Abstract
Background
T-cell factor (TCF) proteins represent key transcription factors that activate Wnt/β-catenin signaling. We have reported that a pair of TCF-4 isoforms (TCF-4C and TCF-4D) exhibits differential TCF transcriptional activity in hepatocellular carcinoma (HCC) cells, although their structure differs by only the presence (TCF-4D) or absence (TCF-4C) of exon 4.
Aim
To demonstrate a regulatory role of exon 4 in HCC development.
Methods
TCF-4C and TCF-4D expression profiles were examined in 27 pairs of human HCC and adjacent liver tissues. The functional role of the TCF-4 isoforms was evaluated in OUMS-29 (an immortalized hepatocyte-derived) and HAK-1A (a well differentiated HCC) cell lines using stable clones overexpressing the TCF-4 isoforms.
Results
TCF-4C was significantly upregulated in HCC tissues compared to corresponding peritumor and normal liver tissues; in contrast, there was no difference of TCF-4D expression. TCF-4C clones derived from both cell lines exhibited increased TCF activity, Wnt-responsive target genes, cell proliferation, cell cycle progression, and resistance to chemotherapeutic drugs compared to TCF-4D clones. Capability of cell migration and colony formation was significantly higher in TCF-4C than TCF-4D clones. In a nude mice xenograft model, the HAK-1A-derived TCF-4C clone rapidly developed tumors compared to the TCF-4D clone. TCF-4C clone-derived tumors exhibited upregulation of Wnt-responsive target genes compared to the slow developing and small TCF-4D-derived tumors.
Conclusion
These results demonstrate that the TCF-4C isoform lacking exon 4 is associated with a malignant phenotype compared to the exon 4-harboring TCF-4D isoform, indicating that exon 4 of TCF-4 plays a prominent role in HCC development.
Keywords: T-cell factor (TCF)-4, isoform, Wnt, hepatocellular carcinoma (HCC), exon 4
INTRODUCTION
Wnt/β-catenin pathway is an evolutionarily conserved signal transduction cascade, which is involved in the regulation of cell proliferation, cell polarity, and cell-fate determination during embryonic development and tissue homeostasis (1). In the absence of Wnt ligands, cytoplasmic β-catenin is constitutively phosphorylated, which leads to ubiquitination of β-catenin and subsequent proteasomal degradation. In the presence of an active Wnt signal, mediated through Frizzled (FZD) receptors, β-catenin accumulates in the cytoplasm, translocates into the nucleus where it forms a transcriptional complex with T-cell factor (TCF)/lymphoid enhancer factor (LEF) proteins, activating Wnt-responsive target genes (2).
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide (3). Aberrant activation of the Wnt/β-catenin signaling pathway has been found in most HCC tumors, which may be associated with genetic and/or epigenetic deregulation involving overexpression of FZD receptors and Wnt ligands (4–6). Previous studies have identified 14 different TCF-4 isoforms that are expressed in HCC cell lines and demonstrated that these alternative spliced isoforms display different TCF transcriptional activity (7). Among such isoforms, a pair designated TCF-4C and D has been further characterized based on the absence (TCF-4C) or presence (TCF-4D) of exon 4 (Fig. 1A). Although TCF-4C and D differ only by exon 4, TCF-4C has higher TCF transcriptional activity than TCF-4D. Accordingly, we hypothesized that exon 4 of TCF-4D may have a repressive function for TCF transcriptional activity. In addition, ectopic expression of TCF-4C (lacking exon 4) exhibited the highest transcriptional activity among the 14 different TCF-4 isoforms in both HEK293 and Huh7 HCC cells (7). Although abnormal activation of the Wnt/β-catenin signaling pathway may be reflected by TCF transcriptional activity and linked functionally to hepatocarcinogenesis (6, 8–10), the biologic role of each TCF-4 isoform containing specific motifs has not been defined during the oncogenic process. In the present study, we explored the concept that exon 4 of a TCF-4 isoform may have a regulatory function during HCC development. Here, we investigated expression profiles of TCF-4C and D mRNA in human HCCs and conducted functional assays using established TCF-4C and D expressing stable clones. The functional consequences of TCF-4C and D expression were assessed in non-transformed hepatocyte-derived cells and HCC cells to define the potential biologic role of exon 4 expression in HCC tumor formation.
