Abstract
Micro (mi)RNAs form a class of short RNAs (~21 nt) that post-transcriptionally regulate partially complementary messenger (m)RNAs. Each miRNA may target tens-to-hundreds of transcripts to control key biological processes. While the biochemical reactions underpinning miRNA biogenesis and activity are relatively well-defined1,2, and the importance of their homeostasis increasingly evident, the processes underlying regulation of miRNA pathways in vivo are still largely elusive3. Autophagy, a degradative process in which cytoplasmic material is targeted into double-membrane vacuoles, is recognized to critically contribute to cellular homeostasis. Here, we show that the miRNA-processing enzyme, DICER, and the major miRNA effector, AGO2, are targeted for degradation as miRNA-free entities by the selective autophagy receptor NDP52. Autophagy establishes a checkpoint required for continued loading of miRNA into AGO2; accordingly, NDP52 and autophagy are required for homeostasis and activity of tested miRNAs. Autophagy also engages post-transcriptional regulation of the DICER mRNA, underscoring the importance of fine-tuned regulation of the miRNA pathway. These findings have implications for human diseases linked to misregulated autophagy, DICER- and miRNA-levels, including cancer.
Keywords: Autophagy, NDP52, DICER, Argonaute, microRNA, post-transcriptional
Following their export to the cytoplasm, stem-loop precursor (pre)-miRNAs are cleaved by Dicer into ~21bp double-stranded miRNA-miRNA* duplexes and transferred into the groove of Argonaute proteins (AGO). Upon miRNA*-strand dissociation, a mature single-stranded miRNA remains loaded into, and stabilized by AGO1. AGO then detaches from DICER to bind TNRC64, an essential co-factor in the miRNA-Induced Silencing Complex (miRISC). Activated miRISC can then repress miRNA-targeted mRNA by inhibiting its translation and accelerating its decay2,5. The mechanisms regulating RNA and protein components of miRNA pathways in vivo has only recently gained attention in mammals. Hence, mammalian miRNA turnover can be enhanced via nucleotide trimming-tailing6 or the exonuclease Xrn27, and limited evidence suggests that a fraction of unloaded Ago may be selectively targeted for degradation8. Interestingly, DICER protein levels undergo fine-tuning mediated both by DICER substrates through competition of pre-miRNA with DICER mRNA for XPO5-dependent nuclear-cytoplasmic export, and by DICER products through repression of the DICER mRNA by specific mature miRNAs in distant organisms including humans9 and plants10. Thus, a requirement for fine-tuned, homeostatic regulation of miRNA pathways appears to be conserved across kingdoms.
Macroautophagy (herein referred to as autophagy) is an intracellular degradation process contributing to cellular homeostasis11. Thirty-five conserved autophagy-related (ATG) proteins control the initiation and elongation of double-membrane autophagosomes, engulfing cargoes that are ultimately degraded upon fusion with lysosomes11. Selective degradation is enabled by autophagy receptors, including p62 (SQSTM1)12 and NDP5213 that bind both cytosolic substrates and Atg8 (i.e. LC3) family proteins inserted into the autophagosome membrane. The detailed molecular mechanisms underpinning cargo selectivity are only emerging. Accordingly, autophagy receptors may confer some cargo selectivity by recognizing conjugated ubiquitin14, although this may also occur via recognition of other modifications or molecules independently of ubiquitin13,15. We and others previously showed that several miRNA pathway components including DICER, AGO and TNRC6, associate with membranes16-18. In this context, we proposed that autophagy might modulate the levels of these or other protein or RNA involved in miRNA biogenesis and action, by promoting their regulated degradation19.
