Abstract
Background
Lipid-mediated delivery of DNA is hindered by extracellular and intracellular barriers that significantly reduce the transfection efficiency of synthetic nonviral vectors.
Results
In this study we investigated the role of the actin and microtubule networks on the uptake and cytoplasmic transport of multicomponent cationic liposome–DNA complexes in CHO-K1 live cells by means of confocal laser scanning microscopy and 3D single particle tracking. Treatment with actin (latrunculin B)- and microtubule-disrupting (nocodazole) reagents indicated that intracellular trafficking of complexes predominantly involves microtubule-dependent active transport. We found that the actin network has a major effect on the initial uptake of complexes, while the microtubule network is mainly responsible for the subsequent active transportation to the lysosomes.
Conclusion
Collectively, a strategy to improve the efficiency of lipid gene vectors can be formulated. We could find a lipid formulation that allows the nanoparticles to avoid the microtubule pathway to lysosomes.
Gene therapy represents a promising approach for the treatment of many fatal diseases [1]. To introduce gene therapeutics in clinical practice, vectors that transfer genes efficiently and safely are needed. Viruses can be effective vectors but their use is hindered by safety considerations. In contrast, nonviral transfection systems are usually safer and do not limit the size of DNA that can be delivered. Nonviral vectors such as cationic liposomes (CLs), polymers, dendrimers and peptides, have gained increasing attention because they are non-immunogenic, not oncogenic, easy to produce in large scale and capable of delivering large genetic material. There have also been attempts to combine the benefits of different nonviral systems into one delivery vehicle. Among nonviral vectors, CLs are commonly used as a transfection reagent for DNA, RNA or proteins and as co-adjuvant of antigens for vaccination [2]. Currently, there are approximately 12 liposome-based drugs approved for clinical use and more are in various stages of clinical trials. Most liposomal drug formulations are approved for intravenous application [2]. For wider clinical use their low transfection efficiency (TE) needs to be improved.
The authors have recently shown that multicomponent (MC) lipoplexes, containing from four to six lipid species within their lipid bilayer, are superior in TE with respect to frequently used binary formulations [3–15]. Since MC systems are highly efficient and completely not cytotoxic, they represent very promising candidates for gene-delivery applications both in vitro and in vivo. In previous investigations, we have provided evidence that MC lipoplexes have intrinsic endosomal rupture properties that allow plasmid DNA to escape from endosomes with extremely high efficiency. Endosomal rupture results in an extraordinarily homogeneous distribution of unbound plasmid DNA throughout the cytoplasm and in the nucleus. Since endosomal escape and intracellular dynamics are two strongly related transfection processes, we asked whether a correlation between the unusual endosomal escape ability of MC lipoplexes and the mechanism for the cytoplasmic transport would exist. The involvement of microtubules in the intracellular dynamics of lipoplexes has been the subject of previous investigations [16–18]. Hasegawa et al. demonstrated that 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-3β[N-(2-hydroxyethylaminoethane)-carbamoyl]–cholestene/DNA (DOPE-HyC–Chol/DNA) lipoplexes are transported to lysosomes by a microtubule-mediated pathway [16]. More recently, Wang and MacDonald found that some microtubule-depolymerizing agents, such as colchicine, vinblastine, vincristine, nocodazole (NCZ) and podophyllotoxin dramatically increase the transfection of O-ethyldioleoylphosphatidylcholinium/DNA (EDOPC/DNA) complexes [17] . Akita et al. have shown that octa-arginine-modified liposome–DNA complexes are directionally transported in their intact form before endosomal escape [18]. Ondrej et al. observed an indispensable but different role of both types of cytoskeleton, actin and microtubular, in the processes of gene delivery [19]. Disruption of either of these networks led to the interruption of transport to the nucleus, and accumulation of large aggregates at the cell periphery. While the contribution of actin and microtubule-dependent transport seems to be clear [1,16–19], the issue of whether and, if so, how actin microfilaments and microtubules affect the internalization of MC lipoplexes remains to be better elucidated.
The goal of this study was to investigate the role of cytoskeleton structure on the uptake and intracellular dynamics of MC lipoplexes in CHO-K1 live cells. This cell line was chosen due to its common use in biological and medical research. Indeed, CHO-K1 cells are among the most widely used mammalian cells for transfection, expression, and large-scale recombinant protein production. We applied 3D single particle tracking (SPT) to study the trafficking of lipoplexes and confocal laser scanning microscopy (CLSM) to identify their intracellular fate. From the 3D trajectories obtained in SPT experiments we calculated 3D mean square displacements (MSD) over time to classify the lipoplex intracellular trafficking as ‘Brownian motion’ or ‘directed motion’. Thus, we used CLSM to visualize the uptake and intracellular trafficking of lipoplexes. In particular, their final fate was investigated by determining the extent of co-localization of fluorescently-tagged complexes and lysosomes.
Experimental
Cationic liposomes preparation
The cationic lipids 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and 3β-[N-(N’,N’-dimethylaminoethane)-carbamoyl]–cholesterol (DC–Chol), and the zwitterionic helper lipids dioleoylphosphocholine (DOPC) and dioleoylphosphatidylethanolamine, were purchased from Avanti Polar Lipids Inc. (AL, USA). They were used without further purification for the preparation of DOTAP–DOPC and DC–Chol–DOPE CLs. In brief, binary lipid mixtures, at molar fractions of neutral lipid in the bilayer ϕ = neutral/(neutral+cationic) (mol/mol) = 0.5, were first dissolved in chloroform and then left to evaporate under vacuum for at least 24 h. The obtained lipid films were hydrated with Nanopure® water until a final concentration of approximately 1 mg/ml. Sonication to clarity of the obtained dispersions was performed to obtain unilamellar CLs.
Lipoplexes preparation
For size and ζ-potential measurements samples were prepared at nine cationic lipid/DNA charge ratio (mol/mol), that is, ρ = (cationic lipid [by mole]/DNA base) = 0.062, 0.125, 0.25, 0.5, 1, 1.5, 2, 2.5 and 3. On the basis of size and ζ-potential results, all the SPT and CLSM experiments were performed at ρ = 2.5. To form quaternary MC lipoplexes (DOTAP–DOPC–DC–Chol–DOPE/pDNA) ready for administration to CHO-K1 cells, 100 μl of phosphate-buffered saline (PBS) were added to a 10-μl dispersion containing binary CLs (5 μl of DOTAP–DOPC and 5 μl of DC–Chol–DOPE) [4]. The same amount of buffer was added to 1 μl of pDNA. These solutions were equilibrated for a few minutes (the final concentration of lipid is, therefore, approximately 47 pg/μl). Then, the pDNA solution was poured in the liposome dispersion and after 20 min the complexes were ready to use. Fluorescein- and Cy3-labeled plasmids DNA (2.7 kbp, double stranded, circular plasmid) were purchased from Mirus Bio Corporation (Madison, WI, USA) and used for SPT and CLSM experiments, respectively. They are both supplied at 1 μg/μl in a 10 mM Tris-HCl and 1 mM EDTA buffer (pH 7.5).
