Abstract
One of the main contributions of Drosophila to the JAK-STAT field is the study of morphogenesis. JAK-STAT signaling controls the formation of many different structures through surprisingly different morphogenetic behaviors that include induction of cell rearrangements, invagination, folding of tissues, modulation of cell shape, and migration. This variability may be explained by the many transcription factors and signaling molecules STAT regulates at early stages of development. But is STAT just acting as an upstream inducer of morphogenesis or does it have a more direct role in controlling cell behaviors? Here we review what is known about how the canonical phosphorylation of STAT contributes to shaping the embryonic and imaginal structures.
Keywords: morphogenesis, Drosophila, JAK, STAT, signaling, development, organogenesis
One of the advantages of the Drosophila signaling pathway is its simplicity in terms of core signaling elements. There is only one receptor (Dome), one JAK kinase (Hop), and one STAT transcription factor protein (Stat92E).1-6 In the embryo these are expressed ubiquitously, with an early maternal contribution that confers all cells the capacity to respond to ligand activation. Cell-specific activation is achieved by the localized expression of three ligands known as Unpaired (Upd), Unpaired-2 (Upd2), and Unpaired-3 (Upd3). These three ligands can explain almost all cases of pathway activation and thus knowing where they are transcribed provides an indication of where the pathway is active. During embryogenesis Upd and Upd2 are expressed in similar patterns3,4 while Upd3 is expressed at late embryo and larval stages.4,7 The activity of the pathway is modulated by a series of positive and negative feedback loops that result in either signaling reinforcement or in pulses of expression. The detection of such feedbacks has been frequently used as a read-out of pathway activity. For example, Dome is subject to a positive feedback and enhancer trap alleles and certain Dome enhancers are upregulated by Stat92E activation and can be used to detect pathway activity. Another read-out of pathway activity is the expression of the negative JAK-STAT regulator Suppressor of cytokine signaling 36E (Socs36E).8 Socs36E is a Stat92E target that is activated in a pattern similar to that of upd although reaching more cells due to ligand diffusion. Socs36E introns contain multiple high affinity STAT binding sites.8 Two of these sites have been multimerized and cloned upstream of GFP proteins creating reporters for STAT activation (×STAT-GFP reporter9). These reporters have become very useful tools to uncover cells where the JAK-STAT pathway is active. Using the expression of this reporter and upd transcription as a guide (Fig. 1) we will focus on the different organs where the pathway is active and describe what is known about Stat92E requirement in their morphogenesis.
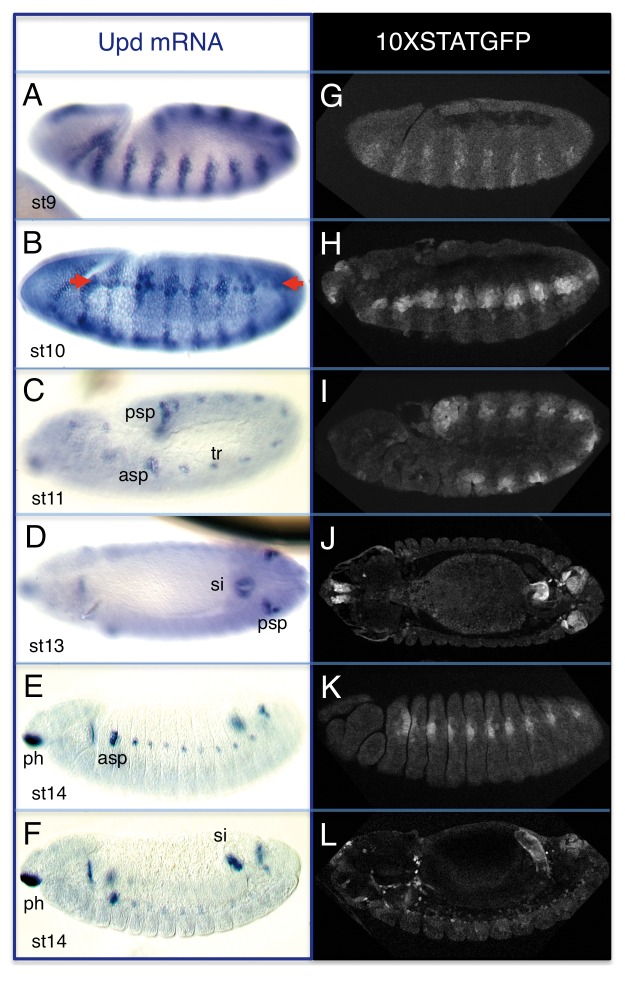
Figure 1.upd expression and JAK-STAT pathway activation during embryogenesis. Embryonic upd RNA expression (A–F) and activity of the ×STAT-GFP reporter at comparable stages (G–L). upd has a very dynamic pattern of expression ranging from a striped pattern at st9 (A) that develops a transient anterior-posterior stripe ([B], arrows), and then restricts to organ primordia (C–F). Note the similarity of the upd expression and the ×STAT-GFP reporter patterns. Anterior is left and dorsal up except in (D and J), which show dorsal views. asp, anterior spiracle; psp, posterior spiracle; tr, trachea; si, small intestine; ph, pharynx.
