Abstract
8-Oxo-7,8 dihydroguanine (8-oxoG) accumulates in the genome over time and is believed to contribute to the development of aging characteristics of skeletal muscle and various aging-related diseases. Here, we show a significantly increased level of intrahelical 8-oxoG and 8-oxoguanine DNA glycosylase (OGG1) expression in aged human skeletal muscle compared to that of young individuals. In response to exercise, the 8-oxoG level was found to be lastingly elevated in sedentary young and old subjects, but returned rapidly to pre-exercise levels in the DNA of physically active individuals independent of age. 8-OxoG levels in DNA were inversely correlated with the abundance of acetylated OGG1 (Ac-OGG1), but not with total OGG1, apurinic/apyrimidinic endonuclease (AP)-1 or Ac-APE1. The actual Ac-OGG1 level was linked to exercise-induced oxidative stress, as shown by changes in lipid peroxide levels and expression of Cu,Zn-SOD, Mn-SOD and SIRT3, as well as the balance between acetyl transferase p300/CBP and the deacetylase SIRT1, but not SIRT6 expression. Together these data suggest that that acetylated form of OGG1, and not OGGl itself, correlates inversely with the 8-oxoG level in the DNA of human skeletal muscle, and the Ac-OGG1 level is dependent on adaptive cellular responses to physical activity, but is age independent.
Introduction
Age-associated increases in levels of reactive oxygen species (ROS), especially during the last quarter of life, result in excessive oxidative damage to macromolecules, including DNA [1-5]. Among DNA and RNA bases, guanine is predominantly prone to oxidation due to its lowest reduction potential [6]. It is modified primarily by hydroxyl radicals at or near diffusion-controlled rates (reviewed in [7-9]). More than 20 oxidation products of guanine base have been identified [10] and among them one of the most abundant is 8-oxo-7,8 dihydroguanine (8-oxoG) [7, 8, 9]. In DNA, the 8-oxoG level increases upon radiation, ischemia/reperfusion, acute exercise and aging [4, 11-14]. 8-OxoG is excised from DNA by formamidopyrimidine-DNA glycosylase (Fpg) in E. coli and by its functional homolog 8-oxoguanine DNA glycosylase (OGG1) in mammals during the base excision repair (BER) pathway [15-18]. While Fpg is well known to excise 4,6-diamino-5-formamidopyrimidine (FapyA), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 8-oxoG with nearly similar excision kinetics [18, 19], the mammalian and yeast OGG1 is specific for 8-oxoG and FapyG, but not for FapyA [20, 21]. When 8-oxoG is not repaired, it is mutagenic, as it has been shown to pair with adenine (A) instead of cytosine and thereby (C) induces G:C→T:A transversions [15, 22].
It is documented that in covalent modifications of DNA repair proteins, e.g., by acetylation, phosphorylation plays a significant role, particularly in their repair activity which consists of the removal/repair of oxidative base lesions [23, 24]. In fact, it has been shown that OGG1 and the human AP-endonuclease (APE1) activities are primarily regulated by p300/CBP-mediated acetylation reactions, processes that significantly influence their repair activities and hence cell fate [23-25]. The role of sirtuin family deacetylases has gathered considerable attention [26], as SIRT1 and SIRT6 have been shown to be involved in DNA repair [27-29]. An increased deacetylase activity of sirtuins may lead to a decrease in acetylation levels of proteins, which, in turn, would result in a decline in enzymatic activities, including those of OGG1 and APE1.
Although it is well-documented that acetylation increases OGG1 activity in cell cultures and in vitro assays, the existence of acetylated OGG1 (Ac-OGG1) and APE1 (Ac-APE) in in vivo conditions is still unknown. The goal of the present investigation was a) to determine changes in Ac-OGG1 and Ac-APE1 in human skeletal muscle; b) to study the effects of aging and acute as well as regular physical conditioning on acetylation levels of these DNA repair enzymes; c) and to evaluate the possible role of SIRT1, SIRT3, and SIRT6 in the adaptability of human skeletal muscle. This report shows that the level of acetylated OGG1 changes as a function of age, and exercise training increases this post-translational modification independent from age in human muscles.
Materials and Methods
Subjects
Forty-eight healthy men volunteered to participate in the present study. A written informed consent was signed by all participants regarding their participation after they were told of all risks, discomforts and benefits involved in the study. Procedures were in accordance with the Helsinki Declaration of 1975 and were approved by the ethical committee of the University of Thessaly.
Participants were assigned to one of four groups according to cross-over, repeated-measures design: a) young sedentary (YS, 26.0 ± 4.5 yrs), b) young physically active (YA, 30.2 ± 7.9 yrs), c) old sedentary (OS, 63.4 ± 4.7 yrs), and d) old physically active (OA, 62.4 ± 2.9 yrs). Subjects were exposed to a single bout of exercise protocol and muscle biopsies were taken. Participants were assigned to the young or old sedentary group based upon the following criteria: a) maximal oxygen uptake (VO2max) was below 25 ml/kg/min for old participants and below 35 ml/kg/min and young or old physically active group were based upon the description of ACSM [30], b) VO2max was over 45 ml/kg/min for old participants and over 35 ml/kg/min (YS: 35.9 ± 4.7, OS. 25.1 ± 3.0, YA: 51.8 ± 7.9, OA: 37.1 ± 2.9 ml/kg/min).
