Abstract
Glioblastoma Multiforme (GBM) is an aggressive brain tumor for which there is no cure. Overexpression of wild-type EGFR and loss of the tumor suppressor genes Ink4a/Arf and PTEN are salient features of this deadly cancer. Surprisingly, targeted inhibition of EGFR has been clinically disappointing, demonstrating an innate ability for GBM to develop resistance. Efforts at modeling GBM in mice using wild-type EGFR have proven unsuccessful to date, hampering endeavors at understanding molecular mechanisms of therapeutic resistance. Here, we describe a unique genetically engineered mouse model of EGFR-driven gliomagenesis that uses a somatic conditional overexpression and chronic activation of wild-type EGFR in cooperation with deletions in the Ink4a/Arf and PTEN genes in adult brains. Using this model, we establish that chronic activation of wild-type EGFR with a ligand is necessary for generating tumors with histopathological and molecular characteristics of GBMs. We show that these GBMs are resistant to EGFR kinase inhibition and we define this resistance molecularly. Inhibition of EGFR kinase activity using tyrosine kinase inhibitors in GBM tumor cells generates a cytostatic response characterized by a cell cycle arrest, which is accompanied by a substantial change in global gene expression levels. We demonstrate that a key component of this pattern is the transcriptional activation of the MET receptor tyrosine kinase and that pharmacological inhibition of MET overcomes the resistance to EGFR inhibition in these cells. These findings provide important new insights into mechanisms of resistance to EGFR inhibition and suggest that inhibition of multiple targets will be necessary to provide therapeutic benefit for GBM patients.
Keywords: Glioblastoma, genetically engineered mouse model, EGFR, PTEN, c-MET
Introduction
GBM is a highly malignant primary brain tumor with poor prognosis. The five-year relative survival rate (~3%) has not improved significantly over the last five decades. This is because GBM tumors are characterized by an uncontrolled cellular proliferation and a robust infiltrative capacity and are notoriously resistant to all therapeutic interventions attempted so far. This results in fatal recurrence in nearly all patients. Therefore, there is an urgent need to develop and utilize appropriate experimental systems to understand mechanisms of resistance, discover novel targets and test therapeutic agents.
The EGFR signaling pathway plays a crucial role in GBM pathogenesis, initiating the early stages of tumor development, sustaining tumor growth and promoting infiltration. Expression of EGFR and its ligands are observed in >65% of GBMs and are almost exclusively associated with loss of the Cdkn2a (p16INK4A/p19ARF) tumor suppressor locus and to a lesser extent with loss of the PTEN tumor suppressor (McLendon et al., 2008; Verhaak et al., 2010). Given the importance of EGFR signaling in GBM, it is surprising then that the use of EGFR tyrosine kinase inhibitors (TKIs) for GBM treatment has proven inadequate (reviewed in (Huang et al., 2009)). The most significant challenge in using EGFR TKIs clinically has been the intrinsic ability for GBMs to develop resistance to receptor inhibition.
Loss of PTEN in EGFR-positive GBMs is thought to uncouple phosphatidylinositol 3-kinase (PI3K) signaling from the control of EGFR (Friedman and Bigner, 2005; Mellinghoff et al., 2007; Mellinghoff et al., 2005), an event that was considered predictive of TKI inefficiency. However, subsequent work reported that the concomitant expression of activated EGFR and PTEN does not correspond to improved survival in patients treated with an EGFR TKI (van den Bent et al., 2009). These contradicting findings suggest that more complex molecular events are associated with EGFR inhibition resistance and that there is a pressing need to expand our understanding of these events to tackle resistance to therapeutic interventions in GBM.
Recently, several groups have molecularly redefined the classification of GBM tumors based on cataloged recurrent genomic abnormalities, robust gene expression-based molecular classification and signaling network profiles (Brennan et al., 2009; Phillips et al., 2006; Verhaak et al., 2010). GBMs are now categorized as classical, mesenchymal and proneural with aberrant gene copy number, mutation and expression of EGFR, NF1, and PDGFRA/IDH1 defining each subtypes respectively. A fourth category referred to as neural is less well defined molecularly. These efforts now provide a clinically relevant molecular framework for stratification of GBM tumors with important clinical ramifications. For example, response to standard of care therapy was shown to differ by subtype, with the greatest benefit being observed in classical GBMs and no benefit in proneural GBMs. Not only does this work demonstrate that distinct molecular subtypes exist within GBMs, it also raises the possibility that response to specific treatments may vary based on the molecular subtype.
Our knowledge of EGFR signaling comes mostly from in vitro studies of acute and transient ligand-stimulated activation of the receptor. This pattern is disparate from the clinical setting where EGFR is chronically active in GBM as a result of autocrine and paracrine expression of EGFR and its ligands (Ekstrand et al., 1991; Mishima et al., 1998; Schlegel et al., 1990; Tang et al., 1997). Therefore, studies of mechanisms of EGFR signaling and resistance to treatment in clinically relevant contexts remain largely unexplored, particularly in germane in vivo model systems.
Here, we describe a novel genetically engineered mouse model of EGFR-driven GBM based on co-expression of wild-type EGFR (EGFRWT) and TGFα, an EGFR ligand expressed in GBM. We established that a strict spatiotemporal expression and activation of EGFRWT with loss of tumor suppressor genes p16Ink4a/p19Arf and PTEN efficiently induce gliomagenesis in adults. Using these mice, we reveal a new and distinctive mechanism of resistance to EGFR TKI treatment. EGFR inhibition causes a global change in the transcription profile of GBM tumor cells, including expression and activation of the MET tyrosine kinase receptor. The acquired MET activity results in the persistent activation of downstream signaling pathways and pharmacological inactivation of MET reverses its resistance function. Our results demonstrate that multi target inhibition is necessary to overcome resistance in GBM.
Results
Sustained activation of EGFRWT and loss of tumor suppressor genes in mice form GBM tumors
Ligand-receptor autocrine and paracrine loops are commonly observed between EGFR and its ligands in GBMs (Ekstrand et al., 1991; Mishima et al., 1998; Tang et al., 1997). Having established that somatic overexpression of EGFRWT in the CNS of adult mice is insufficient to induce gliomagenesis (Zhu et al., 2009), we hypothesized that, similar to the clinical setting, the presence of an EGFR ligand is necessary for receptor activation and cellular transformation in vivo. Therefore, we developed a strategy to co-express EGFRWT and TGFα, an EGFR ligand expressed in human GBMs (Maruno et al., 1991; Ramnarain et al., 2006; Samuels et al., 1989; Schlegel et al., 1990; Tang et al., 1997; van der Valk et al., 1997; Yung et al., 1990) in the adult mouse brain using a conditional lox-stop-lox EGFRWT transgenic mouse strain (CAG-LSL-EGFRWT) in which overexpression of human EGFRWT is Cre-dependent (Zhu et al., 2009). This strain was crossed to the InkΔ2/3−/− constitutive Cdkn2a locus (p16Ink4a/p19Arf) knock out line (Serrano et al., 1996) and/or to a conditional PTEN2lox knock out strain (Lesche et al., 2002) to generate genetically accurate compound animals.
