Summary
Differential gene expression is a key strategy adopted by the Lyme disease spirochaete, Borrelia burgdorferi, for adaptation and survival in the mammalian host and the tick vector. Many B. burgdorferi surface lipoproteins fall into two distinct groups according to their expression patterns: one group primarily expressed in the tick and the other group primarily expressed in the mammal. Here, we show that the Fur homologue in this bacterium, also known as Borrelia oxidative stress regulator (BosR), is required for repression of outer surface protein A (OspA) and OspD in the mammal. Furthermore, BosR binds directly to sequences upstream of the ospAB operon and the ospD gene through recognition of palindromic motifs similar to those recognized by other Fur homologues but with a 1-bp variation in the spacer length. Putative BosR-binding sites have been identified upstream of 156 B. burgdorferi genes. Some of these genes share the same expression pattern as ospA and ospD. Most notably, 12 (67%) of the 18 genes previously identified in a genome-wide microarray study to be most significantly repressed in the mammal are among the putative BosR regulon. These data indicate that BosR may directly repress transcription of many genes that are down-regulated in the mammal.
Keywords: Borrelia burgdorferi, Lyme disease, Borrelia oxidative stress regulator, ferric uptake regulator, gene regulation
Introduction
Lyme disease, the most common vector-borne disease in the northern hemisphere, is caused by infection with Borrelia burgdorferi sensu lato transmitted through a tick bite (Steere, 2001; Stanek et al., 2012). The spirochaetes survive in nature in a complex enzootic cycle consisting of an Ixodes tick vector and a vertebrate reservoir host, often a small mammal (Piesman & Gern, 2004). Differential gene expression has been recognized as a key strategy adopted by B. burgdorferi for survival in these two vastly different environments (Samuels, 2011; Radolf et al., 2012). Many of these differentially expressed genes encode surface lipoproteins that can be divided into two distinct groups according to their expression patterns, one group primarily expressed in the tick and the other group primarily expressed in the mammal. For example, genes encoding outer surface protein A and B (OspAB), OspD, and the 6.6-kDa lipoprotein (Lp6.6) are primarily expressed in the tick, whereas genes encoding OspC, fibronectin-binding protein BBK32, and decorin-binding proteins B and A (DbpBA) are primarily expressed in the mammal.
During its natural life cycle, B. burgdorferi changes its surface lipoproteins at the interface between the tick and the mammal, with the changes in OspA and OspC expression being the most evident (Schwan et al., 1995; Schwan & Piesman, 2000). Although OspA expression is turned off in the mammal, as early as 24 hours after tick attachment, spirochaetes found in the tick are shown to express OspA but not OspC. Likewise, although spirochaetes colonizing the unfed tick express OspA not OspC, during tick feeding, spirochaetes switch from OspA to OspC before migrating from tick midgut to salivary gland and then entering the mammal. Therefore, it appears that the spirochaete reciprocally regulates OspA and OspC expression during tick feeding– turning on OspC and turning off OspA prior to entering the mammal and doing the opposite when entering the tick.
Reciprocal regulation of OspC and OspA expression can be observed in spirochaetes cultivated in vitro in Barbour-Stoenner-Kelly (BSK) medium and its derivatives (Barbour, 1984; Pollack et al., 1993). When grown in BSK medium, the spirochaete abundantly expresses OspA (Barbour et al., 1983). Conditions such as increased temperature, reduced pH, and stationary phase all have been shown to induce OspC expression (Schwan et al., 1995; Carroll et al., 1999; Yang et al., 2000). Repression of OspA, on the other hand, appears to require mammalian host-specific signals, and is the most evident when spirochaetes are cultivated in BSK medium in a dialysis membrane chamber (DMC) implanted inside the peritoneal cavity of a mouse or a rat (Akins et al., 1998; Brooks et al., 2003; Caimano et al., 2005; Caimano et al., 2007).
Norgard and colleagues have demonstrated that alternative sigma factor RpoS (σs) plays a critical role in activating transcription of ospC and several other lipoprotein genes that are primarily expressed in the mammal (Hubner et al., 2001). The rpoS gene can be transcribed from a proximal σ54 promoter (Studholme & Buck, 2000) or a distal σ70 promoter (Lybecker & Samuels, 2007). Transcription from the σ54 promoter not only requires RpoN (σ54, σN), the other alternative sigma factor in B. burgdorferi, but also requires the ATPase activity of response regulator protein 2 (Rrp2) (Yang et al., 2003). Phosphorylation/activation of Rrp2 requires acetyl-phosphate but not its cognate histidine kinase sensor, HK2, as the phosphor donor (Burtnick et al., 2007; Xu et al., 2010). Transcription from the σ70 promoter results in an rpoS transcript with an extended 5’ non-coding region, which allows temperature-dependent control of RpoS translation by a small regulatory RNA, DsrA (Lybecker & Samuels, 2007).
The regulatory pathway(s) for ospA and other lipoprotein genes that are primarily expressed in the tick and repressed in the mammal remains poorly defined. Early on, Margolis and Samuels detected direct protein binding to the ospAB promoter using B. burgdorferi whole-cell lysate, but the identity of a 23-kDa protein that co-eluted with the binding activity remained unknown (Margolis & Samuels, 1995). Recent studies indicated that RpoS was required for OspA repression in the mammal (Caimano et al., 2005), and the Borrelia oxidative stress regulator (BosR) was required for the activation of the RpoS regulon (Hyde et al., 2009; Ouyang et al., 2009; Samuels & Radolf, 2009). These observations predict that BosR, a 20-kDa DNA-binding protein belonging to the ferric uptake regulator (Fur) family, would be required for OspA repression.
Fur was first identified in the model organism Escherichia coli as a transcriptional regulator that represses transcription of multiple genes involved in iron uptake (Bagg & Neilands, 1987; Hantke, 1981). The Fur family is widely distributed in bacteria (Lee & Helmann, 2007; Carpenter et al., 2009), and some genomes encode multiple Fur homologues (orthologues or paralogues). Besides Fur, the E. coli genome encodes Zur, a Fur paralogue involved in regulating zinc uptake (Patzer & Hantke, 1998). The Bacillus subtilis genome encodes three Fur homologues, including a Fur orthologue, a Zur orthologue, and Per, which regulates peroxide stress response (Bsat et al., 1998; Gaballa & Helmann, 1998). DNA-binding activity of the Fur family is regulated either directly by metal ions or indirectly by metal ion-dependent sensing of other signals (Lee & Helmann, 2007). Crystal structures of Fur and several homologues indicate that they are all homodimers with up to three metal-binding sites (Pohl et al., 2003; Traore et al., 2006; Lucarelli et al., 2007; An et al., 2009; Sheikh & Taylor, 2009; Dian et al., 2011; Shin et al., 2011; Butcher et al., 2012). While the Fur family of metalloregulators is known for their role in regulating homeostasis of transition metals, some members have been shown to regulate other cellular functions such as energy metabolism, oxidative stress response, and virulence (Carpenter et al., 2009). Recent studies in Pseudomonas syringae, Vibrio cholera, and Campylobacter jejuni revealed 312, 57, and 95 Fur binding sites in their respective genomes of 6.5, 4, and 1.7 Mb (Butcher et al., 2011; Davies et al., 2011; Butcher et al., 2012).
The 1.5-Mb genome of B. burgdorferi encodes only one Fur homologue. Despite of its naming and apparent sequence homology to Fur, whether BosR plays any role in regulating oxidative stress response or transition metal homeostasis in B. burgdorferi remains uncertain (Samuels & Radolf, 2009). Currently, BosR-dependent repression of OspA expression in B. burgdorferi is thought to be indirect. BosR first activates transcription of the rpoS gene (Ouyang et al., 2011), and RpoS in turn represses OspA expression through an unknown mechanism (Caimano et al., 2007). Given that Fur and its homologues are best known for their function as transcriptional repressors, we examine here whether BosR directly represses OspA expression.
Results
BosR is required for repression of OspA and OspD in the mammal
Recent studies showed that BosR-deficient B. burgdorferi mutants were non-infectious in the mammal, probably due to their inability to activate OspC expression (Ouyang et al., 2009; Hyde et al., 2009). To determine whether B. burgdorferi lacking BosR is also defective in OspA repression, we cultivated a bosR mutant and the isogenic complemented bosR mutant in DMCs implanted in the rat peritoneal cavity. These strains were constructed previously from B. burgdorferi type strain B31 (Ouyang et al., 2009), and were kindly provided by M.V. Norgard at University of Texas Southwestern Medical Center.
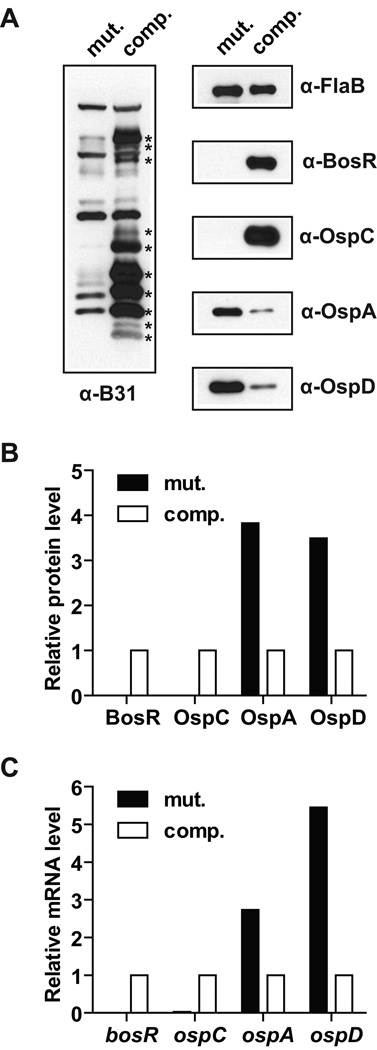
Consistent with previous reports (Ouyang et al., 2009; Hyde et al., 2009), the bosR mutant did not express OspC or several other lipoproteins that elicit strong antibody responses in mice infected by B. burgdorferi through tick infestation, but the levels of these proteins were restored in the complemented bosR mutant (Fig. 1A). Other significant differences between these two strains were in their OspA and OspD protein levels. While the wild-type B31 strain is known to repress OspA and OspD expression during adaption in DMC (Akins et al., 1998; Brooks et al., 2003), the bosR mutant failed to do so, and complementation of the mutant with BosR was able to restore OspA and OspD repression (Fig. 1A). Quantification of the immunoblot bands indicated that, after normalization against the FlaB protein level, the relative OspA and OspD protein levels were 3.8-fold and 3.5-fold higher, respectively, in the bosR mutant than in the complemented bosR mutant (Fig. 1B).
