Preface
Cellular signal transduction occurs in complex and redundant interaction networks that are best examined at the level of single cells by simultaneously monitoring the activation dynamics of multiple components. Recent advances in biosensor technology have made it possible to visualize and quantify the activation of multiple network nodes in the same living cell. The precision and scope of this approach has been greatly extended by novel computational approaches to determine the relationships between different networks, studied in separate cells.
Introduction
Methods such as phospho-proteomics1, protein–protein interaction studies2 and gene expression profiling3 allow analysis of signaling networks via the simultaneous observation of every pathway component. However, these methods average information over many cells and do not take into account cell-to-cell heterogeneity, which has been shown to play key roles in the function of signaling networks4–6. Flow cytometry allows the measurement of signaling at the single cell level7 but, similar to proteomic methods, temporal information is available only as a many-cell average, and spatial information is either completely absent or at very coarse levels, such as membrane and organelle compartmentalization.
A tool chest of fluorescent proteins emitting from the ultra violet (UV) to the near infrared have made it possible to simultaneously visualize the subcellular dynamics of multiple proteins in the same living cell — a technique referred to as experimental multiplexing. Such ‘multiplexed imaging’, which can reveal the coordination of two or more subcellular structures, is now routinely exploited to study, for example, the interactions of cytoskeletal fibers8, 9, the transient coupling of adhesion molecules and cytoskeleton flows10, and the choreography of protein recruitment to endocytic sites at the plasma membrane11. In contrast, the subcellular coordination of the signaling events that regulate these dynamics has remained considerably more obscure. In some cases, a single signaling activity can be visualized together with the dynamics of subcellular structures, but this has until recently been restricted largely to second messengers, for example, fluorescent chelators that report ion concentrations12, 13, or fluorescently tagged domains that bind accumulations of lipid second messengers14, 15 or mark cellular structures16. The study of the coordinated activation of multiple signaling proteins has been hindered by formidable technical hurdles. These obstacles have been recently overcome by two very different but complementary approaches — the development of powerful new biosensors and the development of computational multiplexing.
To understand the spatiotemporal dynamics of signaling events, it is generally not sufficient to simply follow the changing localization of proteins. Signaling proteins interact with downstream targets only in specific ‘activated’ states. Therefore, to dissect the mechanism of signal transduction it is necessary to monitor protein localization and activation. Biosensors have been devised to report protein activation states, usually relying on Förster resonance energy transfer (FRET) readouts of a conformational change or protein interaction17, 18. Due to the availability of fluorescent proteins spanning much of the visible spectrum, FRET-based biosensors can now be built using fluorescent proteins with orthogonal wavelengths, enabling the imaging of two biosensors in the same cell. Furthermore, a deeper understanding of fluorescent proteins has permitted the design of biosensors that respond directly to endogenous signaling molecules, without the need to use two fluorescent proteins. These experimental multiplexing approaches may ultimately enable us to visualize the activities of three and four signaling molecules at the same time.
However, even with these improved designs, only a limited number of biosensors can be introduced into one cell. In addition, multi-spectral imaging and introduction of multiple exogenous proteins is often accompanied by increased phototoxicity and the perturbation of cell physiology. Ideally one could study the dynamics of each protein species separately in different cells and subsequently relate them to each other via correlation with a common fiduciary event that occurs in each cell. Indeed, this approach has been applied, to study the timing of signaling events during phagocytosis and wound closure19–21. However, many cellular functions display stochastic behaviors, resulting in a wide range of heterogeneous, less stereotypical signaling patterns. In this case, establishing the relationships between signaling activities that are not all observed in the same cell is quite challenging. Recent work by several labs has begun to break through this barrier. It has been shown that spatiotemporal relationships between any two co-observed cellular activities can be extracted from their constitutive, basal fluctuations22–24. It has also been shown that, when measuring the relationships between two pairs of activities in different cells, with one activity in common between the pairs, it is possible to predict the spatiotemporal relationships among all of the individual activities, even when they are observed in different cells22. This process has been referred to as computational multiplexing in order to distinguish it from experimental multiplexing. Although computational multiplexing is still in its very early days, it may ultimately become the technology of choice for reconstructing complex pathways with many component activities. Importantly, the concept of computational multiplexing relies on concurrent measurements of activities in pairs, triplets, or even quadruplets. The more relations that can be extracted through direct observation in single experiments, the more robust will be the computationally inferred relations between signaling components. Therefore, advances in experimental multiplexing will lay the groundwork for advances in computational multiplexing. The goal of this Innovation is to outline the current state of both experimental and computational multiplexing, and to project their joint potential for the analysis of signal transduction at the single cell level.
Experimental multiplexing
The great majority of experiments studying multiple signaling nodes in the same living cell have been accomplished by combining genetically encoded FRET biosensors, the most tractable and commonly used type of biosensor. These have been extended to multiplexing primarily through development of new FRET pairs with orthogonal wavelengths, and advances in instrumentation and techniques to enhance sensitivity. Both are important, as the primary limitation to experimental multiplexing appears to be the ability of living cells to tolerate multiple exogenous reporter molecules. A host of other developing approaches show promise, some requiring the use of only one fluorophore to monitor each activity, or with significantly reduced cell perturbation.