JAK-STAT Pathway Requirement in Blastoderm and Early Embryogenesis
upd is expressed in all blastoderm cells excepting the most anterior and posterior ones. Several important decisions are taken at this stage. First, the chromosomal content of the embryo is checked by all cells, so that they define their morphological sexual characteristics. Second, the pole cells become determined. Third, the blastoderm becomes segmented. Fourth, gastrulation begins and the germ band extends.
Determination of sexual characteristics
All JAK-STAT signaling elements with the exception of the ligands are supplied maternally and as a result the pathway has the capacity of becoming activated from very early stages. The pathway is able to influence sexual determination at two levels. First, it is used to reinforce the mechanisms by which the embryo translates the information about its sex chromosome content into the activation of the genetic male or female morphogenetic programs. In Drosophila the ratio of X chromosomes to autosomes (A) determines the activation state of the Sex lethal (Sxl) gene (if the ratio is 1 [2X/2A] the embryo will activate Sxl and develop as a female, but if the ratio is 0.5 [1X/2A] the embryo will not activate Sxl and develop as a male). The upd gene is located on the X chromosome and the JAK-STAT pathway has been found to reinforce the activity of the early X-linked signals with Stat92E boosting an early Sxl enhancer.10 Mutation of the upd ligands, hop, or Stat92E results in lower levels of Sxl expression in the female, perturbing a Sxl autoregulatory loop that affects the way the cells perceive they are female.11
A more direct function occurs in male germ cells where sexual determination is controlled differently. In this case, sexual determination depends not only on germ cell autonomous mechanisms but is also influenced by interactions with the gonadal mesoderm. Upd is absent in the embryonic female gonads, but is expressed in the male gonadal mesoderm at late embryogenesis from where it activates JAK-STAT signaling in the male germ cells.12 Activation of Stat92E is sufficient to induce a male pattern of proliferation in the gonad and the activation of certain male specific germ line genes. However, Stat92E activation in female germ cells cannot completely override the cell autonomous mechanisms, with XX/AA germ cells still capable of forming oocytes.
Pole cell migration
Pole cells form the progenitors of the germ line. They become specified when they get in contact with the pole plasm localized at the posterior end of the egg. During gastrulation, the pole cells migrate from the posterior end of the blastoderm toward the abdominal segments where they join the gonadal mesoderm and constitute the gonads. This complex migration proceeds through several steps. First the pole cells are carried into the invaginating posterior midgut, from here they traverse the midgut epithelium getting into the embryo cavity where they migrate anteriorly along the mesoderm until they reach the gonadal mesoderm with which they aggregate at stage 16 (st16) forming the spherical gonad primordium. Stat92E activation is required at several steps during this migration. Stat92E is first phosphorylated at the anterior and posterior blastoderm ends (including in the pole cells where Stat92E stabilizes in the nucleus) by the Torso tyrosine kinase, independently of the canonical pathway.13 Mutants lacking Stat92E have slow migrating pole cells with some not being able to abandon the posterior midgut pocket. Conversely, ectopic activation of Stat92E results in earlier migratory movements. After crossing the midgut epithelium, pole cells still show activated Stat92E but now this requires the canonical pathway as mutations for the Upd ligands, the receptor or the kinase affect their migration toward the gonad.7 Upd is unlikely to act as a chemoattractant of the pole cells toward the gonadal mesoderm as abnormal phenotypes can be observed before the upd ligands are expressed in the gonad. At this stage Stat92E controls the filopodial dynamics.7
Embryonic segmentation
One of the most conspicuous phenotypes of JAK-STAT pathway mutants is the formation of embryos with fusions and deletion of segments. This is caused by the abnormal activation of pair-rule genes, like even skipped (eve) and runt, required for embryonic segmentation.5,6,14 Stat92E controls eve through a 511 bp enhancer that drives eve expression of stripes 3 and 7.14 Mutation of Stat92E or the STAT binding sites in this enhancer results in the loss of expression in both stripes. In this case, Stat92E acts as a global transcriptional activator in collaboration with other early transcription factors like Zelda, with the exact eve stripe localization established by GAP gene repression. Knirps represses the enhancer between the 3 and 7 stripes and Hunchback represses enhancer activity anterior to stripe 3 and posterior to stripe 7.15 Thus, the segmentation morphogenetic defects are caused by the requirement of Stat92E as an upstream transcriptional regulator of patterning genes. In fact, it has been proposed that, similar to the Zelda protein, Stat92E could be acting as a general transcription factor boosting the expression of several early patterning genes during the maternal to zygotic transcriptional transition.16
Cell intercalation during gastrulation
Germ band extension is caused by cell intercalation in the ventro-lateral cells of the embryo. During this process, ventro-lateral cells accumulate myosin-II (Myo-II) in the apical cortex. JAK-STAT mutants have defective germ band extension, where instead of intercalating, the ventro-lateral cells go through an apical constriction process.17 Observation of Myo-II in ventro-lateral cells of JAK-STAT mutants shows that besides accumulating in the cortex, Myo-II is mislocalized in a medial meshwork similar to its localization in invaginating mesodermal cells. The medial meshwork localization is responsible for the tensile forces causing the apical constriction of Stat92E mutant cells. This phenotype is ameliorated by reducing Rho1 activity or the amount of the myosin regulatory light chain (MRLC) squash, indicating that in JAK-STAT mutant embryos, Rho1 activation of Myo-II causes the cell constriction phenotypes observed during germ band elongation. However, Stat92E does not regulate Myo-II concentration or activation, but acts through Wasp protein regulation. Bertet and collaborators show that Wasp normally disappears from the apical cortex in rearranging cells but this does not occur in JAK-STAT mutants, explaining why Myo-II appears in a central meshwork. Thus Stat92E regulates cell rearrangement vs. cell constriction by keeping Myo-II away from the medial meshwork and in the cell cortex. JAK-STAT pathway prevents actin meshwork formation through the posttranscriptional downregulation of Wasp. The direct targets in this process are unknown, but the effect is not mediated by the regulation of segmentation genes.17
JAK-STAT Pathway Requirement during Organogenesis
During embryogenesis, the JAK-STAT pathway plays an important role in the morphogenesis of the respiratory system, where it is required for the trachea and the spiracle development; as well as in the endocrine organs; in the gut and in various mesodermal derivatives.
