Abstract
Colorectal cancer (CRC) is a major cause of cancer mortality. Whereas some patients respond well to therapy, others do not, and thus more precise, individualized treatment strategies are needed. To that end, we analyzed gene expression profiles from 1,290 CRC tumors using consensus-based unsupervised clustering. The resultant clusters were then associated with therapeutic response data to the epidermal growth factor receptor–targeted drug cetuximab in 80 patients. The results of these studies define six clinically relevant CRC subtypes. Each subtype shares similarities to distinct cell types within the normal colon crypt and shows differing degrees of ‘stemness’ and Wnt signaling. Subtype-specific gene signatures are proposed to identify these subtypes. Three subtypes have markedly better disease-free survival (DFS) after surgical resection, suggesting these patients might be spared from the adverse effects of chemotherapy when they have localized disease. One of these three subtypes, identified by filamin A expression, does not respond to cetuximab but may respond to cMET receptor tyrosine kinase inhibitors in the metastatic setting. Two other subtypes, with poor and intermediate DFS, associate with improved response to the chemotherapy regimen FOLFIRI1 in adjuvant or metastatic settings. Development of clinically deployable assays for these subtypes and of subtype-specific therapies may contribute to more effective management of this challenging disease.
Previous studies have identified molecular subtypes of various human cancers by gene expression profiling2–8, including CRC subtypes9,10. However, these subtypes have not been associated with outcomes in patients treated with specific therapeutic interventions. Therefore, we sought to refine the approach of molecular classification of CRC by associating gene expression profiles of CRC tumors with corresponding clinical response to cetuximab. We first used consensus-based non-negative matrix factorization (NMF)11 to cluster two published gene expression data sets (GSE13294 (ref. 12) and GSE14333 (ref. 13)) derived from resected primary CRCs (core data sets, n = 445). These data were corrected for batch effects and merged using the distance-weighted discrimination method5,14 before clustering. This analysis defined five distinct high-consensus molecular subtypes of CRC (Supplementary Fig. 1a–e and Supplementary Results and Discussion). We used silhouette width2,15 to identify samples that most closely represent one of these five molecular subtypes, and this analysis yielded a ‘core’ set of 387 CRC tumors (Supplementary Results and Discussion and Supplementary Fig. 1f). We identified markers associated with the five subtypes using significance analysis of microarrays (SAM16, false discovery rate (FDR) = 0), followed by prediction analysis for microarrays (PAM17, nearest shrunken centroids–based method) to identify 786 subtype-specific signature genes (a collection dubbed CRCassigner-786; Fig. 1a, Supplementary Results and Discussion, Supplementary Data and Supplementary Table 1b) with the lowest prediction error.
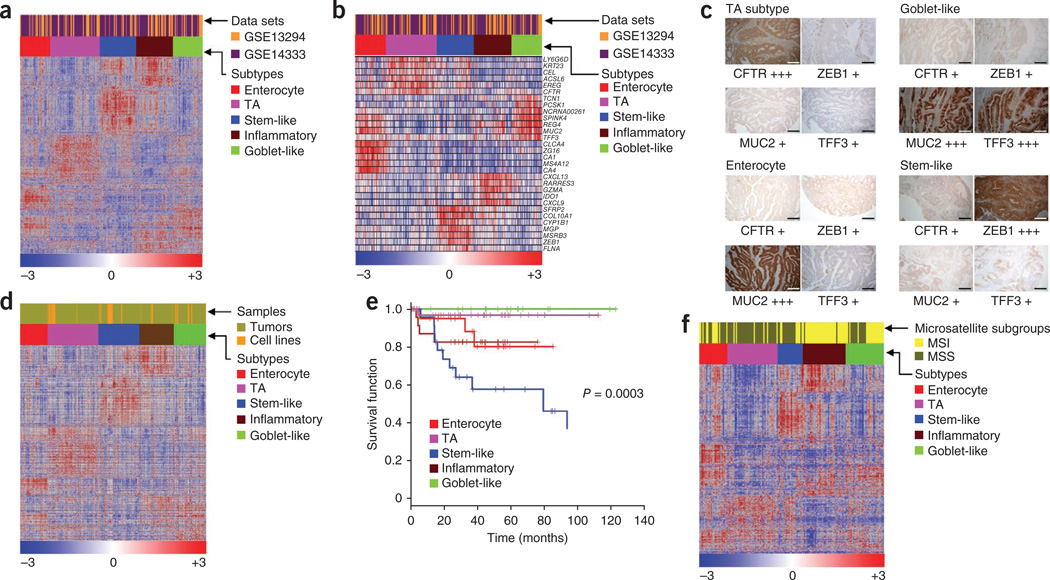
Figure 1.
Classification of colorectal tumors and cell lines into subtypes. (a,b) Heatmaps showing CRC subtypes in tumors (from two merged core data sets: GSE13294 and GSE14333) defined by CRCassigner-786 (a) and CRCassigner-30 gene signatures (b). (c) Immunohistochemistry assays of patient CRC samples using candidate CRC subtype-specific markers (four from the CRCassigner-7) to assign subtypes. For immunohistochemistry, each subtype-specific marker was scored +, ++ or +++ for weak, moderate or strong intensity of staining, respectively; see Supplementary Methods and Supplementary Table 1d for more information. Scale bar represents 100 µm. (d) Heatmap showing CRC tumor subtypes from the core tumor data sets described in a and b merged with cancer cell line data19,20. (e) Differential DFS amongst the CRC subtypes from untreated patients from the GSE14333 data set plotted as Kaplan-Meier survival curves. (f) Heatmap depicting known MSI or MSS status for each of the colorectal tumor subtype samples from the data set GSE13294. CFTR, cystic fibrosis transmembrane conductance regulator; MUC2, mucin 2; TA, transit amplifying; TFF3, trefoil factor 3; ZEB1, zing finger E-box–binding homeobox-1.