Multiplexing with genetically encoded FRET
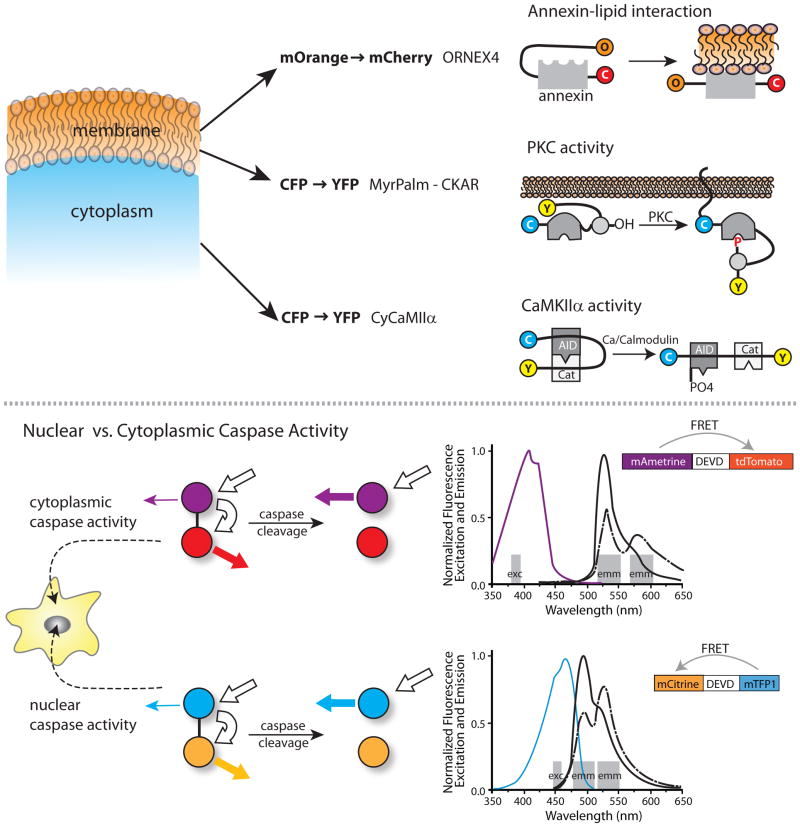
Although several different FRET pairs have been used to construct biosensors (for example, BFP/CFP, EGFP/DsRed and CFP/YFP), the CFP/YFP pair has proven most useful in multiplexing, due to its superior fluorescence properties and excitation/emission wavelengths that are compatible with other fluorophores. The development of orange and red proteins spectrally distinct from CFP and YFP25, and the subsequent improvement in their photostability and brightness26, 27, have provided compatible FRET pairs for multiplexing (See Figure 1 and Table 1). In an impressive example of multiparameter imaging, Piljic et al. studied two, three, and four activities simultaneously28. These authors used two CFP/YFP FRET probes (a Ca2+/Calmodulin-dependent kinase IIα sensor and a membrane-bound PKC sensor) together with an orthogonal mOrange/mCherry FRET pair in a sensor for the assembly of Annexin4 (a calcium-dependent phospholipid binding protein). This enabled ratiometric, spatially resolved FRET imaging. By restricting themselves to monitoring only the FRET intensity changes of the Annexin4 biosensor, the kinetics of annexin assembly into a trimer, which is facilitated by calcium influx, could be followed together with Fura Red to sense Ca2+ level changes, and these two activities could then be correlated with the activities of CaMKII and PKC signaling in live cells to determine the time sequence of Ca2+ influx, cellular signaling, and the resultant Annexin4 assembly.
Figure 1. Examples of Experimental Multiplexing.
Top) Pilijik and Schultz discriminate signals from three biosensors by using both orthogonal FRET pairs and differences in subcellular localization. They examine the relative kinetics of lipid modification, PCK activity and Cam Kinase IIa activity by using a biosensor with mOrange to mCherry FRET, combined with two biosensors using CFP to YFP FRET. One CFP to YFP biosensor is restricted to the cytosol and the other to the plasma membrane. Bottom) Ai et al. developed a novel fluorescent protein, mAmetrine, with an unusually large Stokes shift. This facilitated separation of wavelengths to image two biosensors independently in the same cell. The Stokes shift enabled fluorophore excitation and monitoring of emission at the orthogonal wavelength bands shown by the grey bars. For each FRET pair a single excitation band is used, leading to emission due to either direct excition of the donor (mAmetrine or mTFP) or FRET emission from the acceptor (mTomato or mTFP). Excitation and emission spectra are shown for the FRET pair in each biosensor, with the color of the excitation spectra corresponding to the donor fluorophores in the biosensors. Bold white arrows show the excitation and FRET of the donor fluorophore, while the colored arrows show the level of emission from the acceptors before and after cleavage. These orthogonal FRET biosensors were used to differentiate caspase activities monitored simultaneously in the nucleus and cytoplasm.
Rather than use FRET pairs compatible with CFP/YFP, Ai et al.29 generated new fluorophores and combined mTeal/YFP with mAmetrine/tdTomato to image both nucleus-targeted and nucleus-excluded caspase-3 activity within the same cell. mAmetrine is a unique fluorophore with a large Stokes shift and a violet-shifted excitation spectrum; this allows its effective separation from mTeal. Similarly, T-Sapphire/DsRed has also been used in combination with a CFP/YFP FRET pair, to detect cGMP concentrations30. But, since T-Sapphire has an excitation spectrum similar to CFP and a large Stokes shift that allows it to FRET with DsRed, this fluorophore permits simultaneous excitation of two spectrally separable FRET pairs. Recently, “ultramarine” fluorophores with excitation/emission in the violet have emerged31, 32. These require cell irradiation with more ‘toxic’ UV light, but may enable simultaneous imaging of three FRET biosensors when paired with another violet-shifted fluorophore such as BFP.