Participants visited the laboratory on three occasions. During their first visit, participants were examined by a trained physician for limiting health complications; in their second visit, participants had their body height/weight and skin-folds measured and underwent a GXT to evaluate their VO2max. During their third visit, a week later, participants underwent a sub maximal exercise bout to exhaustion on the treadmill, while muscle biopsies were collected before and following exercise.
Measurement of peak oxygen uptake (VO2peak)
VO2peak was determined during a GXT on a treadmill to voluntary exhaustion as previously described [31].
Exercise protocol
A single bout of exercise included initially 45 min of running on a treadmill at 70–75% of their VO2max. After 45 min, speed increased to 90% of VO2max, and exercise was terminated at exhaustion [32].
Muscle biopsy sampling
Participants had been instructed to refrain from physical activity and caffeine consumption for 48 hours prior to exercise. Both muscle specimens (pre- and post-exercise), of approximately 100-120 mg each were obtained from the vastus lateralis of the same leg of each participant by using the needle biopsy technique [33]. The first biopsy was obtained approximately 20 cm away from the mid patella of the right (dominant) leg with the application of suction [34].
Assessment of Malondialdehyde levels
Blood samples were collected from an antecubital arm vein into evacuated tubes containing ethylenediaminetetraacetic acid (EDTA). Plasma was separated by centrifugation (1500 g, 4°C, for 15 min). Samples were stored at -80°C. Malondialdehyde (MDA) levels were measured by reverse-phase, high-performance liquid chromatography (rp-HPLC) with fluorimetric detection (excitation 532 nm and emission 550 nm) as described [35].
Real time quantitative RT-PCR
Total RNA from skeletal muscle samples (~30 mg) was extracted with NucleoSpin® RNA/Protein (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. Analyses of the real-time quantitative PCR data were performed by using the comparative threshold cycle [Ct] method, as suggested by Applied Biosystems (User Bulletin #2). The following primers were used:
| Reference genes | ||
| β-actin | 5’ GCT CGT CGT CGA CAA CGG CTC 3’ | Forward |
| 5’ CAA ACA TGA TCT GGG TCA TCT TCT 3’ | Reverse | |
| RP28S | 5’ AGC CGA TCC ATC ATC CGC AAT G 3’ | Forward |
| 5’ CAG CCA AGC TCA GCG CAA C 3’ | Reverse | |
| Target genes | ||
| OGG1 | 5’ GTG GAC TCC CAC TTC CAA GA 3’ | Forward |
| 5’ GAG ATG AGC CTC CAC CTC TG 3’ | Reverse | |
| EP300 | 5’ TCA TCT CCG GCC CTC TCG GC 3’ | Forward |
| 5’ GCT CTG TTG GGC CTG GCT GG 3’ | Reverse | |
| SIRT1 | 5’ TGC GGG AAT CCA AAG GAT AAT TCA GTG TC 3’ | Forward |
| 5’ CTT CAT CTT TGT CAT ACT TCA TGG CTC TAT G | Reverse | |
| SIRT3 | 5’ GTC GGG CAT CCC TGC CTC AAA GC 3’ | Forward |
| 5’ GGA ACC CTG TCT GCC ATC ACG TCA G 3’ | Reverse | |
| SIRT6 | 5’ GAG GAG CTG GAG CGG AAG GTG TG 3’ | Forward |
| 5’ GGC CAG ACC TCG CTC CTC CAT GG 3’ | Reverse | |
| SOD1 | 5’ AGG GCA TCA TCA ATT TCG AG 3’ | Forward |
| 5’ ACA TTG CCC AAG TCT CCA AC 3’ | Reverse | |
| SOD2 | 5’ GCA GAA GCA CAG CCT CCC CG 3’ | Forward |
| 5’ CCT TGG CCA ACG CCT CCT GG 3’ | Reverse | |
| XRCC6 (Ku70) | 5’ CTG TCC AAG TTG GTC GCT TC 3’ | Forward |
| 5’ CTG CCC CTT AAA CTG GTC AA 3’ | Reverse |
Fluorescence imaging and quantification
At optimal cutting, temperature–fixed, paraffin-embedded muscles were sectioned into 5-μm sections. The measurement of 8-oxoG levels in nuclear DNA of muscles was assessed by quantitative microscopic imaging, as we previously described [23, 36]. Briefly, sections were de-parafinized, air-dried, and fixed in acetone–methanol (1:1), rehydrated in PBS for 15 min, then sequentially treated with RNAase (100 μg/ml) for 15 min followed by 100 μg/ml pepsin in the presence of 0.1N HCl for 30 min at 37°C. The sections were washed and then incubated with affinity-purified, non-immune IgG (100 μg/ml) for 30 min and washed in PBS containing 0.5% bovine serum albumin, and 0.1% Tween 20 (PBS-T). After incubation with anti-8-oxoG antibody (Trevigen, Gaithersburg, MD; 1:300 dilution) [37] for 30 min, the sections were washed for 15 min three times with PBS-T and then binding of primary antibody was detected with conjugated secondary antibody.