We named the five subtypes by the genes preferentially expressed in each (Fig. 1a,b and Supplementary Fig. 2): (i) goblet-like, defined by high mRNA expression of goblet-specific MUC2 and TFF3 (ref. 18); (ii) enterocyte, defined by high expression of enterocyte-specific genes18; (iii) stem-like, with high expression of Wnt signaling targets plus stem cell, myoepithelial and mesenchymal genes and low expression of differentiation markers; (iv) inflammatory, marked by comparatively high expression of chemokines and interferon-related genes; and (v) transit-amplifying, a heterogeneous collection of samples with variable expression of stem cell and Wnt-target genes. We then condensed the 786-gene signature into two smaller sub-signatures. One, dubbed as CRCassigner-30, has 30 genes with high PAM scores and characteristics of specific subtypes that might be used clinically for robust definition of these subtypes (Fig. 1b and Supplementary Table 1b). The second comprises a reduced feature set of seven genes (CRCassigner-7) that we explored for possible development as a classification assay using either quantitative RT-PCR (qRT-PCR) or immunohistochemistry. Six of these seven markers could be used to classify 50% of 72 patient-derived tumors (10 out of 19 samples using qRT-PCR and 26 out of 53 samples using immunohistochemistry) into one of the five CRC subtypes (Fig. 1c, Supplementary Fig. 2g, Supplementary Table 1c,d and Supplementary Methods). The use of the seventh CRCassigner-7 gene is discussed below and in the Supplementary Results and Discussion. The inflammatory subtype currently cannot be defined using an immunohistochemistry assay owing to the lack of antibodies that identify the markers for this subtype. Development of a clinically deployable qRT-PCR assay will require identification of reference genes and precisely defined decision thresholds.
We further validated the colon cancer subtypes in seven independent patient gene expression profile data sets (n = 744), including a recent The Cancer Genome Atlas study9, by projecting the CRCassigner-786 genes onto the data sets and then performing NMF consensus clustering (Supplementary Fig. 3, Supplementary Table 2 and Supplementary Results and Discussion). Four of our five subtypes were also represented in a panel of human CRC cell lines19,20 (n = 51; see Supplementary Results and Discussion for identification of subtypes in cell lines, Fig. 1d, Supplementary Fig. 4a–c and Supplementary Table 2c). In three cases, we showed that the subtype signature was stably maintained when subtyped CRC lines were grown as xenograft tumors in mice and analyzed for marker expression by qRT-PCR (Supplementary Fig. 4d,e).
We next examined the association of CRC subtypes with DFS after surgery for 197 patients in one of the core CRC data sets, GSE14333 (ref. 13), for which reported follow-up data were available. We first evaluated DFS for all the samples irrespective of stage or treatment (adjuvant chemotherapy or chemoradiotherapy21). This did not reveal a significant association between subtype and DFS (P = 0.12; log-rank test; Supplementary Results and Discussion, Supplementary Fig. 5b and Supplementary Table 3). We did, however, detect significant associations of subtypes with DFS within treatment subgroups. In untreated patients, stem-like–subtype tumors had the shortest DFS, inflammatory and enterocyte subtypes had intermediate DFS, and transit-amplifying and goblet-like subtypes showed a good prognosis (P = 0.0003; n = 120; log-rank test, Fig. 1e). However, there was no significant association between subtype and DFS in the treated patients (n = 77, Supplementary Fig. 5c). There was a trend suggesting that adjuvant chemotherapy or chemo-radiotherapy preferentially improved DFS in patients with stem-like–subtype tumors, whereas both treatments were associated with a detrimental effect in the transit-amplifying and goblet-like subtypes (Supplementary Fig. 5a–h). These results suggest that stem-like tumors might be preferentially responsive to adjuvant chemotherapy or chemoradiotherapy, whereas transit-amplifying– and goblet-like–subtype tumors might not benefit from these treatments. As there were only 43 events of tumor recurrence among the treated and untreated samples, additional studies involving larger patient data sets will be needed to test the validity of the suggested relationships between subtype, treatment and DFS.
We next compared our subtypes with the well-established micro-satellite stability (MSS) or instability (MSI) phenotypes using GSE13294 (n = 155)12. We observed that 94% (n = 36) of the inflammatory-subtype samples showed MSI, whereas 86% (n = 42) of the transit-amplifying and 67% (n = 21) of the stem-like subtypes were MSS (Fig. 1f). We obtained consistent results by associating MSI status for tumors previously classified with our subtype signature and querying their core data sets for MSI status using a published MSI gene signature22 and the nearest template prediction (NTP) algorithm23 (Supplementary Fig. 5i–k). Although there are clear associations between MSI or MSS status and specific subtypes, the transcriptional signatures and subtype definitions allow refinement beyond what can be achieved by annotating microsatellite stability status.
