SUMMARY
The Type II Secretion System (T2SS), a multi-protein machinery spanning two membranes in Gram-negative bacteria, is responsible for the secretion of folded proteins from the periplasm across the outer membrane. The critical multi-domain T2SS assembly ATPase GspEEpsE had so far not been structurally characterized as a hexamer. Here, four hexamers of Vibrio cholerae GspEEpsE are obtained when fused to Hcp1 as an assistant hexamer, as shown by native mass spectrometry. The enzymatic activity of the GspEEpsE-Hcp1 fusions is ~20 times higher than that of a GspEEpsE monomer indicating that increasing the local concentration of GspEEpsE by the fusion strategy was successful. Crystal structures of GspEEpsE-Hcp1 fusions with different linker lengths reveal regular and elongated hexamers of GspEEpsE with major differences in domain orientation within subunits, and in subunit assembly. SAXS studies on GspEEpsE-Hcp1 fusions suggest that even further variability in GspEEpsE hexamer architecture is likely.
INTRODUCTION
The type II secretion system (T2SS) is a complex, two-membrane spanning machinery occurring in a wide variety of pathogenic and non-pathogenic Gram-negative bacteria, where it plays a crucial role in the secretion of folded proteins from the periplasm across the outer membrane into the extracellular milieu (Korotkov et al., 2012; Douzi et al., 2012; McLaughlin et al., 2012). For instance, the Vibrio cholerae T2SS is responsible for secreting the major virulence factor cholera toxin across the outer membrane in such a manner that the delicate AB5 heterohexamer architecture of the toxin (O’Neal et al., 2004) is maintained without unfolding (Hirst et al., 1984). Key sub-assemblies of the T2SS machinery are: an outer membrane channel formed by the secretin GspD; a filamentous pseudopilus with, as major component, the pseudopilin GspG; a central multi-protein inner membrane complex which interacts with all other subassemblies; and a “secretion ATPase” GspE in the cytosol which interacts with the inner membrane protein GspL. The T2SS GspE is an ATPase that enables formation of the pseudopilus, which likely acts as a piston during exoprotein translocation across the outer membrane.
There are several other important machineries with components that are closely or distantly related to the T2SS, including the type IV pilus systems, the filamentous phage assembly system in Gram-negative bacteria, the archaella (also called the archaeal flagella) assembly system in Archaea, and the bacterial transformation system in Gram-positives (Korotkov et al., 2011; Korotkov et al., 2012). The closest relative of the T2SS is the type IV pilus system (T4PS), which also spans the inner and outer membranes in Gram-negative bacteria. The T4PS and T2SS share several features, yet there are also major differences (Ayers et al., 2010). One distinct difference is that the type IV pilus assembled by a T4PS often contains thousands of pilin subunits and extends far into the milieu surrounding the bacterium, while the pseudopilus assembled by a T2SS consists probably of 5–20 subunits, and this pseudopilus remains in the periplasm under physiological conditions. Moreover, most T4PS machineries have two or even three ATPases including an assembly ATPase and one or more retraction ATPases ( Craig and Li, 2008; Giltner et al., 2012). In contrast, T2SS machineries have one ATPase, GspE. Here we focus on the architecture and activity of the T2SS ATPase and its relatives, in particular from the T4PS.
The ~56 kDa T2SS secretion ATPase GspE belongs to the family of Type II/IV secretion ATPases (Peabody et al., 2003; Planet et al., 2001). Almost all T2SS ATPases have three major domains: two N-terminal domains, N1D and N2D, and a C-terminal domain (CTD), although, in a few species, an additional N-terminal N0 domain occurs (Chen et al., 2005). The N1D and N2D are connected by a linker of ~20 residues, and the N2D by a ~14 residue linker to the CTD. The N1D interacts with the cytoplasmic domain of the inner membrane protein GspL (Abendroth et al., 2005; Sandkvist et al., 1995; Shiue et al., 2006). The CTD consists of three subdomains: the major nucleotide binding domain (C1D), a four-helical domain (C2D), and a metal-binding domain (CMD) (Figure 1A). The metal binding domain occurs in almost all T2SS ATPases from Gram-negative bacteria and contains an essential tetra-cysteine motif which coordinates zinc ions (Camberg et al., 2007; Robien et al., 2003). While the assembly ATPase of the T4PS that is required for polymerization of pilins into pili has an identical domain organization to the T2SS ATPase, the retraction ATPase that promotes depolymerization of pili lacks the N1D and CMD domains (Figure 1A).
Figure 1. Characterization of Vibrio cholerae GspEEpsE fused to Hcp1 in solution.
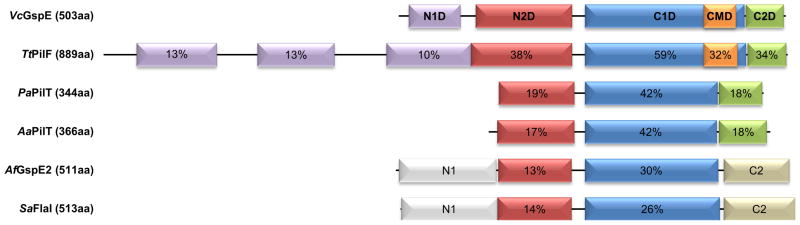
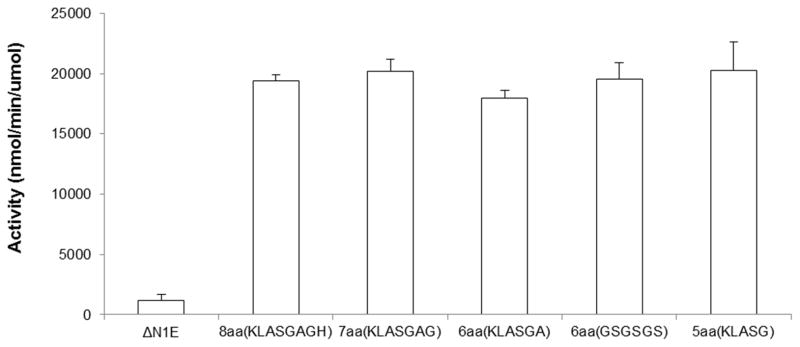
Figure 1A. Domain bar diagram of GspEEpsE and homologous ATPases with electron microscopy reconstructions or crystal structures.
VcGspE =GspE, the assembly ATPase from the T2SS in the Gram-negative Vibrio cholerae; TtPilF = PilF, an assembly ATPase from the T4PS in the Gram-negative eubacterium Thermus thermopilus; PaPilT = PilT, a retraction ATPase from the T4PS in the Gram-negative eubacterium Pseudomonas aeruginosa; AaPilT = PilT, a retraction ATPase from the T4PS in the Gramnegative eubacterium Aquifex aeolicus; AfGspE2 = one of three related ATPases in the Archaeon Archaeoglobus fulgidus; SaFlaI = FlaI, an ATPase from the archaellum assembly system in the Archaeon Sulfolobus acidocaldarius. The percentages of amino acid sequence identity given are from the comparison with GspEEpsE. Where domains are homologous in structures to GspEEpsE, the percentage sequence identity is given compared to GspEEpsE.
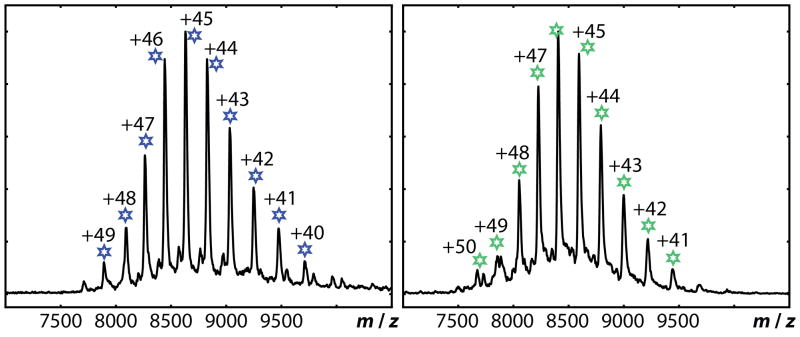
Figure 1B. Native mass spectra of ΔN1GspEEpsE-Hcp1 fusion proteins.
