Abstract
The micro-viscosity and molecular crowding experienced by specific proteins can regulate dynamics and function within live cells. Taking advantage of the emerging TMP-tag technology, we present the design, synthesis and application of a hybrid genetic-chemical molecular rotor probe whose fluorescence lifetime can report on protein-specific micro-environments in live cells.
The micro-mechanical environment of proteins inside live cells and its effect on key biochemical processes are important yet unresolved issues in cell biology.1 Intracellular viscosity plays an important role in biochemical processes such as signal transduction, nuclear envelope function, chromatin localization, ribonucleoprotein assembly and diffusion of reactive oxygen species.2–4 Changes in viscosity at a sub-cellular level have been related to a number of diseases and pathologies.5 The determination of local viscosities and other micro-environmental parameters within the nucleus and other critical cellular organelles in live cells is of great interest.2–13
Experimentally, intracellular viscosity has been measured by tracking fluorophore diffusion, fluorescence correlation spectroscopy, and fluorescence recovery after photobleaching.6–9 These methods, however, suffer from long acquisition times (~100 sec) and cannot report on the spatial variation of micro-viscosities in a rapid imaging mode compatible with live cell imaging. Recently, fluorescent molecular rotors have emerged as novel, environmentally-sensitive probes capable of generating high-resolution images of the spatial distribution of micro-viscosities in a biological sample.10–13 However, the current molecular rotor approach has low organelle specificity and does not allow for protein-specific micro-environment measurements.10–13 This missing protein-specific information, if obtainable, would enhance our understanding of the dynamics and function of proteins inside cells. The fluorescence properties (intensity, lifetime and quantum yield) of genetically-encoded fluorescent proteins (such as GFP) are, unfortunately, insensitive to the medium viscosity because of shielding and isolation of the chromophore from the surroundings by the protein β-barrels.14 In addition, fluorescence anisotropy cannot report on the viscosity experienced by stationary proteins (e.g., H2B) inside cells. In the present study, we explored the emerging chemical tagging technology and developed a hybrid genetic-chemical eDHFR-TMP-Cy3 rotor tag to report protein-specific micro-viscosity by fluorescence lifetime imaging microscopy (FLIM).
The emerging chemical tagging technology has offered a route to selectively label a protein of interest in vivo with an organic fluorescent dye in a genetically encoded manner.15 The TMP-tag, which is designed around the nano-molar affinity (and recent covalent) interaction between E. coli dihydrofolate reductase (eDHFR) and trimethoprim (TMP), stands out as one of the few tags that work inside living cells with a high labeling specificity and efficiency.16 The DNA sequence that encodes the small (18kDa) and monomeric eDHFR is genetically fused to a protein of interest, and then a highly cell-permeable TMP-dye conjugate is introduced. The TMP-dye conjugate diffuses into the cell and recognizes the eDHFR fusion protein. By incorporating bright organic fluorophores such as Atto dyes, the TMP-tag has demonstrated its utility in super-resolution microscopy and in single-molecule biophysics.17, 18
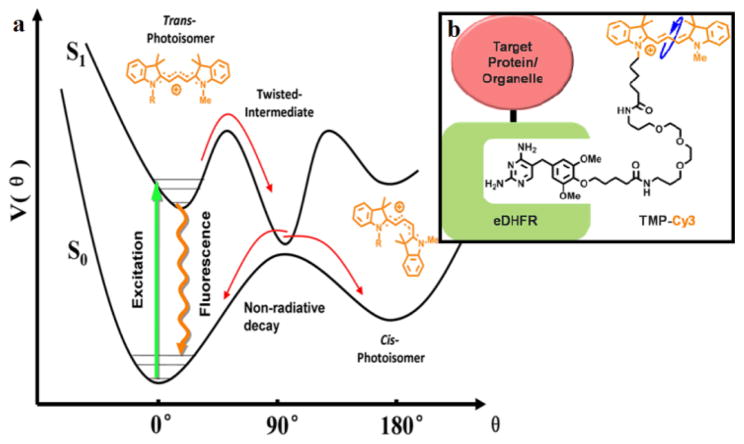
We selected Cy3 as the conjugation probe because of its environment-sensitive fluorescence lifetime. The photophysical properties of Cy3 have been well studied.19–22 As shown in Figure 1a, after excitation, in addition to the radiative decay pathway generating fluorescence, Cy3 can also isomerize from the trans-to the cis- configuration through a torsional motion, bringing Cy3 back to its ground state without photon emission. As a result, the measurable fluorescence lifetime, τ,
Figure 1.