Formation of the trachea
The trachea of Drosophila is a segmentally repeated ectodermal organ. It develops from the invagination of ten placodes present from the second thoracic (T2) to the eighth abdominal (A8) segments. At st10 the placodes invaginate forming the tracheal pits that bud in different directions at st11 to make the main tracheal branches. Early specification of the trachea occurs when the primordia activate the transcription factors trachealess (trh) and ventral veinless (vvl).18,19 After their early activation, Trh and Vvl establish a mutual autoregulatory feedback that maintains the expression of each other during development.20,21 In the tracheal pits, the Trh and Vvl proteins activate elements of the FGF and EGF pathways required for the correct branching patterns, as well as transcription factors that specify the primary branches.18,19,21,22 Mutation of trh results in the tracheal cells remaining at the ectodermal surface, while mutation of vvl allows the tracheal pits to invaginate but they are unable to branch. Mutations for JAK-STAT pathway elements result in strong tracheal defects due to the absence of trh and vvl expression from the trachea primordia.2,20 Analysis of genomic conserved regions with accumulation of STAT binding sites around the trh and vvl loci allowed the isolation of early tracheal enhancers.20 Some of these enhancers are not activated in JAK-STAT mutant embryos suggesting they are direct Stat92E targets. Mutation of the putative STAT sites in vvl1+2, the only enhancer that has been directly tested, results in lack of activity. The requirement of JAK-STAT signaling for trachea specification difficults the analysis of later requirements in tracheal morphogenesis; however, a later function in the trachea is suggested by the activation of upd transcription in the tracheal pits downstream of Trh.23
Formation of the spiracles
The spiracles are the external respiratory organs connecting the trachea to the outside. In the larva, the anterior spiracles form in the first thoracic segment (T1) and the posterior spiracles in A8. The anterior spiracle is not functional until the second instar larva (L2), while the posterior spiracle is functional as the first instar larva (L1) hatches. The posterior spiracles are composed of an internal spiracular chamber that forms the tubular connection to the trachea; and the external stigmatophore that forms a protruding structure lodging the internal tube.24 The posterior spiracle forms in the dorsal region of the A8 segment from cells posterior to the last tracheal pit. The connexion between spiracle and trachea results from the concerted invagination of the tracheal pit and the spiracular chamber primordium, with the tracheal pit invaginating slightly earlier (st10) than the spiracular chamber (st11). The cells of the spiracular chamber invaginate at st11 by constricting their apical membranes through Rho1 activation of actin/myosin.25 This is followed at st13 by a process of apico-basal elongation that in some of the invaginated cells can result in a 4-fold elongation. As the cells of the internal spiracular chamber invaginate, the ectodermal cells surrounding them become specified to form the stigmatophore and activate a process of cell rearrangement. This rearrangement results in a circumferential convergent extension that closes the spiracle opening while creating the proximo-distal axis of the stigmatophore.24 upd is expressed from st11 in the cells of the spiracular chamber primordium before they start invaginating. In upd mutant spiracles cell elongation does not occur and the stigmatophore is flattened.26 The activation of the 10×STAT-GFP reporter in the spiracular chamber and the stigmatophore suggests that Upd can diffuse from the spiracular chamber influencing the development of the stigmatophore. Thus JAK-STAT signaling regulates several distinct morphogenetic behaviors in the spiracles including cell invagination, cell elongation and cell rearrangements.
The only confirmed direct target of Stat92E described in the posterior spiracles is the crumbs (crb) gene. Crb is a subapical transmembrane protein that functions as one of the main cell polarity regulators. Crb is expressed in all ectodermal derivatives but is upregulated in many invaginating tissues probably because it is required to reinforce the polarity of the invaginating cells as they rearrange their apical actin cytoskeleton.27 In the posterior spiracles, crb upregulation is mediated by the JAK-STAT pathway through a specific enhancer localized in the first intron. The regulation of this enhancer depends on JAK-STAT signaling as in upd, hop, or Stat92E mutants, the crb spiracle enhancer is severely downregulated. In addition, mutation of the putative STAT binding sites results in a lack of enhancer expression. Mutants for crb have shorter spiracles, indicating that Stat92E upregulation of crb is required for spiracle morphogenesis.26
Despite the delayed development of the anterior spiracle, the anterior and posterior spiracles express in the embryo some common genes like the homeodomain protein Cut and the Upd ligand; however, the morphogenesis of this organ is not well studied.
