Background: Cyclin D1/Cdk4 and NDR1/2 are critical kinases in cell cycle regulation.
Results: We demonstrate that cyclin D1 interacts with NDR1 and NDR2 and enhances their kinase activity.
Conclusion: Cyclin D1 promotes cell cycle progression partly through up-regulating NDR1/2 activity.
Significance: We revealed the novel function of cyclin D1 in cell cycle progression in a Cdk4-independent manner.
Keywords: Cell Cycle, Cyclins, Protein Phosphorylation, Protein-Protein Interactions, Serine/Threonine Protein Kinase
Abstract
Cyclin/cyclin-dependent kinases (Cdks) are critical protein kinases in regulating cell cycle progression. Among them, cyclin D1/Cdk4 exerts its function mainly in the G1 phase. By using the tandem affinity purification tag approach, we identified a set of proteins interacting with Cdk4, including NDR1/2. Interestingly, confirming the interactions between NDR1/2 and cyclin D1/Cdk4, we observed that NDR1/2 interacted with cyclin D1 independent of Cdk4, but NDR1/2 and cyclin D1/Cdk4 did not phosphorylate each other. In addition, we found that NDR1/2 did not affect the kinase activity of cyclin D1/Cdk4 upon phosphorylation of GST-Rb. However, cyclin D1 but not Cdk4 promoted the kinase activity of NDR1/2. We also demonstrated that cyclin D1 K112E, which could not bind Cdk4, enhanced the kinase activity of NDR1/2. To test whether cyclin D1 promotes G1/S transition though enhancing NDR1/2 kinase activity, we performed flow cytometry analysis using cyclin D1 and cyclin D1 K112E Tet-On inducible cell lines. The data show that both cyclin D1 and cyclin D1 K112E promoted G1/S transition. Importantly, knockdown of NDR1/2 almost completely abolished the function of cyclin D1 K112E in promoting G1/S transition. Consistently, we found that the protein level of p21 was reduced in cells overexpressing cyclin D1 K112E but not when NDR1/2 was knocked down. Taken together, these results reveal a novel function of cyclin D1 in promoting cell cycle progression by enhancing NDR kinase activity independent of Cdk4.
Introduction
Cyclin/cyclin-dependent kinases (Cdks)2 are key regulators during cell cycle progression by phosphorylating downstream substrates. Different cyclin/Cdks execute their functions in different phases of the cell cycle. The cyclin D1-Cdk4 complex plays an important role in G1 phase. It can phosphorylate Rb family proteins to disable their function as transcriptional suppressors and allow activation of E2F-dependent transcription to promote S phase entry and initiation of DNA synthesis (1). Also, cyclin D1/Cdk4 phosphorylates Smad3 to inhibit its transcriptional activity and antiproliferative function (2). Recently, it has been revealed that cyclin D1/Cdk4 can phosphorylate the transcriptional factor FOXM1, which up-regulated its stability and activity in melanoma cells (3).
To further explore the mechanism of cyclin D1/Cdk4 in regulating cell cycle progression, we used the tandem affinity purification (TAP) tag approach to identify Cdk4-associated proteins and found that NDR1/2 (mammalian nuclear Dbf2-related protein kinase 1/2; also named STK38 and STK38-like, respectively) interacts with Cdk4. The functions of NDR1 and NDR2 have been revealed in many aspects of cell cycle progression, such as cell proliferation, centrosome duplication, chromosome alignment, and apoptosis (4–8). Recently, it was reported that NDR1 and NDR2 promote cell cycle progression by influencing the stability of p21 and ubiquitination of c-Myc (5). The kinase activity of NDR1/2 is strictly regulated and controlled by multiple components. MOB1A (Mps one binder 1A) or S100B (S100 calcium-binding protein B) binds to the N-terminal region of NDR1 and NDR2 to stimulate their autophosphorylation at Ser-281/Ser-282 and promote their kinase activity (9, 10), whereas MOB2 competitively binds to the N-terminal region of NDR1 and NDR2 to inhibit their activity (11). MST (mammalian Ste20-like kinase) family kinases phosphorylate NDR1/2 at Thr-444/Thr-442 to enhance NDR kinase activity (8). MICAL-1 (molecules interacting with CasL protein 1) blocks the interaction of NDR1/2 with MST1 and functions as a negative regulator of NDR1/2 in the apoptosis signaling pathway (12). The activity of NDR1/2 peaks in G1 phase and persists at certain levels in S phase (5). However, whether NDR1/2 activated in G1 phase relates to cyclin/Cdks following cell cycle signal transduction is not clear.
Here, we demonstrate that cyclin D1/Cdk4 interacts with NDR1/2. Importantly, we found that cyclin D1 enhances the activity of NDR1/2 to promote G1/S transition independent of Cdk4. Our findings reveal the novel function of cyclin D1 through regulating NDR kinase activity during cell cycle progression.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
The NDR1/2 and Cdk4 cDNAs were amplified by RT-PCR from a human embryonic kidney cDNA library. Full-length NDR1 and NDR2 were cloned into pFLAG-CMV2, pET41b, and pET30a (Novagen). Full-length Cdk4 was cloned into pCMV-Myc, pET41b, and pET30a. The cyclin D1 construct (kindly provided by Dr. Xavier Mayol) was cloned into pCMV-Myc, pET41b, and pET30a. The cDNA encoding Rb(773–832) was cloned into pET41b. Full-length p21 was cloned into pET41b. pCMV5-MOB1A and pCMV-FLAG-MST1 were provided by Xiaolong Yang and Jonathan Chernoff, respectively. The Myc-cyclin D1 K112E, Myc-Cdk4 D158N, FLAG-NDR1 K118A, and FLAG-NDR2 K119A expression plasmids were generated as described previously (13–15). The NDR2 mutant (NDR2 IM) was generated using primers 5′-GCAGAAACTTGGGCAGCAAACAGGAGACAACTGGCA-3′ and 5′-CCAAGTTTCTGCTGCTGCTGCTGAGTTCATGTTCTG-3′ and cloned into pFLAG-CMV2 (9). pET41b-NDR2-PIFtide was generated as described previously (15).
The polyclonal antibodies against NDR1/2 phosphorylated at Ser-281/Ser-282 or Thr-444/Thr-442 were generated as described previously (10). Anti-FLAG antibody (M2) and anti-c-Myc antibody (9E10) were purchased from Sigma. Anti-NDR1 (YJ-7), anti-NDR2 (K-22), anti-Cdk4 (C-22), anti-cyclin D1 (H-295), anti-p21 (F-5), and anti-β-actin (C-11) antibodies were purchased from Santa Cruz Biotechnology.
TAP Tag Purification
The detailed protocol of TAP tag purification was described previously (16). Briefly, pMSCV-C-FLAG-HA-Cdk4 was transfected into the viral packaging cell line Phoenix. The viruses were harvested and used to infect 293T cells, followed by selection with puromycin. Cell lysates were incubated with anti-FLAG beads, and bound proteins were eluted with FLAG peptide. The elution was incubated with anti-HA beads and eluted with HA peptide. The elution was run on 4–12% NuPAGE (Invitrogen) and subjected to mass spectrometry analysis.
