Abstract
Molecular dynamics force field development and assessment requires a reliable means for obtaining a well-converged conformational ensemble of a molecule in both a time-efficient and cost-effective manner. This remains a challenge for RNA because its rugged energy landscape results in slow conformational sampling and accurate results typically require explicit solvent which increases computational cost. To address this, we performed both traditional and modified replica exchange molecular dynamics simulations on a test system (alanine dipeptide) and an RNA tetramer known to populate A-form-like conformations in solution (single-stranded rGACC). A key focus is on providing the means to demonstrate that convergence is obtained, for example by investigating replica RMSD profiles and/or detailed ensemble analysis through clustering. We found that traditional replica exchange simulations still require prohibitive time and resource expenditures, even when using GPU accelerated hardware, and our results are not well converged even at 2 microseconds of simulation time per replica. In contrast, a modified version of replica exchange, reservoir replica exchange in explicit solvent, showed much better convergence and proved to be both a cost-effective and reliable alternative to the traditional approach. We expect this method will be attractive for future research that requires quantitative conformational analysis from explicitly solvated simulations.
Keywords: enhanced sampling, RNA, force field, AMBER, replica exchange, reservoir REMD, TIGER2, convergence
Introduction
RNA takes on many essential roles in biology, from information encoding to catalysis to regulatory functions.1 Molecular dynamics (MD) simulations provide a critical connection between structural theory and experimental data for RNA as well as other biomolecules.2-5 Unfortunately, the force fields which underlie the physical representation of RNA tend to be less reliable than those used for proteins and have required numerous refinements over the past several years.6-11 This is likely due to a variety of factors, for example the highly charged and highly flexible nature of the RNA backbone. As we and others continue efforts at force field development, it has become apparent that a time-efficient and cost-effective method is necessary for producing biomolecular simulation data, from which quantitative results defining the ensemble of sampled conformations can be obtained and the underlying force field evaluated. Conventional MD simulations, typically performed at laboratory temperatures in order to match experiment, often require cost-prohibitive timescales and/or special purpose hardware to produce converged results.12 Additionally, cost-saving approaches such as implicit solvation, which reduces the total degrees of freedom in the system, tend to result in major conformational distortions for many RNA systems13,14 (as is demonstrated in the results of this work). To address the need for efficient simulation methods for generating well-converged conformational ensembles required for force field development and assessment, we turned to approaches from the widely studied field of enhanced sampling.15-17
Replica exchange MD (REMD) is a commonly used method for enhanced sampling which deploys an ensemble of independent system replicas, or MD simulations, that exchange various properties.18,19 By allowing a system replica at target conditions (such as the laboratory temperature) to exchange with system replicas at conditions which favor sampling (such as higher temperatures), REMD enhances conformational sampling on a rugged landscape while maintaining a Boltzmann-weighted ensemble at each target condition.17 Due to the computational cost of simulating multiple system replicas, temperature REMD is often used with implicit solvent which reduces the number of degrees of freedom in the system, requiring fewer replicas to span a given temperature regime while maintaining acceptable exchange ratios. Unfortunately, as noted above, RNA simulations in implicit solvent tend to produce poor results. There are a variety of small systems that have been studied using REMD with explicit solvent, including proteins,20-28 DNA,29,30 and RNA.31-34 In most cases, convergence of the REMD ensemble is discussed in qualitative terms, if at all, and typically only one REMD simulation is performed for each condition of interest. In this work we aim to provide a more detailed understanding of convergence in explicit solvent. Due to our interest in obtaining quantitative results, we chose to study two small systems in order to reduce computational cost: alanine dipeptide and a tetranucleotide RNA, rGACC (Figure 1). Alanine dipeptide is a frequently used test molecule due to its simplicity 35-38 and rGACC is optimal because it is small, has detailed NMR data published regarding its structure, and has been studied previously using conventional MD.39
Figure 1.
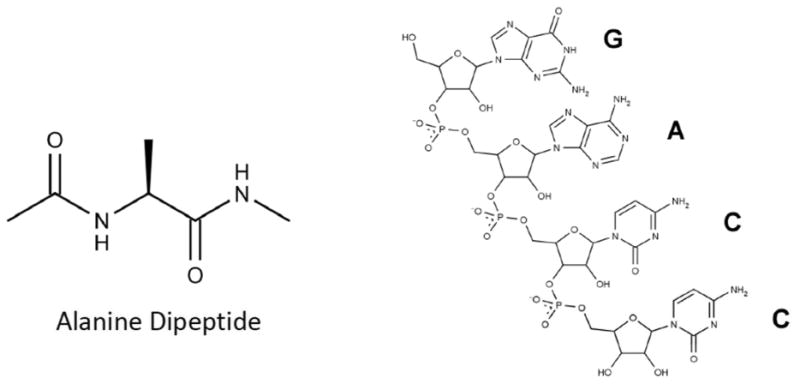
Molecules investigated in this research: alanine dipeptide (left), RNA tetranucleotide rGACC (right).
Finally, in addition to traditional REMD, we also investigate two methods which were previously reported to offer the benefits of REMD at reduced resource cost: TIGER2 38 and reservoir REMD (R-REMD).40 The TIGER2 method reduces the number of replicas required for the ensemble by incorporating a velocity rescaling and thermal equilibration step both prior to and immediately following the exchange attempt. This added step shifts the potential energy distribution of high temperature replicas nearer to the distribution of the baseline temperature replica and allows exchanges to occur which would otherwise be improbable due to the large potential energy distribution spacing. The other method, R-REMD, enhances convergence by adding a high-temperature structure reservoir to the top of the REMD temperature ensemble. Thus, a replica at the highest target temperature can exchange with the pre-generated reservoir and the costly wait associated with traversing energy barriers is overcome by exchange. In effect, the reservoir drives convergence of the entire REMD ensemble. This approach has been reported previously with implicit solvent,40,41 and in explicit solvent with multiple reservoirs for the disordered Abeta(21-30) peptide.42 Here, we apply the single reservoir approach and explore convergence for RNA. Quite surprisingly, even with a relatively small RNA system like rGACC, the results suggest that standard REMD methods may require over 2 μs per replica for convergence, and even with reasonable starting reservoirs, R-REMD still requires 20 – 100 nanoseconds per replica, depending on the required accuracy measures of the conformational ensemble populations for convergence.
Methods
System Building
Two explicitly solvated systems were studied in this research: alanine dipeptide and the RNA tetranucleotide rGACC (Figure 1). Alanine dipeptide was built from three residues, ACE-ALA-NME, using AMBER's LEaP program and parameterized with the AMBER ff12SB force field.43,44 To solvate the system, 544 TIP3P water molecules45 were added to a cubic periodic box around the solute. The system size was chosen to match and be consistent with the systems used in previous studies with the TIGER2 protocol.38
The initial conformation for the RNA tetranucleotide, rGACC, was taken from an A-form portion of a RNA crystal structure (PDB: 3G6E, residues 2623-2626). The RNA, parameterized with the ff12SB force field (note, for nucleic acids this is identical to ff10 which was released with AMBER11), was solvated with 2500 TIP3P water molecules in a truncated octahedron periodic box and the total system charge was neutralized with three sodium ions. Additional salt was not added to be consistent with previous computational studies by Turner and co-workers.39 This RNA was previously studied with AMBER ff99 force field as well as a modified variant by Turner and co-workers,9 which includes corrections to the χ torsion parameters. We chose to use the ff12SB force field for the following reasons: it includes backbone torsion corrections,6 higher accuracy χ torsion corrections (in comparison to the Turner variant),8 and has been tested on larger, more representative RNA structures.7,46,47
System Heating and Equilibration
All simulations were performed with AMBER12 43 using either the PMEMD or SANDER programs. PMEMD was used when possible due to its superior computational performance and parallel efficiency. However, PMEMD does not (yet) support R-REMD simulations and thus SANDER was used for these simulations. Additionally, TIGER2 simulations are not implemented in AMBER and thus an in-house script was used as a wrapper to implement the simulation cycle (details discussed later). Prior to production simulations, both the alanine dipeptide and RNA systems were energy minimized and equilibrated. Energy minimization was performed with 25 kcal/mol-Å2 atomic positional restraints on the solute and consisted of 1000 steps with the steepest descent algorithm followed by 1000 steps with the conjugate gradient algorithm. Following minimization, the systems were heated from 10 K to 150 K at constant volume with 25 kcal/mol-Å2 atomic positional restraints on the solute using 1 fs timestep. This heating step was accomplished with 100 ps of MD simulation for the alanine dipeptide system and 1 ns for the RNA system. During heating, the temperature was controlled using a Langevin thermostat with a collision frequency of 2.0 ps-1. Further heating to the target temperature (300 K in most cases) was performed at constant pressure using a weak-coupling algorithm,48 a pressure relaxation time of 1 ps, 5 kcal/mol-Å2 atomic positional restraints on the solute, and the same thermostat, timestep, and duration settings as the previous heating step. After reaching the target temperature, the system was further equilibrated (1 ns for alanine dipeptide, 5 ns for the RNA) with 0.5 kcal/mol-Å2 positional restraints on the solute using weaker-coupling constant pressure relaxation time of 5.0 ps. The integration time step during this equilibration step was 2 fs and temperature regulation the same as previous steps. For all explicit solvent MD in this research, the non-bonded direct space cutoff was set to 8.0 Å and the default AMBER12 particle mesh Ewald49 settings were used to control reciprocal space calculations. SHAKE constraints50 with a tolerance of 0.00001 Å were used to eliminate short timescale bond vibrations between hydrogen atoms and heavy atoms.
Production MD
Table 1 provides a list of the production simulations discussed in this research. In the text below, we provide details on the settings for each type of simulation: Conventional MD, TIGER2, REMD, and R-REMD.
Table 1.