Formation of the ring gland
The ring gland is the main endocrine organ of the larva. It is formed by the fusion of three independent glands that migrate dorsally from their place of specification toward the anterior aorta. The ring structure is formed ventrally by the corpora cardiaca, dorsally by the corpora allata, and both are linked laterally by the two prothoracic glands. The corpora allata and the prothoracic glands have a similar migratory behavior to that observed in the border cells of the ovary. Mutants for the JAK-STAT pathway lack a ring gland, and there is evidence that this is due to the lack of specification of the corpora allata and the prothoracic gland primordia. As for the trachea, the involvement of JAK-STAT signaling in ring gland specification obscures any possible involvement of the pathway in regulating the migratory behavior of these primordia (Sánchez-Higeras C, SS, and CGH, submitted).
Formation of the gut
The Drosophila gut is formed by an anterior ectodermal foregut, an intermediate endodermal midgut and a posterior ectodermal hindgut. In the foregut, upd is expressed in the pharynx and in the region that will form the proventriculus. In the hindgut, upd is expressed in the anterior region that will form the small intestine (Fig. 2).
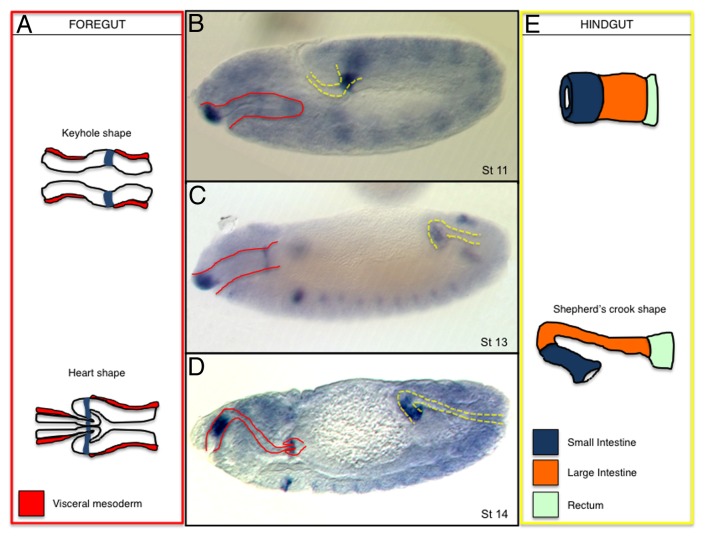
Figure 2. JAK-STAT function during embryonic gut development. Schematic representation of the embryonic foregut (A) and hindgut (E) and their localization at different embryonic stages (B–D). In the schemes, upd expression is shadowed in blue. Expression of upd transcripts in embryos at st11 (B), st13 (C), and st14 (D). In (B–D) red lines mark the position of the foregut and yellow lines of the hindgut. (A) upd expression is restricted to a group of ectoderm cells from where it diffuses to control the proventriculus folding (the enveloping visceral mesoderm is labeled in red). (E) In the hindgut, upd expression is restricted to the small intestine from where it coordinates the cell rearrangements and gut bending (blue, small intestine; orange, large intestine; light green, rectum).
The proventriculus is a valve-like structure separating the foregut from the midgut.28,29 At early st13, the connection between the foregut and the hindgut consists of a monolayered tube. This tube is surrounded by mesoderm except in a gap around the most posterior hindgut (Fig. 2A). In this mesoderm free area, the foregut broadens up forming a small evagination that gives the structure a keyhole appearance in longitudinal sections. The proventriculus morphogenesis occurs when this expanded end of the foregut folds at st15 and inserts into the adjacent endodermal tube forming a heart-shaped valve (Fig. 2A). Upd is expressed in a ring of foregut cells abutting the endodermal tube (Fig. 2A, blue). Mutations in any element of the pathway result in the abnormal formation of the valve. Upd activation in the foregut is regulated by the Drm/Lin/Bowl relief of repression cascade.28 Stat92E is activated at a distance from the upd expressing cells suggesting that the ligand can diffuse a few cell diameters.28,29 The proventriculus morphogenesis defects seen in JAK-STAT mutants are probably due to its function on the ectodermal or endodermal cells as mutations affecting muscle development (twist) have a rather normal shape.29 It is unclear how JAK-STAT signaling affects proventriculus morphogenesis at the cellular level, but it has been observed that the Notch target gene short stop that encodes a cytoskeletal linker protein of the spectraplaking family is not activated properly in JAK-STAT mutants.29
The hindgut can be subdivided into three morphologically distinct regions. The most anterior region connecting to the midgut is the small intestine, the intermediate region is the large intestine, and the most posterior region forms the rectum (Fig. 2E). upd is expressed from st10 to the end of embryogenesis in what will become the small intestine (ref. 30 and Fig. 2B–D). The expression of dome and Stat92E increases in the area where upd is expressed and at a distance, suggesting that the pathway is autoregulated at the level of these two elements. From st10 to st14 the intestine transforms from a short and wide tube into a long and narrow tube due to cell rearrangements causing convergent extension (Fig. 2E). Mutants for any of the JAK-STAT signaling elements result in a short and wide hindgut consistent with absence of convergent extension and suggesting that Stat92E is involved in cell rearrangements along the hindgut. Analysis of Stat92E localization indicates that the protein accumulates in the nucleus at a distance from the upd source. This indicates that Upd is diffusing along the ectoderm. Surprisingly, ectopic expression of upd or of an activated JAK kinase also