The normal colon is composed of cell types with varying degrees of differentiation potential and specialized functions24. Although colonic stem cells are thought to be the cell of origin for CRC, more differentiated cells may also be susceptible to transformation18,25,26. We assigned cell of origin or phenotypes to the transcriptional CRC subtypes defined here using a published gene signature that discriminates between the normal colon crypt top (where terminally differentiated cells are transiently located) and the normal crypt base (where stem cells and their partially differentiated derivatives reside)27. We used the NTP algorithm23 (Supplementary Results and Discussion and Fig. 2a) to show that 98% (n = 44) of the stem-like subtype tumors were significantly (FDR < 0.2) associated with the crypt base signature. In addition, we found that several published stem cell–specific gene and pathway signatures were significantly associated with the stem-like subtype (Supplementary Fig. 6a,b). In contrast, 92% (n = 52) and 82% (n = 33) of samples from the enterocyte and goblet-like subtype tumors, respectively, were associated with crypt top by their concordant gene signatures. The inflammatory subtype was not associated with either the crypt base or top (about 75% of the samples were undetermined with FDR > 0.2). Notably, 59% (n = 59) of the transit-amplifying–subtype tumor samples had a crypt-top signature with low expression of the Wnt signaling targets LGR5 and ASCL2 (ref. 18). In contrast, the remaining transit-amplifying–subtype tumors were significantly associated with the crypt base (Fig. 2a) and showed high mRNA expression of the stem and progenitor markers LGR5 and ASCL2 (Supplementary Fig. 6c). This observation suggests that the transit-amplifying subtype can be further subdivided.
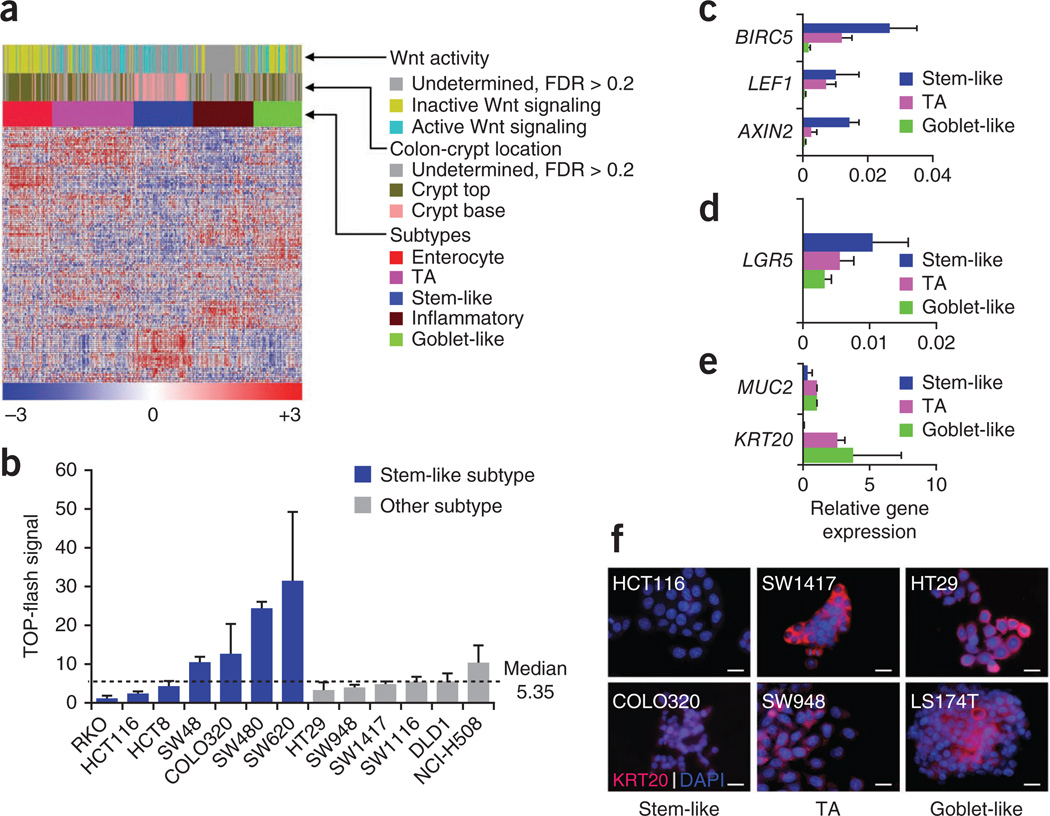
Figure 2.
Cellular phenotype and Wnt signaling in the CRC subtypes. (a) Heatmap showing association of colon-crypt location (top or base) and Wnt activity in the patient colorectal tumors from the core data sets revealed by applying specific signatures27,28 using the NTP algorithm. In these analyses, statistics include only those samples that were predicted with FDR < 0.2 (see Supplementary Results and Discussion for statistics from all samples including those with FDR > 0.2). (b) TOP-flash assay depicting Wnt activity in CRC cell lines. (c–e) qRT-PCR analysis depicting the average expression of Wnt signaling pathway (c), stem cell (d) and differentiation-specific (e) markers in a set of CRC subtype-representative cancer cell lines (HT29 and LS174T for goblet-like; LS1034, NCI-H508 and SW948 for transit-amplifying; and SW48, HCT8 and SW620 for the stem-like subtype). The qRT-PCR data is plotted relative to the housekeeping gene RPL13A. Error bars represent the s.d. of technical replicates from a representative experiment. (f) Immunostaining analyses for the differentiation markers KRT20 are presented in red, and nuclei are counterstained with DAPI (blue). HCT116 and COLO320 belong to the stem-like, SW1417 and SW948 belong to transit-amplifying, and HT29 and LS174T belong to the goblet-like subtypes. Scale bar represents 15 µm.