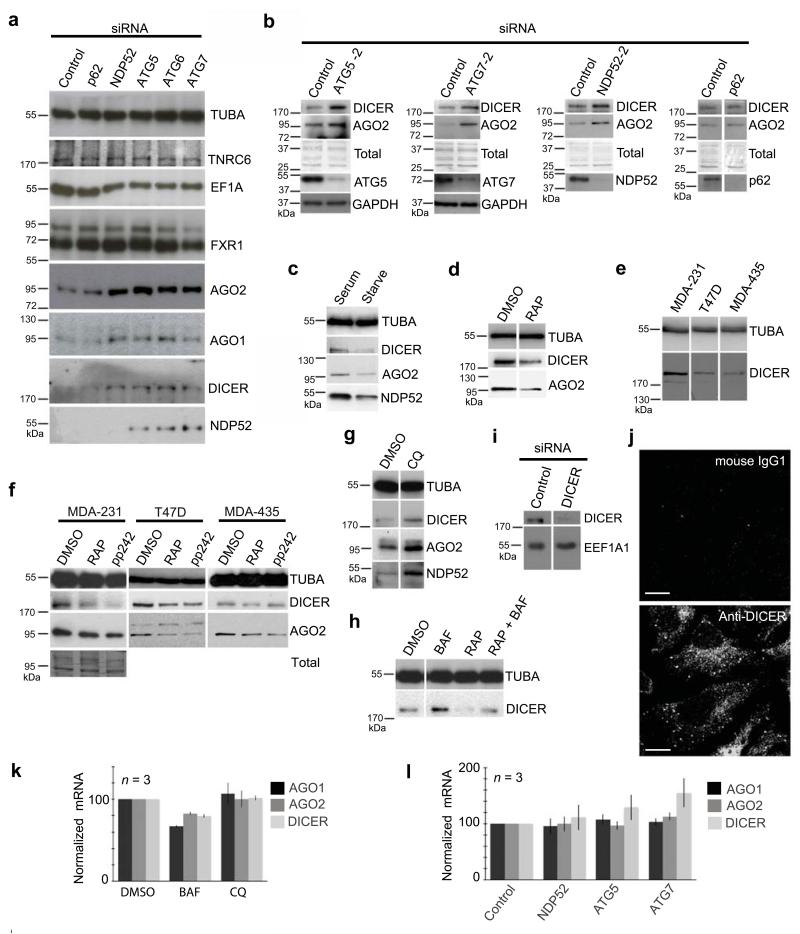
To investigate the potential contribution of autophagy to the degradation of cytosolic miRNA-containing complexes, HeLa cells were depleted of the key autophagy components ATG5, ATG6 or ATG7, or of the selective autophagy receptors NDP52 or p62 using siRNAs. AGO2, AGO1 and DICER accumulated in cells depleted of ATG5, ATG6, ATG7 or NDP52, but not of p62 (Fig.1a-b). In contrast, siRNA-mediated depletion of the autophagy machinery did not result in elevated levels of the key miRISC component TNRC64, of the AGO-interacting protein, FXR1, or of EF1A, an AGO-binding protein that mediates AGO-dependent inhibition of translational elongation20 (Fig.1a). Activating autophagy by serum starvation or with an mTOR inhibitor (rapamycin, [RAP]) decreased DICER and AGO2 levels (Fig.1c-d). DICER levels vary among representative cell lines (Fig. 1e), and DICER levels decreased in each line tested upon treatment with mTOR inhibitors (RAP, pp242, Fig. 1f). Conversely, DICER and AGO2 levels increased in HeLa cells treated with inhibitors of lysosomal acidification (BafilomycinA1 [BAF], chloroquine [CQ]), known to block autophagy (Fig.1g-h). Moreover, DICER levels decreased by RAP treatment were rescued by co-treatment with BAF (Fig.1h). The protein detected with anti-DICER mAb was confirmed to be DICER by its selective depletion with siRNA targeting Dicer mRNA and specific labeling of HeLa cells by confocal microscopy, which was consistent with the uneven cytoplasmic distribution of endogenous DICER previously reported21 (Fig. 1i-j). Effects of autophagy-modulating treatments on DICER, AGO1 and AGO2 were at the protein level, as their mRNA levels remained unchanged (Fig.1k-l).
Figure 1.
Levels of DICER and AGO proteins are regulated by autophagy. (a) Western blot analysis of HeLa cells treated with siRNA for 84 h, including alpha-tubulin loading control (TUBA). (b) Western blot analysis of HeLa cells treated with a second independent siRNA targeting ATG5, ATG7, or NDP52. GAPDH and total protein stained with Coomassie blue serve as loading controls. (c) Western blot analysis of HeLa cells cultured in Hank’s buffered salt solution (starved) or DMEM + 10% FBS (serum). (d) Western blot analysis of HeLa cells treated (48 h) with an inhibitor of mTORC1 (RAP, 20 nM) or control (DMSO). (e) Western blot analysis comparing the levels of DICER in MDA-231, T47D and MDA-435 cells. (f) Western blot analysis of MDA-231, T47D and MDA-435 cells treated (48 h) with inhibitors of mTORC1 (RAP, 20 nM), mTORC1 and 2 (pp242, 100 nM) or DMSO (control). (g) Western blot analysis of HeLa cells treated with CQ or DMSO for 12 h. (h) Western blot analysis of HeLa cells treated with BAF (200 nM, 12 h), RAP (20 nM, 24 h), or RAP and BAF. (i) Western blot analysis of DICER (mAb 13D6) or control (EEF1A1) in HeLa cells treated with control siRNA or siRNA targeting DICER (j) Confocal analysis of signal from anti-DICER mAb (clone 13D6) or an isotype control mAb in HeLa cells. Results in (a-h) are representative of three experiments and use α-tubulin (TUBA) as a loading control. (k-l) RT-qPCR analysis of AGO1, AGO2, and DICER mRNA in HeLa cells treated for 12 h with DMSO, BAF, or CQ (k) or siRNA for 84 h (l). Error bars show SEM.