OGG1 and Ac-OGG1 levels were also determined via quantitative microscopic imaging [36, 38]. Purified mouse anti-OGG1 antibody (human OGG1 reactive) generated against a synthetic peptide (C-DLRQSRHAQEPPAK-N), representing the C-terminus of OGG1 was acquired from Antibodies-Online GmbH (Atlanta, GA, USA). The immunogen affinity-purified, human-reactive rabbit polyclonal antibody to Ac-OGG1 was generated against an AcLys-containing peptide (PAKRRAcKGGAcKGPEC) [23] obtained from AbCam (Cat # ab93670) [23, 36]. Antibody reactive with human APE1 [39] and rabbit anti-Ac-APE1 antibody was characterized previously [40]. Binding of primary antibodies were visualized with flourochrom-labeled secondary antibodies. Confocal microscopic evaluations were performed on a Zeiss LSM510 META System by using the 488-nm line of the Argon-laser for excitation of FITC and Helium-Neon 543-nm line excitation of Rd, combined with appropriate dichroic mirrors and emission band filters to discriminate between green and red fluorescence. Images were captured at a magnification of 60 (60x oil immersion objective; numerical aperture 1.4). To objectively quantify fluorescence intensities morphometric analyses were done by using Metamorph™ software Version 9.0r (Universal Imaging Corp, Downingtown, PA) as we described [38]. Specifically, images were obtained from >15 fields per muscle section containing 160-180 nuclei and reassembled by using the montage stage stitching algorithm of the Metamorph™ software [41]. Co-localization was visualized by superimposition of green and red images using MetaMorph software version 9.0r.
Statistical analyses
Statistical significance was assessed by three-way ANOVA (age X physical activity status X time), followed by Tukey’s post hoc test. The significance level was set at p < 0.05.
Results
Changes in 8-oxoG level in DNA as a function of age and physical activity in human skeletal muscle
DNA glycosylase/AP lyase activity of OGG1 declines with age [42-44]. Here, first we investigated the association between abundance of 8-oxoG in DNA and OGG1, as well as Ac-OGG1 in nuclei of skeletal muscle of sedentary old (OS) and young (YS) individuals. Results from quantitative fluorescence intensity analysis showed that there was a significant (p<0.01) increase in genomic 8-oxoG (8-oxodG; Fig. 1A) and total OGG1 (p<0.01) levels in skeletal muscle (muscle) of elderly compared to young participants (Fig. 1B). This paradoxical observation suggests an increase in oxidative stress and/or decrease in OGG1 activity, the latter may be due to altered OGG1’s post-translational modification(s) such as acetylation [23]. The acetylated form of OGG1, when compared to the unacetylated form, shows an approximately 10-fold increase in repair activity [23]. Immunohistochemical analysis shows that the level of Ac-OGG1 was significantly higher in the skeletal muscle of young individuals (Fig. 1C, upper and lower panels) compared to that of older subjects. Ac-OGG1 was nearly undetectable in the skeletal muscle of the elderly (Fig. 1D, upper and lower panels). As calculated from fluorescence intensities, only 5.1 ± 2.5% of total OGG1 was acetylated in old, while 24.5 ± 6% of total OGG1 reacted with anti-Ac-OGG1 antibody in young individuals (Fig. 1E). APE1 is a multi functional and abundant protein [39] and has been shown to stimulate 8-oxoG repair initiated by OGG1 during BER [45]. Due to APE1’s abundance, it was not surprising to observe that its level was not different in muscle of young and old groups (data not shown). Ac-APE1 [46] levels were only substantially higher in skeletal muscle of YS individuals compared to that in OS subjects (Fig. 2A), not the APE1 level but the Ac-APE1, together with Ac-OGG1, play a role in the repair of 8-oxoG. These results support the hypothesis that an increase in the genomic 8-oxoG level is associated with an inability of aged SkM to post-translationally modify OGG1 [25].
Figure 1. 8-oxoG, OGG1 and Ac-OGG1 levels in SkM prior and after SEB.
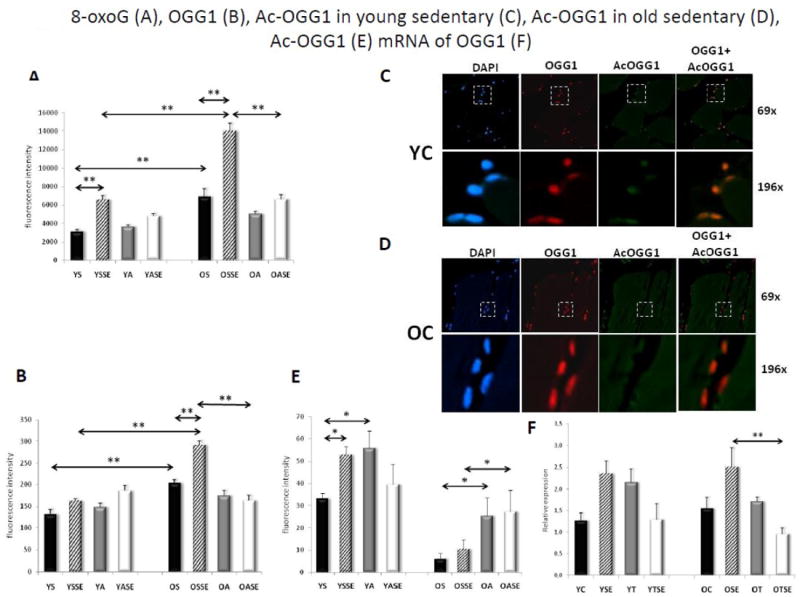
A, Increase in 8-oxoG level in genomic DNA of aged muscles and in response to SEB. B, Total OGG1 level in SkM of sedentary and physically active subjects. In A and B, sections were stained and fluorescence intensities were analyzed by using a montage stage stitching algorithm of the Metamorph™ software (Materials and Methods). C shows representative fluorescent images of OGG1 and Ac-OGG1 in sections from the muscles of young individuals. Upper panels: magnification 69x; lower panels: magnification, 196x. Most left panels are DAPI, the most right panels are the super imposition of OGG1- and Ac-OGG1-mediated fluorescent images. D shows representative fluorescent images of OGG1 and Ac-OGG1 in muscle sections of old volunteers. Upper panels, magnification: 69x; lower panels, magnification: 196x. Most left panels are DAPI-stained, the most right panels are the super imposition of OGG1- and Ac-OGG1-mediated fluorescent images. E, Changes in Ac-OGG1 level of skeletal muscle of young and elderly subjects in response to SEB. DAPI, 4’,6’ diamino-2-phenylindole; YS, young sedentary; YSSE, young sedentary after a single bout exercise; YA, young active; YASE, young active with a single bout of exercise, OS, old sedentary; OSSE, old sedentary with single bout of exercise; OA, old active and OASE, old active with a single bout of exercise. Values are means ± SE for six subjects per group. *p<0.05, **p<0.01.