The colon-crypt base is composed predominantly of stem and progenitor cells that are known to have high Wnt activity28, and we identified several canonical Wnt gene targets as components of our stem-like–subtype signature (Supplementary Table 1b). The majority of the stem-like–subtype tumors from the core CRC data set were associated with high Wnt activity signature28, as observed in the colon crypt top or base gene signature comparison, whereas the enterocyte and goblet-like subtypes were not (Fig. 2a). We tested this association by performing an in vitro Wnt activity assay (TOP-flash) on subtype-specific CRC cell lines. We observed that 57% (n = 7) of stem-like–subtype cell lines showed high Wnt activity (above the median TOP-flash signal), as compared to 17% (n = 6) among cell lines from the other subtypes (Fig. 2b). We further tested this observation by performing qRT-PCR and immunofluorescence assays on a panel of CRC cell lines using known CRC markers of differentiation, Wnt signaling or stemness28. This analysis confirmed that the stem-like subtype was the least differentiated and had the highest expression of Wnt signaling and stem cell markers. The goblet-like subtype, in contrast, had a well-differentiated gene expression pattern with comparatively low expression of the stem cell and Wnt markers (Fig. 2c–f and Supplementary Fig. 2). These results provided further evidence that the stem-like subtype indeed has a stem or progenitor cell phenotype, whereas the goblet-like and enterocyte subtypes have a more differentiated phenotype.
The epidermal growth factor receptor (EGFR)-specific monoclonal antibody cetuximab, which is a mainstay of treatment for metastatic CRC with wild-type KRAS29,30, has failed to show significant benefit in the adjuvant setting, irrespective of KRAS genotype31. We correlated subtypes with cetuximab response using a CRC liver metastases microarray (Khambata-Ford) data set32 annotated with therapeutic responses to cetuximab in 80 patients. NMF consensus clustering with the CRCassigner-786 genes showed that three of our five subtypes were present in this collection of 80 CRC samples (Fig. 3a and Supplementary Fig. 7a). We identified a subgroup of samples (n = 26) (termed ‘unknown’) with a gene expression profile that was highly similar to that of normal liver (Fig. 3a and Supplementary Table 4a). These samples were not analyzed further. Within the remaining metastatic CRCs, only 23% (out of 22 samples with known cetuximab response) from the goblet-like and stem-like subtypes responded to cetuximab. However, 54% (n = 26) of patients with transit-amplifying–subtype cancer benefitted from cetuximab, whereas the other 46% of patients had progressive disease. In this case, complete response, partial response and stable disease were considered as beneficial. These data suggest that the transit-amplifying subtype designation includes two populations that differ in cetuximab sensitivity (Fig. 3a).
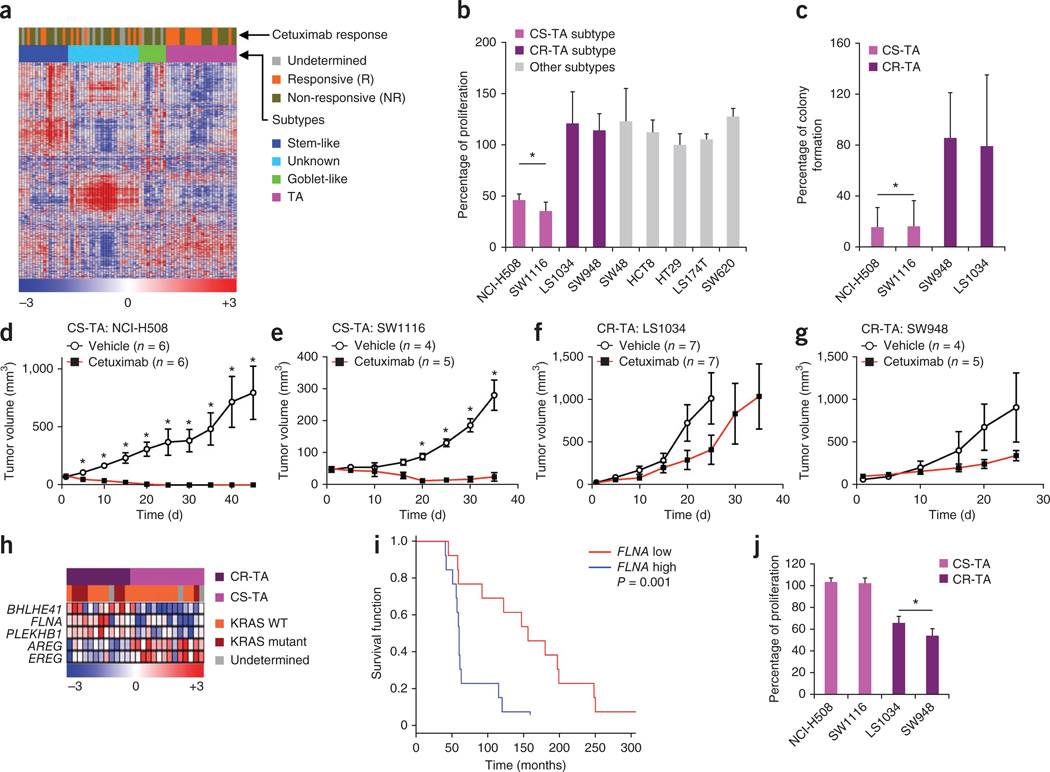
Figure 3.