Simulations performed in this work.
| Simulation ID | Simulation Details | Temp. | Rep. | Length |
|---|---|---|---|---|
|
| ||||
| ALA-NVT | NVT, dt=2fs, cwi=1ps | 300 | 1 | 100 |
| ALA-NPT | NPT, dt=2fs, cwi=1ps | 300 | 1 | 314 |
| ALA-398 | NVT, dt=2fs, cwi=10ps | 398 | 1 | 47 |
| ALA-10REMD | REMD, NVT, dt=2fs, eai=0.2ps, cwi=1ps | 300 - 398 | 10 | 138 |
| ALA-24REMD | REMD, NVT, dt=1fs, eai=1ps, cwi=1ps | 300 - 600 | 24 | 96 |
| ALA-R-REMD-S | R-REMD, NVT, dt=2fs, eai=0.2ps, cwi=1ps, r=538 | 300 - 398 | 10 | 240 |
| ALA-R-REMD-L | R-REMD, NVT, dt=2fs, eai=0.2ps, cwi=1ps, r=4764 | 300 - 398 | 10 | 84 |
| ALA-TIG-1 | TIGER2, NVT, dt=1fs, eai=3ps, cwi=1ps | 300 - 600 | 4 | 75 |
| ALA-TIG-2 | TIGER2, NVT, dt=1fs, eai=4ps, cwi=1ps | 300 - 600 | 4 | 100 |
| ALA-TIG-3 | TIGER2, NVT, dt=1fs, eai=7ps, cwi=1ps | 300 - 600 | 4 | 105 |
| RNA-NPT | NPT, dt=2fs, cwi=2ps | 300 | 1 | 5000 |
| RNA-398 | NVT, dt=2fs, cwi=10ps | 398 | 1 | 1405 |
| RNA-REMD-GB | REMD, dt=2fs, eai=0.2ps, cwi=1ps | 277 - 463 | 6 | 500 |
| RNA-REMD-1 | REMD, NVT, dt=2fs, eai=1ps, cwi=1ps | 277 - 396 | 24 | 2010 |
| RNA-REMD-2 | REMD, NVT, dt=2fs, eai=0.2ps, cwi=1ps | 277 - 396 | 24 | 500 |
| RNA-REMD-3 | REMD, NVT, dt=2fs, eai=0.2ps, cwi=1ps | 277 - 396 | 24 | 400 |
| RNA-R-REMD-S | R-REMD, NVT, dt=2fs, eai=1ps, cwi=1ps, r= 10000 | 277 - 396 | 24 | 46 |
| RNA-R-REMD-1 | R-REMD, NVT, dt=2fs, eai=1ps, cwi=1ps, r= 140510 | 277 - 396 | 24 | 205 |
| RNA-R-REMD-2 | R-REMD, NVT, dt=2fs, eai=1ps, cwi=1ps, r= 140510 | 277 - 396 | 24 | 160 |
| RNA-R-REMD-3 | R-REMD, NVT, dt=2fs, eai=1ps, cwi=1ps, r= 140510 | 277 - 396 | 24 | 250 |
Simulation IDs beginning with “ALA-” contained one alanine dipeptide molecule solvated in TIP3P water. Those beginning with “RNA-” contained one rGACC molecule, three Na+ counterions, and TIP3P water (with the exception of RNA-REMD-GB which used implicit solvent). Column titles and abbreviations are as follows: Temp.: the temperature range covered in the simulations, Rep.: the number of replicas used, Length: the per replica simulation time length in nanoseconds, NVT: constant volume/temperature, NPT: constant pressure/temperature, REMD: replica exchange molecular dynamics, R-REMD: reservoir replica exchange molecular dynamics, dt: the simulation time step, cwi: the trajectory coordinate writing interval, eai: the replica exchange attempt interval, r: number of frames in the reservoir.
Conventional MD
Both constant pressure (NPT) and constant volume simulations (NVT) were performed in the conventional manner. The constant pressure simulations (ALA-NPT and RNA-NPT in Table 1) were intended to match typical simulation protocols. The time step was set to 2 fs and a weak coupling algorithm governed both the temperature and pressure regulation with a relaxation time of 10 ps. Constant volume simulations (ALA-NVT, ALA-398, RNA-398) used 2 fs time step and the Langevin thermostat with the collision frequency set to 10 ps-1 for alanine dipeptide, to be consistent with previous work,38 and 2 ps-1 for the RNA.
TIGER2
A method for enhanced sampling, named TIGER2 38 has been previously described and offers the benefits of temperature REMD with reduced computational cost. The TIGER2 algorithm is not implemented in AMBER, but can be achieved fairly easily with a simple wrapper script. Our implementation, which closely followed the published description, uses repeated cycles of PMEMD calculations for the MD steps and a Perl script to perform the velocity rescaling and Metropolis selection steps. Four replicas were used at the following temperatures: 300, 377, 476, and 600 K. A typical cycle consists of four steps starting from initial systems all at 300K: 1) velocity rescaling to one of the four assigned temperatures followed by thermal equilibration, 2) dynamics sampling at the assigned temperature, 3) velocity rescaling of all replicas back to 300 K followed by thermal equilibration, and 4) exchange attempt and temperature reassignment. The last step consists of the following substeps: a candidate system from the three highest temperatures is randomly selected (using Perl's rand function) for an exchange attempt with the baseline temperature replica (300 K), the exchange probability is determined by the Metropolis criterion, the result assigns one of the two considered replicas to the baseline temperature, and the remaining systems are assigned to the higher temperatures according to their system potential energies (higher energies are given a higher temperature). For all TIGER2 simulations, the heating and production sampling time periods were 1 ps each. The cooling period was varied between three time periods, 1ps, 2ps, and 5ps, as this time period had a large affect on exchange acceptance (ALA-TIG-1, ALA-TIG-2, and ALA-TIG-3 in Table 1, respectively). Similar to what was used in the reference publication, we used a Langevin thermostat collision frequency of 25 ps-1 during the heating and quenching portions of the cycle and 10 ps-1 during the sampling portion.
REMD
Traditional REMD calculations were performed using AMBER's PMEMD program. The temperature intervals between replicas were estimated using an online generator at http://folding.bmc.uu.se/remd/ 51 and are listed in Table S1. These distributions led to exchange rates between 0.15-0.30 for the ten replica simulations and 0.15-0.40 for the twenty-four replica simulations. All REMD simulations were performed at constant volume as AMBER does not support constant pressure for REMD. It has been noted that the hydrophobic effect will be increased at high temperatures for constant volume simulations.52,53 The extent of this phenomenon at high temperatures for the systems studied here and its influence on lower temperature conformational results remains unclear. Prior to production simulations, a 200 ps heating period was employed to equilibrate each replica to the assigned initial temperature. Temperature was controlled with the Langevin thermostat set to a collision frequency of 10 ps-1 for alanine dipeptide and 2 ps-1 for the RNA. A time step of either 1 or 2 fs was used and is noted in Table 1. The 2 fs timestep is typically the preferred value for computational performance reasons, but 1 fs was used for simulations with high temperatures (∼600 K) out of caution. The exchange attempt interval was set to either 0.2 or 1.0 ps and is noted in Table 1. An exchange attempt interval of 1 ps is more commonly used, however the use of a shorter interval may improve convergence.54,55 The value we chose was arbitrary in some cases but dependent on the hardware being used in others. For instance, the GPU simulations are fast enough that we were concerned about a performance drop if the exchange attempt interval was too small. To be consistent with the GPU simulation (RNA-REMD-1), we used a 1 ps exchange attempt interval for the R-REMD simulations of the RNA system even though they were performed on CPUs. In the case of alanine dipeptide, a 0.2 ps exchange attempt interval was used for all REMD simulations except ALA-24REMD, which was intended to mimic a more traditional/conservative approach (1 ps exchange attempt interval and 1 fs timestep). In addition to explicit solvent REMD simulations, we also performed one implicit solvent REMD simulation (RNA-REMD-GB). Implicit solvation was implemented using the Hawkins, Cramer, and Truhlar generalized Born (GB) model56,57 with a surface area contribution to the solvation term computed by the LCPO model.58 A salt concentration of 200 mM was approximated using Debye-Hückel screening. The Langevin thermostat was used to control temperature with a collision frequency of 2 ps-1. An infinite cutoff was employed for the non-bonded cutoff and SHAKE constraints were used to eliminate high frequency bond vibrations between hydrogen atoms and heavy atoms. All analysis of the traditional REMD simulations with RNA discarded the first 50 ns as equilibration (the initial structure of each replica was identical so we use the term equilibrate here to mean that a variety of structures are being sampled at each temperature level as determined by inspecting the RMSD plot (Figure S1)).
R-REMD
In contrast to traditional REMD, the highest temperature replica in R-REMD simulations exchanges with a pre-generated reservoir of structures. This method was previously introduced by Okur et. al.40,41 and tested in implicit solvent. In the current work, reservoirs were generated at 398 K using conventional MD simulations and consisted of high precision coordinate/velocity frames saved every 10 ps. The potential energy of each frame in the reservoir was computed and used in the exchange calculation during the R-REMD simulation with a 1/N non-Boltzmann weighting assumed for each frame. Exchange between highest temperature replica and the reservoir was ∼0.47 for the alanine dipeptide simulations and ∼0.33 for RNA simulations. All other simulation settings were identical to those used in the traditional REMD simulations. Note, the “-S” and “-L” suffixes stand for small and large reservoirs, respectively, in Table 1.
Hardware
All simulations were performed using traditional CPU clusters at a variety of resource locations with the exception of the RNA-NPT, RNA-398, and RNA-REMD-1 simulations, which were performed on GPUs. The CPU clusters include NICS Kraken, SDSC Gordon, and the University of Utah CHPC clusters. The GPU simulations were performed using the CUDA enabled PMEMD code59 on GPU accelerated nodes (NVIDIA Tesla M2090 GPUs) at either NICS Keeneland or the University of Utah CHPC.