(a) Cy3 photophysics (b) Design of eDHFR-TMP-Cy3 probe
is the inverse sum of the radiative decay rate constant of spontaneous emission ksp, the non-radiative decay rate, knr, which is sensitive to the medium viscosity, η, and direct internal conversion from the excited state, ki.c..23 In a low-viscosity environment such as in water, the non-radiative photo-isomerization pathway of Cy3 is dominant and the fluorescence lifetime is short (<0.2 ns). A viscous medium will hinder the rate of torsional motion on the potential surface of the excited state and prolong the fluorescence lifetime, as explained by the Kramers theory on barrier crossing.24 Although cyanine derivatives have been widely used in applications from in vitro protein labeling to in vivo animal diagnostic imaging,25 the environmentally-sensitive photo-physical properties of cyanine derivatives have not been explored until recently. Notably, in two recent in vitro single-molecule studies, Cy3 was exploited as a novel local reporter to probe real-time protein binding upon DNA.26,27 These emerging results encourage us to harness the environmental sensitivity of Cy3 in live cells for imaging.
We developed a TMP-Cy3 probe for live cell imaging (Figure 1b). A cell-permeable TMP-Cy3 conjugate was synthesized by modularized conjugation of TMP-NH2 with a sulfonate-free version of Cy3 (Scheme S1, Figure S1). A flexible polyethylene glycol (PEG) spacer was introduced between TMP and Cy3 to minimize the potential influence of the eDHFR protein on the nearby Cy3 probe.
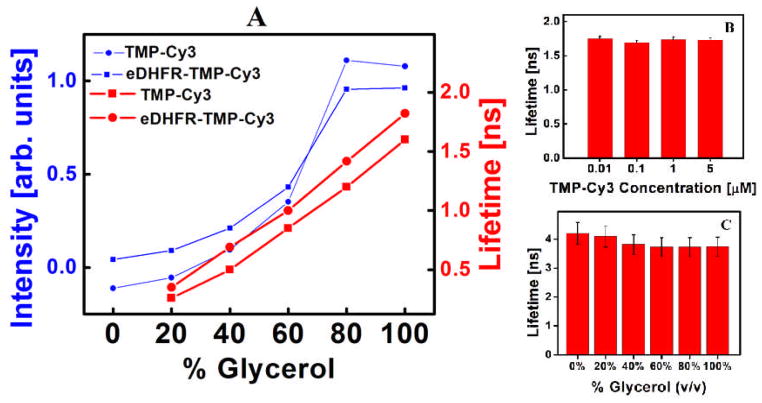
TMP-Cy3 was tested as an environment sensor in vitro before its use in cellular experiments (Figure 2). Fluorescence intensity and lifetime measurements on TMP-Cy3 were carried out in glycerol/water solutions with different viscosities. All measurements were taken on a home-built frequency-domain FLIM microscope (Supporting Information). The fluorescence lifetime of TMP-Cy3 increases from 0.2 ± 0.1 ns to 1.6 ± 0.2 ns with increasing glycerol volume fraction from 20% to 100%, consistent with a restriction of torsional motion and a hindered non-radiative decay. The fluorescence of TMP-Cy3 and eDHFR-TMP-Cy3 were compared in order to examine the potential perturbation effect of the protein on the nearby fluorophore. As shown in Figure 2A, a minimal effect of the eDHFR binding on the lifetime and brightness of TMP-Cy3 was observed, most likely attributed to the long PEG spacer designed between TMP and Cy3.
Figure 2.
Fluorescence intensity and lifetime of TMP-Cy3 and eDHFR-TMP-Cy3 (A). Concentration independence of TMP-Cy3 (B), Rhodamine insensitivity to viscosity (C).
For use as a specific micro-environment sensor inside cells, TMP-Cy3 should have negligible interactions with DNA, proteins and ions. Indeed, this is the case, as confirmed by in vitro spectroscopy experiments of TMP-Cy3 in solutions of varying DNA, bovine serum albumin (BSA) and NaCl concentrations (Supplementary Information). TMP-Cy3, however, does interact weakly interact with micelles formed by SDS (Sodium Dodecyl Sulfate). The fluorescence lifetime of TMP-Cy3 is independent from the fluorophore concentration (Figure 2B), which is an advantage in quantitative FLIM studies as the concentration of fluorophore within live cells isn’t well controlled. At concentrations from 10nm to 5μM, the fluorescence lifetime of TMP-Cy3 is constant (Figure 2B). Rhodamine 6G, a common non-rotor dye, has a constant fluorescence lifetime with varying glycerol concentrations (Figure 2C), underscoring the necessity of flexible rotor tags for viscosity imaging.