Differential sensitivity among CRC subtypes to cetuximab. (a) Heatmap showing individual responses of patients with metastatic CRC (Khambata-Ford data set32) to cetuximab treatment and their association with subtypes. (b) Cetuximab response in CRC subtype–specific cell lines is plotted as percentage of proliferation of treated cells (cetuximab, 62.5 µg ml−1) normalized to vehicle-treated control. (c) Cetuximab response in transit-amplifying sub-subtype specific cell lines are plotted as percentage colony formation of treated cells (cetuximab, 15.6 µg ml−1) normalized to vehicle-treated cells. (d–g) Cetuximab response in transit-amplifying sub-subtype–specific xenograft tumors using the CS-TA cell lines NCI-H508 (d) and SW1116 (e) and the CR-TA cell lines LS1034 (f) and SW948 (g). (h) Heatmap depicting differential gene expression patterns and the KRAS mutation status between CR-TA and CS-TA subtypes (Khambata-Ford data set). (i) Kaplan-Meier curve of differential DFS based on FLNA expression in transit-amplifying subtype samples (Khambata-Ford data set). The expression of FLNA was median-centered across all the 80 samples that belong to different subtypes; those above median were considered as ‘FLNA high’, and those below the median were considered as ‘FLNA low’. (j) Differential response to the cMET inhibitor PHA-665752 (625 nM) in CR-TA and CS-TA subtype-specific cell lines, plotted relative to vehicle-treated cells. Error bars represent the s.d. of technical replicates (triplicates in b,c and j, where as the sample sizes are indicated for d–g in the figure) from a representative experiment. *P < 0.05.
We explored this segregation in responsiveness by assessing cell proliferation and colony-forming potential in cultured CRC cell lines representing different subtypes and then analyzing their growth as xenograft tumors, with and without cetuximab treatment. We found that a subset of transit-amplifying–subtype cell lines was selectively sensitive to treatment (Fig. 3b–g and Supplementary Fig. 7b,c). Specifically, the proliferation of two transit-amplifying–subtype cell lines (NCI-H508 and SW1116) was significantly impaired by cetuximab both in vitro and in xenograft tumors, compared to vehicle controls (Fig. 3b–g and Supplementary Fig. 7b,c). Notably, tumors from the NCI-H508 cell line had not recurred 45 d after the conclusion of treatment. In contrast, two other transit-amplifying cell lines showed resistance to cetuximab in vitro (LS1034 and SW948), and both showed progressive growth as xenograft tumors during treatment with cetuximab (Fig. 3b–g). The clinical and experimental data collectively support the division of the transit-amplifying–subtype tumors and cell lines into two sub-subtypes: cetuximab-sensitive transit-amplifying (CS-TA) and cetuximab-resistant transit-amplifying (CR-TA). This delineation increases the number of CRC subtypes to six.
We next performed SAM-based differential gene expression analysis on the transit-amplifying subtype tumors from the Khambata-Ford data set32 (transit-amplifying signature; FDR = 0.1). This revealed that CS-TA tumors expressed significantly higher levels of the EGFR ligands epiregulin (EREG) and amphiregulin (AREG), which are known to be positive predictors of cetuximab response32, as compared to CR-TA tumors (Fig. 3h and Supplementary Fig. 7d). In contrast, filamin A (FLNA), which regulates the expression and signaling of the cMET receptor33, was overexpressed in CR-TA compared to CS-TA (Fig. 3h and Supplementary Fig. 7e). This correlation was further confirmed using receiver operating curve analysis (Supplementary Fig. 7f,g). High FLNA expression was significantly (P = 0.001; log-rank test; n = 26, Fig. 3i) associated with shorter progression-free survival only within the transit-amplifying–subtype tumors. However, FLNA expression did not show prognostic differences when samples from all the subtypes were included or when all samples were segregated by KRAS status (Supplementary Fig. 7h–k). Our observation of elevated FLNA expression in CR-TA tumors then led us to examine the effects on proliferation of pharmacologically inhibiting cMET using the selective small-molecule inhibitor PFA-665752 in a panel of transit-amplifying cell lines. We found that CR-TA cell lines were more sensitive to cMET inhibition than CS-TA cell lines (Fig. 3j). Moreover, we found that three transit-amplifying–subtype samples from Supplementary Figure 2g could be assigned to CR-TA or CS-TA sub-subtypes (Supplementary Fig. 7l,m) using a qRT-PCR assay for FLNA (one of the seven genes of CRCassigner-7 signature) expression. We did not find a significant association between the transit-amplifying subtype and KRAS mutation status (P = 0.1; chi-square test; Supplementary Results and Discussion Fig. 3h and Supplementary Fig. 7n–q). Collectively, these results suggest that screening first for the transit-amplifying subtype with CFTR expression followed by FLNA expression using qRT-PCR assays (Supplementary Fig. 7l,m) to subdivide the transit-amplifying subtype into two sub-subtypes could provide an effective means to predict sensitivity to either cetuximab (low FLNA) or to a cMET inhibitor (high FLNA) in patients with metastatic, transit-amplifying cancer.
We next examined the possibility that the subtypes might show differential responses to a chemotherapy regimen deployed in first-line treatment of patients with metastatic CRC (FOLFIRI, a combination of irinotecan, 5-fluorouracil, and leukovorin)1. This evaluation was performed by NMF consensus clustering using a gene expression profile data set (Del Rio data set) of primary CRC samples from patients with metastatic disease with matched FOLFIRI response data34. We found that 71% of stem-like–subtype tumors (n = 7) in this data set were associated with clinical benefit to FOLFIRI treatment, whereas only 29% (n = 14) of tumors from the other subtypes were associated with the treatment benefit (Fig. 4a and Supplementary Fig. 8a–c). We further tested this association by showing that stem-like samples (100%, n = 18) were significantly (FDR < 0.2) associated with the FOLFIRI response signature35 in the patients with meta-static disease (Fig. 4b) in the Khambata-Ford data set32, using the NTP algorithm23.