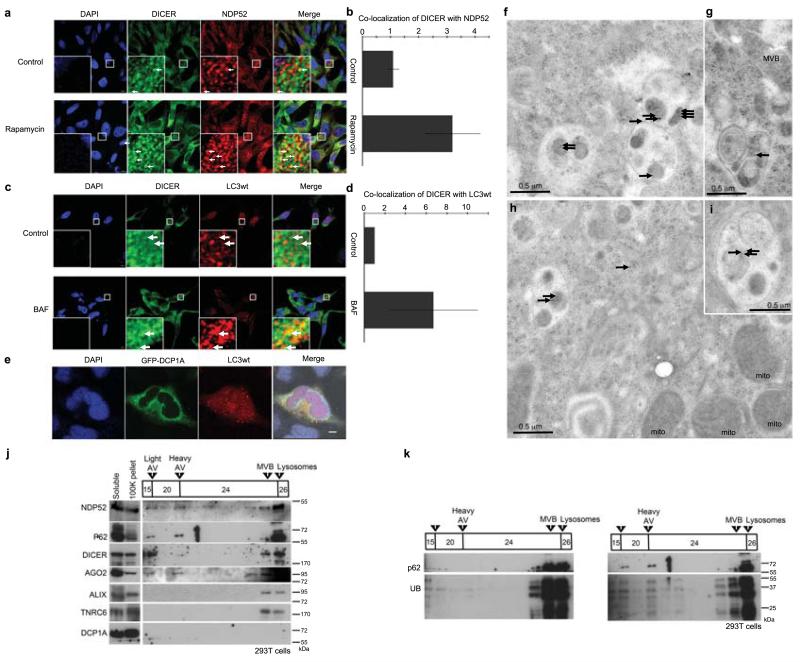
By confocal microscopy, DICER significantly co-localized with a fraction of the autophagy receptor NDP52 in HeLa cells (using Costes’, Fay’s or van Steensel’s tests for non-random co-localization, p<0.0001); moreover, this co-localization increased 3.2-fold in cells treated with RAP (Fig.2a-b, Costes’, Fay’s and van Steensel’s tests, p<0.0001), consistent with targeting of DICER to NDP52-dependent autophagy (Fig.1a-b). Also consistent with targeting of DICER for autophagic degradation, DICER co-localized significantly with the autophagolysosome marker HcRed-LC3 and this co-localization increased 6.7-fold in cells treated with BAF (Fig.2c-d, Costes’, Fay’s or van Steensel’s test, p<0.0001, BAF and control treated). In contrast, no co-localization was observed between autophagy markers and GFP-labeled DCP1A, an AGO-associated mRNA decapping factor that localizes to P-bodies22 (Fig.2e).
Figure 2.
DICER co-localizes and associates with autophagosomes. (a) Localization using confocal microscopy of endogenous DICER (mAb 13D6) and NDP52 in HeLa cells treated with RAP or control. Arrows highlight some examples of co-localization. Scale bar = 5 μm (b) Quantification of DICER co-localization with NDP52. Total fluorescence intensity of DICER co-localized with punctae (>0.2 μm3) of NDP52 was quantified with Volocity software in RAP-treated cells / control-treated cells over several Z-stacked microscope fields per experiment (n=4, error bars SEM). (c) Localization using confocal microscopy of endogenous DICER (mAb 13D6) and Hc-Red-LC3 in HeLa cells treated with bafilomycin or control. Arrows highlight some examples of co-localization. Scale bar = 5 μm (d) Quantification of DICER co-localization with HcRed-LC3 was performed as for NDP52 in (b). (e) Confocal microscopy analysis of GFP-DCP1A and HcRed-LC3wt in HeLa cells. Scale bar = 10 μm (f-i) Localization of DICER (mAb 13D6) detected with anti-mouse IgG (10 nm gold beads) by electron microscopy in CQ (20 μM, 12 h) treated HeLa cells. Mito (mitochondria), MVB (multivesicular body). Black arrows highlight all gold beads. (j) Western blot analysis of fractions from a discontinuous gradient of Histodenz™ (15%, 20%, 24%, 26%) described to enrich autophagosomes (light AV) and autophagolysosomes (heavy AV)24. 293T cells were treated with CQ (20 μM, 16 h). Fractions enriched in MVB and lysosomal markers are indicated. Material that was - pelleted by centrifugation at 100,000 g (100K pellet) or that remained in solution after the 100,000 g spin (soluble) was not added to the gradient. (k) Western blot analysis of UB and p62 in discontinuous Histodenz™ gradients (as in j) of 293T cells treated with control (DMSO) or CQ.
We then sought evidence for physical association of DICER and AGO2 with autophagosomes. First, using electron microcopy in CQ-treated cells, we confirmed accumulation of membrane-bound structures containing irregular electron dense, vesicular and organellar content, hallmark features of autophagolysosomes23, in which DICER was enriched compared to the surrounding cytoplasm (Fig.2f-i). Second, we isolated autophagosome- and autophagolysosome-enriched cell fractions using density gradients24. As expected, ubiquitin and the autophagy receptors NDP52 and p62 accumulated in fractions containing autophagosomes and autophagolysosomes in CQ-treated cells (Fig.2j-k). By contrast, ALIX, a marker of multivesicular bodies that associate with TNRC616, did not accumulate in these fractions, nor did the miRISC component TNRC6 or the P-body associated DCP1A (Fig.2j). By contrast, DICER and AGO2 were detected in autophagosome and autophagolysosome fractions from CQ-treated cells (Fig.2j). Taken together, the data presented in Fig.1-2 strongly support the idea that autophagy degrades DICER and AGO2 in an NDP52-selective manner. These results further suggest that autophagy-mediated regulation occurs upstream of the formation of miRISC (probed with TNRC6) and miRISC activity (probed with EF1A and DCP1a).
