Abstract
OBJECTIVE
To examine the ability of various postoperative nomograms to predict prostate cancer-specific mortality (PCSM) and to validate that they could predict aggressive biochemical recurrence (BCR). Prostate-specific antigen (PSA), grade, and stage are the classic triad used to predict BCR after radical prostatectomy (RP). Multiple nomograms use these to predict risk of BCR. A previous study showed that several nomograms could predict aggressive BCR (prostate-specific antigen doubling time [PSADT] <9 months) more accurately than BCR. However, it remains unknown if they can predict more definitive endpoints, such as PCSM.
METHODS
We performed Cox analyses to examine the ability of 4 postoperative nomograms, the Duke Prostate Center (DPC) nomogram, the Kattan postoperative nomogram, the Johns Hopkins Hospital (JHH) nomogram, and the joint Center for Prostate Disease Research(CPDR)/Cancer of the Prostate Strategic Urologic Research Endeavor (CaPSURE) nomogram to predict BCR and PCSM among 1778 men in the Shared Equal Access Regional Cancer Hospital (SEARCH) database who underwent RP between 1990 and 2009. We also compared their ability to predict BCR and aggressive BCR in a subset of men. We calculated the c-index for each nomogram to determine its predictive accuracy for estimating actual outcomes.
RESULTS
We found that each nomogram could predict aggressive BCR and PCSM in a statistically significant manner and that they all predicted PCSM more accurately than they predicted BCR (ie, with higher c-index values).
CONCLUSION
Currently available nomograms used to predict BCR accurately predict PCSM and other more clinically relevant endpoints. Moreover, not only do they significantly predict PCSM, but do so with generally greater accuracy than BCR.
Because prostate cancer (PCa) is heterogenous, many models exist to help risk-stratify men by their risk of developing various endpoints to help patients receive the appropriate intensity of treatment.1–7 These endpoints occur years after initial treatment and correlate to varying degrees with mortality risk. Most models include the classic triad of prostate-specific antigen (PSA), grade, and stage and were developed to predict biochemical recurrence (BCR) after radical prostatectomy (RP). However, BCR is not always a clinically meaningful endpoint. Many men with BCR live for years ultimately dying of unrelated causes. Others progress rapidly and die of PCa.
Although predicting BCR is useful, it would be more useful to predict distant outcomes, such as aggressive BCR(ie, recurrence with a PSA doubling time [PSADT], <9 months)8 or PCa-specific mortality (PCSM). One approach is to develop new nomograms that predict these outcomes. However, we hypothesize that currently available nomograms designed to predict BCR may accurately predict BCR and PCSM. Previously, we showed that several nomograms could predict aggressive BCR (PSADT <9 months) more accurately than they predicted overall BCR.9 However, no study has examined the ability of these nomograms to predict PCSM. Therefore, we examined the ability of various post-operative nomograms to predict PCSM and to redemonstrate their ability to predict aggressive BCR in the multicenter Shared Equal Access Regional Cancer Hospital (SEARCH) database, which includes clinicopathological information on men treated with RP at 4 Veterans Affairs medical centers.10
MATERIAL AND METHODS
Clinical and Pathological Variables
After obtaining institutional review board approval from each institution to abstract and combine data, data from patients undergoing RP at the Veterans Affairs Medical Centers in West Los Angeles and Palo Alto, California, Augusta, Georgia, and Durham, North Carolina, were entered into the SEARCH database.10 This database includes information on patient age at surgery, race, height, weight, clinical stage, grade of cancer on diagnostic biopsies, preoperative PSA, surgical specimen pathology (specimen weight, tumor grade, stage, and surgical margin status), and follow-up PSA data. Body mass index was calculated as weight in kilograms divided by height in meters squared (kg/m2). The prostatectomy specimens were sectioned per each institution’s protocol.
BCR was defined as a single PSA >0.2 ng/mL, 2 concentrations at 0.2 ng/mL, or secondary treatment for an elevated postoperative PSA. Aggressive BCR was defined as recurrence with a PSADT <9 months based on a previous study that showed that men who recurred with a PSADT >9 months were not at increased risk of mortality, whereas those men who recurred with a PSADT <9 months were at an increased risk of all-cause mortality and PCSM.11 PSADT, calculated using the log-slope method,12 was calculated for all patients meeting the recurrence definition who had a minimum of 2 PSA values, separated by at least 3 months, within 2 years after recurrence. All PSAs within the first 2 years after recurrence were used to calculate PSADT. For patients starting salvage hormone or radiation therapy within this timeframe, only PSAs before salvage therapy were used. Patients with a PSADT ≤0 (ie, decline/no change) or very long PSADT (>100 months) were assigned a value of 100 months for ease of calculations (n = 61). In determining PCSM, men who died with metastatic, progressive, castrate-resistant PCa were considered to have died of PCa.