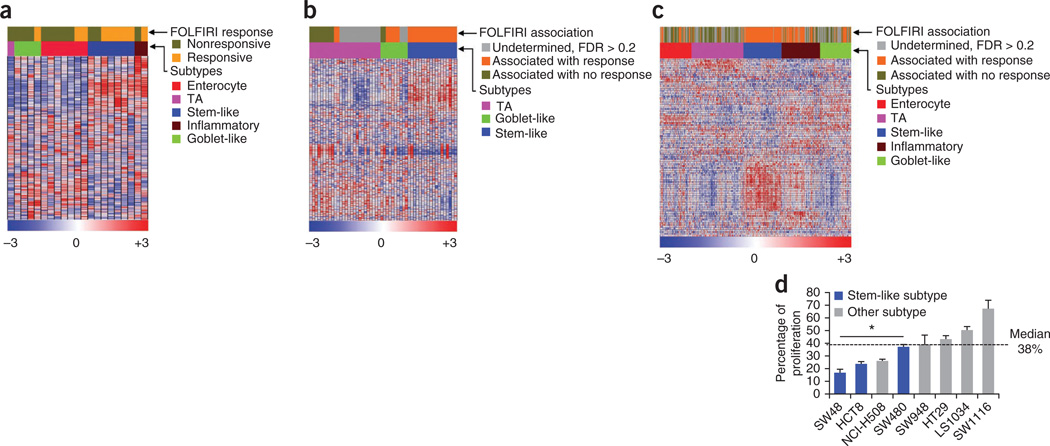
Figure 4.
Specific response to chemotherapy in CRC subtypes. (a) Heatmap showing individual responses of patients with primary CRC (Del Rio data set34, n = 21) to FOLFIRI treatment and their association with subtypes. The subtypes in the Del Rio data set were identified after merging the data set with the core CRC data sets (Supplementary Fig. 8c). Complete and partial responses and stable disease were considered as beneficial response, whereas progressive disease was deemed as no response. (b,c) Heatmaps showing association of individual patient CRC responses in the Khambata-Ford data set (metastases) (b) and in the core data sets (includes samples from all of the Dukes’ stages) (c) to FOLFIRI by applying published FOLFIRI response signatures35 using the NTP algorithm. In these analyses, statistics include only those samples that were predicted with FDR < 0.2. (d) CRC subtype–specific cell line response to FOLFIRI components. Namely, the combination of 5-FU (239 µM) and irinotecan (22.5 µM), plotted as percentage cellular proliferation and normalized to vehicle-treated cells. Error bars represent the s.d. of technical replicates from a representative experiment. *P < 0.05. TA, transit amplifying.
Similarly, the FOLFIRI response signature35 was significantly (FDR < 0.2) associated with 100% (n = 74) of the stem-like– and 75% (n = 53) of the inflammatory-subtype samples, as compared to only 14% (n = 56) of the transit-amplifying–, 39% (n = 33) of the goblet-like– and 38% (n = 40) of the enterocyte–subtype tumors in the core CRC data sets (comprised of all Dukes’ stage samples; Fig. 4c and Supplementary Fig. 8d–f) as assessed using the NTP algorithm. We experimentally assessed the association of the stem-like CRC subtype with sensitivity to FOLFIRI in a panel of eight CRC cell lines representing different transcriptional subtypes. We treated these cell lines with 5-fluorouracil (5-FU) plus irinotecan (the two chemotherapy components of FOLFIRI). Three of the four most sensitive cell lines were of the stem-like subtype (Fig. 4d). These results are consistent with the data presented in Figure 1e and Supplementary Figure 5a–d, demonstrating that patients with stem-like tumors have improved DFS when treated with chemotherapy or chemoradiotherapy in the adjuvant setting. This finding is also consistent with data from poor-prognosis subtypes identified in other cancer types, such as basal and claudin-low breast cancer36 and quasi-mesenchymal pancreatic ductal adenocarcinoma5, which are comparatively more responsive to chemotherapy than other subtypes.
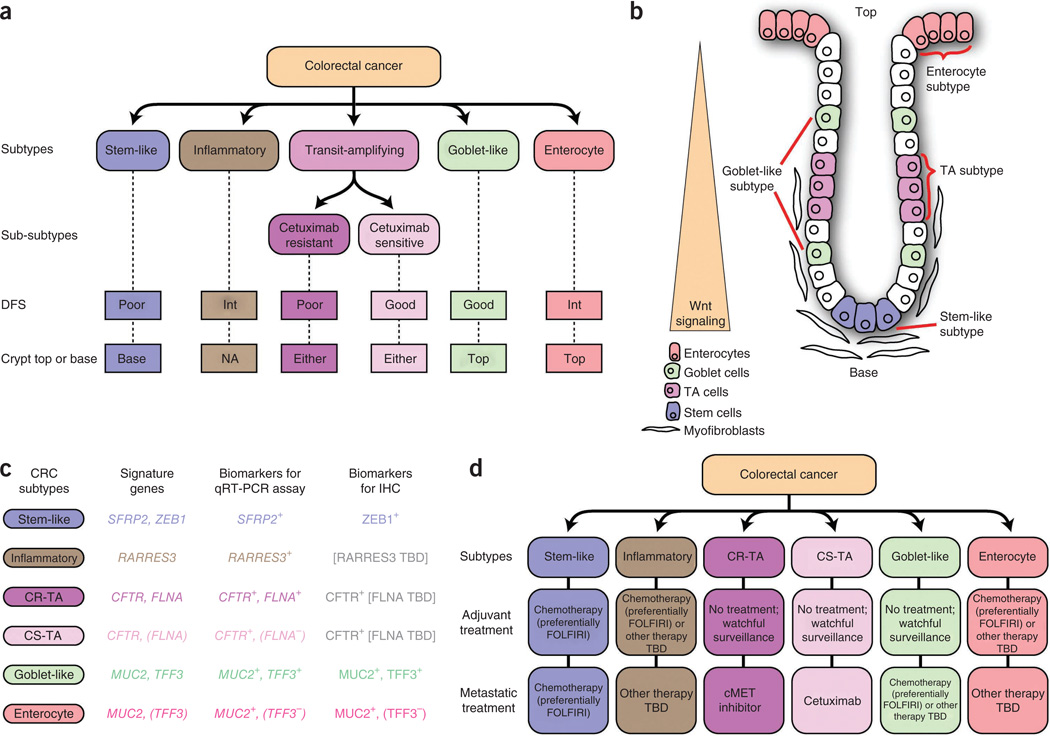
In summary, we document the existence of six subtypes of CRC based on the combined analysis of gene expression profiles and differential response to cetuximab. These subtypes are phenotypically distinct in their DFS (Fig. 5a) and vary in degree of response to cetuximab and standard-of-care chemotherapy. We also have shown that these CRC subtypes are associated with distinctive anatomical regions of the colon crypts (phenotype) and with location-dependent differentiation states and Wnt signaling activity (Fig. 5b). We identified candidate biomarkers that might be developed into clinical qRT-PCR or immunohistochemical assays to classify CRC tumors into one of six subtypes (Fig. 5c) as a guide to assignment of subtype-specific therapeutic agents (Fig. 5d). With regard to first-line chemotherapy, we infer that particular subtypes might show beneficial responses to FOLFIRI in either adjuvant or metastatic settings (Fig. 5d), whereas in unselected CRC this treatment did not improve survival in the adjuvant setting37 Our analyses suggest that stem-like–subtype tumors, both in the adjuvant and metastatic settings, as well as inflammatory-subtype tumors in the adjuvant setting, may best be treated with FOLFIRI. Additionally, the transit-amplifying sub-subtypes and the goblet-like subtype will probably not respond to FOLFIRI in the