adams JC. Cooperative function of Tbx1 and Brn4 in the periotic mesenchyme is necessary for cochlea formation. J Assoc Res Otolaryngol. 2008;9:33–43. doi: 10.1007/s10162-008-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuskin I, Bodson M, Thelen N, Thiry M, Borgs L, Nguyen L, Lefebvre PP, Malgrange B. Sox10 promotes the survival of cochlear progenitors during the establishment of the organ of Corti. Dev Biol. 2009;335:327–339. doi: 10.1016/j.ydbio.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Bronner ME. Formation and migration of neural crest cells in the vertebrate embryo. Histochem Cell Biol. 2012;138:179–186. doi: 10.1007/s00418-012-0999-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Q, Cole LK, Mulheisen M, Chang W, Wu DK. The role of Pax2 in mouse inner ear development. Dev Biol. 2004;272:161–175. doi: 10.1016/j.ydbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Chan WY, Tam PP. A morphological and experimental study of the mesencephalic neural crest cells in the mouse embryo using wheat germ agglutinin-gold conjugate as the cell marker. Development. 1988;102:427–442. doi: 10.1242/dev.102.2.427. [DOI] [PubMed] [Google Scholar]
- Choo D. The role of the hindbrain in patterning of the otocyst. Dev Biol. 2007;308:257–265. doi: 10.1016/j.ydbio.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elms P, Siggers P, Napper D, Greenfield A, Arkell R. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev Biol. 2003;264:391–406. doi: 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Fujimi TJ, Hatayama M, Aruga J. Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/beta-catenin signaling pathway. Dev Biol. 2012;361:220–231. doi: 10.1016/j.ydbio.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Gong TW, Hegeman AD, Shin JJ, Adler HJ, Raphael Y, Lomax MI. Identification of genes expressed after noise exposure in the chick basilar papilla. Hear Res. 1996;96:20–32. doi: 10.1016/0378-5955(96)00013-5. [DOI] [PubMed] [Google Scholar]
- Grinberg I, Millen KJ. The ZIC gene family in development and disease. Clin Genet. 2005;67:290–296. doi: 10.1111/j.1399-0004.2005.00418.x. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Herman GE, El-Hodiri HM. The role of ZIC3 in vertebrate development. Cytogenet Genome Res. 2002;99:229–235. doi: 10.1159/000071598. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Alvarado-Mallart R, Alvarez IS. Pax2, Otx2, Gbx2 and Fgf8 expression in early otic vesicle development. Mech Dev. 2000;95:225–229. doi: 10.1016/s0925-4773(00)00332-4. [DOI] [PubMed] [Google Scholar]
- Hutson MR, Lewis JE, Nguyen-Luu D, Lindberg KH, Barald KF. Expression of Pax2 and patterning of the chick inner ear. J Neurocytol. 1999;28:795–807. doi: 10.1023/a:1007057719025. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hatayama M, Tohmonda T, Itohara S, Aruga J, Mikoshiba K. Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev Biol. 2004;270:146–162. doi: 10.1016/j.ydbio.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil SH, Streit A, Brown ST, Agrawal N, Collazo A, Zile MH, Groves AK. Distinct roles for hindbrain and paraxial mesoderm in the induction and patterning of the inner ear revealed by a study of vitamin-A-deficient quail. Dev Biol. 2005;285:252–271. doi: 10.1016/j.ydbio.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Klootwijk R, Franke B, van der Zee CE, de Boer RT, Wilms W, Hol FA, Mariman EC. A deletion encompassing Zic3 in bent tail, a mouse model for X-linked neural tube defects. Hum Mol Genet. 2000;9:1615–1622. doi: 10.1093/hmg/9.11.1615. [DOI] [PubMed] [Google Scholar]
- Koyabu Y, Nakata K, Mizugishi K, Aruga J, Mikoshiba K. Physical and functional interactions between Zic and Gli proteins. J Biol Chem. 2001;276:6889–6892. doi: 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- Lawoko-Kerali G, Rivolta MN, Holley M. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J Comp Neurol. 2002;442:378–391. doi: 10.1002/cne.10088. [DOI] [PubMed] [Google Scholar]
- Li H, Liu H, Corrales CE, Mutai H, Heller S. Correlation of Pax-2 expression with cell proliferation in the developing chicken inner ear. J Neurobiol. 2004;60:61–70. doi: 10.1002/neu.20013. [DOI] [PubMed] [Google Scholar]
- Liang JK, Bok J, Wu DK. Distinct contributions from the hindbrain and mesenchyme to inner ear morphogenesis. Dev Biol. 2010;337:324–334. doi: 10.1016/j.ydbio.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Maurus D, Harris WA. Zic-associated holoprosencephaly: zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, Hedgehog, and retinoic acid signaling. Genes Dev. 2009;23:1461–1473. doi: 10.1101/gad.517009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AR, Merzdorf CS. Expression of the zic1, zic2, zic3, and zic4 genes in early chick embryos. BMC Res Notes. 2010;3:167. doi: 10.1186/1756-0500-3-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- Merzdorf CS, Sive HL. The zic1 gene is an activator of Wnt signaling. Int J Dev Biol. 2006;50:611–617. doi: 10.1387/ijdb.052110cm. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem. 2001;276:2180–2188. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Hatayama M, Tohmonda T, Ogawa M, Inoue T, Mikoshiba K, Aruga J. Myogenic repressor I-mfa interferes with the function of Zic family proteins. Biochem Biophys Res Commun. 