results in a shorter and wider hindgut, which has been interpreted as a requirement of a graded Stat92E activation to orient the rearrangements more than a requirement for the induction of cell rearrangement. upd expression in the hindgut also depends on the Drm/Lin/Bowl cascade.28 Mutants for bowl have a strong defect on hindgut elongation that can be almost rescued by upd expression in the small intestine. Although it is likely that Bowl may be required in other ways, the activity of the JAK-STAT pathway mediates most of the cell rearrangements.30 Upd is required non-autonomously in the hindgut, as can be shown by the activation of Stat92E at a distance. Independent confirmation for direct activation of the pathway at a distance in the large intestine comes from the regulation of the vvl gene in the hindgut. In upd or Stat92E mutants vvl is not expressed in the hindgut. Although a vvl hindgut enhancer that is likely to be directly regulated by the pathway has been isolated,20 it is not yet known if vvl is required for hindgut elongation.
Work by Wells et al.31 shows that, apart from the cell rearrangement processes, JAK-STAT function is required for the hindgut curvature to achieve its typical shepherd’s crook morphology. Lack of function or overactivation of the JAK-STAT pathway results in a straightened hindgut. Analyses of cell polarity markers show that although most apical and lateral ectodermal proteins are expressed at similar levels in the hindgut, Fasciclin III (FasIII) is upregulated asymmetrically at st12 in the internal, most curved side of the intestine. This upregulation requires JAK-STAT function and is important for hindgut curvature. FasIII is a homophilic adhesion molecule normally associated with the septate junctions. In the hindgut, the direct upregulation of FasIII or the activation of the JAK-STAT pathway, results in the spreading of FasIII along the lateral plasma membrane. Mutant embryos for FasIII have a less curved hindgut, suggesting that FasIII upregulation by JAK-STAT signaling contributes to the curvature of the hindgut. Mathematical modeling suggests that the asymmetry of FasIII expression in the hindgut is important for its shape, a suggestion that is backed by the phenotypes observed after ectopic activation of FasIII in the outer side of the hindgut. It is still unclear what is responsible for the asymmetric FasIII expression, but analysis of ×STAT-GFP and Socs36E expression indicates that Stat92E may be more active in the internal than in the external half of the hindgut curvature.
Mesoderm requirement
Except in the gonads, upd is not expressed in mesoderm cells. However, there is ample evidence that Upd from the overlying ectoderm activates the pathway in the mesoderm.
Tinman (Tin), an important mesoderm transcription factor required for heart, visceral mesoderm and for the development of some somatic muscles, upregulates Stat92E transcription through binding to a Stat92E intronic enhancer.32 Null Stat92E mutant embryos show a disorganized somatic musculature with many muscles missing and most of the remaining muscles having altered morphology and structure. Although more work should be done to find out what part of these anomalies may be secondary to the ectoderm defects, the generalized muscle aberrations may indicate a requirement of Stat92E for muscle morphogenesis. By comparison to the strong somatic musculature defects, visceral muscles form relatively normal in Stat92E mutant embryos.
JAK-STAT signaling is also required for heart development where it regulates cardiac morphology and heart precursor diversification.33 The source of Upd is supposed to come from a brief ventral segmental expression at st9 (Fig. 1B, arrows) from where Upd regulates restriction of tin expression in the mesoderm. In Stat92E null embryos tin expression is not restricted. This phenotype can be rescued by providing Stat92E exclusively in the mesoderm demonstrating its direct requirement in the mesoderm cells. Stat92E mediates tin downregulation indirectly through the transcriptional activation of HLHm5, one of the Enhancer of split complex genes [E(spl)]. Mutation of Stat92E results in the downregulation of the HLHm5 transcript. A partial deletion of the E(spl) complex results in the lack of tin restriction in the dorsal mesoderm causing a phenotype similar to that observed in Stat92E mutants. Careful analysis of different heart cell types indicates that Stat92E is required to regulate the number of pericardial cells and is probably dispensable for the formation of the cardioblasts.33 It is interesting to note that this is a sophisticated delayed autoinhibitory circuit where Tin upregulates Stat92E transcription in the mesoderm and Stat92E contributes indirectly to downregulate tin expression at later stages.