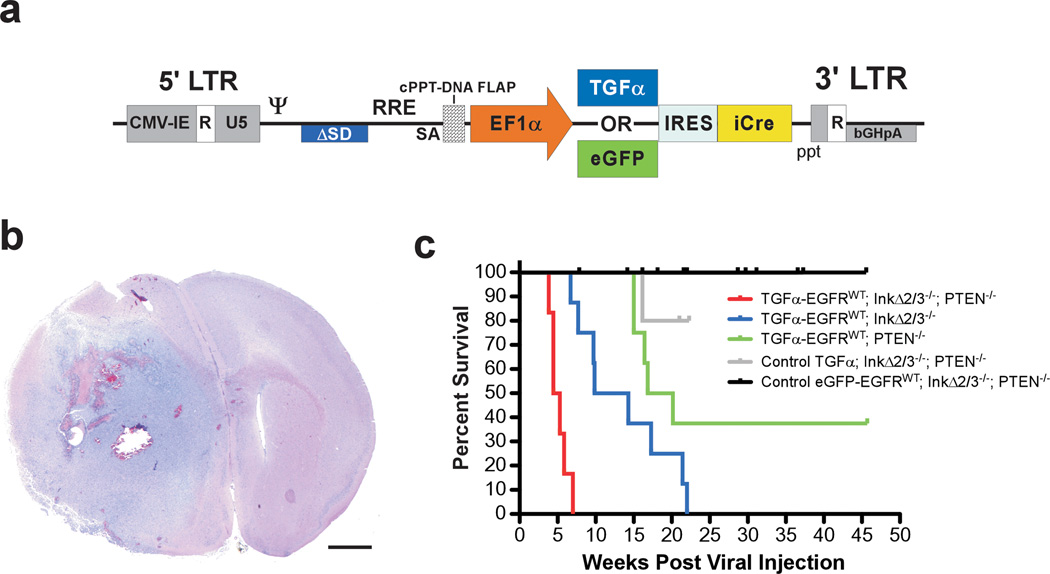
We created bicistronic lentiviral vectors designed to express TGFα and Cre recombinase (Figure 1a). For control experiments, we used eGFP in lieu of TGFα. We somatically induced the expression of EGFRWT with stereotactic intracranial injections of matched titers of the TGFα and eGFP viruses in cohorts of CAG-LSL-EGFRWT;InkΔ2/3−/−, CAG-LSL-EGFRWT;InkΔ2/3−/−;PTEN2lox and CAG-LSL-EGFRWT;PTEN2lox compound animals and monitored tumor formation and survival over time (Figure 1b,c). Mice co-expressing EGFRWT and TGFα in the context of p16Ink4a/p19Arf null conditions developed brain tumors with a median survival of 10 weeks post injection (Figure 1c). Additional loss of PTEN expression exacerbated this latency by half, suggesting a more aggressive tumor progression in the absence of the PTEN tumor suppressor gene (Figure 1c). Expression of TGFα-EGFRWT in the absence of PTEN also resulted in the formation of tumors albeit at a much-reduced penetrance and longer latency when compared to either p16Ink4a/p19Arf null and p16Ink4a/p19Arf;PTEN null backgrounds (Figure 1c). Finally, expression of EGFRWT in the absence of a ligand (eGFP- EGFRWT) under p16Ink4a/p19Arf;PTEN null conditions did not induce tumor formation in mice reaffirming the need for expression of an EGFR ligand for gliomagenesis.
Figure 1.
EGFRWT cooperates with loss of tumor suppressor genes to form brain tumors. (a) Schematic representation of the self-inactivating bicistronic lentiviral transducing vectors. These vectors carry a cassette composed of either the human TGFα cDNA or enhanced green fluorescent protein (eGFP) gene (for control experiments) followed by a human poliovirus1 internal ribosomal entry site (IRES) and improved Cre recombinase (iCre) cDNA (Shimshek et al., 2002) driven by the human elongation factor-1α (EF1α) promoter. The message is stabilized by the presence of the bovine growth hormone poly adenylation signal sequence. These lentiviral vectors were derived from a previously described vector (Coleman et al., 2003). The presence of a central polypurine tract (cPPT)-DNA FLAP element upstream of the multiple cloning site significantly improves the transduction efficiency in CNS tissues (Follenzi et al., 2000; Zennou et al., 2001). LTR; long terminal repeat. (b) Photomicrograph of an H&E-stained coronal section of a representative TGFα-EGFRWT;InkΔ2/3−/−;PTENlox brain tumor. Scale bar; 1.0 mm. (c) Survival (Kaplan-Meier) analysis of conditional EGFRWT mice. Cohorts of mice of the indicated genotypes were stereotactically injected in the striatum with titer-matched pTyf-TGFα-IRES-iCre or pTyf-eGFP-IRES-iCre viruses and monitored for survival over time.
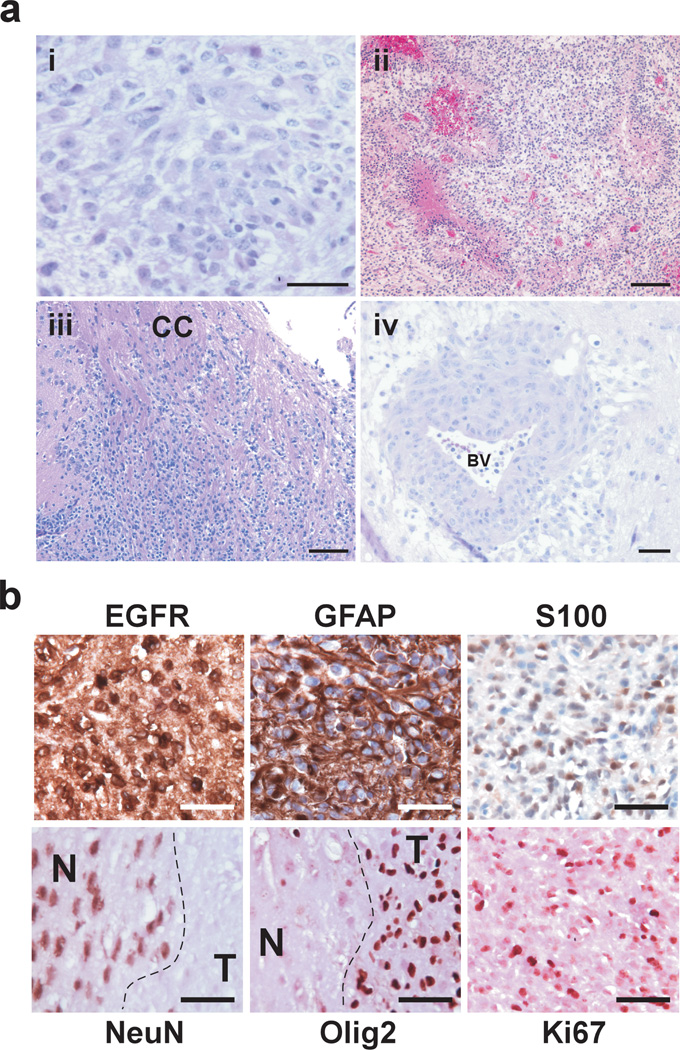
InkΔ2/3 null and InkΔ2/3;PTEN null TGFα-EGFRWT tumors resemble human GBMs histopathologically. All TGFα-EGFRWT tumors are composed of cells growing on a fibrillary background with prominent pleomorphic nuclei (Figure 2a–i). Tumors invariably display areas of pseudopallisading necrosis (Figure 2a–ii). In addition, tumor cells are highly infiltrative, showing invasion along white matter tracks (Figure 2a–iii) and the perivascular space distant from the bulk mass (Figure 2a–iv). Immunohistochemical staining of TGFα-EGFRWT tumors for human EGFR protein reveals a robust membrane expression. Staining for markers associated with astrocytic (GFAP and S100) and neuronal (NeuN) differentiation reveals that these tumor cells express markers of astrocytic lineage only (Figure 2b).
Figure 2.
Representative photomicrographs of H&E-stained histological sections of TGFα-EGFRWT tumors. (a) (i) Tumor cells are set on a fibrillary background and contain densely packed cells featuring pleomorphic nuclei with prominent nucleoli. (ii) Tumors exhibit marked pseudo pallisading necrosis. (iii-iv) The highly infiltrative nature of TGFα-EGFRWT tumor cells is depicted. (iii) Tumor cells migrate and infiltrate parenchyma along white matter tracks (corpus callosum CC) and (iv) migrate along blood vessels (BV) and invade the perivascular space. Scale bars; 50 µm (i,iv), 125 µm (ii, iii). (b) TGFα-EGFRWT tumors express markers of astrocytic differentiation. Representative photomicrographs of TGFα-EGFRWT tumors stained for the indicated cell lineage markers using immunohistochemistry. Tumors stain positive for the astrocytic lineage markers glial fibrillary acidic protein (GFAP) and S100 and negative for the neuronal lineage marker NeuN. Tumors also stain positive for human EGFR, the proliferation marker Ki67 and for Olig2. EGFR, GFAP and S100 sections were counterstained with hematoxylin and sections for the nuclear NeuN, Olig2 and Ki67 markers were counterstained with eosin. N, normal brain; T, tumor. Scale bars 50 µm.