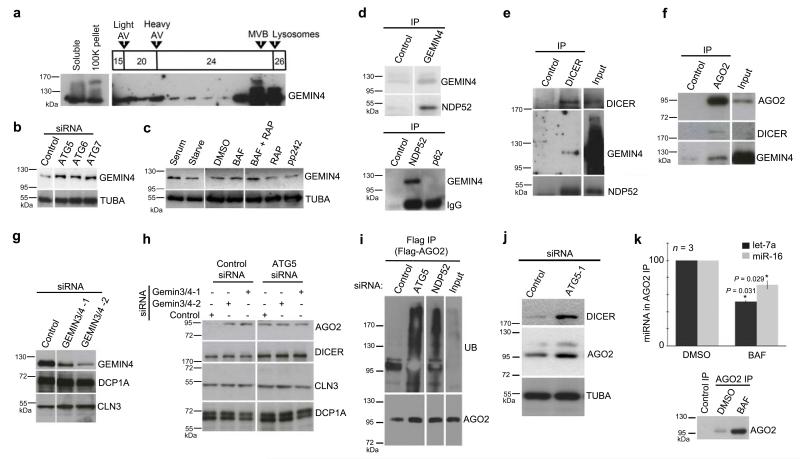
DICER and AGO2 localize to NDP52-labelled autophagosomes (Fig.2a-b,j) and are subject to NDP52-dependent autophagy (Fig.1a-b) suggesting that they may interact with NDP52. Previous studies showed that NDP52 binds GEMIN425, a component of the multi-protein SMN (Survival of Motor Neuron) complex involved in mRNA splicing, that also localizes to the cytoplasm. GEMIN4 and GEMIN3 are routinely found associated with AGO proteins, in complexes also containing DICER26, albeit for reasons not yet understood. We thus hypothesized that GEMIN4 may link AGO2 and DICER to NDP52 and promote their autophagic degradation. GEMIN4 was detected in autophagosome-enriched fractions of CQ-treated cells and GEMIN4 levels were sensitive to depletion of ATG5, ATG6 or ATG7, and to modulation of autophagy by serum starvation or with drugs (Fig.3a-c), suggesting that GEMIN4, like DICER or AGO2, is degraded by autophagy. We confirmed that GEMIN4 immunoprecipitates with NDP52 but not with the distinct autophagy receptor, p62 (Fig.3d). Agreeing with the previously reported co-enrichment of DICER and GEMIN4 in AGO complexes26, DICER immunoprecipitates contained GEMIN4 (Fig.3e) and, conversely, AGO2 immunoprecipitates contained GEMIN4 and DICER (Fig.3f). Furthermore, NDP52 co-immunoprecipitated with DICER (Fig.3e). Collectively, these data suggest that DICER, AGO2 and GEMIN4 are targeted to autophagic degradation via their association with NDP52.
Figure 3.
DICER associates with NDP52 and is targeted to autophagy independent of an RNA cargo. (a) Western blot analysis of GEMIN4 in fractions from a discontinuous gradient of Histodenz™ (15%, 20%, 24%, 26%) described to enrich autophagosomes (light AV) and autophagolysosomes (heavy AV). 293T cells were treated with CQ (20 μM, 16 h). Fractions enriched in MVB and lysosomal markers are indicated. (b) Western blot of GEMIN4 or loading control (TUBA) in HeLa cells treated with control siRNA or siRNA targeting ATG5, ATG6 or ATG7. (c) Western blot analysis of GEMIN4 or control (TUBA) in HeLa cells cultured in DMEM 10% FBS (serum), HBSS (starve) for 2 h, or treated for 16 h with control (DMSO), BAF (200 nM), RAP (20 nM), RAP + BAF, or pp242 (100 nM). (d) Top, Western blot of GEMIN4 immunoprecipitates from HeLa. Bottom, Western blot analysis of GEMIN4 in immunoprecipitates of anti-NDP52, -p62 or control antibody from HeLa cells. (e) Western blot of DICER immunoprecipitates from HeLa cells. (f) Western blot of AGO2 immunoprecipites from HeLa cells. (g) Western blot analysis of GEMIN4, compared to loading controls (DCP1A, CLN3) in HeLa cells treated with control siRNA or siRNA targeting GEMIN3 and GEMIN4. (h) Western blot analysis of HeLa cells treated with indicated combinations of control siRNA or siRNA targeting GEMIN3, GEMIN4 and ATG5. Western blot analysis of CLN3 and DCP1A are used to demonstrate equal loading of wells. (i) Western blot analysis with anti-Ub mAb (mono- and poly-ubiquitinated proteins, FK2 clone) in Flag immunoprecipitates performed under stringent denaturing conditions. Flag-AGO2 was induced for 24 hwith tetracycline in 293T cells with Tet-inducible Flag-AGO2 stably integrated that had been treated 24 h previous with indicated siRNA. (j) Western blot analysis of HeLa cells treated with control siRNA or siRNA targeting ATG5 for 24 h. TUBA, alpha-tubulin loading control. (k) Top, RT-qPCR analysis of miR-16 or let-7a levels in AGO2 immunoprecipitates from DMSO and BAF treated HeLa cells. Bottom, a representative western blot of AGO2 immunoprecipitates used in one replicate of these experiments.
