Abstract
Superfamily 2 helicases are involved in all aspects of RNA metabolism, and many steps in DNA metabolism. This review focuses on the basic mechanistic, structural and biological properties of each of the families of helicases within superfamily 2. There are ten separate families of helicases within superfamily 2, each playing specific roles in nucleic acid metabolism. The mechanisms of action are diverse, as well as the effect on the nucleic acid. Some families translocate on single-stranded nucleic acid and unwind duplexes, some unwind double-stranded nucleic acids without translocation, and some translocate on double-stranded or single-stranded nucleic acids without unwinding.
Keywords: Helicase, Superfamily 2, DEAD-box, DExH, DEAH
2. INTRODUCTION
Helicases are a ubiquitous group of enzymes that use the energy of nucleoside triphosphate (NTP) hydrolysis to catalyze the separation of double-stranded nucleic acids (dsNA). The resulting single-stranded nucleic acids (ssNA) are substrates for numerous cellular reactions. Consequently, helicases are involved in essentially every step in DNA and RNA metabolism, including replication, DNA repair, recombination, transcription, translation, chromatin rearrangement, ribosome synthesis, RNA maturation and splicing, nuclear export, Holliday junction movement, and displacement of proteins from DNA and RNA (reviewed in (1, 2, 3, 4). Helicases were originally identified as proteins that could separate double-stranded nucleic acids based on 7 conserved sequence motifs (5) and were later classified into superfamilies (SF) (6, 4). However, only a subset of these enzymes have dsNA unwinding activity. The helicase motifs, instead, are characteristic of all nucleic acid dependent NTPases, of which helicases are a subset (7, 1). Some helicases have been shown to be translocases (ie: utilize the energy of NTP hydrolysis for directional translocation on NA), including PcrA (8, 9, 10), NS3 (11, 12), NS3h (13), Rep (14), and UvrD (15). However, not all translocases have helicase activity, including EcoR124I (16) RIG-I (17), SWI/SNF (18), and ISW2 (19), and not all helicases have translocase activity, such as Ded1 (20, 21). Therefore, not all proteins possessing the helicase motifs and the helicase core structure are helicases (able to unwind dsDNA or dsRNA in an ATP dependent manner); examples are the Swi/Snf family and the type I restriction enzymes (Table 1).
Table 1.
Members and enzymatic activities of superfamily 2 helicase families.
| Family | Family members discussed in this review | Activities |
|---|---|---|
| DEAD-box | Mammalian eIF4A(37) S. cerevisiae Mss116p (41, 164, 37) S. cerevisiae Ded1p (37) E. coli DbpA (50, 49) E. coli DbpA (50, 49) Neurospora crassa CYP-19 (44) |
dsRNA unwinding |
| DEAH/RHA |
S. cerevisiae Prp16p(77) S. cerevisiae Prp22p(78) S. cerevisiae Prp43p (74, 79, 81) |
ssRNA translocase dsRNA unwinding |
| RecQ-like | human BLM (90) human WRN (91, 24) human RecQ1 (23, 161, 165) human RecQ4 (92) |
ssDNA translocase dsDNA unwinding |
| Rad3/XPD | archaeal XPD (97) human XPD (100, 95) |
ssDNA translocase dsDNA unwinding |
| Swi/Snf | human CSB (111) human ATRX (116, 114) S. cerevisiae INO80(108) S. cerevisiae ISWI (19) S. cerevisiae Rad54(113) S. solfataricus SWI2/SNF2 (166) |
dsDNA translocase |
| RIG-I-like | archaeal Hef (167) human Dicer (123, 124) human RIG-I(17, 126) |
dsRNA translocase |
| Type I Restriction Enzyme | E. coli EcoR124I(168, 132) | dsDNA translocase |
| Ski2-like |
S. cerevisiae Ski2p (52) S. cerevisiae Mtr4 (56) archaeal Hel308 (85) |
ssRNA translocase dsRNA unwinding |
| RecG-like |
E. coli RecG (152, 147, 155) T. maritima RecG (162) E. coli PriA (158, 147, 160) |
dsDNA translocase branched DNA unwinding |
| NS3/NPH-II | Dengue Virus NS3(86, 62) Vaccinia Virus NPH-II(59, 60) Hepatitis C Virus NS3 (63, 71, 69, 58) |
ssRNA translocase dsRNA unwinding |
Of the conserved sequence motifs, only the Walker A and B motifs are common to all helicases (22). These are involved in NTP binding and hydrolysis. Superfamily (SF) 1 and SF2 are the largest of the superfamilies and the conserved motifs are similar (6). The SF1 and SF2 helicases appear to function as monomers or dimers (3) for unwinding. Some specialized activities such as Holliday junction resolution and strand annealing require larger oligomers (23, 24). Each monomer contains two RecA-like domains (4). SF3-SF6 helicases show little similarity to SF1 and SF2 helicases. They have fewer conserved motifs (6, 4), contain one RecA-like domain per monomer (4), and function as hexamers or double hexamers (4, 25). These hexameric rings encircle the DNA, resulting in a highly processive helicase. The replicative helicases involved in chromosomal DNA replication are members of these superfamilies (26, 4).
3. SUPERFAMILY 2 HELICASES
Superfamily 2 is the largest and most diverse of the helicase superfamilies. It has been further divided into families including RecQ-like, RecG-like, Rad3/XPD, Ski2-like, type I restriction enzyme, RIG-I-like, NS3/NPH-II, DEAH/RHA, DEAD-box, and Swi/Snf families based on sequence homology (Table 1) (1, 27, 25). It also includes smaller groups, such as type III restriction enzymes and Suv3 (1). Although they are classified as helicases, some have not been shown to separate the strands of duplex NA or translocate on nucleic acids (16, 17, 18, 19, 28). Some unwind DNA or RNA while translocating on the NA, some unwind without translocation, and some translocate without unwinding. All have nucleic acid stimulated ATPase activity (25). SF2 helicases are involved in transcription, DNA repair, and chromatin rearrangement (29, 30, 31) and all aspects of RNA metabolism (25, 3). Since SF2 helicases function in diverse parts of nucleic acid metabolism, defects are associated with a variety of diseases including predisposition to cancer, premature aging, immunodeficiency, and mental retardation (32, 33).
4. FAMILIES
Based on recent work by the Jankowsky lab there are 10 families within SF2, in addition to the smaller type III restriction enzyme group and helicases such as Suv3 which belong to SF2, but not to any of the families or groups (1, 27, 25). They are collectively referred to as DExH/D helicases. However, the structures, mechanisms, and biological functions of the members of each of the families within this SF vary widely. This review will summarize available information about each of the families within SF2.
4.1 DEAD-box
The DEAD-box family is the largest in SF2 and is conserved from bacteria to humans (25). They are required for all aspects of RNA metabolism including transcription, splicing, transport, ribosome biogenesis, translation, RNA/protein complex assembly, and degradation (34, 25, 7). These enzymes are named for the conserved sequence of amino acids in the Walker B motif. They function exclusively on RNA as ATP-dependent chaperones that reconfigure the RNA. Diverse substrates are utilized by these enzymes, but they are most efficient when the RNA binding domain, separate from the unwinding active site, is also bound to RNA (35, 20). It can be ssRNA, dsRNA, or even structured RNA (36). DEAD-box proteins bind to RNA in an ATP dependent manner, but they don’t translocate. Instead, they manipulate structured RNAs and RNA protein complexes (RNPs) by disrupting local secondary and tertiary structures and RNA-protein interactions (34). ATP-dependent unwinding of RNA by DEAD-box proteins occurs by binding to the dsRNA and prying the strands apart instead of by translocation on nucleic acid (21). This allows unwinding to occur without directionality in some cases (37). It also limits the DEAD-box helicases to unwinding short duplexes, normally less than 2 helical turns (21).
The helicase motifs are clustered in the cleft between the two RecA-like domains (Figure 1). They form the ATP and RNA binding sites. The Q motif (38) contacts the nucleotide base, providing a specificity for ATP (7). The N-terminal RecA-like domain of all DEAD box helicases contains motifs I-III. Motifs IV-VI are in the C-terminal domain (34). When ATP and RNA are absent, the helicase is in an open conformation and the two RecA-like domains do not interact (39). Upon binding of RNA and ATP, the helicases domains close (40). In the structures of DEAD-box proteins bound to ssRNA, the phosphate backbone is kinked (Figure 1) (41, 40, 42, 43). This may aid in duplex destabilization, thereby providing a mechanism for unwinding short duplexes. Many contacts are made between the DEAD-box proteins and the RNA backbone, but little or no interactions with the bases are observed in the structures (40, 42, 43). Hydrogen bonding between the ribose 2′-OH and helicase motifs Ia, Ib, and IV confers specificity for RNA (42, 43).