JAK-STAT signaling is also activated in the pharyngeal mesoderm and in the visceral mesoderm surrounding the small intestine. The source of upd is the ectodermal part of the pharynx and the ectodermal small intestine. Here, there is evidence that the pathway is active as Stat92E directly upregulates domeless transcription in the pharynx and small intestine mesoderm.34 In Stat92E mutants the pharyngeal muscles are abnormal. In the hindgut mesoderm no obvious phenotypes are observed in Stat92E mutant embryos but no studies have analyzed visceral mesoderm regionalization.
JAK-STAT Requirement during Imaginal Disc Development
JAK-STAT signaling plays important roles in imaginal disc development at all larval stages. Early Stat92E function serves for the patterning and specification of broad imaginal disc areas. At later stages JAK-STAT activity is refined to smaller regions where the pathway has more subtle effects on morphogenesis (Fig. 3). In many cases upd imaginal disc expression has been inferred genetically, either by analyzing the cells expressing an upd-Gal4 enhancer trap line,35 or by using the G-TRACE system.36 G-TRACE allows detecting the cells that at any point during development have expressed upd-Gal4 even if at the moment when it is assayed they have stopped expressing it. In this second approach upd-GAL4 expresses the FLP recombinase that will induce clones of permanently labeled cells. Both tests give consistent results; however, caution has to be taken as the quality of the results depends on the reliability of the reporter line.
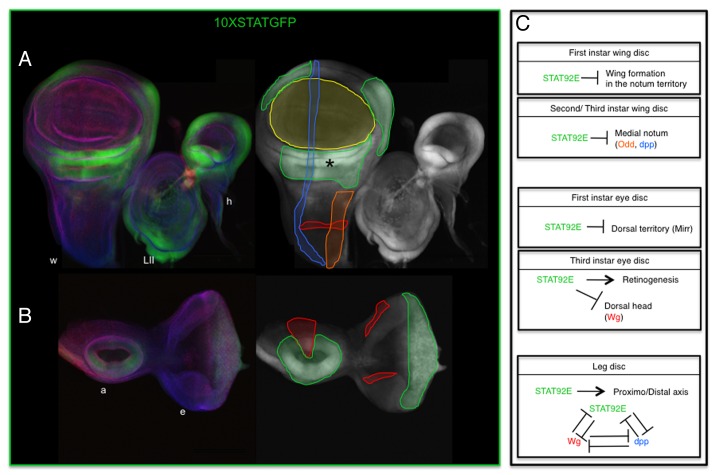
Figure 3. JAK-STAT activity in imaginal discs. Activation of the pathway visualized using the ×STAT-GFP reporter in the wing disc (left disc in [A]) and eye-antennal disc (B). (A and B) Color images show the imaginal discs stained for GFP (green), aPKC (red), and Discs large (blue). The corresponding GFP single channel image by its side shows schematically the extent of GFP (green line) and the approximate expression patterns of Dpp (blue lines), Odd (orange line), and Wingless (red lines). The localization of the wing blade is highlighted by a yellow line. Wingless expression in the wing blade and hinge is omitted. (C) Schematic representation of JAK-STAT functions at different larval stages and imaginal discs. w, wing; h, haltere; LII, mesothorathic leg; e, eye; a, antenna. Asterisk in (A) marks the hinge region. ×STAT-GFP reporter expression in the leg disc has been omitted since it does not correctly recapitulate JAK-STAT activation.9
The eye-antennal disc will be described focusing first on the eye development, while the antenna will be discussed in the leg disc section due to the similar function JAK-STAT signaling has in both cases.
Function on the eye imaginal primordium
Lack of upd expression causes smaller eyes, while Upd overexpression induces larger ones, a phenotype that was interpreted as due to control of cell proliferation.35,37 Further work has showed that the interpretation of these phenotypes is more complex as upd controls early eye patterning. Upd is expressed at the first larval instar (L1) in the ventral eye primordium. Eye cells activating the JAK-STAT pathway repress wingless (wg),38,39 and as a result, the Iroquois-complex (Iro-C) genes that respond to Wg signaling are only activated dorsally. This subdivides the eye into a dorsal half expressing the Iro-C and a ventral region expressing the Sloppy paired transcription factor. The confrontation of dorsal and ventral regions, induces the activation of a Notch organizer along the equator of the eye. This organizer is responsible for the eye growth and the activation of Eyegone (Eyg). Ectopic expression of upd in the eye primordium results in bigger eyes due to more proliferation on the dorsal side,37 which has been attributed to the repression of Wg and the creation of a second organizer on the dorsal eye.39
Ventral eye expression is transient, with upd becoming restricted to the most posterior edge of the eye. This refined expression is controlled by Eyg, which is activated at the eye’s equator by Notch.40,41 The site of upd expression is called the firing point as it is the point from where the eye morphogenetic furrow begins its anterior wave-like movement. Photoreceptor cell specification occurs sequentially just behind the morphogenetic furrow. Although this source of Upd has been suggested to be required for cell proliferation, the effect of JAK-STAT signaling on proliferation may be indirect through its earlier effect on Notch activation. In fact, overexpression of the JAK-STAT pathway is unable to restore eye size when Notch signaling is attenuated.39 Several experiments indicate that Upd diffusion from the firing point is only required for wg repression. Mutant Stat92E clones cause ectopic wg expression and impede the progression of the morphogenetic furrow in the dorsal but not in the ventral eye. Conversely, ectopic Upd represses wg expression and induces precocious furrow initiation in the dorsal eye. This is prevented if wg and upd are coexpressed, suggesting that the main JAK-STAT pathway function is to regulate wg.38,42 However, it is still possible that JAK-STAT signaling has more direct functions in eye morphogenesis.39
Function on the antenna and leg discs