Figure 1.
OspA and OspD repression in DMC requires BosR. The bosR mutant (mut.) and the complemented bosR mutant (comp.) were cultivated for 4 weeks in DMCs implanted in rat peritoneal cavities and then subjected to protein and RNA analyses. (A) Immunoblot analyses were performed using pooled sera from mice infected with B. burgdorferi type strain B31 through tick infestation (α-B31) as well as antibodies specific to FlaB, BosR, OspC, OspA, and OspD. (B) Relative protein levels of BosR, OspC, OspA, and OspD (after normalization against the FlaB level) in the bosR mutant as compared to those in the complemented bosR mutant. Data were based on quantification of the immunoblots shown in panel A. (C) The mRNA levels of bosR, ospA, ospC, and rpoS (after normalization against the flaB mRNA level sample) in the bosR mutant as relative their respective levels in the complemented bosR mutant. Data were based on triplicate q RT-PCR analysis of the RNA samples obtained from the same DMC-adapted bacterial samples that were subjected to immunoblot analyses in panel A.
To determine whether these differences at the protein levels could be attributed to differences at the mRNA levels, quantitative reverse transcription-polymerase chain reaction (q RT-PCR) was performed using primers targeting specific genes (Table S1). After normalization against the flaB mRNA level, the relative ospA and ospD mRNA levels were 2.7-fold and 5.5-fold higher, respectively, in the bosR mutant than in the complemented bosR mutant (Fig. 1C). These data indicated that in the absence of BosR, there were significant increases in the ospA and ospD mRNA levels, which could contribute to the increases in the OspA and OspD protein levels.
As mentioned above, the experimental results shown here are consistent with previous studies suggesting that BosR is required for activation of RpoS (Hyde et al., 2009; Ouyang et al., 2009; Ouyang et al., 2011), and RpoS is required for in vivo-specific repression of OspA (Caimano et al., 2005). However, the Fur family of transcriptional regulators is best known for their function as repressors (Carpenter et al., 2009; Lee & Helmann, 2007) and sigma factors are best known for their ability to activate transcription from specific promoters. An alternative interpretation for the increased ospA and ospD mRNA levels in the bosR mutant could be that BosR directly represses transcription of these two genes.
BosR binds directly to sequences upstream of the ospAB operon and the ospD gene
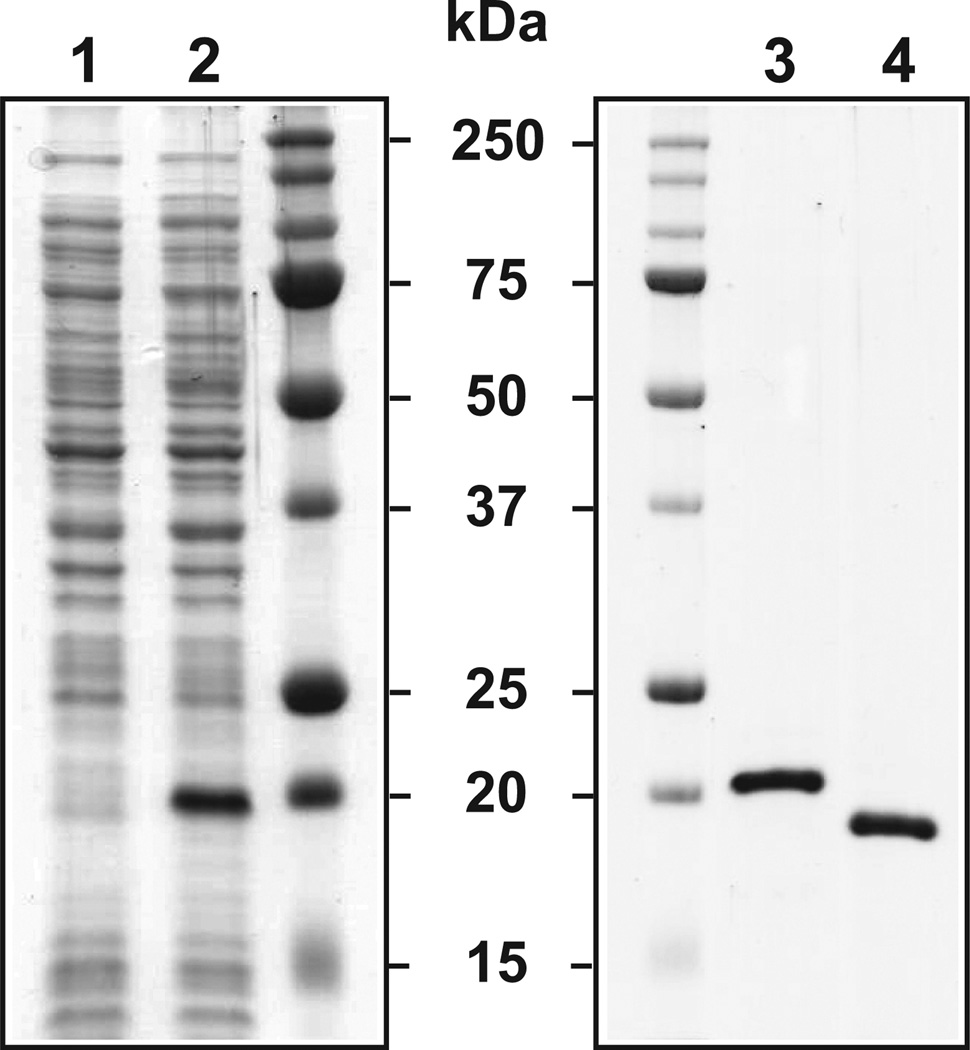
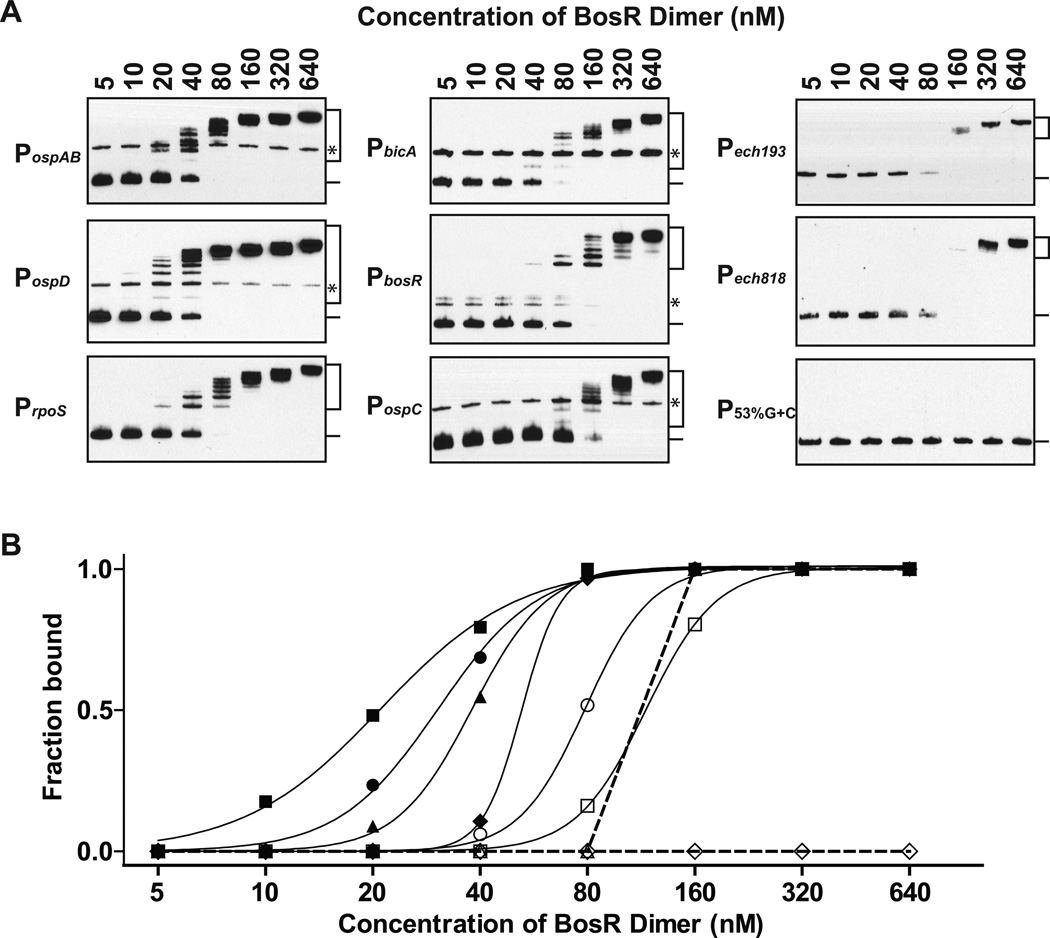
We utilized electrophoretic mobility shift assay (EMSA) to examine whether BosR binds directly to sequences upstream of the ospAB operon and the ospD gene. BosR was affinity-purified from E. coli as a soluble recombinant protein (Fig. 2, see Experimental Procedures for details). Although both His6-tagged and tag-free BosR were active, all EMSA data shown in Fig. 3 and Fig. 4 were obtained using the His6-tagged BosR. DNA fragments spanning 330 bps upstream and 20 bps downstream of the start of the ospA gene or 231 bps upstream and 68 bps downstream of the start of the ospD gene (Fig. S1) were used as probes. Dose-dependent BosR binding to PospAB and PospD was readily detected when the probes were incubated with 5- to 640-fold molar excess of BosR dimers (Fig. 3A).