Co-immunoprecipitation and GST Pulldown
293T cells were transfected with the indicated plasmids and then lysed in lysis buffer (50 mm Tris (pH 8.0), 150 mm NaCl, and 0.5% Triton X-100) with protease inhibitor (Roche Applied Science) at 4 °C for 15 min. The cell lysates were incubated with antibodies at 4 °C for 2 h, followed by the addition of protein A-agarose beads. After 2 h of incubation at 4 °C, the beads were washed with lysis buffer. The immunoprecipitates were separated by SDS-PAGE and immunoblotted with the indicated antibodies. For GST pulldown assay, GST-NDR1/2 or GST (used as a negative control) was incubated with His-Cdk4 or His-cyclin D1 for 1 h at 4 °C. The glutathione beads were then added and incubated for 1 h at 4 °C. The bound proteins were eluted with sample loading buffer and analyzed by immunoblotting with anti-cyclin D1 or anti-Cdk4 antibody.
Cell Culture and Synchronization
HEK293T cells were cultured in DMEM containing 10% fetal calf serum. The human normal hepatocyte cell line HL77-02 (purchased from the Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) was cultured in RPMI 1640 medium containing 20% fetal calf serum. T-RExTM-HeLa cell lines (Invitrogen) were maintained in DMEM containing 10% fetal calf serum plus 5 μg/ml blasticidin. To synchronize the cells to M phase, the cells were treated with 2 mm thymidine for 12 h, released for 6 h, and then treated with 100 ng/ml nocodazole for 6 h. The mitotic cells were collected by the mitotic shake-off method.
Immunofluorescence Assays
For BrdU incorporation, cells were pulse-labeled with BrdU (10 μm) for 10 min, fixed and incubated in 3 n HCl for 10 min, and stained with Alexa Fluor 594-conjugated mouse anti-BrdU antibody (Molecular Probes). Nuclear DNA was revealed by DAPI staining. The cells were observed under a Leica microscope.
In Vitro Kinase Assay
GST-fused NDR2-PIFtide, p21, cyclin D1, Cdk4, and Rb(773–832) proteins were purified by standard procedures (16). For in vitro kinase assay, 1 μg of each protein as indicated was incubated in kinase buffer (50 mm Tris (pH 7.5), 10 mm MgCl2, 0.02% BSA, and 0.04 mm ATP) in the presence of 0.5 μCi of [γ-32P]ATP for 30 min at 30 °C. For detection of the Cdk4 or NDR1/2 kinase activities in cells, 293T cells were transfected with pCMV-Myc-Cdk4 or pCMV-FLAG-NDR1/2 together with the indicated plasmids. For Cdk4, the cell lysates were immunoprecipitated with anti-Cdk4 antibody and protein A beads. For NDR1/2, the cell lysates were immunoprecipitated with FLAG beads. The beads were then resuspended in kinase buffer and subjected to kinase assay with the indicated substrates. Samples were resolved by 10% SDS-PAGE and autoradiographed on x-ray film.
RNA Interference
HeLa cells were seeded on a 24-well plate and transfected with the indicated siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions for 48 h. siRNAs were synthesized as duplexes with dTdT overhangs (RiboBio, Guangzhou, China). The sense sequences of NDR1 and NDR2 siRNAs were 5′-GGACAUGAUGACCUUGUUGAU-3′ and 5′-GUUACGUCGAUCACAACAC-3′, respectively. The sense sequence of p21 siRNA was 5′-CUUCGACUUUGUCACCGAG-3′.
Flow Cytometry
For DNA content analysis, cells were harvested and fixed in ice-cold 70% ethanol at −20 °C, washed with PBS and 1% BSA, and then incubated with PBS and 1% BSA containing 20 μg/ml propidium iodide and 100 μg/ml RNase A for 30 min at 4 °C. All samples were analyzed on a FACSCalibur cytometer (BD Biosciences). The percentage of cells in each phase of the cell cycle was estimated with ModFit.
Generation of Tet-On Stable Cell Lines
FLAG-tagged cyclin D1 or cyclin D1 K112E was cloned into pcDNATM/TO (Invitrogen). The plasmids were transfected into T-RExTM-HeLa cells. 48 h after transfection, the cells were selected with 5 μg/ml blasticidin and 250 μg/ml Zeocin for 3 weeks. The individual clones were picked and expanded. Cyclin D1 expression was analyzed by immunoblotting for the cells treated with tetracycline (1 μg/ml).
RESULTS
NDR1 and NDR2 Interact with Cyclin D1/Cdk4
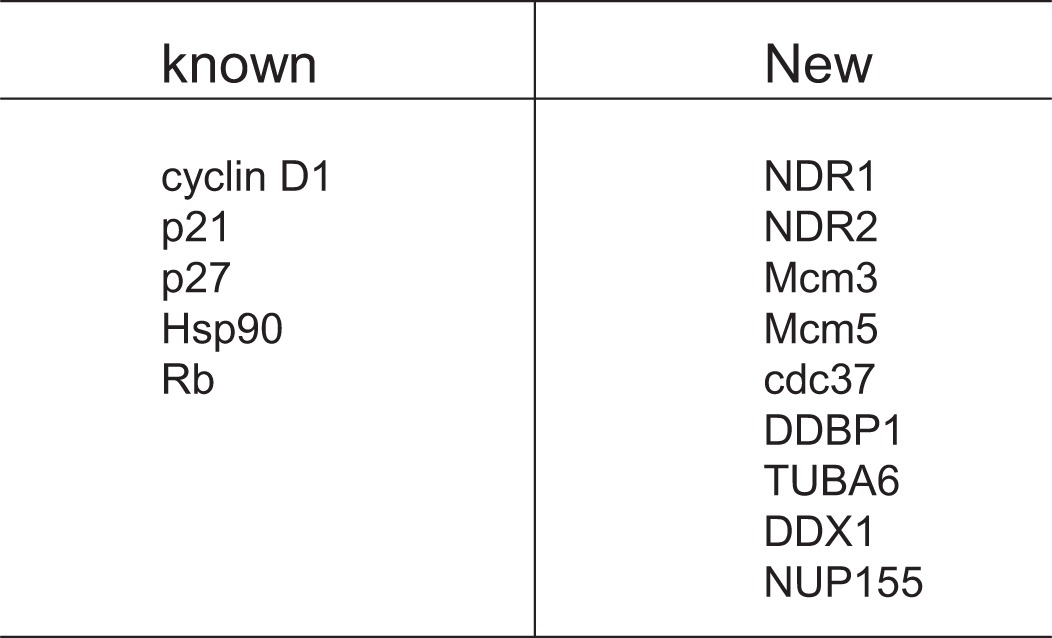
To identify potential Cdk4-associated proteins, we performed a TAP tag purification experiment with Cdk4 as the bait. As shown in Table 1, we identified a set of proteins as potential novel Cdk4-associated partners, in addition to some proteins that are known as Cdk4-interacting proteins, such as cyclin D1, Hsp90, p21, p27, and pRb. Two of the novel proteins are the serine/threonine protein kinases NDR1 and NDR2.
TABLE 1.