Conformational Analysis
The REMD algorithm implemented in AMBER specifies that when an exchange attempt is successful, replicas will exchange thermostat temperatures (the alternative being the exchange of system coordinates). This results in each replica containing simulation data from a variety of temperatures. Often, the researcher is most interested data for a specific temperature and thus the REMD trajectory ensemble must be sorted such that contiguous data is obtained for each temperature level. In this paper, we refer to such a process as “sort by temperature”. Alternatively, it is occasionally of interest to study the data directly for each replica without sorting by temperature (as we do for our ensemble RMSD profile analysis). We will refer to this as “sort by replica”. In order to sort by temperature we used a development version of AMBER's Cpptraj. Conformational analysis, including RMSD profiles, clustering, principal component analysis, and torsion and distance calculations, were performed using a combination of AMBER's Ptraj and Cpptraj programs and in-house Perl scripts. All RMSD analysis was made using a common reference structure, which ensured that results could be compared. The common reference structure for both alanine dipeptide and rGACC was generated by GB energy minimization of initial build structure for each molecule. RMSD analysis was performed with mass-weighting using only heavy atoms for alanine dipeptide and all atoms for the RNA. Clustering of the RNA simulations was performed with Ptraj using the “averagelinkage” agglomerative algorithm, a critical distance epsilon value of 2.3 Å, and a variable sieve value which ensured that the initial clustering pass contained ∼5000 frames. The sieve, which uses an initial subset of randomly chosen frames to define the clustering divisions, was required because memory limitations do not allow complete clustering of a large number of frames. Thus five independent cluster analyses were performed for each simulation of interest and an average and standard deviation was reported. Animation of the PCA eigenvectors was performed using PCAsuite.60 Molecular graphics images were generated using UCSF Chimera.61
Results And Discussion
Alanine Dipeptide Conformations and Convergence
Due to its small size and simple structure, a solvated alanine dipeptide system is convenient for quick testing and demonstrating proof of principle. Thus we began by studying the feasibility of various explicitly solvated REMD simulations with this system. The primary purpose of these initial investigations was to demonstrate that a given simulation can reliably sample the conformational space of the solute of interest. One convenient method for observing the sampled space is to generate a histogram profile of the atomic RMSD values with respect to a common reference structure. We found that conventional MD (both constant volume and constant pressure), traditional REMD, and R-REMD produce nearly indistinguishable RMSD profiles (Figure 2). We note here that RMSD profiles represent a necessary but not sufficient test of convergence. Thus they are useful as an initial test to compare whether ensembles from two independent simulations overlap. For complex molecules, more detailed conformational analysis is required (and provided later) to confirm this in situations where multiple conformations may occupy the same RMSD space.
Figure 2.
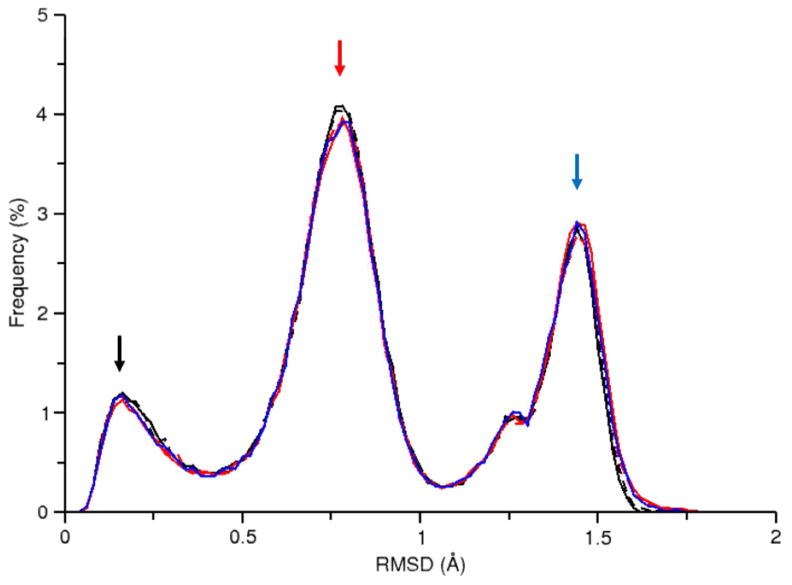
RMSD profile for the 300 K simulation data from the following alanine dipeptide simulations: ALA-NVT (solid black), ALA-NPT (dashed black), ALA-10REMD (solid red), ALA-24REMD (dashed red), ALA-R-REMD-L (solid blue). Tight overlap in the profiles suggests that these simulations explore very similar RMSD space. The colored arrows indicate RMSD profile points studied for the time convergence displayed in Figure 3. The points were chosen due to their variability as observed in Figure S2.
In order to gauge the convergence of these simulations, a more in-depth analysis is required. By plotting the cumulative RMSD profile over simulation time (Figure S2), it is possible to observe the time convergence at any point along the profile, such as the frequency maxima (Figure 3). From this convergence plot we find that REMD calculations require significantly less time to reach convergence than conventional MD and R-REMD calculations are nearly converged from the beginning of the simulation (the latter observation was also made by Okur et. al40). A comparison of the ten replica REMD simulation (ALA-10REMD) with the twenty-four replica REMD simulation (ALA-24REMD) suggests that the larger ensemble takes longer to converge to the same precision as the smaller ensemble, although near quantitative results are obtained fairly quickly (Figure S3).
Figure 3.
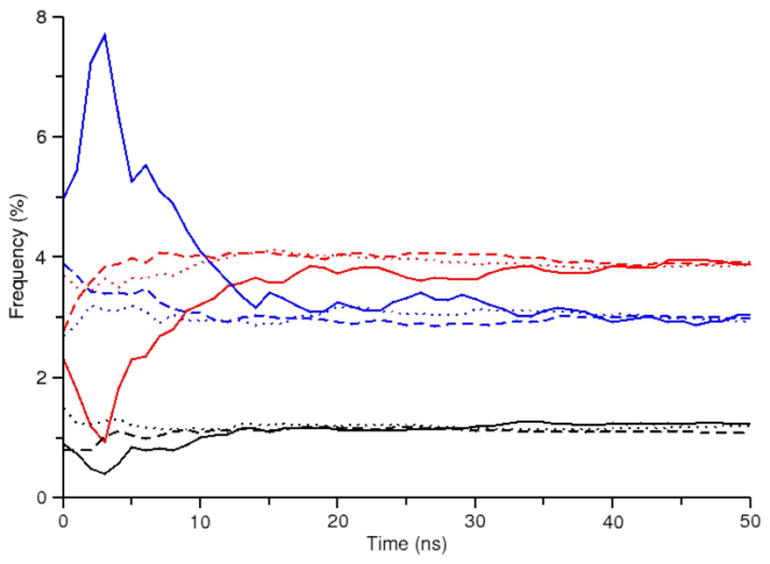
Convergence of alanine dipeptide RMSD profile maxima versus simulation time at 300 K for the following simulations: ALA-NVT (solid), ALA-10REMD (dashed), ALA-R-REMD-L (dotted). Colors correspond to the three RMSD profile maxima observed in Figure 2 at the following values: 0.16 (black), 0.78 (red), 1.44 Å (blue). Only the first 50 ns of each simulation are shown to emphasize the initial convergence period.
Unfortunately, cumulative-type plots of convergence don't always indicate that the ensemble has converged to the true value dictated by the force field if for some reason an individual replica becomes improperly trapped in a given conformation. A potentially more revealing test of convergence is found by plotting the RMSD profile of each replica trajectory prior to sorting the data by temperature. In an ideal scenario, a converged REMD ensemble would consist of replicas which each traverse temperature space many times and therefore, over the course of a long simulation, would sample identical conformational space. Thus, the RMSD profile of each replica would be identical to one another. Nearly complete convergence of the replica RMSD profiles can be qualitatively demonstrated for the REMD simulations of alanine dipeptide (Figure S4) and we suggest such plots should be regularly included in publications. As we show later in the results for the RNA system, replicas which become conformationally trapped are easily spotted using this method.
To quantify the conformational distribution of the alanine dipeptide simulations, the phi/psi torsion space of alanine dipeptide was divided into six regions (Figure 4, left) and the percent occupancy was calculated (Table 2). The regions were identified by locating the minima (which correspond to regions of highest density) in a population based free energy plot of the phi/psi space (Figure 4, right). A comparison of the conformational populations between the conventional MD simulations (ALA-NVT, ALA-NPT) and the traditional REMD simulations (ALA-10REMD, ALA-24REMD) suggests that the two approaches yield very similar conformational results (Table 2). Also, the use of constant pressure or constant volume does not seem to significantly affect the results for conventional MD. Additionally, it appears that REMD with ten replicas and a maximum temperature of 398 K (ALA-10REMD) yields similar results to the twenty-four replica REMD with a maximum temperature of 600 K (ALA- 24REMD). It is interesting to note that the REMD methods appear to sample the rare F conformation more frequently than conventional MD. This may suggest that that the conventional MD simulations require more simulation time than was collected to adequately sample the F conformation and that REMD efficiently traverses the energy barriers to this conformation.
Figure 4.
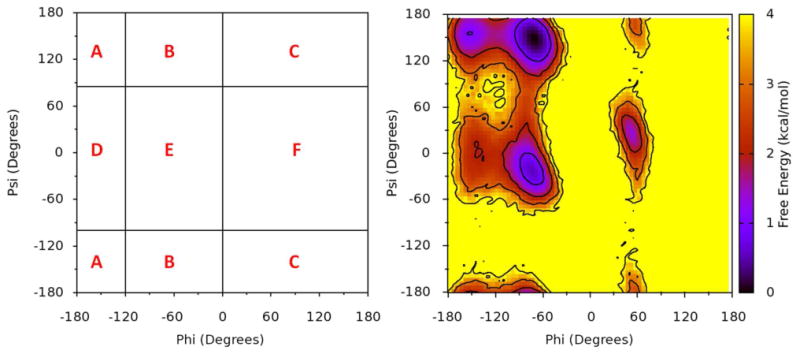
Identification of alanine dipeptide phi/psi conformational divisions. (Left) Six regions were identified and labeled A-F. (Right) Population based free energy plot based on phi/psi dihedral angles for the ALA-10REMD simulation at 300 K. Free energy estimates are calculated using the following equation: Gi = -kB T ln(Ni/NtOt).
Table 2.