Size & ζ-potential
The size and size distribution of liposomes and lipoplexes were measured at a temperature of 25°C by a Malvern ZetaSizer Nanospectrometer equipped with a 5-mW HeNe laser (wavelength λ = 632.8 nm) and a digital logarithmic correlator. The normalized intensity autocorrelation functions were detected at a 90° by a logarithmic digital correlator and were analyzed using the CONTIN algorithm; an inverse Laplace transform method to obtain the distribution of the diffusion coefficient (D) of the particles [20,21]. Each of these coefficients is then converted into an effective hydrodynamic radius RH using the Stokes–Einstein relationship RH = KBT/(6πηD), where KBT is the thermal energy and η the solvent viscosity. Both liposomes and lipoplexes invariably showed a size distribution, and the values of the radii reported here correspond to the socalled ‘intensity weighted’ average [20]. The electrophoretic mobility measurements were carried out by means of the laser Doppler electrophoresis technique, with the same apparatus used for size measurements. The mobility (u) was converted into the ζ-potential using the Smoluchowski relation ζ = uη/ε, where η and ε are the viscosity and the permittivity of the solvent phase, respectively (η = 0.88 centipoise (cP); ε=78.5).
Cell culture & transfection
CHO-K1 cells were purchased from American Type Culture Collection (CCL-61 ATCC) and were grown in Ham’s F12K medium supplemented with 10% of fetal bovine serum (FBS) at 37°C and in 5% CO2. Cells were split every 2–4 days to maintain monolayer coverage. The day before transfection, cells were seeded in single-well plates (150,000 cells/well, typical working volume of ~2 ml) using a medium without antibiotics. On the day of transfection, the culture medium was removed, and cells were washed three times with PBS before adding MC lipoplexes in a total medium volume of approximately 2 ml (the final DNA concentration is therefore 0.5 μg/ml). After administration, 1–4 h of incubation allowed complete internalization of the lipoplexes (data not reported). In SPT experiments, the normal medium was then replaced with a medium containing 200 μl of DiIC18(3) (Invitrogen, CA, USA), a lipophilic membrane marker that aided the localization of a single lipoplex inside the cell during the tracking procedure (DiIC18[3] working solution prepared at 1 μM). In CLSM experiments, instead, the normal medium was then replaced with a medium containing 1 μl of Lyso Sensor (a lysosomal marker, purchased from Sigma-Aldrich [MO, USA]) from a working solution at 50 mM. After 15 min, in both SPT and CLSM experiments, the aforementioned media were at last replaced with a measurement medium depleted of phenol red. In the case of Latrunculin B (LAT) or NCZ treatment, both inhibitors purchased from Sigma-Aldrich, the transfection procedure was slightly different. Before lipoplex administration, cells were first incubated for no more than 15 min with a 20-μM LAT or NCZ medium and then washed three-times with approximately 2 ml of PBS. Such drug concentrations were the highest possible concentrations that did not significantly reduce cell viability (Supplementary Figure 1).
SPT experiments
Two-photon microscopy experiments were carried out using a Zeiss Axiovert 135 TV microscope. The excitation source was a mode-locked Ti:Sapphire laser (Mai Tai HP with integrated Millennia, Spectra-Physics [CA, USA]) tuned at 790 nm. More details about the experimental setup can be found elsewhere [22–25]. The experiments were controlled by a custom-made data acquisition and analysis software (SimFCS, Laboratory for Fluorescence Dynamics – LFD [CA, USA]), downloadable from the LFD website [101]. The complete theory of 3D single-particle tracking based on the orbital method can be found elsewhere [26–28]. In brief, the particle position is determined on the fly by analyzing the fast Fourier transform of the integrated fluorescence intensity. During each tracking cycle, the intensity signal is integrated at different points along the circular orbits traced around the particle by the excitation beam. Before the next cycle, the center of scanning is moved to the position determined for the particle in the previous cycle, allowing to be always ‘on top’ of the particle.
SPT data analysis
From the discrete experimental trajectory (xn = x(nδt), yn = y(nδt), zn = z(nδt), n = 0,1,2… N-1) with δt being the data acquisition time interval, the 3D MSD was calculated averaging over all pairs (of points that are nδt time steps apart) (Equation 1).
| (EQUATION 1) |
where N is the total number of trajectory points and nδt is the time lag τ.
Fitting the calculated MSDs with theoretical expressions allows to classify the lipoplex intracellular trafficking and to obtain the dynamic parameters (i.e., the diffusion coefficient and the velocity). Equations 2 & 3 represent the theoretical 3D MSD when lipoplexes undergo Brownian and directed diffusion, respectively.
| (EQUATION 2) |
| (EQUATION 3) |
where D and v are the 3D diffusion coefficient and the velocity, respectively.
In order to enhance the statistic accuracy of the derived dynamic parameters, it is mandatory to set a proper maximum time lag (mδt) before the fitting procedure [29]. Taking into account the wide range of trajectory lengths, we decided to set m = 49 that following Qian et al. yields to a relative error less than 27% [30].
CLSM experiments
Confocal microscopy experiments were carried out using a Fluoview FV-1000 (Olympus, Tokyo, Japan) microscope. The excitation sources were a Multiline Argon laser tuned at 488 nm and HeNe laser tuned at 543 nm. More details regarding the experimental setup can be found elsewhere [31]. Following the guideline by Bolte and Cordelieres [32], the experiments were controlled by a data acquisition software (FV10-ASW, Olympus, Tokyo, Japan). Instead, Fiji was used to analyze the data. It is a distribution of ImageJ containing several plugins and downloadable from the Fiji website [102,103].
