Abstract
Affinity purification coupled with mass spectrometry (AP-MS) is now a widely used approach for the identification of protein-protein interactions. However, for any given protein of interest, determining which of the identified polypeptides represent bona fide interactors versus those that are background contaminants (e.g. proteins that interact with the solid-phase support, affinity reagent or epitope tag) is a challenging task. While the standard approach is to identify nonspecific interactions using one or more negative controls, most small-scale AP-MS studies do not capture a complete, accurate background protein set. Fortunately, negative controls are largely bait-independent. Hence, aggregating negative controls from multiple AP-MS studies can increase coverage and improve the characterization of background associated with a given experimental protocol. Here we present the Contaminant Repository for Affinity Purification (the CRAPome) and describe the use of this resource to score protein-protein interactions. The repository (currently available for Homo sapiens and Saccharomyces cerevisiae) and computational tools are freely available online at www.crapome.org.
Introduction
Affinity purification (AP) coupled with mass spectrometry (MS) has become a ubiquitous approach for the identification of protein-protein interactions1. In most cases, however, a large number of nonspecific interactors (here referred to as “background contaminants”, or “contaminants”) are co-purified with bait proteins and identified by MS. Methods to discern bona fide interacting partners from background contaminants are thus essential. In the case of affinity purification using epitope-tagged proteins, this is often aided by the inclusion of ‘negative control’ purifications, typically consisting of one or more “mock” purifications using the same support resin and cell line, but without expression of the polypeptide(s) of interest (referred to here as “bait” protein(s)). These controls (when not using isotope labeling2–5_ENREF_2) can be considered as “universal”, meaning that they are useful for filtering the background from any bait protein subjected to the same purification scheme3, 6–10.
A question that arises when designing and performing AP-MS experiments is how to use previous knowledge regarding background contaminants to best score interaction data. Small variations in the sample or sample preparation may influence the recovery of proteins, including contaminants. It is therefore not uncommon for a negative control experiment to fail to capture a complete set of contaminants, due to undetected variations at one or more experimental steps. This issue is compounded by the fact that low abundance peptides (and hence proteins) may not be reliably detected in a given MS analysis. Analyzing one or a few negative control samples will thus generally not allow for a comprehensive characterization of background contaminants for a given purification regime.
Here we present the Contaminant Repository for Affinity Purification, a web-accessible resource that stores and annotates negative controls generated by the proteomics research community, and enables their use for scoring AP-MS data. Users employ an intuitive graphical user interface to explore the database, by either querying one protein at a time, downloading background contaminant lists for selected experimental conditions, or uploading their own data (alongside their own negative controls when available) and performing data analysis. We also describe database structure and composition, provide examples of the use of this resource to filter contaminants with properly chosen controls, and demonstrate the utility of the scoring scheme for identifying bona fide interaction partners. The CRAPome accommodates a variety of purification schemes and, while it currently contains only H. sapiens and S. cerevisiae data, will be expanded to other species.
Results
Creation of the CRAPome repository
The CRAPome database is a web-accessible (www.crapome.org) repository of negative control AP-MS experiments (both published7, 9–27 and unpublished) associated with detailed protocols and controlled vocabularies (CVs) used to organize the data. Data contributors first submit raw MS files (Fig. 1a; database architecture in Supplementary Fig. 1) which are processed using a uniform data analysis pipeline followed by several quality control checks (see Methods), prior to association of metadata (CVs and text-based protocols; see Supplementary Note). These annotated negative control runs form the core of the repository. Currently (version 1.0, March 2013), 360 experiments contributed by 12 laboratories are available in the repository, of which the bulk of the data (343 experiments) were generated using human cell lines. This large dataset covers many of the most commonly used AP-MS protocols (see Supplementary Table 1 for CVs and the download section of the CRAPome for the current list of all experiments). For each experiment, mapping of the protein identifiers to NCBI Gene IDs is performed, and spectral count information is parsed to the relational database (see Methods). The database is expandable and new data are added to the CRAPome using the same deposition and annotation process. New protocols and CVs will adapt the database to new experimental workflows.
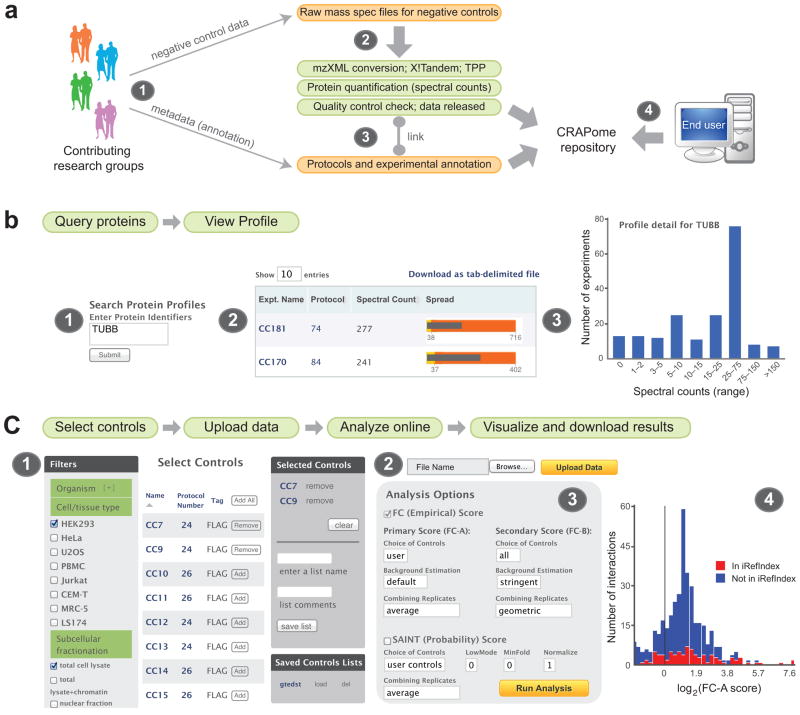
Figure 1.
The CRAPome at a glance. (a) Creation of the CRAPome. (1) Contributors to the CRAPome submit raw MS files for negative control runs, detailed experimental protocols and mapping information. (2) Raw MS files are first converted to mzXML and analyzed by X!Tandem and the Trans-Proteomic Pipeline; counts are extracted for protein quantification and the CRAPome administrator performs a quality control check (see Methods). (3) Released high quality runs (data) are associated with experimental descriptions and protocols (metadata) by the CRAPome administrator in consultation with the data provider. (4) Query of the CRAPome database by external users via the web interface. (b) Overview of the first CRAPome workflow. (1) Proteins are queried against the CRAPome by inputting one of several identifiers (Supplementary Note) which enable mapping to Gene ID. Different views enable exploration of the contaminant profile of each queried protein, either as a summary table (2) or in graphical formats (3). (c) Overview of the third CRAPome workflow (note that the second workflow is similar, except that no user data is uploaded; the second workflow generates lists of contaminant proteins). (1) Desired controls are selected, with the help of CVs. (2) Users upload their own data (test experiments and controls if available) to the CRAPome and (3) select parameters for data analysis. Data is displayed in a table format and in different graphical formats, which include the detection of a given interaction in the public repository iRefIndex (4).