adjuvant setting. Watchful surveillance might spare patients with these forms of disease from the harmful side effects of debilitating and ineffective FOLFIRI treatment. Moreover, and in contrast to the adjuvant setting, the CS-TA or CR-TA subtype might be effectively treated with cetuximab or a cMET inhibitor, respectively, in the metastatic setting (Fig. 5d). These associations warrant further retrospective and prospective validation. Lastly, we demonstrated that subtype-specific CRC cell lines and xenograft tumors can serve as surrogates for assessing subtype-specific treatment responses. Recognition of these subtypes may prove applicable to the assessment of new investigational drugs in preclinical trials. The outcomes could in turn guide ‘personalized’ therapeutic trial designs that target subtype-selective sensitivities in those patients with CRC who are most likely to see clinical benefit, much as is becoming standard of care in non– small-cell lung cancer38.
Figure 5.
Summary, including clinically deployable markers and potential subtype-guided therapies for CRC. (a) Salient characteristics of the six CRC subtypes. Int, intermediate DFS; Either, either crypt top or base; NA, no clear association. (b) CRC subtype phenotypes correlated with colon-crypt location and Wnt signaling. (c) Summary of subtype-specific candidate biomarkers (CRCassigner-7) that were tested using qRT-PCR and immunohistochemistry (IHC), and (d) subtype guided therapeutic strategies suggested by the association studies. ZEB1 was not useful for qRT-PCR because ZEB1 is expressed both by fibroblast and tumor cells. The expression of ZEB1 specifically in tumor cells was evaluated using immunohistochemistry as in Figure 1c. TBD, to be determined.
ONLINE METHODS
Combining different microarray data sets
Microarray data sets from different published studies were screened separately for variable genes using s.d. cut off greater than 0.8. The screened data sets were column (sample) normalized to N(0,1) and row (gene) normalized and then merged using Java-based distance-weighted discrimination14. Finally, the rows were median centered before further downstream analysis, as described5. Additional methodological details can be found in the Supplementary Methods and Supplementary Results and Discussion.
NMF, SAM and PAM analysis
The stable subtypes were identified using consensus clustering-based NMF11 followed by SAM16 (using classes defined by NMF analysis) and PAM17 (using significant genes defined by SAM) analysis to identify gene signature specific to each of the subtypes with modified methods described for glioblastoma classification2. Additional methodological details can be found in the Supplementary Methods and Supplementary Results and Discussion.
Drug response in cell lines using proliferation assay
Cells were added (5 × 103) into 96-well plates on day 0 and treated with cetuximab (Merck Serono, Geneva, Switzerland), cMET inhibitor (PHA-665752, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), a combination of 5-FU (Sigma-Aldrich, Buchs SG, Switzerland) and irinotecan (Pfizer AG, Zurich, Switzerland) or vehicle control (medium alone or DMSO) in the presence of fetal bovine serum on day 1. Proliferation was monitored using CellTiter-Glo assay kit according to the manufacturer’s instructions (Promega, Dubendorf, Switzerland) on day 3 (72 h).
TOP flash assay
The TOP/FOP-flash assay was performed as instructed by the manufacturer (Upstate, USA). Briefly, colon cancer cell lines were plated into 24-well dishes in biological triplicate at 10,000 cells/well in full growth medium (RPMI + 10% FBS). The next day, the medium was changed to that containing 3 µL of polyethylenimine (stock, 1 mg mL−1), TOP or FOP-flash DNA (0.25 µg/well) and a plasmid encoding constitutive expression of Renilla luciferase (to normalize for transfection efficiency). Two days later, the cells were assayed. Samples were prepared in biological triplicate (n = 3) and the experiment was repeated twice.