Molecular classification of TGFα-EGFRWT GBM tumors defines two subtypes
To gain insight into molecular similarities between our model and the different subtypes of human GBMs, we carried out gene expression profiling on TGFα-EGFRWT;InkΔ2/3−/− and TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells and performed a gene set enrichment analysis (GSEA; (Subramanian et al., 2005)) using gene set lists defining the different human GBM subtypes. We also used hierarchical clustering (pvclust; (Suzuki and Shimodaira, 2006)) to further characterize the relationship between the mouse and human gene expression data. GSEA was used to examine the distribution of the markers for each human subtype in a list of genes arranged according to differential expression between TGFα-EGFRWT;InkΔ2/3−/− and TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor types. The most significant association was observed between the markers of the human classical subtype and high expression in TGFα-EGFRWT;InkΔ2/3−/− tumors (Supplementary Figure 1a and Supplementary table 3). Clustering analysis further supports this association (Supplementary Figure 1b). Gene expression data from our mouse GBMs were merged to the unified TCGA GBM data and the data were summarized to a single value per gene, per tumor group using averaging. The TGFα-EGFRWT;InkΔ2/3−/− tumor data clusters in a highly supported clade with human classical tumors. In addition, our results classify the TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumors as most similar to the mesenchymal subtype (Supplementary Figure 1b).
EGFR kinase inhibitor treatment of TGFα-EGFRWT GBM cells
EGFR kinase inhibitors are inefficient at conferring clinical benefits to GBM patients. To better understand the molecular basis for TKI resistance in GBM, we evaluated tumor cell growth and signaling events under EGFR kinase inhibition in our GBM mouse model. We derived a series of primary tumor cultures from InkΔ2/3 null and InkΔ2/3;PTEN null TGFα-EGFRWT tumors. These primary cultures are stable, can proliferate in low serum concentrations and are capable of unrestricted growth when orthotopically implanted as allografts (data not shown).
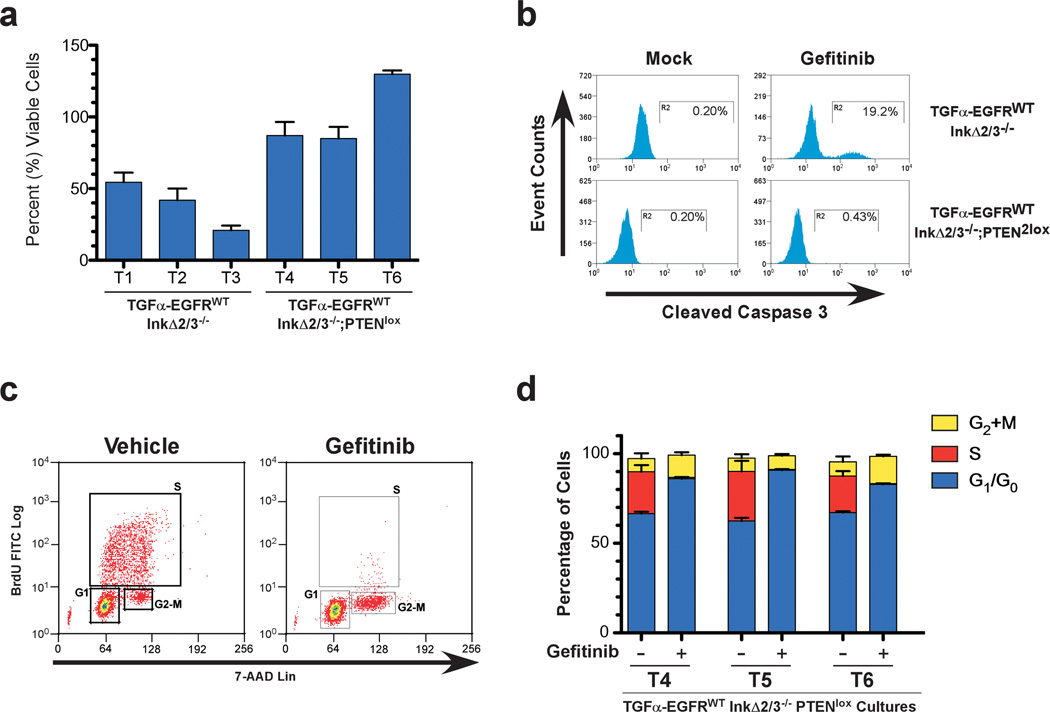
Primary cultures of GBMs were incubated in low serum (0.1% FBS) media and treated either with vehicle or with the EGFR kinase inhibitor gefitinib and analyzed for viable cell growth (Figure 3a). After 24 hours of treatment, the TGFα-EGFRWT;InkΔ2/3−/− tumor cultures (T1, T2, and T3) displayed a 50–80% reduction in number of cells whereas cells derived from TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumors (T4, T5, and T6) did not show significant reductions in viability (Figure 3a). To better understand these effects, we analyzed gefitinib-treated cells for apoptosis with the apoptotic marker cleaved caspase-3 using flow cytometry. The levels of cleaved caspase-3 increased dramatically after 24 hours of gefitinib treatment in TGFα-EGFRWT;InkΔ2/3−/− cells and remained low in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cultures (Figure 3b,c). We validated these results with the EGFR TKI erlotinib and observed similar results (data not shown). These results demonstrate that EGFR TKI treatment of TGFα-EGFRWT;InkΔ2/3−/− tumor cells is cytotoxic whereas identical treatment in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cultures is cytostatic.
Figure 3.
Cytostaticity of EGFR kinase inhibition in TGFα-EGFRWT GBM tumor cells. (a) TGFα-EGFRWT;InkΔ2/3−/− tumor cells are sensitive to gefitinib treatment whereas TGFα-EGFRWT;InkΔ2/3−/−;PTENlox cells are resistant. Viability assays of three independent tumor cell cultures from the indicated genotypes after vehicle or gefitinib treatment (10 µM) for 24 hours. Data is plotted as percentage of viable cells of treated over vehicle treatment (mean ± SD; n=3 in each group). (b) Representative flow-cytometric analysis of vehicle- and gefitinib-treated TGFα-EGFRWT;InkΔ2/3−/− and TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM tumor cells indicating an increase in cleaved caspase-3 positive cells upon EGFR kinase inhibitor treatment. (c) Primary tumor cultures from TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBMs were incubated overnight in low serum (0.1% FBS) media then challenged with 10 µM of gefitinib for 24 hours. BrdU incorporation and 7-AAD levels were measured by flow cytometry to determine cell cycle profiles for treated and untreated cultures. Representative plots for BrdU incorporation vs. 7-AAD levels for determination of cell cycle profiles in vehicle and gefitinib-treated cells are shown. (d) Graphical representation of the distribution (percentage) of cells in G2+M, S, or G1/G0 for TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM cultures T4, T5, and T6. The small percentage of cells in sub-G1 is not included in this graph. (mean ± SD; n=3 in each group).
We characterized this cytostatic response by performing a cell cycle analysis on gefitinib-treated TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells. Cells were treated as above and flow cytometry was used to assess the extent of BrdU incorporation and the levels of 7-AAD staining. EGFR inhibition induces a dramatic decrease in the percentage of cells in S phase after 24 hour treatment, which is accompanied by an increase in the percentage of cells in G1/G0 as well as a smaller increase in the percentage of cells in G2+M phases (Figure 3d). Thus in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM cells, inhibition of EGFR mostly induces a G1 arrest. We further analyzed TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cultures treated with gefitinib for the activation status of canonical MAPK (Mek1/2 and Erk1/2), PI3K/Akt and mTOR pathways by immunoblot analysis (Supplementary Figure 2). Treatment with gefitinib resulted in a robust attenuation of the MAPK signaling pathway and to a much lesser extent that of the PI3K/Akt and mTORC1 signaling pathways.