Figure 2.
Expression and purification of recombinant BosR. SDS-PAGE analysis of whole-cell lysates of E. coli obtained before (lane 1) and after (lane 2) addition of IPTG indicated that expression of the His6-tagged BosR was induced by IPTG. SDS-PAGE analysis of the His6-tagged (lane 3) and the tag-free (lane 4) BosR indicated that these proteins were purified to apparent homogeneity. Sizes of protein markers were indicated in kDa.
Figure 3.
BosR directly binds to sequences upstream of the ospA and the ospD genes. (A) BosR binding to PospAB, PospD, PrpoS, PbicA, PbosR, PospC, Pech193, Pech818, and P53%G+C was analyzed by EMSA. Probes (1 nM) were first incubated with BosR (5 – 640 nM of dimer) and then separated on 6% polyacrylamide gels in 0.5× TBE. (B) BosR binding curves for PospAB (closed circle), PospD (closed square), PrpoS (closed triangle), PbicA (closed diamond), PbosR (open circle), PospC (open square), Pech193 or Pech818 (open triangle), and P53%G+C (open diamond) were generated based on quantification of the EMSA data shown in panel A. Fraction of probe bound by BosR in each reaction was calculated by dividing the intensity of all shifted bands (indicated with a bracket) with the total intensity of both the shifted bands and the unbound band (indicated with a line). In some probes, there were minor isomers (indicated with asterisks) that migrated slower and were often not recognized by BosR. These isomers were excluded in the quantitative analysis. For clarity, curves for the B. burgdorferi probes were indicated with solid lines, and curves for the non-B. burgdorferi probes were indicated with dashed lines.
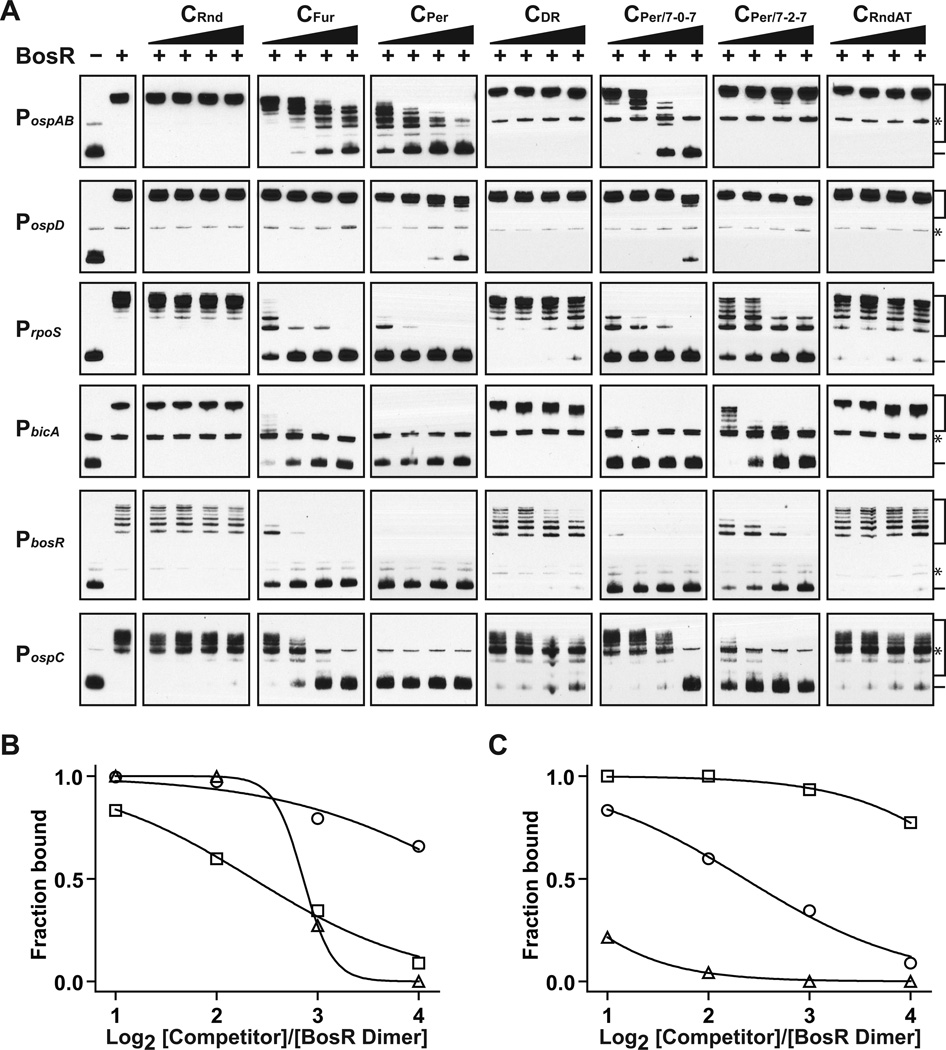
Figure 4.
BosR recognizes both the Per box and the Fur box, and it tolerates 1-bp variation in the spacer length. (A) Seven different competitor oligos, CRnd, CFur, CPer , CDR , CPer/7-0-7, CPer/7-2–7, and CRndAT were tested for their ability to block BosR binding to six different probes, PospAB, PospD, PrpoS, PbicA, PbosR, and PospC. BosR (320 nM of dimer) was incubated with a competitor at molar ratios of 1:2, 1:4, 1:8, and 1:16 prior to incubation with a probe (1 nM). For each probe, a reaction that did not contain BosR or any competitor was included as a negative control, and a reaction that contained BosR but not any competitor was included as a positive control. (B) Inhibition curves were generated for CFur (open circle), CPer (open square), and CPer/7-0–7 (open triangle) based on quantification of their effect on BosR binding to PospAB. Curves were not plotted for any of the other four competitors because none had any inhibitory effect on BosR binding to PospAB. (C) Inhibition curves were generated for CPer based on quantification of its effect on BosR binding to PospAB (open circle), PospD (open square), and PrpoS (open triangle). Curves were not plotted for any of the other three promoters because BosR binding to these promoters was completely blocked by CPer even at the lowest concentration. For panels B and C, fraction of probe bound by BosR in each reaction was calculated as described above.
To date, BosR has been shown to recognize sequences upstream of at least three other genes, rpoS, bosR itself, and the gene encoding neutrophil activating peptide A (NapA) (Boylan et al., 2003; Katona et al., 2004; Ouyang et al., 2011). We recently showed that NapA was a novel fusion between a ferritin-like Dps molecule and a copper thionein and thus renamed it BicA for Borrelia iron- and copper-binding protein A (Wang et al., 2012). For comparison with PospAB and PospD, we examined BosR-binding to PrpoS, PbosR, and PbicA (Fig. S1, Fig. 3A). Binding curves were generated based on intensity measurements of bound and unbound bands in each reaction (Fig. 3B), which allowed ranking of the strength of BosR binding to various probes as the following: PospD > PospAB > PrpoS > PbicA > PbosR (Fig. 3B). Sequences upstream of the ospC gene and two unrelated genes from Ehrlichia chaffeensis (ech193 and ech818) were included as negative controls in our analysis. It is well established the ospC gene is transcriptionally activated by RpoS, not a direct target of BosR. Weaker but nonetheless dose-dependent BosR binding to PospC, Pech193, and Pech818 was detected (Fig. 3A and 3B). Since all sequences tested so far are AT-rich (15~28% G+C) (Fig. S1), we then tested a synthetic DNA fragment with 53% G+C and found that BosR did not bind to this probe at all (Fig. 3A). Collectively, these data indicate that despite having a propensity for AT-rich sequences, BosR binds to sequences upstream of the ospAB operon and the ospD gene more strongly than to other known targets of BosR, and much more strongly than to sequence upstream of the ospC gene (Fig. 3B).
BosR recognizes the Fur and the Per boxes but tolerates variation in the spacer length
The Fur family of DNA-binding proteins is known to recognize 15-bp palindromic sequences that follow the 7-1–7 rule (Lee & Helmann, 2007). For example, E. coli Fur recognizes the sequence TGATAAT-n-ATTATCA, known as the Fur box, whereas Bacillus subtilis PerR recognizes the sequence TTATAAT-n-ATTATAA, known as the Per box. Although several studies have showed and we have further confirmed here that BosR bind to sequences upstream of bicA, bosR, and rpoS, there has not been a consensus on what sequence BosR recognizes (Boylan et al., 2003; Katona et al., 2004; Ouyang et al., 2011). One study suggested that BosR bound to sequences upstream of the bosR and the bicA genes through recognition of putative Per boxes (Katona et al., 2004). Another recent study, however, suggested that BosR bound to the σ54 promoter of the rpoS gene through recognition of the direct repeats (DR) TAAATTAAAT (Ouyang et al., 2011).
We used a competitor EMSA to determine the sequence specificity of BosR binding to PospAB and PospD. At first, we tested four competitors, one with a random sequence (CRnd) and the other three containing consensus sequence of the Fur box (CFur), the Per box (CPer), or the DR (CDR) (Table S1). To compensate for its shorter length, two copies of DR (i.e. four copies of the pentanucleotide) were included in CDR (Table S1). Pre-incubation with either CFur or CPer blocked BosR binding to PospAB in a dose dependent manner, but neither CDR nor CRnd had any adverse effect on the binding (Fig. 4A). BosR-binding to PospD appeared to be much stronger than that to PospAB, because it was not affected by at all by CFur, CDR, or CRnd and the effect of CPer was only evident at the highest concentration tested. In comparison, BosR binding to PrpoS, PbosR, PbicA, or PospC was much weaker than that to PospAB, because it was more severely affected by CPer and CFur. Overall, these data support an earlier conclusion by Benach and colleagues that BosR recognizes the Fur box and the Per box (Katona et al., 2004), and demonstrate again that BosR binds more strongly to PospAB and PospD than to the other probes.