Imaging fluorescence lifetime rather than fluorescence intensity provides important advantages for FRET multiplexing. FRET quenches donor fluorescence, thus affecting donor fluorescence lifetime. Unlike intensity measurements, the donor lifetime is insensitive to sources of artifacts such as uneven illumination, subcellular variation in probe concentration, or cell geometry (reviewed in45–47 and elsewhere). Fluorescence lifetime imaging (FLIM) expands the number of usable FRET pairs for multiplexing because only effects on donor fluorescence are measured. Thus, the same acceptor can be used for two different donors48, and the choice of available proteins can be extended to include non-fluorescent acceptors, such as dkYFP49. With current technology, FLIM does incur either reduced temporal or spatial resolution relative to widefield imaging, and not all donor fluorophores are useful for FLIM imaging. However, donor fluorophores and fluorophore combinations with potential for FLIM multiplexing have been described50–54: mRFP/GFP, mStrawberry/GFP, mRFP/Venus, mStrawberry/Venus, and mDarkVenus/mGFP were all found to be comparable for FLIM imaging of CaMKII using variants of the Camui sensor, a FRET-based sensor of CaMKII activity.
On the horizon
FRET can also be accomplished using organic dyes, quantum dots, and other inorganic fluorophores. Compared to fluorescent proteins these fluorescent markers provide superior brightness and photostability33, 34, but they are not genetically encoded. Brightness and photostability are important for multiplexing, as they determine how much biosensor must be used in a cell to generate a robust signal. This is especially important when multiple biosensors are loaded in the same cell, as more biosensors increase cellular perturbation. In a recent study, a FRET biosensor for RhoA was combined with a Cdc42 biosensor based on an environment-sensing dye. The dye was attached to a protein domain that bound selectively to activate Cdc4222. Upon binding activated Cdc42 the dye on the biosensor underwent a shift in fluorescence, revealing the localization of Cdc42 activity. This design substantially reduced the concentration of biosensor needed, as it enabled the sensing of endogenous Cdc42, and used direct excitation of a bright dye. However, despite their greater sensitivity and ability to operate at lower intracellular concentrations, non-genetic biosensors are rarely used because they have to be loaded into cells via cumbersome methods such as microinjection or electroporation. Recent approaches to attach dyes to proteins in vivo show promise for harnessing the advantages of non-genetic fluorophores 35.
Signaling behaviors have also been reported by using only a single fluorescent protein, rather than by FRET. This facilitates experimental multiplexing as each biosensor uses a smaller portion of the wavelength spectrum. These approaches include fluorescent proteins that respond to ions, pH, voltage, or reactive oxygen species18. In bimolecular fluorescence complementation (BiFc), a fluorescent protein is split into two nonfluorescent halves, each linked to a different signaling protein. When the two signaling proteins bind, they bring the two halves of the fluorescent protein together, generating fluorescence. In some applications, one half of the fluorescent protein can combine with two different second halves36–40, each generating a unique color, including a far red version of the split protein, mLumin41. This technique is very sensitive because the signal is created over a ‘dark’ background. However, the interaction of the two halves is currently irreversible and the fluorescence forms with relatively slow kinetics. FRET between identical fluorophores, observed through effects on fluorescence polarization anisotropy, has been used to study receptor oligomerization, or protein aggregation, without the need for two different fluorophores42–44.
Important strides in experimental multiplexing will have to come from improved sensitivity in biosensor imaging, as toxicity and cell perturbation are important issues when studying low abundance signaling proteins. Several papers have addressed how the relative concentrations of biosensors and endogenous proteins or molecules affect cell behavior55–61; some have proposed that sensors can be applied to specific, well selected proteins to reveal the behavior of larger networks, ideally proteins of high concentration less likely to be perturbed by introducing biosensors62–64.
Computational multiplexing
Progress in biosensor design and the spectral decomposition of images, and improvements in filters and instrumentation, will steadily increase the number of simultaneously observable molecular activities. However, deriving a complete analysis of pathway states by studying all of the components in the same cell is unlikely in the foreseeable future. Thus, methods are needed that complement the direct imaging of multiplexed biosensors with the ability to integrate data from independent experiments into a comprehensive pathway model. These methods are collectively referred to as ‘computational multiplexing (Figure 2). Several challenges must be overcome when implementing computational multiplexing approaches. First, the integration of data from multiple experiments implies that the properties of the studied pathway, that is, the hierarchy of pathway components and the kinetics of information transfer between them, do not change between experiments. Practically, this means that the cell state and the environmental conditions are held identical between experiments, for example, by using clonal cell populations. Second, even then, the activation dynamics of the pathway will vary from cell to cell. Therefore, methods for data integration are required that identify from variable cell responses a canonical, cell-invariant representation of the pathway. Third, the spatiotemporal coupling of biosensor activities, whether observed in the same or separate experiments, usually results from nonlinear pathways, the feedback and feedforward relations of which are unknown a priori. Therefore, data integration must be combined with mathematical approaches for network inference. In this section we discuss strategies towards fulfilling these requirements, with reference to studies where the computational multiplexing of biosensors has provided insights into signaling pathways with sub-cellular resolution.