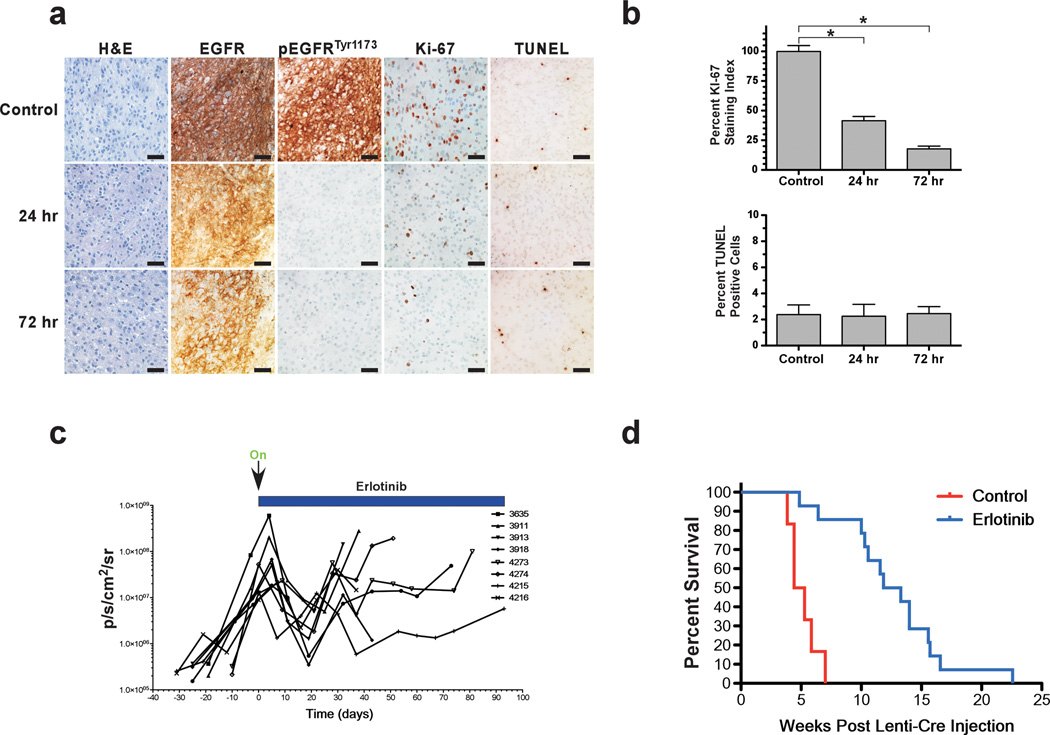
We then ascertained the effect of EGFR inhibition treatment on TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumors in vivo. To allow for real-time monitoring of tumor initiation and growth, we retrogressed a conditional luciferase reporter transgene (Woolfenden et al., 2009) into our TGFα-EGFRWT;InkΔ2/3−/−;PTEN2lox strain. GBM tumors were initiated as above and monitored for growth using bioluminescence imaging (BLI). Once GBM tumors were established, animals were administered with vehicle or erlotinib 150 mg/kg p.o. and sacrificed 24 and 72 hours later and processed for histology to determine the extent of EGFR inhibition in vivo. Figure 4a shows that 24 hours after a single dose of erlotinib, the levels of activated EGFR (as measured by the levels of phosphotyrosine 1173) decrease to undetectable levels when compared to control treatment whereas the levels of total EGFR remained identical. This inhibition continued for another 72 hours and is accompanied by a decrease in proliferation as measured by lower levels of Ki-67 positive cells with no change in apoptosis as measured by TUNEL staining (Figure 4a,b).
Figure 4.
Inhibition of EGFR prolongs survival in mice. (a) Treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox mice with the EGFR TKI erlotinib inhibits EGFR activity. Mice were treated with erlotinib formulated in Ora-Plus (Paddock Lab) and administered by oral gavage daily Monday to Friday at 150mg/kg dose and sacrificed at the indicated times and processed for histology. IHC of representative GBM tumors for total human EGFR expression, activated EGFR (pTyr1173), the proliferation marker Ki-67 and for apoptosis by TUNEL are shown. (b) Graphical representation of the quantification of proliferation assayed by Ki-67 staining and for the percentage of apoptotic cells as measured by the number of TUNEL positive cells. The Ki-67 staining data is presented as percentage of Ki-67 positive cells in treated tumors over control tumors (*p<0.001, two-tailed t-test) and quantification of apoptosis is presented as percentage of TUNEL positive cells per field of view. (c) Bioluminescence imaging monitoring of treatment course. EGFRWT;InkΔ2/3−/−;PTEN2lox mice were injected with pTyf-TGFα-IRES-iCre lentivirus and 14 days later were imaged for presence of tumor. Once tumors were detected, animals were imaged once a week. Initiation of treatment with erlotinib commenced once BLI values surpassed 1.0 × 107 photons/sec/cm2/sr. Animals were treated with erlotinib on a schedule of five days on 2 days off and monitored for response using BLI (d) Animals remained on this treatment until moribund to establish overall survival. Kaplan-Meier survival curves of control (red, n=6) and erlotinib-treated (blue, n=14) tumor bearing animals. The difference in survival is significant, p<0.0001, Log-rank (Mantel-Cox) test.
To determine the clinical outcome of EGFR TKI therapy, we established cohorts of tumor bearing animals as above and initiated treatment once GBM development was detected by BLI. Animals treated with erlotinib initially responded to the drug as visualized by a decrease in BLI output (Figure 4c). The response subsided and tumor growth eventually resumed and proved to be fatal. This initial response translated into a prolongation in the overall survival of the treated cohort (Figure 4d). To control for the responsiveness of the system, we initiated and then suspended erlotinib treatment in a TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor bearing mouse and monitored tumor response by BLI (Supplementary Figure 3). Withdrawal of erlotinib treatment resulted in a rapid resumption of tumor growth that could be attenuated by recommencement of treatment (Supplementary Figure 3).