As both GEMIN3 and -4 associate with AGO26,27, the two proteins were co-depleted in subsequent investigations to prevent potential functional redundancy (Fig.3g). AGO2 accumulated in HeLa cells depleted of GEMINs (Fig.3h) but did not accumulate significantly further in cells depleted of both GEMINs and ATG5 (Fig.3h) suggesting that GEMINs operate in the same pathway as ATG5, which mediates autophagy-dependent AGO2 degradation. NDP52 recognizes both ubiquitinated and ubiquitin-independent cargoes for delivery to autophagosomes13. In cells depleted of NDP52 or ATG5, ubiquitinated AGO2 or an ubiquitinated protein tightly associated to AGO2, accumulated to high levels (Fig.3i). Thus, NDP52 may selectively direct AGO2 to autophagy using non-mutually exclusive and parallel recognition events including binding of AGO2 by GEMIN4 and post-translational modification of AGO2 or associated partner(s) by ubiquitin. In contrast, DICER levels were not significantly affected by depletion of GEMINs (Fig.3h), even though DICER associates with GEMIN4 and NDP52 (Fig.3e), co-localizes with NDP52 (Fig.2a) and is degraded in an NDP52-dependent manner (Fig.1-b). Dicer may thus be recruited to NDP52-dependent autophagic degradation via multiple recognition events potentially compensating for the absence of GEMINs.
Because lysosomes contain RNA-degrading nucleases, we tested if autophagy might degrade pre-miRNA or miRNA cargoes along with DICER or AGO2. The levels of ubiquituous miRNAs (e.g. miR-16 and let-7a) and of their corresponding miRNA* and pre-miRNA were, however, unperturbed in cells treated with BAF or ATG5-targeting siRNAs (24 h, Fig. 3j, Supplementary Fig.S1a). Likewise, mTOR inhibitors RAP or pp242 did not affect the levels of tested miRNAs (Supplementary Fig.S1b). DICER and AGO2 levels were clearly altered by these treatments (Fig.1c-h, 3j), strongly suggesting that neither pre-miRNA, miRNA* nor miRNA are degraded by autophagy in association with DICER or AGO2. This predicts that miRNA-free (i.e. unloaded) AGO2 should specifically accumulate in autophagy-compromised cells. Indeed, significantly less miR-16 and let-7a was detected in AGO2 immunoprecipitates from BAF-treated cells (Fig.3k). Taken together, these data suggest that a complex of DICER and AGO unattached to miRNA-related cargoes is selectively targeted for autophagy.
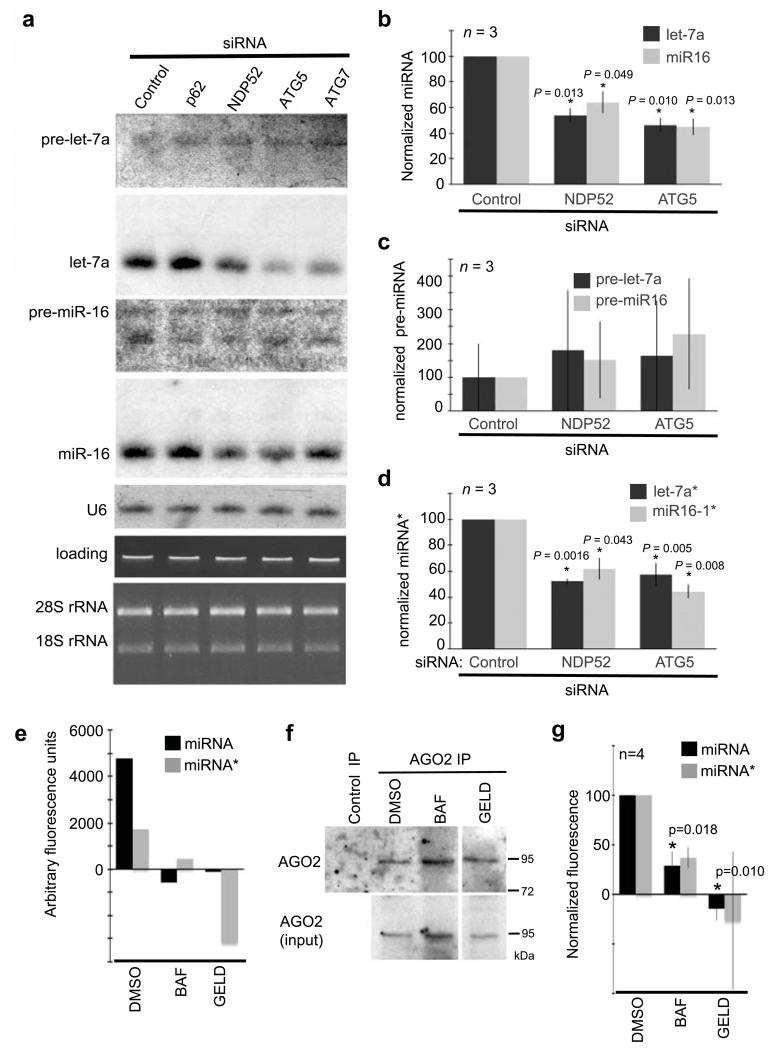
miRNA levels are not affected by short-term alterations to autophagy (Supplementary Fig.S1a-b), consistent with their avoidance of direct autophagic degradation. However, miRNA have surprisingly long half-lives (~5 days)28, suggesting that more extended periods might be required to destabilize miRNA levels as a consequence of perturbed homeostasis of key proteins involved in their biogenesis or stability, including DICER or AGO. Levels of miR-16 and let-7a, measured by Northern blot or RT-qPCR, indeed decreased significantly in cells treated with siRNAs targeting NDP52, ATG5 or ATG7, but not p62, for an extended period of 4 days (Fig.4a-b). In contrast, pre-miR-16 and pre-let-7a levels were not overtly modified (Fig.4a-c) suggesting that prolonged defects in autophagy specifically affect miRNA stability after pre-miRNA processing, but before miRISC formation and activity (Fig.1-2).