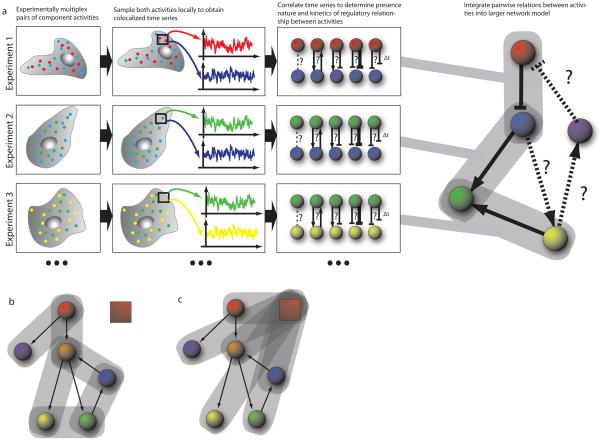
Figure 2. Workflow of Computational Multiplexing.
a) In each experiment, a sub-set of the signaling network components (‘nodes’) are experimentally multiplexed (two in the illustrated example). Component activities are sampled locally to extract co-localized time-series for each experimentally multiplexed node, in each region of the cell. The sampling windows are defined in a cell-centered frame of reference (see Fig. 4) in order to generate time series that are independent of the cell-cell to variation in shape. These time series are then analyzed by correlation methods, testing first the presence of an interaction between the activities and then determining the direction and sign (stimulating or inhibiting) as well as the timing of the interaction. This process is repeated for other activity pairs in the network, with each subsequent experiment sharing a common node with any of the previous experiments – that is in this illustration experiments 1 and 2 share observations from the blue node, while experiments 2 and 3 share observations from the green node. Continuing this process allows traversal of the network, without requiring each node to be observed simultaneously. Mathematical tools allow integration of the pairwise and possibly redundant observations into a larger network model. b, c) Two strategies for traversing the network. Experiment establishes pairwise relations bewteen activities throughout the network, b); or each experiment established the relation of one activities relative to a common fiduciary activity, depicted by red box, c).
A need for conserved experimental conditions
The conservation of the fundamental pathway properties is an implicit assumption made by every investigator who repeats an experiment to average the responses of multiple cells. This is no different with computational multiplexing. However, when combining data from experiments probing different aspects of a pathway with different biosensors, this assumption must be thoroughly validated. The expression or microinjection of a biosensor can shift the pathway properties. To control for such effects, the reproducibility of a cell function related to the studied pathway must be tested for each biosensor that is expressed, over a range of concentrations. For example, for pathways that regulate cell morphogenesis, cell morphology or morphodynamics are excellent measures against which to identify probe-induced biases. These measurements can be accomplished either by manual inspection or, ideally, by quantitative morphological65 or morphodynamic66 analyses.
A common fiduciary mark between experiments
To determine the spatiotemporal relations between activities monitored in independent experiments, spatial and temporal fiduciaries are required that are common between experiments and invariant to changes in cell morphology and cell behavior. Temporal fiduciaries can be defined by the acute stimulation or inhibition of a pathway67. By applying the same intervention protocol across multiple experiments, different biosensor activities can be aligned in time to deduce a response hierarchy. Similarly, spatiotemporal fiduciaries can be defined by spatially limiting the acute stimulation or inhibition of a pathway. Numerous approaches have been proposed to achieve this while monitoring downstream signaling responses, including mechano-stimulation of cells by micro-spheres68, 69 or photo-switchable reagents70, 71.
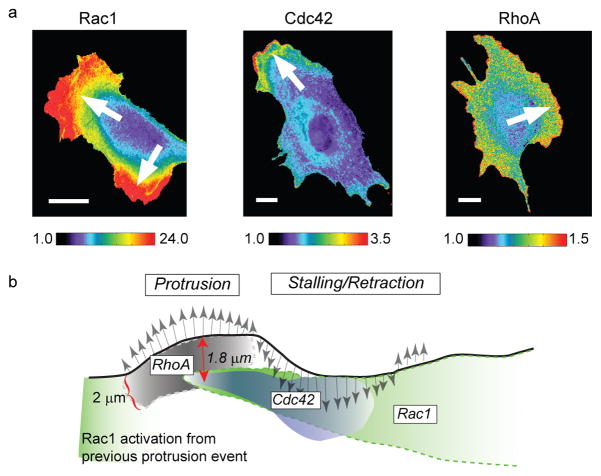
Recently, two studies independently established the concept of inferring a response hierarchy from constitutive fluctuations in the activity of biosensors. Time shifts between signaling events were determined by temporal and spatial cross-correlation between the activation of two biosensors, and/or between the activation of one biosensor and spontaneous fluctuations in cell morphology22, 24 (Fig. 3). There are several key advantages to this strategy as compared to the acute stimulation or inhibition of a pathway. First, acute interventions may over stimulate or over perturb pathways. As a result, the kinetics and response hierarchies deduced from these experiments may reflect a pathway that operates far outside the range of normal physiology. As well as avoiding this problem, the second advantage of analyzing constitutive fluctuations is that this permits a dense mapping of spatial gradients in signaling hierarchies. For example, the study by Machacek et al.22, of the coordination of Rho GTPases during cell protrusion, suggested that not only is the activation of Rac1 delayed by ~40 s relative to the activation of RhoA, but it is also shifted in space by ~2 microns. While RhoA is activated at the cell edge, Rac1 is activated behind the cell edge, possibly in growing nascent adhesions. It would be very tedious to derive such predictions of the spatiotemporal organization of pathways from a few scarcely distributed interventions. Third, in concept fluctuation analysis is readily scalable to pathways involving a very large number of components. To make use of this potential, pairwise cross-correlation analysis must be replaced by more sophisticated mathematical tools that integrate, from multiple experiments, the image fluctuation sequences of partially overlapping sets of component activities.