Re-expression of PTEN in established tumor cells has no therapeutic consequences
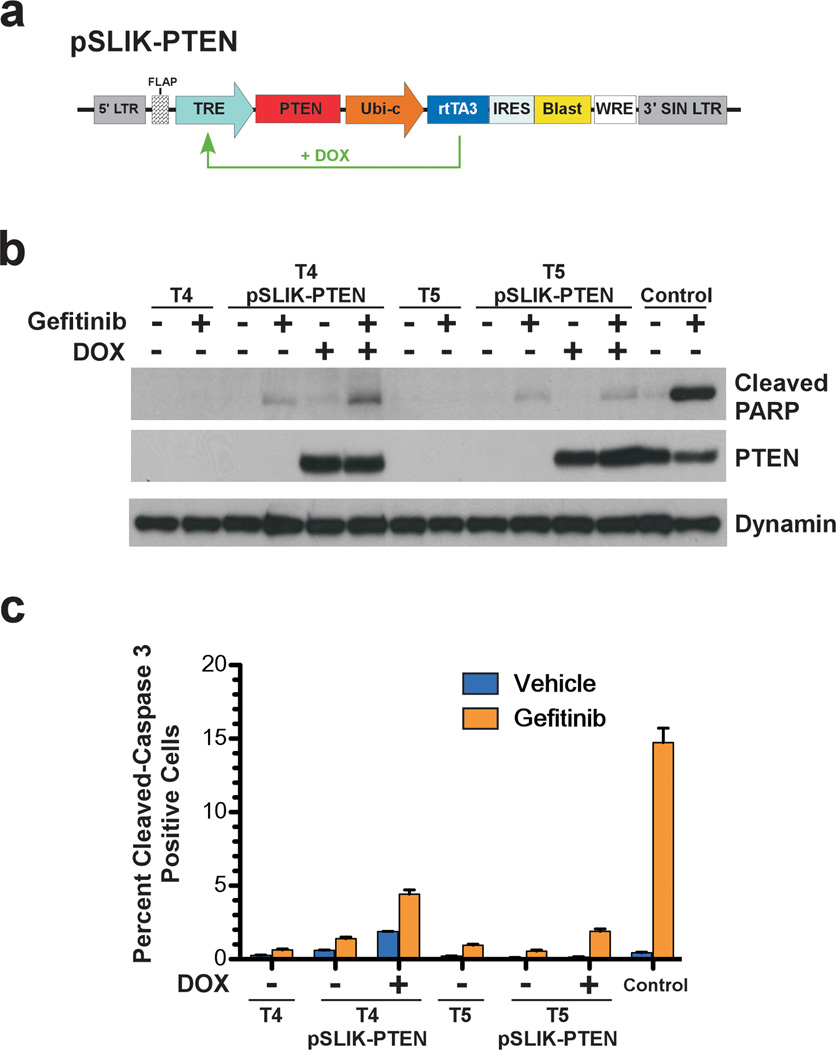
Loss of PTEN in GBM is thought to confer resistance to EGFR TKI treatment by uncoupling the dependence of PI3K activity from EGFR (Mellinghoff et al., 2007; Mellinghoff et al., 2005). Similar to these clinical observations, we demonstrated that our PTEN-null GBM tumor cells are resistant to EGFR TKI treatment. To determine if PTEN directly functions in this resistance, we used a tetracycline-inducible lentiviral vector system (Shin et al., 2006; Zhu et al., 2007) to conditionally re-express PTEN in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells and assayed for sensitivity to EGFR TKI treatment (Figure 5a). Parental and daughter cultures were treated with doxycycline to induce PTEN expression and treated with gefitinib as described above. Immunoblot analysis of total cell lysates for cleaved PARP demonstrates that doxycycline-induced PTEN expression does not confer sensitivity to EGFR kinase inhibitor-induced apoptosis (Figure 5b) and flow cytometry analysis of cleaved caspase-3 further confirms these results (Figure 5c). This demonstrates that re-expression of PTEN in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBMs, post tumor formation, does not sensitize them to gefitinib-induced apoptosis.
Figure 5.
PTEN re-expression in GBM tumor cells does not confer sensitivity to EGFR kinase inhibition. (a) Schematic representation of the pSLIK-PTEN lentiviral vector for Tet-inducible expression of PTEN. The original pSLIK vector (Shin et al., 2006; Zhu et al., 2007) was modified to confer blasticidine resistance as described in Supplementary Information. (b) Immunoblot analysis of total cell lysates from PTEN deficient GBM tumor cell cultures (T4 & T5) re-expressing PTEN probed for cleaved PARP. Control cultured cells sensitive to gefitinib-induced apoptosis are derived from a TGFα-EGFRWT;InkΔ2/3−/− tumor. (c) Graphical representation of flow cytometry analysis of cleaved caspase-3 in PTEN deficient and isogenic PTEN re-expressing GBM tumor cell cultures (T4 & T5) (mean ± SD; n=3 in each group).
EGFR kinase inhibition induces gene expression profile changes in InkΔ2/3;PTEN-null GBMs
To gain insight into the observed cytostaticity, we performed gene expression profiles of vehicle- and gefitinib-treated TGFα-EGFRWT;InkΔ2/3−/− and TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells. Unsupervised hierarchical clustering of a 1000 random genes demonstrated that global gene expression remained unchanged in control versus gefitinib-treated TGFα-EGFRWT;InkΔ2/3−/− tumor cells (Supplementary Figure 4a). Pathway analysis from the gene profiling data failed to identify apoptosis-specific signature pathway utilization (data not shown) suggesting that the rapid onset of apoptosis observed in TGFα-EGFRWT;InkΔ2/3−/− tumor cells is less likely to be transcriptionally mediated. Conversely, treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells with gefitinib triggered significant changes in global gene expression (Supplementary Figure 4a). All three gefitinib-treated TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cultures clustered together rather than clustering pair-wise with their vehicle treated controls. These extensive changes in gene expression allowed us to hypothesize that the mechanism(s) responsible for the observed gefitinib-induced cytostaticity might be reflected at the level of transcription of one or several genes. First, we validated the array results on a subset of 24 of those genes (10 downregulated and 14 upregulated, Supplementary Table 4) by qRT-PCR assays performed on newly isolated RNA from gefitinib- and vehicle-treated cells (Supplementary Figure 5a,b). Then, we focused our attention on genes whose expression is most significantly up or down regulated upon gefitinib treatment (Supplementary Figure 4b).
Activation of c-met expression in response to EGFR inhibition leads to survival of GBM tumor cells
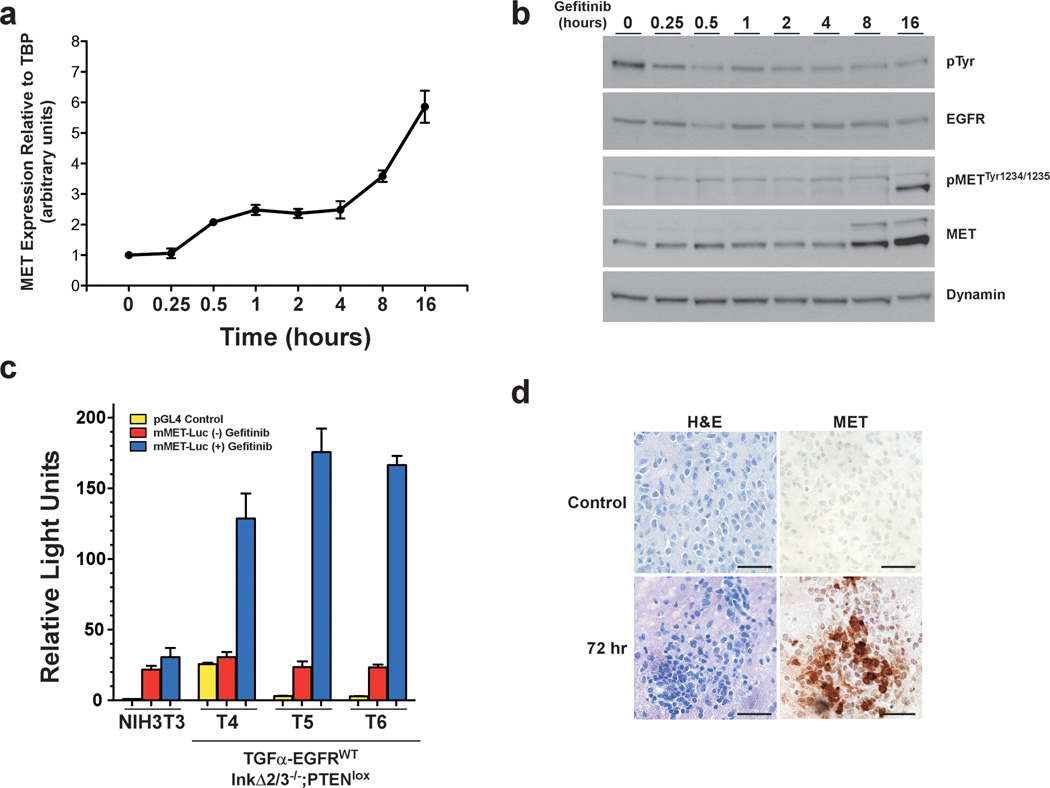
We surveyed the genes with the most significant changes in expression levels upon gefitinib treatment and concentrated on the c-met receptor tyrosine kinase (RTK) gene. We treated TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cell cultures with gefitinib for 16 hours and harvested total RNA at different times and performed qRT-PCR to measure the relative expression levels of c-met mRNA over time (Figure 6a). Our results demonstrate a biphasic increase in the c-met mRNA levels upon gefitinib treatment. Within 30 min of treatment the levels of c-met mRNA doubled and stayed constant for 3.5 hours, after which the levels increased to over 5 folds after 16 hours. This latter increase in c-met mRNA levels corresponded to the appearance of detectable levels of activated MET receptors (increase in MET autophosphorylation sites Tyr1234/1235 levels) (Figure 6b). We also determined that this induction in MET expression upon gefitinib treatment is irrespective of PTEN status (Supplementary Figure 6).