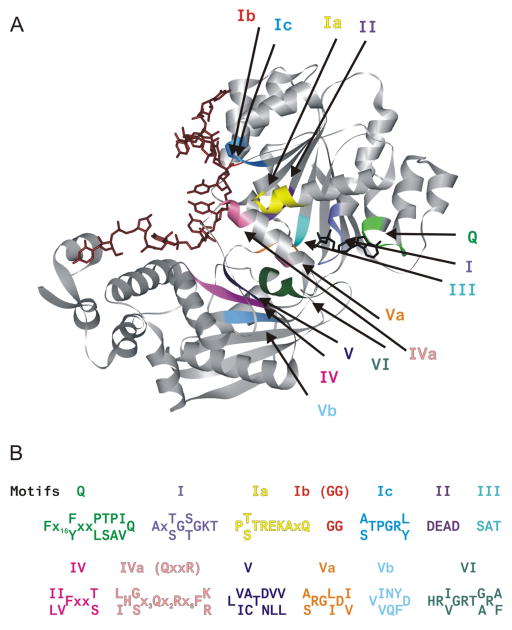
Figure 1.
Structure of DEAD-box helicase Mss116p. A. The crystal structure (Protein Data Bank code 3I61 (41)) of the DEAD-box helicase Mss116p bound to ssRNA and the ATP transition state analog, ADP-BeF3−, illustrates the bending of the nucleic acid substrate by DEAD-box helicases. The helicase motifs are in the cleft formed between the two RecA-like domains. B. The consensus sequence for the conserved helicase motifs (1, 34) in DEAD-box family members is shown.
ATP binding, but not hydrolysis, is required for strand separation (37, 44). S. cerevisiae DEAD-box proteins Ded1p and Mss116, and mammalian eIF4A can unwind dsRNA with the nonhydrolyzable ATP analog ADP-BeF3, but not with or ADPNP (37). These results suggest that ATP hydrolysis is needed for protein recycling, not strand separation. When bound to ATP, the helicase has a high affinity for RNA, but the ADP bound and free enzymes have low affinity for RNA (7, 45), resulting in release of the RNA after hydrolysis of ATP. Interestingly, although Mss116p functions differently with various ATP analogs, the structures of Mss116p bound to AMP-PNP, ADP-BeF3, and are all similar (41). Comparisons of the kinetic parameters for ATPase and unwinding reactions also suggest that ATP binding results in unwinding (44). ATP hydrolysis is not required for unwinding, but it is possible that ATP hydrolysis occurs after the dsRNA is melted due to ATP binding, but before the strands have completely separated. Helicase dissociation increases after ATP hydrolysis, but unwinding is faster than helicase dissociation, resulting in strand separation upon ATP hydrolysis. This could explain the reduced unwinding rates with ADP-BeF3 compared to ATP (37). Local unwinding, whether or not it is accompanied by ATP hydrolysis allows the remainder of short duplexes to spontaneously melt. Due to this unwinding mechanism, the unwinding rate constants decrease as duplex length and stability increase (21). With longer duplexes, the enzyme is likely to dissociate following ATP hydrolysis before the strands are completely separated, resulting in nonproductive ATP hydrolysis (44).
DbpA appears to be an exception. ATP and RNA bind cooperatively to most DEAD-box proteins (45, 46, 47, 48), and this has been accepted as a characteristic of DEAD-box proteins (34). However, kinetic studies on DbpA indicate no cooperativity in binding (49). Also, when the equilibrium and rate constants of each step in the ATPase cycle of DbpA were measured, it was found that the high affinity RNA binding state is ADP-bound (50), in contrast to other DEAD-box proteins which have high affinity for RNA when bound to ATP. Upon release of phosphate from the ADP-Pi bound state, the DbpA loses its high affinity for RNA and returns to the low affinity state. This results in unwinding of an 8 nucleotide nucleic acid strand after ATP hydrolysis, but before phosphate release (50). This is in contrast to Ded1p, Mss116p, and eIF4A which all unwind dsRNA before ATP hydrolysis, as shown in Figure 2 (37).
Figure 2.
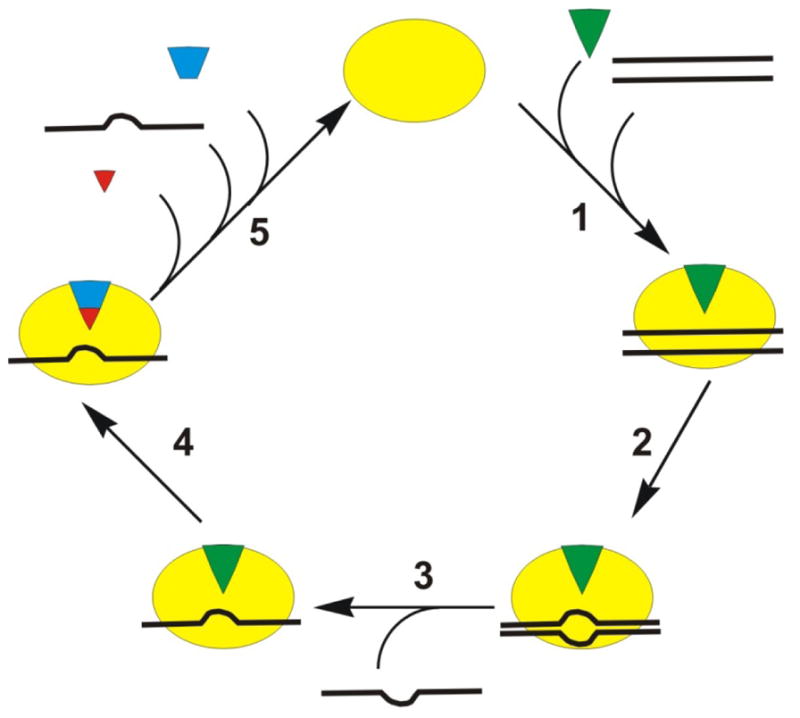
Proposed destabilization mechanism for unwinding by DEAD-box helicases (34, 37). The helicase (yellow oval) binds to the ATP (green triangle) and dsRNA (black lines) (step 1). Following a rearrangement (step 2) one strand of RNA can dissociate (step 3). ATP hydrolysis occurs after unwinding (step 4), and the cycle is completed with the release of phosphate (red triangle), ssRNA, and ADP (blue trapezoid) (step 5).
4.1.1. Destabilization mechanism
Since unwinding can occur in the absence of ATP, a destabilization mechanism has been suggested (Figure 2) (7, 34). ADP-bound and the free protein have low affinity for RNA for most DEAD-box proteins (37, 44). Helicase bound to ADP and Pi (49) or bound to non-hydrolyzable ATP analogs (37) has high affinity for RNA. The rate limiting step in unwinding is phosphate release (49). This is in agreement with unwinding occurring in the presence of ADP-BeF3 for some DEAD-box proteins (37). The kink in the ssRNA bound in the active site disrupts base pairing and causes separation of a few basepairs of the dsRNA substrate (step 2 in Figure 2), which has been suggested to be the initial step in unwinding (43). Strand separation occurs before ATP hydrolysis, but ATP hydrolysis is required for enzyme recycling (37) (step 5 in Figure 2). Since the substrates unwound are short duplexes, destabilization of a small portion of the duplex may be sufficient for unwinding as the remainder of the duplex may spontaneously melt.
Since AMP-PNP and binding also produce a kink in the RNA, but not unwinding, a rearrangement of the protein/ATP complex before hydrolysis has been suggested (34). This is consistent with two phase nucleotide binding (49). Based on these data, a model where the protein binds to ATP and dsRNA rearranges to a hydrolysis competent form has been proposed (7, 34, 41). Rearrangement can occur when ADP-BeF3 is bound, so unwinding can occur. Rearrangement does not take place when or AMP-PNP is bound, so no unwinding occurs. ATP hydrolysis (step 4 in Figure 2) occurs after dissociation (unwinding) of the first RNA strand (step 3 in Figure 2) (37). The hydrolysis of one ATP molecule either nearly or completely unwinds a short helix due to a kink that distorts the duplex RNA. For longer duplexes, the dsRNA may not be separated by the local distortion of the duplex, resulting in ATP hydrolysis while duplex RNA is bound to the enzyme (no unwinding) (44). After ATP hydrolysis, the enzyme is bound to ssRNA (or dsRNA if unwinding did not occur due to long or stable duplex), ADP, and Pi (step 4 in Figure 2). The ADP-Pi bound helicase has high affinity for RNA, so phosphate likely dissociates before RNA. This causes the enzyme to return to the open conformation, where ADP and RNA are released (step 5 in Figure 2)(49, 45).
4.2. Ski2-like
Ski2 family members are RNA helicases essential for removal of polyadenylated RNA from the cell (51, 52), and are thought to unwind secondary structure in and displace proteins from the RNA targeted to the exosome (53). mRNA is degraded in eukaryotes in the 5′-to-3′ direction by the exonuclease XRN1 and in the 3′-to-5′ direction by the exosome, a complex of exonuclases and the Ski complex which includes a Ski2 helicase (54, 55). Ski2 helicases translocate on ssRNA and unwind dsRNA in the 3′-to-5′ direction (56). Eukaryotes have nuclear and cytosolic exosomes. XRN1, the cytosolic exosome, and the Ski complex are also a part of the nonsense-mediated decay (NMD) pathway that degrades mRNAs with premature translation termination codons and the nonstop decay (NSD) pathway that eliminated mRNAs without stop codons (55). The nuclear exosome interacts with the TRAMP (Trf4-Air1-Mtr4) complex which recognizes polyadenylated RNA substrates. Trf4 is a poly(A) polymerase, Air1 is a Zn knuckle protein, and Mtr4 is a member of the Ski2-like RNA helicase family (53).