Figure 3. Assessing the activity of Rho GTPases using computational multiplexing.
a) Three distinct members of the family of Rho GTPases (Rac1, Cdc42 and RhoA) were imaged in different cells using different biosensors. To monitor Rac1 activation, a bimolecular FRET reporter was used, which produces a high signal when a Rac-binding fragment of the Rac1-effector Pak conjugated to cyPet associates the activated form of Rac1 conjugated to YPet. To monitor Cdc42 activation, an environmentally-sensitive dye, mero87, was coupled to a Cdc42-binding fragment of the Cdc42-effector WASP. Upon association of this labeled fragment with the activated form of Cdc42 the dye undergoes a change in its fluorescence spectrum that is observed as a strong increase in fluorescence at a particular wavelength. To monitor RhoA activation, an intramolecular FRET reporter was used. In contrast to the Rac1 reporter, here RhoA and the RhoA-binding fragment are in a single chain, which undergoes conformational changes upon activation of RhoA. These changes are detected by changes in the FRET intensity between cyPet-/YPet-fluorescent proteins inserted into the single chain sensor. Pseudo-color scales indicate the dynamic range of the biosensor responses (1, no significant response; maximum value, strongest response throughout the time-lapse sequence. Arrows indicate the direction of cell protrusion. b) Cartoon illustrating the spatially and temporally differentiated activation of the three Rho GTPases during protrusion (gray arrows pointing north) and retraction/stalling (gray arrows pointing south) events at the cell edge. The shading in the activation clouds indicates how the signaling strength decreases after initial induction. The relations between the signaling activities were predicted first indirectly by spatiotemporal cross-correlation analysis of each biosensor in a separate cell to the velocity of edge movement as a common fiduciary (strategy Fig. 2c). The inferred relations were then confirmed by direct observation of two of the biosensors (Cdc42 and RhoA) in the same cell (strategy Fig. 2b). Images reproduced from22 with permission from Nature.
Appropriate spatiotemporal sampling
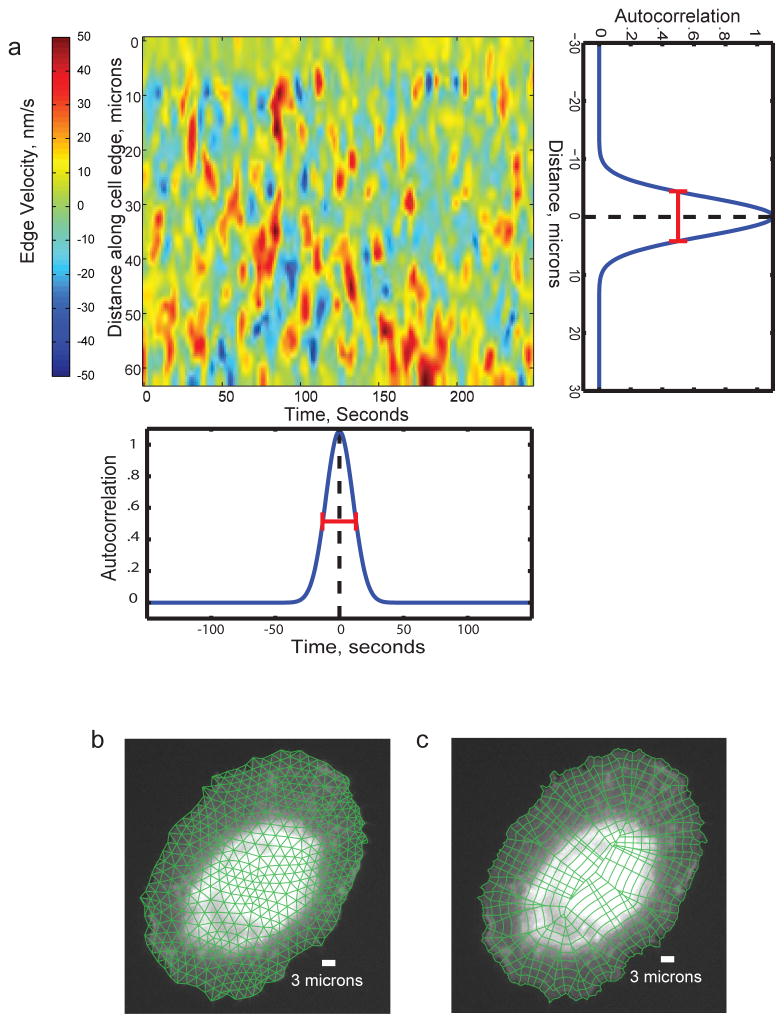
The possibility of exploiting constitutive fluctuations to infer the response hierarchy and kinetics of pathways relies on the proper sampling of biosensor activities. The sampling resolution in time must be a few times faster than the time scale of information transmission through the pathway. Otherwise, the fluctuation sequences of different pathway components are decoupled. The sampling resolution in space must be high enough to preclude the averaging of adjacent yet independent pathway events. On the other hand, some averaging may be desirable to reduce measurement noise in the fluctuations. As illustrated in Fig. 4, the required time and length scales for sampling signaling activities can be deduced from the autocorrelation functions of the cellular outputs of the pathway; that is, in the example shown, cell protrusion dynamics.