Study Population
To be included in the study population, men had to have information available regarding all of the clinical variables that were included in each nomogram. Of the 2211 men in SEARCH, we excluded 76 men who were missing follow-up data and 233 men who were missing clinicopathological variables (62, 28, 30, 4, and 233 men were dropped because they were missing preoperative PSA, postoperative Gleason sum, pathological stage characteristics, race, and prostate weight). This resulted in a final study population of 1778 men who underwent RP between 1990 and 2009. In a secondary analysis that compared the ability of these nomograms to predict overall vs aggressive BCR, we excluded an additional 272 men who were missing additional data pertaining to this endpoint, yielding a population of 1506 men.
Statistical Analysis
Our primary goal was to test the ability of nomograms that were designed to predict BCR and PCSM to determine if they could do so more accurately than they could predict BCR. In a secondary analysis, we sought to validate our group’s previous findings9 that these nomograms could more accurately predict aggressive BCR (ie, recurrence with a PSADT <9 months) than overall BCR. To do so, we chose 4 representative previously published postoperative nomograms, the Duke Prostate Center (DPC) nomogram,1 the Kattan postoperative nomogram,7 the Johns Hopkins Hospital (JHH) nomogram,13 and the joint Center for Prostate Disease Research (CPDR)/Cancer of the Prostate Strategic Urologic Research Endeavor (CaPSURE) nomogram,5 and tested them for their ability to predict the aforementioned outcomes.
We first calculated the linear predictor scores for each nomogram based on the equations that the authors used, which are listed below.
DPC: 108.36634 + 0.43664285 * log (PSA) + 0.71718536 * (positive surgical margins) + 0.54469524 * (seminal vesicle invasion) + 0.20156322 * (extraprostatic extension) + 0.32334319 * (pathological Gleason sum of 2–6) + 0.80299179 * (pathological Gleason sum of 3 + 4) + 0.98773794 * (pathological Gleason sum ≥4 + 3) − 0.37209739 * log (prostate weight) + 0.31395694 * (black race) - 0.054236003 * (year of surgery) − 1.6355449 * (adjuvant radiation) + 0 * (pathological Gleason sum) * (race) * (surgical margin status) * (extraprostatic extension status) * (seminal vesicle invasion status).
Kattan postoperative nomogram: 110.52302 - 0.056105835 * (year of surgery) + 0.3931648 * (surgical margin status) + 0.8748934 * (extracapsular extension) + 0.94251512 * (seminal vesicle invasion) + 0.43389125 * (lymph node metastasis) + 1.3792909 * (primary pathological Gleason sum >3) + 0.57039863 * (secondary pathological Gleason sum >3) − 1.4998377 * (adjuvant radiation) + 0.39237815 * log (PSA) − 0.032117323 * max (log (PSA) − 1.0650555, 0) ‸ 3 +0.059594888 * max (log (PSA) − 1.8407084, 0) ‸ 3 − 0.027477565 * max (log (PSA) − 2.747335, 0) ‸ 3 + 0 * (primary pathological Gleason sum) * (secondary pathological Gleason sum) * (lymph node involvement status).
JHH: 1.43 * (lymph node metastasis) + 1.15 * (surgical margin status) + 0.71 * (pathological Gleason sum ≥5) + 0.51 * (seminal vesicle invasion status).
CPDR/CaPSURE: exponent (0.54 × race) + (0.05 × sigmoidal transformation of PSA) + (0.23 * pathological Gleason sum) + (0.69 * pathological stage).
We then performed a series of Cox proportional hazard analyses to determine the ability of all of these nomograms to predict BCR and PCSM in the SEARCH database. We then calculated the c-index for each nomogram for its ability to predict the aforementioned PCa outcomes and compared their respective predictive abilities. We first compared each nomogram’s ability to predict BCR with its ability to predict PCSM. Because men have to have a minimum of 3 years of follow-up in order to determine whether or not they will have an aggressive BCR, we examined each nomogram’s ability to predict BCR, aggressive BCR, and PCSM in a subset of men who had the appropriate follow-up such that each endpoint could be predicted.