Alanine dipeptide conformational distribution at 300 K for simulations in this work.
| Alanine Dipeptide Conformational Frequency (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Simulation | A | B | C | D | E | F | ||||||
|
| ||||||||||||
| ALA-NVT | 12.3 | (0.2) | 56.6 | (1.9) | 0.0 | (0.0) | 4.1 | (0.1) | 27.0 | (1.7) | 0.0 | (0.0) |
| ALA-NPT | 12.2 | (0.0) | 56.9 | (1.2) | 0.2 | (0.1) | 4.0 | (0.1) | 25.1 | (0.6) | 1.7 | (0.3) |
| ALA-10REMD | 11.3 | (0.2) | 54.2 | (0.1) | 0.6 | (0.1) | 3.8 | (0.0) | 26.0 | (0.3) | 4.1 | (0.7) |
| ALA-24REMD | 11.8 | (0.5) | 55.6 | (0.1) | 0.3 | (0.1) | 3.9 | (0.0) | 25.5 | (0.3) | 3.0 | (0.7) |
| ALA-R-REMD-S | 13.3 | (0.2) | 58.9 | (0.1) | 0.1 | (0.0) | 3.4 | (0.0) | 23.0 | (0.1) | 1.2 | (0.2) |
| ALA-R-REMD-L | 11.6 | (0.0) | 54.9 | (1.0) | 0.3 | (0.0) | 4.1 | (0.2) | 26.3 | (1.2) | 2.9 | (0.5) |
| ALA-TIG-1 | 11.3 | (0.7) | 49.5 | (2.9) | 0.9 | (0.3) | 4.2 | (0.2) | 28.9 | (1.5) | 5.2 | (2.1) |
| ALA-TIG-2 | 12.3 | (0.8) | 45.9 | (0.1) | 1.3 | (0.2) | 4.8 | (0.4) | 28.6 | (1.9) | 7.1 | (2.7) |
| ALA-TIG-3 | 11.3 | (0.2) | 46.0 | (2.0) | 2.0 | (0.5) | 4.7 | (0.1) | 27.8 | (0.2) | 8.2 | (2.0) |
Letters A-F indicate regions of the phi/psi space indicated in Figure 4, left. Given in parentheses is a crude approximation of error obtained by comparing the average frequency values from the first and second halves of a simulation according to the following equation: Error = abs(FirstHalf -SecondHalf)/2
When performing R-REMD simulations, in which a predefined reservoir quickly drives convergence of the REMD ensemble, the size and conformational diversity of the reservoir is an important concern. To understand how the composition of the reservoir affects the results, we examined a variety of reservoir sizes and compositions and report two examples here. The first, used in the ALA-R-REMD-L simulation, was a larger reservoir and consisted of 4764 frames collected every 10 ps from a 47 ns simulation at 398 K. Comparison of the RMSD profile of this reservoir with the 397.7 K temperature replica from the ALA-10REMD simulation suggests that the reservoir is a reasonable approximation of a converged ensemble at 398 K, although its profile is not smooth due to the relatively small number of frames used (Figure S5). Using this reservoir, the ALA-R-REMD-L simulation generates a conformational ensemble at 300 K very similar to those observed using traditional REMD (Table 2).
We also studied a much smaller reservoir (ALA-R-REMD-S), containing 538 artificially selected frames from the larger reservoir, in which structures from each of the three RMSD profile maxima are present but not at the correct relative frequencies (Figure S5). The resulting conformational distribution at 300 K from the ALA-R-REMD-S simulation is perturbed slightly relative to the other REMD and R-REMD simulations (Table 2), yet the small difference suggests that the R-REMD method is fairly robust at recovering the correct conformational frequencies even when the reservoir is sparse and non-Boltzmann weighted.
The R-REMD method offers significant savings in computational resources over traditional REMD. An even greater savings can potentially be found in the previously published TIGER2 method.38 Briefly, the TIGER2 method involves a small number of replicas for which a subset are rapidly heated from the baseline temperature to a higher sampling temperature and after a brief period rapidly quenched and equilibrated back to the baseline temperature. Following this heating and quenching cycle, the replicas are exchanged with the baseline replica with a probability based on the Metropolis criterion. The heating step allows the system to rapidly overcome energy barriers at high temperatures and the quenching step allows for replica exchange to occur at a reasonable rate at the baseline temperature. The TIGER2 algorithm is not implemented in AMBER, yet its simplicity makes using a wrapper script feasible, which is the approach we took. After testing our implementation of TIGER2, it was immediately clear that the equilibration period following the quenching step had a large effect on the exchange probability. We tested three quench-equilibration periods, 1, 2, and 5 ps, which resulted in exchange success rates of 0.066, 0.277, and 0.475 respectively (corresponding to ALA-TIG-1, ALA-TIG-2, and ALA-TIG-3). The conformational distribution of the TIGER2 simulations is similar in the overall trend to the other methods tested, yet there is a noticeable difference in the frequency of the B and F conformations (Table 2). It is not clear what causes this difference and further investigation will be necessary to understand the discrepancy. For this reason the TIGER2 method was not used to investigate the larger and more complex RNA system.
rGACC RNA conformational analysis
As part of an overall effort to improve RNA force fields, we wanted to focus computational efforts on a minimal RNA system for which conformational convergence could be quickly obtained and experimental data was available. The rGACC RNA has previously been studied in solution using NMR and in simulation using other force fields.39 Although it is very small, it still populates (at least two) A- form-like conformations and is therefore an ideal test case. The difficulty with using conventional MD simulations is that conformational convergence at the temperature of interest, even with the latest accelerated hardware, is difficult to achieve. We performed a 5 μs simulation at 300 K (RNA-NPT) and the atomic RMSD versus time plot indicates that only a few transitions occur between the major conformations on that timescale (Figure 5, top).
Figure 5.
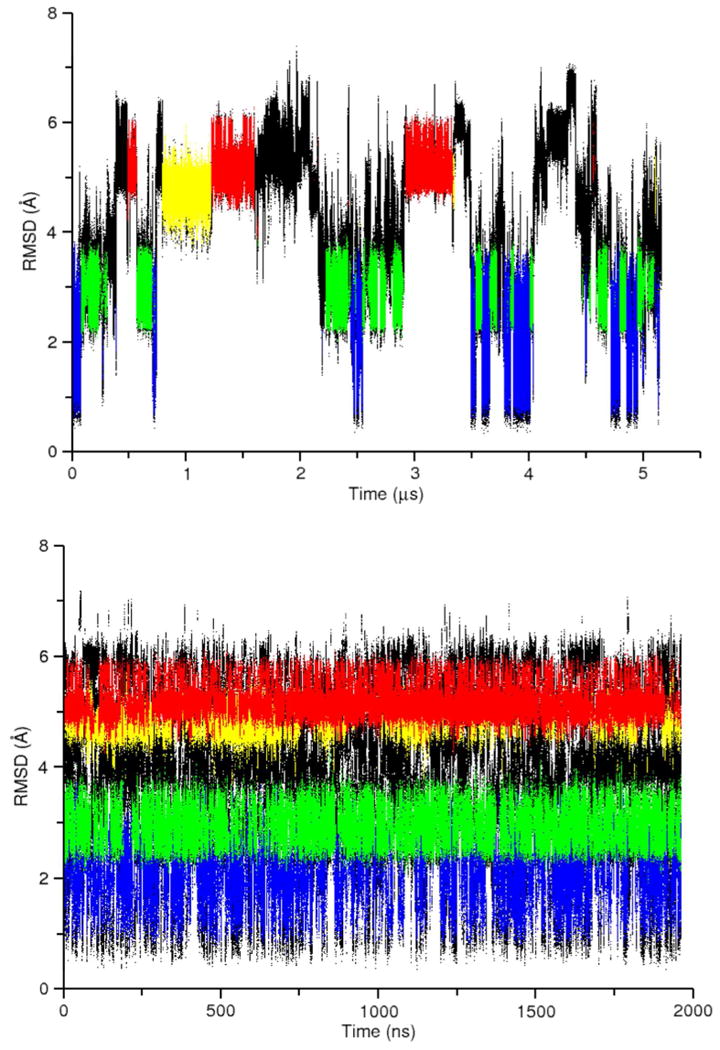
RMSD analysis of the RNA-NPT (top) and the RNA-REMD-1 (bottom) simulations. The data for RNA-REMD-1 is taken exclusively from the 277 K temperature level. The colored regions correspond to the four most populated conformational clusters depicted in Figure 6 as follows: Intercalated (red), NMR Minor (green), NMR Major (blue), Inverted (yellow), other conformations (black). (Right) RMSD profile for the data at left.
Identification of four major conformations was performed by clustering and a representative structure of each is shown in Figure 6 in both molecular graphics and simplifying cartoon. Additional conformations were identified other than the four described here, but due to their population frequencies being 3% or less, they were not studied further in great detail. Quantitative frequencies of the four conformations are given in Table 3. The most populated conformation in the RNA-NPT simulation, a non-A-form conformation which we term the “Intercalated structure”, does not fit the NMR data and was also observed to dominate a microsecond timescale simulation using the modified force field by Yildirim et. al.39 The next most populated conformations, termed the “NMR Minor” and “NMR Major”, are consistent with the NMR data as their names suggest and are essentially A-form structures. However, Yildirim et. al. concluded that the NMR Major structure should dominate in solution with a small fraction of the NMR Minor structure present as well.39 In contrast to the experimental results, we observed a greater frequency of the NMR Minor structure compared to the NMR Major structure. The fourth structure, which was not described in the previous computational study of rGACC but was reported in a recent study of rCCCC,62 is termed the “Inverted” structure. It is a non-A-form conformation and is also not consistent with the observed NMR data.
Figure 6.
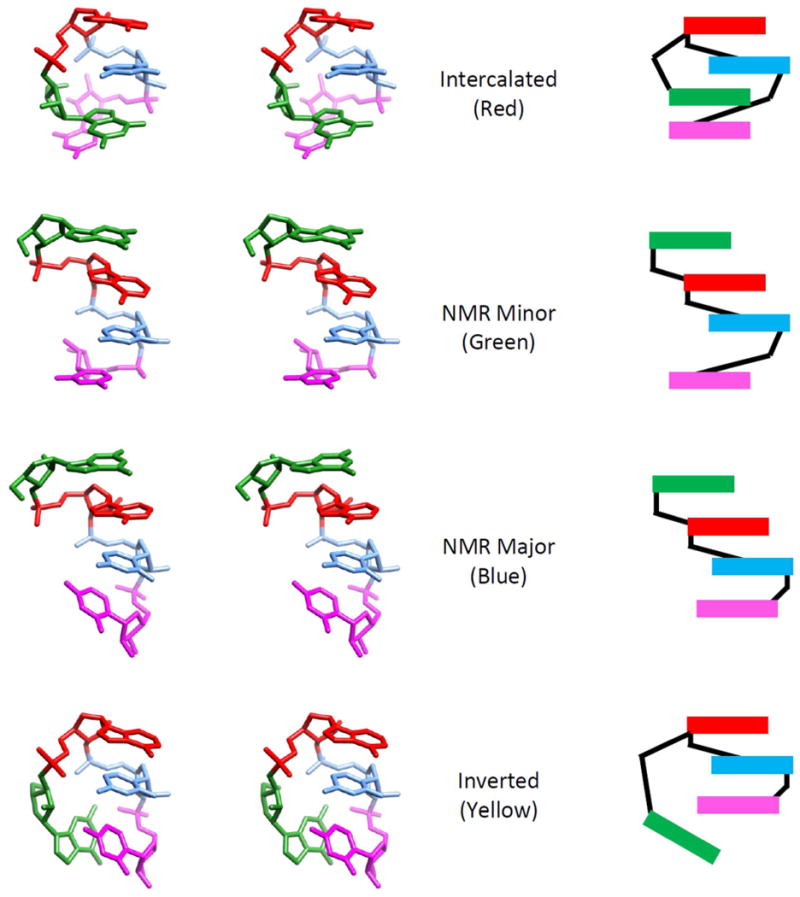
Stereoview and cartoon representation of the representative structures for the four most populated conformational clusters of the RNA-NPT simulation. Each structure is labeled with a reference name and color which is used in other plots to indicate to the conformation.