4.3. Viral NS3/NPH-II
Viral NS3/NPH-II helicases are DExH helicases encoded by many positive strand RNA viruses that unwind substrates with a 3′-ssNA overhang (57, 58). NPH-II requires a RNA loading strand (59). It translocates along the loading strand through an interaction with the sugar-phosphate backbone (60). NPH-II has been proposed to unwind DNA by strand exclusion (wire-stripper or wedge mechanism) (60). Nicks in the displaced strand do not affect unwinding by NPH-II or NS3, but both are halted by nicks in the loading strand (60, 61), indicating that these enzymes track along one of the strands.
A number of the Flaviviridae family of viruses encode a SF2 helicase, including hepatitis C, dengue virus, West Nile virus, yellow fever virus, and Japanese encephalitis virus (62). Hepatitis C virus (HCV) nonstructural protein 3 (NS3) is a dual function helicase/protease. The C-terminal portion forms a SF2 helicase (3, 1) while the N-terminal domain forms the protease. The HCV nonstructural proteins replicate and package the viral genome, and NS3 is required for viral replication (58).
Several structures of the helicase domain of NS3 (NS3h) (63, 64) and full length NS3 (65) from Hepatitis C have been solved. The structures suggest that translocation of 1 nucleotide per ATP hydrolyzed occurs in a ratcheting or inchworm mechanism due to several conformational changes that occur upon ATP binding and hydrolysis (64). NS3 has been reported to have a large kinetic step size of 11–18 base pairs (66, 67, 68) composed of smaller steps of 3 base pairs (69) and 1 base pair (63, 64, 69). The large kinetic step size has been proposed to be due to slow dissociation of the displaced strand from an as yet unidentified site on the enzyme (70). Using single-molecule FRET, Myong et al (69) observed unwinding in three base pair steps, with smaller translocation steps of one nucleotide (69). A spring-loaded inchworm mechanism was proposed based on this data in which three translocation steps of one nucleotide each produce strain in the enzyme’s structure. Release of the strain occurs when the trailing edge of the enzyme springs forward to melt three basepairs. This model received support from recent crystal structures (71) which indicated that the leading edge of NS3 could translocate one nucleotide while the trailing edge remained bound to the same RNA bases of the 3′-end of the RNA (71). It is possible that some form of ‘scrunching’ of RNA might also account for the single molecule FRET data as has been reported for the SF1 helicase UvrD (15).
4.4 DEAH/RHA
The DEAH family of RNA helicases is found in eukaryotes (72) and prokaryotes (73). It is distinct from the NS3/NPH-II (viral DExH) family of helicases (74). It is named for human RNA helicase A (RHA) and for the conserved sequence of the Walker B box. DEAH/RHA helicases have two conserved domains C-terminal to the two RecA-like helicase domains (72). One is a helicase-associated domain and the other has unknown function. These domains are required (73) and are specific to DEAH/RHA helicases; viral NS3/NPH-II helicases do not have these conserved domains (74).
DEAH/RHA helicases are involved in ribosome biogenesis (Dhr1p and Drh2p in yeast, and DHX32 and DHX37 in humans) and mRNA splicing (Prp2p, Prp16p, Prp22p, and Prp43p in yeast and DHX8, DHX16, and DHX38 in humans) (72). In bacteria, DEAH/RHA helicases are also involved in RNA processing (73). Helicases in the human RHA subfamily (DHX9, DHX29, DHX30, DHX36, and DHX57) are involved in nuclear import and export, RNA localization, translational regulation, splicing (75), and RHA knockout mice were not viable (76). During splicing, DEAH/RHA helicases appear to remodel structural RNAs. Prp16p melts the U2/U6 helix in the spliceosome after cleavage of the 5′-exon (77). Prp22p catalyzes mRNA release from the spliceosome by translocating on the mRNA (78). Prp43p is required for synthesis of both subunits of the ribosome (79, 80) and releases the lariat intron from the spliceosome in an ATP dependent process (81, 82, 83, 84).
The structure of Prp43p is similar to the Ski2-like DNA helicase Hel308 (74). Like Hel308, Prp43p has a ratchet domain. Stacking interactions with amino acids of the helix and the nucleic acid are proposed to pull the ssNA into the active site and allow the helicase to translocate along the NA in a processive manner (85, 74). However, unlike Hel308, C-terminal to the RecA-like domains, Prp43p has an oligonucleotide/oligosaccharide-binding (OB)-fold that is characteristic of DEAH/RHA proteins. The OB-fold is required for ATPase activity, interaction with protein partners, and increases its affinity for RNA (74). The helicase interacts with the nucleic acid through the phosphate backbone. ATP is bound in a conformation similar to that of NS3/NPH-II helicases (86), not like it is typically bound in DEAD-box helicases (43) or Ski2-like helicases (85). Since DEAH/RHA and Ski2-like helicases share a similar structure, it suggests that the catalytic mechanisms are similar between these two families. Ski-2-like helicases translocate in the 3′-to-5′ direction along the ssNA, with a beta-hairpin (Figure 3) causing strand separation (74).
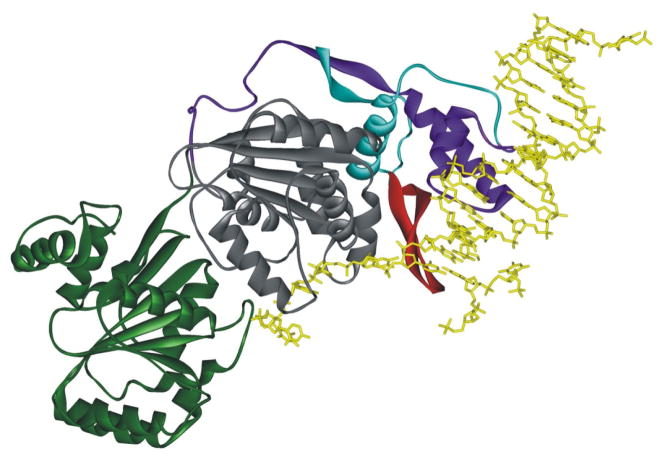
Figure 3.
The crystal structure of the human RecQ1 (Protein Data Bank code 2WWY (23)) shows the beta-hairpin separating the strands of duplex DNA. The two RecA-like domains are colored in green and gray. The Zn domain is purple. The winged helix domain is cyan, and the beta-hairpin is red.
4.5 RecQ-like
RecQ-like helicases are involved in DNA recombination, telomere maintenance, and DNA damage signaling (87). They reduce illegitimate recombination by unwinding branched DNA structures (88). Cells lacking RecQ proteins have increased recombination and chromosome missegregation, in addition to defects in meiosis. There are five human RecQ-like helicases (BLM, WRN, RecQ1, RecQ4, and RecQ5) (29). Mutations in three are associated with predispositions to cancer and premature aging (33, 89, 87, 32). Mutations in BLM are associated with Bloom’s syndrome (90), WRN with Werner’s syndrome (91), and RecQ4 with Rothmund-Thomson syndrome (92). RecQ helicases interact with the recombination and replication complexes to remove intermediates during recombination and to stabilize replication forks, thereby aiding in maintenance of genomic integrity (29).
RecQ family members are 3′-to 5′ DNA helicases. In addition to duplexes, they can unwind triplexes, quadruplexes, and 3- and 4-way junctions (29, 93). The beta-hairpin (Figure 3) is required for unwinding, HJ resolution, dimer and tetramer formation. The oligomeric properties may regulate some functions of these helicases. Monomers unwind DNA, although dimers are preferred; tetramers are required for HJ resolution by RecQ1 (23) and hexamers of WRN are needed for strand annealing (24).
4.6. Rad3/XPD
Rad3 family members in humans include: XPD, involved in nucleotide excision repair (NER) and RNA transcription, FancJ, involved in recombinational repair, Rtel1, involved in homologous recombination (HR) by unwinding toxic recombinational intermediates, and ChlR1, involved in sister chromatid cohesion (94). XPD is part of the transcription factor II H (TFIIH) complex which initiates transcription of genes regulated by RNA polymerase II promoters. The helicase activity of XPD is required in NER but only its presence, not its activity, is required for transcription initiation (95). Mutations in XPD affect nucleic excision repair, resulting in photosensitivity and an increased risk of skin cancer (32). Three distinct disorders can result, Xeroderma pigmentosum (XP), a predisposition to cancer, and the aging disorders: trichothiodystrophy (TTD) and Cockayne Syndrome (CS), depending on whether the helicase activity, interactions with TFIIH or both are affected (96). All have photosensitivity, but only XP has increased risk of skin cancer. CS and TTD have developmental disorders and premature aging. CS is more severe with patients exhibiting mental retardation (96)
All members of the Rad3 family that have been studies are 5′-to-3′ DNA helicases involved in DNA repair, and genome maintenance. They translocate on ssDNA and unwind dsDNA (97). Two insertions into helicase domain 1 distinguish Rad3 family helicases from other SF2 helicases: an Arch domain (98, 99, 100) and an iron-sulfer (FeS) cluster (101). A mutation in the FeS domain of XPD causes TTD (102). A mutation in the FeS domain of FancJ, causes Fanconi anemia (103) and a predisposition to early-onset breast cancer (102, 104). The FeS domain forms a secondary ssDNA binding site and couples ATP hydrolysis to translocation on ssDNA (97, 101).