Cdk4-interacting proteins identified by mass spectrometry
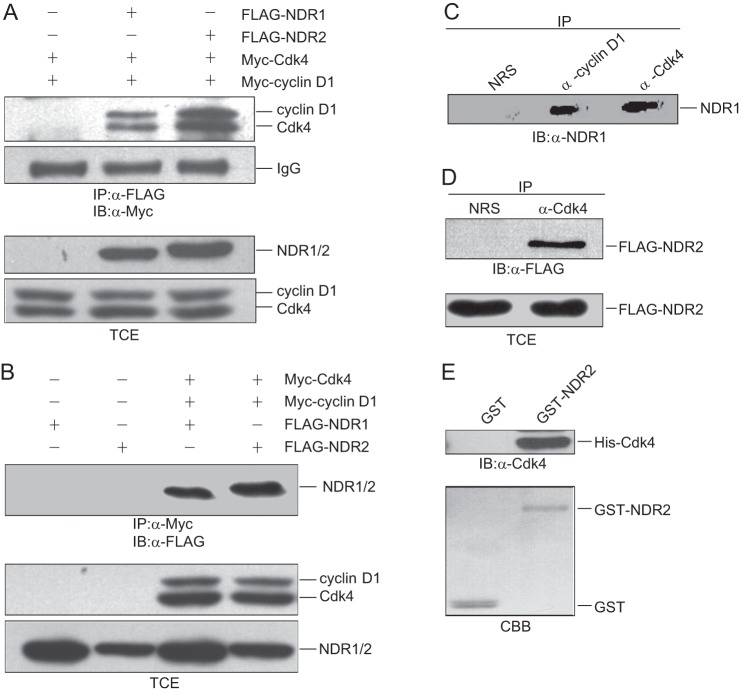
To confirm the interaction of NDR1/2 with cyclin D1/Cdk4, immunoprecipitation experiments were performed using 293T cells coexpressing FLAG-NDR1 or FLAG-NDR2 with Myc-cyclin D1 and Myc-Cdk4. As shown in Fig. 1 (A and B), NDR1/2 interacted with cyclin D1 and Cdk4. To detect the interaction of NDR1/2 with endogenous cyclin D1 and Cdk4, we performed a similar experiment with anti-cyclin D1 and anti-Cdk4 antibodies for co-immunoprecipitation. As shown in Fig. 1C, NDR1 interacted with endogenous cyclin D1 and Cdk4. Also, we found that NDR2 interacted with endogenous Cdk4 as well (Fig. 1D). To confirm the direct interaction of NDR with Cdk4, we purified the GST-NDR2 and His-Cdk4 proteins for GST pulldown assay. The data show that NDR2 interacted with Cdk4 directly (Fig. 1E). These results indicate that NDR interacts with cyclin D1/Cdk4 in vivo and in vitro.
FIGURE 1.
Cyclin D1/Cdk4 interact with NDR1/2. A, 293T cells were cotransfected with pCMV-FLAG-NDR1/2 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-Cdk4. The cell lysates were harvested and subjected to co-immunoprecipitation (IP) with anti-FLAG antibody and then immunoblotted (IB) with anti-Myc antibody. B, 293T cells were cotransfected with pCMV-FLAG-NDR1/2 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-Cdk4. The cell lysates were harvested and subjected to co-immunoprecipitation with anti-Myc antibody, followed by immunoblotting with anti-FLAG antibody. C, 293T cell lysates were harvested and subjected to co-immunoprecipitation with anti-cyclin D1 or anti-Cdk4 antibody with normal rabbit serum (NRS) as a control and then immunoblotted with anti-NDR1 antibody. D, 293T cells were transfected with pCMV-FLAG-tagged NDR2. The cell lysates were harvested and subjected to co-immunoprecipitation with anti-Cdk4 antibody, followed by immunoblotting with anti-FLAG antibody. E, GST protein or GST-NDR2 immobilized on glutathione beads was incubated with His-Cdk4 in lysis buffer. The associated protein was eluted with SDS loading buffer and immunoblotted with anti-Cdk4 antibody. TCE, total cell extract; CBB, Coomassie Brilliant Blue.
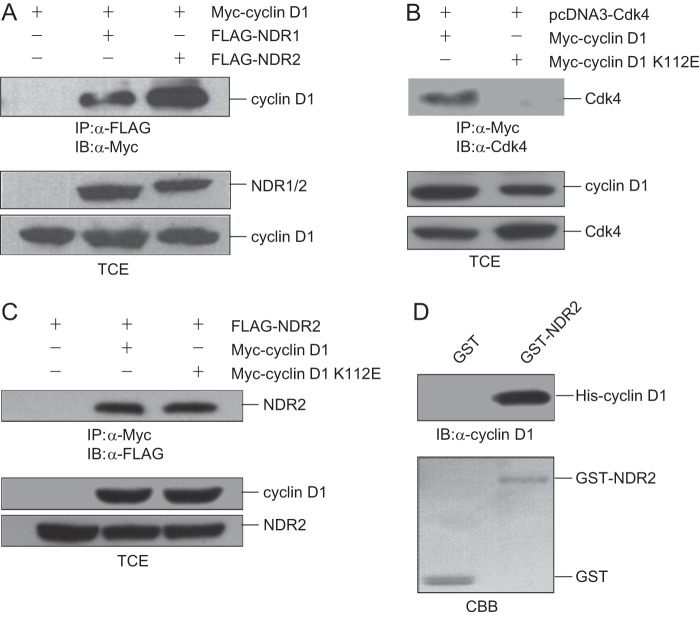
We noticed that NDR1/2 interacted with cyclin D1 and Cdk4 in our experiment, which suggested that cyclin D1 may interact with NDR1/2 independently instead of as a subunit of the cyclin D1-Cdk4 complex. To verify this assumption, we transfected 293T cells with pCMV-FLAG-NDR1/2 and pCMV-Myc-cyclin D1 and performed co-immunoprecipitation assay. As shown in Fig. 2A, NDR1/2 interacted with cyclin D1 independently as expected. To exclude the possibility of endogenous Cdk4 as an adaptor, we generated the cyclin D1 K112E mutant, which could not bind to Cdk4 (Fig. 2B) or activate Cdk4, as reported previously (14), and performed co-immunoprecipitation assay. The data indicate that cyclin D1 K112E interacted with NDR2 distinctly (Fig. 2C). Furthermore, GST pulldown assay using purified GST-NDR2 and His-cyclin D1 showed that NDR2 interacted with cyclin D1 directly (Fig. 2D). These results indicate that cyclin D1 interacts with NDR1/2 independent of Cdk4.
FIGURE 2.
Cyclin D1 interacts with NDR1/2 independent of Cdk4. A, 293T cells were cotransfected with pCMV-Myc-cyclin D1 alone or with pCMV-FLAG-NDR1 or pCMV-FLAG-NDR2. The cell lysates were immunoprecipitated (IP) with anti-FLAG antibody and then immunoblotted (IB) with anti-Myc antibody. B, 293T cells were cotransfected with pcDNA3-Cdk4 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-cyclin D1 K112E. The cell lysates were immunoprecipitated with anti-Myc antibody and then immunoblotted with anti-Cdk4 antibody. C, 293T cells were cotransfected with pCMV-FLAG-NDR2 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-cyclin D1 K112E. The cell lysates were immunoprecipitated with anti-Myc antibody and then immunoblotted with anti-FLAG antibody. D, GST protein or GST-NDR2 immobilized on glutathione beads was incubated with His-cyclin D1 in lysis buffer. The associated protein was eluted with SDS loading buffer and immunoblotted with anti-cyclin D1 antibody. TCE, total cell extract; CBB, Coomassie Brilliant Blue.