P values <.05 were considered significant. Data were similar among all sites and were combined for analysis. All statistical analyses were performed using STATA 10 (Stata Corp., College Station, TX).
RESULTS
Patient Characteristics
The mean age of the men included in this study was 61.5 years (SD 6.5). The cohort was racially diverse (51% of patients were white, 42% were African American, and 7% were of other races). Overall, this cohort was intermediate-risk (39% of men had pathological Gleason sums of ≤6, 39% had Gleason 7 disease, and 18% had Gleason 8–10 disease). The mean and median preoperative PSA values of this cohort were relatively low at 9.2 (SD 9.2) and 6.8 (interquartile range [IQR] 4.9–10.3), respectively. A total of 84% of men had clinical stage T1c or T2a disease (Table 1). Among men who died of PCa, the median time to PCa death was 94.9 months (IQR 63.0–136.2). Among men who died of non-PCa causes, the median time to death was 74.9 months (IQR 44.6–111.7).
Table 1.
Demographic, clinical, and pathologic characteristics of included patients (n = 1778)
| Variables | |
| Mean age at surgery ± SD (y) | 61.5 ± 6.5 |
| Median y of surgery | 2002 |
| Race | |
| White | 904 (51%) |
| African American | 751 (42%) |
| Other | 123 (7%) |
| BMI | |
| <25 kg/m2 | 395 (24%) |
| 25–29.9 kg/m2 | 765 (47%) |
| ≥30 kg/m2 | 478 (29%) |
| PSA (ng/mL) | |
| Mean ± SD | 9.2 ± 9.2 |
| Median (IQR) | 6.8 (4.9–10.3) |
| Biopsy Gleason Sum | |
| 2–6 | 1069 (61%) |
| 7 | 376 (21%) |
| 8–10 | 310 (18%) |
| Pathological Gleason Sum | |
| 2–6 | 713 (40%) |
| 7 | 686 (39%) |
| 8–10 | 379 (21%) |
| Percentage positive cores | |
| Mean ± SD | 35.3 ± 22.9 |
| Median (IQR) | 33.3 (16.7–50.0) |
| Prostate weight (grams) | |
| Mean ± SD | 44.2 ± 21.8 |
| Median (IQR) | 40.0 (31.0–50.7) |
| Extraprostatic extension | 378 (21%) |
| Positive surgical margins | 806 (45%) |
| Seminal vesicle invasion | 171 (10%) |
| Clinical stage | |
| T1a | 21 (1%) |
| T1b | 11 (1%) |
| T1c | 949 (58%) |
| T2X | 66 (4%) |
| T2a | 431 (26%) |
| T2b | 107 (7%) |
| T2c | 57 (3%) |
| T3 | 1 (0.1%) |
| Pathologic stage grouping | |
| Organ-confined, margin negative | 835 (47%) |
| EPE and/or positive margins | 761 (43%) |
| Seminal vesicle invasion | 157 (8%) |
| Lymph node involvement | 25 (1%) |
BMI, body mass index; EPE, extraprostatic extension; IQR, interquartile range; PSA, prostate-specific antigen.