Figure 2. Ac-APE1 level and expression of p300/CPB, SIRT1, 3 and 6 prior and after physical exercise in skeletal muscle.
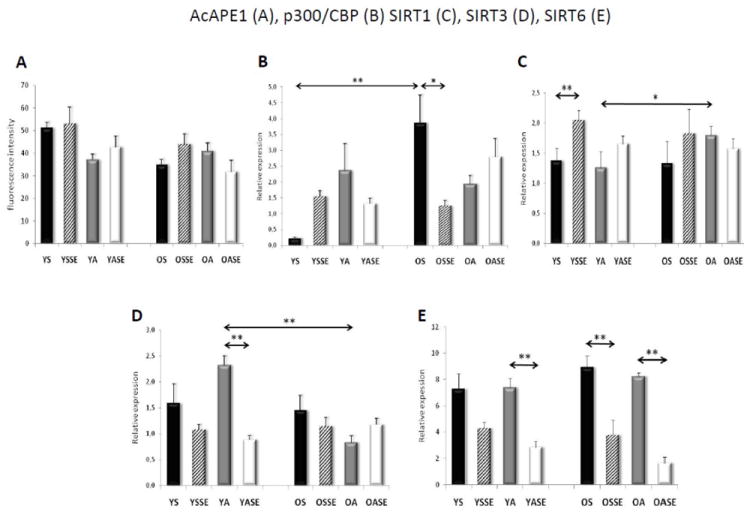
A, Level of Ac-APE1 as assessed by fluorescent imaging (analyzed as in Fig. 1A). B, C and D, Expressions at mRNA levels of SIRT1 (B), p300/CBP (C) and SIRT3 (D). RNA was isolated from muscle biopsies excised prior to and 24 h after SEB. Quantitative Rt-PCR was undertaken as described in Materials and Methods. YS, young sedentary; YSSE, young sedentary after a single bout exercise; YA, young active; YASE, young active with a single bout of exercise, OS, old sedentary; OSSE, old sedentary with single bout of exercise; OA, old active and OASE, old active with a single bout of exercise. Values are means ± SE for six subjects per group. *p<0.05, **p<0.01.
OGG1’s acetylation level is altered by the activity of acetyl transferase p300/CBP [23, 25] and deacetylases such as sirtuins [27]. Our results show that expression of p300/CBP is increased (p<0.01) in skeletal muscle of OS subjects compared to that in younger counterparts (Fig. 2B). On the other hand, expression of SIRT1 and SIRT6 (Fig. 2C, D and 3A) was not affected by age, while SIRT3 expression is significantly lower in the OS compared to the YS group (Fig. 3D). In controls, there were no differences in the expression of Ku70 (binds directly to free DNA ends) in the muscles of young and old individuals (Fig. 3B), an indication that repair efficiency of 8-oxoG is unaffected by age and level of unrepaired AP sites, and DNA single-strand breaks are not sufficient to alter the expression of Ku70.
Figure 3. Effect of SEB on expressions of Ku70, Cu,Zn- and Mn-superoxide dismutase.
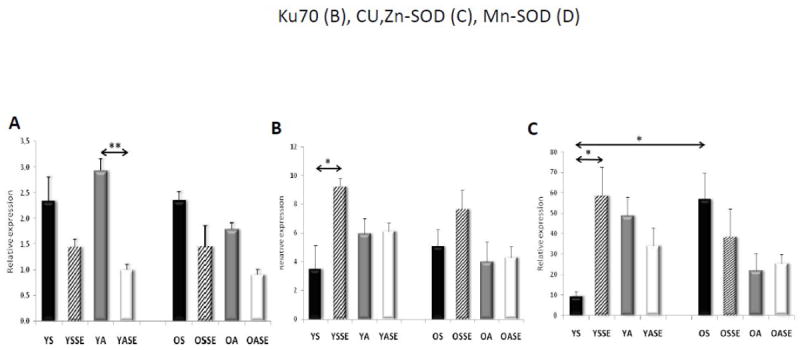
RNA was isolated from muscle biopsies excised prior to and 24 h after SEB. QRt-PCR was carried out as described in Materials and Methods. A, Ku70; B, Cu,Zn-SOD, C, MnSOD. YS, young sedentary; YSSE, young sedentary after a single bout exercise; YA, young active; YASE, young active with a single bout of exercise, OS, old sedentary; OSSE, old sedentary with single bout of exercise; OA, old active and OASE, old active with a single bout of exercise. Values are means ± SE for six subjects per group. *p<0.05, **p<0.01.