The antenna and the legs respond similarly to Upd. One of the most important factors to generate the leg proximo-distal (PD) axis is the activation of Wg and Decapentaplegic (Dpp) expression in opposing sectors of the imaginal disc. Interaction between these two pathways is necessary for the activation of Distalless (Dll) in the center of the disc and the creation of the PD axis. In both the antenna and the leg imaginal discs Upd is expressed in a pattern complementary to that of Wg and Dpp.43 Stat92E mutant clones cause ectopic wg expression and induce the duplication of leg and antenna structures by creating a secondary PD axis that activates Dll. Accordingly, ectopic upd expression results in both wg and dpp repression. Not only activation of Stat92E represses wg and dpp in the disc but Wg and Dpp can restrict upd expression indicating that the cross-inhibition between the three signaling pathways is important for leg and antenna patterning.43
Function on the wing disc
The wing disc forms the notum, wing hinge, pleura, and wing blade. At L2 the wing disc has an almost global activation of upd. At this stage Stat92E is required to repress the formation of a secondary wing field.44 However, at early L3 the expression of ×STAT-GFP is restricted to the hinge region (Fig. 3A, asterisk). This downregulation from the wing pouch and the notum coincides with the activation in the pouch of the wing marker Nubbin and of Eyg in the notum suggesting a causal effect. JAK-STAT signaling downregulation is required for the normal growth of the disc and proximo-distal and medio-lateral pattern elaboration. This is mediated on the one hand by reducing the scope of Odd-skipped (Odd) function in the notum, which is important to organize the notum’s AP axis; and by antagonizing Dpp that is important to pattern the notum’s medio-lateral axis. Maintained upd expression in the whole disc results in gene expression anomalies and aberrant development of the disc.44
At later stages JAK-STAT signaling is required in the hinge. In Stat92E mutants there is an abnormal development of the hinge that results in the characteristic outstretched wing posture of the adult flies.45 Downregulation of Stat92E results in abnormal hinge folds in the imaginal disc probably caused by the abnormal localization of FasIII in the lateral membrane of the cells forming the hinge wing disc folds.31 Supporting this, the hinge fold phenotype observed in Stat92E mutants can be mimicked by expression of a fasIII RNAi. These defects in fold formation are reminiscent of those described for hindgut curvature indicating a possible common cause.31
In summary, the JAK-STAT pathway has different functions during morphogenesis: initially contributes to organ specification and patterning and later controls molecules directly modulating cell behavior. The characterization of direct Stat92E targets should open exciting avenues of research.
Acknowledgments
This work was supported by the Spanish Ministerio de Investigación Ciencia e Innovación, Consolider, the European Regional Development Fund, and Junta de Andalucía.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/26089
References
- 1.Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–12. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- 2.Brown S, Hu N, Hombría JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–5. doi: 10.1016/S0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 3.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–63. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hombría JC, Brown S, Häder S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–33. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 5.Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–9. doi: 10.1016/S0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 6.Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–30. doi: 10.1016/S0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 7.Brown S, Zeidler MP, Hombría JE. JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev Dyn. 2006;235:958–66. doi: 10.1002/dvdy.20709. [DOI] [PubMed] [Google Scholar]
- 8.Karsten P, Häder S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–6. doi: 10.1016/S0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 9.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–31. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Sefton L, Timmer JR, Zhang Y, Béranger F, Cline TW. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405:970–3. doi: 10.1038/35016119. [DOI] [PubMed] [Google Scholar]
- 11.Avila FW, Erickson JW. Drosophila JAK/STAT pathway reveals distinct initiation and reinforcement steps in early transcription of Sxl. Curr Biol. 2007;17:643–8. doi: 10.1016/j.cub.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 12.Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, Van Doren M. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 2005;436:563–7. doi: 10.1038/nature03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Xia F, Li WX. Coactivation of STAT and Ras is required for germ cell proliferation and invasive migration in Drosophila. Dev Cell. 2003;5:787–98. doi: 10.1016/S1534-5807(03)00328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–24. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 15.Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, Small S. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development. 2011;138:4291–9. doi: 10.1242/dev.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsurumi A, Xia F, Li J, Larson K, LaFrance R, Li WX. STAT is an essential activator of the zygotic genome in the early Drosophila embryo. PLoS Genet. 2011;7:e1002086. doi: 10.1371/journal.pgen.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertet C, Rauzi M, Lecuit T. Repression of Wasp by JAK/STAT signalling inhibits medial actomyosin network assembly and apical cell constriction in intercalating epithelial cells. Development. 2009;136:4199–212. doi: 10.1242/dev.040402. [DOI] [PubMed] [Google Scholar]