BCR, Aggressive BCR, and PCSM Rates of Included Patients
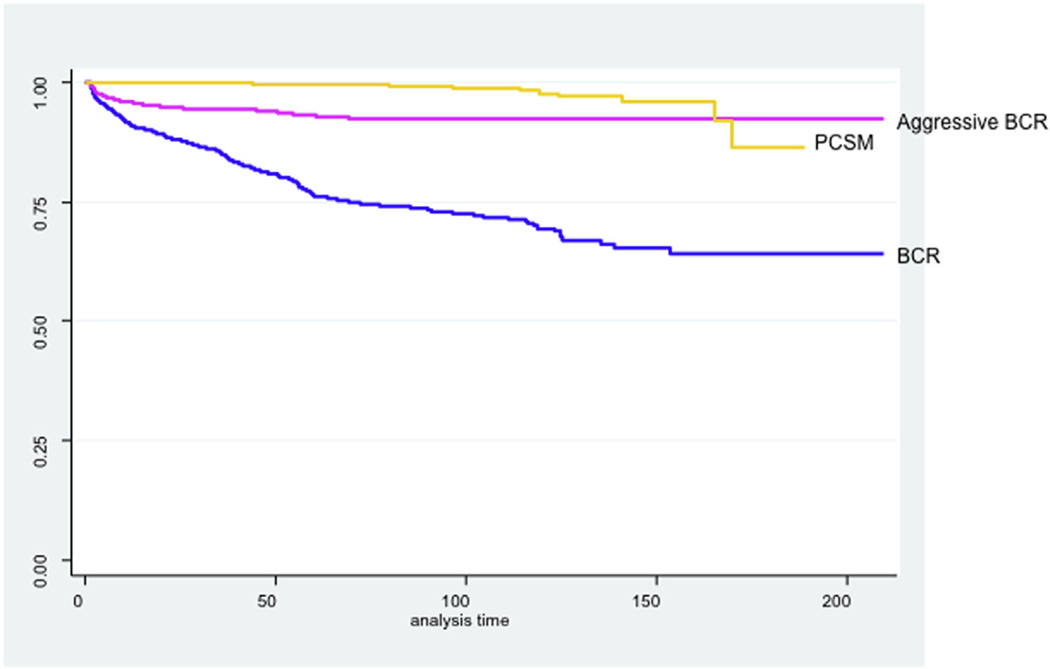
In this cohort, 435 patients (24%) experienced BCR and 27 patients (2%) died of PCa. The median time to BCR among men in this cohort was 15.4 months (IQR 3.7– 40.9). Among the subset of men who experienced aggressive BCR, the median time to recurrence was 6.1 months (IQR 2.1–17.2). The median time to PCa death was 94.9 months (IQR 63.0–136.2, Fig. 1).
Figure 1.
Kaplan-Meier curves for biochemical recurrence (BCR), aggressive BCR, and prostate cancer-specific mortality (PCSM). (Color version available online.)
Ability of Nomograms to Predict BCR, Aggressive BCR, and PCSM
Overall, the DPC, Kattan postoperative, JHH, and CPDR/CaPSURE nomograms predicted PCSM with a higher degree of accuracy than they predicted overall BCR (c-index: 0.83 vs 0.75, 0.86 vs 0.74, 0.87 vs 0.74, and 0.82 vs 0.72, respectively, Table 2). When we sought to validate the findings of the previous SEARCH study9 by comparing the ability of the models to predict BCR vs aggressive BCR in a subset of men, we again found that the models were able to predict aggressive BCR more accurately than overall BCR (0.82 vs 0.77, 0.81 vs 0.75, 0.79 vs 0.71, and 0.77 vs 0.70 for the DPC, Kattan postoperative, JHH, and CPDR/CaPSURE nomograms, respectively, Table 3).
Table 2.
Ability of various postoperative nomograms to predict biochemical recurrence and prostate cancer-specific mortality (n = 1778)
| BCR | PCSM | |||||||
|---|---|---|---|---|---|---|---|---|
| Nomogram | HR | 95% CI | P Value | c-index | HR | 95% CI | P Value | c-index |
| DPC | 2.74 | 2.50–3.01 | <.001 | 0.75 | 2.81 | 1.90–4.14 | <.001 | 0.83 |
| Kattan postoperative | 2.02 | 1.89–2.16 | <.001 | 0.74 | 2.57 | 1.92–3.45 | <.001 | 0.86 |
| JHH | 2.09 | 1.93–2.24 | <.001 | 0.74 | 2.97 | 2.12–4.18 | <.001 | 0.87 |
| CPDR/CaPSURE | 1.09 | 1.07–1.10 | <.001 | 0.72 | 1.10 | 1.07–1.14 | <.001 | 0.82 |
BCR, biochemical recurrence; CI, confidence interval; CPDR/CaPSURE, joint Center for Prostate Disease Research/Cancer of the Prostate Strategic Urologic Research Endeavor; DPC, Duke Prostate Center; HR, hazard ratio; JHH, Johns Hopkins Hospital; PCSM, prostate cancer-specific mortality.
Table 3.