Figure 4. Determining the requirements for spatiotemporal sampling to allow computational multiplexing based on constitutive fluctuations.
a) Map of the spatial (vertical axis) and temporal (horizontal axis) activity of an experimental fiduciary (here cell edge velocity). The activity map displays typical stochastic oscillations, as seen for many cellular activities. The horizontal autocorrelation of the map indicates the characteristic time scale of the oscillations (~40 s in this example, as deduced from the full width at half maximum (FWHM) of the function, red bar). This time scale defines the requirement for temporal sampling. The vertical autocorrelation of the map indicates the characteristic length scale of the oscillations (~15 μm in this example, as deduced from the FWHM). This length scale defines the requirement for spatial sampling. For both, time and space domain 2 – 4 fold oversampling should be achieved. That is, movies should be acquired at 10 s frame intervals or faster and biosensor intensities should be sampled in probing windows of max. 4 microns side length. b) Definition of a triangular mesh of appropriately-sized probing windows. c) Definition of a polygonal mesh of appropriately-sized probing windows. In contrast to the triangular mesh in b), each window inside the cell perimeter has a unique relation to a window at the cell periphery. Thus, this mode of windowing is preferred for studies that relate intracellular signals to cell protrusion and retraction events.
Reconstructing the pathway hierarchy
Until recently, studies involving multiple biosensor activities have used pairwise cross-correlation to infer the relations between pathway components22, 72. Component pairs, the correlation of which does not reach a defined significance level are considered disconnected in the pathway, whereas pairs with a significant correlation are considered directly or indirectly coupled. These pairwise analysis methods, though simple, have major limitations. First, as mentioned above, they do not provide the means for a rigorous integration of multiple experiments in one model. Second, cross-correlation approaches break down in the presence of strong feedback and/or feedforward interactions between pathway components. Third, although temporal cross-correlation generates some evidence for the hierarchy of activation, by determining what comes first and what comes second, it cannot define causal relations between pathway components. Two unconnected activities may be correlated with each other simply because they are downstream of the same activity. Much work is underway to identify causal networks from common fluctuations in gene expression profiles or proteomic data7, 73, 74. Some of these frameworks even have been expanded to non-simultaneously observed data 75. In contrast to genomic and proteomic data sets, however, biosensor images offer more finely resolved information about the dynamics and sub-cellular localization of pathway activities. While this additional information will greatly enhance the inference of the pathway hierarchy, it introduces new challenges for the design of algorithms that integrate data from a sequence of different experiments.
Conclusion
In summary, new biosensor designs with extended spectral properties and vastly increased sensitivity have opened exciting opportunities for signal transduction research. It has become possible to simultaneously monitor two or three signaling activities, and enhanced sensitivity permits the analysis of highly localized constitutive pathway fluctuations. This in turn enables the use of powerful statistical formalisms for the integration of multiple experiments and for the inference of hierarchy in signaling networks. Conceptually, this has brought us to the point where complex pathways with many components and redundant and nonlinear cascades can be studied in living cells under fairly physiological conditions. However, to turn this concept into a routine tool for the cell biologist, developments on the horizon will play a major role. To apply computational multiplexing by relying on combinations of biosensors in the same cell, issues of cell perturbation will become even more important in biosensor design. To overcome these issues, biosensors must be developed with enhanced sensitivity that enables their effective use at lower concentrations; this may stem from brighter fluorophores, direct excitation, and improvements in instrumentation. Designs responding to endogenous proteins will also reduce cell perturbation. Novel approaches to decrease the spectral bandwidth of each biosensor will increase the number of activities that can be imaged simultaneously, and might include the development of FLIM with an enhanced signal to noise ratio, biosensors that are based on single fluorophores, and spectral deconvolution imaging. As computational multiplexing provides us with an ability to multiplex an essentially unlimited number of signaling molecules, our ability to produce biosensors will become a bottleneck. Soon, biosensors will not need to be based on naturally occurring domains or substrates with the desired specificity, but can be generated via high throughput screening of engineered scaffolds76.