Graphical User Interface
End users access the database via a web interface (Fig. 1b, 1c, Supplementary Note; www.crapome.org). After selecting the organism of interest (currently H. sapiens or S. cerevisiae), the database can be queried in three ways (called “user workflows”).
1) Query selected proteins
In the first workflow, users submit queries consisting of protein or gene identifiers and retrieve summaries of the occurrence of queried entries. An expanded view summarizes the conditions and protocols in which the protein has been identified, associated with quantitative information (Fig. 1b).
2) Create contaminant lists
The second user workflow generates background lists from a subset of the CRAPome controls. In this case, the user simply selects the list of desired controls (filtered using CVs and protocol details; Fig. 1c) and downloads the resulting tables of contaminants. Quantitative parameters, including the occurrence of identification across selected controls and the average spectral counts across selected controls in which the protein was detected, are included (a maximum of 30 experiments can be viewed online and included for analysis in workflow 3 below; the entire dataset can be downloaded as a tab delimited file from www.crapome.org/Download). Registered users can also save the selected list of controls for future use.
3) Analyze user data
The third workflow allows users to analyze their own data, using selected CRAPome controls and/or their own controls. The input data consists of one or multiple AP-MS experiments, ideally including biological replicates, along with user controls (optional, but recommended for better discrimination of true interactors from contaminants). Preparation of the data for upload to the CRAPome is described in Methods. In a first step, the user selects relevant controls from the CRAPome database (using the same interface as for workflow 2; Fig. 1c), or chooses previously saved selected lists of controls. The user then uploads their data in the specified format (or uses previously uploaded data). Upon selection of baits and controls, analysis is performed with the Significance Analysis of INTeractome (SAINT) score28–30 and/or a simpler Fold Change calculation (detailed below). These scoring tools create lists of interacting partners, ranked by confidence. Previously reported interactions documented in the interaction database aggregator iRefIndex (version 9.031) are also mapped onto user data. The results are presented in a tabular format and can be downloaded as a tab delimited file. Additionally, summary graphical views of the data are provided for each bait protein (Fig. 1c), or for all baits combined, enabling the user to view their data at a glance.
Characterization of the CRAPome
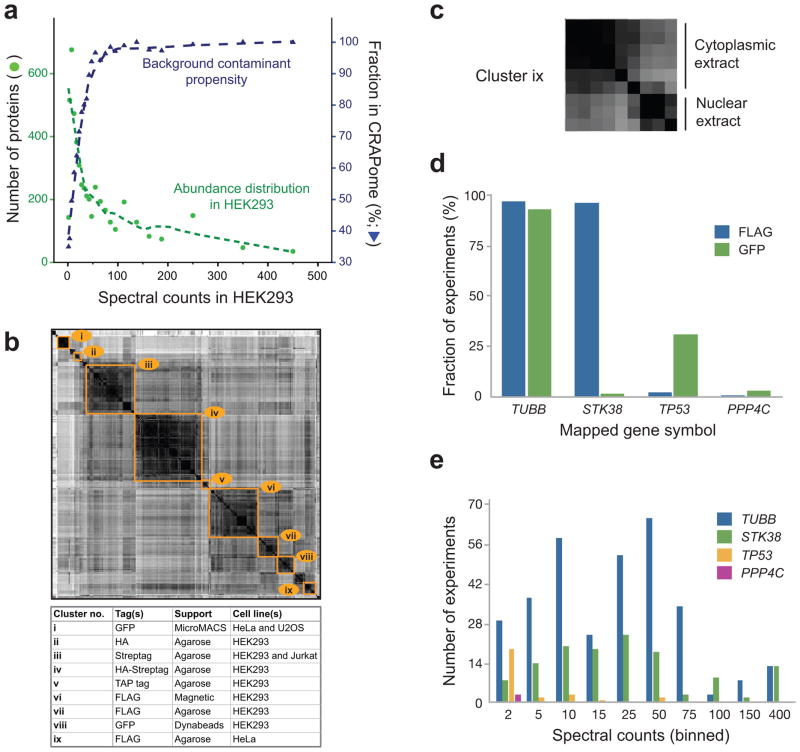
We mined the database to determine: (i) which proteins have a higher propensity to be contaminants, and (ii) how background proteins differ based on experimental conditions. First, to understand whether the abundance level of a protein in a sample increases the propensity of the protein to be a contaminant, we plotted the proteins reported in the CRAPome repository (restricting the analysis to HEK293 cells, by far the most common human cell line in the CRAPome) against a list of proteins ranked by their abundance estimates based on whole proteome analysis of HEK293 cell lysate32. There is a clear relationship between the abundance of a protein in HEK293 and its detection in at least one of the HEK293 experiments in the CRAPome (Fig. 2a). We next analyzed the frequency of detection of individual proteins in the CRAPome (mapped to gene names, as throughout this manuscript). Using stringent filtering (protein FDR < 1%), 4449 non-redundant protein groups (or 7782 gene names without compression of the data, see Methods for details) were identified (see Supplementary Tables 2–3 for most frequently detected proteins and the “Supplementary data” section of the CRAPome website for complete lists). Of these, 14 proteins were detected in >90% of all experiments, and 89 in >50% of the experiments, qualifying them as ubiquitous contaminants (Table 1). Not surprisingly, these include keratins, cytoskeletal proteins such as tubulins and actins, and high abundance proteins including translation elongation factors and histones (Table 2). Other proteins were not detected consistently across all purifications, but were abundant (in terms of total spectral counts) across the database: these were notably enriched for several functional categories, most predominantly associated with RNA functions (see Supplementary Tables 4–6 for most enriched GO BP, MF, and CC categories). However, a large fraction of the proteins present in the CRAPome were detected in only a small fraction of experiments: 3571, or 80% of the proteins in the CRAPome, were found in ≤10% of the experiments.
Figure 2.