Oxidative stress induced by physical activity mediates an adaptive response for efficient oxidative DNA damage repair
Old and young physically inactive and active individuals were subjected to a single exercise bout (SEB). SEB-induced changes in oxidative stress levels were determined indirectly by measuring the levels of lipid peroxidation product malondialdehyde (MDA) in plasma (YS: 0.176 ± 0.02, YSSE: 0.262 ± 0.03*, YA: 0.143 ± 0.01, YASE: 0.181 ± 0.02, OS: 0.254 ± 0.04, OSSE: 0.338 ± 0.06*, OA: 0.188 ± 0.03, OASE: 0.233 ± 0.03 μmol/L). It is obvious that the MDA level was significantly increased only in the plasma of physically inactive old and young subjects. Although, we recognize the limitation of MDA measurements [47], the strong match between MDA and 8-oxoG (p=0.001) levels suggests that indeed aging and SEB elevate the level of oxidative damage. These results are supported by the observed increase in the expression of Cu,ZnSOD (Fig. 3B) in the muscle of physically inactive (old and young) subjects. MnSOD expression is increased in response to SEB only in young subjects (Fig. 3C). Surprisingly, MnSOD expression was not affected by SEB in active/trained old and young individuals (Fig. 3C). Together these data imply an adaptive response of the skeletal muscle to SEB in trained/active individuals.
An increase in MDA level predicts enhanced genomic 8-oxoG levels upon exercise. Thus we asked if regular physical exercise-induced antioxidant responses protect guanine from oxidation in the DNA from muscle biopsies of sedentary vs. trained and young vs. old subjects. In response to a SEB, the 8-oxoG level was doubled in the muscle of all individuals regardless of whether they were sedentary or physically active. Importantly, while 8-oxoG levels returned to pre-exercise levels in physically active individuals, both OA and YA, its level in DNA remained high in sedentary young and old subjects after a 24-h recovery period (Fig. 1A). For example, 8-oxoG levels were approximately 4-times higher in untrained older (Fig. 1A) compared to younger individuals without SEB (Fig. 1A). Importantly, there was no change in genomic 8-oxoG levels in muscle biopsies of OA individuals after SEB (Fig. 1A).
The sub-physiological level of genomic 8-oxoG in physically active subjects suggested an efficient repair of DNA. We observed that OGG1 levels did not significantly change in younger subjects, while it increased in the older subjects in response to SEB (Fig. 1B). In contrast, Ac-OGG1 levels were significantly increased in younger individuals, while in the older subjects no significant change was observed in response to SEB. Ac-OGG1 level was ~3-fold higher in active compared to older, sedentary individuals (Fig. 1E and C). SEB did not change Ac-APE1 (Fig. 2A), which was similar to APE1 levels (data not shown), suggesting that neither Ac-APE1 nor APE1 is limiting in the repair of 8-oxoG.
In response to SEB, the expression of p300/CBP increased approximately 5-fold in the younger subjects, while unexpectedly, significantly decreased in older subjects (Fig. 3A). If indeed p300/CBP is the acetyl transferase in muscle, these results are in line with levels of Ac-OGG1 (Fig. 1C,E). In physically active subjects SEB did not significantly alter p300/CBP levels (Fig. 2B). Expression of the deacetylase SIRT1 showed a significant increase only in younger sedentary subjects in response to SEB (Fig. 2C). SIRT3’ expression that has no deacetylase activity was the highest in muscle biopsies of active, younger subjects (Fig. 2D), while its expression did change upon SEB (Fig. 2D). SIRT6 expression (Fig. 2E) along with Ku70 (Fig. 3A) decreased in both young and old muscles following SEB. Together these data suggest that a physically active life-style induces an adaptive response by generating mild oxidative stress, and prevented the age-associated increase in genomic 8-oxoG levels possibly due to the age-independent increase in OGG1’s acetylation.
Discussion
Age-related and physical exercise-associated changes in DNA damage levels in skeletal muscle of experimental animals have been reported previously [13, 14, 48]. The present study analyzed levels of 8-oxoG in DNA and abundance of rate-limiting BER enzymes in human muscle biopsies prior to and after a single exercise bout. We also examined expression of acetyl-transferases, and deacetylases linked to DNA repair pathways and antioxidant genes that could reflect on cellular redox conditions. We show that the genomic 8-oxoG level is lastingly elevated in sedentary young and old subjects, but it returned rapidly to pre-exercise levels in physically active individuals independent of age upon a single exercise bout. The 8-oxoG level in DNA inversely correlated with the abundance of Ac-OGG1, but not with total OGG1, APE1 or Ac-APE1. Importantly, our data also demonstrate a physical activity-dependent increase in the acetylated forms of OGG1 in human skeletal muscle. Accordingly, it is possible that an exercise-induced acetylation pathway would enhance OGG1 activity, not only in muscles, but in other tissues, and thereby exercise may decrease the incidence of various pathological conditions, such as inflammation, that have been linked to carcinogenesis, cardiovascular diseases, strokes or Alzheimer’s disease.