CLSM data analysis
A qualitative co-localization analysis involves a simple pixel-by-pixel overlay of different channels, pseudo-colored using a color look-up table. For instance, in our experiments we chose ‘green’ for LysoSensor-labeled lysosomes and ‘red’ for Cy3-labeled lipoplexes, which gives ‘yellow’ hotspots where the two fluorescent labels are present in the same pixels (i.e., co-localized). However, this method is highly dependent on the relative signal intensity collected in both channels; in fact, the overlay image yields a reliable degree of co-localization only if both images exhibit similar gray level dynamics, for example, when the histograms of each channel are similar. This is rarely the case when imaging two channels characterized by different signal strength. A more quantitative analysis, instead, might rely on the so-called object-based approach. The basic idea is to employ image segmentation methods for object identification and delineation. After that, it is possible to calculate centroids and centers of mass (intensity centers) from the structures. A measure of co-localization is then achieved by comparing the position of the 3D centroids or centers of mass of the respective subcellular structures in the two channels. In the instance of objects larger than the optical resolution or different in size between the two channels, Lachmanovich et al. developed an overlap approach: objects in the two channels are classified as ‘co-localized’ if the center of mass (and not centroids to avoid possible under-estimation due to anisotropic intensity distributions) of the object in the green channel falls into the area covered by the object in the red channel [33]. The overall degree of co-localization of red (green) objects C1 (C2) is then given by the percentage of red (green) objects co-localizing with green (red) objects in the area of interest:
| (EQUATION 4) |
where Nr, Ng and NC represent the number of red objects, green objects and the co-localized (red or green) objects, respectively. Please note: in our system, ‘object’ refers to ‘fluorescent vesicle’.
We obtained the centers of mass using a simple custom-made modification of the 3D objects counter plugin described in Bolte and Cordelieres [32], and contained in Fiji [102]. Before running the segmentation analysis, images were corrected with a simple background subtraction. The presence of cell auto-fluorescence in the green channel made a further intensity subtraction necessary; in fact, selecting by visual inspection different (not less than five) small region of interests in the portions of cytoplasm not containing particles, it has been possible to calculate an average intensity, afterwards set as a threshold.
Results
Size & ζ-potential results
MC liposomes were found to be positively charged vesicles (ζP ~57 mV) with an average hydrodynamic radius RH approximately 70 nm. Upon addition of DNA to CLs, MC lipoplexes formed in a self-assembled manner. Size and ζ-potential of lipoplexes are reported in Table 1. As Table 1 clearly shows, both reentrant condensation and charge inversion occurred [34]. To choose the proper lipoplex formulation for SPT and CLSM experiments, the following general considerations were made:
▪ Lipoplexes must be positively charged to associate electrostatically with cells;
▪ Charge ratio, ρ, must be minimized due to potential cytotoxicity of cationic lipids (Supplementary Figure 2);
▪ Small-size complexes are better internalized and processed by cells. As a result, lipoplexes at ρ = 2.5 were selected because they exhibited the lowest charge ratio ensuring positive ζP (~40 mV) and the lowest colloidal dimensions.
Table 1.
Hydrodynamic radius, RH, and values of ζ-potential, ζP, of multi-component lipoplexes as a function of increasing charge ratio, ρ.
| ρ | RH [nm] | ζp [mV] |
|---|---|---|
| 0.062 | 114 ± 2 | −40.1 ± 0.9 |
| 0.125 | 134 ± 3 | −39.2 ± 0.8 |
| 0.250 | 159 ± 4 | −25.3 ± 0.5 |
| 0.500 | 189 ± 5 | −10.2 ± 0.5 |
| 1.000 | 300 ± 8 | 5.1 ± 0.7 |
| 1.500 | 212 ± 4 | 13.4 ± 0.6 |
| 2.000 | 183 ± 4 | 25.1 ± 0.5 |
| 2.500 | 125 ± 3 | 42.4 ± 0.3 |
| 3.000 | 120 ± 3 | 46.7 ± 0.5 |
SPT results
The 3D SPT technique allowed to obtain 100 single particle (3D) trajectories, divided into three different categories:
▪ Fifty-six trajectories of lipoplexes tracked in CHO-K1 cells not treated (NT) with inhibitors;
▪ Twenty-five trajectories of lipoplexes tracked in CHO-K1 cells treated with Latrunculin B;
▪ Nineteen trajectories of lipoplexes tracked in CHO-K1 cells treated with NCZ.
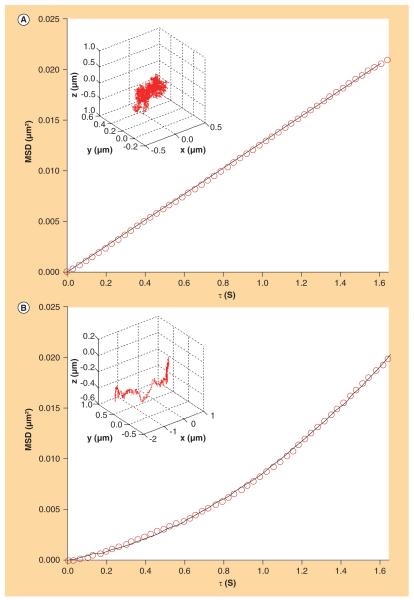
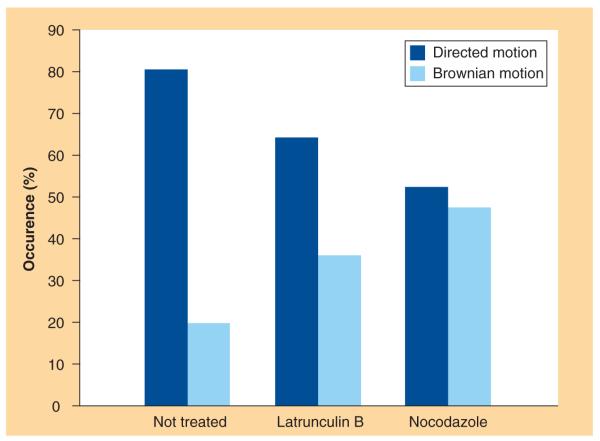
As described in the ‘Materials and Methods’ section, we calculated the 3D MSD directly from the discrete trajectory coordinates using Equation 1. Fittings of all the 100 MSD curves following the theoretical expressions (Equations 2 & 3) led to the classification of the involved modes of motion and to the derivation of dynamic parameters of the lipoplex intracellular trafficking. The first qualitative result is the occurrence of either Brownian motion or directed motion. Figure 1 shows two representative MSD plots as well as the corresponding fitting curves and 3D trajectories (inset in the top left corner of each panel). Figure 1A & 1B are representative of Brownian motion and directed motion, respectively. By matching the MSD curves with the corresponding category of motion we obtain the occurrence percentage of each mode of motion for NT, LAT-treated and NCZ-treated cells. This is shown in the histogram of Figure 2, where a specific trend emerges. In fact, the percentage of directed motion (dark gray bars) decreases in both LAT- and NCZ-treated cells with respect to NT cells. In a complementary way, the percentage of Brownian motion (light gray bars) evidently increases in treated cells with respect to control cells. According to the literature [22–26], some tenths of trajectories are absolutely sufficient for the categorization to be accurate. Thus, the classification reported in Figure 2, which is related to 56 NT-, 25 LAT- and 19 NCZ-trajectories, can be considered as statistically meaningful. The (unweighted) MSD curve fittings allowed deriving the 3D diffusion coefficients and velocities. Mean values and corresponding standard deviations are listed in Table 2. No specific trend for the mean diffusion coefficients emerges if we consider the large standard deviation. According to the Stoke–Einstein equation, the diffusion coefficient and the hydrodynamic radius of a particle in a diffusing medium are inversely related. So, the absence of any specific trend is not due to the number of particles followed by the tracking procedure, but might reflect the high polydispersity in size of MC lipoplexes, the larger the size distribution of lipoplexes the larger the distribution of diffusion coefficients. In this sense, it does not seem to be a limit of our data analysis but it is rather an intrinsic feature of lipofection experiments. In a similar way, the mean velocities for NT, LAT-treated and NCZ-treated cells lay within the same interval of values. The most reliable explanation is that the directed motion along either actin filaments or microtubule is nonspecific and mainly dependent on the nanocarrier properties [18].