Composition of the CRAPome (human data). (a) Relationship between the detection of a given protein in the CRAPome and its protein abundance (all entries are mapped to official gene identification numbers and displayed as corresponding gene symbols). The abundance distribution in HEK293 cells was calculated from shotgun mass spectrometry data (see Methods). The left axis indicates the number of proteins identified at each of the spectral count abundances (green circles; green dashed line shows fit to data); the right axis indicates the fraction of the proteins at a given binned abundance in the CRAPome database (blue triangles). (b) Similarity clusters of all experiments. All experiments in the CRAPome were scored for similarity in their contaminant profiles based on a cosine function: the size of the clusters represents the number of the experiments with strong similarity. Selected similarity clusters are indicated, alongside their composition. (c) Cluster ix, described in b as FLAG agarose in HeLa cells, can be further defined as two sub-clusters based on subcellular fractionation performed prior to the affinity purification (cytosolic and nuclear fractions); other clusters can also be further refined. (d) Example of epitope-tag specificity for selected proteins/genes. (e) Spectral count distribution of the proteins shown in d across the entire dataset. Spectral count bins are shown for all non-zero experiments. The highest spectral count boundary for each bin is shown.
Table 1.
General overview of the frequency of detection across the CRAPome (H. sapiens data). The two numbers are computed at different frequencies: (i) “Redundant” gene counts are based on a generous estimation of shared peptides: in this case, each protein/gene to which a given peptide is matched is counted as a contaminant (ii) “Reduced” gene counts are based on a more stringent definition of protein/gene parsimony, as described in Methods.
| Frequency in CRAPome | Redundant gene counts | Reduced gene counts |
|---|---|---|
| > 90% | 15 | 14 |
| > 75% | 37 | 30 |
| > 50% | 110 | 89 |
| > 20% | 504 | 463 |
| > 10% | 898 | 878 |
| ≤ 10% | 6884 | 3571 |
| TOTAL | 7782 | 4449 |
Table 2.
List of the most frequently detected protein families across the CRAPome, alongside some of the most frequently detected representative genes (H. sapiens data).
| Gene family | Example gene symbols |
|---|---|
| Heat shock proteins | HSPA1A, HSPA8, HSPA2 |
| Keratins | KRT1, KRT10, KRT2 |
| Tubulins | TUBA1B, TUBA3C, TUBB |
| Actins | ACTB, ACTA2, ACTBL2 |
| Elongation factors | EEF1A, EEF1A2 |
| Histones | HIST1H1C, H2AFX, HIST2H2B |
| Ribonucleo proteins | HNRNPK, HNRNPU, HNRNPH1 |
| Ribosomal proteins | RPS3, RPS18, RPL23 |
To further explore the contaminant propensity of the proteins in the CRAPome, we computed the similarity of all experiments (restricting the analysis to human data only), generating the heat map displayed in Fig. 2b (see Methods). The data clustered primarily according to experimental conditions (though there was a bias in the type of background detected across different laboratories). Several of the clusters could be further separated into subclusters, as exemplified by the “FLAG HeLa agarose” cluster, showing a clear separation based on subcellular fractionation (cytoplasmic or nuclear) performed prior to AP-MS (Fig. 2c). Based on our analysis of the most important determinants of background behavior, we annotated all experiments in the CRAPome using 14 categories of CVs (Supplementary Table 1), which can be used to select experiments that are most similar to those in a query set. More complete protocol descriptions of the experiments are provided via a free text form.
To illustrate the different contaminant propensities of individual proteins, and the need to take into account not only the overall frequency of detection in the dataset, but also the experimental conditions, we analyzed the frequency distribution of four proteins with two types of epitope tags, FLAG and GFP (Fig. 2d). TUBB (tubulin beta) was detected across nearly all of the experiments, irrespective of the epitope tag. By contrast, STK38 (a serine/threonine kinase) co-purified in nearly all FLAG experiments, but not in GFP experiments, while TP53 (the tumor suppressor protein p53) was detected predominantly in GFP-based AP protocols. The serine/threonine phosphatase PPP4C was not detected at a high frequency in experiments performed with either of these epitope tags (it was identified in 3/343 experiments across the entire database). Frequency and experimental conditions are also clearly not sufficient to describe contaminant propensity: abundance measures are also critical. For instance, if a protein is detected at a high frequency but low abundance (i.e. a low number of spectral counts in a high number of MS runs) in the CRAPome, but is detected with a high spectral count in bait purifications performed by a user, it is more likely to be a true interactor than if it is always detected with high abundance in the CRAPome. To illustrate this concept, we compared the non-zero values for the four proteins in Fig. 2d, but specifically examined spectral count distributions (binned values). This analysis revealed that while TUBB and STK38 were often present in very high counts in the CRAPome, TP53 was usually detected with much lower spectral counts (Fig. 2e). These comparisons are easily accessed via the CRAPome user interface (see Supplementary Note). They also provide the basis for statistical or empirical scoring of interactions, as described below.
Using the CRAPome to score interactions
The CRAPome can be used for the analysis of diverse AP-MS datasets, and most importantly for relatively small datasets, where eliminating background contaminants computationally is a difficult task. The CRAPome implements two complementary scoring strategies, both based on quantitative comparisons of prey abundance levels (estimated using spectral counts) in purifications with bait proteins against the distribution of prey abundances across a set of negative controls (see Methods). SAINT, described previously10, 28–30, allows advanced statistical modeling of the input bait-prey spectral count data and reports a posterior probability of true interaction. A simpler Fold Change (FC) calculation is based on computing the ratio of average normalized spectral counts in bait purifications versus negative controls. FC scoring is customizable and, in addition to the calculation of the standard FC score (referred to as primary score, or FC-A), computes a secondary, more stringent score (FC-B, see next section). Both FC and SAINT calculations are run in parallel using the facile CRAPome interface (allowing specification of key model parameters30), and comparison of their relative performances for each of the tested baits can be assessed by a Receiver Operating Characteristic (ROC) analysis provided via the CRAPome interface.
The use of the analytical pipeline within the CRAPome is illustrated here by a small dataset consisting of two biological replicates of each of the following four baits: RAF1, EIF4A2, WASL and MEPCE. In addition, six matching controls (user controls) were generated and processed together with the four baits to generate the input data (see Methods for detail). MEPCE and EIF4A2 have many documented interactors31 while WASL and RAF1 have fewer known interactors; all proteins provide challenges for background definition based on their association with polypeptides with contaminant-like behavior (chaperones, cytoskeletal proteins, RNA binding proteins, etc.; Table 2).