We searched for palindromic sequences similar to the Per or the Fur box and found two in PospAB and seven in PospD. One sequence in PospAB, TTATAAT-ATAATTA, completely covers the −10 region of the ospAB promoter, and the other, TTATATT-AATATAA, is situated further downstream (Fig. S1). The seven sequences found in PospD are identical (TGATATT-aa-AATATAA), each contained in one of seven 17-bp direct repeats previously identified upstream of the ospD gene (Norris et al., 1992). The number of 17-bp direct repeats upstream of ospD varies significantly among strains, ranging from 1 to 12, as reported in one study (Marconi et al., 1994). Notably, the two sequences found in PospAB both lack the 1-bp spacer defined by the 7-1–7 rule, whereas the seven sequences found in PospD all have a 2-bp spacer.
To determine whether BosR has a less stringent requirement for the spacer length, we tested two competitors, each containing a modified Per box, one with no spacer (CPer/7-0–7), and the other with a 2-bp spacer (CPer/7-2–7) (Table S1). Given that both the Per box and the DR are AT-rich, we also tested a competitor oligo with a random sequence of A and T (CRndAT) (Table S1). As shown in Fig. 4A, pre-incubation of BosR with CPer/7-0–7 or CPer/7-2–7 adversely affected its subsequent binding to some but not all probes. In general, the inhibitory effect of CPer/7-0–7 was slightly weaker than that of CPer, and the inhibitory effect of CPer/7-2–7 was weaker than that of CPer/7-0–7. The only exception was that BosR binding to PospC was more effectively blocked by CPer/7-2–7 than by CPer/7-0–7. Like CDR, CRndAT had no effect on BosR binding to PospAB or PospD, but it had some adverse effect on BosR binding to the other probes.
We further quantified the EMSA data to compare the inhibitory effects of all competitors on BosR binding to PospAB, and the results indicated that CPer was the strongest competitor, followed in order by CPer/7-0–7 and CFur (Fig. 4B). CPer/7-2–7 did not affect BosR binding to either PospAB or PospD, but it did inhibit BosR binding to the other probes to various degrees. The remaining three competitors, CDR, CRndAT, and CRnd, had no (or minimal) effect on BosR binding to all probes. By and large, each of these competitors affected BosR binding to all probes similarly, with the overall strengths of these competitors ranked in the following order: CPer > CPer/7-0–7 > CFur > CPer/7-2–7 > CDR = CRndAT > CRnd.
We also quantified the inhibitory effects of CPer on BosR binding to all probes, and the data indicated that BosR binding to PospD was the least affected by CPer, which was followed in order by BosR binding to PospAB and to PrpoS (Fig. 4C). BosR binding to the remaining three probes, PbicA, PbosR, and PospC, was completely blocked by CPer even at the lowest concentration tested. However, three other competitors, CPer/7-0–7, CFur, and CPer/7-2–7, all had varying degrees of inhibitory effect of on BosR binding to these three probes (Fig. 4A). Given that a stronger inhibitory effect implies a weaker binding, these data allowed ranking of the strengths of BosR binding to these various probes as the following: PospD > PospAB > PrpoS > PbicA > PbosR > PospC. Notably, this is the same order as shown in Fig. 3B.
BosR recognizes the palindromic sequences in the ospAB and the ospD promoters
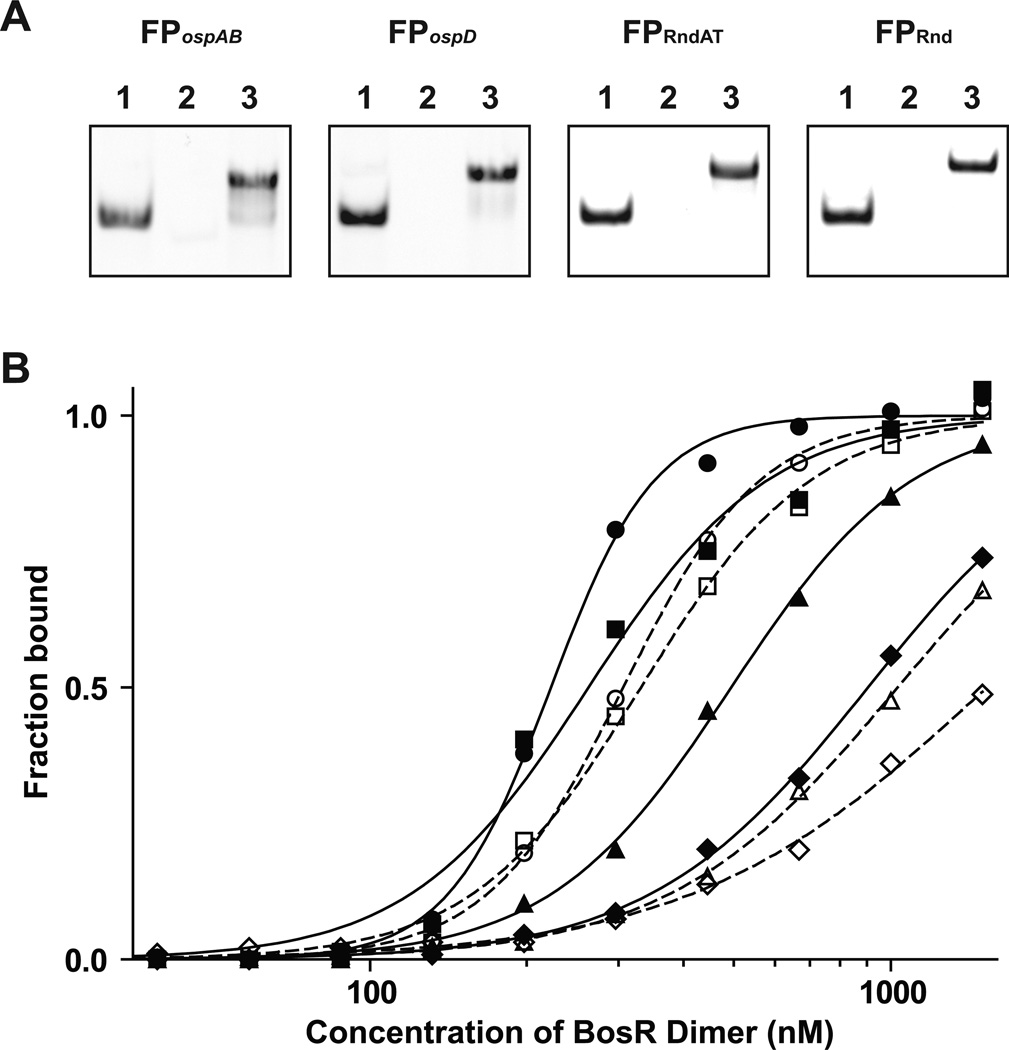
We developed a fluorescence anisotropy (FA)-based assay to determine whether BosR indeed recognizes the palindromic sequences identified in the ospAB and the ospD promoters. Double-stranded fluorescent probes (FP) were prepared by annealing two synthetic oligos with complementary sequences, one of which were labeled at the 5’ end with 6-FAM (see Experimental Procedures for details). For comparison, we also tested oligos with either a random sequence or a random sequence of A and T, which were used as competitors in EMSA, the only difference being that the oligos used here were labeled with 6-FAM on one strand. All four FPs were visualized as single fluorescent bands after electrophoresis on 18% polyacrylamide gels (Fig. 5A; Experiment Procedures).
Figure 5.
BosR recognizes the palindromic sequences near the ospAB and the ospD promoters. (A) Electrophoretic analysis of the fluorescent probes FPospAB, FPospD, FPRndAT, and FPRnd. For each probe, the 6-FAM-labeled single-stranded oligo (lane 1), the unlabeled single-stranded oligo (lane 2), and the double-stranded oligo (lane 3) were analyzed. (B) BosR binding curves to FPospAB (open or closed circle), FPospD (open or closed square), FPRndAT (open or closed triangle) and FPRnd (open or closed diamond) were generated using a FA-based assay. Data obtained using the His6-tagged BosR were indicated with closed symbols and solid lines, and data obtained using the tag-free BosR were indicated with open symbols and dashed lines.
BosR binding to FPospAB, FPospD, FPRndAT, and FPRnd were tested using both the His6-tagged and the tag-free proteins (Fig. 5B). The respective dissociation constants (Kd) for binding to FPospAB, FPospD, FPRndAT, and FPRnd were 222, 262, 494, and 904 nM for the His6-tagged BosR, and 307, 332, 1,032, and 1,531 nM for the tag-free BosR. These data indicated that the His6-tagged protein was slightly more active than the tag-free protein, which could be attributed to BosR losing activity during the extra steps of enzymatic digestion and purification required for preparation of the tag-free protein. Nevertheless, both sets of data showed that the strength of BosR binding to various probes followed the same order: FPospAB > FPospD > FPRndAT > FPRnd. Again, these FA data are consistent with the EMSA data. Despite of having a propensity for AT-rich sequence, BosR recognizes the palindromic motifs identified in the ospAB and the ospD promoters in a sequence-specific manner.
BosR binding protects regions of B. burgdorferi genome from DNase I digestion
The B. burgdorferi genome itself is AT-rich, with the G+C content of its linear chromosome and the 22 linear or circular plasmids ranging from 21% to 32%. The AT-rich sequences in B. burgdorferi genome could potentially function as competitors to ensure that BosR binds more selectively to its targeting sequences. To determine what regions of B. burgdorferi genome are targeted by BosR, we performed, in essence, a genome footprint analysis. Purified B. burgdorferi genomic DNA was first incubated with either BSA or BosR and then digested with DNase I. Quantitative PCR using primers targeting specific regions of B. burgdorferi genome was performed to determine whether these regions were protected by BosR from DNase I digestion (see details in Experimental Procedures).