Honda et al. (97) used a single molecule approach to simultaneously monitor translocation of the XPD helicase from Ferroplasma acidarmanus in the presence of ssDNA binding proteins (SSB) RPA1 and RPA2. Both SSBs have similar DNA binding affinities, but had different effects on XPD translocation. RPA1 competed with XPD for binding, and RPA2 did not interfere with XPD binding to ssDNA, but slowed down XPD translocation. RPA2 facilitates XPD binding to DNA by bending the DNA. RPA1 and competes with XPD for DNA binding by extending the DNA. When XPD encountered RPA1, it displaced it from the DNA. However, RPA2 was not displaced from ssDNA during translocation. XPD bypassed RPA2 without displacing it from DNA. Simultaneous visualization of both the helicase and its obstacle brought them to the conclusion that XPD can translocate on the protein-coated ssDNA without disrupting the protein-nucleic acid complex. SF2 helicases translocate along the phosphodiester backbone of nucleic acids (4), and RPA binding to ssDNA primarily involves the bases (105, 106). Since the helicases contacts the DNA backbone while the SSB interacts with bases, this allows both proteins to be bound simultaneously to the same region of ssDNA and allows the helicase to move by the SSB. Translocation over the bound protein and displacement of the protein both provide mechanisms for these helicases to bypass ssDNA binding proteins (97).
4.7. Swi/Snf
Swi/Snf complexes are involved in diverse processes in the cell, including replication, DNA repair, regulation of RNA polymerase II and III, cell signaling, cell cycle progression, metamorphosis, and tumor suppression (107, 108). Many of these processes involve their ability to remodel chromatin. This occurs by a variety of mechanisms, including repositioning histone octamers, unwrapping the DNA around an octamer, ejection of a histone octamer, or dimer, or exchange of a histone dimer (108, 107).
Chromatin remodeling complexes are large, multisubunit complexes that change the structure of nucleosomes to vary the accessibility of the DNA (109). They all contain a member of the Swi/Snf family of helicases, but the other components of the complex vary, and can be used to subdivide the family (31, 110). Swi/Snf helicases are subdivided into Snf2, ISWI, CHD, INO80, CSB, RAD54, and DDM1 subfamilies of chromatin remodeling proteins (109, 108). The INO80 subfamily is involved in DNA repair and activation of transcription (108). It is the only member of the family which has been shown to have helicase (unwinding) activity (31). Snf2 subfamily members disrupt nucleosomes and ISWI subfamily members can assemble nucleosomes, resulting in repression of transcription (31). CDH subfamily members have been implicated in chromatin remodeling and transcription activation (108, 31). Cockayne Syndrome protein B (CSB) is involved in NER and remodels chromatin (111). DDM1 is required for maintaining proper DNA methylation in the plant Arabidopsis thaliana (112). Rad54, along with Rad51, is involved in HR (31, 113). Mutations in ATRX, a Rad54 family member, result in ATR-X syndrome, characterized by alpha-thalassaemia and mental retardation (114). The majority of the mutations that have been linked to the disease occur in the helicase domains and the histone H3 binding domain (115). ATRX localizes to telomeres and other repetitive DNA sequences, in particular sequences that have the potential to form quadruplexes (116, 114).
Swi/Snf helicases have a high affinity for nucleosomes and are able to recognize histone modifications. The translocation activity of the helicase can reposition or eject histones from the DNA. This activity is modulated by other associated subunits of the remodeling complexes (108, 107). Several mechanisms have been proposed for nucleosome sliding (117). In the twist diffusion model (118), a single base pair is shifted from the linker to wrapped around the nucleoseome core with an accompanying twist to accommodate an extra basepair. Because a rotation of ~35° would be required for each basepair, this is not likely to be the mechanism of histone repositioning by Swi/Snf (117). Another model is the histone core swiveling model. It proposes that the nucleosome rotates relative to the DNA (117). Two variations of the loop or bulge propogation model have been proposed. In one, the remodeling protein pulls the DNA from the nucleosome entry or exit site, resulting in a bulge. In the second, the protein binds to an internal site and pulls DNA from the linker. They propose that the helicase binds to the DNA and upon ATP hydrolysis, the dsDNA is translocated, without being unwound, while the remodeling complex remains bound to the nucleosome (108, 109). This results in formation of a loop of DNA on the surface of the nucleosome where the histone-DNA contacts are broken (19, 119, 120, 18). This loop moves around the nucleosome, repositioning the nucleosome. Loops are visible in single molecule experiments, and the direction of translocation has been observed to switch (18, 121). It is unknown whether the helicases can translocate in either direction on DNA or whether the nucleosome rebinds in the opposite orientation. In addition to repositioning of nucleosomes, small DNA loops can also allow replacement of dimer with a modified dimer or ejection of dimers or nucleosomes by remodelers or localized unwrapping (108).
4.8. RIG-I-like
RIG-I-like helicases are involved in the antiviral immune response (122). Dicer has RNase activity for dsRNA (123) and a RIG-I like helicase domain. The helicase is required for efficient processing of pre-miRNA, and may allow Dicer to cleave structured viral RNAs (124, 125). RIG-I serves a sensor of viral RNA to initiate an immune response that leads to production of interferon (IFN). (126). Activation of two N-terminal caspase activation and recruitment domains (CARDs) occurs by recognition of 5′-triphosphate (127, 128) or dsRNA (126). 5′-triphosphates and dsRNA are present during viral replication but not during most cellular processes (129). In the presence of viral RNA, CARD ubiquitination results in IFN expression (130). RIG-I also has a C-terminal regulatory domain (RD) and is a DExH box RNA helicase. The RD inhibits signaling when viral RNA is absent. The ATPase activity of the helicase is required for signaling (126). The helicase has dsRNA translocase activity, but does not unwind the RNA (17). The CARDS prevent translocation of RIG-I in the absence of 5′-triphosphate recognition by the RD (17). In the presence of viral triggers, RIG-I tranlsocates on the RNA strand in a RNA/DNA heteroduplex or on dsRNA (17).
4.9. Type I Restriction enzymes
Type I restriction enzymes or endonucleases (T1RE) are part of the restriction-modification system in bacteria (131, 132). They protect the bacterial genome against cleavage by methylating target sequences and restriction of foreign DNA. They are classified into three groups (types I-III) based on their recognition sequence, subunit composition, cleavage position, and cofactor requirements (133, 134). T1RE are pentamers made up of three different subunits: specificity (S), methylase (M), and restriction (R) with R2M2S stoichiometry (135, 131). They are encoded by the Host Specificity for DNA (hsdS, hsdM, and hsdR genes (136). A dimer of two R2M2S holoenzymes, with each bound to a target sequence, is the active form of the enzyme for restriction of the DNA (132).
Bacterial DNA is methylated, and T1RE dissociate upon binding to fully methylated DNA (137); therefore T1RE have no effect on host DNA. Hemi-methylated DNA is methylated at the target sequence (137). HsdM and HsdS are sufficient for methylation (136). When the target sequence is unmethylated, HsdR restricts the foreign DNA (132).
All T1RE utilize S-adenosyl methionine (SAM) as the methyl donor for the methylation reaction, catalyzed by HsdM (138). It is also required for DNA restriction for most T1RE, EcoR124I being the exception (138, 139). The HdsS subunit recognizes and binds to a non-methylated DNA sequence containing two 3–5 base pair domains separated by a 6–8 base pair spacer (138). After binding, the enzyme translocates dsDNA in an ATP dependent manner toward the enzyme while it remains bound to the recognition sequence (140), resulting in formation of dsDNA loops (141). HsdR contains both a helicase domain, and an endonuclease domain (138). T1RE are not known to unwind dsNA (138) but appear to function as a dsDNA translocases (142). After translocation of 400 to 7000 basepairs (138), while remaining bound to the target sequence, the DNA is cleaved by nicking of each strand by one HsdR subunit (143). The cleaved DNA can then be degraded by exonucleases (ie: RecBCD).
A model describing T1RE was proposed by Szczelkun (144). The enzyme binds to the DNA at the target sequence, upon binding of ATP. Two holoenzymes bind to non-methylated target sequences and associate to form a dimer. ATP dependent dsDNA translocation occurs, while remaining bound to the target sequence, resulting in formation of loops. Eventually, translocation is impeded, possibly due to a collision with another protein, and the DNA is cleaved, resulting in enzyme dissociation.