Cyclin D1 but Not Cdk4 Positively Regulates NDR Kinase Activity
To determine whether NDR1/2 kinases are phosphorylated by Cdk4, we performed in vitro kinase assay with Cdk4 or Cdk4 D158N (kinase-dead mutant) (13) immunoprecipitated from transfected 293T cells as the kinase and with GST-NDR1 K118A, GST-NDR2 K119A, and GST-Rb(773–832) as the substrates. As shown in Fig. 3A, neither GST-NDR1 K118A nor GST-NDR2 K119A was phosphorylated by Cdk4, whereas phosphorylation of GST-Rb(773–832) indicated that Cdk4 was activated in the assay. These results indicate that NDR1 and NDR2 are not substrates of cyclin D1/Cdk4.
FIGURE 3.
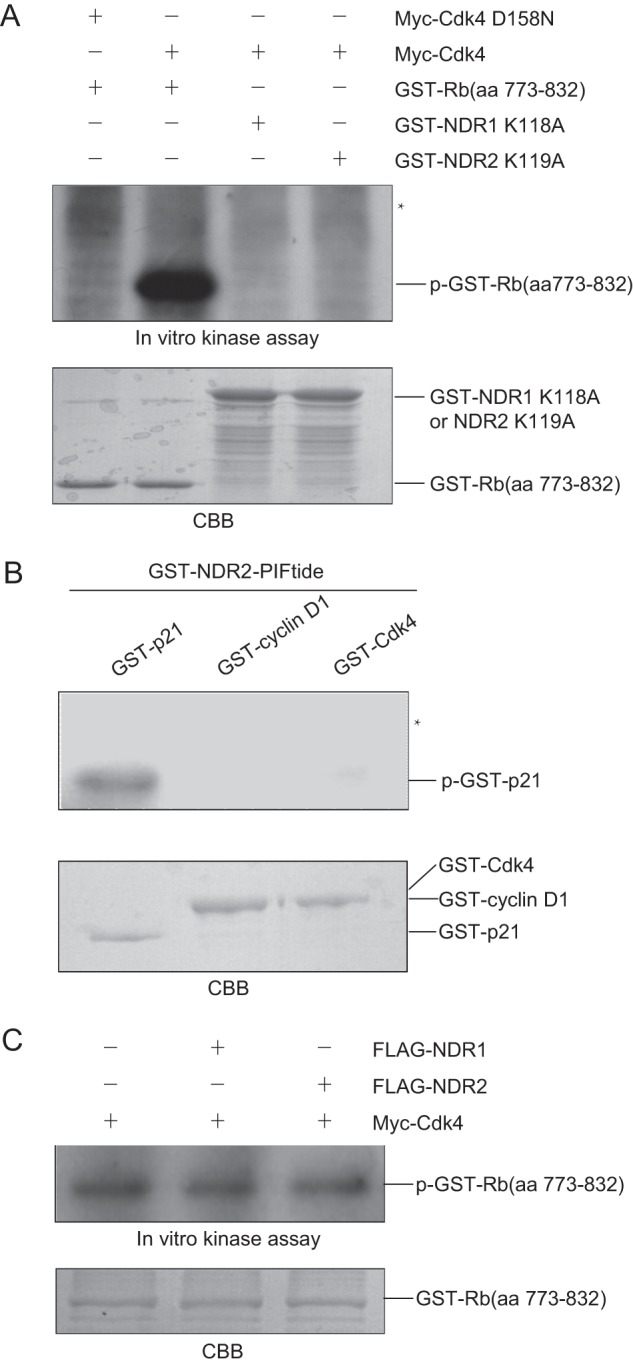
NDR1/2 and cyclin D1/Cdk4 cannot phosphorylate each other in vitro. A, 293T cells were transfected with pCMV-Myc-Cdk4 or pCMV-Myc-Cdk4 D158N. The cell lysates were immunoprecipitated with anti-Cdk4 antibody. The beads were incubated with 1 μg of GST-Rb(773–832), GST-NDR1 K118A, or GST-NDR2 K119A in the presence of [γ-32P]ATP and then assessed by SDS-PAGE, followed by autoradiography of the gel. The asterisk indicates the expected position of GST-NDR1 K118A and GST-NDR2 K119A. B, 1 μg of GST-p21, GST-cyclin D1, or GST-Cdk4 was incubated with GST-NDR2-PIFtide in the presence of [γ-32P]ATP for in vitro kinase assay. The samples were assessed by SDS-PAGE, followed by autoradiography. The asterisk indicates the expected position of GST-cyclin D1 and GST-Cdk4. C, 293T cells were transfected with pCMV-Myc-cyclin D1/Cdk4 alone or with pCMV-FLAG-NDR1/2. The cell lysates were immunoprecipitated with anti-Cdk4 antibody. The beads were incubated with 1 μg of GST-Rb(773–832) in the presence of [γ-32P]ATP for in vitro kinase assay. aa, amino acids; CBB, Coomassie Brilliant Blue.
We next wondered whether cyclin D1 or Cdk4 is phosphorylated by NDR kinases. To ensure the activity of NDR kinases in the assay, we generated the constitutively active form of NDR2, GST-NDR2-PIFtide, to perform in vitro kinase assay with GST-cyclin D1 or GST-Cdk4 as the substrate and GST-p21 as the positive control. As shown in Fig. 3B, GST-cyclin D1 or GST-Cdk4 was not phosphorylated by NDR2-PIFtide, whereas GST-p21 was phosphorylated by active NDR2. These results indicate that cyclin D1 and Cdk4 are not substrates of NDR1/2.
Because NDR1/2 interacts with cyclin D1/Cdk4, NDR1/2 may influence the activity of cyclin D1/Cdk4. To investigate the possible effect of NDR1/2 on the kinase activity of Cdk4, we transfected 293T cells with pCMV-Myc-cyclin D1/Cdk4 alone or with pCMV-FLAG-NDR1/2, and Cdk4 was isolated by immunoprecipitation and subjected to in vitro kinase assay with GST-Rb(773–832) as the substrate. As shown in Fig. 3C, NDR1/2 displayed no effect on the kinase activity of Cdk4 toward GST-Rb(773–832).
We next examined whether cyclin D1 or Cdk4 influences the activity of NDR1/2. Because the autophosphorylation of NDR1 and NDR2 at Ser-281/Ser-282 and the phosphorylation at Thr-444/Thr-442 are required for their kinase activity (10), we first examined whether cyclin D1 or Cdk4 affects the autophosphorylation of NDR2. 293T cells were cotransfected with pCMV-FLAG-NDR2 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-Cdk4. Cell lysates were harvested for immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 antibody. As shown in Fig. 4A, the phosphorylation of NDR2 at Ser-282 was elevated in cells cotransfected with cyclin D1 but not Cdk4, indicating that cyclin D1 positively regulates the activity of NDR kinases. To investigate whether cyclin D1 regulates NDR2 phosphorylation independent of Cdk4, we cotransfected 293T cells with pCMV-FLAG-NDR2 and pCMV-Myc-cyclin D1 or pCMV-Myc-cyclin D1 K112E and performed immunoblotting with the indicated antibodies. As shown in Fig. 4B, cyclin D1 K112E also enhanced the phosphorylation of NDR2 at Ser-282 and Thr-442 as well as wild-type cyclin D1. The above cell lysates were also immunoprecipitated with anti-FLAG antibody and subjected to in vitro kinase assay for NDR2 with GST-p21 as the substrate. As shown in Fig. 4C, the phosphorylation of GST-p21 by NDR2 was enhanced in cells cotransfected with NDR2 and cyclin D1 or cyclin D1 K112E. These results indicate that cyclin D1 promotes NDR kinase activity independent of Cdk4.
FIGURE 4.