Left: ΔN1GspEEpsE-8aa-Hcp1 Right: ΔN1GspEEpsE-6aa-Hcp1. The data show that these proteins each assemble as hexamers in solution. The mass measured for each complex is only slightly greater than that expected assuming that each protein subunit in the hexamer contains one Zn and one nucleotide. The native mass spectra for ΔN1GspEEpsE-5aa-Hcp1 and ΔN1GspEEpsE-7aa-Hcp1, fragmentation spectra for ΔN1GspEEpsE-8aa-Hcp1, and a table of observed and expected masses are shown in Figure S1.
Figure 1C. ATPase activities of V. cholerae ΔN1GspEEpsE-linker-Hcp1 variants.
From left to right the activities for: ΔN1GspEEpsE (control monomer), ΔN1GspEEpsE-8aa-Hcp1, ΔN1GspEEpsE-7aa-Hcp1, ΔN1GspEEpsE-6aa(KLASGA)-Hcp1, ΔN1GspEEpsE-6aa(GSGSGS)-Hcp1 and ΔN1GspEEpsE-5aa-Hcp1. Two variant linkers were tested in the case of ΔN1GspEEpsE-6aa-Hcp1 fusions to evaluate the effect of linker sequence. As shown, both types of linkers gave the same increase in activity. The crystal structure was determined of ΔN1GspEEpsE-6aa(GSGSGS)-Hcp1 which is called ΔN1GspEEpsE-6aa-Hcp1 throughout the paper.
Many non-T2SS GspE homologs, such as the retraction ATPase PilT and archaeal flagellar ATPases, form hexamers with various point group symmetries (Misic et al., 2010; Patrick et al., 2011; Reindl et al., 2013; Satyshur et al., 2007; Yamagata and Tainer, 2007). In contrast, obtaining T2SS ATPases as homogeneous hexamers in solution, or as hexamers in crystals, has not been possible so far. The crystal structure of a truncated form of Vibrio cholerae GspE (also called EpsE (Sandkvist et al., 1995)) has been elucidated. This variant of GspEEpsE lacked the N1D and crystallized in a helical arrangement with 61 symmetry (Robien et al., 2003). Solution studies indicate hexamer formation of the GspEXpsE from the Xanthomonas campestris T2SS upon the addition of nucleotides (Shiue et al., 2007). Activity measurements of V. cholerae GspEEpsE showed that oligomers of an approximately hexameric size according to gel filtration analysis possess much higher activity than monomers, but hexamers are only present in small amounts (Camberg and Sandkvist, 2005). Full length V. cholerae GspEEpsE is able to form oligomers, possibly hexamers, in the presence of the cytoplasmic domain of GspL and acidic phospholipids, specifically cardiolipin, with a concomitant considerable increase in ATPase activity (Camberg et al., 2007). Mutations interrupting putative inter-subunit interfaces of V. cholerae GspEEpsE result in activity loss, indicating that a precise interaction between adjoining subunits is essential for the function of GspEEpsE (Patrick et al., 2011). High resolution structures of retraction T4PS ATPase hexamers are known (Misic et al., 2010; Satyshur et al., 2007) and recently an electron microscopy reconstruction of a hexamer of the T4PS assembly ATPase PilF from Thermus thermopilus, a eukaryotic extremophile, has been reported (Collins et al., 2013) So far, however, no hexamers of T2SS ATPases have been characterized structurally.
To overcome the reluctance of GspE to form hexamers, we embarked on a strategy of fusing GspEEpsE to another protein known to form hexamers by itself. The rationale behind this approach is to increase the local concentration of GspEEpsE subunits, thereby increasing the probability of GspEEpsE hexamer formation. Here we selected Hcp1 (Mougous et al., 2006) as a fusion partner for V. cholerae ΔN1GspEEpsE, and observe that the fusion protein forms hexamers in solution with a concomitant ~20-fold increase in ATPase activity with respect to the monomeric enzyme. The application of the assistant hexamer strategy was clearly successful. In crystallographic studies of the GspEEpsE-Hcp1 fusion proteins, V. cholerae ΔN1GspEEpsE hexameric assemblies with two different symmetries, and distinctly different subunit conformations were observed. SAXS studies of ΔN1GspEEpsE-Hcp1 fusions, when combined with the new ΔN1GspEEpsE-Hcp1 hexamer crystal structures, indicate that additional types of GspEEpsE hexamers are likely. These results show considerable interdomain flexibility, consistent with large dynamic motions expected to occur in the T2SS while in action. Comparison of the new structures with related ATPases indicates that a construction unit consisting of two domains from adjacent subunits is a hallmark of this large family of secretion ATPases, as also reported in a recent study of the archaellum assembly ATPase FlaI (Reindl, et al., 2013). The global general principle of these ATPases appears to be that construction units are largely fixed and subunits are highly flexible.
RESULTS
Characterization of GspE-Hcp1 Fusions
Four fusion proteins were successfully expressed, purified, characterized and crystallized: ΔN1GspEEpsE-KLASG-Hcp1, ΔN1GspEEpsE-GSGSGS-Hcp1, ΔN1GspEEpsE-KLASGAG-Hcp1 and ΔN1GspEEpsE-KLASGAGH-Hcp1, called hereafter ΔN1GspEEpsE-5aa-Hcp1, ΔN1GspEEpsE-6aa-Hcp1, ΔN1GspEEpsE-7aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1, respectively. Homogeneous hexamer formation of these four V. cholerae ΔN1GspEEpsE-linker-Hcp1 fusions was determined by native mass spectrometry (Figures 1B and S1). The expected and observed molecular masses differed by less than ~1 kDa, which is in the expected range of mass differences for such ~390 kDa complexes. In the case of the 5 amino acid linker, the mass spectrometry data also provided evidence for the presence of pentamers. The hexamerization greatly enhanced the ATPase activity of these four ΔN1GspEEpsE-linker-Hcp1 variants compared to monomeric ΔN1GspEEpsE: in all cases, an increased activity by a factor of approximately twenty was observed (Figure 1C). This increase in activity might be an underestimate since there is some tendency for the control ΔN1GspEEpsE protein to dimerize according to dynamic light scattering and size exclusion experiments, but not according to native mass spectrometry (data not shown). In addition, the activity of the oligomeric forms was 2–4 fold greater than the cardiolipin-stimulated activity of full length GspEEpsE when in complex with the cytoplasmic domain of GspLEpsL (Camberg et al., 2007; Patrick et al., 2011).
Crystal Structure of Two GspE Hexamers
Crystals of all four V. cholerae ΔN1GspEEpsE-linker-Hcp1 variants were obtained and the structures could be solved by molecular replacement (see Methods and Table 1)). The tetragonal crystals of ΔN1GspEEpsE-5aa-Hcp1 and ΔN1GspEEpsE-6aa-Hcp1 exhibited space group P42212 with essentially the same cell dimensions and diffracted to about 7.6 Å resolution. The asymmetric unit contains one Hcp1 hexamer and a quite regular arrangement of six ΔN1GspEEpsE subunits with quasi C6 point group symmetry (Figure 2A), hereafter called the qC6 hexamer. The 7aa- and 8aa-linked fusions crystallized in space group P22121 with similar cell dimensions. The crystals of ΔN1GspEEpsE-7aa-Hcp1 protein reach a resolution of about 7 Å, while those of ΔN1GspEEpsE-8aa-Hcp1 diffracted highly anisotropically with a resolution of ~5 Å along the c axis and ~4.1 Å in the other two directions. The asymmetric unit contains three ΔN1GspEEpsE-8aa-Hcp1 subunits, which are each different as described below. A crystallographic two-fold axis creates an elongated hexamer with C2 point group symmetry from the three crystallographically independent ΔN1GspEEpsE subunits (Figure 2A), hereafter called the C2 hexamer. Below, we describe only the ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 structures since these are essentially the same as those of, respectively, the 5aa- and 7aa-linked proteins.
TABLE 1.