Figure 1. Representative 3D mean square displacements curves (circles).
(A) Brownian motion and (B) directed motion. The best curve fittings are also shown (-). In the top left corner of each panel, a 3D plot of the corresponding experimental trajectory is shown.
MSD: Mean square displacements.
Figure 2.
Occurrence percentage of directed motion and Brownian motion for each experimental condition.
Table 2.
Physical parameters from unweighted fits of 3D mean square displacements curves.
| Not treated cells | Latrunculin B- treated cells |
Nocodazole- treated cells |
|
|---|---|---|---|
| Directed motion | |||
| D† [10−3 μm2/s] | 1.54 ± 1.53 | 3.61 ± 2.12 | 1.80 ± 1.19 |
| v† [μm/s] | 0.054 ± 0.028 | 0.060 ± 0.032 | 0.055 ± 0.034 |
| Brownian motion | |||
| D† [10−3 μm2/s] | 1.44 ± 0.77 | 4.68 ± 3.47 | 1.82 ± 0.66 |
Represents the average value.
CLSM results
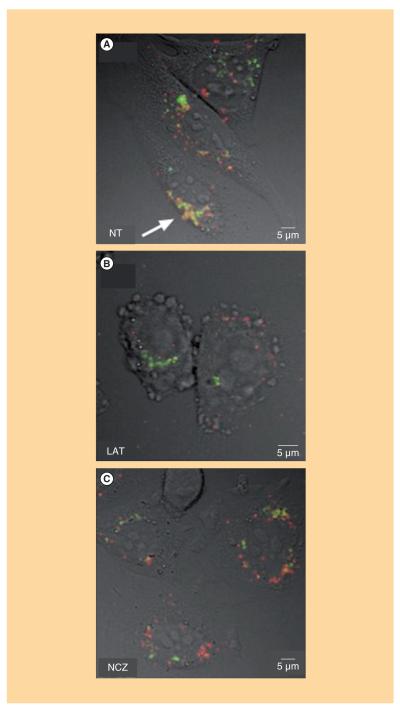
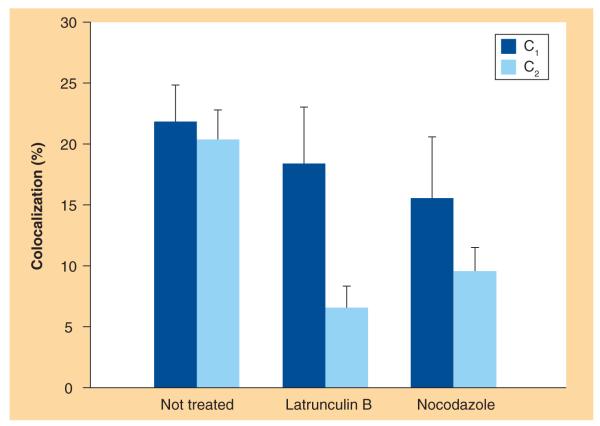
Similarly to the SPT results, confocal images were also divided into the same study groups: images of NT cells, LAT-treated cells and NCZ-treated cells. A qualitative co-localization was obtained by overlaying the two channels (pseudo-colored using red and green look-up tables) pixel by pixel. In Figure 3, representative overlays of fluorescence and Nomarski images of NT, LAT-treated and NCZ-treated cells are shown. In NT cells (Figure 3A), lipoplexes (red) are largely internalized within the cells. They are found both at the cell periphery and in the perinuclear region. Lipoplexes approaching the cell nucleus largely co-localize with lysosomes (green), as denoted by the yellow hotspots (white arrows). Notably, in LAT-treated cells (Figure 3B), the largest amount of lipoplexes accumulates at the plasma membrane, while only a minor part seems to be effectively uptaken by the cell. Accordingly, the internalized fraction co-localizes much less with lysosomes, compared with NT cells. Finally, in NCZ-treated cells (Figure 3C), lipoplexes even less clearly co-localize with lysosomes. As explained in the Materials and Methods section, an object-based co-localization approach allowed us to quantitatively investigate the degree of co-localization, for example, the percentages C1 (C2) of lipoplexes (lysosomes) co-localized with lysosomes (lipoplexes). Average values of C1 and C2 for each experimental condition are shown in the histogram of Figure 4. To better interpret and discuss these data, one more comment on the co-localization coefficient is needed. While C1 is proportional to the fraction of lipoplexes that have reached the lysosomes, C2 is directly related to the percentage of lysosomes that have been reached by internalized lipoplexes. This means that, within the same study group, the value of these parameters are definitely independent from each other. For instance, if all the lipoplexes co-localized with lysosomes, C1 would tend to its maximal value (C1 = 1). Nevertheless, this would not assure that all the lysosomes would be reached by lipoplexes. Indeed, even though one assumes that all the lypoplexes are shuttled to lysosomes for degradation, it could be that a minor fraction of lysosomes would be enough to accommodate all the internalized lipoplexes. This would result in a low C2 value (C2<1). On the other hand, comparing the value of the same co-localization coefficient within different study groups (i.e., NT, LAT and NCZ) is a meaningful operation. As Figure 4 shows, a specific trend emerges for both of the co-localization coefficients. With respect to NT cells, the average value of C1 (dark gray bars) sensibly decreases in LAT-treated cells and even more in NCZ-treated cells. With respect to NT cells, the average values of C2 (light gray bars) decrease in LAT- and NCZ-treated cells.