Table 3.
Conformational frequency of the RNA simulations determined by cluster analysis.
| rGACC Conformational Frequency (%) | ||||
|---|---|---|---|---|
| Simulation ID | Intercalated | NMR Minor | NMR Major | Inverted |
|
| ||||
| RNA-NPT1 | 16.0 (0.3) | 12.9 (0.7) | 9.2 (0.4) | 8.4 (0.1) |
| RNA-3982 | 6.2 (0.4) | 3.5 (0.3) | 3.1 (0.1) | 7.1 (0.5) |
| RNA-REMD-GB | -- -- | -- -- | -- -- | 92.9 (0.7) |
| RNA-REMD-1 | 24.5 (0.9) | 15.9 (0.7) | 11.8 (0.6) | 7.6 (0.0) |
| RNA-REMD-2 | 24.2 (1.2) | 10.5 (1.0) | 8.8 (0.5) | 9.9 (0.1) |
| RNA-REMD-3 | 18.8 (0.9) | 16.3 (1.0) | 13.1 (0.5) | 7.3 (0.1) |
| RNA-rREMD-S | 29.4 (0.1) | 28.3 (1.1) | 12.0 (0.2) | -- -- |
| RNA-rREMD-1 | 18.7 (0.3) | 15.5 (0.7) | 13.1 (0.4) | 11.3 (0.1) |
| RNA-rREMD-2 | 18.5 (0.1) | 15.3 (1.0) | 13.6 (0.1) | 10.9 (0.0) |
| RNA-rREMD-3 | 18.7 (0.5) | 14.6 (0.7) | 14.0 (0.4) | 10.2 (0.1) |
The four conformational categories are structurally depicted in Figure 6. Data shown is for the 277 K temperature level, except for the RNA-NPT and RNA-398 simulations, for which the temperature is indicated (below). Data at 299 K and 328 K are given in Table S2. An error estimate is given in parentheses which corresponds to the standard deviation of five independent clustering calculations.1 Simulation performed at 300 K.2 Simulation performed at 398 K. The “-- --” indicates that the conformation was not observed in the top fifteen most populated clusters.
At 5 μs of conventional MD, the RNA-NPT simulation is still not converged enough for reliable force field comparison purposes. Even at 100 ns/day simulation speeds, attainable through use of GPU accelerated simulations, this simulation required 50 days to complete. Thus a more convenient, reliable, and cost effective method for studying the conformational landscape of complex structures is of interest. REMD simulations provide an attractive alternative. By coupling high temperature simulations which traverse energy barriers quickly with low temperatures simulations at experimentally relevant conditions, a converged conformational ensemble is expected to be obtained quicker than by conventional MD alone.63 REMD is typically performed with implicit solvent because the number of replicas increases with the size of the system. Unfortunately, as of present, RNA simulations generally do not behave well in implicit solvent. To demonstrate this, we performed a 500 ns per replica REMD simulation in GB solvent using six replicas spanning a temperature range from 277 – 463 K. At lower temperatures, the RNA nearly exclusively adopts the Inverted conformation whereas at the maximum temperature the RNA is largely unstructured (Figure S6). These results are consistent with our previous experience with RNA simulations in implicit solvent (unpublished) and suggest that explicit solvent is necessary for (even crudely accurate) simulations in the foreseeable future.
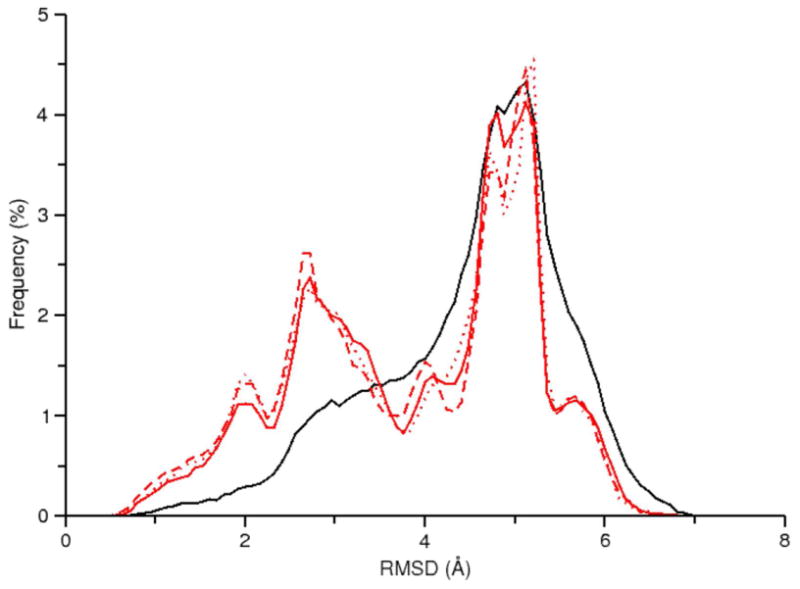
To evaluate the performance of explicitly solvated REMD simulations, we performed three simulations: one 2 μs per replica timescale simulation using GPU acceleration (RNA-REMD-1) and two shorter, 400-500 ns per replica simulations using traditional CPU hardware (RNA-REMD-2 and RNA-REMD-3). Inspection of the RMSD versus time plot for RNA-REMD-1 suggests that frequent conversion between the four major conformations occurs over the course of the simulation (Figure 5, bottom). Comparison of the RMSD profile at 277 K for the three traditional REMD simulations reveal the difficulty in obtaining converged results (Figure 7). For instance a large peak at an RMSD value of 4.0 Å is observed in the RNA-REMD-2 simulation but is much smaller in the other two simulations. The corresponding structure to this peak (Figure S7) is not one of the previously mentioned conformations. Although it is present in low frequencies in the other REMD simulations it seems to dominate the RNA-REMD-2 simulation. A small number of replicas are the main contributor to the overpopulation of this conformation in the ensemble and those replicas remain trapped for significant time periods despite regularly traversing the complete temperature space (Figure S8). This problem may be exacerbated by a short exchange attempt interval which will tend to reduce the average duration a replica stays at a given temperature. If a conformational transition requires some minimum time to proceed and also requires a high temperature to make traversal of an energy barrier more likely, a short exchange attempt interval may cause the replica to stay conformationally trapped. Extending the exchange attempt interval and/or increasing the number of target temperatures intervals above 398 K may alleviate this problem.
Figure 7.
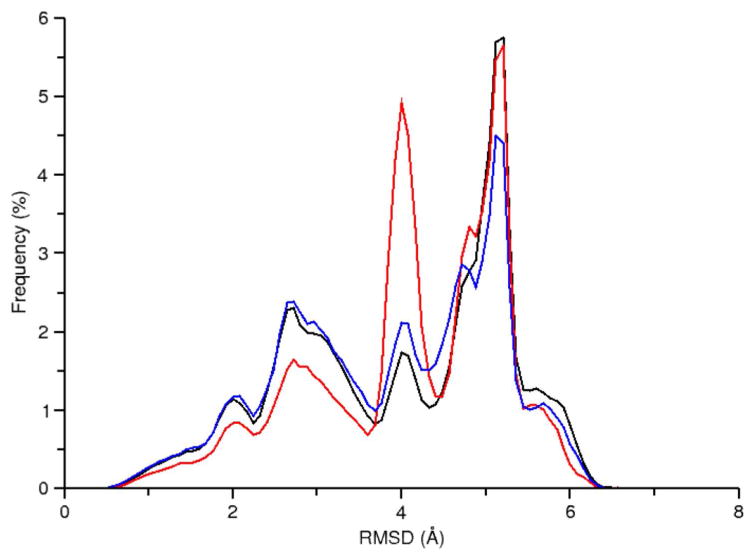
RMSD profile for data collected at 277 K from the following simulations: RNA-REMD-1 (black), RNA-REMD-2 (red), and RNA-REMD-3 (blue).
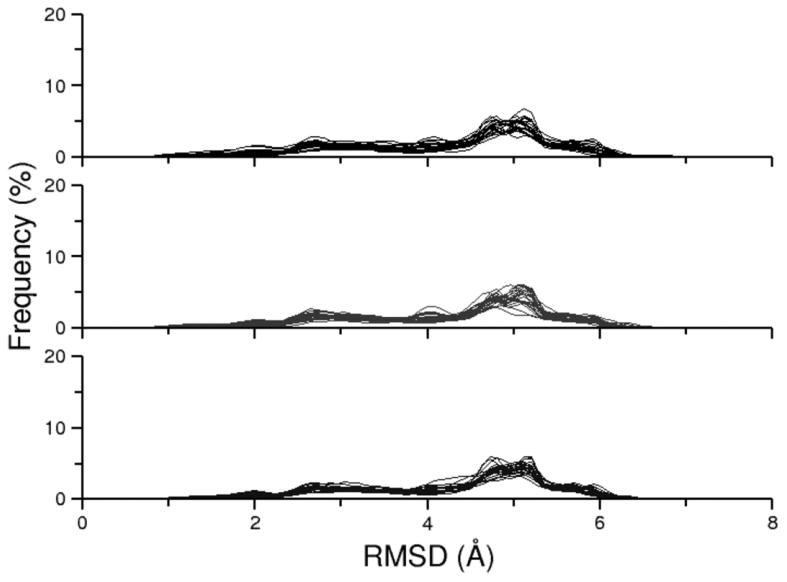
As we mentioned in the results for alanine dipeptide, a convenient method to examine REMD ensemble convergence is to study the replica RMSD profiles for the ensemble data sorted by replica rather than sorted by temperature. In the ideal case, a converged REMD ensemble will generate an identical curve for each replica. We demonstrate the tendency towards a converged RMSD profile in Figure S9. At 200 ns, the replica RMSD profiles are dispersed and a consensus profile is difficult to distinguish. By the end of the 2 μs simulation, the RMSD profiles are much more consistent although a few outliers remain, suggesting that longer simulation is required for complete convergence. A comparison can also be made between the three traditional REMD simulations (Figure 8). These results also suggest that the 2 μs simulation (RNA-REMD-1) is closer to convergence than the shorter simulations (RNA-REMD-2 and RNA-REMD-3), although a single consensus profile is still not achieved.