Crystallographic data collection and refinement1
| ΔN1GspE-6aa-Hcp1 | ΔN1GspE-8aa-Hcp1 | |
|---|---|---|
| Data Collection | ||
| Space Group | P 42 21 2 | P 2 21 21 |
| Unit cell dimensions a,b,c (Å) | 205.1, 205.1, 235.0 | 112.5, 132.9, 142.8 |
| Mosaicity (°) | 0.53 | 0.23 |
| Resolution range (Å) | 60.0–7.6 (7.87-7.6) | 38.7, 4.09 (4.31-4.09) |
| Total no. of reflections | 45,135 | 111,225 |
| Unique reflections | 6,587 | 17,346 |
| Average redundancy | 6.9 (7.0) | 6.4 (6.1) |
| % completeness | 99.6 (99.9) | 99.2 (97.5) |
| Rmerge | 0.25 (>1) | 0.083 (0.959) |
| <I/σ(I)> | 10.9 (2.0) | 7.1 (0.8) |
| Refinement | ||
| Resolution (Å) | 50.0–7.6 (7.9-7.6) | 38.7–4.2 (4.38-4.2) |
| No. reflections | 6265 (432) | 15196 (1014) |
| Rwork & Rfree | 0.363/0.368 | 0.341 & 0.371 |
| Number of protein chains per a.u. | Six ΔN1EpsE-6aa-Hcp1 chains. Total Mw 383 kDa. | Three ΔN1EpsE-8aa-Hcp1 chains. Total Mw 193 kDa. |
Numbers in parentheses refer to outer resolution shell
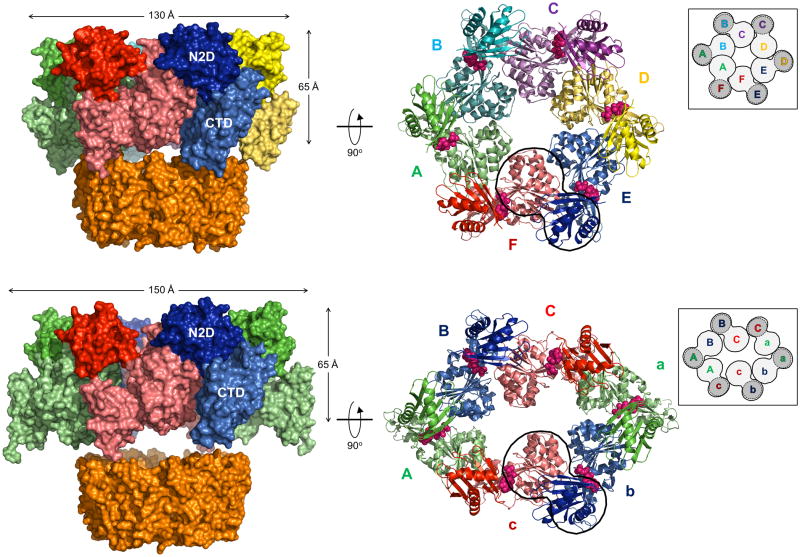
Figure 2. Crystal structures of hexameric ΔN1GspEEpsE from Vibrio cholerae fused to Hcp1.
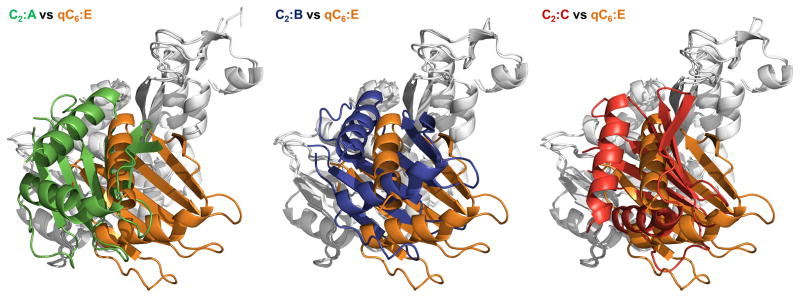
Insets: schematic view of the hexamer outlining all six CTD•N2D’ construction units. CTD domains in light grey, N2D’ domains in dark grey.
Figure 2A. V. cholerae ΔN1GspEEpsE-6aa-Hcp1, with a quasi-C6 ΔN1GspEEpsE hexamer and V. cholerae ΔN1GspEEpsE-8aa-Hcp1 containing a C2 hexamer.
Upper panel: The quasi-C6 hexamer of V. cholerae ΔN1GspEEpsE-6aa-Hcp1.
This fusion forms a ΔN1GspEEpsE hexamer with quasi-C6 point group symmetry. Shown are subunits A (green), B (cyan), C (purple), D (yellow), E (blue) and F (red). The CTDs are shown in a lighter shade of the same color as the N2Ds of the same subunit. The Hcp1 assistant hexamer is shown in orange. Left: view perpendicular to the quasi sixfold depicting also the Hcp1 hexamer. Right: view along the quasi-sixfold axis of the ΔN1GspEEpsE hexamer, with the Hcp1 hexamer omitted. The nucleotides shown in spheres are AMPPNP superposed from the helical Δ90GspEEpsE structure (PDB: 1p9w (Robien et al., 2003)). The shape of the hexamer in this view is very regular. One CTD•N2D’ construction unit is outlined.
Lower panel: The C2 hexamer of V. cholerae ΔN1GspEEpsE-8aa-Hcp1.
This fusion forms a ΔN1GspEEpsE hexamer with C2 point group symmetry. Shown are subunits A (green), B (blue), C (red) – each occurring twice in the hexamer. The CTDs are shown in a lighter shade of the same color as the N2Ds of the same subunit. The Hcp1 assistant hexamer is shown in orange. Left: view perpendicular to the twofold depicting also the Hcp1 hexamer. Right: view along the twofold axis of the ΔN1GspEEpsE hexamer, with the Hcp1 hexamer omitted. The nucleotides shown in pink spheres are AMPPNP superposed from the helical Δ90GspEEpsE structure (PDB: 1p9w (Robien et al., 2003)). The shape of the hexamer in this view is an approximate ellipsoid of 105 Å by 150 Å. One CTD•N2D’ construction unit is outlined. See also Figure S2, S3, S4 and S7.
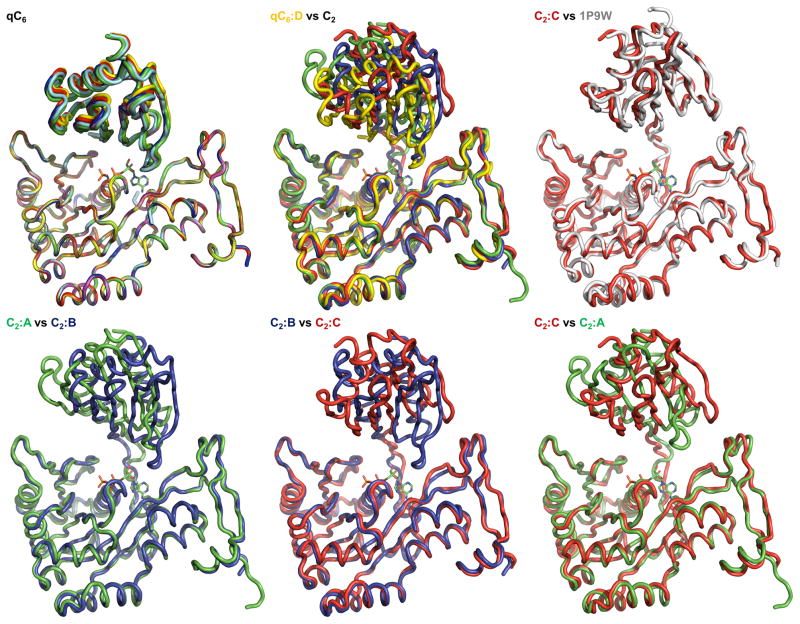
Figure 2B. The variability of the N2D-vs-CTD orientations in V. cholerae GspEEpsE.
Shown are superimposed subunits in the “canonical view” with the CTDs superimposed below and the N2Ds on top (colored as Figure 2A). For a different “orthogonal view,” see Figure 2C. The nucleotide shown for reference is AMPPNP from the helical Δ90GspEEpsE structure (PDB: 1p9w (Robien et al)). For N2D-vs-CTD orientations see also Table S1.
Top left: Superposition of the six subunits of the qC6 ΔN1GspEEpsE hexamer from the ΔN1GspEEpsE-6aa-Hcp1 structure, revealing only small differences, by one to five degrees, in N2D-vs-CTD orientations.
Top middle: Superposition of subunit D (yellow) from of the qC6 ΔN1GspEEpsE hexamer and the three subunits of the C2 ΔN1GspEEpsE hexamer from the ΔN1GspEEpsE-8aa-Hcp1 structure. N2D-vs-CTD orientations vary by 16 to 41 degrees.
Top right: Superposition of subunit C (red) from the ΔN1C2 GspEEpsE hexamer and ΔN1GspEEpsE (grey) from the helical Δ90GspEEpsE structure (PDB: 1p9w (Robien et al)). The difference in N2Dvs-CTD orientation is only 2 degrees.