2004;320:233–240. doi: 10.1016/j.bbrc.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Minowa O, Sugimoto T, Ohno Y, Noda T, Mikoshiba K. Zic2 regulates the kinetics of neurulation. Proc Natl Acad Sci U S A. 2000;97:1618–1623. doi: 10.1073/pnas.97.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nornes HO, Dressler GR, Knapik EW, Deutsch U, Gruss P. Spatially and temporally restricted expression of Pax2 during murine neurogenesis. Development. 1990;109:797–809. doi: 10.1242/dev.109.4.797. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Pan H, Gustafsson MK, Aruga J, Tiedken JJ, Chen JC, Emerson CP., Jr A role for Zic1 and Zic2 in Myf5 regulation and somite myogenesis. Dev Biol. 2011;351:120–127. doi: 10.1016/j.ydbio.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippard D, Heydemann A, Lechner M, Lu L, Lee D, Kyin T, Crenshaw EB., 3rd Changes in the subcellular localization of the Brn4 gene product precede mesenchymal remodeling of the otic capsule. Hear Res. 1998;120:77–85. doi: 10.1016/s0378-5955(98)00059-8. [DOI] [PubMed] [Google Scholar]
- Pourebrahim R, Houtmeyers R, Ghogomu S, Janssens S, Thelie A, Tran HT, Langenberg T, Vleminckx K, Bellefroid E, Cassiman JJ, Tejpar S. Transcription factor Zic2 inhibits Wnt/beta-catenin protein signaling. J Biol Chem. 2011;286:37732–37740. doi: 10.1074/jbc.M111.242826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschel AW, Westerfield M, Dressler GR. Comparative analysis of Pax-2 protein distributions during neurulation in mice and zebrafish. Mech Dev. 1992;38:197–208. doi: 10.1016/0925-4773(92)90053-m. [DOI] [PubMed] [Google Scholar]
- Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–1812. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkwitz-Brandt S, Arnold HH, Bober E. Regionalized expression of Nkx5-1, Nkx5-2, Pax2 and sek genes during mouse inner ear development. Hear Res. 1996;99:129–138. doi: 10.1016/s0378-5955(96)00093-7. [DOI] [PubMed] [Google Scholar]
- Rinkwitz-Brandt S, Justus M, Oldenettel I, Arnold HH, Bober E. Distinct temporal expression of mouse Nkx-5.1 and Nkx-5.2 homeobox genes during brain and ear development. Mech Dev. 1995;52:371–381. doi: 10.1016/0925-4773(95)00414-v. [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon H, Martin-Partido G, Hidalgo-Sanchez M. Differential expression of Otx2, Gbx2, Pax2, and Fgf8 in the developing vestibular and auditory sensory organs. Brain Res Bull. 2002;57:321–323. doi: 10.1016/s0361-9230(01)00725-0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon H, Martin-Partido G, Hidalgo-Sanchez M. Pax2 expression patterns in the developing chick inner ear. Gene Expr Patterns. 2005;5:763–773. doi: 10.1016/j.modgep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Sanek NA, Taylor AA, Nyholm MK, Grinblat Y. Zebrafish zic2a patterns the forebrain through modulation of Hedgehog-activated gene expression. Development. 2009;136:3791–3800. doi: 10.1242/dev.037820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul SM, Brzezinski JAt, Altschuler RA, Shore SE, Rudolph DD, Kabara LL, Halsey KE, Hufnagel RB, Zhou J, Dolan DF, Glaser T. Math5 expression and function in the central auditory system. Mol Cell Neurosci. 2008;37:153–169. doi: 10.1016/j.mcn.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechrist J, Serbedzija GN, Scherson T, Fraser SE, Bronner-Fraser M. Segmental migration of the hindbrain neural crest does not arise from its segmental generation. Development. 1993;118:691–703. doi: 10.1242/dev.118.3.691. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116:297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Sun Rhodes LS, Merzdorf CS. The zic1 gene is expressed in chick somites but not in migratory neural crest. Gene Expr Patterns. 2006;6:539–545. doi: 10.1016/j.modgep.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Tosney KW. The segregation and early migration of cranial neural crest cells in the avian embryo. Dev Biol. 1982;89:13–24. doi: 10.1016/0012-1606(82)90289-5. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Sobieszczuk D, Wilkinson D, Krumlauf R. Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development. 2002;129:433–442. doi: 10.1242/dev.129.2.433. [DOI] [PubMed] [Google Scholar]
- Vazquez-Echeverria C, Dominguez-Frutos E, Charnay P, Schimmang T, Pujades C. Analysis of mouse kreisler mutants reveals new roles of hindbrain-derived signals in the establishment of the otic neurogenic domain. Dev Biol. 2008;322:167–178. doi: 10.1016/j.ydbio.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Viola A, Morishima M, Pramparo T, Baldini A, Lindsay E. TBX1 is required for inner ear morphogenesis. Hum Mol Genet. 2003;12:2041–2048. doi: 10.1093/hmg/ddg216. [DOI] [PubMed] [Google Scholar]
- Warner SJ, Hutson MR, Oh SH, Gerlach-Bank LM, Lomax MI, Barald KF. Expression of ZIC genes in the development of the chick inner ear and nervous system. Dev Dyn. 2003;226:702–712. doi: 10.1002/dvdy.10262. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Takeda K, Katori Y, Ikeda K, Oshima T, Yasumoto K, Saito H, Takasaka T, Shibahara S. Expression of the Sox10 gene during mouse inner ear development. Brain Res Mol Brain Res. 2000;84:141–145. doi: 10.1016/s0169-328x(00)00236-9. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]