Figure 8.
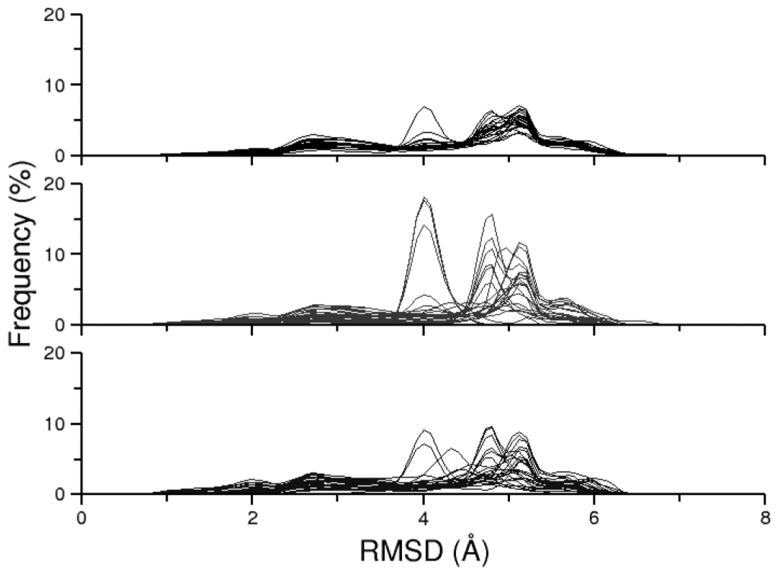
Overlay of the twenty-four replica RMSD profiles for the following simulations: RNA-REMD-1 (top), RNA-REMD-2 (middle), RNA-REMD-3 (bottom). A fully converged REMD ensemble should produce an identical curve for each replica and thus disorder in the plot indicates that the simulation has not yet fully converged.
The conformational landscape of the rGACC structure is much more complicated than alanine dipeptide and cannot easily be represented by the equivalent of a phi/psi plot. Instead we used principal component analysis to identify the primary motional modes of the RNA in the RNA-REMD-1 simulation (Figure 9A,B). The division of clustering along these PCA axes can be visualized (Figure 9C) and a population based free energy plot reveals the relative minima (Figure S10). These plots show that the two conformations consistent with NMR data, NMR Major and NMR Minor, are nearly overlapping in the primary motional axes whereas the two non-A-form conformations are distant from the NMR structures, suggesting a force field problem which leads to incorrect population sampling.
Figure 9.
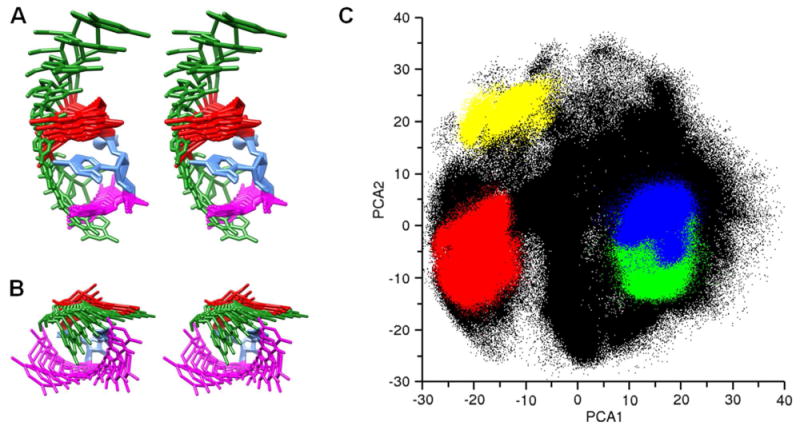
PCA analysis of the RNA-REMD-1 simulation at 277 K. Stereoview depiction of the motion described by the first (A) and second (B) eigenvectors determined by principal component analysis. (C) Distribution of the simulation data along the first two eigenvectors identified by PCA. Colored regions correspond to the four most populated conformational clusters depicted in Figure 6 as follows: Intercalated (red), NMR Minor (green), NMR Major (blue), Inverted (yellow), other conformations (black).
The results of traditional REMD suggest that even on fairly long timescales by current standards and at significant resource cost, conformational convergence is not achieved. Thus, this is not a very feasible method for force field development which requires multiple such simulations run in a linear fashion. Given the success we found with R-REMD simulations of alanine dipeptide, we decided to investigate whether the method was also feasible for larger more complicated systems like rGACC. To generate the reservoir, a 1.4 μs simulation of the RNA at 398 K was performed saving frames every 10 ps (RNA-398). A plot of the RMSD versus simulation time reveals that all four conformations are sampled many times during the simulation and a smooth RMSD profile is generated (Figure S11). This is an important observation because it shows that conventional MD at 398 K does not exhibit the conformational trapping behavior which was observed for the traditional REMD approach. As we demonstrate below, the R-REMD method also does not suffer from conformational trapping due to the frequent exchange with this high temperature reservoir.
Three independent R-REMD simulations were run using this reservoir: RNA-R-REMD-1 (205 ns/replica), RNA-R-REMD-2 (160 ns/replica), and RNA-R-REMD-3 (250 ns/replica). Analysis of the RMSD profile for these simulations shows that the conformational detail at 277 K emerges from the smooth, distinctly different reservoir profile at 398 K (Figure 10). Inspection of the replica RMSD profiles (sorted by replica, not temperature) for the three R-REMD simulations suggest improved convergence (Figure 11) as do the similar quantitative results obtained from cluster analysis (Table 3). Comparison of the quantitative clustering results for the R-REMD method at three temperatures, 277, 299, and 328 K, reveals an interesting trend (Table 3 and Table S2). Increasing the temperature significantly reduces the frequency of the two NMR consistent structures but not the non-A-form structures. Only a small decrease is seen for the Intercalated structure and an increase is observed for the Inverted structure. This suggests that the entropic component of the free energy favors the non-A-form structures more than the NMR structures and thus this preference increases with temperature.
Figure 10.
RMSD profile at the 277 K temperature level for RNA-R-REMD-1 (solid red), RNA-R-REMD-2 (dashed red), RNA-R-REMD-3 (dotted red) as well as the profile for the structural reservoir used in these simulations (which was generated by conventional MD at 398 K).
Figure 11.
Overlay of the twenty-four replica RMSD profiles for the following simulations: RNA-R-REMD-1 (top), RNA-R-REMD-2 (middle), RNA-R-REMD-3 (bottom). The replica profiles for these R-REMD simulations seem to be closer to convergence than the traditional REMD simulations (see Figure 8).
In order to test how the composition of the reservoir affects the R-REMD results for the RNA system, we performed an additional simulation with a much smaller reservoir. This simulation, RNA-R-REMD-S, contained only 10,000 frames from the first 100 ns of the RNA-398 simulation. In this time period, three of the four major conformations were sampled including the Intercalated, NMR Minor, and NMR Major (Figure S11). The Inverted conformation was not sampled until ∼150 ns and thus was not present in this reservoir. As expected, quantitative conformational analysis of the RNA-R-REMD-S simulation shows that the Inverted conformation is not present in any significant amount at the three listed temperatures (Table 3 and Table S2). This underscores the importance of having representatives from each significantly populated structural class present in the R-REMD reservoir.
Comparison of rGACC RNA simulation data with NMR data
Given that the R-REMD simulations appear to be a reliable method for obtaining an accurate conformational data about rGACC in simulation, we decided to make a deeper comparison between the published NMR data and the simulation data (specifically for RNA-R-REMD-3). Two easy comparisons include the sugar pucker and base orientation preferences. Values at three different temperatures for these metrics are given in Table 4.
Table 4.
Sugar pucker and base orientation at three temperatures from the RNA-R-REMD-3 simulation.
| 277 K | 299 K | 328 K | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | % C3′-endo | % Anti | % C3′-endo | % Anti | % C3′-endo | % Anti | |||
|
| |||||||||
| G1 | 91 | (80-90) | 64 | 89 | (60-70) | 57 | 84 | (50-60) | 48 |
| A2 | 81 | (80-90) | 96 | 79 | (80-90) | 94 | 76 | (70-80) | 92 |
| C3 | 87 | (80-90) | 98 | 84 | (80-90) | 98 | 80 | (70-80) | 97 |
| C4 | 52 | (70-80) | 99 | 50 | (70-80) | 99 | 49 | (60-70) | 99 |
In parentheses are the experimentally determined ranges for the sugar pucker determined by NMR at the following temperatures: 278, 298, and 328 K.39 The base orientation, determined by NMR, was anti for all four residues at 275 K. Values discussed in the main text are underlined.
The sugar pucker values observed in simulation are consistent with the NMR measurements for the first three residues, however residue C4 significantly underpopulates the C3′-endo conformation compared to experiment at 277 K. At 328 K, both residues G1 and C4 are out of the experimental range, with G1 overpopulating C3′-endo and C4 still underpopulating C3′-endo. The deviation of the simulation sugar pucker from experiment at C4 is likely due to the overpopulation of the NMR Minor structure, which prefers C2′-endo at C4. Quantitative values for the other metric of interest, base orientation, were not obtained in the experimental publication; however the NOESY spectrum indicated that all four residues preferred the anti conformation at 275 K. Simulation values are consistent with this observation, with the possible exception of residue G1. It seems likely that a stronger NOE would have been observed if the anti/syn ratio was 64/36, as was observed in simulation. Taken together, these two metrics offer only a weak indication that the force field is incorrectly modeling the solution structure, primarily by highlighting the under-population of the C3′-endo sugar pucker state by residue C4.
In order to make a more detailed comparison with the NMR data, we investigated whether any of experimentally observed NOEs were violated. To study this, the r6-averaged distances for all possible NOE pairs were calculated for the RNA-R-REMD-3 simulation at 277 K. Of the 21 experimentally supported atom distance restraints, only one pair was in violation: atoms G1 H8 and A2 H8. The experimental restraint lower and upper bounds are 2.0 – 5.0 Å, while the simulation r6-average is 5.3 Å. (Note: the lower/upper bounds listed for this restraint in the publication SI were written as 2.0 – 10.0 Å, but this was done to “search broader conformational space” during annealing. In fact, the NOESY data indicate that the upper bound should be less than 5.0 Å.39) A violation rate of 1/21 would ordinarily be fairly high; however, in this case it leaves us with just one data point. Thus it is important to inspect atom pairs for which the simulation results would predict an NOE signal but where none is observed experimentally. This approach was noted in the previous computational study when discussing the non-A-form Intercalated structure.39 We identified fifteen atom pairs for which the r6-average value was 5.0 Å or less, but for which no NOE signal was identified by NMR (Table 5). A visual depiction of these predicted atom pairs, overlaid on both the NMR Major and Intercalated conformations, as well as the single restraint violation are shown in Figure 12.