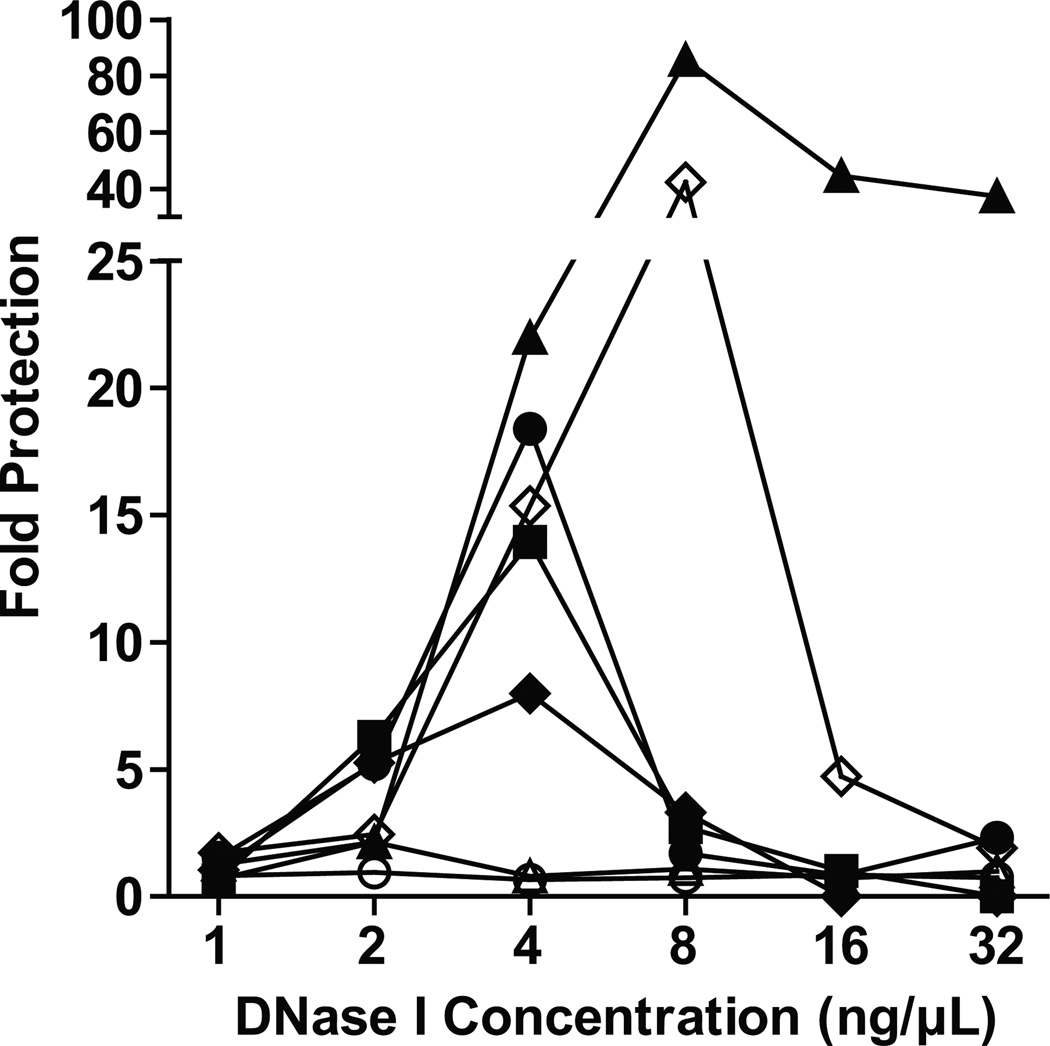
As expected, either too little or too much DNase I digestion resulted in little protection by BosR-binding, but with an optimal concentration of DNase I, BosR-mediated protection became evident for some genomic regions (Fig. 6). Regions upstream of the ospA gene (−330 ~ +20), the ospD gene (−231 ~ +68), the bicA gene (−170 ~ +156), and the bosR gene (−183 ~ +10), were protected by as much as 18-, 14-, 8-, and 86-fold, respectively. The relatively higher degree of protection for the bosR region is likely due to its smaller size. When the targeted bicA region was reduced from 329 bps to 170 bps, the fold of protection increased from 8 to 42. In contrast, a coding region of the ospA gene (+166 ~ +295) was not protected by BosR, even though it was only 130-bp long. A region upstream of the rpoS gene (−277 ~ +60) was also not protected, even though it was comparable in length to the ospA (−330 ~ +20) and the ospD (−231 ~ +68) regions targeted for analysis. In summary, these data showed that BosR protects specific regions of B. burgdorferi genome from DNase I digestion, suggesting that despite its propensity for AT-rich sequences, BosR selectively targets specific regions of the AT-rich genome.
Figure 6.
BosR protects specific regions of B. burgdorferi genome from DNase I digestion. Fold of protection by BosR was determined by q PCR analysis for the following regions of B. burgdorferi genome: ospA (−330 ~ +20) (closed circle), ospA (+166 ~ +295) (open circle), ospD (−231 ~ +68) (closed square), rpoS (−277 ~ +60) (open triangle), bicA (−170 ~ +159) (closed diamond), bicA (−170 ~ −1) (open diamond), bosR (−183 ~ +10) (closed triangle). The ends of each targeted region are indicated by their distances to the start of the gene, with a negative value indicating a position upstream and a positive value indicating a position downstream.
Putative BosR regulon contains genes that are prominently repressed in the mammal
Given that BosR recognizes both Per and Fur boxes and appears to tolerate a 1-bp variation in spacer length, we utilized the Regulatory Sequence Analysis Tools (RSAT) (Thomas-Chollier et al., 2011) to search the intergenic regions of the B. burgdorferi genome for consensus sequence T(T/G)ATAAT-N{0,2}-ATTAT(A/C)A, allowing 2 mismatches. Our search revealed a total of 184 putative BosR-binding sites at 124 intergenic loci, and among those loci, 45 (36%) had 2 or more BosR-binding sites (Table S2). Since 32 (26%) of the 124 loci situate in between two divergent genes, a total of 156 B. burgdorferi genes have at least one BosR-binding site identified within 250-bp of their upstream intergenic region. We termed these genes collectively the putative BosR regulon, which includes ospA, bosR, and bicA, but not rpoS or ospC (Table S2).
Interestingly, this putative BosR regulon contains many genes that had been previously identified by Akins and colleagues in a genome-wide microarray analysis to be prominently down-regulated by mammalian host-specific signals (Brooks et al., 2003). Of the 18 genes whose mRNA levels were the most significantly repressed in host-adapted spirochaetes compared to those grown at 37°C, 12 (67%) belong to the putative BosR regulon (Table 1). One of the remaining six genes is ospB, which is known to be co-transcribed with ospA. Notably, the gene that is the most significantly repressed in the mammal, ospD, also has the most number of BosR-binding sites identified in its upstream intergenic region.
Table 1.
Putative BosR-binding sites are found upstream of many genes that are greatly repressed in the mammal.
| Gene Description a | Fold Repression a | No. of BosR-binding Site b |
|---|---|---|
| BBJ09, outer surface protein D (ospD) | −18.77 | 7 |
| BBH16, hypothetical protein | −12.81 | |
| BBI02, conserved hypothetical protein | −11.78 | 1 |
| BBA62, lipoprotein | −11.73 | 3 |
| BBI39, hypothetical protein | −11.56 | 1 |
| BBD18, hypothetical protein | −11.17 | |
| BBJ41, antigen P35, putative | −10.8 | 1 |
| BBI36, antigen P35, putative | −9.53 | 2 |
| BBA16, outer surface protein B (ospB) | −8.66 | |
| BBI38, hypothetical protein | −7.54 | 2 |
| BBA69 , hypothetical protein | −6.65 | 3 |
| BBA68, hypothetical protein | −6.57 | 1 |
| BBH29, conserved hypothetical protein | −5.9 | |
| BBA15, outer surface protein A (ospA) | −5.85 | 2 |
| BBA74, outer membrane porin (oms28) | −5.79 | |
| BBK45, immunogenic protein P37, putative | −5.68 | |
| BBA38, hypothetical protein | −5.4 | 2 |
| BBA61, conserved hypothetical protein | −5.18 | 3 |
From Table 3 of the referenced study (Brooks et al., 2003).
From this study (see Table S2 for details).
Discussion
BosR has emerged as a key virulence regulator in B. burgdorferi (Samuels & Radolf, 2009). However, there has not been a consensus on what sequence or motif BosR recognizes. Data presented in this study demonstrate that like other Fur family members, BosR recognizes palindromic motifs similar to the Per or Fur box, but with an apparent tolerance for a 1-bp variation in the spacer length. With this simple modification, putative BosR-binding sites were identified upstream of many genes known to be primarily expressed in the tick and prominently repressed in the mammal. Footprint analysis using B. burgdorferi genomic DNA showed that several regions containing putative BosR-binding sites were indeed protected by BosR from DNase I digestion. Therefore, these data strongly suggest that BosR directly represses transcription of these genes in the mammal.