Ability of various postoperative nomograms to predict overall and aggressive biochemical recurrence (n = 1506)
| BCR | Aggressive BCR | |||||||
|---|---|---|---|---|---|---|---|---|
| Nomogram | HR | 95% CI | P Value | c-index | HR | 95% CI | P Value | c-index |
| DPC | 3.06 | 2.69–3.48 | <.001 | 0.77 | 3.80 | 3.02–4.76 | <.001 | 0.82 |
| Kattan postoperative | 2.12 | 1.93–2.32 | <.001 | 0.75 | 2.42 | 2.06–2.85 | <.001 | 0.81 |
| JHH | 1.98 | 1.72–2.05 | <.001 | 0.71 | 2.49 | 2.06–3.00 | <.001 | 0.79 |
| CPDR/CaPSURE | 1.09 | 1.08–1.11 | <.001 | 0.70 | 1.11 | 1.08–1.13 | <.001 | 0.77 |
Abbreviations as in Table 2.
DISCUSSION
Many nomograms and predictive models have been developed to help predict outcome after treatment. However, typically, these nomograms were developed to predict BCR.1–3,5–7,13 BCR is an important endpoint that can indicate recurrent PCa up to 8 years before clinical evidence of metastases.12 However, experiencing BCR does not mean the patient will die of PCa or even that he will ever experience symptoms of recurrent disease. Because many currently available nomograms include the classic triad of PSA, stage, and grade and have been shown to predict aggressive BCR, which is a more relevant clinical endpoint than overall BCR,8 we determined whether 4 postoperative nomograms originally designed to predict BCR (ie, the DPC, Kattan postoperative, JHH, and CPDR/CaPSURE nomograms) could predict the more definitive clinical outcome of PCSM with a similar or higher degree of accuracy than they predicted BCR. We also sought to validate the findings of our prior SEARCH study that these models could more accurately predict aggressive BCR than overall BCR in a secondary analysis.9 Encouragingly, we found that all 4 nomograms predicted both aggressive BCR and PCSM more accurately than BCR.
As mentioned above, although BCR is clearly an important endpoint, it is not necessarily a definitive clinical endpoint in all patients. For example, in several previous studies, the group at JHH showed that of men who experience BCR after RP and have a calculable PSADT, only those with a PSADT <9 months are at increased risk of all-cause mortality and PCSM.11,14 Men who recurred with a PSADT ≥9 months were at no higher risk of all-cause death than men who did not recur, highlighting the limitations of BCR. This study was recently validated by our group using a cohort of men from the SEARCH database.15 Because this subgroup of men who recur with a short (ie, <9 months) PSADT are at increased risk of death, BCR with a PSADT <9 months can be termed an “aggressive” BCR as it confers a worse prognosis than a BCR with a longer PSADT. We have previously shown using the SEARCH database that aggressive BCR can be predicted with a high degree of accuracy using a model that, like the 4 nomograms analyzed in this article, include the aforementioned classic triad of predictor variables (ie, PSA, stage, and grade).8 This finding was later validated in a cohort of men from the DPC database.16 Furthermore, we have also shown using the SEARCH database that nomograms originally designed to predict BCR are able to predict aggressive BCR with a higher degree of accuracy than they are able to predict overall BCR (c-index = 0.756 vs 0.702).9 However, this prior study did not examine their ability to predict PCSM, the primary analysis of the current study.
Although the 4 nomograms included in this study were all designed to predict BCR, the variables included in the equations used to generate the predictions, as well as the development cohorts, differed from one another to a certain degree and from SEARCH. The DPC nomogram was developed in a cohort of 3194 patients who underwent RP at Duke University between 1988 and 2007.1 The authors examined whether or not race and prostate weight were independent predictors of BCR. In a multivariate Cox proportional hazards regression model, they added these parameters to a nomogram that included preoperative PSA, pathological Gleason sum, pathological stage parameters (ie, surgical margin status, seminal vesicle invasion, and extraprostatic extension), year of surgery, and whether or not a patient received adjuvant radiotherapy and were able to predict BCR with a c-index of 0.75.
The Kattan postoperative nomogram was developed using a cohort including 1881 men who underwent RP at Memorial Sloan-Kettering Cancer Center or Baylor College of Medicine between 1983 and 2003.7 The nomogram includes preoperative PSA, pathological Gleason sum, adjuvant radiation, and pathological stage parameters (the same factors that were included in the DPC nomogram minus race plus lymph node status). The concordance index of the nomogram when applied to independent validation sets was 0.81 and 0.79.