Bottom left: Superposition of subunits A (green) and B (blue) of the C2 ΔN1GspEEpsE hexamer. The difference in N2D-vs-CTD orientation is 32 degrees.
Bottom middle: Superposition of subunits B (blue) and C (red) of the C2 ΔN1GspEEpsE hexamer. The difference in N2D-vs-CTD orientation is 48 degrees.
Bottom right: Superposition of subunits C (red) and A (green) of the C2 ΔN1GspEEpsE hexamer. The difference in N2D-vs-CTD orientation is 47 degrees.
Figure 2C. “Orthogonal views” of the subunits in V. cholerae GspEEpsE.
Pairwise comparison of ΔN1GspEEpsE subunits after superposition of the CTDs (grey, as background)viewed in a direction approximately perpendicular to the “canonical view” in Figure 2B. The N2D of subunit E from the qC6 ΔN1GspEEpsE hexamer (orange) is used as reference for each case.
Comparing the two ΔN1GspEEpsE hexamers reveals striking differences such as very different shapes of the outside envelopes and also of the central cavities (Figures 2A). The regular qC6 hexamer has a cylindrical shape with an exterior diameter of ~130 Å and an inner diameter of ~25 Å. The C2 hexamer, in contrast, has an elongated appearance of ~ 105 by 150 Å. The inner diameter of the C2 hexamer is quite irregular measuring ~14 Å at the narrowest point and ~50 Å at the widest point. The six C1Ds in the C2 hexamer are less tightly packed together than in the qC6 hexamer. In both hexamers, the Hcp1 rings are positioned at the expected position, near the C-termini of the ΔN1GspEEpsE rings. While the six-fold axis of the Hcp1 hexamer in the ΔN1GspEEpsE-8aa-Hcp1 structure coincides with the crystallographic two-fold axis which is shared with the two-fold axis of the C2 hexamer, in the ΔN1GspEEpsE-6aa-Hcp1 structure the six-fold axis of the Hcp1 hexamer does not align perfectly with the quasi six-fold axis of the qC6 hexamer (Figure 2A).
Including the Δ90GspEEpsE structure (Robien et al., 2003), we can now compare ΔN1GspEEpsE subunits in three different crystal forms. In the ΔN1GspEEpsE-6aa-Hcp1 structure with ΔN1GspEEpsE hexamers with qC6 symmetry, the N2D-vs-CTD domain orientation within one subunit is essentially the same for all six subunits, differing by only 2.0 to 5.2 degrees. In contrast, in the ΔN1GspEEpsE-8aa-Hcp1 structure with ΔN1GspEEpsE hexamers with C2 symmetry, the N2D-vs-CTD orientation is highly variable, differing by 32 to 48 degrees when compared pairwise (Table S1). The N2D-vs-CTD orientation observed in the three independent subunits in the C2 hexamer deviates by 16 to 41 degrees from the average orientation in the qC6 hexamer (Figures 2B and 2C). Interestingly, the N2D-vs-CTD orientations in subunit C of the C2 hexamer and the subunit in the helical structure (Robien et al., 2003) differ by only 2 degrees and hence are essentially the same (Figure 2B). Hence, the three available ΔN1GspEEpsE crystal structures, with ten (i.e. 6+3+1) independent views of ΔN1GspEEpsE subunits, reveal essentially four different N2D-vs-CTD domain orientations (Figure 2).
Despite the large freedom of domain motion within one subunit revealed by this comparison, the qC6 and C2 hexamers of ΔN1GspEEpsE share the manner in which the CTD of one subunit interacts with the N2D of an adjacent subunit (N2D’) (Figure S2). The same CTD•N2D’ arrangement is also observed in the helical structure of Δ90GspEEpsE ((Robien et al., 2003) Figure S2). Apparently, the new ΔN1GspEEpsE hexamers reported here, and the Δ90GspEEpsE helix with 61 symmetry, are all built from essentially the same CTD•N2D’ construction unit. The different ΔN1GspEEpsE hexamers are obtained by different conformations of the N2D-to-CTD linkers connecting six such construction units. This results in strikingly different orientations and positions of all domains in the two hexamers. For instance, the distances between the Cα of residue Ile458, located near the end of the first helix α10 in the C2D, is 35.5 ± 2.5 Å in nonadjacent subunits in the qC6 hexamer but ranges from 39 to 70 Å in the C2 hexamer. Such variations suggest that large dynamic motions of the subunits during functioning of this assembly ATPase in the T2SS may occur. A similar architectural principle, with essentially the same CTD•N2D’ building block, has recently been reported for the distantly related FlaI ATPase of the archaellum assembly system from the crenarchaeon Sulfolobus acidocaldarius (Reindl et al., 2013).
Crystals of the four V. cholerae ΔN1GspEEpsE-linker-Hcp1 variants were obtained in the presence of 5 mM nucleotide(s) and 5 mM MgCl2. Electron density at the expected position of the nucleotide, close to the CTD of ΔN1GspEEpsE subunits, could be observed in the three crystallographically independent subunits of the C2 hexamer, and also in the six subunits of the qC6 hexamer (Figure S3). The densities for the phosphoryl groups of the nucleotides occur at approximately the same position as Mg•AMPPNP in the helical Δ90GspEEpsE structure (Robien et al., 2003). The ΔN1GspEEpsE-8aa-Hcp1 structure was solved with Mg•ADP plus AlCl3 and NaF but no clear evidence for density beyond the β-phosphoryl group was present. The densities vary in the nine crystallographically independent subunits which might indicate that the nucleotide is present with different occupancies, but given the uncertainties due to the low resolution of the structures it seems best not to discuss this point here in detail. Higher resolution structures will be required to reveal the mode of nucleotide binding more precisely.
The metal binding domains are located on the periphery of both ΔN1GspEEpsE hexamers. Despite the ~7 Å resolution of the data, the presence of zinc ions in the CMDs was unambiguously confirmed by six peaks in the anomalous difference density at the expected positions in the ΔN1GspEEpsE-6aa-Hcp1 crystal structure (Figure S4). The positions of zinc ions of non-adjacent subunits are 114 ± 8 Å apart in the qC6 hexamer. These inter-zinc distances range from 91 to 140 Å in the C2 hexamer, again showing that the two hexamers are dramatically different.
SAXS studies
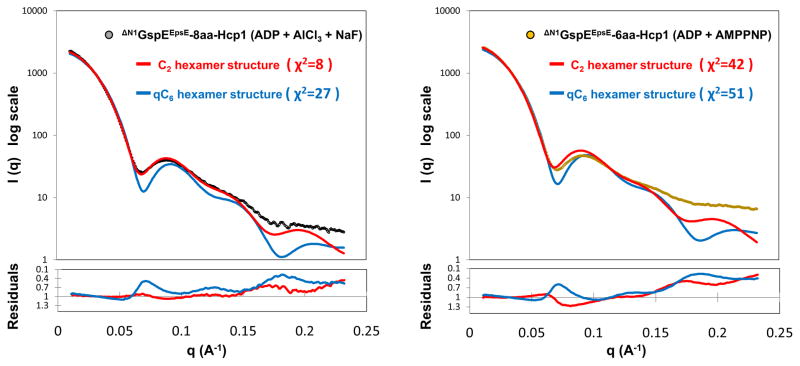
SAXS studies were carried out to investigate whether the crystal structures correspond with the ΔN1GspEEpsE hexamers in solution and also if the presence of different nucleotides affects the structure of the ΔN1GspEEpsE hexamer of the fusion proteins. The observed scattering curve of ΔN1GspEEpsE-8aa-Hcp1 in the presence of 5 mM ADP, 5 mM MgCl2, 5 mM AlCl3 and 15 mM NaF follows closely the calculated scattering curve based on the crystal structure of ΔN1GspEEpsE-8aa-Hcp1 elucidated in the presence of the same compounds and containing the elongated C2 hexamer (Figure 3, Left panel). In contrast, the calculated scattering curve using the crystal structure of ΔN1GspEEpsE-6aa-Hcp1 with the qC6 ΔN1GspEEpsE hexamer matches the observed scattering curve of ΔN1GspEEpsE-8aa-Hcp1 poorly. Hence it appears that in solution the structure of ΔN1GspEEpsE-8aa-Hcp1 is close to that observed in the crystal structure.
Figure 3. SAXS studies on ΔN1GspEEpsE-8aa-Hcp1 and ΔN1GspEEpsE-6aa-Hcp1.