4.10. RecG-like
RecG-like helicases are involved in resolution of recombination intermediates through translocation on dsDNA. In prokaryotes, Mfd removes RNA polymerase from stalled replication forks by destabilizing its interaction with the transcription complex during translocation (145, 146). PriA recognizes and binds to D-loops formed during recombination and loads the replisome onto the branched DNA to allow restart of replication (147, 148). RecG limits origin independent DNA replication (149, 150).
Until recently, RecG was believed to catalyze Holliday junction intermediate branch migration in a pathway that overlaps functionally with the RuvABC pathway (151). However, it now seems likely that RecG may function to limit replication at sites remote from oriC initiated by PriA (152). RecG translocates on dsDNA and catalyzes Holliday junction branch migration (153), in addition to unwinding various branched DNA substrates, in vitro (154). RecG decreases replication by unwinding D (147) and R (155) loops, which prevents PriA from loading DnaB helicase at the branch, leading to assembly of the replisome and replication (156, 157). By resolving these loops, RecG limits replication to oriC (152).
PriA, on the other hand, binds to and stabilizes stalled replication forks, eventually leading to replication restart (158). PriA has a 3′-terminus binding pocket that binds specifically to the 3′-end of the invading strand in the D loop (147) or the nascent leading strand (159) and displaces the nascent lagging strand (160). Primasome assembly at stalled replication forks is dependent upon PriA (148).
5. CONCLUSIONS
Superfamily 2 is a diverse group of helicases. Although some sequence motifs are conserved among all families, there are motifs that are unique to separate families (1, 25). All SF2 helicases are capable of binding nucleic acids and have nucleic acid stimulated NTPase activity (3). However, some families, such as type I restriction enzymes and RIG-I do not perform the canonical helicase reaction of unwinding duplex NA (132)). DEAD-box family members unwind RNA, but without translocation (34). In some cases, DEAD-box helicases even unwind dsRNA without ATP hydrolysis (44).
Crystal structures of Hel308 (85), an archaeal Ski2-like helicase, and RecQ1 (23, 161), a RecQ-like helicase, complexed with a single-strand/double-strand junction contain a beta-hairpin loop in a position where it could separate the strands of dsNA (Figures 3 and 4). Interestingly, HCV NS3 and S. cerevisiae Prp43p, which also unwind duplex nucleic acids in a translocation dependent process, also contain a prominent beta-hairpin (Figure 4A). XPD also unwinds dsNA and translocates on NA, but it contains a wedge made of two alpha-helices instead (Figure 4B). Another way that helicases couple translocation to unwinding is through a formal wedge domain, such as in the case of T. maritima RecG. It has a beta-hairpin on one side of the three-way or four-way junction and a wedge domain on the other which separate the dsDNA on each side of the junction simultaneously (162). The DEAD-box helicase mss116p unwinds dsRNA without translocation, and its structure complexed with a single-strand/double-strand junction does not appear to contain a beta-hairpin (Figure 4C). Structures of SWI2/SNF2, RIG-I-like Hef, and the type I restriction enzyme EcoR124I also do not appear to have a beta-hairpin (Figure 4C). These enzymes are known to translocate on nucleic acids, but not unwind dsNA. Thus, the beta-hairpin may be a conserved feature among helicases that translocate on dsNA to unwind the duplex. Pin-like structures have been proposed to serve the purpose of splitting the duplex in SF1 helicases PcrA (161) and UvrD (85) and RecD2 (163). The lack of such a structure in some SF2 helicases may serve to further distinguish structure-function relationships among this large, diverse class of enzymes.
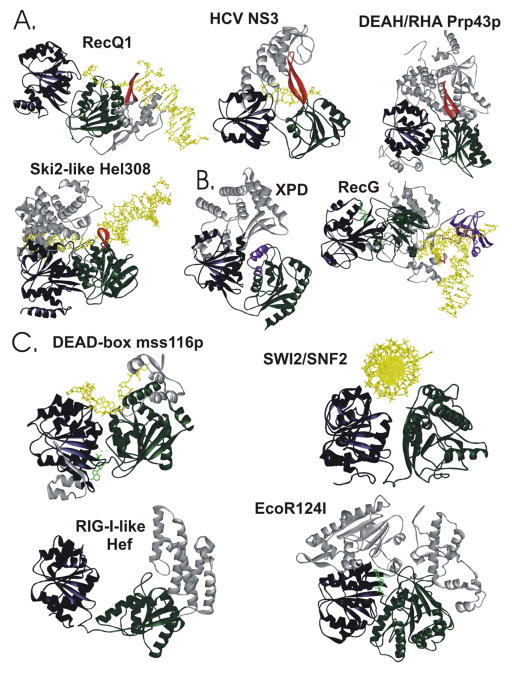
Figure 4.
A. Families containing helicases that unwind duplex nucleic acids using a mechanism dependent on translocation upon the substrate contain a beta-hairpin. The Protein Data Bank codes are RecQ1: 2WWY (23), NS3: 1A1V (63), Hel308: 2P6R (85), and Prp43p: 2XAU (74). B. XPD (Protein Data Bank code 2VSF (100)) has translocase and helicase activity, but does not have a beta-hairpin. Instead, the wedge appears to be formed from two alpha-helices (100). RecG (Protein Data Bank code 1GM5 (162)) has a beta-hairpin and a wedge domain which simultaneously separate two arms of a DNA fork. C. Families which either do not unwind nucleic acids, or unwind without translocating, do not have the beta-hairpin. The Protein Data Bank codes are Mss116p: 3I61 (41), SWI2/SNF2: 1Z63 (166), Hef: 1WP9 (167), and EcoR124I: 2W00 (168). In each of the structures, the N-terminal RecA-like domain is navy and the C-terminal RecA-like domain is green. Accessory domains are gray, except the beta-hairpin, which is red and wedge domains are purple. Nucleic acid is yellow and nucleotide is green.
Acknowledgments
This work was supported by NIH grant R01 GM089001. “This is an, un-copyedited, author manuscript that has been accepted for publication in the Frontiers in Bioscience”. Cite this article as appearing in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (http://bioscience.org/search/authors/htm/search.htm) following publication and at PubMed(http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience. From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at http://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the un-copyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.”
Abbreviations
- NTP
nucleoside triphosphate
- ss
single-stranded
- NA
nucleic acid
- ds
double-stranded
- SF
superfamily
- RNP
RNA-protein complexes
- Pi
inorganic phosphate
- HCV
hepatitis C virus
- DENV
dengue virus
- SAM
S-adenosyl methionine
- T1RE
type I restriction enzyme
- HJ
Holliday junction
- TFIIH
transcription factor IIH
- NER
nucleotide excision repair
- HR
homologous recombination
- XP
Xeroderma pigmentosum
- TDD
trichothiodystrophy
- CS
Cockayne’s Syndrome
- Pi
inorganic phosphate
- RHA
RNA helicase A
- CARD
caspase activation and recruitment domains
- RD
regulatory domain
References
- 1.Fairman-Williams Margaret E, Guenther Ulf Peter, Jankowsky Eckhard. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lohman Timothy M, Tomko Eric J, Wu Colin G. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 3.Pyle Anna Marie. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biochem. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 4.Singleton Martin R, Dillingham Mark S, Wigley Dale B. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya Alexander E, Koonin Eugene V, Donchenko Alexi P, Blinov Vladimir M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorbalenya Alexander E, Koonin Eugene V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. doi: 10.1016/S0959-440X(05)80116-2. [DOI] [Google Scholar]
- 7.Cordin Olivier, Banroques Josette, Kyle Tanner N, Linder Patrick. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Dillingham Mark S, Wigley Dale B, Webb Martin R. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–51. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 9.Dillingham Mark S, Wigley Dale B, Webb Martin R. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 10.Soultanas Panos, Dillingham Mark S, Wiley Paul, Webb Martin R, Wigley Dale B. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopal Vaishnavi, Gurjar Madhura, Levin Mikhail K, Patel Smita S. The Protease Domain Increases the Translocation Stepping Efficiency of the Hepatitis C Virus NS3-4A Helicase. J Biol Chem. 2010;285:17821–17832. doi: 10.1074/jbc.M110.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Jin, Cheng Wei, Bustamante Carlos, Oster George. Coupling Translocation with Nucleic Acid Unwinding by NS3 Helicase. J Mol Biol. 2010;404:439–455. doi: 10.1016/j.jmb.2010.09.047. [DOI] [PubMed] [Google Scholar]