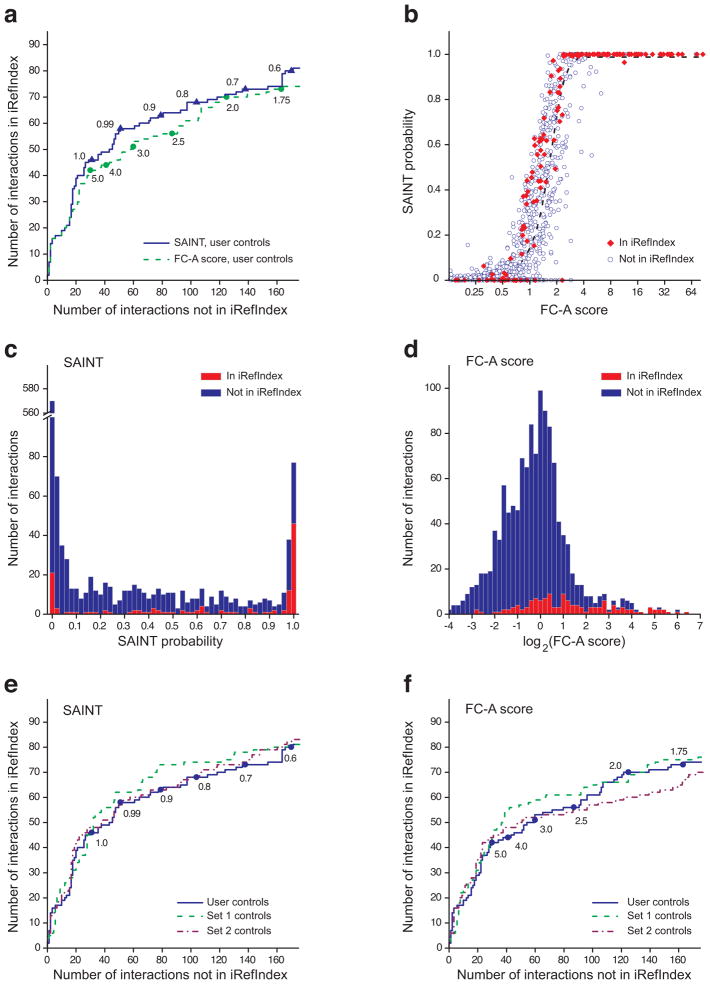
The results were first evaluated by plotting ROC curves based on the information extracted from iRefIndex31. The protein interaction list (all four baits combined) was sorted based either on the SAINT probability or the primary FC score computed using the six user controls (Fig. 3a). While SAINT did outperform the FC score on this dataset, both scoring schemes were able to efficiently recapitulate known interactions from the literature. Both scores also tracked very similarly for most of the proteins analyzed (Fig. 3b), with SAINT essentially providing a statistical conversion of the fold change onto the probability scale via the mixture model analysis of the underlying spectral count distributions. The performance of the interaction scores was further visualized by plotting the distribution of scores (histograms) separately based on iRefIndex annotation, showing that high scoring interactions (SAINT probability above 0.9, FC score above 4) are clearly enriched for previously reported interactions (Fig. 3c – d). The CRAPome interface provides (both separately for each analyzed bait and for all baits combined) a ROC and a histogram view (with mouse-over function), which enables the user to explore the reported interactions at different scores for SAINT or FC, and assists in establishing appropriate thresholds.
Figure 3.
Scoring functions in the CRAPome illustrated on a four bait dataset (MEPCE, EIF4A2, WASL, RAF1; 8 experiments). (a) Comparison between the primary Fold Change score (FC-A) and SAINT for scoring known interactions using negative control runs (n = 6) provided by the user; ROC based on the interactions in iRefIndex. Note that when SAINT scores are identical, ties are broken by the FC-A score. Selected SAINT probability or FC-A score thresholds are represented by triangles and circles, respectively. (b) The relationship between SAINT probability and FC score is well represented by a sigmoid function (dashed curve). (c – d) Histogram visualization of the data presented in (b) can help with data exploration and threshold selection. (e – f) Scoring protein interactions using controls from the CRAPome with SAINT (e) and FC-A (f): User controls (n = 6) are compared to two sets of controls from the CRAPome, selected based on the CVs (Set 1 = 10 controls; Set 2 = 11 controls).
We next tested whether the controls deposited in the CRAPome could be used for scoring interactions in the absence of user controls. While we recommend always using at least some user controls for scoring interactions, there are certainly cases where such controls do not appropriately model the background. Controls from the repository were thus selected on the basis of the CVs and protocols. We identified two relevant control sets from two different laboratories that fulfilled our criteria (HEK293 cells, FLAG tag, single step purification on M2 agarose) which contained 10 (Set 1; CRAPome protocol #56) and 11 (Set 2; CRAPome protocol #26) experiments, respectively. Using ROC analysis, we showed that each of these sets of controls performed very similarly to the user controls both in SAINT (Fig. 3e) and FC (Fig. 3f) calculations.
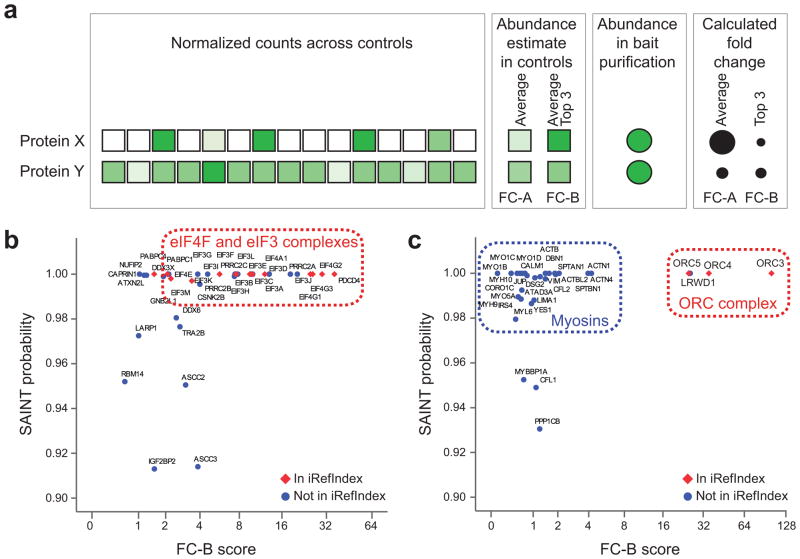
One issue affecting scoring of AP-MS data is the existence of contaminants (e.g. myosin and the proteins that co-purify with it) that are usually present in small amounts across most controls, but can spike to high abundance in some controls (or across batches of purifications), making detection of the true interactors much more difficult. Such contaminants are normally “diluted out” when multiple experiments are used for FC calculation, or even SAINT analysis (Fig. 4a). To assist in the identification of these “rare” contaminants, we implemented a secondary, more conservative FC score (FC-B) that is automatically calculated to supplement normal scoring using SAINT or the primary FC score (see Methods and Fig. 4a). We applied this more stringent scoring scheme to two biological replicates of the bait ORC2L, which, through visual inspection of the results, were found to contain large quantities of myosin contamination. While SAINT is capable of identifying true interactors in successful experiments as exemplified by EIF4A2 (Fig. 4b; see the relatively good agreement between SAINT score and FC-B score), it assigned a high probability to myosins and associated proteins in the ORC2L samples (Fig. 4c). By contrast, the conservative FC scores readily distinguished between these contaminants and true interaction partners (ORC3, ORC4 and ORC5 are in iRefIndex31, and LRWD1 is reported in PubMed33). Importantly, the CRAPome interface enables rapid visualization of the samples likely affected by this type of low frequency contaminants, by providing comparisons between FC-B and SAINT or FC-A.