Experimental (black and yellow circles) and calculated (red and blue) SAXS scattering curves for ΔN1GspEEpsE-8aa-Hcp1 and ΔN1GspEEpsE-6aa-Hcp1 with the same nucleotides as in the mother liquor of the crystal structures (See the right upper corner of each panel, and for specific details see methods). The calculated SAXS curve based on the ΔN1GspEEpsE-6aa-Hcp1 structure with the qC6 hexamer of ΔN1GspEEpsE shown in blue, and for the ΔN1GspEEpsE-8aa-Hcp1 structure with the C2 hexamer of ΔN1GspEEpsE shown in red. Residual plots are shown in the lower panels. See also Figure S9.
Surprisingly, the observed scattering curve of ΔN1GspEEpsE-6aa-Hcp1 deviates from the calculated scattering curve based on the crystal structure of ΔN1GspEEpsE-6aa-Hcp1. The calculated scattering curve using the crystal structure of ΔN1GspEEpsE-8aa-Hcp1 also resulted in a poor fit with the observed curve of ΔN1GspEEpsE-6aa-Hcp1 (Figure 3, Right panel). It is therefore likely that the architecture of the ΔN1GspEEpsE hexamer of the ΔN1GspEEpsE-6aa-Hcp1 complex in solution is neither that of the qC6 nor of the C2 hexamer observed in the crystals.
Comparison of T2SS and T4PS ATPase Hexamers
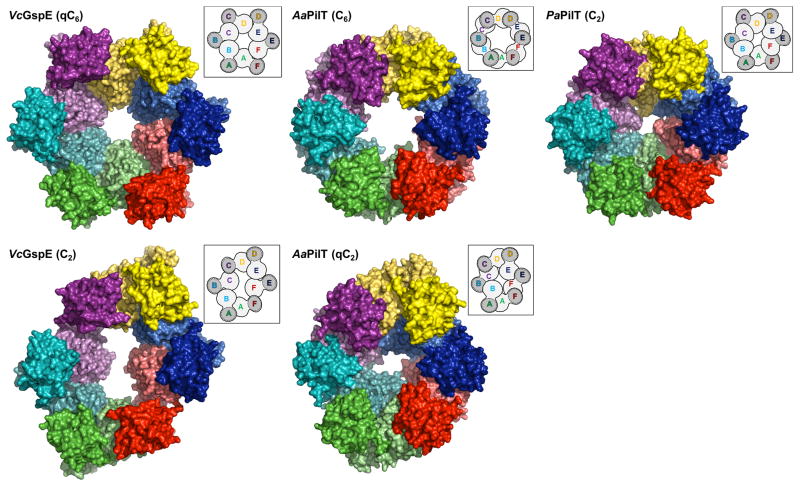
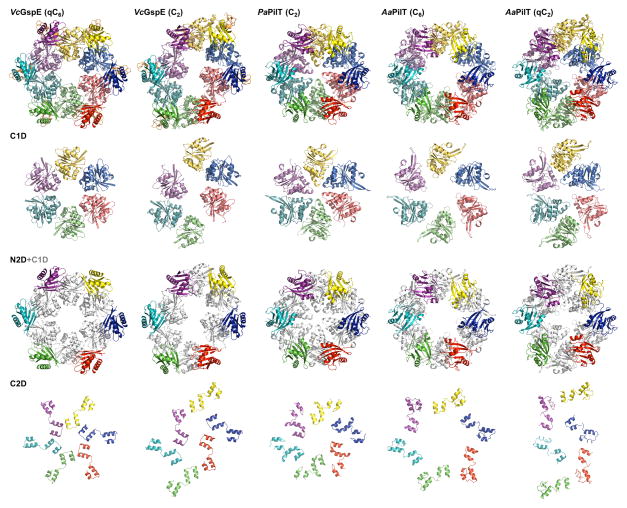
Crystal structures of hexamers with various symmetries have been reported for the T4PS ATPases PilT (Misic et al., 2010; Satyshur et al., 2007) from the Gram-negative bacteria Aquifex aeolicus and Pseudomonas aeruginosa (hereafter called AaPilT and PaPilT, respectively). These retraction ATPases differ substantially from the T2SS ATPases since they lack two domains: the N1D and CMD. Regarding the common domains, the amino acid sequence identities of AaPilT and PaPilT compared to V. cholerae GspEEpsE are ~18 % for the N2Ds, ~42 % for the C1Ds, and ~18 % for the C2Ds, respectively (Figure 1A). While the ATPase hexamers of V. cholerae ΔN1GspEEpsE-6aa-Hcp1 and of AaPilT, with qC6 and C6 symmetry respectively, have approximately the same overall dimensions (Figure 4A), the N2D-vs-CTD orientations within subunits are surprisingly different in these two cases; varying by ~40 degrees in pairwise comparisons (Table S1; Figure 4B and S6). As a consequence, the N2Ds of the qC6 ΔN1GspEEpsE hexamer move further away from the central C1D domains than the N2Ds of the AaPilT C6 hexamer and the PaPilT C2 hexamer (Figure 4C). The elongated ΔN1GspEEpsE C2 hexamer is in turn very dissimilar from the irregular qC2 hexamer of AaPilT (Satyshur et al., 2007): in this case the N2D arrangement of the ΔN1GspEEpsE C2 hexamer is distinctly different from that in the AaPilT qC2 hexamer (Figure 4C). Yet, despite these major differences in hexamer shape, the two-domain CTD•N2D’ construction unit observed in the two new ΔN1GspEEpsE hexamers is essentially the same as in AaPilT and in PaPilT (Figure S5).
Figure 4. Comparison of T2SS GspEEpsE and T4PS secretion ATPases.
Figure 4A. The regular and irregular GspEEpsE and PilT hexamers.
NOTE: The N2D and CTD domains of the same subunit are represented with darker and lighter shades of the same color. Insets: Hexamers with the CTD•N2D’ construction units outlined
Top: comparison of regular hexamers. Left: V. cholerae ΔN1GspEEpsE qC6 hexamer. Middle: A. aeolicus PilT C6 hexamer (PDB: 2EWV). Right: P. aeruginosa PilT C2 hexamer (PDB: 3JVV).
Bottom: comparison of irregular hexamers. Left: V. cholerae ΔN1GspEEpsE C2 hexamer. Middle: A. aeolicus PilT qC2 hexamer (PDB: 2GSZ). See also Figure S5.
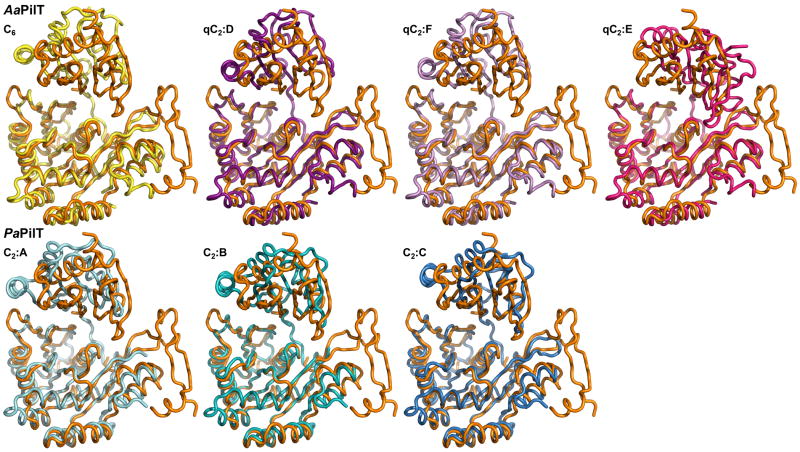
Figure 4B. The variability of the N2D-vs-CTD orientations in GspEEpsE and PilT hexamers.
Shown are superimposed subunits in the “canonical view” with the CTDs superimposed below and the N2Ds on top. For a different, “orthogonal view,” see Figure S6. For N2D-vs-CTD orientations see Table S1.
Top: superposition of the CTD from the subunit in the AaPilT C6 hexamer (yellow; PDB: 2EWV) and the three independent subunits of the AaPilT qC2 hexamer (different shades of purple; PDB: 2GSZ) onto subunit E from the ΔN1GspEEpsE qC6 hexamer (orange).
Bottom: superposition of the CTDs of PaPilT (different shades of blue; PDB: 3JVV) onto subunit E from the ΔN1GspEEpsE qC6 hexamer (orange). See also Figure S6 and S8.