Table 5.
NOEs predicted by the RNA-R-REMD-3 simulation at 277 K but not observed experimentally by NMR.
| Atom 1 | Atom 2 | r6 Avg. | Int. | Min. | Maj. | Inv. |
|---|---|---|---|---|---|---|
|
| ||||||
| :3@H3′ | :4@H3′ | 2.9 | 2.6 | 2.8 | 4.8 | 5.0 |
| :3@H3′ | :4@H2′ | 2.9 | 4.8 | 2.9 | 6.1 | 6.5 |
| :1@H2′ | :3@H5 | 3.2 | 3.6 | 6.6 | 5.8 | 3.1 |
| :1@H5′ | :4@H2′ | 3.4 | 2.7 | 12.6 | 14.1 | 11.2 |
| :1@H8 | :4@H3′ | 3.8 | 2.9 | 12.5 | 14.4 | 7.5 |
| : 1@H2′ | :4@H5 | 3.8 | 6.3 | 13.6 | 8.4 | 2.6 |
| :1@H8 | :4@H2′ | 3.9 | 3.2 | 12.1 | 13.9 | 6.7 |
| :1@H8 | :3@H3′ | 4.5 | 3.2 | 12.2 | 11.7 | 8.5 |
| :1@H3′ | :3@H6 | 4.6 | 4.0 | 7.3 | 6.5 | 4.3 |
| :1@H8 | :4@H6 | 4.7 | 3.5 | 14.1 | 11.9 | 6.7 |
| :1@H5′ | :4@H3′ | 4.7 | 4.5 | 12.9 | 14.6 | 10.6 |
| :1@H8 | :4@H5 | 4.8 | 4.0 | 14.9 | 10.1 | 5.0 |
| :1@H8 | :3@H2′ | 4.9 | 4.0 | 12.6 | 12.3 | 8.7 |
| :1@H8 | :3@H6 | 4.9 | 4.2 | 9.6 | 8.9 | 8.5 |
| :1@H5′′ | :4@H2′ | 5.0 | 4.0 | 11.0 | 12.7 | 11.0 |
The first three columns indicate the atom pairs and the r6-averaged distances obtained from the simulation. The last four columns list the corresponding distance observed in the most populous representative structures which are depicted in Figure 6. Conformational abbreviations: Int. (Intercalated). Min. (NMR Minor). Maj. (NMR Major). Inv. (Inverted).
Figure 12.
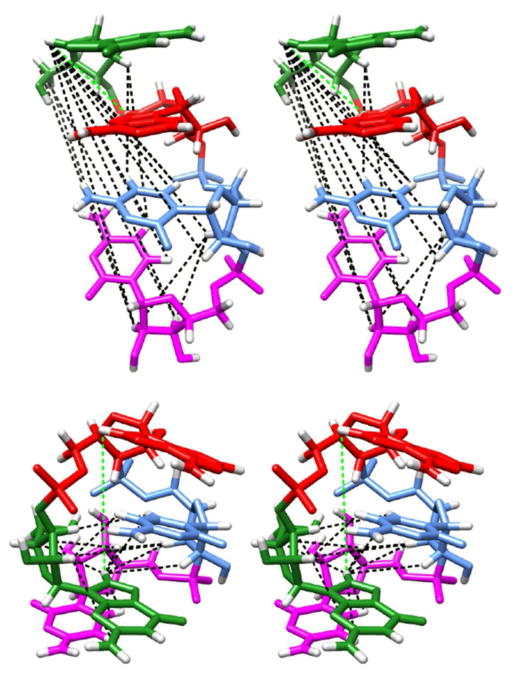
Stereo views of the simulation predicted NOEs (black lines) for RNA-R-REMD-3 at 277 K mapped onto the NMR Major conformation (top) and the Intercalated conformation (bottom). Only the NOEs for which the Intercalated conformation is the primary contributor are shown. The single NOE violation between G1 H8 and A2 H8 is also depicted (green line).
This data strongly suggests that the force field overpopulates the non-A-form structures. We acknowledge that even if the non-A-form structures were truly present in solution, it is unlikely that NOE signals would be observable for all fifteen identified atom pairs due to the limitations of experimental studies. However, we do show that there exist many atom pairs for which an NOE signal should arise if the non-A-form conformation is really present. Taken together, our comparison to the published NMR data suggests that the ff12SB force field requires further improvements in order to adequately model this RNA structure.
Conclusion
In this work we have demonstrated a tractable method for reliable generation of conformational ensembles from explicitly solvated simulations. This represents a necessary step towards cost-effective force field development of RNA. Conventional MD, even with advances in accelerated hardware, is still not a feasible method for research that requires both quick turnaround and extensive sampling of a rugged conformational landscape. REMD, which has been one of the traditional solutions to this problem, is not nearly as cost effective when explicit solvent is required as is the case for most RNA simulations. In addition, we have shown that even REMD simulations of significant length (2 μs) may be slow to converge for certain systems. In other words, although traditional REMD has improved convergence with respect to conventional MD, that does not mean it is guaranteed to converge. Thus we have turned to the R-REMD method, in which a pre-generated, high temperature reservoir drives the convergence of a REMD ensemble. At high temperatures, the rugged conformational landscape is much more easily traversed than at lower temperatures (compare Figure 5, top and S11) and thus the reservoir is quickly and relatively cheaply generated. Use of the reservoir reduces the simulation time required for the resource intensive REMD step and thus leads to converged results which are cheaper than traditional REMD. It should also be mentioned that the aggregate simulation time for all replicas is far less than traditional REMD and comparable to that of the brute force conventional MD, and thus is a more efficient use of computational resources.
Care must be used when employing R-REMD method. For example, the reservoir must sufficiently sample the energy landscape in order to include all of the major conformations. We observed that a deliberately small reservoir completely eliminated one of the major rGACC conformations. Despite this, a perfectly converged reservoir does not seem to be necessary. We showed that a reservoir with skewed populations (albeit one with that included all major conformational classes) only slightly modified the conformational distribution at the baseline temperature. Finally, we suggest it is necessary to plot RMSD profiles for the data sorted by replica, rather than temperature, in order to determine how much simulation time is required for convergence. Significant discrepancies between these profiles can indicate that the ensemble has not yet converged and might contain trapped replicas.
After we determined that the R-REMD method was a reliable method for generating consistent data regarding the conformational landscape of rGACC, we were able to make specific comparisons with the published NMR data. These comparisons suggest that the ff12SB force field overpopulates non-A-form conformations for the rGACC tetramer. The source of this error is not immediately clear, although we suspect that refinements to the sugar pucker torsions or backbone torsions may improve the results. In addition to studying force field refinements, we can also easily study the effect of various water models, salt composition, and salt concentration on the results. The R-REMD method also holds promise for studying flexible portions of much larger molecules. For instance, a non-canonical region of a larger RNA could be studied by generating a high temperature reservoir in which base pair restraints are used to prevent complete unfolding. The resulting R-REMD simulation would allow a focus on the conformational landscape of regions where the current force field is flawed (i.e., non-canonical regions), while limiting unfolding in helical regions where the force field behaves better. This approach may also be useful in studying ligand binding modes in which receptors which undergo significant rearrangement.
Supplementary Material
Acknowledgments
The authors would like to acknowledge funding from the NIH (R01-GM081411, TEC), the Eccles Foundation (NMH), and the American Foundation for Pharmaceutical Education (NMH), as well as generous computing allocations from NSF XSEDE MCA01S027, extensive friendly user time on the KIDS and KFS Keeneland systems at GATech/NICS, and the University of Utah Center for High Performance Computing.