- 13.Matlock Dennis L, Yeruva Laxmi, Byrd Alicia K, Mackintosh Samuel G, Langston Clint, Brown Carrie, Cameron Craig E, Fischer Christopher J, Raney Kevin D. Investigation of Translocation, DNA Unwinding, and Protein Displacement by NS3h, the Helicase Domain from the Hepatitis C Virus Helicase. Biochemistry. 2010;49:2097–2109. doi: 10.1021/bi901977k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myong Sua, Rasnik Ivan, Joo Chirlmin, Lohman Timopthy M, Ha Taekjip. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 15.Tomko Eric J, Fischer Christopher J, Niedziela-Majka Anita, Lohman Timothy M. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidel Ralf, Bloom Joost G, Dekker Cees, Szczelkun Mark D. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myong Sua, Cui Sheng, Cornish Peter V, Kirchhofer Axel, Gack Michaela U, Jung Jae U, Hopfner Karl-Peter, Ha Taekjip. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Yongli, Smith Corey L, Saha Anjanabha, Grill Stephan W, Mihardja Shirley, Smith Steven B, Cairns Bradley R, Peterson Craig L, Bustamante Carlos. DNA Translocation and Loop Formation Mechanism of Chromatin Remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zofall Martin, Persinger Jim, Kassabov Stefan R, Bartholomew Blaine. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 20.Yang Quansheng, Jankowsky Eckhard. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 21.Yang Quansheng, Campo Mark Del, Lambowitz Alan M, Jankowsky Eckhard. DEAD-Box Proteins Unwind Duplexes by Local Strand Separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucic Bojana, Zhang Ying, King Oliver, Mendoza-Maldonado Ramiro, Berti Matteo, Niesen Frank H, Burgess-Brown Nicola A, Pike Ashley CW, Cooper Christopher DO, Gileadi Opher, Vindigni Alessandro. A prominent beta-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 2011;39:1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muftuoglu Meltem, Kulikowicz Tomasz, Beck Gad, Lee Jae Wan, Piotrowski Jason, Bohr Vilhelm A. Intrinsic ssDNA Annealing Activity in the C-Terminal Region of WRN. Biochemistry. 2008;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankowsky Eckhard, Fairman Margaret E. RNA helicases - one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Patel Smita S, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 27.Jankowsky Eckhard, Fairman Margaret E. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairman Margaret E, Maroney Patricia A, Wang Wen, Bowers Heath A, Gollnick Paul, Nilsen Timothy W, Jankowsky Eckhard. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 29.Bennett Richard J, Keck James L. Structure and Function of RecQ DNA Helicases. Crit Rev Biochem Mol Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 30.Fuller-Pace Frances V. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2008;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexandra Lusser, Kadonaga James T. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 32.van Brabant Anja J, Stan Rodica, Ellis Nathan A. DNA HELICASES, GENOMIC INSTABILITY, AND HUMAN GENETIC DISEASE. Annu Rev Genom Human Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 33.Ellis Nathan A. DNA helicases in inherited human disorders. Curr Opin Gen Dev. 1997;7:354–363. doi: 10.1016/S0959-437X(97)80149-9. [DOI] [PubMed] [Google Scholar]
- 34.Hilbert Manuel, Karow Anne R, Klostermeier Dagmar. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 35.Grohman Jacob K, Campo Mark Del, Bhaskaran Hari, Tijerina Pilar, Lambowitz Alan M, Russell Rick. Probing the Mechanisms of DEAD-Box Proteins as General RNA Chaperones: The C-Terminal Domain of CYT-19 Mediates General Recognition of RNAG. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tijerina Pilar, Bhaskaran Hari, Russell Rick. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proceedings of the National Academy of Sciences. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Fei, Putnam Andrea, Jankowsky Eckhard. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyle Tanner N, Cordin Olivier, Banroques Josette, Doere Monique, Linder Patrick. The Q Motif: A Newly Identified Motif in DEAD Box Helicases May Regulate ATP Binding and Hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 39.Collins Ruairi, Karlberg Tobias, Lehtio Lari, Schutz Patrick, van den Berg Susanne, Dahlgren Lars Goran, Hammarstrom Martin, Weigelt Johan, Schuler Herwig. The DEXD/H-box RNA Helicase DDX19 Is Regulated by an alpha-Helical Switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen Christian BF, Ballut Lionel, Johansen Jesper S, Chamieh Hala, Nielsen Klaus H, Oliveira Cristiano LP, Pedersen Jan Skov, Seraphin Bertrand, Hir Herve Le, Andersen Gregers Rom. Structure of the Exon Junction Core Complex with a Trapped DEAD-Box ATPase Bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 41.Campo Mark Del, Lambowitz Alan M. Structure of the Yeast DEAD Box Protein Mss116p Reveals Two Wedges that Crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bono Fulvia, Ebert Judith, Lorentzen Esben, Conti Elena. The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Sengoku Toru, Nureki Osamu, Nakamura Akira, Kobayashi Satoru, Yokoyama Shigeyuki. Structural Basis for RNA Unwinding by the DEAD-Box Protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 44.Chen Yingfeng, Potratz Jeffrey P, Tijerina Pilar, Campo Mark Del, Lambowitz Alan M, Russell Rick. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorsch Jon R, Herschlag Daniel. The DEAD Box Protein eIF4A. 1. A Minimal Kinetic and Thermodynamic Framework Reveals Coupled Binding of RNA and Nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 46.Polach Kevin J, Uhlenbeck Olke C. Cooperative Binding of ATP and RNA Substrates to the DEAD/H Protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 47.Cordin Olivier, Kyle Tanner N, Doere Monique, Linder Patrick, Banroques Josette. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iost Isabelle, Dreyfus Marc, Linder Patrick. Ded1p, a DEAD-box Protein Required for Translation Initiation in Saccharomyces cerevisiae, Is an RNA Helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 49.Henn Arnon, Cao Wenxiang, Hackney David D, De La Cruz Enrique M. The ATPase Cycle Mechanism of the DEAD-box rRNA Helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henn Arnon, Cao Wenxiang, Licciardello N, Heitkamp SE, Hackney David D, De La Cruz Enrique M. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci U S A. 2010;107:4046–4050. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Xuying, Jia Huijue, Jankowsky Eckhard, Anderson James T. Degradation of hypomodified tRNAiMet in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widner William R, Wickner Reed B. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson Ryan N, Alejandra Klauer A, Hintze Bradley J, Robinson Howard, van Hoof Ambro, Johnson Sean J. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown Justin T, Bai Xinxue, Johnson Arlen W. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/S1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ORBAN TAMASI, Izaurralde Elisa. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weir John R, Bonneau Fabien, Hentschel Jendrik, Conti Elena. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci U S A. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jankowsky Eckhard, Gross Christian H, Shuman Stewart, Pyle Anna Marie. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 58.Raney Kevin D, Sharma Suresh D, Moustafa Ibrahim M, Cameron Craig E. Hepatitis C Virus Non-structural Protein 3 (HCV NS3): A Multifunctional Antiviral Target. J Biol Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawaoka Jane, Pyle Anna M. Choosing beteen DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 2005;33:644–649. doi: 10.1093/nar/gki208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawaoka Jane, Jankowsky Eckhard, Pyle Anna M. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 61.Beran Rudolf KF, Bruno Michael M, Bowers Heath A, Jankowsky Eckhard, Pyle Anna Marie. Robust Translocation Along a Molecular Monorail: the NS3 Helicase from Hepatitis C Virus Traverses Unusually Large Disruptions in its Track. J Mol Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 62.Wang Chun Chung, Huang Zhi Shun, Chiang Pei Ling, Chen Chien Tsun, Wu Huey Nan. Analysis of the nucleoside triphosphatase, RNA triphosphatase, and unwinding activities of the helicase domain of dengue virus NS3 protein. FEBS Letters. 2009;583:691–696. doi: 10.1016/j.febslet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 63.Kim Joseph L, Morgenstern Kurt A, Griffith James P, Dwyer Maureen D, Thomson John A, Murcko Mark A, Lin Chao, Caron Paul R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/S0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 64.Gu Meigang, Rice Charles M. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao Nanhua, Reichert Paul, Shane Taremi S, Prosise Winifred W, Weber Patricia C. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;7:1353–1363. doi: 10.1016/S0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 66.Dumont Sophie, Cheng Wei, Serebrov Victor, Beran Rudolf K, Tinoco Ignacio, Pyle Anna Marie, Bustamante Carlos. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tackett Alan J, Chen Yingfeng, Cameron Craig E, Raney Kevin D. Multiple Full-length NS3 Molecules Are Required for Optimal Unwinding of Oligonucleotide DNA in Vitro. J Biol Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 68.Serebrov Victor, Pyle Anna Marie. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 69.Myong Sua, Bruno Michael M, Pyle Anna Marie, Ha Taekjip. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Serebrov Victor, Beran Rudolf K, Pyle Anna Marie. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J Biol Chem. 2009;284:2512–2521. doi: 10.1074/jbc.M805460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Appleby Todd C, Anderson Robert, Fedorova Olga, Pyle Anna M, Wang Ruth, Liu Xiaohong, Brendza Katherine M, Somoza John R. Visualizing ATP-Dependent RNA Translocation by the NS3 Helicase from HCV. J Mol Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanjuan Rafael, Marin Ignacio. Tracing the Origin of the Compensasome: Evolutionary History of DEAH Helicase and MYST Acetyltransferase Gene Families. Mol Biol Evol. 2001;18:330–343. doi: 10.1093/oxfordjournals.molbev.a003809. [DOI] [PubMed] [Google Scholar]