Figure 6.
Gefitinib treatment increases expression and activation of c-Met in PTEN deficient GBM tumor cells. (a) Representative qRT-PCR from total RNA isolated from a TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor culture (T5) treated with gefitinib (10 µM) for the indicated time. (b) Immunoblot of total cell lysates isolated from cells as in (a) and probed using antibodies against the indicated proteins. (c) Graphical representation of luciferase reporter assay results. A 3.5 kb fragment of the mouse c-Met promoter was used to drive the expression of firefly luciferase. Control plasmid (pGL4.10[luc2]) was used as a negative control. Firefly luciferase plasmids were co-transfected with TK-renilla luciferase internal transfection efficiency control plasmid. Cells were treated with gefitinib (10 µM) for 24 hours and luciferase outputs measured using a luminometer. Relative Light Units represent the ratio of firefly/renilla luc (mean ± SD; n=3 in each group). (d) Representative H&E and IHC against MET on tumor brain sections from treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor-bearing mice with EGFR TKI (erlotinib, 150 mg/kg).
Next, we investigated whether the increase in c-met mRNA levels upon gefitinib treatment resulted from an enhanced transcription of the c-met gene by using a 3.5 kilobase (kb) fragment of the c-met promoter region (Liang et al., 2004; Seol and Zarnegar, 1998) in a firefly luciferase reporter gene assay. c-met promoter and control luciferase vectors were transiently expressed in control NIH3T3 cells and in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cell cultures (T4–6), treated with vehicle or gefitinib and assayed for luciferase activity 24 hours after transfection. Treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cell cultures with gefitinib lead to a significant increase in the activity of the c-met promoter (Figure 6c). Finally, we validated these observations in vivo by performing IHC against MET on GBM tumor sections from mice that have been treated with erlotinib. Figure 6d demonstrates that treatment of GBM tumor-bearing mice with the EGFR TKI erlotinib resulted in the expression of MET tyrosine kinase 72 hours post treatment.
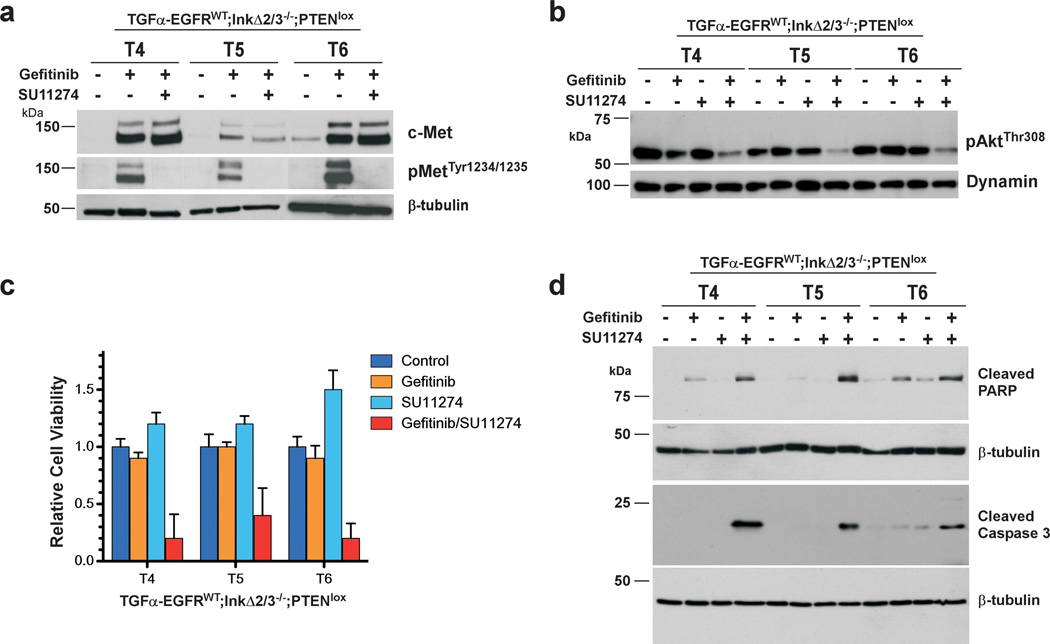
Our work indicated that this gefitinib-induced increase in MET expression and activity is responsible for sustaining a pro-survival Akt-based signaling. As such, we reasoned that co-treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells with gefitinib and the MET inhibitor SU11274 may sensitize these cells to apoptosis. Treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cell cultures with both gefitinib and SU11274 robustly abrogated the levels of phospho MET Tyr1234/1235, indicating a complete inhibition of MET activity (Figure 7a). Inhibition of MET activity paralleled a reduction in the activity of Akt as measured by a decline in the levels of phospho Akt Thr308 (Figure 7b). To directly address the role of MET activation on survival of gefitinib-treated TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells, we evaluated the effects of co-inhibition of EGFR and MET kinase on cell viability. Dual treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox tumor cells with gefitinib and SU11274 effectively decreased cell viability compared to gefitinib or SU11274 treatment alone (Figure 7c). This decrease in viability was accompanied by an increase in the levels of cleaved caspase-3 and cleaved PARP suggesting that dual EGFR and MET inhibition resulted in the induction of apoptosis (Figure 7d). Together, these results demonstrate that inhibition of EGFR kinase activity channels signaling pathways to induce MET kinase expression and activation, which in turn triggers the activation of survival signaling events that counteract the actions of EGFR kinase inhibition.
Figure 7.
Inhibition of c-MET sensitizes PTEN deficient tumor cells to EGFR TKI-induced apoptosis. PTEN deficient TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM tumor cells were treated with gefitinib (10 µM) and/or the c-MET kinase inhibitor SU11274 (10 µM). (a) Immunoblot analysis of total cell lysates from TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM tumor cells treated with the indicated inhibitors using c-MET and c-MET phosphotyrosine 1234,1235 antibodies. Anti β-tubulin probing is used as an internal loading control. (b) Activation of Akt is attenuated by inhibition of c-MET. Immunoblot analysis of total cell lysates against phospho Akt (Thr308) in TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM tumor cells. Anti dynamin probing is used as an internal loading control. (c,d) Inhibition of c-MET induces apoptosis. Co-treatment of TGFα-EGFRWT;InkΔ2/3−/−;PTENlox GBM tumor cells with gefitinib and SU11274 causes a marked reduction in the relative viability of these cells (c). This reduced viability corresponds to an increase in the rate of drug-induced apoptosis (d) as demonstrated by immunoblot analysis of cleaved caspase 3 and cleaved PARP.
Discussion
Several in vitro studies have unequivocally established that overexpression of wild-type EGFR results in cellular transformation solely in the presence of ligands (Ciardiello et al., 1990; Di Fiore et al., 1987; Di Marco et al., 1990; Di Marco et al., 1989; Rosenthal et al., 1986; Shankar et al., 1989; Watanabe et al., 1987). This dual requirement for receptor and ligand expression to elicit oncogenic transformation by EGFR is further exemplified in animal models where receptor overexpression alone in various tissues, including glia, does not result in tumor formation (Andl et al., 2003; Cheng et al., 2002; Ding et al., 2003). The evidence thus suggests that physiologically relevant overexpression of wild-type EGFR is an oncogenic event only in the presence of ligands. Here we demonstrate that sustained, TGFα-mediated activation of EGFR is necessary for gliomagenesis in vivo, functionally substantiating the clinical observation that EGFR-positive GBMs also express EGFR ligands (Maruno et al., 1991; Ramnarain et al., 2006; Samuels et al., 1989; Schlegel et al., 1990; Tang et al., 1997; van der Valk et al., 1997; Yung et al., 1990).