Figure 3. Representative overlays of fluorescence and Nomarski images of (A) non-, (B) latrunculin B- and (C) nocodazole-treated cells.
A qualitative co-localization was obtained simply overlaying pixel-by-pixel different channels, pseudo-colored using red and green look-up tables. We used green for LysoSensor-labeled lysosomes while red for Cy3-labeled lipoplexes. The presence of the two fluorescent labels in the same pixels (i.e., co-localization) produces yellow hotspots (indicated by white arrows).
LAT: Latrunculin B; NCZ: Nocodazole; NT: Not treated.
Figure 4.
Average co-localization percentages C1 and C2 for each experimental condition (10 not-treated, 25 latrunculin B-treated and 23 nocodazole-treated cells).
Discussion
MC lipoplexes have emerged as promising nonviral gene carriers due to high TE and low cytotoxicity in several cell lines [3–15]. Their key feature seems to be their uncommon endosomal escape ability. Indeed, entry of the complexes into the lysosomal compartment has been suggested to cause massive DNA degradation and to prevent transfection. Since intracellular trafficking and endosomal escape are strongly related to each other, novel insights into the cytoplasmic transport of MC lipoplexes could help to explain their unusual tendency to escape from endosomes and to prevent lysosomal degradation. The aim of the present work was to gain novel quantitative insights into the role played by both actin microfilaments and microtubules in the intracellular dynamics of MC lipoplexes. SPT experiments allowed us to follow the 3D lipoplex journey inside the cell. Data analysis allowed us to categorize the intracellular trafficking of MC lipoplexes by their mode of motion (Figure 2). In NT cells, lipoplex intracellular motion was found to be mainly directed (~80%), while treatment with cytoskeleton inhibitors resulted in a pronounced reduction of directional movement. This finding means that both the cytoskeleton structures are involved in the active transport of MC lipoplexes. In LAT-treated cells, the reduction in directional movement may indicate that some lipoplexes attach to the actin microfilaments and ‘walk’ along them before attaching to the microtubule network [19]. The simultaneous increase of Brownian motion may also be due to the tracking of lipoplexes accumulated at the plasma membrane whose diffusion is, at least locally, Brownian [35,36]. Figure 2 indicates a preferential role played by the microtubule network in regulating the lipoplex intracellular trafficking. This finding confirms that microtubules are used as a preferential route for cytoplasmic transport of vesicular structures [16–19]. The occurrence of relatively high percentages (35–45%) of direct motion in LAT and NCZ cells deserves further consideration. In LAT-treated cells such percentage is most likely to reflect the direct motion of MC lipoplexes along the intact microtubule network, while, in NCZ-treated cells, it could arise from the active transportation along the actin microfilaments. In principle, such percentages of direct motion could be due, at least in part, to the reorganization of the actin and microtubule networks. We tested this hypothesis by staining the cytoskeleton with actin-GFP and tubulin-YFP and by following its temporal recovery by CLSM (Supplementary Figure 3). Upon the time range of SPT experiments (i.e., 4–8 h after lipoplex administration) a gradual, but not complete, restructuring of cytoskeleton was clearly found. Confocal images, therefore, corroborate our suggestion that the percentages of directed motion in inhibitors-treated cells are partly due to active transport along newly reformed cytoskeleton filaments. Given the results of Supplementary Figure 3, an alternative approach would consist in adding inhibitors just before starting SPT experiments; for example, 4 h after lipoplex administration. By doing so, the effect of inhibitors would be definitely stronger and probably more detectable. However, 4 h after administration, lipolexes are mainly inside the cell so that any information about the effect of cytoskeleton on the cellular uptake would not be possible to obtain. Thus, the followed experimental strategy seems to be the best compromise to examine the effect of cytoskeleton on the intracellular dynamics without leaving to investigate its influence on the cellular uptake. Furthermore, comparable dynamical parameters after different cell treatments might be reliably explained by assuming that the directed motion along either actin filaments or microtubule is nonspecific and mainly dependent on the nanocarrier properties. The lipid formulation might influence the motion if MC lipoplexes are uptaken by either endocytosis or fusion with the plasma membrane. The former mechanism does not hamper interactions between inner and outer lipid bilayers of endosomes, granting to the engulfed lipoplex surface characteristics similar to those of bare vesicles. The latter mechanism has been found to occur as well [4] and, according to the present understanding, fusion of multilamellar lipoplexes results in the internalization of vesicular structures retaining the largest part of the gene cargo with just a number of lipid layers peeled off by interaction with the plasma membrane.
In light of the large involvement of the microtubule network in the cytoplasmic transport of MC lipoplexes (Figure 2), a massive degradation at the lysosomal level would be expected. Clearly, the opposite would stand true in cells pretreated with NCZ. We put to test this hypothesis by CLSM co-localization experiments. Confocal images (Figure 3) allowed us to visualize fluorescently labeled lipoplexes (red) in the presence of Lyso Sensor (green), a primary lysosome marker. At a first glance, it is clear that lipoplexes are rapidly destined to degradation in NT cells, while both LAT and NCZ treatments largely affected their intracellular localization. However, the pixel-by-pixel overlay of confocal images is merely qualitative and highly dependent on the relative fluorescence intensity. Co-localization analysis [32] reported in Figure 4 was, therefore, decisive to support our visual interpretation of CLSM images. After treatment with LAT, the fraction of lysosomes reached by lipoplexes (C2) decreased of more than 65% with respect to NT cells. This finding is most likely to indicate that a lower number of MC lipoplexes was internalized within LAT-treated cells. On the other side, once inside the cell, lipoplexes could move towards the nucleus as efficiently as in NT cells (Supplementary Figure 4). This is quantitatively demonstrated by observing that the fraction of internalized lipoplexes that reached lysosomes was similar to its counterpart in NT cells (C1LAT~C1NT). The combined analysis of C1 and C2 suggests that actin filaments most likely play a role in the uptake of MC lipoplexes rather than in their following intracellular trafficking towards the nucleus. This conclusion is in very good agreement with recent findings showing that lipoplexes are taken up in living cells mainly through fluid-phase macropinocytosis [37,38]. Macropinocytosis relies on the formation of large endocytic vesicles generated by actin-driven invaginations of the plasma membrane, called membrane ruffles [39]. Thus, it is conceivable that changes in actin organization as those induced by treatment with LAT do promote the destabilization of actin-rich membrane ruffles with the result that the macropinocytosis pathway is largely inhibited. This interpretation is further confirmed by the slow transport rate (Table 2) typical of the trafficking of vesicles uptaken by macropinocytosis [18]. In summary, our findings suggest that the actin network plays an active role in the cellular uptake of MC lipolexes so that a lower number of lysosomal compartments is involved (C2LAT<C2NT), but it has a minor involvement, if any, in the cytoplasmic transport and final fate of internalized lipoplexes (C1LAT~C1NT). Our conclusions are supported by the recent results reported by Dos Santos et al. who showed that the uptake of nanoparticles in HeLa and 1321N1 cells was strongly affected by actin depolymerisation [40].