The JHH nomogram development cohort included 904 men who underwent RP between 1982 and 1999 at Johns Hopkins University.13 The nomogram included PSA, pathological Gleason sum, and the same pathological stage characteristics used in the Kattan postoperative nomogram. The authors also cross-validated the model in a cohort of 901 men who underwent RP at the Mayo Clinic matched for Gleason score with the JHH patients. The authors did not specifically comment on the predictive accuracy of the model.
Although the first 3 nomograms discussed included patients treated at tertiary care centers and who were predominately white, the CPDR/CaPSURE cohort had several characteristics that made it slightly different than the other development cohorts. The CPDR database includes men treated at Walter Reed Army Medical Center. All of the men in CPDR were active military members, and is a more racially diverse cohort than the 3 development cohorts discussed above. CaPSURE is a large database that includes information on men with PCa who received treatment at 1 of many academic practices, community practices, or Veterans Affairs hospitals across the United States.17 The characteristics of the patients in these 2 databases, particularly CPDR, make them more similar to SEARCH than the other nomogram cohorts discussed above. This particular study included 1012 men from CaPSURE who underwent RP between 1995 and 2000 and 503 patients from CPDR who underwent RP between 1988 and 1998.5 The authors validated a previously developed nomogram that included preoperative PSA, pathological stage, pathological Gleason sum, and ethnicity and revised it using data for their cohort. The authors did not specifically comment on the performance characteristics of the nomogram in the development cohort. The 4 nomograms that we examined, therefore, contained the same core predictor variables (PSA, grade, and stage) but differed slightly from one another in other included variables. The study populations included men treated at tertiary care academic hospitals, Veterans Affairs hospitals, active military hospitals, and community practices. Although each development cohort had certain unique characteristics, overall, these 4 nomograms represent a diversity of populations, making them ideal to examine for their ability to predict aggressive BCR and PCSM. Of note, we do not claim these models are the most predictive models. Rather, we selected these to present a spectrum of populations and are used to test the general hypothesis that nomograms designed to predict BCR can actually predict PCSM with greater accuracy.
We found all 4 models (DPC, Kattan postoperative, JHH, and the CPDR/CaPSURE nomogram) predicted PCSM more accurately than they predicted BCR and did so with similar c-index values (although the JHH nomogram had a slightly higher c-index than the other nomograms, overall they were quite similar). This is inline with a prior study that showed that PCSM is a highly predictable endpoint.18 Additionally, in a secondary analysis, we again showed these 4 nomograms could predict aggressive BCR more accurately than BCR. Thus, these widely used nomograms can be used to predict more clinically meaningful PCa endpoints with a higher degree of accuracy than they can predict the endpoint that they were originally designed to predict. This has clinical utility because it means that new models incorporating the same clinicopathological characteristics do not need to be designed to predict these later endpoints because the models that we already have can do so quite accurately. If these models are used to predict aggressive BCR and PCSM, however, the model calibration will differ given that the frequency of occurrence of these endpoints are lower than for overall BCR. One way to deal with this would be to recalibrate the preexisting nomograms, which our group has previously shown can be successfully done.19 Additionally, as new markers are developed that improve PCa prognostication, these models would likely benefit from modification.
Our study had several limitations. First, data were missing for several key variables included in the various nomograms on some of the otherwise eligible patients in SEARCH. Second, the number of PCa deaths was limited. Therefore, although we found these nomograms were accurately able to predict PCSM in the SEARCH cohort, there is a possibility our results were impacted by type I error. However, given the consistency of our findings with previous studies, this is unlikely. It would also be helpful to repeat this analysis in a cohort with a higher rate of PCSM. Ultimately, whether nomograms designed to predict BCR after other treatment modalities (ie, radiation) can also predict PCSM with improved accuracy requires further testing.
CONCLUSIONS
We found 4 commonly used postoperative nomograms designed to predict BCR after RP (ie, the Kattan postoperative, JHH, DPC, and CPDR/CaPSURE nomograms) more accurately predicted distant PCa endpoints and specifically PCSM than they were able to predict BCR. If validated, our findings suggest currently available nomograms could be recalibrated to predict long-term, definitive clinical outcomes.
Acknowledgments
Funding Support: This study was supported by the Department of Veterans Affairs, Department of Defense, National Institutes of Health, the Georgia Cancer Coalition, the American Urological Association (AUA) Foundation/Astellas Rising Star in Urology Award, and Duke University’s CTSA grant UL1RR024128 (NCRR/NIH).