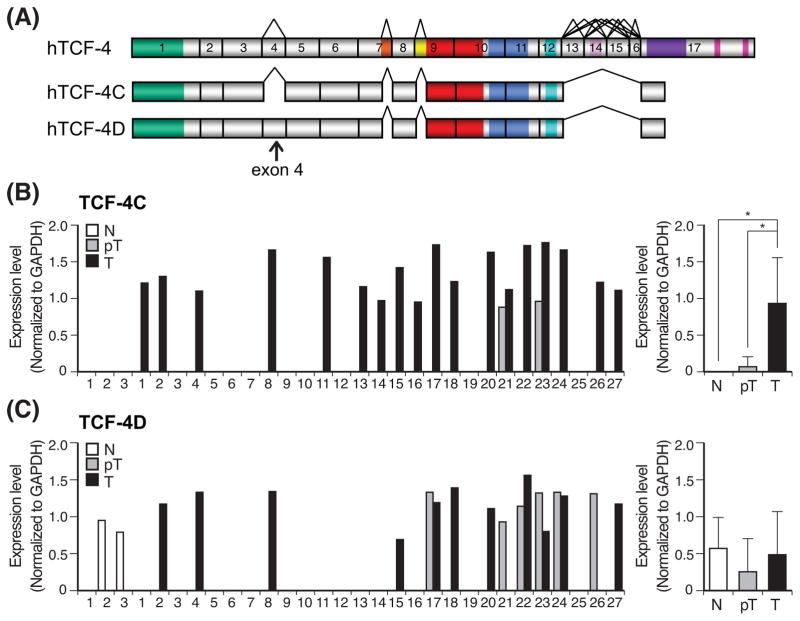
Fig. 1. Expression profiles of TCF-4C and D mRNA in human HCC samples.
(A) A schematic representation of human TCF7L2 gene compromising 17 exons based on the reported cDNA sequences (36) and structural organization of human TCF-4C and TCF-4D isoforms. The alternatively spliced sites, shown with a tee-pee shape, are exon 4, 7L (LVPQ), 9L (SxxSS), 13, 14, 15, 16, and 17L. (B, C) Expression level of TCF-4C (B) and TCF-4D (C) measured by semi-quantitative RT-PCR in human HCC tumors (T), adjacent peritumor tissue (pT) and normal liver (N). Left panel shows expression level in each sample, and right panel depicts as mean ± SD. The expression level of TCF-4C and TCF-4D was evaluated by semi-quantitative RT-PCR and the values were normalized by GAPDH. *, P < 0.05.
Materials and methods
Cell lines and cultures
The immortalized fetal liver-derived cell line OUMS-29 was kindly provided by Dr. Namba and Kobayashi (Okayama University, Japan) (11). The HAK-1A HCC cell line was a generous gift from Dr. Yano (Kurume University, Japan) (12). These cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). To generate stable transfectants overexpressing TCF-4C, TCF-4D, or EV, OUMS-29 and HAK-1A cells were transfected with each plasmid construct and selected by G418 (Invitrogen, Carlsbad, CA).
Plasmids and transfection
Human TCF-4C-myc, TCF-4D-myc, and EV plasmids have been previously described (7). For transient expression, plasmids were transfected by a TransIT-LT1 reagent (Mirus Bio Co., Madison, WI) according to manufacturers’ instructions.
Human HCC tissues
There were 27 pairs of HCC tumors and matched peritumoral liver tissues including 3 normal livers used in this study. The clinical and pathological features of these tumors have been previously described (13).
Semi-quantitative RT-PCR analysis
Semi-quantitative RT-PCR analysis was performed as described (7). Expression levels of TCF-4C and TCF-4D were normalized to mRNA levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 18S rRNA, or ribosomal protein S9 (RPS9). Primers used in this study are listed in a previous paper (7) and Table S1.
Quantitative real-time PCR analysis
Total RNA was extracted from cells using the TRIzol Reagent (Invitrogen), and reverse transcription was performed with First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche Diagnostics, Indianapolis, IN) according to manufacturers’ instruction. Quantitative real-time PCR analysis was carried out on a Mastercycler ep realplex instrument and software (Eppendorf AG, Hamburg, Germany), using SYBR Green PCR reagents. Relative quantification was done using ΔΔCt method, normalizing to 18S rRNA. Dissociation curves were generated to evaluate PCR product specificity and purity. Primers used are listed in Table S2.