Supplementary Material
Glossary
- Orthogonal wavelengths
Emission or excitation wavelengths of two fluorescent probes are sufficiently different to be imaged separately
- Ratiometric imaging
Biosensors can be designed so that the ratio of emission or excitation at two different wavelengths reflects the biological activity being measured. This ratio is independent of the biosensor’s fluorescence intensity, so eliminates effects of cell thickness, uneven biosensor distribution, uneven illumination etc
- Stokes shift
Difference between the excitation and emission wavelengths of a fluorescent probe
- Quantum dots
Small semiconductor crystals that emit light of a longer wavelength upon excitation with a shorter wavelength, akin fluorophores
- Fluorescence polarization anisotropy
This technique measures the rotational diffusion of fluorophores by measuring the difference in the polarization of excitation and emission light. Changes in fluorescence polarization anisotropy indicate changes in the rotational diffusion of molecules induced by their interactions with other molecules
- Spectral image decomposition
Mathematical technique to separate the contribution of multiple fluorophore species to the image signal at a certain wavelength. This allows the separation of the signals of fluorophores with overlapping emission spectra
References
- 1.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 2.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 3.Margolin AA, et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. Bmc Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–U144. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U. Dynamics and Variability of ERK2 Response to EGF in Individual Living Cells. Molecular Cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal Protein-Signaling Networks Derived from Multiparameter Single-Cell Data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 8.Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–7. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer AW, et al. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev Cell. 2008;15:146–62. doi: 10.1016/j.devcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–5. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 11.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbeck MC, et al. Simultaneous optical measurements of cytosolic Ca2+ and cAMP in single cells. Sci STKE. 2006:pl6. doi: 10.1126/stke.3532006pl6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wier WG, Rizzo MA, Raina H, Zacharia J. A technique for simultaneous measurement of Ca2+, FRET fluorescence and force in intact mouse small arteries. J Physiol. 2008;586:2437–43. doi: 10.1113/jphysiol.2008.151522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tengholm A, Teruel MN, Meyer T. Single cell imaging of PI3K activity and glucose transporter insertion into the plasma membrane by dual color evanescent wave microscopy. Sci STKE. 2003:PL4. doi: 10.1126/stke.2003.169.pl4. [DOI] [PubMed] [Google Scholar]
- 15.Tanimura A, Nezu A, Morita T, Turner RJ, Tojyo Y. Fluorescent biosensor for quantitative real-time measurements of inositol 1,4,5-trisphosphate in single living cells. J Biol Chem. 2004;279:38095–8. doi: 10.1074/jbc.C400312200. [DOI] [PubMed] [Google Scholar]
- 16.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–5. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 17.VanEngelenburg SB, Palmer AE. Fluorescent biosensors of protein function. Curr Opin Chem Biol. 2008;12:60–5. doi: 10.1016/j.cbpa.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 111:3614–66. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoppe AD, Swanson JA. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004;15:3509–19. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson JA, Hoppe AD. The coordination of signaling during Fc receptor-mediated phagocytosis. J Leukoc Biol. 2004;76:1093–103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- 21.Vaughan EM, Miller AL, Yu HY, Bement WM. Control of local Rho GTPase crosstalk by Abr. Curr Biol. 21:270–7. doi: 10.1016/j.cub.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nature Cell Biology. 2008;10:1393–U38. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukada Y, et al. Quantification of local morphodynamics and local GTPase activity by edge evolution tracking. PLoS Comput Biol. 2008;4:e1000223. doi: 10.1371/journal.pcbi.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matz MV, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–73. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 26.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 27.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem Biol. 2008;3:156–60. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 29.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–3. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 30.Niino Y, Hotta K, Oka K. Blue fluorescent cGMP sensor for multiparameter fluorescence imaging. PLoS One. 2010;5:e9164. doi: 10.1371/journal.pone.0009164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niino Y, Hotta K, Oka K. Simultaneous live cell imaging using dual FRET sensors with a single excitation light. PLoS One. 2009;4:e6036. doi: 10.1371/journal.pone.0006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomosugi W, et al. An ultramarine fluorescent protein with increased photostability and pH insensitivity. Nat Methods. 2009;6:351–3. doi: 10.1038/nmeth.1317. [DOI] [PubMed] [Google Scholar]
- 33.Sapsford KE, Berti L, Medintz IL. Materials for fluorescence resonance energy transfer analysis: beyond traditional donor-acceptor combinations. Angew Chem Int Ed Engl. 2006;45:4562–89. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 34.Pertz O, Hahn KM. Designing biosensors for Rho family proteins--deciphering the dynamics of Rho family GTPase activation in living cells. Journal of Cell Science. 2004;117:1313–1318. doi: 10.1242/jcs.01117. [DOI] [PubMed] [Google Scholar]
- 35.Hinner MJ, Johnsson K. How to obtain labeled proteins and what to do with them. Curr Opin Biotechnol. 21:766–76. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nature Biotechnology. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodama Y, Wada M. Simultaneous visualization of two protein complexes in a single plant cell using multicolor fluorescence complementation analysis. Plant Mol Biol. 2009;70:211–7. doi: 10.1007/s11103-009-9467-0. [DOI] [PubMed] [Google Scholar]
- 38.Vidi PA, Chemel BR, Hu CD, Watts VJ. Ligand-dependent oligomerization of dopamine D(2) and adenosine A(2A) receptors in living neuronal cells. Mol Pharmacol. 2008;74:544–51. doi: 10.1124/mol.108.047472. [DOI] [PubMed] [Google Scholar]