Figure 4.
Use of a more stringent Fold Change score (FC-B) to recover true interacting partners for ORC2L. (a) Schematic illustration of the consequences of averaging all spectral counts as opposed to selecting the top three maximal values for scoring protein-protein interactions. Here, protein X represents a contaminant in the purification scheme that is detected with variable counts across the 15 selected controls (the intensity of shading is proportional to the spectral counts). By contrast, protein Y is a contaminant detected with similar counts across all selected controls. The standard primary Fold Change calculation (FC-A) averages the counts across all controls while the more stringent secondary Fold Change score (FC-B) takes the average of the top 3 highest spectral counts for the abundance estimate. The resulting FC-A and FC-B scores are represented schematically where a larger circle indicates a higher fold change, with FC-A and FC-B assigning a similar score to protein Y, but not to protein X. (b) Comparison of SAINT scoring and stringent FC-B with good bait samples. Note here that only the top of the map (the interactions with SAINT probability ≥ 0.9) are displayed. (c) Same as c for bait samples (ORC2L) contaminated with myosin: the more stringent fold change score FC-B helps in discriminating between true interaction partners (labeled “ORC complex”) and contaminants (labeled “myosins”).
CONCLUDING REMARKS
While lists of contaminating proteins have been reported in the past3, 34, 35, there was no central repository for this type of data, or freely available software tools for their utilization. The CRAPome facilitates access to a standardized (in terms of protein identification pipeline, ID mapping, abundance measures, etc.) set of negative control experiments, organized via CVs based on experimental considerations. The freely accessible user interface is intuitive and informative, even for those who may be new to mass spectrometry.
While we are currently using spectral counts as the sole quantification tool within the repository, extension of the system to other types of quantification (especially MS1, which is becoming possible as high mass resolution instruments are increasingly being used for AP-MS experiments) may help to further discriminate between background contaminants and true interactors. We expect a constant stream of data to be deposited in the CRAPome, which would partly fulfill the mandate from journals to make data publicly available. While we have restricted the release of the first version of the CRAPome to H. sapiens and S. cerevisiae data, the system is ready to accommodate data from other species, which will further increase the usefulness of the system. As contributors continue depositing their data in the repository, robustness in scoring will increase, and in-depth characterization of contaminant behavior will be possible. The CRAPome can be used as a retrospective tool to analyze AP-MS data, and will be instrumental to curators of protein-protein interaction databases. It should also assist with establishing guidelines regarding the scoring and annotation of such data. Widespread adoption of the CRAPome (by experimentalists, computational biologists, database curators, and reviewers alike) will improve the overall quality of AP-MS protein interaction data, addressing one of the key challenges in proteomics research.
Online Methods
Design and architecture of the CRAPome
The CRAPome interface was developed using Drupal, an open source PHP-based web framework, and MySQL and SQLite relational databases. The processing pipeline for adding data to the database, processing user input data, extracting data from the database, computing Fold Change scores, and preparing summary reports was developed using Python and a SQLite database. SAINT analysis29, 30 is computationally intensive and is executed on a set of dedicated compute nodes. SAINT jobs are managed using Torque, an open source computing resource management system. All SAINT analysis requests are queued and executed on a first come, first served basis. The entire infrastructure is currently hosted on FLUX, the university-wide shared high-performance computing service at the University of Michigan. In addition to professional data backup and system management, its allocation based system allows adding computing nodes to the system if additional computing nodes are needed for running SAINT or other computation-heavy steps that may be added in the future.
The actual data for each experiment (‘data’; Supplementary Fig. 1), such as the protein/gene accession numbers, the sequences of the identified peptides, peptide probabilities, and the spectral counts are stored in a SQLite database. The attributes used to annotate the experimental conditions (‘meta-data’) are stored in a separate MySQL database. The separation of data and meta-data is performed for the convenience of developing the web interface, which allows annotation of experiments (management of meta-data) directly by data contributors, while the processing and management of the data itself is performed by the database administrator.
In order to keep annotation of data consistent, the attributes and values that describe the experimental conditions are predefined. The corpus of these attributes (and their values) is referred to as the “controlled vocabularies”, or CVs (Supplementary Table 1). In addition to the CVs, each experiment deposited in the CRAPome repository is also annotated with a detailed description of the experimental protocol that enables users to obtain additional details about the experiments.
Processing of mass spectrometry data and population of the CRAPome database
Datasets were obtained from the contributing laboratories in the .raw or .mgf file format. The files were converted to the open mzXML file format, and further processed using the X! Tandem/Trans-Proteomic Pipeline (TPP) suite of tools36–38. MS/MS spectra were searched against RefSeq protein sequence database version 4739 (H. sapiens) or SGD ORF protein sequence database orf_trans.20100105.fasta (S. cerevisiae), appended with an equal number of decoy sequences, using X!Tandem40 with k-score plug-in. For the purposes of simplicity and uniformity, we developed two standard parameter templates for processing using X! Tandem and TPP, which were applied to data generated on low or high mass accuracy instruments, respectively. MS/MS spectra were searched using a precursor ion mass tolerance of 100 ppm (monoisotopic mass), or using −1 to +4 Da (average mass) window for high and low mass accuracy instruments, respectively. All other database search parameters were identical: cysteine carbamylation (C + 57.0215) and methionine oxidation (M + 15.9949) were specified as variable modifications. The search results were processed using PeptideProphet (high mass accuracy data was analyzed using high mass accuracy binning option), and then further processed using ProteinProphet to create protein summary files. For each experiment, all contributing data (multiple gel band fractions, technical replicates, etc.) were combined to generate a single set of PeptideProphet and ProteinProphet output files (pepXML and protXML files, respectively). One of the submitted datasets16 consisted of a very large number (300) negative controls in which proteins were separated using 1D SDS-PAGE. In a fraction of these experiments, only selected bands were analyzed using MS. To avoid the problem of data inconsistency due to missing MS data for a subset of gel fractions, and to reduce the total number of entries in the CRAPome representing this dataset, the individual experiments from this dataset were combined to generate 10 composite experiments (protocol #66; experiments CC185 – CC194).