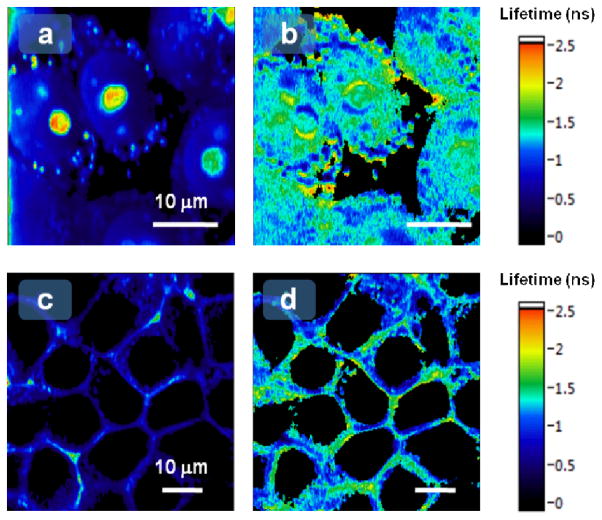
Encouraged by the in vitro experiments, we moved on to cellular experiments to evaluate the ability of TMP-Cy3 to map local micro-environments. To target the molecular rotor to the cell nucleus, eDHFR was fused to histone H2B protein. We also studied the cytosol environment in the vicinity of the cell membrane by anchoring eDHFR with a plasma membrane localization signal (PMLS). We transiently transfected HEK 293T cells with plasmids encoding H2B-eDHFR or PMLS-eDHFR fusion protein, then incubated the cells with our synthesized, cell-permeable TMP-Cy3 for 10 min, and imaged them. The H2B-eDHFR images show minor degrees of non-specific granular-shaped binding of TMP-Cy3 to lipid-rich organelles.28 Nevertheless, oval-shaped nuclei with distinct nucleoli were clearly observed (Figure 3a). Characteristic plasma membrane patterns were observed in cells expressing PMLS-eDHFR (Figure 3c). Therefore, this approach would be suitable for monitoring the nuclear and plasma membrane environment as non-specific staining was minor inside the nucleus and near the plasma membrane.
Figure 3.
Fluorescence intensity (a, c) and fluorescence lifetime (b, d) images of H2B-eDHFR-TMP-Cy3 (a & b) and PMLS-eDHFR-TMP-Cy3 (c & d) in live cells.
FLIM microscopy was used to image the distributions of local environments experienced by H2B within the cell nuclei and cytosol viscosities near the cell membrane. FLIM imaging (Figure 3a) revealed that the nuclear viscosity is high, comparable to glycerol solutions from 50% volume fraction (~1 ns) up to considerably higher values in certain regions (>2.0 ns). The average lifetime over the two brightest nuclei with clearly visible nucleoli to the left-of-center in Figure 3a is 1.4 ± 0.3ns, indicative of a viscous and crowded environment. Our lifetime measurements correspond to average viscosities of around 60–70cP within the nucleus, comparable to other reports.13 There is also remarkable heterogeneity in the distributions of the measured micro-environment, offering information not available from the confocal fluorescence intensity image. This heterogeneity may be related to the recently reported heterogeneous level of chromatin compaction detected by fluorescence anisotropy imaging of H2B-EGFP.29 FLIM images were also captured for TMP-Cy3 labeled PMLS-eDHFR in the vicinity of cell membranes (Figure 3d). In this scenario, the average lifetime is typically 0.9 ± 0.2 ns, indicating a relatively less viscous and crowded area of the cell cytoplasm compared to the nucleus experienced by H2B.
Conclusions
In conclusion, we developed a hybrid genetic-chemical molecular rotor tag (eDHFR-TMP-Cy3) to measure protein-specific local environments in live cells using FLIM. Although we only used the TMP-tag, the same Cy3 rotor moiety can be readily applied to other chemical tagging techniques such as the SNAP, CLIP, and HaloTag.15, 30 This methodology, with its good genetically-encoded specificity, high spatial-temporal resolution and simple interpretation, could provide valuable mechanistic information about protein function in the complex and constantly changing cellular environment. For example, the observed heterogeneous micro-environment could have broader implications in understanding chromatin condensation and transcription control within live cells.31 Chemical tags can be engineered with arbitrary open-structure biophysical probes that are exposed to their surroundings and can sense the local environment more sensitively than regular fluorescent proteins. To our knowledge, this advantageous aspect of chemical tags has been largely unexplored. This protein/organelle specific FLIM technique should be useful for evaluating a wider variety of protein or organelle-specific cellular micro-environments.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (U54 GM087519 and RC1GM091804 to V. W. C) and by the start up funds from Columbia University (to W. M.). We thank Dr. Steffen Jockusch for experimental assistance.