The inhibition of lipoplex uptake in LAT-treated cells could also explain the reduction in TE usually observed after LAT treatment [6] and confirmed in the case of MC lipoplexes (data not reported).
By contrast, in NCZ-treated cells, MC lipoplexes were located diffusely throughout the cell 1 h after transfection, indicating that disruption of the microtubule network played a minor role, if any, in the uptake process (Figure 3C). Our observation is in good agreement with recent results showing that NCZ does not affect the internalization of lipoplexes [41]. CLSM images indicated that most lipoplexesdo not co-localize with lysosomes (Figure 3C). Figure 4 shows that the fraction of lipoplexes that avoid the lysosomal degradationis larger than that of NT cells (C1NCZ<C1NT). Likewise, the portion of lysosomes reached by lipoplexesis lower than that of NT cells (C2NCZ<C2NT). Collectively, CLSM findings denote that, in NCZ-treated cells, the ability of MC lipoplexes to enter cells is not severely compromised, while their intracellular motion does. At this stage, it is worth considering that the effectiveness of gene vectors is proportional to the expression levels of the transgene [42]. It appears that, in the case of lipoplexes such as those used here, the transfection efficiency is highly hampered by the vector entrapment within lysosomes, with the subsequent degradation of the DNA payload. In this light, the reported increase in transfection efficiency upon inhibition of microtubule-dependent vesicular transport [16,17] appears an obvious outcome (Supplementary Figure 5). Our data also explains why lipoplexes that enter the cytoplasm in the immediate vicinity of the nucleus can efficiently reach it, while those entering at locations far from the nucleus may fail to traffic in the absence of microtubules [43]. However, even if active transport is inhibited due to microtubules disruption, MC lipoplexes can still move towards the nucleus undergoing Brownian diffusion. It is proved by the results shown in Figure 2 and in Supplementary Figure 5.
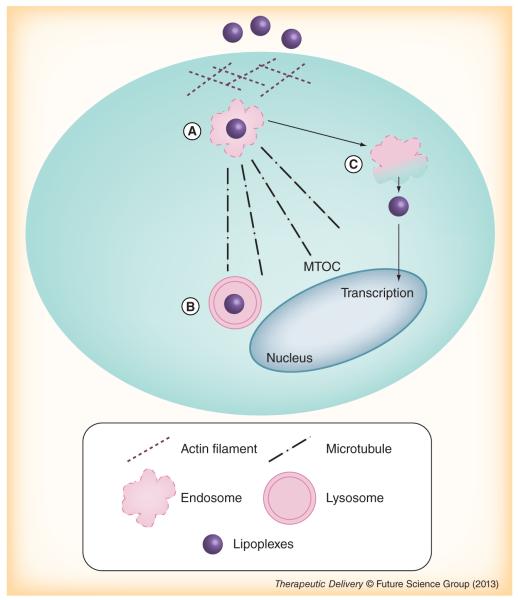
Figure 5 summarizes our present understanding of the role of cytoskeleton structure on the uptake and intracellular trafficking of lipoplexes. Cholesterol-sensitive macropinocytosis is the major pathway responsible for lipoplex internalization and is regulated by formation of large endocytic vesicles of irregular size and shape, generated by actin-driven invaginations of the plasma membrane (Figure 5A) [37,38]. Accordingly, if the actin network is destroyed, the ability of lipoplexes to penetrate within cells is dramatically reduced and they accumulate at the plasma membrane. Instead, once inside the cell, complexes are actively transported, preferentially along the microtubules, from endosomes to lysosomes (close to microtubule organizing center) for degradation (Figure 5B) [42]. Only a few of the DNA molecules can escape from endosomes to the cytoplasm before reaching the lysosomes and even less will enter the nucleus for transcription (Figure 5C). In this connection, if microtubules are disrupted, the complexes are less frequently destined to lysosomes, thus, increasing their chances to escape from vesicles and promote transcription of the target gene.
Figure 5. Present understanding of the role of cytoskeleton structure on the uptake and intracellular trafficking of lipoplexes.
(A) Cholesterol-sensitive macropinocytosis is the major pathway responsible for lipoplex internalization and is regulated by formation of large endocytic vesicles of irregular size and shape. (B) Once inside the cell, the complexes are actively transported from endosomes to lysosomes, preferentially, along the microtubules, near the MTOC for degradation. (C) Only a few of the DNA molecules can escape from endosomes to cytoplasm before entering lysosomes and even less will enter the nucleus for transcription.
MTOC: microtubule organizing center.
Conclusion
Despite the increased application of nanomaterials in diagnostics and therapeutics, the interactions of nanoparticles with subcellular structures in living cells remain relatively undeveloped [44]. Thus, quantitative investigations that allows for the precise tracking of cell-associated nanoparticles in a living cell are highly desirable [44]. In this work, we have investigated the role of the cytoskeleton architecture in both the cellular uptake and intracellular trafficking of MC lipoplexes by combining 3D SPT and CLSM. We have shown that the intracellular uptake of MC lipoplexesis is largely affected by the actin network, while cytoplasmic trafficking predominantly involves microtubule-dependent transport. This mechanism of action of CLs should be considered for the rational design of novel delivery systems with superior transfection efficiency. We could find a lipid formulation that allows the nanoparticles to contribute more efficiently to the transfection process; for example, release the gene payload for nucleus entry and consequent transcription, by avoiding the microtubule pathway to lysosomes. It requires a better understanding of the molecular mechanisms involved in the trafficking. This crucial aspect of nanodelivery is currently being investigated in our laboratory. It has also recently been shown that uptake and cellular processing of nanoparticles can be largely affected by the cell cycle [45]. Experiments aimed at evaluating the role of cell cycle and type on the uptake and final fate of MC lipoplexes will be addressed in future investigations.