CNS expression of TGFα-EGFRWT and loss of Cdkn2a in mice lead to the formation of neoplasms with molecular and histopathological characteristics of the classical subtype of GBMs. The additional loss of PTEN resulted in tumor with mesenchymal molecular features. Although the majority of EGFR positive GBMs belong to the classical subtype, there are a considerable number of mesenchymal GBMs that are EGFR positive (Verhaak et al., 2010). Our model suggests that those are likely to be PTEN deficient. The newly established molecular stratification of GBMs reveals that response to treatments varies based on tumor subtypes. We show that the GBM tumors from our models behave similarly, where classical GBMs are sensitive to EGFR TKI therapy and mesenchymal tumors are not. Our genetically-defined pre-clinical model systems thus offer a powerful means to study responses to new therapies according to GBM subtypes.
We show that p16Ink4a/p19Arf null TGFα-EGFRWT tumor cells are sensitive to EGFR inhibition, undergoing rapid apoptosis upon gefitinib treatment. This is a response that is reminiscent of EGFR-addicted non-small cell lung cancers (NSCLC) (reviewed in (Engelman and Settleman, 2008)) and suggests that under Cdkn2a null conditions, GBMs are addicted to EGFR signaling. TGFα-EGFR;InkΔ2/3−/−;PTEN null tumors on the other hand displayed a cytostatic response both in vitro and in vivo under the same treatment. Cytostatic responses resulting from EGFR inhibition in cancer have been reported to be mediated by a p27KIP1-dependent G1 arrest (Busse et al., 2000; Ling et al., 2007; Peng et al., 1996). Although the molecular mechanisms that link EGFR inhibition to induction of p27KIP1 remain ill defined, it is conceivable that the combination of the sudden loss in the proliferative MAPK signaling and a sustained Akt pro-survival activity resulting from the PTEN null status induces p27KIP1 in our system.
On the surface, the cytostatic response to EGFR inhibition in our GBMs is analogous to the resistance to TKI therapy displayed by PTEN null GBM patients (Fan et al., 2007; Guillamo et al., 2009; Mellinghoff et al., 2007; Mellinghoff et al., 2005; Sarkaria et al., 2007). This is not the case since re-expressing PTEN in GBM cells did not change their response to EGFR inhibition nor did it sensitize them to gefitinib-induced apoptosis. Instead, our gene expression profiling experiments revealed that the cytostatic effect we observed was characterized by global changes in gene expression of which, MET was identified as an integral component.
The transcriptional induction of the c-met gene as a result of growth factor inhibition has never been described and it remains to be observed in GBM patients who underwent EGFR TKI treatments. The paucity and difficult access to these samples makes such studies difficult to carry out. However, co-expression of MET and EGFR has been reported in GBM, NSCLC and breast cancer and suggested to play an important role in these cancers’ ability to escape single agent targeted therapy (Engelman et al., 2007; Huang et al., 2007; Mueller et al., 2008; Mueller et al., 2010; Stommel et al., 2007; Turke et al., 2010).
In NSCLC, recent studies demonstrated that erlotinib resistance arises from acquisition of MET expression after prolonged exposure to EGFR TKI thus defining a mode of secondary or acquired resistance to TKI treatment (Engelman et al., 2007; Turke et al., 2010). In contrast, breast cancer and glioma studies define an upfront or primary mode of resistance to EGFR TKI treatment where MET expression and activation is already present prior to EGFR TKI exposure (Mueller et al., 2008; Mueller et al., 2010; Stommel et al., 2007). Although the mechanistic details of the resistance described herein differ from those examples, together they nevertheless underscore a potent selection pressure for RTK switching during the development of resistance to targeted therapeutic treatment in tumors.
This study reports a novel mode of resistance to EGFR targeted therapy in GBM. We demonstrated that in a genetically accurate mouse model of GBM, resistance to EGFR kinase inhibition is a dynamic process involving rapid and drastic changes in gene expression profiles of which, induction of the c-met gene bears most of the resistance phenotype. It remains to be determined whether a subset of patients would benefit from dual MET and EGFR TKI treatment. Our work supports the notion that up front, multi-target therapy represents a compelling approach towards the treatment of GBM and that the pre-clinical mouse models presented here offer a unique platform to decipher and exploit the mechanisms of action of therapeutic agents.
Materials and Methods
EGFR conditional mice and procedures
All mouse procedures were performed in accordance with Tufts University’s recommendations for the care and use of animals and were maintained and handled under protocols approved by the Institutional Animal Care and Use Committee. Cre/Lox-mediated conditional expression of the human EGFRWT was described elsewhere (Zhu et al., 2009). Stereotactic intracranial injections of viruses and BLI monitoring are described in details in the Supplementary Materials and Methods section.
Histology and immunodetection
Brains were excised and post-fixed and processed for histology as described in Supplementary Materials and Methods. Immunodetection of markers of astrocytic and neuronal differentiation was performed by IHC and immunoblots were performed using standard protocols using antibodies listed in Supplementary Tables 1 and 2 and as described in details in Supplementary Materials and Methods.
Survival assays and inhibitor treatments
Cell viability was measured by trypan blue exclusion assay as described in Supplementary Materials and Methods. Drug (gefitinib (LC Labs) and SU11274 (Selleck chemicals LLC)) were added at the indicated final concentrations and incubated for 24 hours before the total number of viable cells remaining post-treatment was determined by counting and reported as percent of control treated.
Gene promoter reporter assays
The relative activity of the c-met promoter was assayed by transient co-transfection of mMet-Luc and TK-renilla Luc plasmids in cells, incubated overnight followed by 24 hours of vehicle or gefitinib. Cell lysates were harvested and assayed in a luminometer using a dual glow luciferase kit (Promega). Promoter activity was calculated as the ratio of firefly luciferase over renilla luciferase and is represented as Relative Light Units.
Supplementary Material
Acknowledgments
We thank Drs. Bill Chiu and Ken Hung for helpful comments and Dr. Robert R. Langley (MD Anderson Cancer Center, Houston, Texas) for human TGFα cDNA. Dr. Jason Coleman (MIT, Cambridge, MA) for the pTyf vector and iCre cDNA. Dr. John Alberta (DFCI, Boston, MA) for the Olig2 antibody. This work was supported by NIH grant NCI U01 CA141556 (A.C. and F.W.), by American Cancer Society Research Scholar Award 117409 (A.C.), by NIH grant U54 CA119349 (D.H. and A.C.), by NINDS P30 NS047243 (L.I.) and NCI T32 CA009429 (D.C.).
Footnotes
The authors have declared that no conflict of interest exists.