To build the CRAPome database, spectral counts were extracted from protXML files using an in house-built software tool. For each protein in the protXML file, peptide to spectrum matches with a probability greater than or equal to 0.9 were extracted. The cumulative sum of the spectral assignments for these peptides constituted the spectral count for the corresponding protein. The spectral count was computed for each protein in the output file regardless of whether peptides mapping to a given protein could also map to other proteins. We note that this represents a deviation from the conventional approach of performing stringent false discovery rate (FDR) filtering and removing redundant or inconclusive, i.e. not supported by unique peptides, protein identifications41 (the results of such stringent filtering are described below, see Global analysis and reduced gene counts section). The liberal approach for creating protein summaries for each experiment taken here in fact enables a conservative approach for scoring protein interactions. As discussed in8, it ensures that the spectral counts of proteins from homologous families such as keratins, tubulins, and actins are not underestimated due to the ambiguities related to the identification of shared peptides. Finally, RefSeq protein accession numbers were mapped to official gene identification numbers using Ensembl Biomart tools and displayed as corresponding gene symbols (entries with NP accession numbers only; proteins with XP numbers and those with NP accession numbers that cannot be mapped to gene symbols are presently not visible in the database). When multiple proteins mapped to the same gene entry, the maximum spectral count among these proteins was selected as the spectral count for that gene. These data provided the basis of the CRAPome as accessible online, and were used to calculate ‘redundant gene counts’ shown in Table 1.
Quality control
As part of the process of creating the database, the CRAPome administrator performs a quality control check of the database search results. Experiments containing only a few identifications (less than 10 gene symbols with non-zero counts) are removed automatically, and experiments with less than 50 gene symbols are inspected in more detail. Furthermore, all negative control experiments generated using the same protocol (biological replicates) are inspected for consistency, and inconsistent samples are removed. Lastly, possible carry-over issues are identified and referred to the data depositors for further inspection. From the 402 experiments submitted to the CRAPome, 42 experiments were excluded based on these quality control steps.
Global analysis and reduced gene counts
To allow a more informative analysis of the contaminant profiles and comparison with other data, all pepXML and protXML files generated as described above were processed using a more conventional set of filtering thresholds. All pepXML files used to generate the CRAPome repository (human data subset, 343 files) were processed together using ProteinProphet to generate a single protein summary file (protXML file). This combined protXML file, as well as the pepXML and protXML files for each individual experiment, were then processed using ABACUS42 to generate a combined spectral count matrix using default parameters (accepting proteins with at least one peptide having PeptideProphet probability of 0.99 or greater, and protein probability as computed by ProteinProphet of 0.9 or greater). Each row in the filtered ABACUS file represented a protein group based on the combined protXML file, with a single accession number selected among indistinguishable protein entries forming that group. Spectral counts for the representative proteins were extracted from pepXML files for each individual experiment. The false discovery rate (FDR) for the combined protein list was less than 1% as estimated using decoy counts. The resulting spectral count matrix was used to compute similarity scores to generate the clustergram (see below), and to analyze the global properties of the data such as frequency of identification across the entire dataset (Table 1, ‘reduced gene count’).
Gene Ontology (GO) enrichment analysis was performed on the reduced list, and considering the top 25% most abundant proteins in each experiment only (1427 genes in total). The analysis was done using the online DAVID tool43, restricting the analysis to level 3 biological process (BP), molecular function (MF) or cellular component (CC).
To generate the clustergram (Fig. 2b), we first computed experiment-experiment similarity scores using cosine function from square root transformed spectral counts (data from protocol #6616, was excluded from this analysis; see above). For computing the final clustergram, we required that each experiment had at least 2 additional experiments with a similarity score of 0.7 or higher. The final clustergram was generated using Cluster 3.0 software44, with single linkage clustering using Pearson correlation (uncentered) as the similarity measure. The clustergram was visualized using TreeView software45.
Background contaminant propensity as a function of protein abundance in HEK293 cells
To generate the list of proteins and protein abundances in the HEK293 whole cell lysate, we used publicly available data taken from32. Raw mass spectrometry data for this cell line were downloaded from the Tranche data exchange system (https://proteomecommons.org) using the hash specified in the original manuscript. Data were processed as described above (Global analysis and reduced gene counts). For each identified protein (representative protein per group, see above) in the filtered ABACUS file the summed spectral count across the 4 biological replicates was taken as a measure of the protein abundance in the cell line. A global histogram of protein abundances was then generated by binning (Fig. 2a). The background contaminant propensity was then calculated as a fraction of HEK293 cell line identified proteins in each spectral count bin that were also detected in at least one HEK293 experiment in the CRAPome. For this comparison, we selected CRAPome experiments having the ‘Cell Line’ attribute value ‘HEK293’ only and queried protein accession numbers identified in the HEK293 whole cell lysate against the CRAPome HEK293 identified proteins. We then plotted the “fraction in CRAPome” as a function of protein abundance (binned spectral counts).
Data formats
When querying the database to view contaminant profiles for selected proteins of interest (workflow 1), proteins can be referenced using a variety of identifiers: RefSeq protein ID, Ensembl protein ID, NCBI Gene ID, Uniprot entry name, Uniprot entry ID, HGNC gene symbol (human) or SGD ID (S. cerevisiae). All input identifiers are internally mapped to official gene identification numbers using Ensembl Biomart46 tools and displayed as corresponding gene HGNC symbols (SGD ID for S. cerevisiae). Input data for uploading to the CRAPome for analysis in workflow 3 can be formatted using any of the accession schemes references above. The input file needs to be formatted as to contain four columns: Bait Name, AP Name, Prey Name, and Spectral Count. Each row in this file lists the spectral count (Spectral Count column) for each protein (referenced in Prey Name column) in purification with a particular bait protein (bait protein/gene identifier is referenced in the Bait Name column). When multiple biological replicates for the same bait are available, they are distinguished using different text strings in the AP Name column (e.g. ‘R1’, ‘R2’, etc.). The negative controls runs are specified as text string ‘CONTROL’ in the Bait Name column (and named differently in the AP Name column, e.g. ‘UC1’, ‘UC2’, etc.).
AP-MS test data
The analytical pipeline is illustrated using two biological replicates of each of the following four baits. RAF1 is a serine/threonine kinase that binds to Ras, several chaperones, and 14-3-3 proteins47, 48. EIF4A2 is a translation initiation factor that is part of the EIF4F complex, which bridges the mRNA cap structure to the ribosome via the EIF3 complex49. WASL (also known as N-WASP) belongs to the Wiskott-Aldrich syndrome (WAS) family of proteins, involved in transduction of signals from receptors on the cell surface to the actin cytoskeleton50. Finally, MEPCE, the 7SK snRNA methylphosphate capping enzyme, interacts with numerous transcriptional and RNA processing proteins51.