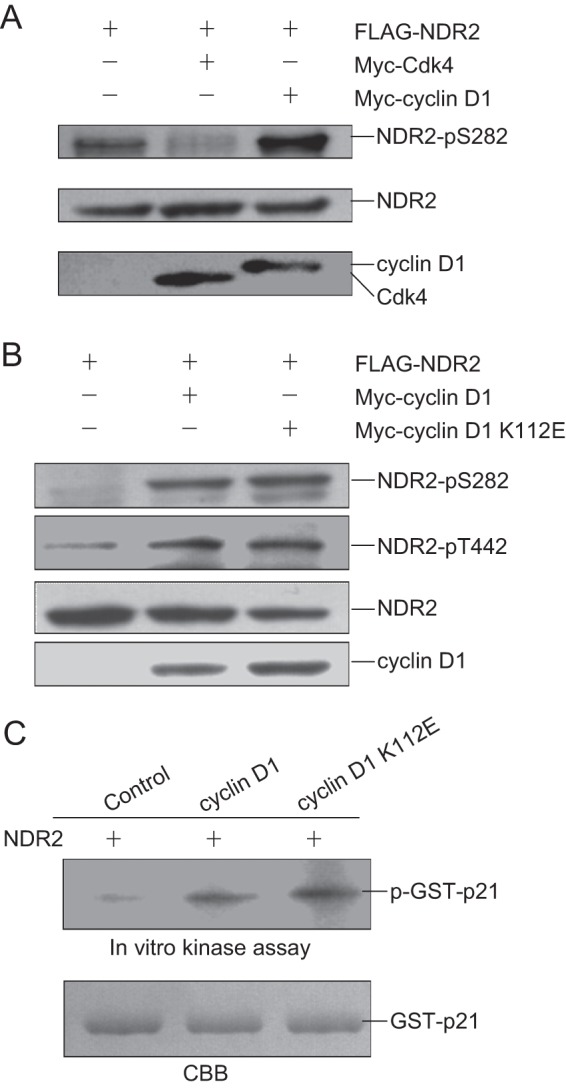
Cyclin D1 promotes the activity of NDR kinase independent of Cdk4. A, 293T cells were cotransfected with pCMV-FLAG-NDR2 alone or with pCMV-Myc-Cdk4 or pCMV-Myc-cyclin D1. The cell lysates were harvested and subjected to immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 antibody. B, 293T cells were cotransfected with pCMV-FLAG-NDR2 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-cyclin D1 K112E. The cell lysates were harvested and subjected to immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 or anti-NDR phospho-Thr-444/phospho-Thr-442 antibody. C, 293T cells were cotransfected with pCMV-FLAG-NDR2 alone or with pCMV-Myc-cyclin D1 or pCMV-Myc-cyclin D1 K112E. The cell lysates were immunoprecipitated with anti-FLAG antibody. The beads were incubated with 1 μg of GST-p21 in the presence of [γ-32P]ATP for in vitro kinase assay. The samples were assessed by SDS-PAGE, followed by autoradiography of the gel. CBB, Coomassie Brilliant Blue.
Cyclin D1 Regulates Activation of NDR Kinases by Releasing Its Self-inhibition Motif
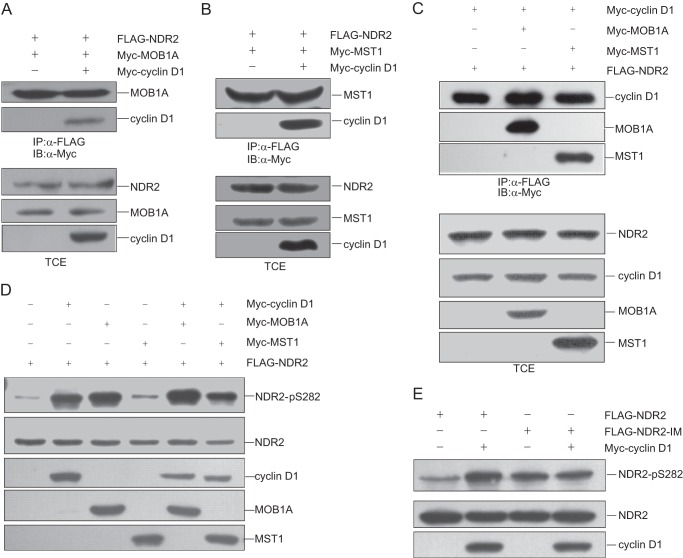
It is known that the kinase activity of NDR1/2 is regulated by MOB1A and MST1. We asked whether cyclin D1 regulates the kinase activity of NDR1 and NDR2 by influencing their association with MOB1A or MST1. To address this question, we transfected 293T cells with pCMV-FLAG-NDR2 and pCMV-Myc-MOB1A, or pCMV-FLAG-NDR2 and pCMV-Myc-MST1 in the absence or presence of pCMV-Myc-cyclin D1 followed by co-immunoprecipitation using anti-FLAG antibody. As shown in Fig. 5 (A and B), cyclin D1 exhibited no effect on the binding of MOB1A or MST1 to NDR2. Also we found that MOB1A or MST1 did not affect the binding of cyclin D1 to NDR2 (Fig. 5C). We then asked whether cyclin D1 and MOB1A or cyclin D1 and MST1 can co-stimulate the phosphorylation of NDR2 at Ser-282. We transfected 293T cells with pCMV-FLAG-NDR2 alone or with pCMV-Myc-MOB1A, pCMV-Myc-MST1, or pCMV-Myc-cyclin D1 as indicated. The cell lysates were harvested for immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 antibody. As shown in Fig. 5D, cyclin D1 further enhanced NDR2 phosphorylation stimulated by MOB1A or MST1.
FIGURE 5.
Cyclin D1 enhances NDR activity by releasing its self-inhibition motif. A, 293T cells were cotransfected with pCMV-FLAG-NDR2 and pCMV-Myc-MOB1A with or without pCMV-Myc-cyclin D1. The cell lysates were immunoprecipitated (IP) with anti-FLAG antibody and then immunoblotted (IB) with anti-Myc antibody. B, 293T cells were cotransfected with pCMV-FLAG-NDR2 and pCMV-Myc-MST1 and with or without pCMV-Myc-cyclin D1. The cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with anti-Myc antibody. C, 293T cells were transfected with pCMV-FLAG-NDR2 and pCMV-Myc-cyclin D1 with or without pCMV-Myc-MOB1A or pCMV-Myc-MST1. The cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with anti-Myc antibody. D, 293T cells were transfected with the indicated plasmids. The cell lysates were harvested and subjected to immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 antibody or other antibodies as shown. E, 293T cells were transfected with pCMV-FLAG-NDR2 or pCMV-FLAG-NDR2 IM alone or with pCMV-Myc-cyclin D1. The cell lysates were harvested and subjected to immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 antibody. TCE, total cell extract.
Because the phosphorylation of NDR1/2 at Ser-281/Ser-282 is inhibited by the insert sequence between the catalytic domains of NDR kinases (9), we wondered whether the binding of cyclin D1 to NDR affects the autoinhibitory insert sequence of NDR1/2. Therefore, we generated the insert mutant of NDR2 (NDR2 IM) to address this question. 293T cells were cotransfected with pCMV-FLAG-NDR2 or pCMV-FLAG-NDR2 IM alone or with pCMV-Myc-cyclin D1. Cell lysates were harvested for immunoblotting with anti-NDR phospho-Ser-281/phospho-Ser-282 antibody. As shown in Fig. 5E, cyclin D1 enhanced the phosphorylation of NDR2 but not NDR2 IM. These results demonstrate that cyclin D1 enhances the kinase activity of NDR1/2 by releasing the autoinhibitory effect of its insert sequence.