Figure 4C. Domain rearrangements of T2SS GspEEpsE and T4PS secretion ATPases.
Rows from top to bottom: hexamers; the C1Ds only; the N2Ds in color with C1Ds in grey as background; the C2Ds only. Note how different the domain positions are in the various hexamers.
The five available crystal structures of PilT hexamers provide seven independent views of AaPilT and PaPilT subunits. Two of these, subunits D and F of AaPilT qC2 (PDB: 2GSZ) (Satyshur et al., 2007), are very similar to each other (Table S1). Hence, six different N2D-vs-CTD PilT orientations are available for comparison with the four orientations in GspEEpsE subunits in the two new hexamers. For 24 mutual comparisons of four T2SS GspEEpsE versus six T4PS PilT ATPase subunits, the smallest difference in N2D-vs-CTD orientation is 14.7 degrees and the largest as much as 73.4 degrees. Clearly, none of these six PilT orientations are similar to the four GspE orientation observed in the current study (Figure 4B and Table S1). This is particularly clear when viewed in a direction (Figure S6) which is approximately orthogonal with respect to the “classical” direction of view (Figure 4B). Hence, despite considerable sequence similarity of the N2D and C1D domains of GspEEpsE and PilT, there is no similarity in the N2D-vs-CTD orientation among the subunits in the known structures of these two ATPases.
DISCUSSION
We have succeeded in obtaining ΔN1GspEEpsE hexamers in solution by fusion to an “assistant hexamer” as revealed by native mass spectrometry (Figures 1B and S1). In crystals, depending on linker length, two types of ΔN1GspEEpsE hexamers are observed (Figures 2A). Importantly, the four ΔN1GspEEpsE-Hcp1 hexamers of ~380 kDa obtained show an approximately 20-fold increase in activity with respect to ΔN1GspEEpsE not fused to an assistant hexamer (Figure 1C). The strategy to bring GspEEpsE subunits in close proximity by linking to an assistant hexamer has resulted for the first time in structures of T2SS ATPase hexamers, and observing a substantial increase in ATPase activity as a result of the hexamerization of ΔN1GspEEpsE.
Linking domains, peptides and proteins has in the past been applied mainly to cases where two proteins, or a peptide and a protein, with weak affinity for each other were linked together (see e.g. (Reddy Chichili et al., 2013)) although sometimes also up to four domains from different proteins have been connected (Park and Hol, 2012). Another interesting example of protein crosslinking in structural studies is the hexameric CA building block of the HIV capsid. While CA hexamers had been observed at ~9 Å resolution by cryo-electron microscopy (Ganser-Pornillos et al., 2004), fusing CA to the assistant hexamer CcmK4 resulted in a higher resolution crystal structure of CA hexamers (Pornillos et al., 2009). Intriguingly, in spite of a linker of only two residues in this case, the CcmK4 assistant hexamer could not be localized in the crystals of the CA-2aa-CcmK4 fusion protein. Both crystal forms we obtained of ΔN1GspEEpsE-Hcp1 fusions did reveal ΔN1GspEEpsE and Hcp1 hexamers, though the crystal packing differs. In particular in the case of the C2 hexamers alternating layers of ΔN1GspEEpsE and Hcp1 hexamers occur (Figure S7).
The fact that subunit C of the ΔN1GspEEpsE-8aa-Hcp1 structure is highly similar to the Δ90GspEEpsE helical structure suggests that subunit conformation to be an important one. An interesting remaining question is what the total number of major conformations is which T2SS GspEEpsE subunits adopt under physiological conditions. Are the four N2D-vs-C2D orientations observed in the current study the sole conformations available to GspEEpsE subunits? Are the two hexamers observed in the present study the only hexameric arrangements possible for GspEEpsE? The SAXS data reveals that the elongated C2 hexamer of ΔN1GspEEpsE occurs in solution as part of the ΔN1GspEEpsE-8aa-Hcp1 hexamer. In contrast, in the conditions tested, neither the regular qC6 nor the elongated C2 hexamer appeared to correspond with the SAXS data of the ΔN1GspEEpsE-6aa-Hcp1 hexamer (Figure 3). This suggests that GspEEpsE in the assembled T2SS might form more hexamer variants than the two observed in our current structures. Perhaps GspEEpsE in the ΔN1GspEEpsE-6aa-Hcp1 fusion exists in multiple conformations but only one of these, which Is not predominant in solution, is seen in the crystals. Interestingly, the ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 assemblies have essentially the same ATPase activity (Figure 1B). This suggests that in both hexamers the subunits of ΔN1GspEEpsE can adopt with equal frequency the conformations capable of ATP hydrolysis.
Possibly the six different PilT N2D-vs-CTD orientations (Figures 4C), provide an additional set of subunit conformations available to GspEEpsE. The N2D and CTD could then differ by up to 73 degrees in mutual orientation (Table S1). An even larger set of possible hexamer arrangements is obtained when considering distant assembly systems occurring in Archaea. These prokaryotes lack an outer membrane but contain machineries which assemble flagellins, relatives of type IV pilins and T2SS pseudopilins, into archaella. When compared with GspEEpsE, the archaellum assembly system ATPases have an amino acid sequence identity of ~28% for the C1Ds and of ~13% for the N2Ds, while the N1D and C2D domains have different folds, and the CMD is absent (Figure 1A). In the subunits of archaellum hexameric ATPases (Reindl et al., 2013; Yamagata and Tainer, 2007), about three additional N2D-vs-CTD orientations occur which differ from the N2D-vs-CTD orientations observed so far in GspEEpsE and in PilT subunits (Figure S8). If some or all of the known N2D-vs-C2D orientations in these ATPases would also be available to the T2SS GspE ATPases, then the ensemble of possible hexameric T2SS GspE conformations would become truly large. Alternatively, the T2SS, T4PS and archaellum assembly systems might each have a specific, more limited, set of N2D-vs-CTD orientations at their disposal to perform their respective functions. Interestingly, the CTD•N2D’ construction unit of the FlaI ATPase from the archaellum assembly system in S. acidocaldarius (Reindl et al., 2013) differs from the CTD•N2D’ construction unit of V. cholerae GspEEpsE with an r.m.s.d. of only about ~ 2.5 Å. Clearly, this large family of secretion ATPases is using quite inflexible construction units and very flexible subunits.
Recently, a cryo-electron microscopy study of the assembly ATPase PilF of the T4PS involved in DNA transformation by the Gram-negative Thermus thermophilus has been reported (Collins et al., 2013). This T4PS assembly ATPase contains the zinc-binding CMD, like the GspE T2SS assembly ATPases, but has three, not one, N1 domains which are homologous to each other (Figure 1A). In these reconstructions of TtPilF, with imposed six-fold symmetry, the ATPase rings have distinctly different shapes in the absence and presence of AMPPNP. Whether or not TtPilF can also form hexamers with lower symmetry will require further investigations. Possibly the N-terminal domains of TtPilF assist in the hexamerization of the TtPilF assembly ATPase domains, analogous to the way the Hcp1 hexamer induces hexamer formation of GspEEpsE in our fusion proteins (Figures 2A).
While the assistant-hexamer approach has been successful in the current in vitro study, a crucial question for understanding the mechanism of T2SS secretion is which factors are responsible for hexamerization of GspEEpsE in vivo. It is obviously important to avoid futile ATP hydrolysis by the T2SS ATPase in the absence of exoproteins in the periplasm. Hence, full activation, almost certainly by hexamerization, of the T2SS GspE is likely to occur only under specific conditions. It has been proposed that the inner membrane platform depends on the assembly of the secretin GspD in the outer membrane (Howard, 2013). Hence, T2SS GspE hexamerization might only occur after the GspD dodecamer is formed in the outer membrane, and the inner membrane platform is assembled with GspL likely being a key factor in the hexamerization of GspE. Maybe an exoprotein catalyzes the GspD-assisted assembly process of the inner membrane platform, as has been suggested for the T2SS in Xanthomonas campestris. (Chen and Hu, 2013). However, there are studies which show that in the related T4PS, the type IV pilus can be formed when the T4PS secretin PilQ is not expressed (Wolfgang et al., 2000). Given that the T2SS and the T4PS share several features, this might suggest that also in the T2SS GspD is not absolutely required for pseudopilin assembly. Evidently, despite major progress made in recent years, profound mysteries regarding the mechanism of action of the T2SS remain.