Footnotes
Financial Disclosure: The authors declare that they have no relevant financial interests.
References
- 1.Schroeck FR, Sun L, Freedland SJ, et al. Race and prostate weight as independent predictors for biochemical recurrence after radical prostatectomy. Prostate Cancer Prostatic Dis. 2008;11:371–376. doi: 10.1038/pcan.2008.18. [DOI] [PubMed] [Google Scholar]
- 2.Kattan MW, Eastham JA, Stapleton AM, et al. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 3.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 4.Han M, Partin AW, Zahurak M, et al. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169:517–523. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- 5.Moul JW, Connelly RR, Lubeck DP, et al. Predicting risk of prostate specific antigen recurrence after radical prostatectomy with the Center for Prostate Disease Research and Cancer of the Prostate Strategic Urologic Research Endeavor databases. J Urol. 2001;166:1322–1327. [PubMed] [Google Scholar]
- 6.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 7.Stephenson AJ, Scardino PT, Eastham JA, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2005;23:7005–7012. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teeter AE, Bañez LL, Presti JC, Jr, et al. What are the factors associated with short prostate specific antigen doubling time after radical prostatectomy? A report from the SEARCH database group. J Urol. 2008;180:1980–1984. doi: 10.1016/j.juro.2008.07.031. [discussion: 1985]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroeck FR, Aronson WJ, Presti JC, Jr, et al. Do nomograms predict aggressive recurrence after radical prostatectomy more accurately than biochemical recurrence alone? BJU Int. 2009;103:603–608. doi: 10.1111/j.1464-410X.2008.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bañez LL, Loftis RM, Freedland SJ, et al. The influence of hepatic function on prostate cancer outcomes after radical prostatectomy. Prostate Cancer Prostatic Dis. 2010;13:173–177. doi: 10.1038/pcan.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedland SJ, Humphreys EB, Mangold LA, et al. Death in patients with recurrent prostate cancer after radical prostatectomy: prostate-specific antigen doubling time subgroups and their associated contributions to all-cause mortality. J Clin Oncol. 2007;25:1765–1771. doi: 10.1200/JCO.2006.08.0572. [DOI] [PubMed] [Google Scholar]
- 12.Pound CR, Partin AW, Eisenberger MA, et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 13.Roberts WW, Bergstralh EJ, Blute ML, et al. Contemporary identification of patients at high risk of early prostate cancer recurrence after radical retropubic prostatectomy. Urology. 2001;57:1033–1037. doi: 10.1016/s0090-4295(01)00978-5. [DOI] [PubMed] [Google Scholar]
- 14.Freedland SJ, Humphreys EB, Mangold LA, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294:433–439. doi: 10.1001/jama.294.4.433. [DOI] [PubMed] [Google Scholar]
- 15.Teeter AE, Presti JC, Jr, Aronson WJ, et al. Does PSADT after radical prostatectomy correlate with overall survival? A report from the SEARCH database group. Urology. 2011;77:149–153. doi: 10.1016/j.urology.2010.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teeter AE, Sun L, Moul JW, Freedland SJ. External validation of the SEARCH model for predicting aggressive recurrence after radical prostatectomy: results from the Duke Prostate Center Database. BJU Int. 2010;106:796–800. doi: 10.1111/j.1464-410X.2010.09214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubeck DP, Litwin MS, Henning JM, et al. The CaPSURE database: a methodology for clinical practice and research in prostate cancer. CaPSURE Research Panel. Cancer of the Prostate Strategic Urologic Research Endeavor. Urology. 1996;48:773–777. doi: 10.1016/s0090-4295(96)00226-9. [DOI] [PubMed] [Google Scholar]
- 18.Stephenson AJ, Klein EA, Kattan MW, et al. Predicting the long-term risk of prostate cancer-specific mortality after radical prostatectomy. J Clin Oncol. 2009;(suppl):27. doi: 10.1200/JCO.2008.18.2501. abstr 5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeck FR, Kattan MW, Moul JW, et al. Re-calibration and external validation of an existing nomogram to predict aggressive recurrences after radical prostatectomy. BJU Int. 2010;105:1654–1659. doi: 10.1111/j.1464-410X.2009.09060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]