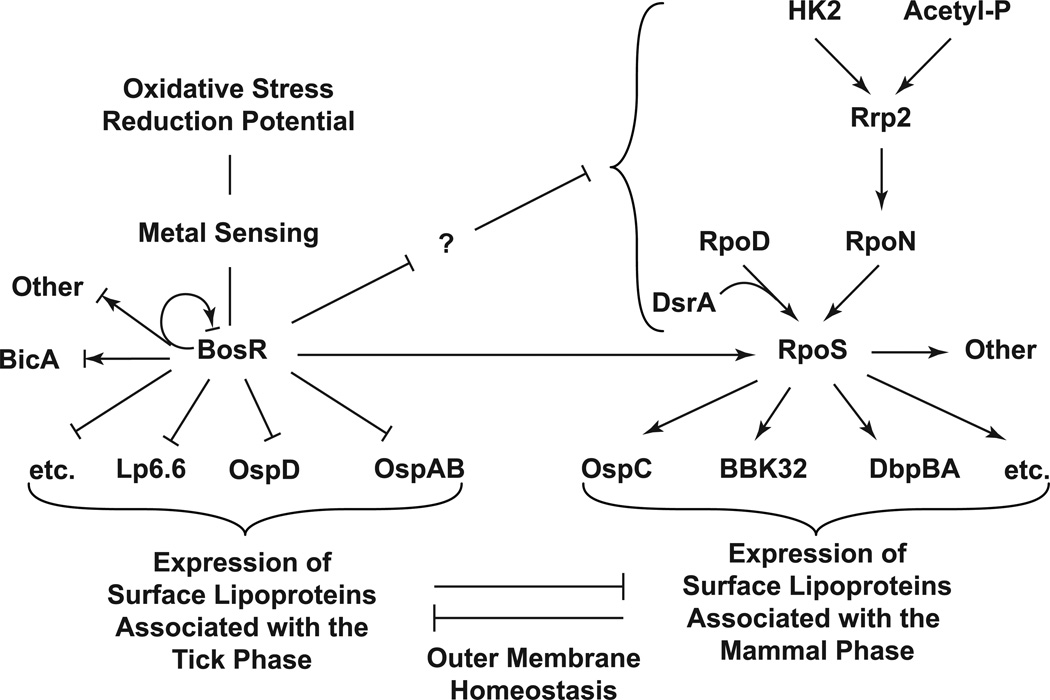
We propose a model where BosR regulates transcription of surface lipoproteins that are primarily expressed in the tick, and the Rrp2-RpoN-RpoS pathway regulates transcription of surface lipoproteins that are primarily expressed in the mammal (Fig. 7). These two pathways work in concert to drive spirochaetal adaptation in the mammal, with RpoS turning on lipoproteins associated with the mammal phase and BosR turning off the tick-phase lipoproteins. These two seemingly parallel pathways are dependent on each other because a deficiency in either RpoS or BosR results in the spirochaete being locked in the tick phase. While the underlying molecular mechanisms remain to be defined (see discussion below), such interdependence of these two pathways is consistent with the biphasic nature of the spirochaete’s life cycle. In a sense, the tick phase – with OspA on and OspC off – appears to be the default state for the Lyme disease spirochaete because while OspC expression requires both BosR and RpoS, OspA expression requires neither and appears to be constitutive. Given the nutrient limitation in the tick, perhaps the spirochaete can only afford default expression of OspA, a key virulence factor required for its survival in the tick.
Figure 7.
A model for reciprocal regulation of B. burgdorferi surface lipoproteins by BosR and RpoS. During transition from the tick to the mammal, B. burgdorferi surface lipoproteins that are primarily expressed in the tick are repressed by BosR whereas those primarily expressed in the mammal are activated by RpoS. The interdependence of these two seemingly parallel pathways may be dictated, in part, by outer membrane homeostasis (see text for details).
A deficiency in BosR resulted in a significant reduction in the RpoS protein level and also a slight reduction in the rpoS mRNA level, suggesting that BosR has a positive effect on RpoS. It has been suggested that BosR directly activates transcription of rpoS through recognition of DR motifs overlapping with the σ54 promoter (Ouyang et al., 2011). Here, we confirmed that BosR did bind to the sequence upstream of the rpoS gene, but showed that this binding was more effectively blocked by CPer and CFer than by CDR. We found a putative BosR-binding site within one of the BosR footprints previously identified upstream of the rpoS gene, BS1 (Ouyang et al., 2011). This binding site, TTACAAT-ga-ATTACAA, has a 2-bp spacer and deviates from the consensus BosR-binding sequence by two nucleotides. It was not identified in our search for intergenic BosR-binding sites (Table S2) because it is situated within the upstream gene bb0771a and therefore not considered intergenic. Binding of BosR to this site could potentially activate rpoS transcription from the downstream σ54 promoter (see discussion below). The lack of BosR protection for this region in the genome footprint analysis does not rule out that BosR may bind to this site in a manner that could not offer protection for the entire region targeted by q PCR analysis. In addition to transcriptional activation of rpoS, BosR could exert a positive influence on RpoS protein level by repressing a negative regulator of the Rrp2-RpoN-RpoS pathway (Fig. 7), which may not be a transcriptional regulator because activation of this pathway is largely dependent on phosphor-relay (Xu et al., 2010), and because the RpoS protein level can also be regulated by proteolysis (Hengge-Aronis, 2002).
A deficiency in RpoS did not affect BosR expression (data not shown), but nonetheless resulted in a lack of repression of OspA in the mammal (Caimano et al., 2007), suggesting that although BosR is required, BosR alone is not sufficient for OspA repression. This may be dictated by lipoprotein homeostasis on the outer surface of the spirochaete (Fig. 7). The phenotypes of several B. burgdorferi mutants suggest that there is a need for the spirochaete to maintain lipoprotein homeostasis on its outer surface. When the ospA gene was abolished from the B. burgdorferi genome, the spirochaete invariably increased the expression of other lipoproteins. In one study, the deletion of the ospAB operon in strain B31 or 297 resulted in constitutive activation of the Rrp2-RpoN-RpoS pathway and thus constitutive expression of OspC (He et al., 2008). In another study, although for an unknown reason, neither the parental B31 strain nor its isogenic ospA mutant expressed OspC when the spirochaetes were cultivated in vitro at 35°C, the mutant appeared to have increased expression of several proteins that reacted strongly with sera from infected mice (Fig. 4 from the reference) (Battisti et al., 2008). In another ospAB mutant constructed from an infectious clone of B31, it was two lipoproteins belonging to the BosR regulon (OspD and BBI39) that were significantly up-regulated due to loss of OspA (F.T. Liang, unpublished data). Similarly, in an ospC mutant constructed from strain 297 (Pal et al., 2004), several other lipoproteins in the RpoS regulon, including BBK32, became up-regulated (Fig. S2). In each of these cases, the spirochaete appeared to compensate the loss of one lipoprotein with increased expression of other lipoproteins. Therefore, it is possible that in the absence of RpoS-mediated activation of lipoproteins associated with the mammalian phase, the spirochaete could not afford to turn off those associated with the tick phase. Notably, in its natural life cycle through the tick and the mammal, the spirochaete never turns off both sets of these lipoproteins.
BicA has long been thought to be the primary target of BosR. We have identified two putative BosR-binding sites in the intergenic region upstream of the bicA gene, and showed that BosR protected this region of the B. burgdorferi genome from DNase I digestion. These data further support the consensus that BosR regulates bicA transcription (Boylan et al., 2003; Katona et al., 2004; Ouyang et al., 2011). However, to date, there have been both evidences for BosR activating bicA transcription and evidences for BosR repressing bicA transcription. Using a reporter system in E. coli, Boylan et al. showed that BosR activated transcription from the bicA promoter (Boylan et al., 2003). Katona et al., however, showed that BosR binding to the upstream region of bicA was adversely affected by peroxide, and thus proposed that BosR functioned as a repressor of bicA transcription (Katona et al., 2004). When the bosR gene was abolished, Hyde et al. reported a reduction in the BicA protein level, but Ouyang et al. reported an increase in the bicA mRNA level (Hyde et al., 2009; Ouyang et al., 2009). In our hands, the bosR mutant constructed by Ouyang et al. also had a significant reduction in BicA protein level when cultivated in vitro (Fig. S3). However, when the spirochaetes were cultivated in the DMCs, presence of BosR was associated with a slight reduction in BicA protein level as well as a slight reduction in the bicA mRNA level (Fig. S3). The BosR-dependent repression of BicA in the DMC is consistent with at least two other pieces of evidence suggesting that BicA expression is relatively repressed in the mammal. A genome-wide microarray study comparing spirochaetes grown in the DMC with those grown in vitro at 37°C showed a slight reduction of the bicA mRNA level in the host-adapted spirochaetes (Brooks et al., 2003). Another study showed that the bicA mRNA level was lower in the mammal compared to that in the tick (Li et al., 2007). Collectively, these data suggest that while BicA expression may be modestly repressed by BosR in the mammal, there are other situations where BicA expression could be activated by BosR.
Gene regulation by Fur is more complex than a binary mode based on the presence or absence of the regulator. Although Fur homologues are best known as repressors, there have been reports of Fur homologues directly activating gene transcription (Lee & Helmann, 2007; Carpenter et al., 2009). It has been noted that Fur boxes overlapping with a promoter often result in repression whereas those further upstream usually result in activation (Lee & Helmann, 2007). The BosR-binding sites found upstream of ospAB and ospD clearly overlap with their respectively promoters (Table S1, Fig. S1). The tandem arrangement of seven BosR-binding sites upstream of ospD implies extensive BosR coverage of this region. A footprint analysis also showed that BosR coverage of PospAB extended beyond the putative BosR-binding sites (Fig. S4). By and large, these data support that BosR represses transcription of ospAB and ospD.
While it is intuitive that Fur binding to a promoter could block or interfere with its access by the RNA polymerase, it is less clear how Fur binding to regions upstream of a promoter could activate transcription. It has been shown that Fur binding to a DNA molecule results in increased rigidity at the bound region (Le Cam et al., 1994). Perhaps, increased rigidity at regions upstream of a promoter would force the RNA polymerase to go the other direction and initiate transcription of the downstream gene. This mechanism would allow activation of transcription from either a σ70 or a σ54 promoter, so long as the ATPase required for activation of the σ54 promoter is present. It has also been shown that Fur is capable of polymerizing along a DNA molecule and covering regions beyond the initial binding site (Le Cam et al., 1994). Thus, the farther upstream a binding-site is from a promoter, the more protein would be needed to extend coverage to the promoter to block its access by the RNA polymerase. This could potentially allow the action of Fur at a site upstream of a promoter to be determined by the concentration of Fur, activating at a lower concentration and repressing at a higher concentration. Boylan et al. had previously identified a large BosR footprint 173–222 bps upstream of the bicA gene (Boylan et al., 2003). Whether BosR-binding to this region plays any role in activating bicA transcription remains to be investigated.
The DNA-binding activity of Fur homologues is known to be regulated by metals and/or metal-dependent sensing of other signals such as oxidative stress (Lee & Helmann, 2007). We have recently shown that the DNA-binding activity of BosR is negatively regulated by copper, and BosR also regulates copper homeostasis in B. burgdorferi (X. Li, unpublished data). We have shown in another study that BicA plays an important role in detoxifying copper, and the carboxyl cysteine-rich domain of BicA functions as a copper thionein (Wang et al., 2012). Given that the two BosR-bindings sites identified upstream of bicA are very close to its start codon (Table S2), we propose that BosR-binding to these sites may repress bicA transcription, and copper could activate bicA transcription by inactivate the DNA-binding activity of BosR.