Additional methods
Detailed methodology is described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Schulz (Charité, Universitätsmedizin) for providing RNA from xenograft tumors and for comments of the manuscript. We thank R.A. Du Pasquier (CHUV) for providing the HT29 cell line, P. Depeille (University of California– San Francisco) for the SW480, SW48, HCT8, LS174T and SW948 cell lines, and H. Ying (MD Anderson Medical Center) for the NCI-H508, LS1034, SW620, COLO320, SW1417, HCT116, RKO and DLD1 cell lines. The TOP/FOP-flash and Renilla constructs were a generous gift from S. Kobayashi (Beth Israel Deaconess Medical Center). We particularly acknowledge G. Poulogiannis for insightful feedback and assistance with statistical analysis of survival data. We also thank C. Fuerer, S.S. Sidhu, J. Yun and N. Divorne-Formenton for advice on the experimental design, C.R. Thomas for help with editing the manuscript, the Histology Core Facility of EPFL for help with immunohistochemistry. A.S. was partially supported by a US Department of Defense Postdoctoral Fellowship (BC087768). C.A.L. is the Amgen Fellow of the Damon Runyon Cancer Research Foundation (DRG-2056-10). J.W.G. is supported by the US National Institutes of Health grant U54 CA 112970 and by the Stand Up To Cancer–AACR Dream Team Translational Cancer Research Grant SU2C–AACR-DT0409. This work was supported by a Swiss National Science Foundation project grant awarded to D.H.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
A.S. conceived of the hypothesis, designed and performed experiments, interpreted results and co-wrote the manuscript. C.A.L., K.H., S.W., L.C.G.O., W.A.L. and C.G. performed experiments. M.D.R. provided CRC microarray data with FOLFIRI response data. B.L. provided pathology expertise, and A.B.O. provided statistical expertise. C.A.L., K.H., E.A.C., W.J.G., L.C.C. and B.W. participated in critical discussions and helped edit the manuscript. J.W.G. interpreted results, helped edit the manuscript, and co-supervised the project. D.H. co-supervised the project, interpreted results and co-wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Andre T, et al. CPT-11 (irinotecan) addition to bimonthly, high-dose leucovorin and bolus and continuous-infusion 5-fuorouracil (FOLFIRI) for pretreated metastatic colorectal cancer. GERCOR. Eur. J. Cancer. 1999;35:1343–1347. doi: 10.1016/s0959-8049(99)00150-1. [DOI] [PubMed] [Google Scholar]
- 2.Verhaak RG, et al. Integrated genomic analysis identifes clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classifcation of prostate cancer using curated expression signatures. Proc. Natl. Acad. Sci. USA. 2011;108:21276–21281. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identifed by gene expression profling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 8.Tothill RW, et al. Novel molecular subtypes of serous and endometrioid cancer linked to clinical outcome. Clin. Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez Villamil B, et al. Colon cancer molecular subtypes identifed by expression profling and associated to stroma, mucinous type and different clinical behavior. BMC Cancer. 2012;12:260. doi: 10.1186/1471-2407-12-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorissen RN, et al. DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin. Cancer Res. 2008;14:8061–8069. doi: 10.1158/1078-0432.CCR-08-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorissen RN, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin. Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benito M, et al. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20:105–114. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 15.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Signifcance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greshock J, et al. Molecular target class is predictive of in vitro response profle. Cancer Res. 2010;70:3677–3686. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 20.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Cutsem E, Oliveira J. Primary colon cancer: ESMO clinical recommendations for diagnosis, adjuvant treatment and follow-up. Ann. Oncol. 2009;20(suppl. 4):49–50. doi: 10.1093/annonc/mdp126. [DOI] [PubMed] [Google Scholar]
- 22.Banerjea A, et al. Colorectal cancers with microsatellite instability display mRNA expression signatures characteristic of increased immunogenicity. Mol. Cancer. 2004;3:21. doi: 10.1186/1476-4598-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshida Y. Nearest template prediction: a single-sample-based fexible class prediction with confdence assessment. PLoS ONE. 2010;5:e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 25.Shih IM, et al. Top-down morphogenesis of colorectal tumors. Proc. Natl. Acad. Sci. USA. 2001;98:2640–2645. doi: 10.1073/pnas.051629398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Kosinski C, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl. Acad. Sci. USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermeulen L, et al. Wnt activity defnes colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham D, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N. Engl. J. Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 30.De Roock W, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann. Oncol. 2008;19:508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 31.Ogino S, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin. Cancer Res. 2009;15:7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khambata-Ford S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J. Clin. Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 33.Zhou AX, et al. Filamin a mediates HGF/c-MET signaling in tumor cell migration. Int. J. Cancer. 2011;128:839–846. doi: 10.1002/ijc.25417. [DOI] [PubMed] [Google Scholar]
- 34.Del Rio M, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fuorouracil, and irinotecan. J. Clin. Oncol. 2007;25:773–780. doi: 10.1200/JCO.2006.07.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graudens E, et al. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heiser LM, et al. Subtype and pathway specifc responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Cutsem E, et al. Randomized phase III trial comparing biweekly infusional fuorouracil/leucovorin alone or with irinotecan in the adjuvant treatment of stage III colon cancer: PETACC-3. J. Clin. Oncol. 2009;27:3117–3125. doi: 10.1200/JCO.2008.21.6663. [DOI] [PubMed] [Google Scholar]
- 38.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.