- 73.Koo Jovanka T, Choe Juno, Moseley Steve L. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol Microbiol. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- 74.Walbott Helene, Mouffok Saida, Capeyrou Regine, Lebaron Simon, Humbert Odile, van Tilbeurgh Herman, Henry Yves, Leulliot Nicolas. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toretsky Jeffrey A, Erkizan Verda, Levenson Amy, Abaan Ogan D, Parvin Jeffrey D, Cripe Timothy P, Rice Anna M, Lee Sean Bong, Üren Aykut. Oncoprotein EWS-FLI1 Activity Is Enhanced by RNA Helicase A. Cancer Res. 2006;66:5574–5581. doi: 10.1158/0008-5472.CAN-05-3293. [DOI] [PubMed] [Google Scholar]
- 76.Lee Chee Gun, da Costa Soares Vera, Newberger Carol, Manova Katia, Lacy Elizabeth, Hurwitz Jerard. RNA helicase A is essential for normal gastrulation. Proc Natl Acad Sci U S A. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mefford Melissa A, Staley Jonathan P. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009;15:1386–1397. doi: 10.1261/rna.1582609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwer Beate. A Conformational Rearrangement in the Spliceosome Sets the Stage for Prp22-Dependent mRNA Release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lebaron Simon, Froment Carine, Fromont-Racine Micheline, Rain Jean Christophe, Monsarrat Bernard, Caizergues-Ferrer Michele, Henry Yves. The Splicing ATPase Prp43p Is a Component of Multiple Preribosomal Particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leeds Nina B, Small Eliza C, Hiley Shawna L, Hughes Timothy R, Staley Jonathan P. The Splicing Factor Prp43p, a DEAH Box ATPase, Functions in Ribosome Biogenesis. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai Rong Tzong, Fu Ru Huei, Yeh Fu Lung, Tseng Chi Kang, Lin Yu Chieh, Huang Yu hsin, Cheng Soo Chen. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanaka Naoko, Aronova Anna, Schwer Beate. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes & Development. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin Arnold, Schneider Susanne, Schwer Beate. Prp43 Is an Essential RNA-dependent ATPase Required for Release of Lariat-Intron from the Spliceosome. Journal of Biological Chemistry. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 84.Arenas Jaime E, Abelson John N. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proceedings of the National Academy of Sciences. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buttner Katharina, Nehring Sebastian, Hopfner Karl Peter. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 86.Luo Dahai, Xu Ting, Watson Randall P, Scherer-Becker Daniella, Sampath Aruna, Jahnke Wolfgang, Yeong Sui Sum, Wang Chern Hoe, Lim Siew Pheng, Strongin Alex, Vasudevan Subhash G, Lescar Julien. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bohr Vilhelm A. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGlynn Peter, Lloyd Robert G. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 89.Brosh Robert M, Jr, ohr Vilhelm A. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ellis Nathan A, Groden Joanna, Ye Tian Zhang, Straughen Joel, Lennon David J, Ciocci Susan, Proytcheva Maria, German James. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 91.Yu Chang En, Oshima Junko, Fu Ying Hui, Wijsman Ellen M, Hisama Fuki, Alisch Reid, Matthews Shellie, Nakura Jun, Miki Tetsuro, Ouais Samir, Martin George M, Mulligan John, Schellenberg Gerard D. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 92.Kitao Saori, Shimamoto Akira, Goto Makoto, Miller Robert W, Smithson William A, Lindor Noralane M, Furuichi Yasuhiro. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 93.Mohaghegh Payam, Karow Julia K, Brosh Robert M, Jr, Bohr Vilhelm A, Hickson Ian D. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu Y, Suhasini A, Brosh R. Welcome the Family of FANCJ-like Helicases to the Block of Genome Stability Maintenance Proteins. Cell Mol Life Sci. 2009;66:1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rudolf Jana, Rouillon Christophe, Schwarz-Linek Ulrich, White Malcolm F. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucleic Acids Res. 2010;38:931–941. doi: 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lehmann Alan R. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 97.Honda Masayoshi, Park Jeehae, Pugh Robert A, Ha Taekjip, Spies Maria. Single-Molecule Analysis Reveals Differential Effect of ssDNA-Binding Proteins on DNA Translocation by XPD Helicase. Mol Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fan Li, Fuss Jill O, Cheng Quen J, Arvai Andrew S, Hammel Michal, Roberts Victoria A, Cooper Priscilla K, Tainer John A. XPD Helicase Structures and Activities: Insights into the Cancer and Aging Phenotypes from XPD Mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Huanting, Rudolf Jana, Johnson Kenneth A, McMahon Stephen A, Oke Muse, Carter Lester, McRobbie Anne Marie, Brown Sara E, Naismith James H, White Malcolm F. Structure of the DNA Repair Helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolski Stephanie C, Kuper Jochen, Hanzelmann Petra, Truglio James J, Croteau Deborah L, Van Houten Bennett Kisker Caroline. Crystal structure of the FeS cluster-containing necleotide excision repair helicase XPD. PLoS Biol. 2008;6:1332–1342. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pugh Robert A, Honda Masayoshi, Leesley Haley, Thomas Alvin, Lin Yuyen, Nilges Mark J, Cann Isaac KO, Spies Maria. The Iron-containing Domain Is Essential in Rad3 Helicases for Coupling of ATP Hydrolysis to DNA Translocation and for Targeting the Helicase to the Single-stranded DNA-Double-stranded DNA Junction. J Biol Chem. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- 102.Rudolf Jana, Makrantoni Vasso, Ingledew W John, Stark Michael JR, White Malcolm F. The DNA Repair Helicases XPD and FancJ Have Essential Iron-Sulfur Domains. Mol Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 103.Litman Rachel, Peng Min, Jin Zhe, Zhang Fan, Zhang Junran, Powell Simon, Andreassen Paul R, Cantor Sharon B. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 104.Cantor Sharon B, Bell Daphne W, Ganesan Shridar, Kass Elizabeth M, Drapkin Ronny, Grossman Steven, Wahrer Doke CR, Sgroi Dennis C, Lane William S, Haber Daniel A, Livingston David M. BACH1, a Novel Helicase-like Protein, Interacts Directly with BRCA1 and Contributes to Its DNA Repair Function. Cell. 2001;105:149–160. doi: 10.1016/S0092-8674(01)00304-X. [DOI] [PubMed] [Google Scholar]
- 105.Bochkarev Alexey, Bochkareva Elena, Frappier Lori, Edwards Aled M. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kerr Iain D, Wadsworth Ross IM, Cubeddu Liza, Blankenfeldt Wulf, Naismith James H, White Malcolm F. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kasten Margaret M, Clapier Cedric R, Cairns Bradley R. SnapShot: Chromatin Remodeling: SWI/SNF. Cell. 2011;144:310–310. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 108.Clapier Cedric R, Cairns Bradley R. The Biology of Chromatin Remodeling Complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 109.Tang Liling, Nogales Eva, Ciferri Claudio. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102:122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hargreaves Diana C, Crabtree Gerald R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Citterio Elisabetta, Van Den Boom Vincent, Schnitzler Gavin, Kanaar Roland, Bonte Edgar, Kingston Robert E, Hoeijmakers Jan HJ, Vermeulen Wim. ATP-Dependent Chromatin Remodeling by the Cockayne Syndrome B DNA Repair-Transcription-Coupling Factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/MCB.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeddeloh Jeffrey A, Stokes Trevor L, Richards Eric J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 113.Alexeev Andrei, Mazin Alexander, Kowalczykowski Stephen C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Mol Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 114.Law Martin J, Lower Karen M, Voon Hsiao PJ, Hughes Jim R, Garrick David, Viprakasit Vip, Mitson Matthew, De Gobbi Marco, Marra Marco, Morris Andrew, Abbott Aaron, Wilder Steven P, Taylor Stephen, Santos Guilherme M, Cross Joe, Ayyub Helena, Jones Steven, Ragoussis Jiannis, Rhodes Daniela, Dunham Ian, Higgs Douglas R, Gibbons Richard J. ATR-X Syndrome Protein Targets Tandem Repeats and Influences Allele-Specific Expression in a Size-Dependent Manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 115.Berube Nathalie G, Smeenk Cecilia A, Picketts David J. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 116.Wong Lee H, McGhie James D, Sim Marcus, Anderson Melissa A, Ahn Soyeon, Hannan Ross D, George Amee J, Morgan Kylie A, Mann Jeffrey R, Choo KHA. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bowman Gregory D. Mechanisms of ATP-dependent nucleosome sliding. Current Opinion in Structural Biology. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Holde KE, Yager TD. Models for chromatin remodeling: a critical comparison. Biochem Cell Biol. 2003;81:169–172. doi: 10.1139/o03-038. [DOI] [PubMed] [Google Scholar]