References
- Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse D, Doughty RS, Ramsey TT, Russell WE, Price JO, Flanagan WM, et al. Reversible G(1) arrest induced by inhibition of the epidermal growth factor receptor tyrosine kinase requires up-regulation of p27(KIP1) independent of MAPK activity. J Biol Chem. 2000;275:6987–6995. doi: 10.1074/jbc.275.10.6987. [DOI] [PubMed] [Google Scholar]
- Cheng J, Huang H, Zhang ZT, Shapiro E, Pellicer A, Sun TT, et al. Overexpression of epidermal growth factor receptor in urothelium elicits urothelial hyperplasia and promotes bladder tumor growth. Cancer Res. 2002;62:4157–4163. [PubMed] [Google Scholar]
- Ciardiello F, McGeady ML, Kim N, Basolo F, Hynes N, Langton BC, et al. Transforming growth factor-alpha expression is enhanced in human mammary epithelial cells transformed by an activated c-Ha-ras protooncogene but not by the c-neu protooncogene, and overexpression of the transforming growth factor-alpha complementary DNA leads to transformation. Cell Growth Differ. 1990;1:407–420. [PubMed] [Google Scholar]
- Coleman JE, Huentelman MJ, Kasparov S, Metcalfe BL, Paton JF, Katovich MJ, et al. Efficient large-scale production and concentration of HIV-1-based lentiviral vectors for use in vivo. Physiol Genomics. 2003;12:221–228. doi: 10.1152/physiolgenomics.00135.2002. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Pierce JH, Fleming TP, Hazan R, Ullrich A, King CR, et al. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 1987;51:1063–1070. doi: 10.1016/0092-8674(87)90592-7. [DOI] [PubMed] [Google Scholar]
- Di Marco E, Pierce JH, Aaronson SA, Di Fiore PP. Mechanisms by which EGF receptor and TGF alpha contribute to malignant transformation. Nat Immun Cell Growth Regul. 1990;9:209–221. [PubMed] [Google Scholar]
- Di Marco E, Pierce JH, Fleming TP, Kraus MH, Molloy CJ, Aaronson SA, et al. Autocrine interaction between TGF alpha and the EGF-receptor: quantitative requirements for induction of the malignant phenotype. Oncogene. 1989;4:831–838. [PubMed] [Google Scholar]
- Ding H, Shannon P, Lau N, Wu X, Roncari L, Baldwin RL, et al. Oligodendrogliomas result from the expression of an activated mutant epidermal growth factor receptor in a RAS transgenic mouse astrocytoma model. Cancer Res. 2003;63:1106–1113. [PubMed] [Google Scholar]
- Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Fan QW, Cheng CK, Nicolaides TP, Hackett CS, Knight ZA, Shokat KM, et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res. 2007;67:7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- Friedman HS, Bigner DD. Glioblastoma multiforme and the epidermal growth factor receptor. N Engl J Med. 2005;353:1997–1999. doi: 10.1056/NEJMp058186. [DOI] [PubMed] [Google Scholar]
- Guillamo JS, de Bouard S, Valable S, Marteau L, Leuraud P, Marie Y, et al. Molecular mechanisms underlying effects of epidermal growth factor receptor inhibition on invasion, proliferation, and angiogenesis in experimental glioma. Clin Cancer Res. 2009;15:3697–3704. doi: 10.1158/1078-0432.CCR-08-2042. [DOI] [PubMed] [Google Scholar]
- Huang PH, Cavenee WK, Furnari FB, White FM. Uncovering therapeutic targets for glioblastoma: a systems biology approach. Cell Cycle. 2007;6:2750–2754. doi: 10.4161/cc.6.22.4922. [DOI] [PubMed] [Google Scholar]
- Huang TT, Sarkaria SM, Cloughesy TF, Mischel PS. Targeted therapy for malignant glioma patients: lessons learned and the road ahead. Neurotherapeutics. 2009;6:500–512. doi: 10.1016/j.nurt.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- Liang H, O'Reilly S, Liu Y, Abounader R, Laterra J, Maher VM, et al. Sp1 regulates expression of MET, and ribozyme-induced down-regulation of MET in fibrosarcoma-derived human cells reduces or eliminates their tumorigenicity. Int J Oncol. 2004;24:1057–1067. [PubMed] [Google Scholar]
- Ling YH, Li T, Yuan Z, Haigentz M, Jr, Weber TK, Perez-Soler R. Erlotinib, an effective epidermal growth factor receptor tyrosine kinase inhibitor, induces p27KIP1 up-regulation and nuclear translocation in association with cell growth inhibition and G1/S phase arrest in human non-small-cell lung cancer cell lines. Mol Pharmacol. 2007;72:248–258. doi: 10.1124/mol.107.034827. [DOI] [PubMed] [Google Scholar]
- Maruno M, Kovach JS, Kelly PJ, Yanagihara T. Transforming growth factor-alpha, epidermal growth factor receptor, and proliferating potential in benign and malignant gliomas. J Neurosurg. 1991;75:97–102. doi: 10.3171/jns.1991.75.1.0097. [DOI] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Cloughesy TF, Mischel PS. PTEN-mediated resistance to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res. 2007;13:378–381. doi: 10.1158/1078-0432.CCR-06-1992. [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Mishima K, Higashiyama S, Asai A, Yamaoka K, Nagashima Y, Taniguchi N, et al. Heparin-binding epidermal growth factor-like growth factor stimulates mitogenic signaling and is highly expressed in human malignant gliomas. Acta Neuropathol. 1998;96:322–328. doi: 10.1007/s004010050901. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KL, Yang ZQ, Haddad R, Ethier SP, Boerner JL. EGFR/Met association regulates EGFR TKI resistance in breast cancer. J Mol Signal. 2010;5:8. doi: 10.1186/1750-2187-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Fan Z, Lu Y, DeBlasio T, Scher H, Mendelsohn J. Anti-epidermal growth factor receptor monoclonal antibody 225 up-regulates p27KIP1 and induces G1 arrest in prostatic cancer cell line DU145. Cancer Res. 1996;56:3666–3669. [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- Rosenthal A, Lindquist PB, Bringman TS, Goeddel DV, Derynck R. Expression in rat fibroblasts of a human transforming growth factor-alpha cDNA results in transformation. Cell. 1986;46:301–309. doi: 10.1016/0092-8674(86)90747-6. [DOI] [PubMed] [Google Scholar]
- Samuels V, Barrett JM, Bockman S, Pantazis CG, Allen MB., Jr Immunocytochemical study of transforming growth factor expression in benign and malignant gliomas. Am J Pathol. 1989;134:894–902. [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- Schlegel U, Moots PL, Rosenblum MK, Thaler HT, Furneaux HM. Expression of transforming growth factor alpha in human gliomas. Oncogene. 1990;5:1839–1842. [PubMed] [Google Scholar]
- Seol DW, Zarnegar R. Structural and functional characterization of the mouse c-met proto-oncogene (hepatocyte growth factor receptor) promoter. Biochim Biophys Acta. 1998;1395:252–258. doi: 10.1016/s0167-4781(97)00202-9. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Shankar V, Ciardiello F, Kim N, Derynck R, Liscia DS, Merlo G, et al. Transformation of an established mouse mammary epithelial cell line following transfection with a human transforming growth factor alpha cDNA. Mol Carcinog. 1989;2:1–11. doi: 10.1002/mc.2940020102. [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Tang P, Steck PA, Yung WK. The autocrine loop of TGF-alpha/EGFR and brain tumors. J Neurooncol. 1997;35:303–314. doi: 10.1023/a:1005824802617. [DOI] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk P, Lindeman J, Kamphorst W. Growth factor profiles of human gliomas Do non-tumour cells contribute to tumour growth in glioma. Ann Oncol. 1997;8:1023–1029. doi: 10.1023/a:1008265905505. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Lazar E, Sporn MB. Transformation of normal rat kidney (NRK) cells by an infectious retrovirus carrying a synthetic rat type alpha transforming growth factor gene. Proc Natl Acad Sci U S A. 1987;84:1258–1262. doi: 10.1073/pnas.84.5.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfenden S, Zhu H, Charest A. A Cre/LoxP conditional luciferase reporter transgenic mouse for bioluminescence monitoring of tumorigenesis. Genesis. 2009;47:659–666. doi: 10.1002/dvg.20545. [DOI] [PubMed] [Google Scholar]
- Yung WK, Zhang X, Steck PA, Hung MC. Differential amplification of the TGF-alpha gene in human gliomas. Cancer Commun. 1990;2:201–205. doi: 10.3727/095535490820874416. [DOI] [PubMed] [Google Scholar]
- Zennou V, Serguera C, Sarkis C, Colin P, Perret E, Mallet J, et al. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat Biotechnol. 2001;19:446–450. doi: 10.1038/88115. [DOI] [PubMed] [Google Scholar]
- Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Santat LA, Chang MS, Liu J, Zavzavadjian JR, Wall EA, et al. A versatile approach to multiple gene RNA interference using microRNA-based short hairpin RNAs. BMC Mol Biol. 2007;8:98. doi: 10.1186/1471-2199-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.