Figure 4.
MiRNA and miRNA* levels decrease when autophagy is inhibited for extended periods. (a) Northern blot of miR-16 and let-7a in HeLa cells treated with siRNA for 84 h. Levels of U6, and ethidium bromide stained total RNA on northern membrane (loading), and 28S and 18S rRNA on 1% agarose gel are shown as controls for equal loading. (b-d) RT-qPCR analysis of let-7a and miR-16 (b) pre-let-7a and pre-miR-16 (c) and let-7a* and mIR-16* (d) in HeLa cells depleted of NDP52 or ATG5 with siRNA (84 h). n = 3 independent experiments, error bars represent SEM. (e) Quantification of fluorescence in AGO2 immunoprecipitates from HeLa cell extracts incubated with duplexes of let-7a-let-7a* labeled on the 3′ end of either let-7 or let-7a* with FITC. Cells were treated 16 h prior with DMSO (control), BAF (200 nm), or geldanamycin (10 μM). Representative results of one experiment are shown. (f) Western blot analysis of AGO2 immunoprecipitates used in (e). (g) Normalized results of four independent experiments (n = 4) identical to (e). Error bars represent SEM.
We considered the possibility that the miRNA-degrading exonuclease XRN2 might be degraded by autophagy; its excess triggered by autophagy inhibition could then cause miRNA levels to decrease. Arguing against this, XRN2 was not detectable in autophagosome-enriched fractions and XRN2 levels were unaffected by ATG7 siRNA treatments (Supplementary Fig.S1c-d). miRNA are also destabilized by trimming-tailing which generates diagnostic ‘ladder’ patterns on Northern blots6, however, we did not observe such patterns (Fig.4a). This suggests that in autophagy-deficient cells, miRNA levels decrease independently of known pathways of miRNA degradation. Others have demonstrated that impediments to AGO-loading cause a drop in miRNA levels16,28-30. Because DICER and AGO both bind miRNA-miRNA* duplexes and promote their loading into AGO17,29-31, DICER and AGO2 degradation via autophagy may modulate loading of AGO2 and, consequently, miRNA levels. To test directly for effects on AGO-loading, miRNA-miRNA* complexes were incubated with lysates of Hela cells subjected to various treatments. In lysate from untreated cells, miRNA and miRNA* strands were detected in UV-cross-linked AGO2 complexes consistent with their loading into AGO2 (Supplementary Fig.S2). As expected, miRNA strands were more efficiently loaded into AGO2, retrieved by immunoprecipitation, than miRNA* strands (Fig.4e-g, Supplementary Fig.S2a) and incorporation of both small RNA species was impeded by geldanamycin, an hsp90 inhibitor that blocks AGO loading with small RNAs32 (Fig.4e-g). Less miRNA and miRNA* strands were also loaded into AGO2 in lysates from BAF-treated cells (Fig.4e-g), demonstrating that active autophagy is required for continued transfer of miRNA-miRNA* duplexes into AGO2. Consistent with a block in transfer of miRNA-miRNA* duplexes into AGO, the levels of miR16-1* and let-7a* were reduced proportionally to those of miR16-1 and let-7a when NDP52, ATG5, or ATG7 were depleted by siRNAs (Fig.4d).