8-oxoG is arguably one of the important forms of DNA base damage induced by ROS, and it has been proposed to play a role in the aging process and is also linked to age-associated diseases [49-52]. This hypothesis is consistent with the several-fold increase in 8-oxoG (and possibly of other oxidized bases) content in nuclear and mtDNA from aged tissues [49-52]. A single bout of exercise has been shown to cause mild oxidative stress [32, 53, 54], and thus we applied a SEB and determined cellular oxidative states, changes in 8-oxoG levels and abundance of selected repair enzymes. Due to a limited amount of muscle biopsies, we used quantitative fluorescence analysis [36, 38, 41] to assess 8-oxoG levels, as the quantity of DNA isolated from them did not allow us to use HPLC with electrochemical detection [7, 8], which would provide a better estimates. By using highly specific, anti-8-oxodG-specific antibody, we observed significantly higher levels of genomic 8-oxoG in human skeletal muscle of sedentary, older individuals compared to the levels in younger subjects in line with previous observations [13, 14, 43, 44]. In response to SEB-induced ROS, 8-oxoG levels increased further, and were not repaired, even after a 24 h time period in sedentary individuals independent of age. In contrast, 8-oxoG levels returned to pre-exercise levels in physically active individuals, a finding that may mean regular physical activity could prevent accumulation and/or increase repair efficacy of 8-oxoG and possible other bases in DNA human skeletal muscle.
The observed increase in 8-oxoG levels in sedentary individuals points to a possible age-dependent decrease in levels of OGG1. In contrast, our data show a significantly increased OGG1 level in elderly subjects and, interestingly, SEB furthered its level. Unexpectedly, the 8-oxoG level was also enhanced. These paradoxical observations suggested to us that OGG1 may have a low DNA glycosylase/AP lyase activity or that BER activities are significantly lower in aged human muscle. Indeed, a recent publication documents, decreased overall BER activities both in the nuclei and the mitochondrial extracts from skeletal muscles, compared to those in liver or kidneys of the same mice [55]. While decreased overall BER activity could be a possibility, our data also imply that lack/delayed repair of 8-oxoG could be linked to a deficiency in post-translationally modified OGG1 in aged muscles. Indeed, OGG1’s glycosylase/AP-lyase activity is modulated via acetylation, phosphorylation, and redox [23, 25]. For example, OGG1 is acetylated on 338 and 341 lysine and has approximately a 10-fold increase in its 8-oxoG excision activity compared to that in non-acetylated OGG1 [23] To explore the latter possibility we show that ~one-fifth of OGG1 is in an acetylated form in younger individuals and, importantly, Ac-OGG1 was nearly undetectable in the sedentary elderly. This observation is a feasible possibility, as 8-oxoG level in DNA was inversely correlated with levels of Ac-OGG1 in muscles of young and old individuals.
Repair of 8-oxoG is initiated by OGG1 during the BER pathway followed by APE1-mediated cleavage of the DNA strand at the abasic site. After removal of this 3’-blocking group, the single nucleotide gap is filled in by a DNA polymerase, and DNA ligase seals the nick to restore DNA integrity [17]. It has also been shown that OGG1 remains tightly bound to its AP product following base excision, and APE1 prevents its reassociation with its product, thus enhancing OGG1 turnover [45]. Accordingly, APE1 is considered to be rate-limiting in the BER of 8-oxoG [17, 39]. However, neither APE1 nor Ac-APE1 showed significant changes with aging and/or physical activity. Therefore, it may be proposed that the AcOGG1 is limiting in the repair of 8-oxoG lesions in human skeletal muscle during BER processes. As modification by phosphorylation only substantially alters incision activity of OGG1 [24], our earlier observations of an exercise-induced increase in OGG1 activity in skeletal muscles of human and experimental animals [14, 43] may be attributed to Ac-OGG1.
Acetylation levels of OGG1 and APE1 are dependent on level/activity of acetyl transferase p300/CBP [23, 25] and possibly on deacetylase(s) such as some of the sirtuins [56]. Results from our studies show that p300/CBP’s expression was increased in young individuals by SEB, independent of whether they were sedentary or active. However, we were not able show such consistency in the elderly. SIRT1, a NAD-dependent, histone deacetylase [57], has been shown to interact with p300/CBP to regulate its acetyl-transferase activity [56]. SIRT1 levels increased in both young and elderly muscles in response to exercise. These observations are in line with the general role of SIRT1 in DNA damage response and maintenance of genomic integrity, as it promotes proper chromatin structure and DNA damage repair foci formation for repair of DNA base lesions [27, 28]; however, the patterns of changes in SIRT1 expression young vs. old or sedentary vs. physically active suggest an inverse correlation between SIRT1 and level of Ac-OGG1.
Among sirtuins, only SIRT3 expression correlates with the life-span of humans [58]. Interestingly, SIRT3 expression is increased with physical fitness level in only young subjects in this study. SIRT3 has two isoforms with different molecular mass (44 kDa and 28 kDa) which are localized in mitochondria and nucleus, respectively [59]. The translocation of SIRT3 from the nucleus to mitochondria has been shown to be induced by oxidative stress [59]. SIRT3 is also a modulator of apoptosis [60]. Recent findings also indicate that SIRT3 is a downstream target of PGC-1 alpha and one of the regulators of mitochondrial ROS production [61].