Future perspective
An essential requirement for nanocarriers is efficient translocation of DNA to the cell nucleus. To accomplish this mission, nanocarriers must avoid lysosomes, the cellular organelles containing proteases and other enzymes that degrade most biological substances. Our results have demonstrated that accumulation of lipoplexes in lysosomes is a microtubule-mediated process. Thus, we put forth the concept that efficient delivery systems should move to the nucleus by a mechanism other than active transportation along the microtubule pathway. A first step towards that direction is the precise knowledge of all the molecular mechanisms regulating the interaction between nanocarriers and the cytoskeleton network. This crucial aspect of nanodelivery will be the focus of future research in the field of gene delivery.
Supplementary Material
Executive summary.
Single particle tracking results
▪ In control cells, lipoplexes’ intracellular motion is mainly directed.
▪ In cells treated with actin and microtubule-disrupting reagents, the percentage of directed motion decreases for both treatments.
▪ The percentage of Brownian motion increases in treated cells with respect to control cells.
Confocal laser scanning microscopy results
▪ In control cells, lipoplexes are largely internalized within the cells and approaching the cell nucleus largely co-localize with lysosomes.
▪ After latrunculin B treatment we observed that a minor fraction of multicomponent lipoplexescolocalized with lysosomes. This suggests that actin may help lipoplexes to efficiently hop onto the tubulin.
▪ In cells treated with microtubule-disrupting reagents, lipoplexes even less clearly co-localize with lysosomes, with respect to the actin disruption case.
Conclusion
▪ The cytoskeleton structures are involved in the active transport of multicomponent lipoplexes.
▪ The intracellular uptake of multicomponent lipoplexesis is largely affected by the actin network.
▪ The cytoplasmic trafficking predominantly involves microtubule-dependent transport.
Acknowledgments
This work was partially supported by the Italian Minister for University and Research (MIUR) (Futuro in Ricerca, Grant No. RBFR08TLPO; Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale, Grant No. 2009ACFPN9_002). E Gratton, LC Estrada and MA Digman acknowledge support by the National Center for Research Resources (5P41RR003155), the National Institute of General Medical Sciences (8P41GM103540 and 5P50 GM076516) divisions of the NIH. MilkaTitin is gratefully acknowledged for cell cultivation.
No writing assistance was utilized in the production of this manuscript.
Key Term
- Single particle tracking
Powerful fluorescence technique that allows one to follow particles, such as vesicles or proteins, in their 2D or 3D motion with high temporal and spatial resolution.
Footnotes
Supplementary data To view the supplementary data that accompany this paper please visit the journal website at: WWW.FUTURE-SCIENCE.COM/DOI/SUPPL/10.4155/TDE.12.151
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Zuhorn IS, Kalicharan R, Hoekstra D. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J. Biol. Chem. 2002;277(20):18021–18028. doi: 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- 2.Chang H, Yeh M. Clinical development of liposome-based drugs: formulation, characterization, and therapeutic efficacy. Int. J. Nanomed. 2012;7:49–60. doi: 10.2147/IJN.S26766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchini C, Pozzi D, Montani M, et al. Role of temperature-independent lipoplex–cell membrane interactions in the efficiency boost of multicomponent lipoplexes. Cancer Gene Ther. 2011;18(8):543–552. doi: 10.1038/cgt.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caracciolo G, Pozzi D, Caminiti R, Amenitsch H. Two-dimensional lipid mixing entropy regulates the formation of multicomponent lipoplexes. J. Phys. Chem. B. 2006;110(42):20829–20835. doi: 10.1021/jp0620926. [DOI] [PubMed] [Google Scholar]
- 5.Marchini C, Pozzi D, Alfonsi C, et al. Tawiloring lipoplex composition to the lipid composition of plasma membrane: a Trojan horse for cell entry? Langmuir. 2010;26(17):13867–13873. doi: 10.1021/la1023899. [DOI] [PubMed] [Google Scholar]
- 6.Caracciolo G, Caminiti R, Digman MA, Gratton E, Sanchez S. Efficient escape from endosomes determines the superior efficiency of multicomponentlipoplexes. J. Phys. Chem. B. 2009;113(15):4995–4997. doi: 10.1021/jp811423r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caracciolo G, Pozzi D, Caminiti R, et al. Enhanced transfection efficiency of multicomponent lipoplexes in the regime of optimal membrane charge density. J. Phys. Chem. B. 2008;112(36):11298–11304. doi: 10.1021/jp803077n. [DOI] [PubMed] [Google Scholar]
- 8.Caracciolo G, Pozzi D, Caminiti R, et al. Transfection efficiency boost by designer multicomponent lipoplexes. Biochim. Biophys. Acta. 2007;1768(9):2280–2292. doi: 10.1016/j.bbamem.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 9.Caracciolo G, Pozzi D, Amenitsch H, Caminiti R. Multicomponent cationic lipid–DNA complex formation: role of lipid mixing. Langmuir. 2005;21(25):11582–11587. doi: 10.1021/la052077c. [DOI] [PubMed] [Google Scholar]
- 10.Caracciolo G, Pozzi D, Caminiti R, Amenitsch H. Lipid mixing upon deoxyribonucleic acid-induced liposomes fusion investigated by synchrotron small-angle x-ray scattering. Appl. Phys. Lett. 2005;87(13):133901. [Google Scholar]
- 11.Pozzi D, Amenitsch H, Caminiti R, Caracciolo G. How lipid hydration and temperature affect the structure of DC-Chol–DOPE/DNA lipoplexes. Chem. Phys. Lett. 2006;422(4):439–445. [Google Scholar]
- 12.Caracciolo G, Caminiti R. Do DC-Chol/DOPE-DNA complexes really form an inverted hexagonal phase? Chem. Phys. Lett. 2005;411(4):327–332. [Google Scholar]
- 13.Caracciolo G, Pozzi D, Amenitsch H, Caminiti R. Interaction of lipoplexes with anionic lipids resulting in DNA release is a two-stage process. Langmuir. 2007;23(17):8713–8717. doi: 10.1021/la7017665. [DOI] [PubMed] [Google Scholar]
- 14.Caracciolo G, Callipo L, De Sanctis SC, Cavaliere C, Pozzi D, Laganà A. Surface adsorption of protein corona controls the cell internalization mechanism of DC-Chol–DOPE/DNA lipoplexes in serum. Biochim. Biophys. Acta. 2010;1798(3):536–543. doi: 10.1016/j.bbamem.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Caracciolo G, Pozzi D, Candeloro De Sanctis S, et al. Effect of membrane charge density on the protein corona of cationic liposomes: interplay between cationic charge and surface area. Appl. Phys. Lett. 2011;99(3):033702. [Google Scholar]