- 18.Boube M, Llimargas M, Casanova J. Cross-regulatory interactions among tracheal genes support a co-operative model for the induction of tracheal fates in the Drosophila embryo. Mech Dev. 2000;91:271–8. doi: 10.1016/S0925-4773(99)00315-9. [DOI] [PubMed] [Google Scholar]
- 19.Chung S, Chavez C, Andrew DJ. Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis. Dev Biol. 2011;360:160–72. doi: 10.1016/j.ydbio.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sotillos S, Espinosa-Vázquez JM, Foglia F, Hu N, Hombría JC. An efficient approach to isolate STAT regulated enhancers uncovers STAT92E fundamental role in Drosophila tracheal development. Dev Biol. 2010;340:571–82. doi: 10.1016/j.ydbio.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zelzer E, Shilo BZ. Interaction between the bHLH-PAS protein Trachealess and the POU-domain protein Drifter, specifies tracheal cell fates. Mech Dev. 2000;91:163–73. doi: 10.1016/S0925-4773(99)00295-6. [DOI] [PubMed] [Google Scholar]
- 22.Anderson MG, Certel SJ, Certel K, Lee T, Montell DJ, Johnson WA. Function of the Drosophila POU domain transcription factor drifter as an upstream regulator of breathless receptor tyrosine kinase expression in developing trachea. Development. 1996;122:4169–78. doi: 10.1242/dev.122.12.4169. [DOI] [PubMed] [Google Scholar]
- 23.Lovegrove B. Genetic control of cell shape changes and cell rearrangements during Drosophila morphogenesis. Department of Zoology and Darwin College. Cambridge: University of Cambridge, 2004:151. [Google Scholar]
- 24.Hu N, Castelli-Gair J. Study of the posterior spiracles of Drosophila as a model to understand the genetic and cellular mechanisms controlling morphogenesis. Dev Biol. 1999;214:197–210. doi: 10.1006/dbio.1999.9391. [DOI] [PubMed] [Google Scholar]
- 25.Simões S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Hombría JC, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–67. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- 26.Lovegrove B, Simões S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombría JC. Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol. 2006;16:2206–16. doi: 10.1016/j.cub.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 27.Sotillos S, Aguilar M, Hombría JC. Forces shaping a Hox morphogenetic gene network. Proc Natl Acad Sci U S A. 2013;110:4303–8. doi: 10.1073/pnas.1212970110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansen KA, Green RB, Iwaki DD, Hernandez JB, Lengyel JA. The Drm-Bowl-Lin relief-of-repression hierarchy controls fore- and hindgut patterning and morphogenesis. Mech Dev. 2003;120:1139–51. doi: 10.1016/j.mod.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Josten F, Fuss B, Feix M, Meissner T, Hoch M. Cooperation of JAK/STAT and Notch signaling in the Drosophila foregut. Dev Biol. 2004;267:181–9. doi: 10.1016/j.ydbio.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Johansen KA, Iwaki DD, Lengyel JA. Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development. 2003;130:135–45. doi: 10.1242/dev.00202. [DOI] [PubMed] [Google Scholar]
- 31.Wells RE, Barry JD, Cuhlmann S, Evans P, Huber W, Zeidler MP. Control of tissue morphology by Fasciclin III mediated intercellular adhesion. Development. 2013 doi: 10.1242/dev.096214. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu YH, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, Furlong EE. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev Cell. 2009;16:280–91. doi: 10.1016/j.devcel.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Johnson AN, Mokalled MH, Haden TN, Olson EN. JAK/Stat signaling regulates heart precursor diversification in Drosophila. Development. 2011;138:4627–38. doi: 10.1242/dev.071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivas ML, Cobreros L, Zeidler MP, Hombría JC. Plasticity of Drosophila Stat DNA binding shows an evolutionary basis for Stat transcription factor preferences. EMBO Rep. 2008;9:1114–20. doi: 10.1038/embor.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–53. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 36.Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–5. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–66. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekas LA, Baeg GH, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133:4721–9. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- 39.Gutierrez-Aviño FJ, Ferres-Marco D, Dominguez M. The position and function of the Notch-mediated eye growth organizer: the roles of JAK/STAT and four-jointed. EMBO Rep. 2009;10:1051–8. doi: 10.1038/embor.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–47. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 42.Tsai YC, Yao JG, Chen PH, Posakony JW, Barolo S, Kim J, Sun YH. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev Biol. 2007;306:760–71. doi: 10.1016/j.ydbio.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 43.Ayala-Camargo A, Ekas LA, Flaherty MS, Baeg GH, Bach EA. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev Dyn. 2007;236:2721–30. doi: 10.1002/dvdy.21230. [DOI] [PubMed] [Google Scholar]
- 44.Hatini V, Kula-Eversole E, Nusinow D, Del Signore SJ. Essential roles for stat92E in expanding and patterning the proximodistal axis of the Drosophila wing imaginal disc. Dev Biol. 2013;378:38–50. doi: 10.1016/j.ydbio.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnstone K, Wells RE, Strutt D, Zeidler MP. Localised JAK/STAT pathway activation is required for Drosophila wing hinge development. PLoS One. 2013;8:e65076. doi: 10.1371/journal.pone.0065076. [DOI] [PMC free article] [PubMed] [Google Scholar]