Exercise has been shown to cause mild oxidative stress [32, 53, 54, 62]. Although the 8-oxoG level is a documented measure of such an oxidative insult [14], MDA levels and expression of superoxide dismutase(s) were used to evaluate further SEB-induced oxidative stress. An increase in MDA levels in plasma correlated with genomic 8-oxoG level both in young and old subjects in response to SEB. Interestingly, only the expression of Cu,ZnSOD show age independent and exercise-associated changes, while Mn-SOD expression was increased only in the younger sedentary group. Based on these observations, it appears Cu,Zn, SOD expression is a better measure of an adaptive response to ROS than that of mitochondrial MnSOD. These data also imply a decline in adaptive response with age at the level of MnSOD. These observations are in line with those showing that the adaptive capability of an organism to withstand oxidative stress challenge(s) is markedly decreased as a function of age [63, 64]. Based on our data, however, we propose that adaptive responses to ROS are not age dependent, but decided by the physical status of an individual.
In conclusion, the present investigation offers insight into interactions between aging processes, exercise, and regulation of the repair of oxidized DNA base lesions in human skeletal muscle. We show for the first time that 1) acetylated forms of OGG1 and APE1 are present in human tissues, 2) but only Ac-OGG1 seems to be rate-limiting in the BER processes of 8-oxoG. 3) Repair of 8-oxoG seems to be independent of age, 4) but is dependent on the physical state of muscles. Our data also imply that regular exercise induces an adaptive response that involves an improved, more efficient antioxidant and DNA repair machinery.
Acknowledgments
The present work was supported by Hungarian grants from ETT 38388, TéT JAP13/02, JSPS (L-10566), OTKA (K75702) awarded to Z. Radák, and AG 021830 (to I. Boldogh) from US NIH/NIA. We thank Mardelle Susman (Department of Microbiology and Immunology, UTMB) for careful editing our manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 4.Kaneko T, Tahara S, Matsuo M. Non-linear accumulation of 8-hydroxy-2’-deoxyguanosine, a marker of oxidized DNA damage, during aging. Mutat Res. 1996;316:277–285. doi: 10.1016/s0921-8734(96)90010-7. [DOI] [PubMed] [Google Scholar]
- 5.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candeias LP, Steenken S. Reaction of HO* with guanine derivatives in aqueous solution: formation of two different redox-active OH-adduct radicals and their unimolecular transformation reactions. Properties of G(-H)*. Chemistry. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 8.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 9.von Sonntag C. The chemical basis of radiation biology. Taylor and Francis; New York: 1987. [Google Scholar]
- 10.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 11.Dizdaroglu M. Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry. 1985;24:4476–4481. doi: 10.1021/bi00337a032. [DOI] [PubMed] [Google Scholar]
- 12.Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med. 2010;49:587–596. doi: 10.1016/j.freeradbiomed.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radak Z, Kaneko T, Tahara S, Nakamoto H, Ohno H, Sasvari M, Nyakas C, Goto S. The effect of exercise training on oxidative damage of lipids, proteins, and DNA in rat skeletal muscle: evidence for beneficial outcomes. Free Radic Biol Med. 1999;27:69–74. doi: 10.1016/s0891-5849(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 14.Radak Z, Naito H, Kaneko T, Tahara S, Nakamoto H, Takahashi R, Cardozo-Pelaez F, Goto S. Exercise training decreases DNA damage and increases DNA repair and resistance against oxidative stress of proteins in aged rat skeletal muscle. Pflugers Arch. 2002;445:273–278. doi: 10.1007/s00424-002-0918-6. [DOI] [PubMed] [Google Scholar]
- 15.Nakabeppu Y, Tsuchimoto D, Furuichi M, Sakumi K. The defense mechanisms in mammalian cells against oxidative damage in nucleic acids and their involvement in the suppression of mutagenesis and cell death. Free Radic Res. 2004;38:423–429. doi: 10.1080/10715760410001688348. [DOI] [PubMed] [Google Scholar]
- 16.Hazra TK, Hill JW, Izumi T, Mitra S. Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions. Prog Nucleic Acid Res Mol Biol. 2001;68:193–205. doi: 10.1016/s0079-6603(01)68100-5. [DOI] [PubMed] [Google Scholar]
- 17.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 18.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlow-Poehnelt RA, Zharkov DO, Grollman AP, Broyde S. Substrate discrimination by formamidopyrimidine-DNA glycosylase: distinguishing interactions within the active site. Biochemistry. 2004;43:16092–16105. doi: 10.1021/bi048747f. [DOI] [PubMed] [Google Scholar]
- 21.Boiteux S, Gellon L, Guibourt N. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic Biol Med. 2002;32:1244–1253. doi: 10.1016/s0891-5849(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura S. Involvement of mammalian OGG1(MMH) in excision of the 8-hydroxyguanine residue in DNA. Free Radic Biol Med. 2002;32:813–821. doi: 10.1016/s0891-5849(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 23.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Imam SZ, Hashiguchi K, de Souza-Pinto NC, Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase (alpha-OGG1) modulates its function. Nucleic Acids Res. 2005;33:3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szczesny B, Bhakat KK, Mitra S, Boldogh I. Age-dependent modulation of DNA repair enzymes by covalent modification and subcellular distribution. Mech Ageing Dev. 2004;125:755–765. doi: 10.1016/j.mad.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 29.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.American College of Sports Medicine. Lippincott Williams & Wilkins; Philadelphia: 2005. Guidelines for Exercise Testing and Prescription. [Google Scholar]