- 16.Hasegawa S, Hirashima N, Nakanishi M. Microtubule involvement in the intracellular dynamics for gene transfection mediated by cationic liposomes. Gene Ther. 2001;8(21):1669–1673. doi: 10.1038/sj.gt.3301573. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, MacDonald RC. Effects of microtubule-depolymerizing agents on the transfection of cultured vascular smooth muscle cells: enhanced expression with free drug and especially with drug–gene lipoplexes. Mol. Ther. 2004;9(5):729–737. doi: 10.1016/j.ymthe.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Akita H, Enoto K, Masuda T, Mizuguchi H, Tani T, Harashima H. Particle tracking of intracellular trafficking of octaarginine-modified liposomes: a comparative study with adenovirus. Mol. Ther. 2010;18(5):955–964. doi: 10.1038/mt.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ondrej V, Lukasova E, Falk M, Kozubek S. The role of actin and microtubule network in plastid DNA intracellular trafficking. Acta. Biochim. Pol. 2007;54:657–663. [PubMed] [Google Scholar]
- 20.Provencher SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput. Phys. Commun. 1982;27(3):213–227. [Google Scholar]
- 21.Provencher SW. CONTIN: a general purpose constrained regularization program for inverting noisy linear algebraic and integral equations. Comput. Phys. Commun. 1982;27(3):229–242. [Google Scholar]
- 22.Levi V, Serpinskaya AS, Gratton E, Gelfand V. Organelle transport along microtubules in xenopus melanophores: evidence for cooperation between multiple motors. Biophys. J. 2006;90(1):318–327. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levi V, Gelfand VI, Serpinskaya AS, Gratton E. Melanosomes transported by Myosin-V in xenopus melanophores perform slow 35 nm steps. Biophys. J. 2006;90(1):L07–L09. doi: 10.1529/biophysj.105.075556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levi V, Gratton E. Exploring dynamics in living cells by tracking single particles. Cell Biochem. Biophys. 2007;48(1):1–15. doi: 10.1007/s12013-007-0010-0. [DOI] [PubMed] [Google Scholar]
- 25.Levi V, Gratton E. Chromatin dynamics during interphase explored by single-particle tracking. Chromosome Res. 2008;16(3):439–449. doi: 10.1007/s10577-008-1240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kis-Petikova K, Gratton E. Distance measurement by circular scanning of the excitation beam in the two-photon microscope. Microsc. Res. Techniq. 2004;63(1):34–49. doi: 10.1002/jemt.10417. [DOI] [PubMed] [Google Scholar]
- 27.Levi V, Ruan QQ, Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys. J. 2005;88(4):2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levi V, Ruan QQ, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89(6):4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26(1):373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 30.Qian H, Sheetz MP, Elson EL. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 1991;60(4):910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Digman MA, Brown CM, Sengputa P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 2005;89(2):1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224(3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 33.Lachmanovich E, Shvartsman DE, Malka Y, Botvin C, Henis YI, Weiss AM. Co-localization analysis of complex formation among membrane proteins by computerized fluorescence microscopy: application to immunofluorescence co-patching studies. J. Microsc. 2003;212(2):122–131. doi: 10.1046/j.1365-2818.2003.01239.x. [DOI] [PubMed] [Google Scholar]
- 34.Zuzzi S, Cametti C, Onori G. Polyion-induced aggregation of lipidic-coated solid polystyrene spheres: the many facets of complex formation in low-density colloidal suspensions. Langmuir. 2008;24(12):6044–6049. doi: 10.1021/la8005458. [DOI] [PubMed] [Google Scholar]
- 35.Ying W, Huerta G, Steinberg S, Zuniga M. Time series analysis of particle tracking data for molecular motion on the cell membrane. Bull. Math. Biol. 2009;71(8):1967–2024. doi: 10.1007/s11538-009-9434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl Acad. Sci. USA. 2005;102(42):15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardarelli F, Pozzi D, Bifone A, Marchini C, Caracciolo G. Cholesterol-dependent macropinocytosis and endosomal escape control the transfection efficiency of lipoplexes in CHO living cells. Mol. Pharm. 2012;9(2):334–340. doi: 10.1021/mp200374e. [DOI] [PubMed] [Google Scholar]
- 38.Zhang XX, Allen PG, Grinstaff M. Macropinocytosis is the major pathway responsible for DNA transfection in CHO cells by a charge-reversal amphiphile. Mol. Pharm. 2011;8(3):758–766. doi: 10.1021/mp100366h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi Q, Coppolino MG. Automated classification and quantification of F-actin-containing ruffles in confocal micrographs. Biotechniques. 2006;40(6):745–756. doi: 10.2144/000112162. [DOI] [PubMed] [Google Scholar]
- 40.Dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. Effects of transport inhibitors on the cellular uptake of carboxylated polystyrene nanoparticles in different cell lines. PLoS ONE. 2011;6(9):e24438. doi: 10.1371/journal.pone.0024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Li P, Li G, Wang J, Wang E. The effect of nocodazole on the transfection efficiency of lipid-bilayer coated gold nanoparticles. Biomaterials. 2009;30(7):1382–1388. doi: 10.1016/j.biomaterials.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 42.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006;58(1):32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 43.Vaughan EE, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol. Ther. 2006;13(2):422–428. doi: 10.1016/j.ymthe.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JA, Åberg C, Salvati A, Dawson KA. Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population. Nat. Nanotechnol. 2011;7(1):62–68. doi: 10.1038/nnano.2011.191. [DOI] [PubMed] [Google Scholar]
- 45.Sandin P, Fitzpatrick LW, Simpson JC, Dawson KA. High-speed imaging of rab family small GTPases reveals rare events in nanoparticle trafficking in living cells. ACS Nano. 2012;6(2):1513–1521. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
Websites
- 101.LFD website. www.lfd.uci.edu.
- 102.Fiji website. http://fiji.sc/wiki/index.php/Fiji.
- 103.Rasband WS, Image J. US National Institutes of Health. Bethesda, MD, USA: http://imagej.nih.gov/ij. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.