- 39.Grinberg AV, Hu CD, Kerppola TK. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol Cell Biol. 2004;24:4294–308. doi: 10.1128/MCB.24.10.4294-4308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–82. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 41.Chu J, et al. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens Bioelectron. 2009;25:234–9. doi: 10.1016/j.bios.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Altman D, Goswami D, Hasson T, Spudich JA, Mayor S. Precise positioning of myosin VI on endocytic vesicles in vivo. PLoS Biol. 2007;5:e210. doi: 10.1371/journal.pbio.0050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–89. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 44.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 45.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–95. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 46.Jares-Erijman EA, Jovin TM. Imaging molecular interactions in living cells by FRET microscopy. Curr Opin Chem Biol. 2006;10:409–16. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 47.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Peyker A, Rocks O, Bastiaens PI. Imaging activation of two Ras isoforms simultaneously in a single cell. Chembiochem. 2005;6:78–85. doi: 10.1002/cbic.200400280. [DOI] [PubMed] [Google Scholar]
- 49.Murakoshi H, Lee SJ, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwok S, et al. Genetically encoded probe for fluorescence lifetime imaging of CaMKII activity. Biochem Biophys Res Commun. 2008;369:519–25. doi: 10.1016/j.bbrc.2008.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremers GJ, van Munster EB, Goedhart J, Gadella TW., Jr Quantitative lifetime unmixing of multiexponentially decaying fluorophores using single-frequency fluorescence lifetime imaging microscopy. Biophys J. 2008;95:378–89. doi: 10.1529/biophysj.107.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS One. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shcherbo D, et al. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 2009;9:24. doi: 10.1186/1472-6750-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Padilla-Parra S, et al. Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition. Biophys J. 2009;97:2368–76. doi: 10.1016/j.bpj.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–72. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 56.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science (New York, NY) 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 57.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–7. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 58.Berlin JR, Bassani JW, Bers DM. Intrinsic cytosolic calcium buffering properties of single rat cardiac myocytes. Biophys J. 1994;67:1775–87. doi: 10.1016/S0006-3495(94)80652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Helmchen F, Imoto K, Sakmann B. Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys J. 1996;70:1069–81. doi: 10.1016/S0006-3495(96)79653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mochizuki N, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–8. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 61.Sawano A, Takayama S, Matsuda M, Miyawaki A. Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev Cell. 2002;3:245–57. doi: 10.1016/s1534-5807(02)00224-1. [DOI] [PubMed] [Google Scholar]
- 62.O’Rourke NA, Meyer T, Chandy G. Protein localization studies in the age of ‘Omics’. Curr Opin Chem Biol. 2005;9:82–7. doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Schultz C, Schleifenbaum A, Goedhart J, Gadella TW., Jr Multiparameter imaging for the analysis of intracellular signaling. Chembiochem. 2005;6:1323–30. doi: 10.1002/cbic.200500012. [DOI] [PubMed] [Google Scholar]
- 64.Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Developmental Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 65.Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- 66.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophysical Journal. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor RJ, et al. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kiosses WB, Shattil SJ, Pampori N, Schwartz MA. Rac recruits high-affinity integrin alphavbeta3 to lamellipodia in endothelial cell migration. Nat Cell Biol. 2001;3:316–20. doi: 10.1038/35060120. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 70.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–8. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tkachenko E, et al. Protein Kinase A Governs a RhoA-RhoGDI-driven Protrusion-Retraction Pacemaker in Migrating Cells. Nature Cell Biol. doi: 10.1038/ncb2231. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Locasale JW, Wolf-Yadlin A. Maximum Entropy Reconstructions of Dynamic Signaling Networks from Quantitative Proteomics Data. Plos One. 2009;4 doi: 10.1371/journal.pone.0006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perrin BE, et al. Gene networks inference using dynamic Bayesian networks. Bioinformatics. 2003;19:II138–II148. doi: 10.1093/bioinformatics/btg1071. [DOI] [PubMed] [Google Scholar]
- 75.Sachs K, et al. Learning Signaling Network Structures with Sparsely Distributed Data. Journal of Computational Biology. 2009;16:201–212. doi: 10.1089/cmb.2008.07TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gulyani A, et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nature Chemical Biology. 2011 doi: 10.1038/nchembio.585. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galperin E, Verkhusha VV, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods. 2004;1:209–17. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 78.He L, Wu X, Simone J, Hewgill D, Lipsky PE. Determination of tumor necrosis factor receptor-associated factor trimerization in living cells by CFP->YFP->mRFP FRET detected by flow cytometry. Nucleic Acids Res. 2005;33:e61. doi: 10.1093/nar/gni057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X, et al. Measurement of two caspase activities simultaneously in living cells by a novel dual FRET fluorescent indicator probe. Cytometry A. 2006;69:477–86. doi: 10.1002/cyto.a.20300. [DOI] [PubMed] [Google Scholar]
- 80.Kawai H, et al. Simultaneous imaging of initiator/effector caspase activity and mitochondrial membrane potential during cell death in living HeLa cells. Biochim Biophys Acta. 2004;1693:101–10. doi: 10.1016/j.bbamcr.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 81.Kawai H, et al. Simultaneous real-time detection of initiator- and effector-caspase activation by double fluorescence resonance energy transfer analysis. J Pharmacol Sci. 2005;97:361–8. doi: 10.1254/jphs.fp0040592. [DOI] [PubMed] [Google Scholar]
- 82.DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci U S A. 2004;101:16513–8. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brumbaugh J, Schleifenbaum A, Gasch A, Sattler M, Schultz C. A dual parameter FRET probe for measuring PKC and PKA activity in living cells. J Am Chem Soc. 2006;128:24–5. doi: 10.1021/ja0562200. [DOI] [PubMed] [Google Scholar]
- 84.Ouyang M, et al. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res. 2010;70:2204–12. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grant DM, et al. Multiplexed FRET to image multiple signaling events in live cells. Biophys J. 2008;95:L69–71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.