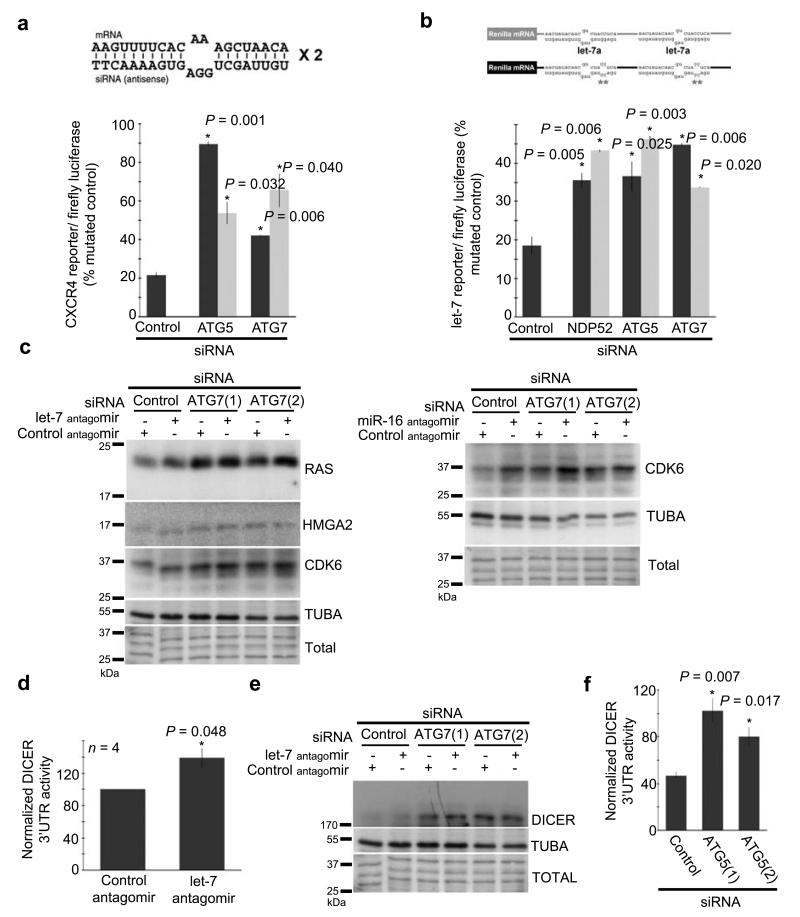
To test the consequence of this defect in AGO-loading on miRNA activity while eliminating any potential impact of autophagy on pre-miRNA processing or upstream steps, we measured the activity of exogenously delivered siRNA-siRNA* duplexes. To also rule out any confounding, autophagy-unrelated effects of mTOR such as generalized suppression of cap-dependent translation33 the following experiments involved anti-ATG5 or -ATG7 siRNAs to block autophagy at the level of autophagosome biogenesis, downstream of mTOR. In autophagy-deficient cells, a siRNA-siRNA* duplex, but not a missense control, silenced a Renilla reporter with partially complementary sites less efficiently than in control cells (Fig.5a). These data demonstrate that autophagy is required for continued loading of miRNA-miRNA* duplexes into AGO and for their durable activity (Fig.4e-g, 5a), consistent with homeostatic degradation of DICER-AGO2 complexes by autophagy (Fig.1-2). To test if autophagy also regulates endogenous miRNA action, we employed a second dual-luciferase reporter34 (Fig.5b, diagram) for let-7 activity, which was indeed less efficiently repressed upon depletion of NDP52, ATG5 or ATG7 (Fig.5b). Furthermore, accumulation of endogenous RAS (targeted by let-735), HMGA2 (targeted by let-736) or Cyclin-Dependent Kinase 6 (CDK6, targeted by let-7a and miR-1636) was increased in ATG7-depleted cells, reaching levels similar to those observed when miRNA activity was directly inhibited with antagomirs (Fig.5c).
Figure 5.
Autophagy is required for miRNA activity and engages post-transcriptional regulation of DICER. (a) Renilla luciferase expression controlled by an exogenous siRNA-siRNA* duplex targeting two partially complementary sites, in HeLa cells depleted of ATG5 or ATG7 with siRNA (black and grey bars correspond to two independent siRNA). Results are normalized to Firefly luciferase and Renilla luciferase expression in cells treated with a non-specific siRNA-siRNA* duplex. n = 2 independent experiments. (b) Renilla luciferase expression controlled by let-7 in HeLa cells depleted of NDP52 or ATG5 with siRNA (black and grey bars are independent siRNA). Results are normalized to firefly luciferase and a version of the Renilla luciferase construct with mutated let-7 binding sites. n = 3 independent experiments. (c) Western blot analysis of endogenous miRNA-repressed proteins (RAS, HMGA2, CDK6 are repressed by let-7, CDK6 is repressed by miR-16 as well [right panel]) or control (TUBA) in HeLa cells treated with control siRNA or either of two independent siRNA targeting ATG7. Cells were treated in parallel with non-specific antagomir or let-7 targeting antagomir. (d) Renilla luciferase expression controlled by the DICER 3′UTR in HeLa cells treated with control antagomir or anti-let-7 antagomir. (e) Western blot analysis of DICER in HeLa cells treated with control siRNA or either of two independent siRNA targeting ATG7. Cells were also treated with non-specific antagomir or let-7 targeting antagomir. (f) Renilla luciferase expression controlled by the DICER 3′UTR in HeLa cells treated with either of two independent siRNA targeting ATG5 or a control siRNA. All error bars are SEM.
Because DICER mRNA is a direct target of let-7a9, we finally investigated the contribution of decreased let-7a activity (Fig.5b-c) to the overall gain in DICER protein levels observed upon autophagy inhibition. Confirming repression of the DICER 3′UTR by let-7a, a let-7 antagomir increased stability of the DICER 3′UTR in a dual luciferase assay (Fig.5d) and caused endogenous DICER protein to over-accumulate (Fig.5e). DICER protein also accumulated in cells depleted of ATG7, reaching levels higher than those attained with the let-7 antagomir alone (Fig.5e), consistent with autophagy regulating DICER by both protein degradation (Fig.1-2) and miRNA-mediated post-transcriptional processes (Fig.5d-e). Moreover, inhibition of autophagy with ATG5-targeting siRNAs also augmented translation controlled by the DICER 3′UTR (Fig.5f). Together, these results suggest that autophagy regulates DICER at two distinct levels: (i) via NDP52-dependent protein degradation (Fig.1-2) and (ii) via post-transcriptional control of the DICER transcript. Thus, autophagy engages DICER, and possibly AGO2, in multilayered regulatory mechanisms impacting miRNA accumulation and activity.