- 119.Strohner Ralf, Wachsmuth Malte, Dachauer Karoline, Mazurkiewicz Jacek, Hochstatter Julia, Rippe Karsten, Langst Gernot. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 120.Saha Anjanabha, Wittmeyer Jacqueline, Cairns Bradley R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lia Giuseppe, Praly Elise, Ferreira Helder, Stockdale Chris, Tse-Dinh Yuk Ching, Dunlap David, Croquette Vincent, Bensimon David, Owen-Hughes Tom. Direct Observation of DNA Distortion by the RSC Complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ding Shou Wei. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 123.Bernstein Emily, Caudy Amy A, Hammond Scott M, Hannon Gregory J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 124.Soifer Harris S, Sano Masayuki, Sakurai Kumi, Chomchan Pritsana, Saetrom Pal, Sherman Mark A, Collingwood Michael A, Behlke Mark A, Rossi John J. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Welker Noah C, Maity Tuhin S, Ye Xuecheng, Joseph Aruscavage P, Krauchuk Ammie A, Liu Qinghua, Bass Brenda L. Dicer’s Helicase Domain Discriminates dsRNA Termini to Promote an Altered Reaction Mode. Molecular Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yoneyama Mitsutoshi, Kikuchi Mika, Natsukawa Takashi, Shinobu Noriaki, Imaizumi Tadaatsu, Miyagishi Makoto, Taira Kazunari, Akira Shizuo, Fujita Takashi. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immun. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 127.Hornung Veit, Ellegast Jana, Kim Sarah, Brzuzka Krzysztof, Jung Andreas, Kato Hiroki, Poeck Hendrik, Akira Shizuo, Conzelmann Karl Klaus, Schlee Martin, Endres Stefan, Hartmann Gunther. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 128.Pichlmair Andreas, Schulz Oliver, Tan Choon Ping, Naslund Tanja I, Liljestrom Peter, Weber Friedemann, Reis e Sousa Caetano. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 129.Shatkin Aaron J, Manley James L. The ends of the affair: Capping and polyadenylation. Nat Struct Mol Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 130.Cui Sheng, Eisenacher Katherina, Kirchhofer Axel, Brzozka Krzysztof, Lammens Alfred, Lammens Katja, Fujita Takashi, Conzelmann Karl-Klaus, Krug Anne, Hopfner Karl-Peter. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 131.Sisakova Eva, Weiserova Marie, Dekker Cees, Seidel Ralf, Szczelkun Mark D. The interrelationship of helicase and nuclease domains during DNA translocation by the molecular motor EcoR124I. J Mol Biol. 2008;384:1273–1286. doi: 10.1016/j.jmb.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bianco Piero R, Xu Cuiling, Chi Min. Type I restriction endonucleases are true catalytic enzymes. Nucleic Acids Res. 2009;37:3377–3390. doi: 10.1093/nar/gkp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roberts Richard J, Vincze Tamas, Posfai Janos, Macelis Dana. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roberts Richard J, Vincze Tamas, Posfai Janos, Macelis Dana. REBASE--enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Davies Graham P, Martin Ina, Sturrock Shane S, Cronshaw Andrew, Murray Noreen E, Dryden David TF. On the structure and operation of type I DNA restriction enzymes. J Mol Biol. 1999;290:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 136.Murray Noreen E. Type I Restriction Systems: Sophisticated Molecular Machines (a Legacy of Bertani and Weigle) Microbiol Mol Biol Rev. 2000;64:412–434. doi: 10.1128/MMBR.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan Robert, Bickle Thomas A, Ebbers Walter, Brack Christine. Multiple steps in DNA recognition by restriction endonuclease from E.coli K. Nature. 1975;256:556–560. doi: 10.1038/256556a0. [DOI] [PubMed] [Google Scholar]
- 138.Sistla Srivani, Rao Desirazu N. S-Adenosyl-L-methionine–Dependent Restriction Enzymes. Crit Rev Biochem Mol Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 139.Janscak Pavel, Abadjieva Agnes, Firman Keith. The Type I Restriction Endonuclease R.EcoR124I: Over-production and Biochemical Properties. J Mol Biol. 1996;257:977–991. doi: 10.1006/jmbi.1996.0217. [DOI] [PubMed] [Google Scholar]
- 140.Firman Keith, Szczelkun Mark D. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rosamond John, Endlich Brian, Linn Stuart. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J Mol Biol. 1979;129:619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- 142.William Studier F, Bandyopadhyay Pradip K. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci U S A. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dreier Jurg, MacWilliams Maria P, Bickle Thomas A. DNA Cleavage by the Type IC Restriction-Modification EnzymeEcoR124II. J Mol Biol. 1996;264:722–733. doi: 10.1006/jmbi.1996.0672. [DOI] [PubMed] [Google Scholar]
- 144.Szczelkun Mark D. Kinetic Models of Translocation, Head-On Collision, and DNA Cleavage by Type I Restriction Endonucleases. Biochemistry. 2002;41:2067–2074. doi: 10.1021/bi011824b. [DOI] [PubMed] [Google Scholar]
- 145.Park Joo Seop, Marr Michael T, Roberts Jeffrey W. E. coli Transcription Repair Coupling Factor (Mfd Protein) Rescues Arrested Complexes by Promoting Forward Translocation. Cell. 2002;109:757–767. doi: 10.1016/S0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 146.Smith Abigail J, Szczelkun Mark D, Savery Nigel J. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McGlynn Peter, Al-Deib Abdulwahab A, Liu Joing, Marians Kenneth J, Lloyd Robert G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 148.Liu Joing, Marians Kenneth J. PriA-directed Assembly of a Primosome on D Loop DNA. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 149.Rudolph Christian J, Upton Amy L, Harris Lynda, Lloyd Robert G. Pathological replication in cells lacking RecG DNA translocase. Mol Microbiol. 2009;73:352–366. doi: 10.1111/j.1365-2958.2009.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rudolph Christian J, Upton Amy L, Briggs Geoffrey S, Lloyd Robert G. Is RecG a general guardian of the bacterial genome? DNA Repair. 2010;9:210–223. doi: 10.1016/j.dnarep.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 151.Lloyd Robert G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Jing, Mahdi Akeel A, Briggs Geoffrey S, Lloyd Robert G. Promoting and Avoiding Recombination: Contrasting Activities of the Escherichia coli RuvABC Holliday Junction Resolvase and RecG DNA Translocase. Genetics. 2010;185:23–37. doi: 10.1534/genetics.110.114413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lloyd Robert G, Sharples GJ. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McGlynn Peter, Lloyd Robert G. Modulation of RNA Polymerase by (p)ppGpp Reveals a RecG-Dependent Mechanism for Replication Fork Progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 155.Vincent Simon D, Mahdi Akeel A, Lloyd Robert G. The RecG Branch Migration Protein of Escherichia coli Dissociates R-loops. J Mol Biol. 1996;264:713–721. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- 156.Asai Tsuneaki, Kogoma Tokio. D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Asai Tsuneaki, Kogoma Tokio. Roles of ruvA, ruvC and recG Gene Functions in Normal and DNA Damage-Inducible Replication of the Escherichia coli Chromosome. Genetics. 1994;137:895–902. doi: 10.1093/genetics/137.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Masai Hisao, Tanaka Taku, Kohda Daisuke. Stalled replication forks: Making ends meet for recognition and stabilization. Bioessays. 2010;32:687–697. doi: 10.1002/bies.200900196. [DOI] [PubMed] [Google Scholar]
- 159.Mizukoshi Toshimi, Tanaka Taku, Arai Ken ichi, Kohda Daisuke, Masai Hisao. A Critical Role of the 3′ Terminus of Nascent DNA Chains in Recognition of Stalled Replication Forks. J Biol Chem. 2003;278:42234–42239. doi: 10.1074/jbc.C300285200. [DOI] [PubMed] [Google Scholar]
- 160.Gregg Amanda V, McGlynn Peter, Jaktaji Razieh P, Lloyd Robert G. Direct Rescue of Stalled DNA Replication Forks via the Combined Action of PriA and RecG Helicase Activities. Mol Cell. 2002;9:241–251. doi: 10.1016/S1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- 161.Pike Ashley CW, Shrestha Binesh, Popuri Venkateswarlu, Burgess-Brown Nicola, Muzzolini Laura, Costantini Silvia, Vindigni Alessandro, Gileadi Opher. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci U S A. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Singleton Martin R, Scaife Sarah, Wigley Dale B. Structural Analysis of DNA Replication Fork Reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/S0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 163.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 164.Halls Coralie, Mohr Sabine, Campo Mark Del, Yang Quansheng, Jankowsky Eckhard, Lambowitz Alan M. Involvement of DEAD-box Proteins in Group I and Group II Intron Splicing. Biochemical Characterization of Mss116p, ATP Hydrolysis-dependent and -independent Mechanisms, and General RNA Chaperone Activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu Yuliang, Brosh Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair. 2010;9:315–324. doi: 10.1016/j.dnarep.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Durr Harald, Korner Christian, Muller Marisa, Hickmann Volker, Hopfner Karl Peter. X-Ray Structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase Core and Its Complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 167.Nishino Tatsuya, Komori Kayoko, Tsuchiya Daisuke, Ishino Yoshizumi, Morikawa Kosuke. Crystal Structure and Functional Implications of Pyrococcus furiosus Hef Helicase Domain Involved in Branched DNA Processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 168.Lapkouski Mikalai, Panjikar Santosh, Janscak Pavel, Smatanova Ivana Kuta, Carey Jannette, Ettrich Rudiger, Csefalvay Eva. Structure of the motor subunit of type I restriction-modification complex EcoR124I. Nat Struct Mol Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]