Cloning and expression of eIF4A2, RAF1 and MEPCE has been previously described15. WASL and ORC2L were amplified by PCR from Mammalian Gene Collection constructs BC052955 and BC014834 respectively, and cloned into pcDNA5-FRT-FLAG (using EcoRI/NotI for WASL, and AscI/NotI for ORC2L), and the junctions sequenced. Primers used were: WASL_5′EcoRI, GATCGAATTCATGAGCTCCGTCCAGCAGC; WASL_3′NotI, GATCGCGGCCGCTCAGTCTTCCCACTCATCATCATC; ORC2L_5′AscI, GATCGGCGCGCCAATGAGTAAACCAGAATTAAAGGAAGAC; ORC2L_3′NotI, GATCGCGGCCGCTCAAGCCTCCTCTTCTTCC. The resulting vectors were stably co-transfected with the Flp-recombinase expressing vector pOG44 into Flp-In T-REx 293 cells (Invitrogen). Selection of stable transformants (single clones), clonal expansion, induction of protein expression and AP-MS were performed essentially as described in15, using FLAG M2 agarose beads (Sigma). Two biological replicate analyses of each bait were performed, alongside six negative controls (cells expressing the tag alone). All samples were analyzed on an LTQ mass spectrometer coupled to an online C18 reversed phase column. The detailed protocol is #48 in the CRAPome. The mass spectrometry data was searched using the X! Tandem/TPP/ABACUS pipeline and settings as described in Global analysis and reduced gene counts. The filtered ABACUS file was formatted for CRAPome as described in Data formats using an in-house tool. Data were uploaded to the CRAPome (workflow 3). Two sets of additional controls (Set 1 and Set 2, see main text for detail) were selected and used alongside the user controls. SAINT and FC scores were generated using different settings (see main text and below). The ORC2L bait was processed in a similar way and uploaded for analysis to the CRAPome separately (it was not used for comparison between SAINT and FC scores shown in Fig. 3). The resulting input data matrices for eIF4A2, RAF1, MEPCE, and WASL baits and the six user controls, as well as for ORC2L and the same user controls, can be downloaded from the CRAPome website.
Interaction scoring: SAINT
SAINT was described in29. Here the data was analyzed using SAINT options LowMode=0, MinFold=0, Normalize=1. In general, SAINT performance varies depending on the choice of options, especially MinFold (requiring a certain minimum fold change as a part of probability calculation) and Normalize (normalization to the total spectral count in each experiment). SAINT run with the options specified above slightly outperform SAINT results with other options in these data (Supplementary Fig. 2). When the bait protein is analyzed in multiple biological replicates, SAINT probabilities computed independently for each bait replicate are averaged, and the average probability (AveP) is reported as the final SAINT score. For in-depth discussion of these options see30. The CRAPome also allows alternative specifications for combining biological replicates (e.g., geometric mean as a more conservative approach).
SAINT has been shown to perform well when using a sufficient number of matching negative controls (ideally at least 3–5 controls) showing a high degree of reproducibility. At the same time, SAINT can be sensitive to changes in the spectral count distributions of a given protein in either the controls or the bait samples, and thus its performance may be affected if the bait sample quality is poor or the negative controls are heterogeneous. SAINT is also computationally intensive.
Interaction scoring: Fold Change
The primary FC score (FC-A, or just FC) can be considered an alternative to SAINT scoring. It is computed for each bait - prey interaction pair (initially separately for each biological replicate of the bait). It is defined as the ratio of the normalized spectral count of protein i in purification with bait j, Ti,j, and the average normalized spectral count of that protein across the negative controls (user controls or selected CRAPome controls), Ci: FCi,j = (Ti,j + α)/(Ci + α). The normalized spectral counts are computed as Ti,j = SCi,j/Nj, where the normalization factor is the sum over all proteins identified in the experiment with bait j, Nj = ΣSCi,j. Similarly, the counts are normalized in each negative control experiment x=1…n, Ci,x = SCi,x/Nx, prior to computing the averaged normalized count across all n controls, Ci = 1/n ΣCi,x. A small background factor α is added to prevent division by zero, calculated as β/ave(Nx), where ave(Nx) is the average normalization factor across all n negative controls. The parameter β is by default set to 1. When the bait protein is analyzed in multiple biological replicates, the FC scores computed independently for each bait replicate are averaged to arrive at the final FC score.
The secondary, more conservative FC score (FC-B) can be used in addition to SAINT or the primary FC-A score for improved detection of several classes of challenging contaminants. It is computed as described above, expect that Ci is computed by averaging the highest 3 normalized spectral counts across all controls (by default, using the combined set of selected CRAPome controls and the user controls, when available). Furthermore, in the case of biological replicates for the bait protein, the final FC-B score is computed by default as the geometric mean of the FC scores for each replicate.
Comparison to literature data
In order to rapidly benchmark scoring performance and to provide users with a view of the new data within the context of previously published results, a mapping of the interactions to those deposited in the iRefIndex repository31 (currently version 9.0) is provided within the interface. iRefIndex was selected based on its comprehensiveness in the number of interactions annotated (it aggregates data from primary curation databases), and the relative ease of download and data mapping. Each entry from the database is mapped to a pair of genes (interacting proteins) using an in-house mapping tool. Entries identified as “complex” are excluded from this mapping. Due to uncertain quality of previously reported interactions involving ribosomal proteins, which are among the most common contaminating proteins in AP-MS experiments, we excluded all RPL and RPS proteins from the computation of ROC curves show in Fig 3.
Access to the database
The CRAPome can be accessed at www.crapome.org. No registration is required to access workflows 1 and 2. Registration is required for users to analyze their own data in workflow 3, and will enhance the functionality of workflow 2. Registration allows the users to save selected lists of controls (Fig. 1c), to use their previously uploaded data, and to access the results of previously performed SAINT and FC analyses.
Supplementary Material
Acknowledgments
We wish to thank G. I. Chen, M. Mullin, M. J. Kean, T. M. Greco, T. Srikumar and Y.-C. Tsai for contributing published data and S. Saha and J.-E. Dazard for constructive comments. We thank A. Stefanovic, M. Planyavsky, A. Stukalov, A. J. Guise, A. C. Müller, A. Pichlmair, B. Larsen, C. Knoll, C. L. Baumann, E. L. Rudashevskaya, F. Grebien, F. P. Breitwieser, H. G. Budayeva, J. W. Bigenzahn, M. Bruckner, M. Licciardello, M. L. Huber, M. Tucholska, N. Venturini, O. Rocks, O. Stein, P. Joshi, R. Giambruno, R. Sacco, S. Zhang, T. Stasyk and V. Nguyen for help with sample analysis.