Previous evidence hinted that miRNA-free AGO2 is degraded by an unknown mechanism: obstructing DICER-dependent AGO2 loading with miRNA-miRNA* duplexes caused AGO2 degradation in a manner only partly rescued with the proteasome inhibitor MG1328. Likewise, plant Ago1 is degraded possibly in an unloaded form37 in an MG132-independent but autophagy-dependent manner38. We have shown that the critical components of miRNA biogenesis complexes, DICER and AGO2, are selectively degraded via NDP52-dependent autophagy while inactive, i.e. without their pre-miRNA, miRNA, or miRNA* cargoes. Removal by autophagy of these inactive DICER-AGO2 complexes from the cellular pool seems required for persistent function of the remaining DICER-AGO2 complexes in stabilizing miRNA-miRNA* duplexes and transferring them into AGO29,30. Indeed, autophagy-deficient cells exhibit decreased AGO-loading with miRNA and miRNA* (Fig.4e-g), diminished miRNA and miRNA* levels (Fig.4a-d), and reduced silencing activity of exogenous siRNA-siRNA* duplexes (Fig.5a). Hence, inactive DICER-AGO2 complexes targeted to autophagy may compete with active complexes for cellular co-factors, substrates or targets. Strikingly, therefore, in both plants38 and metazoans, autophagy may be required for homeostatic clearance of miRNA-free DICER and AGO pools.
NDP52 and p62 can recruit ubiquitinated and ubiquitin-independent cargoes for autophagic degradation13,15. Similarly, our evidence suggests that multiple recognition events might recruit DICER and AGO2 to NDP52-dependent autophagy, including GEMIN4 association and ubiquitination. Provision of selectivity to autophagy is increasingly recognized to require complex and multiple recognition events that will certainly deserve careful attention in the future. Our findings predict that AGO-loading, miRNA abundance and miRNA activity may be perturbed by pathogens, stresses or diseases, known to alter autophagy11. Autophagy regulators, including PIK3CA, ATG6 (Beclin1) and PTEN, are, for instance, frequently mutated in cancers and several core autophagy genes were identified as tumor suppressors39. Consistent with this idea, autophagy is often aberrantly inhibited in cancer cells39. Incidentally, several studies have also reported global down-regulation of miRNAs in most types of tumors, correlating with less differentiated and proliferative states40. Our results thus suggest that autophagy inhibition in cancer cells may well be causal in this global decrease in miRNA levels, providing a potential mechanistic link between these two seemingly disparate phenomena.
Supplementary Material
Acknowledgements
The authors would like to thank Michael Johnston for providing 293T cells stably transfected with Tet-inducible Flag-AGO2 as well as the protocol for detection of ubiquitinated Flag-AGO2. Funding was provided by a core grant from ETH-Z to O.V. S.M. is a Wellcome Trust Research Career Development Fellow. The authors thank Kieran McGourty and Daniel Li for helpful discussions.
Footnotes
Competing Financial Interests:
The Authors declare they have no competing interests.
Supplementary Information:
A Supplementary Information file is attached to this Letter which includes Supplementary Figures 1-2 as well as the corresponding figure legends.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews. Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Molecular cell. 2010;38:323–32. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–53. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 5.Bazzini AA, Lee MT, Giraldez AJ. Ribosome Profiling Shows That miR-430 Reduces Translation Before Causing mRNA Decay in Zebrafish. Science. 2012 doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–9. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–9. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 8.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–9. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & development. 2004;18:1187–97. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 13.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell death and differentiation. 2012 doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y, Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. Journal of cell science. 2011;124:2692–701. doi: 10.1242/jcs.081232. [DOI] [PubMed] [Google Scholar]
- 16.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 17.Haase AD, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–6. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbings D, Voinnet O. Control of RNA silencing and localization by endolysosomes. Trends Cell Biol. 2010;20:491–501. doi: 10.1016/j.tcb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Friend K, et al. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nature structural & molecular biology. 2012;19:176–83. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pare JM, et al. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20:3273–84. doi: 10.1091/mbc.E09-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. The Journal of cell biology. 1982;93:144–54. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Y, et al. HCC-associated protein HCAP1, a variant of GEMIN4, interacts with zinc-finger proteins. J Biochem. 2003;133:713–8. doi: 10.1093/jb/mvg091. [DOI] [PubMed] [Google Scholar]
- 26.Meister G, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–55. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 27.Mourelatos Z, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gantier MP, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic acids research. 2011;39:5692–703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurai K, et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic acids research. 2011;39:1510–25. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Molecular cell. 2011;43:110–21. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwasaki S, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Molecular cell. 2010;39:292–9. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–22. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 34.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slack F. let-7 microRNA reduces tumor growth. Cell Cycle. 2009;8:1823. doi: 10.4161/cc.8.12.8639. [DOI] [PubMed] [Google Scholar]
- 36.Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nature genetics. 2011;43:371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csorba T, Lozsa R, Hutvagner G, Burgyan J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. The Plant journal : for cell and molecular biology. 2010;62:463–72. doi: 10.1111/j.1365-313X.2010.04163.x. [DOI] [PubMed] [Google Scholar]
- 38.Derrien B, et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. The Journal of clinical investigation. 2011;121:1231–41. doi: 10.1172/JCI44145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.