We have also identified a putative BosR-binding site upstream of the bosR gene, and have shown that that this region is protected from DNase I digestion by BosR. These data suggest that BosR may regulate its own transcription. Auto-regulation has been reported for other Fur homologues (Lee & Helmann, 2007; Carpenter et al., 2009). Notably, this BosR-binding site is situated at 120~133 bps upstream of bosR, much farther than those found upstream of ospA, ospD, or bicA (Table S2). As discussed above, BosR-binding to this site could potentially activate transcription. It has been shown that bosR could also be co-transcribed with its immediate upstream and downstream genes (Ouyang et al., 2009). The upstream gene, bb0648, is annotated as a pseudogene, and thus the entire gene is considered intergenic by the RSAT program. The BosR-binding site identified upstream of bb0649 (Table S2), which is divergent from bb0648, is actually situated within bb0648 (+30 ~ +43). BosR binding to this site could potentially inhibit transcription of bb0648-bosR-bb0646. Given that BosR can bind nonspecifically to DNA, it is critically important to maintain the BosR protein level within a certain range so that specific promoters can be targeted. While there may be other regulators involved, concentration-dependent activation or repression of its own transcription by BosR could constitute both positive and negative feedback loops.
Although we have used genome footprint analysis to confirm BosR-binding to several regions containing putative BosR-binding sites, whether BosR binds to the other targets listed in Table S2 remains to be verified. Genome-wide mapping of BosR binding sites can be performed using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq) analysis (Park, 2009). Recently, these methods have been applied to studying Fur regulation in V. cholera (Davies et al., 2011), P. syringae (Butcher et al., 2011), and C. jejuni (Butcher et al., 2012). In each of these studies, the network of genes regulated by Fur has been expanded. Here, our data suggest that BosR, the B. burgdorferi Fur homologue, behaves similarly as those in other pathogenic bacteria, recognizing palindromic sequences and regulating transcription of many genes, including but not limited to those involved in metal homeostasis and virulence.
Experimental procedures
Host adaption of spirochaetes in DMCs
The bosR mutant and the complemented bosR mutant were constructed from B. burgdorferi type strain B31 and kindly provided by M.V. Norgard at the University of Texas Southwestern Medical Center (Ouyang et al., 2009). Host-adapted spirochaetes were prepared in DMCs as previously described (Akins et al., 1998). All animal procedures described here were approved by the Institutional Animal Care and Use Committee at Louisiana State University. The Spectra/Por® 6 Standard Grade Regenerated Cellulose dialysis membrane with molecular weight cut-off of 8 kDa (Spectrum Laboratories Inc., Rancho Dominguez, CA) was treated with 5 mM EDTA and then autoclaved. Using standard aseptic techniques, a sterilized DMC filled with 5 ml of BSK-H complete medium (Sigma-Aldrich) was inoculated with 103 spirochaetes per ml, and implanted into the peritoneum of a Sprague-Dawley rat (6–8 weeks old; Division of Laboratory Animal Medicine at Louisiana State University, Baton rouge, LA). After 4 weeks of adaptation in DMCs, approximately 5×107 of the bosR mutant and 2×107 of the complemented bosR mutant were recovered and subjected to immunoblot and q RT-PCR analyses as described below.
Immunoblot analysis
Host-adapted spirochaetes were suspended in the Laemmli buffer (Laemmli, 1970) to approximately 2×105 spirochaetes per µl, and incubated at 100°C for 10 min. Samples (5 µl per lane) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes for immunoblot analysis using pooled sera from mice infected with B. burgdorferi type strain B31 through tick infestation or antibodies to FlaB, BosR, RpoS, OspC, or OspA. A goat α-mouse IgG-horse radish peroxidase conjugate was purchased from Kirkegaard & Perry Laboratories and used at a 1:10,000 dilution. Signals were developed using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and captured on X-ray films. Adobe Photoshop CS3 was used to compile images and to quantify band intensity.
Quantitative RT-PCR and PCR
Total RNA was extracted from host-adapted spirochaetes using the TRIZOL reagent (Life Technologies), digested with RNase-free DNase I (Roche), and then purified on the RNeasy mini spin column (Qiagen) following the manufacturers’ protocols. For q RT-PCR analysis, RNA was first converted into DNA using Superscript™ III reverse transcriptase (Invitrogen) and random hexamers. Quantitative PCR was performed in 96-well plates using SYBR® green PCR master mix (Applied Biosystems) and primers targeting specific sequences (Table S1). On each plate, a series of 10-fold dilutions of B. burgdorferi genomic DNA with known concentrations were assayed to generate a linear calculation curve, which was used to convert the threshold cycle (Ct) values to gene copy numbers.
Expression and purification of His6-tagged or tag-free BosR
The bosR gene (with omission of the start codon) was amplified from genomic DNA of B. burgdorferi B31 strain using Vent DNA polymerase (New England Biolabs) and specific primers (Table S1). The BamHI and XhoI restrictive sites engineered respectively in the forward and reverse primers allowed directional cloning of the bosR gene into the BamHI- and SalI-digested pQE30 vector, which resulted in an in-frame fusion of BosR with an amino-terminal His6 tag. PCR mutagenesis was applied to modifying the plasmid construct so that the amino-terminal sequence of His6-tagged BosR was changed from Met-Arg-Gly-Ser-His6-Gly-Ser-BosR to Met-Lys-His6-Lys-BosR (changes underlined), which then allowed removal of the His6 tag using the TAGZyme system (Qiagen). Primers used for the first and the second lysine replacement were listed in Table S1. Intended changes were verified by nucleotide sequencing at the Plant-Microbe Genomics Facility of Ohio State University.
Expression of the fusion protein in E. coli strain M15 (Qiagen) was induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG; Sigma). Bacteria were lysed in the B-PER protein extraction buffer containing DNase I, lysozyme, and a cocktail of EDTA-free protease inhibitors, and BosR was affinity-purified from the cell lysate using HisPur cobalt resin following the manufacturer’s protocol (all reagents were from Thermo Scientific). To remove imidazole, protein samples were filtrated using centrifugal filter units with a 10-kDa molecular weight cut off (Millipore). Enzymatic removal of the first eight amino acid residues (including the His6 tag) was carried out using His6-tagged dipeptidyl aminopeptidase I (Qiagen), after which any remaining His6-tagged BosR and the enzyme were removed using HisPur cobalt resin (Thermo Scientific). Tag-free recombinant BosR differs from native BosR only in that it has an arginine instead of methionine at the first residue. Protein concentration was measured using the Bradford reagent (Bio-Rad) according to a standard linear curve generated with a series of BSA solutions of known concentrations. Protein was stored in small aliquots at −80°C until use.
Consistent with a previous report (Ouyang et al., 2011), zinc was the only metal that was detected at a significant level in recombinant BosR, which was measured at approximately 0.9 atoms per monomer by inductively coupled plasma-sector field mass spectrometry as described previously (Wang et al., 2012). Adding excess zinc (10 µM) to binding reactions or removing zinc from BosR did not affect the DNA-binding activity of BosR (unpublished data, X.L.). Therefore, all binding reactions were carried out using recombinant BosR as purified and without addition of excess zinc. For EMSA, some probes were tested multiple times using both His6-tagged and tag-free BosR, whereas others were only tested using His6-tagged BosR. In order to compare BosR binding activities to different probes, all experiments shown in Fig. 3 and Fig. 4 were performed using a single batch of His6-tagged BosR. For the FA-based assay, we tested all probes using both the His6-tagged and the tag-free BosR.
Electrophoretic mobility shift assay (EMSA)
Sequences upstream of B. burgdorferi genes ospA, ospD, rpoS, bicA, bosR, and ospC (Fig. S1) were PCR-amplified from B. burgdorferi type strain B31 using iProof DNA polymerase (Bio-Rad) and pairs of specific primers (Table S1). Either one or both primers were labeled at the 5’-end with biotin. The biotinylated PCR fragments were then purified on a 1.5% SeaPlaque (Lonza) agarose gel using the QIAquick gel extraction kit (Qiagen). DNA concentration was determined by absorbance at 260 nm. The probes (1 nM) were incubated at room temperature for 10 min with 5-, 10-, 20-, 40-, 80-, 160-, 320- or 640-fold molar excess of BosR dimer in 20-µl reactions containing 10 mM Tris (pH7.5), 50 mM KCl, 1mM DTT, 5% glycerol, 50 µg/ml salmon sperm DNA, and 100 µg/ml of BSA. Double-stranded competitors used in EMSA were prepared by annealing pairs of complementary oligos (Table S1) mixed at an equal molar ratio. For competitor EMSA, BosR (320 nM of dimer) was incubated with 2-, 4-, 8-, or 16-fold molar excess of a competitor prior to incubation with a probe (1 nM). All binding reactions were subjected to electrophoresis on 6% polyacrylamide gels prepared in 0.5X TBE (Bio-Rad; 45 mM Tris, pH 8.3, 45 mM boric acid, 1mM EDTA) and then transferred to nylon membranes. The biotin signal was detected using the LightShift chemiluminescent EMSA kit (Thermo Scientific) following the manufacturer’s instructions. The Adobe Photoshop CS3 software was used to compile images captured on X-ray films and to quantify band intensity. Binding and inhibition curves were plotted using the GraphPad Prism 5 software.