Cyclin D1 Promotes Cell Cycle Progression Partly through NDR Kinases
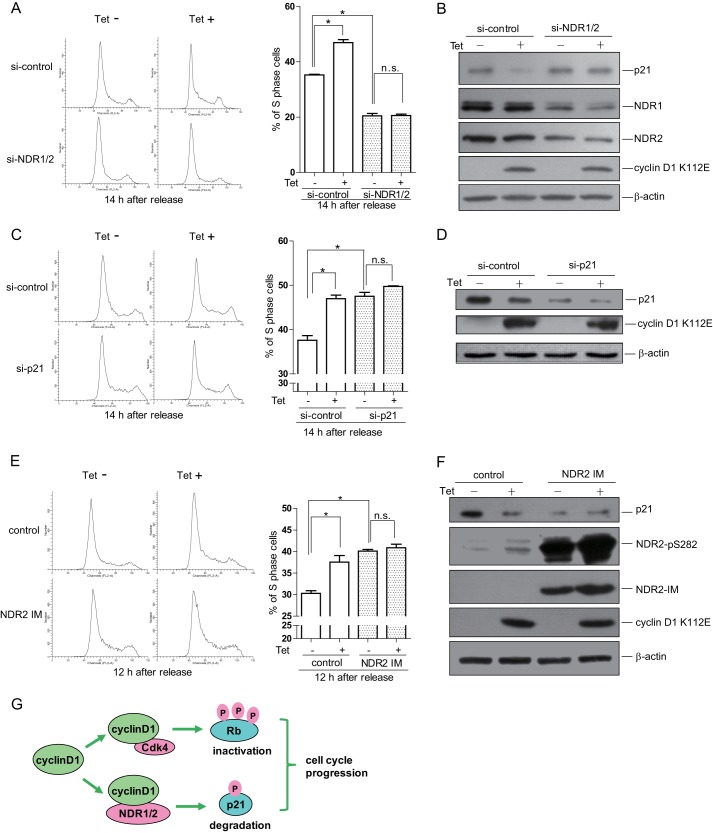
NDR kinases are important regulators in cell cycle progression. To examine whether cyclin D1-promoted NDR kinase activity affects cell cycle progression, we generated the Tet-On inducible HeLa cell lines for cyclin D1 and cyclin D1 K112E. The cells were synchronized in M phase, released for the indicated times, treated with or without tetracycline, and analyzed by flow cytometry (Fig. 6A). The data show that overexpression of cyclin D1 significantly promoted S phase entry, consistent with a previous report (1). Interestingly, overexpression of cyclin D1 K112E also promoted S phase entry, although to a lesser extent compared with wild-type cyclin D1 (Fig. 6, B and C). We also labeled the cells with BrdU, performed immunofluorescence assay, and found that overexpression of cyclin D1 or cyclin D1 K112E promoted DNA synthesis (Fig. 6D). Because cyclin D1 K112E activated NDR but not Cdk4, we wondered if cyclin D1 K112E-promoted S phase entry is dependent on NDR kinase. Because activated NDR kinases will induce p21 degradation in G1 phase to promote G1/S transition (5), we also analyzed the p21 level in cells expressing cyclin D1 or cyclin D1 K112E. As shown in Fig. 6E, the phosphorylation of NDR at Ser-281/Ser-282 was increased, and the protein level of p21 was reduced in cyclin D1- or cyclin D1 K112E-overexpressing cells compared with control cells. We also asked whether the cyclin D1-overexpressing cells accumulate sooner in G2 phase. We synchronized the cells in M phase, released them for 2 h, and then kept them released for 20 h in the presence of nocodazole (Fig. 6F). The cells were collected for flow cytometry analysis. As shown in Fig. 6G, cells overexpressing cyclin D1 or cyclin D1 K112E accumulated in G2 phase sooner compared with control cells. We next asked whether cyclin D1 K112E promotes cell cycle progression in normal cells. To address this question, we transfected the human normal hepatocyte cell line HL77-02 with Myc-tagged cyclin D1 or cyclin D1 K112E. The cells were synchronized by nocodazole in M phase, released for 12 h, and subjected to flow cytometry analysis. As shown in supplemental Fig. S1A, both cyclin D1 and cyclin D1 K112E promoted S phase entry in HL77-02 cells. Consistently, the immunoblotting data show that both cyclin D1 and cyclin D1 K112E promoted the phosphorylation of NDR at Ser-281/Ser-282 and caused the reduction of p21 (supplemental Fig. S1B). Taken together, these results suggest that both cyclin D1 and cyclin D1 K112E promote cell cycle progression, which may be through the NDR-p21 pathway.
FIGURE 6.
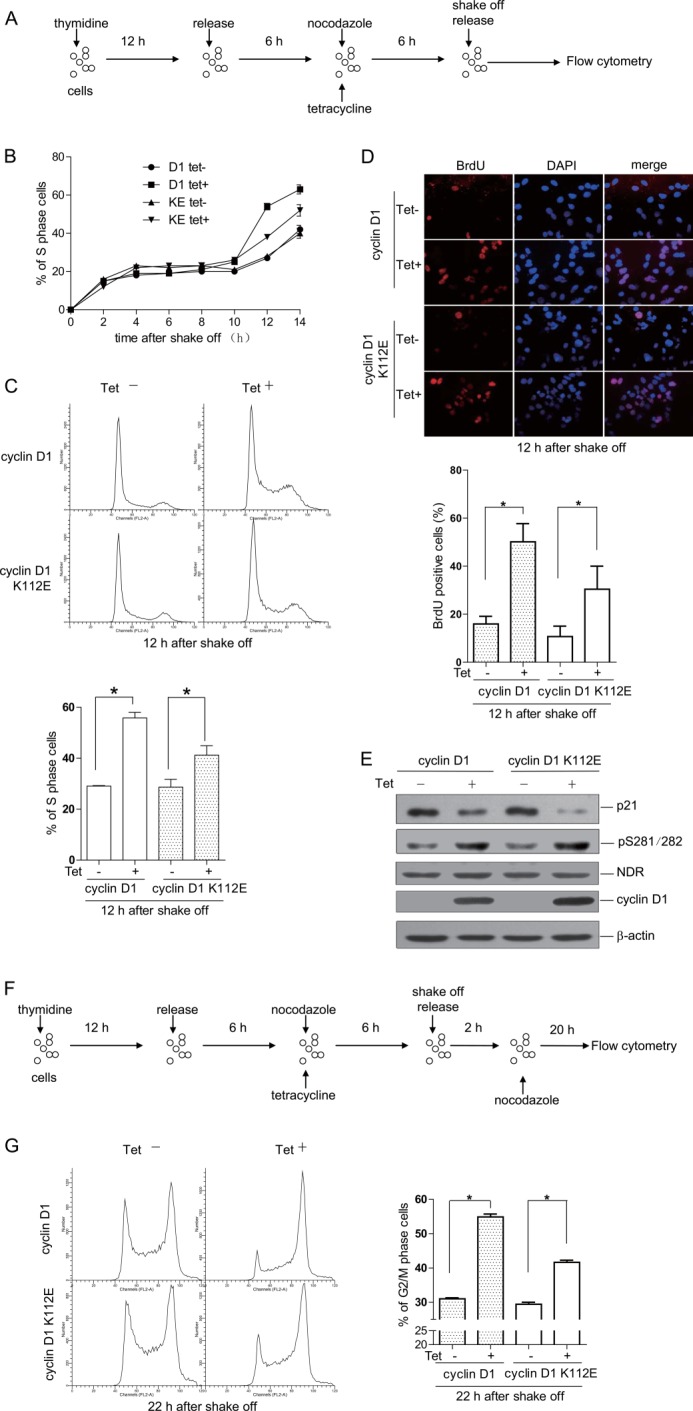
Cyclin D1 K112E promotes G1/S transition. A, schematic view of the experimental design. The cells were arrested at G1/S phase by thymidine treatment for 12 h and then released for 6 h, followed by treatment with nocodazole for 6 h. Cells were shaken off and released for the indicated time points and subjected to flow cytometry. B, cyclin D1 (D1) and cyclin D1 K112E (KE) Tet-On inducible cells were treated as described for A. The percentages of S phase cells in different groups were calculated. Data are presented as means ± S.D. (n = 3). C, cyclin D1 and cyclin D1 K112E Tet-On inducible cells were treated as described for A. The cell cycle profiles at 12 h after release from nocodazole treatment were analyzed by flow cytometry (upper panel), and the percentages of S phase cells in different groups were calculated (lower panel). Data are presented as means ± S.D. (n = 3). D, cyclin D1 and cyclin D1 K112E Tet-On inducible cells were treated as described for A. The cells at 12 h after release from nocodazole treatment were labeled with BrdU and subjected to immunofluorescence (upper panel), and the percentages of BrdU-positive cells in different groups were calculated (lower panel). Data are presented as means ± S.D. (n = 3). E, cyclin D1 and cyclin D1 K112E Tet-On inducible cells were released for 6 h after treatment with nocodazole. Total cell lysates were harvested and subjected to immunoblotting with the indicated antibodies. F, schematic view of experimental design. The cells were arrested at G1/S phase by thymidine treatment for 12 h and then released for 6 h, followed by treatment with nocodazole for 6 h. Cells were shaken off, released for 2 h, treated with nocodazole for 20 h, and then subjected to flow cytometry. G, cyclin D1 and cyclin D1 K112E Tet-On inducible cells were treated as described for F. The cell cycle profiles were analyzed by flow cytometry (left panel), and the percentages of G2/M phase cells in different groups were calculated (right panel). Data are presented as means ± S.D. (n = 3). *, p < 0.05. Tet−, without tetracycline; Tet+, with tetracycline.