EXPERIMENTAL PROCEDURES
Design of ΔN1GspEEpsE-Hcp1 Fusions
P. aeruginosa Hcp1 (PDB ID: 1Y12; (Mougous et al., 2006)), a 165-residue single domain protein that forms stable hexameric rings in solution and structure, was chosen as the assistant hexamer. A hexameric ring model with C6 symmetry of V. cholerae GspEEpsE without its Nterminal 99 residues (ΔN1GspEEpsE) was generated based on the C6 hexameric structure of Aquifex aeolicus PilT bound to ATP (AaPilT; PDB ID: 2EWW) (Satyshur et al., 2007), which shares 25% amino acid sequence identity with ΔN1GspEEpsE. The CTDs of Δ90GspEEpsE were superimposed onto the C-domains of the AaPilT hexamer, and N2D domains were placed in positions to maintain the most extensive and conserved CTD•N2D’ interface observed in the V. cholerae Δ90GspEEpsE helical structure (PDB ID: 1P9W) (Robien et al., 2003). By aligning the six fold axes of the Δ90GspEEpsE hexameric model with that of the Hcp1 hexamer, the two hexamers were placed close to each other without clashes. An approximate distance of 28 Å was obtained between the last observed amino acid at the C-terminus in the Δ90GspEEpsE structure and the first observed amino acid at the N-terminus of the Hcp1 structure. The five C-terminal residues of GspEEpsE, that were not included in the construct of the Δ90GspEEpsE structure (Robien et al., 2003), where maintained in the current constructs. Several variants of ΔN1GspEEpsE-Hcp1 fusions with different linker sequences were constructed and verified by DNA sequencing. The constructs are labeled as ΔN1GspEEpsE-naa-Hcp1, which consists of V. cholerae GspE residues 100–503, a linker with of ‘n’ amino acids, and the full-length Hcp1 sequence. Two “6aa” variants were made to check the effect of amino acids in the linker on ATPase activity: ΔN1GspEEpsE-6aa(KLASGA)-Hcp1 and ΔN1GspEEpsE-6aa(GSGSGS)-Hcp1. The activities were very similar (Figure 1C). The crystal structure was determined of ΔN1GspEEpsE-6aa(GSGSGS)-Hcp1, which is for convenience called ΔN1GspEEpsE-6aa-Hcp1 throughout the paper.
Purification and Characterization
All four variants of ΔN1GspEEpsE -linker-Hcp1, each containing a C-terminal hexa-histidine tag, were cloned into a modified pETDuet vector. Protein was expressed at 12°C in BL21(DE3), by induction with 0.1 mM IPTG at OD600~0.8. Cells were harvested at approximately 18 hours and resuspended in purification buffer consisting of 20 mM Tris-HCl pH 8, 500 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP/HCl, 0.1 mM ATP, 5 mM MgCl2 supplemented with 50 mM imidazole and a protease inhibitor cocktail. All the purification procedures were carried out at 4 °C. The cells were disrupted by sonication. The soluble fraction was loaded onto Ni-NTA resin (Qiagen), washed with 75 mM imidazole added to the purification buffer, and eluted in the purification buffer with 250 mM imidazole. Proteins were then purified by Superose 6 gel-permeation chromatography (Amersham). The final protein solution was concentrated to approximately 5 mg/ml supplemented with 5 mM ADP and 5 mM MgCl2.
ATPase activity measurements
ATPase activities were measured using BIOMOL Green reagent (Enzo Life Sciences). The reaction mixture contained 0.25 μM protein, 5 mM ATP, and 5 mM MgCl2, in 100 mM HEPES, pH 8.5, 65 mM NaCl, 5% glycerol. Reactions were incubated at 37 °C for 0, 5, 10, 15, and 20 minutes and assayed for the release of inorganic phosphate. The amount of phosphate was determined by comparing the absorbance at 650 nm with a phosphate standard curve. Data reported were from three separate samples of the same purified proteins assayed in duplicate.
Native Mass Spectrometry
All native mass spectra were acquired using a hybrid electrospray/quadrupole/ionmobility/time-of-flight mass spectrometer (Waters Synapt G2 HDMS). For each protein, a ~20 μL sample solution was prepared by exchanging 5 μL of 2.5–6.4 mg/mL of the given protein in a storage buffer into a buffer containing 500 mM ammonium acetate buffer (pH 8.0), 50 μM MgCl2, 50 μM ADP using a Corning® Spin-X® UF centrifugal concentrator with a 10K MWCO. Ions were formed by nanospray using borosilicate capillaries that have inner diameters of 0.78 mm and a tip at one end with an inner diameter ~1–3 μm. The capillary was loaded with 2–4 μL of the sample solution and a potential of ~1 kV was applied using a platinum wire electrode inserted into the back of the capillary that makes direct contact with the solution. Mass spectra were calibrated externally using electrospray generated ions from a 50 mg/mL solution of CsI.
Crystallization and Data Collection
ΔN1GspEEpsE-5aa-Hcp1 crystals were grown in sitting drop at 4 °C using 1 μL protein solution in the presence of 5 mM AMPPNP and 5 mM MgCl2, and 1 μL reservoir solution of 12.5% PEG 20,000, 0.1 M Bicine pH 9, and 2% v/v 1,4-dioxane. Crystals were cryo-protected with the above precipitant and nucleotides supplemented with 25% glycerol. Data were collected at the SSRL beamline 12-1. ΔN1GspEEpsE-5aa-Hcp1 data were processed to 7.3 Å in space group P42212, with cell dimensions a = b = 205.1 Å and c = 234.7 Å, using HKL2000 (Otwinowski and Minor, 1997).
ΔN1GspEEpsE-6aa-Hcp1 crystals were grown in sitting drop at 4 °C using 1 μL protein solution in the presence of 5 mM ADP, 5 mM AMPPNP and 5 mM MgCl2, and 1 μL reservoir solution of 7% PEG6000 and 0.1 M Bicine pH 9. Crystals were cryo-protected with the above precipitant and nucleotides supplemented with 20% glycerol. Data were collected in-house using a Saturn 94 CCD detector on a Rigaku Micromax HF-7 rotating anode. The Zn K-edge data were collected at SSRL beamline 12-1. Native ΔN1GspEEpsE-6aa-Hcp1 data were processed to 7.6 Å in space group P42212, with cell dimensions a=b= 205.1 Å and c= 235.0 Å, using HKL2000. The Zn-edge ΔN1GspEEpsE-6aa-Hcp1 data were isomorphous and was processed using XDS (Kabsch, 2010). The solvent content of these tetragonal crystals is ~ 61.9 %.
ΔN1GspEEpsE-7aa-Hcp1 crystals were grown in sitting drop at 4 °C using 1 μL protein solution and 1 μL reservoir solution of 7% PEG3350, 0.12 M ammonium citrate pH 7.0, 5 mM ADP, 5 mM MgCl2, and cryoprotected by the addition of 25% glycerol. Data were collected inhouse using a Saturn 94 CCD detector on a Rigaku Micromax HF-7 generator. The ΔN1GspEEpsE-7aa-Hcp1 data were processed to 6.95 Å in space group P22121 with cell dimension of a= 106.7, b=132.4, c=149.7 Å, using HKL2000.
ΔN1GspEEpsE-8aa-Hcp1 crystals were grown in sitting drops at 4 °C using 1 μL protein solution and 1 μL reservoir solution of 16% PEG300, 0.2 M ammonium sulfate, 0.1 M BisTris pH 6.1, 5 mM ADP, 5 mM MgCl2, 5 mM AlCl3, and 15 mM NaF. Crystals were cryo-protected with the above precipitant with 30% PEG300. Data was collected at the SSRL beamline 12-1. The ΔN1GspEEpsE-8aa-Hcp1 data were processed in space group P22121 with cell dimension of a = 112.5, b = 132.9, c = 142.8 Å, using XDS. The ΔN1GspEEpsE-8aa-Hcp1 data were highly anisotropic, extending to ~4.1 Å in the a and b directions and to only ~5 Å in the c direction. The solvent content of these orthorhombic crystals is ~ 55.6 %.
Structure Determinations
The structure of ΔN1GspEEpsE-5aa-Hcp1 was solved by molecular replacement with Phaser (McCoy, 2007) using the CTD•N2D’ construction unit from a full length structure of GspEEpsE (manuscript in preparation), i.e. employing the same the procedure as used in the structure determination of the unliganded PaPilT structure (Misic et al., 2010). Six CTD•N2D’ construction units and one Hcp1 hexamer were placed in the asymmetric unit. TLS and rigid body refinements were performed by defining each N2D and CTD domain as an independent group, and the Hcp1 hexamer as the thirteenth group.