- 31.Fatouros IG, Jamurtas AZ, Villiotou V, Pouliopoulou S, Fotinakis P, Taxildaris K, Deliconstantinos G. Oxidative stress responses in older men during endurance training and detraining. Med Sci Sports Exerc. 2004;36:2065–2072. doi: 10.1249/01.mss.0000147632.17450.ff. [DOI] [PubMed] [Google Scholar]
- 32.Michailidis Y, Jamurtas AZ, Nikolaidis MG, Fatouros IG, Koutedakis Y, Papassotiriou I, Kouretas D. Sampling time is crucial for measurement of aerobic exercise-induced oxidative stress. Med Sci Sports Exerc. 2007;39:1107–1113. doi: 10.1249/01.mss.0b013e318053e7ba. [DOI] [PubMed] [Google Scholar]
- 33.Bergström J. Muscle electrolytes in man. Scand J Clin Lab Invest. 1962;14:1–110. [Google Scholar]
- 34.Terzis G, Georgiadis G, Stratakos G, Vogiatzis I, Kavouras S, Manta P, Mascher H, Blomstrand E. Resistance exercise-induced increase in muscle mass correlates with p70S6 kinase phosphorylation in human subjects. Eur J Appl Physiol. 2008;102:145–152. doi: 10.1007/s00421-007-0564-y. [DOI] [PubMed] [Google Scholar]
- 35.Lykkesfeldt J. Determination of malondialdehyde as dithiobarbituric acid adduct in biological samples by HPLC with fluorescence detection: comparison with ultraviolet-visible spectrophotometry. Clin Chem. 2001;47:1725–1727. [PubMed] [Google Scholar]
- 36.Bacsi A, Chodaczek G, Hazra TK, Konkel D, Boldogh I. Increased ROS generation in subsets of OGG1 knockout fibroblast cells. Mech Ageing Dev. 2007;128:637–649. doi: 10.1016/j.mad.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bespalov IA, Bond JP, Purmal AA, Wallace SS, Melamede RJ. Fabs specific for 8-oxoguanine: control of DNA binding. J Mol Biol. 1999;293:1085–1095. doi: 10.1006/jmbi.1999.3214. [DOI] [PubMed] [Google Scholar]
- 38.Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, Hazra TK, Mitra S, Goldblum RM, Sur S. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115:2169–2179. doi: 10.1172/JCI24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci U S A. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loo BW, Jr, Meyer-Ilse W, Rothman SS. Automatic image acquisition, calibration and montage assembly for biological X-ray microscopy. J Microsc. 2000;197:185–201. doi: 10.1046/j.1365-2818.2000.00644.x. [DOI] [PubMed] [Google Scholar]
- 42.Xu G, Herzig M, Rotrekl V, Walter CA. Base excision repair, aging and health span. Mech Ageing Dev. 2008;129:366–382. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radak Z, Apor P, Pucsok J, Berkes I, Ogonovszky H, Pavlik G, Nakamoto H, Goto S. Marathon running alters the DNA base excision repair in human skeletal muscle. Life Sci. 2003;72:1627–1633. doi: 10.1016/s0024-3205(02)02476-1. [DOI] [PubMed] [Google Scholar]
- 44.Radak Z, Toldy A, Szabo Z, Siamilis S, Nyakas C, Silye G, Jakus J, Goto S. The effects of training and detraining on memory, neurotrophins and oxidative stress markers in rat brain. Neurochem Int. 2006;49:387–392. doi: 10.1016/j.neuint.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhakat KK, Hazra TK, Mitra S. Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 2004;32:3033–3039. doi: 10.1093/nar/gkh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meagher EA, FitzGerald GA. Indices of lipid peroxidation in vivo: strengths and limitations. Free Radic Biol Med. 2000;28:1745–1750. doi: 10.1016/s0891-5849(00)00232-x. [DOI] [PubMed] [Google Scholar]
- 48.Nakae Y, Stoward PJ, Bespalov IA, Melamede RJ, Wallace SS. A new technique for the quantitative assessment of 8-oxoguanine in nuclear DNA as a marker of oxidative stress. Application to dystrophin-deficient DMD skeletal muscles. Histochem Cell Biol. 2005;124:335–345. doi: 10.1007/s00418-005-0037-5. [DOI] [PubMed] [Google Scholar]
- 49.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 51.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 53.Ji LL, Gomez-Cabrera MC, Vina J. Role of free radicals and antioxidant signaling in skeletal muscle health and pathology. Infect Disord Drug Targets. 2009;9:428–444. doi: 10.2174/187152609788922573. [DOI] [PubMed] [Google Scholar]
- 54.Radak Z, Chung HY, Koltai E, Taylor AW, Goto S. Exercise, oxidative stress and hormesis. Ageing Res Rev. 2008;7:34–42. doi: 10.1016/j.arr.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Szczesny B, Tann AW, Mitra S. Age- and tissue-specific changes in mitochondrial and nuclear DNA base excision repair activity in mice: Susceptibility of skeletal muscles to oxidative injury. Mech Ageing Dev. 2010;131:330–337. doi: 10.1016/j.mad.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 58.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powers SK, Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88:1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji LL. Exercise at old age: does it increase or alleviate oxidative stress? Ann N Y Acad Sci. 2001;928:236–247. doi: 10.1111/j.1749-6632.2001.tb05653.x. [DOI] [PubMed] [Google Scholar]
- 64.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]