To further verify this assumption, we took the RNA interference approach to knock down NDR1/2 and examined the cell cycle profile in cyclin D1 K112E-overexpressing cells. We treated the cells with NDR1 and NDR2 siRNAs, arrested them in M phase with nocodazole, and released them for 14 h. The flow cytometry data show that overexpression of cyclin D1 K112E promoted cell cycle progression in control siRNA-treated cells but not in NDR siRNA-treated cells (Fig. 7A). Consistently, the protein level of p21 was also not affected by overexpression of cyclin D1 K112E if NDR1 and NDR2 were knocked down (Fig. 7B). These results indicate that cyclin D1 K112E promotes the cell cycle dependent upon NDR kinases. We also treated the cells with siRNA against p21 and performed similar experiments as in Fig. 7A. The data show that cyclin D1 K112E promoted cell cycle progression in control cells but not in p21 siRNA-treated cells (Fig. 7, C and D). Because we observed that cyclin D1 did not enhance the phosphorylation of NDR2 IM at Ser-282 (Fig. 5E), we examined whether or not cyclin D1 K112E affects the cell cycle in NDR2 IM-overexpressing cell by taking the same approach as described for Fig. 7A. Fig. 7 (E and F) shows that cyclin D1 K112E did not promote cell cycle progression in NDR2 IM-overexpressing cells. Taken together, we propose that cyclin D1 accelerates cell cycle progression not only by activating Cdk4-mediated signaling but also through the NDR signaling pathway (Fig. 7G).
FIGURE 7.
Cyclin D1 promotes G1/S transition partly dependent of NDR kinase. A, C, and E, cyclin D1 K112E Tet-On inducible cells were transfected with siRNA targeting NDR1/2 (si-NDR1/2) or p21 (si-p21), pCMV-FLAG-NDR2 IM, or the control siRNA (si-control) or plasmid, respectively. The cells were then treated as described in the legend to Fig. 6A and analyzed by flow cytometry (A, C, and E, left panels). The percentage of S phase cells was calculated (A, C, and E, right panel). Data are presented as means ± S.D. (n = 3). B, D, and F, cyclin D1 K112E Tet-On inducible cells were treated as described above. The total cell lysates were harvested and subjected to immunoblotting with the indicated antibodies. G, schematic model showing that cyclin D1 enhances NDR activity to promote G1/S transition. Cyclin D1 can either bind Cdk4 to form an active kinase complex to phosphorylate downstream substrates, such as Rb, or interact with NDR1/2 to enhance its kinase activity for p21. It can activate both pathways to promote cell cycle progression. *, p < 0.05; n.s., not significant. Tet−, without tetracycline; Tet+, with tetracycline.
DISCUSSION
D-type cyclins play a very important role in cell cycle progression, and cyclin D1 has been found to be overexpressed in many types of cancers. How cyclin D1 regulates cell cycle progression is not fully understood. So far, we know only that cyclin D1 binds to Cdk4 or Cdk6 to phosphorylate Rb, Smad3, or FOXM1 to regulate cell cycle progression. Thus, work to identify new cyclin D1/Cdk4-interacting proteins is urgently needed.
Here, we took the TAP tag approach to identify novel cyclin D1/Cdk4-interacting proteins and found that NDR1/2 binds Cdk4 (Table 1). Our results provide evidence showing the novel cross-talk between the cyclin D1/Cdk4 and NDR kinase signaling pathways.
Interestingly, we found that cyclin D1 enhances NDR activation. Cyclin D1 is generally considered to be the Cdk regulatory subunit. However, there was some evidence implying that cyclin D1 exerted its functions independent of Cdk4. For example, it was reported that cyclin D1 inhibits the transcriptional activity of v-Myb, DMP1, BETA2/NeuroD, and STAT3 (18–21). Cyclin D1 has been found to be a positive regulator of estrogen receptors (22). In addition, it has been reported that cyclin D1 is involved in DNA damage repair by directly binding to RAD51 (23). Nevertheless, there is no report revealing that cyclin D1 promotes cell cycle progression independent of Cdk4.
Here, we revealed a new function of cyclin D1 as a NDR activator to regulate cell cycle progression. Cyclin D1 K112E, which could not activate Cdk4, promoted cell cycle progression by enhancing NDR activity and reducing p21 level. Our results indicate that cyclin D1 promotes cell cycle progression not only as a regulatory subunit of Cdk4 but also by activating the NDR pathway by itself.
Our results show that cyclin D1 and cyclin D1 K112E enhanced NDR phosphorylation at Ser-281/Ser-282 and Thr-444/Thr-442, which is a sign of NDR activation. We found that cyclin D1 enhanced phosphorylation of p21 by NDR2 using in vitro kinase assay. Although cyclin D1 did not affect the association of NDR with its positive regulator MOB1A or MST1, it promoted NDR autophosphorylation by releasing its self-inhibition motif between kinase domains, which is the prerequisite of full activation of NDR kinases. We propose that autophosphorylation of NDR can be controlled not only by MOB1A but also by cyclin D1. The in-depth mechanism of how cyclin D1 and MOB1A coordinate with each other to regulate NDR kinase activity needs to be further studied.