This work was supported by grants from the National Institutes of Health (5R01GM94231 to ACG and AIN; DP1DA026192 and HL112618-01 to IMC), the Canadian Institutes of Health Research (MOP-84314 to ACG; MOP-82851 to BC), the government of Ontario via a Global Leadership Round in Genomics and Life Sciences (TP and ACG), the Austrian Academy of Sciences (KLB, JC and GSF), the Austrian Federal Ministry for Science and Research (Gen-Au projects, APP-III and BIN-III; KLB and GSF; No 820965; JC and KLB; No 820962), the European Research Council (GSF; ERC-2009-AdG-250179-i-FIVE), the Austrian Science Fund FWF (GSF, JC and KLB; P24321-B21; P22282-B11), the European Molecular Biology Organisation long-term fellowship (GSF, JC and KLB; ATLF463-2008), The Netherlands Proteomics Center (TYL, VAH, SM AJRH), the European Union 7th Framework Program (PRIME-XS project, grant number 262067, TYL, VAH, SM, RA and AJRH), the Stowers Institute for Medical Research, and the Human Frontier Science Program (RGY0079/2009-C to IMC). ACG is the Canada Research Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics; BR is the Canada Research Chair in Proteomics and Molecular Medicine. RME acknowledges salary support from the Cleveland Foundation, and NIH 1R21 CA16006001A1. JPL was supported by a Canadian Institute for Health Research (CIHR) postdoctoral award. NSD was supported by a TD Bank postdoctoral fellowship, and RDB was supported by a CIHR postdoctoral award.
Abbreviations
- CRAPome
Contaminant Repository for Affinity Purification
- AP
Affinity purification
- AP-MS
Affinity purification followed by mass spectrometry
- FC
Fold Change
- FDR
False Discovery Rate
- SAINT
Significance Analysis of INTeractome
- RefSeq
Reference Sequence from the National Center for Biotechnology Information
- ROC
Receiver Operating Characteristic
- TPP
Trans-Proteomic Pipeline
- GO BP
Gene Ontology Biological Process
- GO MF
Gene Ontology Molecular Function
- GO CC
Gene Ontology Cellular Component
- CV
Controlled vocabulary
Footnotes
CONTRIBUTIONS
D.M., Z.W., A.I.N. and A.-C.G. designed the CRAPome structure and interface; D.M. and Z.W. implemented the system; D.M. and A.I.N. created the scoring scheme with help from H.C. and A.-C.G.; R.M.E., A.-C.G. and A.I.N. initiated the project; D. Fer. helped with processing the data; B.R., A.L.C., N.S.D. and J.P.L. tested the interface and contributed to editing the user manuals; A.L.C., N.S.D., J.P.L., W.H.D., T.L., Y.V.M., S.H., M.E.S., T.Y.L., V.A.H., R.B., N.C.H., A.A.H., A.B., D. Fau., R.M.E., I.M.C., K.L.B. and A.-C.G. provided mass spectrometry data to the CRAPome and/or annotated data in the repository; Z.-Y.L., B.G.B. and M. Gou. contributed the test benchmark dataset; T.P., D.D., B.C., R.A., G.S.F., J.C., A.J.R.H., M. Gst., S.M., I.M.C., K.L.B., M.P.W. and A.-C.G. supervised trainees and were responsible for data generation across the different research groups; H.C., B.R., I.M.C., K.L.B., M.P.W. and R.M.E. provided critical comments throughout the project; A.-C.G. and A.I.N. co-directed the project, analyzed and annotated data; A.I.N., A.-C.G. and D.M. wrote the manuscript and the user manuals with help from B.R.
References
- 1.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 2.Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 3.Trinkle-Mulcahy L, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trinkle-Mulcahy L. Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry. Proteomics. 2012;12:1623–1638. doi: 10.1002/pmic.201100438. [DOI] [PubMed] [Google Scholar]
- 5.Tackett AJ, et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 6.Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 7.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nesvizhskii AI. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics. 2012;12:1639–1655. doi: 10.1002/pmic.201100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skarra DV, et al. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011;11:1508–1516. doi: 10.1002/pmic.201000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Hakim AK, Bashkurov M, Gingras AC, Durocher D, Pelletier L. Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Mol Cell Proteomics. 2012;11:M111 014233. doi: 10.1074/mcp.M111.014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen GI, et al. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem. 2008;283:29273–29284. doi: 10.1074/jbc.M803443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Daniels DL, et al. Examining the complexity of human RNA polymerase complexes using HaloTag technology coupled to label free quantitative proteomics. J Proteome Res. 2012;11:564–575. doi: 10.1021/pr200459c. [DOI] [PubMed] [Google Scholar]
- 15.Dunham WH, et al. A cost-benefit analysis of multidimensional fractionation of affinity purification-mass spectrometry samples. Proteomics. 2011;11:2603–2612. doi: 10.1002/pmic.201000571. [DOI] [PubMed] [Google Scholar]
- 16.Ewing RM, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forget D, et al. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics. 2010;9:2827–2839. doi: 10.1074/mcp.M110.003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goudreault M, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kean MJ, et al. Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. J Biol Chem. 2011;286:25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruiswijk F, et al. Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci Signal. 2012;5:ra40. doi: 10.1126/scisignal.2002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato S, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 22.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 23.Greco TM, Yu F, Guise AJ, Cristea IM. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10:M110 004317. doi: 10.1074/mcp.M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11:M111 015156. doi: 10.1074/mcp.M111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudashevskaya EL, et al. A method to resolve the composition of heterogeneous affinity-purified protein complexes assembled around a common protein by chemical cross-linking, gel electrophoresis and mass spectrometry. Nat Protoc. 2013;8:75–97. doi: 10.1038/nprot.2012.133. [DOI] [PubMed] [Google Scholar]
- 26.Pichlmair A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 27.Varjosalo M, et al. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods. 2013;10:307–314. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 28.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi H, et al. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr Protoc Bioinformatics. 2012;Chapter 8(Unit 8):15. doi: 10.1002/0471250953.bi0815s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakur SS, et al. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol Cell Proteomics. 2011;10:M110 003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Z, et al. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell. 2010;40:99–111. doi: 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen GI, Gingras AC. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 35.Gingras AC, et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 37.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 38.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 41.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11:1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 45.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 46.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database (Oxford) 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzivion G, Luo Z, Avruch J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 48.Wartmann M, Davis RJ. The native structure of the activated Raf protein kinase is a membrane-bound multi-subunit complex. J Biol Chem. 1994;269:6695–6701. [PubMed] [Google Scholar]
- 49.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 50.Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. Embo J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 51.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.