Fluorescence anisotropy (FA)
FPospAB was prepared by annealing the 6-FAM-labeled oligo 5’-taatcTTATAATATAATTAtactt-3’ with an unlabeled oligo of the complementary sequence (Table S1). FPospD was prepared by annealing the 6-FAM-labeled oligo 5’-ataatTGATATTaaAATATAAttgat-3’ with an unlabeled oligo of the complementary sequence (Table S1). Of note, five additional nucleotides were included on each side of the putative BosR binding site (indicated in bold). FPRndAT and FPRnd have same sequences as CRndAT and CRnd, respectively, and are prepared similarly by annealing a 6-FAM-labeled oligo with an unlabeled complementary oligo (Table S1). The 6-FAM-labeled and the unlabeled oligos were mixed at a molar ratio of 1 to 1.1. Using an excess amount of the unlabeled oligos was to ensure that the fluorescently labeled oligos exist mostly in the double-stranded form. To monitor the annealing process of each probe, the two single-stranded oligos used for preparation of the double-stranded probe and the double-stranded probe were electrophoresed on an 18% polyacrylamide gel prepared in 0.5× TBE, and fluorescent signals were captured on a Fujifilm LAS-3000 imager. As expected, signal was not detected from any unlabeled single-stranded oligos, and the double-stranded probes migrated more slowly than their respective 6-FAM-labeled single-stranded oligos (Fig. 5A). Since the amount of 6-FAM-labeled single-stranded oligo remained in each probe preparation was negligible, any change in fluorescence anisotropy can be attributed to the double-stranded probe.
Binding reactions were set up in a Corning 384-well low volume non-binding assay plate. Each 20-µL reaction was set up by adding 2-µL of BosR to 18-µL of probe, both in a buffer containing 10 mM Tris, pH7.5, 100 mM KCl, and 1 mM DTT. The probes (10 nM) were incubated with BosR (2 – 1,500 nM of dimer) for 5 min at room temperature. Fluorescence anisotropy (FA; λex = 470 nM; λex = 520 nM) was measured on a TECAN Infinite M1000Pro instrument equipped the iControl software version 1.9. The plate was read three times each before and after addition of BosR, and the three FA readings were averaged. For each well, the average FA reading before addition of BosR was subtracted as background from the average FA reading obtained after addition of BosR. For each probe, a reaction that contained probe only (with addition of 2 µL of buffer instead of BosR) was set up, and FA reading of this reaction was subtracted as background from reactions containing BosR. Changes in FA (ΔFA) at different concentrations of BosR were fitted with a one site-specific binding curve using the GraphPad Prism 5 software, and the fraction of probe bound by BosR in each reaction was calculated by normalizing against the maximal binding value projected by the curve.
Footprint analysis
Genomic DNA of B. burgdorferi type strain B31 (5 pM) was first incubated with BosR (1 µM) or BSA (1 µM) in the same binding buffer described for EMSA for 30 min at room temperature, and then digested with DNase I (0.5 – 32 ng/µl) for 10 min at room temperature. MgCl2 was added to each reaction to a final concentration of 10 mM prior to DNase I digestion. After DNase I digestion, all reactions were incubated at 75 °C for 10 min to inactivate the enzyme, and then assayed by q PCR to determine the copy number of specific regions of the B. burgdorferi genome. The copy number of a coding region, flaB (+441 ~ +543), was determined in each sample and used for normalization of all other target regions. Fold of protection afforded by BosR for a specific target region was calculated by dividing its relative level in the BosR-treated sample with that in the BSA-treated sample. For example, the fold of protection offered by BosR for the ospA (−330 ~ +20) fragment was calculated by first dividing the copy number of this fragment against the copy number of the flaB (+441~ +543) fragment gene separately in the BSA- and the BosR-treated samples, and then dividing the normalized value of the BosR-treated sample against that of the BSA-treated sample.
Bioinformatic analysis
A search for putative BosR-binding sites was performed using the Regulatory Sequence Analysis Tools (Thomas-Chollier et al., 2011). Since BosR recognizes both the Per box and the Fur box, but allows the spacer length to vary from 0 to 2 bps, we defined the consensus BosR binding sequence as T(T/G)ATAAT-n{0,2}-ATTAT(A/C)A. The intergenic regions of B. burgdorferi type strain B31 genome, limited to within 250-bp upstream of the start codon of a gene, were searched for sequences with up to 2 mismatches.
Supplementary Material
Acknowledgements
We thank M.V. Norgard, X.F. Yang, and U. Pal for providing strains, X.F. Yang, J. Skare and L. Hu for providing reagents, D. Kudryashov, D.B. Heisler, K. Musier-Forsyth, R. Comandur, and P. Upadhyaya for help with the FA assay, and K. Hayes-Ozello for editing the manuscript. We also thank M.V. Norgard, Z. Ouyang, J.D. Radolf, and U. Pal for critical comments and helpful suggestions for our study. X.L. is the recipient of an Arthritis Investigator Award from the Arthritis Foundation. This study is supported by the NIH Grant AI103173 (X.L.) and start-up funds from the Public Health Preparedness for Infectious Diseases Program and the College of Veterinary Medicine of the Ohio State University.
Footnotes
The authors declare no competing financial interests.
References
- Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Invest. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, Cha SS. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 2009;37:3442–3451. doi: 10.1093/nar/gkp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagg A, Neilands JB. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli . Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- Barbour AG. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Tessier SL, Todd WJ. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti JM, Bono JL, Rosa PA, Schrumpf ME, Schwan TG, Policastro PF. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect Immun. 2008;76:5228–5237. doi: 10.1128/IAI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci U S A. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. Insights into the complex regulation of rpoS in Borrelia burgdorferi . Mol Microbiol. 2007;65:277–293. doi: 10.1111/j.1365-2958.2007.05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher BG, Bronstein PA, Myers CR, Stodghill PV, Bolton JJ, Markel EJ, Filiatrault MJ, Swingle B, Gaballa A, Helmann JD, Schneider DJ, Cartinhour SW. Characterization of the Fur regulon in Pseudomonas syringae pv. tomato DC3000. J Bacteriol. 2011;193:4598–4611. doi: 10.1128/JB.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci U S A. 2012;109:10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Gonzalez CA, Radolf JD. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J Bacteriol. 2005;187:7845–7852. doi: 10.1128/JB.187.22.7845-7852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BM, Whitmire JM, Merrell DS. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun. 2009;77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JA, Garon CF, Schwan TG. Effects of environmental pH on membrane proteins in Borrelia burgdorferi . Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Mekalanos JJ. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci U S A. 2011;108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian C, Vitale S, Leonard GA, Bahlawane C, Fauquant C, Leduc D, Muller C, de Reuse H, Michaud-Soret I, Terradot L. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol Microbiol. 2011;79:1260–1275. doi: 10.1111/j.1365-2958.2010.07517.x. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis . J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- He M, Oman T, Xu H, Blevins J, Norgard MV, Yang XF. Abrogation of ospAB constitutively activates the Rrp2-RpoN-RpoS pathway (sigmaN-sigmaS cascade) in Borrelia burgdorferi . Mol Microbiol. 2008;70:1453–1464. doi: 10.1111/j.1365-2958.2008.06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith Iii R, Trzeciakowski JP, Skare JT. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol. 2009;74:1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona LI, Tokarz R, Kuhlow CJ, Benach J, Benach JL. The Fur Homologue in Borrelia burgdorferi . J Bacteriol. 2004;186:6443–6456. doi: 10.1128/JB.186.19.6443-6456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Cam E, Frechon D, Barray M, Fourcade A, Delain E. Observation of binding and polymerization of Fur repressor onto operator-containing DNA with electron and atomic force microscopes. Proc Natl Acad Sci U S A. 1994;91:11816–11820. doi: 10.1073/pnas.91.25.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, Anderson JF, Radolf JD, Fikrig E. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis . J Biol Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi . Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- Marconi RT, Samuels DS, Landry RK, Garon CF. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis N, Samuels DS. Proteins binding to the promoter region of the operon encoding the major outer surface proteins OspA and OspB of Borrelia burgdorferi . Mol Biol Rep. 1995;21:159–164. doi: 10.1007/BF00997238. [DOI] [PubMed] [Google Scholar]
- Norris SJ, Carter CJ, Howell JK, Barbour AG. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Deka RK, Norgard MV. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 2011;7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. BosR (BB0647) governs virulence expression in Borrelia burgdorferi . Mol Microbiol. 2009;74:1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–230. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli . Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Piesman J, Gern L. Lyme borreliosis in Europe and North America. Parasitology. 2004;129(Suppl):S191–S220. doi: 10.1017/s0031182003004694. [DOI] [PubMed] [Google Scholar]
- Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- Pollack RJ, Telford SR, 3rd., Spielman A. Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol. 1993;31:1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS. Gene regulation in Borrelia burgdorferi . Annu Rev Microbiol. 2011;65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- Samuels DS, Radolf JD. Who is the BosR around here anyway? Mol Microbiol. 2009;74:1295–1299. doi: 10.1111/j.1365-2958.2009.06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci U S A. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MA, Taylor GL. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol. 2009;72:1208–1220. doi: 10.1111/j.1365-2958.2009.06718.x. [DOI] [PubMed] [Google Scholar]
- Shin JH, Jung HJ, An YJ, Cho YB, Cha SS, Roe JH. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci U S A. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet. 2012;379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- Steere AC. Lyme disease. N Engl J Med. 2001;345:115–125. doi: 10.1056/NEJM200107123450207. [DOI] [PubMed] [Google Scholar]
- Studholme DJ, Buck M. Novel roles of sigmaN in small genomes. Microbiology. 2000;146(Pt 1):4–5. doi: 10.1099/00221287-146-1-4. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–w91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traore DA, El Ghazouani A, Ilango S, Dupuy J, Jacquamet L, Ferrer JL, Caux-Thang C, Duarte V, Latour JM. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis . Mol Microbiol. 2006;61:1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Lutton A, Olesik J, Vali H, Li X. A novel iron- and copper-binding protein in the Lyme disease spirochaete. Mol Microbiol. 2012;86:1441–1451. doi: 10.1111/mmi.12068. [DOI] [PubMed] [Google Scholar]
- Xu H, Caimano MJ, Lin T, He M, Radolf JD, Norris SJ, Gherardini F, Wolfe AJ, Yang XF. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi . PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi . Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi . Proc Natl Acad Sci U S A. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.