It has been reported that p21CIP1/WAF1 at low protein levels promotes complex formation and nuclear translocation of cyclin D1/Cdk4 but inhibits the kinase activity of Cdk4 at high protein levels (24). In addition, p21 exerts its inhibitory effect on cyclin E/Cdk2 to block G1/S transition (25). It has been reported that cyclin E/Cdk2 can phosphorylate p21 to promote its degradation (26), but no report has revealed that cyclin D1/Cdk4 can phosphorylate p21. Here, we showed that cyclin D1 promoted degradation of p21 by enhancing the activity of NDR kinases. These results demonstrate that the level of p21 is precisely controlled by different kinases to keep the cell cycle progression in order.
Although we did not observe that NDR1/2 was phosphorylated by Cdk4 and vice versa or that NDR affected the activity of Cdk4 by in vitro kinase assay, we noticed that overexpression of Cdk4 reduced the phosphorylation of NDR2 at Ser-282 (Fig. 4A) and that overexpression of Cdk4 inhibited the interaction between NDR2 and MST1 but did not affect the association of NDR2 and MOB1A (supplemental Fig. S2). However, whether Cdk4 regulates cell cycle progression though NDR will be further studied.
In conclusion, we found that cyclin D1/Cdk4 can cross-talk with NDR kinase pathways. We demonstrated that cyclin D1 is a positive regulator of NDR kinase and promotes cell cycle progression independent of Cdk4. These findings extend the role of cyclin D1 as a key regulator in cell cycle regulation and may provide a new avenue of cancer therapy related to cyclin D1.
Acknowledgment
We thank Dr. Y. Li (Institute of Psychology, Chinese Academy of Sciences) for help with flow cytometry.
This work was supported by National Natural Science Foundation of China Grant 81272272, Ministry of Science and Technology of China Grants 2012CB519003 and 2011CB504705, and Chinese Academy of Sciences Innovation Project KSCX2-EW-J-6.

This article contains supplemental Figs. S1 and S2.
- Cdk
- cyclin-dependent kinase
- TAP
- tandem affinity purification
- IM
- insert mutant.
REFERENCES
- 1. Sherr C. J., Roberts J. M. (1999) Cdk inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 2. Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 3. Anders L., Ke N., Hydbring P., Choi Y. J., Widlund H. R., Chick J. M., Zhai H., Vidal M., Gygi S. P., Braun P., Sicinski P. (2011) A systematic screen for Cdk4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 20, 620–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chiba S., Ikeda M., Katsunuma K., Ohashi K., Mizuno K. (2009) MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr. Biol. 19, 675–681 [DOI] [PubMed] [Google Scholar]
- 5. Cornils H., Kohler R. S., Hergovich A., Hemmings B. A. (2011) Human NDR kinases control G1/S cell cycle transition by directly regulating p21 stability. Mol. Cell. Biol. 31, 1382–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hergovich A., Lamla S., Nigg E. A., Hemmings B. A. (2007) Centrosome-associated NDR kinase regulates centrosome duplication. Mol. Cell 25, 625–634 [DOI] [PubMed] [Google Scholar]
- 7. Suzuki A., Ogura T., Esumi H. (2006) NDR2 acts as the upstream kinase of ARK5 during insulin-like growth factor-1 signaling. J. Biol. Chem. 281, 13915–13921 [DOI] [PubMed] [Google Scholar]
- 8. Vichalkovski A., Gresko E., Cornils H., Hergovich A., Schmitz D., Hemmings B. A. (2008) NDR kinase is activated by RASSF1A/MST1 in response to FAS receptor stimulation and promotes apoptosis. Curr. Biol. 18, 1889–1895 [DOI] [PubMed] [Google Scholar]
- 9. Bichsel S. J., Tamaskovic R., Stegert M. R., Hemmings B. A. (2004) Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 279, 35228–35235 [DOI] [PubMed] [Google Scholar]
- 10. Tamaskovic R., Bichsel S. J., Rogniaux H., Stegert M. R., Hemmings B. A. (2003) Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J. Biol. Chem. 278, 6710–6718 [DOI] [PubMed] [Google Scholar]
- 11. Kohler R. S., Schmitz D., Cornils H., Hemmings B. A., Hergovich A. (2010) Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human Ndr kinases. Mol. Cell. Biol. 30, 4507–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou Y., Adolfs Y., Pijnappel W. W., Fuller S. J., Van der Schors R. C., Li K. W., Sugden P. H., Smit A. B., Hergovich A., Pasterkamp R. J. (2011) MICAL-1 is a negative regulator of MST-NDR kinase signaling and apoptosis. Mol. Cell. Biol. 31, 3603–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van den Heuvel S., Harlow E. (1993) Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262, 2050–2054 [DOI] [PubMed] [Google Scholar]
- 14. Hinds P. W., Dowdy S. F., Eaton E. N., Arnold A., Weinberg R. A. (1994) Function of a human cyclin gene as an oncogene. Proc. Natl. Acad. Sci. U.S.A. 91, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stegert M. R., Tamaskovic R., Bichsel S. J., Hergovich A., Hemmings B. A. (2004) Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 279, 23806–23812 [DOI] [PubMed] [Google Scholar]
- 16. Deng M., Li F., Ballif B. A., Li S., Chen X., Guo L., Ye X. (2009) Identification and functional analysis of a novel cyclin E/Cdk2 substrate Ankrd17. J. Biol. Chem. 284, 7875–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deleted in proof [Google Scholar]
- 18. Ratineau C., Petry M. W., Mutoh H., Leiter A. B. (2002) Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J. Biol. Chem. 277, 8847–8853 [DOI] [PubMed] [Google Scholar]
- 19. Inoue K., Sherr C. J. (1998) Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol. Cell. Biol. 18, 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirai H., Sherr C. J. (1996) Interaction of D-type cyclins with a novel Myb-like transcription factor, DMP1. Mol. Cell. Biol. 16, 6457–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bienvenu F., Gascan H., Coqueret O. (2001) Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J. Biol. Chem. 276, 16840–16847 [DOI] [PubMed] [Google Scholar]
- 22. Zwijsen R. M., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R. J. (1997) Cdk-independent activation of estrogen receptor by cyclin D1. Cell 88, 405–415 [DOI] [PubMed] [Google Scholar]
- 23. Jirawatnotai S., Hu Y., Michowski W., Elias J. E., Becks L., Bienvenu F., Zagozdzon A., Goswami T., Wang Y. E., Clark A. B., Kunkel T. A., van Harn T., Xia B., Correll M., Quackenbush J., Livingston D. M., Gygi S. P., Sicinski P. (2011) A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 474, 230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LaBaer J., Garrett M. D., Stevenson L. F., Slingerland J. M., Sandhu C., Chou H. S., Fattaey A., Harlow E. (1997) New functional activities for the p21 family of Cdk inhibitors. Genes Dev. 11, 847–862 [DOI] [PubMed] [Google Scholar]
- 25. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 26. Bornstein G., Bloom J., Sitry-Shevah D., Nakayama K., Pagano M., Hershko A. (2003) Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278, 25752–25757 [DOI] [PubMed] [Google Scholar]