The structures of ΔN1GspEEpsE-6aa-Hcp1 was solved by molecular replacement with Phaser using the procedure as used for the ΔN1GspEEpsE-5aa-Hcp1 structure. Phaser placed six CTD•N2D’ construction units in the asymmetric unit of the ΔN1GspEEpsE-6aa-Hcp1 P42212 crystal. The resultant ΔN1GspEEpsE-5aa-Hcp1 and ΔN1GspEEpsE-6aa-Hcp1 structures are essentially the same.
The structure of ΔN1GspEEpsE-8aa-Hcp1 was solved using Phaser and the same search model as above. Three CTD•N2D’ construction units were placed in the asymmetric unit of the ΔN1GspEEpsE-8aa-Hcp1 P22121 crystal, with a crystallographic two fold generating a hexameric ring of ΔN1GspEEpsE subunits. Subsequently, Phaser was able to position the Hcp1 hexamer (PDB: 1Y12). The ΔN1GspEEpsE-8aa-Hcp1 structure was then used as a model to obtain the structure of ΔN1GspEEpsE-7aa-Hcp1 in the same P22121 space group yielding very similar arrangements of the ΔN1GspEEpsE and Hcp1 hexamers, but at lower resolution.
For the ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 structures further adjustments to the models were performed with REFMAC (Murshudov et al., 1997), initially with each hexameric ring defined as a rigid body. Final rigid body and TLS refinements were carried out for the N2D and CTD domains of each independent ΔN1GspEEpsE subunit and the Hcp1 hexamer. Since the ΔN1GspEEpsE-5aa-Hcp1 structure is very similar to ΔN1GspEEpsE-6aa-Hcp1 structure and the ΔN1GspEEpsE-7aa-Hcp1 variant is very similar to the ΔN1GspEEpsE-8aa-Hcp1 structure but with lower resolution, we focus here on the ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 structure in the structure analysis and description.
SAXS
SAXS data for ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 were collected at the beamline 4-2 at SSRL using a Rayonix MX225-HE detector. ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 were purified in the presence of 5 mM nucleotide analogs and 5 mM MgCl2 in 20 mM Tris-HCl pH 8, 500 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP/HCl, at 4 °C, and then centrifuged to remove the aggregation. 35 μL of purified samples from the size exclusion chromatographic separation, with corresponding matching buffers, were automatically loaded into the capillary flow cell with a Hamilton syringe robot. Measurements were taken at room temperature with a sample-to-detector distance of 1700 mm and an X-ray energy of 11 keV. The SAXS data were measured at three protein concentrations for each sample (Figure S9) and merged together using PRIMUS (Konarev et al., 2003). Scattering curves were calculated from the ΔN1GspEEpsE-8aa-Hcp1 and ΔN1GspEEpsE-6aa-Hcp1 crystal structures, and compared with the experimental scattering curves using FOXS ((Schneidman-Duhovny et al., 2010)). The chi-square value was used to compare calculated and experimental SAXS curves (Svergun et al., 1995).
Supplementary Material
HIGHLIGHTS.
Vibrio cholerae T2SS ATPase GspE forms hexamers when fused to an assistant hexamer
GspE-Hcp1 fusions are twenty times more active than monomeric T2SS GspE
Crystal structures reveal two very different types of T2SS GspE hexamers
SAXS studies indicate that additional T2SS GspE hexamer arrangements may occur
Acknowledgments
We thank the staff of beam line BL12-2 at SSRL for invaluable assistance with crystallographic data collection, and of beam line BL4-2 for assistance with the SAXS scattering experiments. We thank Joseph Mougous for providing the Hcp1 gene, Konstantin Korotkov for important contributions to earlier stages of the project, Wei Mi for stimulating discussions, Miklos Guttman and Natalie Garcia for assistance with the SAXS studies, and Jonathan Kay for technical support. STM and MFB acknowledge support from the University of Washington. Research reported in this publication was supported by NIAID of the National Institutes of Health under award number AI34501 to WGJH and AI49294 to MS.
Footnotes
ACCESSION NUMBERS
The coordinates and structure factors of ΔN1GspEEpsE-6aa-Hcp1 and ΔN1GspEEpsE-8aa-Hcp1 have been deposited with the PDB with accession codes: 4KSR and 4KSS.
Supplemental Information contains one Table and nine Figures and can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J Mol Biol. 2005;348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Ayers M, Howell PL, Burrows LL. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 2010;5:1203–1218. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]
- Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. The EMBO journal. 2007;26:19–27. doi: 10.1038/sj.emboj.7601481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Sandkvist M. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J Bacteriol. 2005;187:249–256. doi: 10.1128/JB.187.1.249-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-L, Hu N-T. Function-Related Positioning of the Type II Secretion ATPase of Xanthomonas campestris pv. campestris. PLoS ONE. 2013:8. doi: 10.1371/journal.pone.0059123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shiue SJ, Huang CW, Chang JL, Chien YL, Hu NT, Chan NL. Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. The Journal of biological chemistry. 2005;280:42356–42363. doi: 10.1074/jbc.M506843200. [DOI] [PubMed] [Google Scholar]
- Collins RF, Hassan D, Karuppiah V, Thistlethwaite A, Derrick JP. Structure and mechanism of the PilF DNA transformation ATPase from Thermus thermophilus. Biochem J. 2013;450:417–425. doi: 10.1042/BJ20121599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Li J. Type IV pilli: paradoxes in form and function. Curr Opin Struc Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzi B, Filloux A, Voulhoux R. On the path to uncover the bacterial type II secretion system. Philos T R Soc B. 2012;367:1059–1072. doi: 10.1098/rstb.2011.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiology and molecular biology reviews : MMBR. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst TR, Sanchez J, Kaper JB, Hardy SJ, Holmgren J. Mechanism of toxin secretion by Vibrio cholerae investigated in strains harboring plasmids that encode heat-labile enterotoxins of Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:7752–7756. doi: 10.1073/pnas.81.24.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SP. Research in Microbiology. 2013. Assembly of the type II secretion system. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- Korotkov KV, Gonen T, Hol WG. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci. 2011;36:433–443. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica Section D-Biological Crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LS, Haft RJF, Forest KT. Structural insights into the Type II secretion nanomachine. Curr Opin Struc Biol. 2012;22:208–216. doi: 10.1016/j.sbi.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J Mol Biol. 2010;400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O’Neal CJ, Amaya EI, Jobling MG, Holmes RK, Hol WG. Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry. 2004;43:3772–3782. doi: 10.1021/bi0360152. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C, Sweet R, editors. Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Park YJ, Hol WG. Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds. J Struct Biol. 2012;180:362–373. doi: 10.1016/j.jsb.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick M, Korotkov KV, Hol WG, Sandkvist M. Oligomerization of EpsE coordinates residues from multiple subunits to facilitate ATPase activity. The Journal of biological chemistry. 2011;286:10378–10386. doi: 10.1074/jbc.M110.167031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua YZ, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-Ray Structures of the Hexameric Building Block of the HIV Capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy Chichili VP, Kumar V, Sivaraman J. Linkers in the structural biology of protein–protein interactions. Protein Sci. 2013;22:153–167. doi: 10.1002/pro.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl S, Ghosh A, Williams GJ, Lassak K, Neiner T, Henche AL, Albers SV, Tainer JA. Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics. Mol Cell. 2013;49:1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J Mol Biol. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- Sandkvist M, Bagdasarian M, Howard SP, DiRita VJ. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. The EMBO journal. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Research. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiue SJ, Chien IL, Chan NL, Leu WM, Hu NT. Mutation of a key residue in the type II secretion system ATPase uncouples ATP hydrolysis from protein translocation. Molecular microbiology. 2007;65:401–412. doi: 10.1111/j.1365-2958.2007.05795.x. [DOI] [PubMed] [Google Scholar]
- Shiue SJ, Kao KM, Leu WM, Chen LY, Chan NL, Hu NT. XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. The EMBO journal. 2006;25:1426–1435. doi: 10.1038/sj.emboj.7601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Wolfgang M, van Putten JPM, Hayes SF, Dorward D, Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 2000:19. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. The EMBO journal. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.