Footnotes
Supporting Information Available: Additional tables and figures which are referenced in the manuscript are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Meister G. RNA Biology: An Introduction. Wiley; 2011. [Google Scholar]
- 2.MacKerell AD, Jr, Nilsson L. Molecular dynamics simulations of nucleic acid-protein complexes. Curr Opin Struct Biol. 2008;18:194. doi: 10.1016/j.sbi.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheatham TE, III, Kollman PA. Molecular Dynamics Simulation of Nucleic Acids. Annu Rev Phys Chem. 2000;51:435. doi: 10.1146/annurev.physchem.51.1.435. [DOI] [PubMed] [Google Scholar]
- 4.Cheatham TE, 3rd, Young MA. Molecular dynamics simulation of nucleic acids: successes, limitations, and promise. Biopolymers. 2000;56:232. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CMT, Onuchic JN, Sanbonmatsu KY. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Natl Acad Sci U S A. 2011;108:18943. doi: 10.1073/pnas.1108363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α/γ Conformers. Biophys J. 2007;92:3817. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banáš P, Hollas D, Zgarbová M, Jurečka P, Orozco M, Cheatham TE, Sponer Ji, Otyepka M. Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins. J Chem Theory Comput. 2010;6:3836. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zgarbová M, Otyepka M, Sponer Ji, Mládek At, Banáš P, Cheatham TE, Jurečka P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J Chem Theory Comput. 2011;7:2886. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildirim I, Stern HA, Kennedy SD, Tubbs JD, Turner DH. Reparameterization of RNA χ Torsion Parameters for the AMBER Force Field and Comparison to NMR Spectra for Cytidine and Uridine. J Chem Theory Comput. 2010;6:1520. doi: 10.1021/ct900604a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yildirim I, Kennedy SD, Stern HA, Hart JM, Kierzek R, Turner DH. Revision of AMBER Torsional Parameters for RNA Improves Free Energy Predictions for Tetramer Duplexes with GC and iGiC Base Pairs. J Chem Theory Comput. 2012;8:172. doi: 10.1021/ct200557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denning EJ, Priyakumar UD, Nilsson L, Mackerell AD., Jr Impact of 2′-hydroxyl sampling on the conformational properties of RNA: update of the CHARMM all-atom additive force field for RNA. J Comput Chem. 2011;32:1929. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Dong F, Wagoner JA, Baker NA. Assessing the performance of implicit solvation models at a nucleic acid surface. Phys Chem Chem Phys. 2008;10:4889. doi: 10.1039/b807384h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong Z, Xiao Y. RNA stability under different combinations of amber force fields and solvation models. J Biomol Struct Dyn. 2010;28:431. doi: 10.1080/07391102.2010.10507372. [DOI] [PubMed] [Google Scholar]
- 15.Kelso C, Simmerling C. In: Computational Studies of RNA and DNA. Šponer J, Lankaš F, editors. Vol. 2. Springer; Netherlands: 2006. p. 147. [Google Scholar]
- 16.Zuckerman DM. Equilibrium Sampling in Biomolecular Simulations. Annu Rev Biophys Biomol Struct. 2011;40:41. doi: 10.1146/annurev-biophys-042910-155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141. [Google Scholar]
- 18.Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60:96. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 19.Nymeyer H, Gnanakaran S, Garcia AE. Atomic simulations of protein folding, using the replica exchange algorithm. Methods Enzymol. 2004;383:119. doi: 10.1016/S0076-6879(04)83006-4. [DOI] [PubMed] [Google Scholar]
- 20.Kannan S, Zacharias M. Folding simulations of Trp-cage mini protein in explicit solvent using biasing potential replica-exchange molecular dynamics simulations. Proteins: Struct., Funct., Bioinf. 2009;76:448. doi: 10.1002/prot.22359. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen PH, Stock G, Mittag E, Hu CK, Li MS. Free energy landscape and folding mechanism of a β-hairpin in explicit water: A replica exchange molecular dynamics study. Proteins: Struct., Funct., Bioinf. 2005;61:795. doi: 10.1002/prot.20696. [DOI] [PubMed] [Google Scholar]
- 22.Paschek D, Nymeyer H, García AE. Replica exchange simulation of reversible folding/unfolding of the Trp-cage miniprotein in explicit solvent: On the structure and possible role of internal water. J Struct Biol. 2007;157:524. doi: 10.1016/j.jsb.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Periole X, Mark AE. Convergence and sampling efficiency in replica exchange simulations of peptide folding in explicit solvent. J Chem Phys. 2007;126:014903. doi: 10.1063/1.2404954. [DOI] [PubMed] [Google Scholar]
- 24.Sanbonmatsu KY, García AE. Structure of Met-enkephalin in explicit aqueous solution using replica exchange molecular dynamics. Proteins: Struct., Funct., Bioinf. 2002;46:225. doi: 10.1002/prot.1167. [DOI] [PubMed] [Google Scholar]
- 25.Sgourakis NG, Merced-Serrano M, Boutsidis C, Drineas P, Du Z, Wang C, Garcia AE. Atomic-Level Characterization of the Ensemble of the Aβ(1-42) Monomer in Water Using Unbiased Molecular Dynamics Simulations and Spectral Algorithms. J Mol Biol. 2011;405:570. doi: 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannan S, Zacharias M. Application of biasing-potential replica-exchange simulations for loop modeling and refinement of proteins in explicit solvent. Proteins: Struct., Funct., Bioinf. 2010;78:2809. doi: 10.1002/prot.22796. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez-Cruz CA, Makhatadze GI, Garcia AE. Protonation/deprotonation effects on the stability of the Trp-cage miniprotein. Phys Chem Chem Phys. 2011;13:17056. doi: 10.1039/c1cp21193e. [DOI] [PubMed] [Google Scholar]
- 28.Paschek D, Day R, Garcia AE. Influence of water-protein hydrogen bonding on the stability of Trp-cage miniprotein. A comparison between the TIP3P and TIP4P-Ew water models. Phys Chem Chem Phys. 2011;13:19840. doi: 10.1039/c1cp22110h. [DOI] [PubMed] [Google Scholar]
- 29.Kannan S, Zacharias M. Folding of a DNA Hairpin Loop Structure in Explicit Solvent Using Replica-Exchange Molecular Dynamics Simulations. Biophys J. 2007;93:3218. doi: 10.1529/biophysj.107.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kannan S, Zacharias M. Role of the closing base pair for d(GCA) hairpin stability: free energy analysis and folding simulations. Nucleic Acids Res. 2011;39:8271. doi: 10.1093/nar/gkr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villa A, Widjajakusuma E, Stock G. Molecular Dynamics Simulation of the Structure, Dynamics, and Thermostability of the RNA Hairpins uCACGg and cUUCGg. J Phys Chem B. 2007;112:134. doi: 10.1021/jp0764337. [DOI] [PubMed] [Google Scholar]
- 32.Zuo G, Li W, Zhang J, Wang J, Wang W. Folding of a Small RNA Hairpin Based on Simulation with Replica Exchange Molecular Dynamics. J Phys Chem B. 2010;114:5835. doi: 10.1021/jp904573r. [DOI] [PubMed] [Google Scholar]
- 33.Garcia AE, Paschek D. Simulation of the Pressure and Temperature Folding/Unfolding Equilibrium of a Small RNA Hairpin. J Am Chem Soc. 2007;130:815. doi: 10.1021/ja074191i. [DOI] [PubMed] [Google Scholar]
- 34.Kirmizialtin S, Elber R. Computational Exploration of Mobile Ion Distributions around RNA Duplex. J Phys Chem B. 2010;114:8207. doi: 10.1021/jp911992t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ioannou F, Archontis G, Leontidis E. Specific Interactions of Sodium Salts with Alanine Dipeptide and Tetrapeptide in Water: Insights from Molecular Dynamics. J Phys Chem B. 2011;115:13389. doi: 10.1021/jp207068m. [DOI] [PubMed] [Google Scholar]
- 36.Kwac K, Lee KK, Han JB, Oh KI, Cho M. Classical and quantum mechanical/molecular mechanical molecular dynamics simulations of alanine dipeptide in water: Comparisons with IR and vibrational circular dichroism spectra. J Chem Phys. 2008;128:105106. doi: 10.1063/1.2837461. [DOI] [PubMed] [Google Scholar]
- 37.Vymetal Ji, Vondrášek Ji. Metadynamics As a Tool for Mapping the Conformational and Free-Energy Space of Peptides — The Alanine Dipeptide Case Study. J Phys Chem B. 2010;114:5632. doi: 10.1021/jp100950w. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Latour RA, Stuart SJ. TIGER2: An improved algorithm for temperature intervals with global exchange of replicas. J Chem Phys. 2009;130:174106. doi: 10.1063/1.3129342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildirim I, Stern HA, Tubbs JD, Kennedy SD, Turner DH. Benchmarking AMBER Force Fields for RNA: Comparisons to NMR Spectra for Single-Stranded r(GACC) Are Improved by Revised χ Torsions. J Phys Chem B. 2011;115:9261. doi: 10.1021/jp2016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okur A, Roe DR, Cui G, Hornak V, Simmerling C. Improving Convergence of Replica-Exchange Simulations through Coupling to a High-Temperature Structure Reservoir. J Chem Theory Comput. 2007;3:557. doi: 10.1021/ct600263e. [DOI] [PubMed] [Google Scholar]
- 41.Roitberg AE, Okur A, Simmerling C. Coupling of replica exchange simulations to a non-Boltzmann structure reservoir. J Phys Chem B. 2007;111:2415. doi: 10.1021/jp068335b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruscio JZ, Fawzi NL, Head-Gordon T. How hot? Systematic convergence of the replica exchange method using multiple reservoirs. J Comput Chem. 2010;31:620. doi: 10.1002/jcc.21355. [DOI] [PubMed] [Google Scholar]
- 43.Case DA, Darden TA, Cheatham TE, I, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 44.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926. [Google Scholar]
- 46.Banas P, Sklenovsky P, Wedekind JE, Sponer J, Otyepka M. Molecular mechanism of preQ1 riboswitch action: a molecular dynamics study. J Phys Chem B. 2012;116:12721. doi: 10.1021/jp309230v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krepl M, Zgarbova M, Stadlbauer P, Otyepka M, Banas P, Koca J, Cheatham TE, 3rd, Jurecka P, Sponer J. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J Chem Theory Comput. 2012;8:2506. doi: 10.1021/ct300275s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak J. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684. [Google Scholar]
- 49.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577. [Google Scholar]
- 50.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327. [Google Scholar]
- 51.Patriksson A, van der Spoel D. A temperature predictor for parallel tempering simulations. Phys Chem Chem Phys. 2008;10:2073. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 52.Hummer G, Garde S, García AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci U S A. 1998;95:1552. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nymeyer H, Gnanakaran S, Garcia AE. Atomic simulations of protein folding, using the replica exchange algorithm. Methods Enzymol. 2004;383:119. doi: 10.1016/S0076-6879(04)83006-4. [DOI] [PubMed] [Google Scholar]
- 54.Sindhikara D, Meng Y, Roitberg AE. Exchange frequency in replica exchange molecular dynamics. J Chem Phys. 2008;128:024103. doi: 10.1063/1.2816560. [DOI] [PubMed] [Google Scholar]
- 55.Sindhikara DJ, Emerson DJ, Roitberg AE. Exchange Often and Properly in Replica Exchange Molecular Dynamics. J Chem Theory Comput. 2010;6:2804. doi: 10.1021/ct100281c. [DOI] [PubMed] [Google Scholar]
- 56.Hawkins GD, Cramer CJ, Truhlar DG. Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett. 1995;246:122. [Google Scholar]
- 57.Hawkins GD, Cramer CJ, Truhlar DG. Parametrized Models of Aqueous Free Energies of Solvation Based on Pairwise Descreening of Solute Atomic Charges from a Dielectric Medium. J Phys Chem. 1996;100:19824. [Google Scholar]
- 58.Weiser J, Shenkin PS, Still WC. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J Comput Chem. 1999;20:217. [Google Scholar]
- 59.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J Chem Theory Comput. 2012;8:1542. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molecular Modelling & Bioinformatics Group; PCAsuite: A tool to compress Molecular Dynamics trajectories using Principal Components Analysis. http://mmb.pcb.ub.edu/software/pcasuite/ [Google Scholar]
- 61.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Tubbs JD, Condon DE, Kennedy SD, Hauser M, Bevilacqua PC, Turner DH. The Nuclear Magnetic Resonance of CCCC RNA Reveals a Right-Handed Helix, and Revised Parameters for AMBER Force Field Torsions Improve Structural Predictions from Molecular Dynamics. Biochemistry. 2013;52:996. doi: 10.1021/bi3010347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosta E, Hummer G. Error and efficiency of replica exchange molecular dynamics simulations. J Chem Phys. 2009;131:165102. doi: 10.1063/1.3249608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