Footnotes
Electronic Supplementary Information (ESI) available: [Synthesis of TMP-Cy3, additional spectroscopic tests, FLIM and microscopy instrumentation]. See DOI: 10.1039/b000000x/
V.W.C holds patents on the TMP-tag technology, and the technology is licensed and commercialized by Active Motif.
Notes and references
- 1.Handwerger KE, Gall JG. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Lang I, Scholz M, Peters R. J Cell Biol. 1986;102:1183–1190. doi: 10.1083/jcb.102.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 4.Kues T, Dickmanns A, Lührmann R, Peters R, Kubitscheck U. Proc Natl Acad Sci USA. 2001;98:12021–12026. doi: 10.1073/pnas.211250098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luby-Phelps K. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 6.Wojcieszyn JW, Schlegel RA, Wu ES, Jacobson KA. Proc Natl Acad Sci USA. 1981;78:4407–4410. doi: 10.1073/pnas.78.7.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang L, Wang X, Xing D, Chen T, Chen WR. J Biomed Opt. 2009;14:024013-1–9. doi: 10.1117/1.3088141. [DOI] [PubMed] [Google Scholar]
- 8.Seksek O, Biwersi J, Verkman AS. J Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grünwald D, Martin RM, Buschmann V, Bazett-Jones DP, Leonhardt H, Kubitscheck U, Cardoso MC. Biophys J. 2008;94:2847–2858. doi: 10.1529/biophysj.107.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haidekker MA, Brady TP, Lichlyter D, Theodorakis EA. J Am Chem Soc. 2006;128:398–399. doi: 10.1021/ja056370a. [DOI] [PubMed] [Google Scholar]
- 11.Kuimova MK, Botchway SW, Parker AW, Balaz M, Collins HA, Anderson HL, Suhling K, Ogilby PR. Nature Chem. 2009;1:69–73. doi: 10.1038/nchem.120. [DOI] [PubMed] [Google Scholar]
- 12.Peng X, Yang Z, Wang J, Fan J, He Y, Song F, Wang B, Sun S, Qu J, Qi J, Yan M. J Am Chem Soc. 2011;133:6626–6635. doi: 10.1021/ja1104014. [DOI] [PubMed] [Google Scholar]
- 13.Kuimova MK, Yahioglu G, Levitt JA, Suhling K. J Am Chem Soc. 2008;130:6672–6673. doi: 10.1021/ja800570d. [DOI] [PubMed] [Google Scholar]
- 14.Suhling K, Siegel J, Phillips D, French PM, Lévêque-Fort S, Webb SE, Davis DM. Biophys J. 2002;83:3589–3595. doi: 10.1016/S0006-3495(02)75359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing C, Cornish VW. Acc Chem Res. 2011;44:784–792. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller LW, Cai Y, Sheetz MP, Cornish VW. Nat Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 17.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish V, Sauer M. Nat Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 18.Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aramendia PF, Negri RM, Roman ES. J Phys Chem. 1994;98:3165–3173. [Google Scholar]
- 20.Sanborn ME, Connolly BK, Gurunathan K, Levitus M. J Phys Chem B. 2007;111:11064–11074. doi: 10.1021/jp072912u. [DOI] [PubMed] [Google Scholar]
- 21.Levitus M, Ranjit S. Q Rev Biophys. 2011;44:123–151. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- 22.Muddana HS, Morgan TT, Adair JH, Butler PJ. Nano Lett. 2009;9:1559–1566. doi: 10.1021/nl803658w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berezin MY, Achilefu S. Chem Reviews. 2010;10:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramers HA. Physica. 1941;7:284–304. [Google Scholar]
- 25.Samanta A, et al. Angew Chem Int Ed. 2011;50:6089–6092. doi: 10.1002/anie.201007841. [DOI] [PubMed] [Google Scholar]
- 26.Luo G, Wang M, Konigsberg WH, Xie XS. Proc Natl Acad Sci USA. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myong S, Cui S, Cornish VP, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson LV, Walsh ML, Bockus BJ, Chen LB. J Cell Biol. 1981;88:526–535. doi: 10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee B, Bhattacharya D, Shivashankar GV. Biophys J. 2006;91:2297–2303. doi: 10.1529/biophysj.105.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinner MJ, Johnsson K. Curr Opin Biotech. 2010;21:766–776. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Nature Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.