TCF transcriptional activity assay
TCF transcriptional activity was performed as previously described (6). In brief, TOPFlash or FOPFlash reporter plasmid was co-transfected with β-galactosidase in the presence of β-catenin expression construct. The β-galactosidase activity was used for normalization of transfection efficiency. After the transfection, the luciferase activity was measured and the relative transcriptional activity was determined by the ratio of TOP to FOP basal activity.
Immunofluorescence
Cells were seeded in Labteck culture chamber slides (Thermo Fisher Scientific, Rockford, IL) and fixed by 4% paraformaldehyde, followed by Protein block serum free solution (DAKO, Glostrup, Denmark) with 0.1% Triton X-100. Cells were incubated with an antibody against Myc-tag (Cell Signaling Technology, Beverly, MA), followed by incubation with secondary antibody (anti-rabbit IgG conjugated with Alexa-594, Invitrogen). Coverslips were mounted using DAPI-containing anti-fade mounting medium (Vector Lab, Berlingame, CA), and the immunofluorescence staining were visualized with Zeiss LSM510 Confocal Laser Scanning Microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY).
Western blot analysis and immunoprecipitation
Western blot analysis was carried out as previously described (6) using primary antibodies against Myc-tag, β-catenin, TLE (Cell Signaling Technology), Axin-2 (AXIN2) (Millipore, Billerrica, MA), CBP, Cyclin D1, PCNA and actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Nuclear proteins or total cell lysates were used as samples in this study. Nuclear proteins were extracted by using NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific). Immunoprecipitation of TCF-4 isoforms was performed using agarose-conjugated antibody against c-Myc (Santa Cruz Biotechnology, Inc.). To evaluate amount of co-factors in the β-catenin/TCF-4 transcriptional complex, the intensity of each protein band in the immunoprecipitated sample was determined with a densitometer and normalized by the intensity of that detected by an anti-Myc-tag antibody.
Immunohistochemistry
Immunochemical staining was performed as described previously (6) using primary antibodies against Axin-2 (AXIN2), WISP2, Osteopontin (SPP1), and CD24 (Abcam, Cambridge, UK). Staining of these proteins was observed in cytoplasm and independently assessed by two investigators.
Cell proliferation assay
Cell proliferation assay was performed as previously described (6). Briefly, cells were seeded in 24-well plates as triplicates. A colorimetric assay (CellTiter 96® Aqueous One Solution Cell Proliferation Assay; Promega, Madison, WI) was carried out at indicated days, and the signals were measured using Spectra Max M5 (Molecular Devices, Sunnyvale, CA).
Cell cycle analysis
Cell cycle progression was evaluated by flow cytometric analysis. In brief, cells were washed and then fixed in 70% cold ethanol. After the fixation, cells were incubated with propidium iodide (Sigma-Aldrich, St. Louis, MO) and RNase (Wako Chemicals USA, Richmond, VA) for 30 min at 37°C and data were acquired on the FACS Calibur (BD Biosciences, Franklin Lakes, NJ). The cell cycle analysis was carried out using ModFIT software (BD Biosciences).
Cell growth inhibition assay
5-Fluorouracil (5-FU) (Sigma-Aldrich) and doxorubicin (DXR) (Sigma-Aldrich) were used in this assay. Cells were seeded to 96-well microplate, and drugs with indicated concentrations were added after 24 hrs. Cells suspended in medium without drugs were used as a control for cell viability. Seventy-two hrs after incubation, the numbers of viable cells were measured by 3-(4-,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Sigma-Aldrich) assay. Briefly, MTT reagents were added to each well and incubated for 4 hrs at 37°C. Unreacted MTT was then removed, leaving the resultant formazan crystals at the bottom of the well. Then, acid-isopropanol was added to dissolve the crystal. The absorbance of the plate was measured using Spectra Max M5 (Molecular Devices).
Wound healing assay
Wound healing assay was carried out as previously described (6). In brief, a scratch was made using a sterile micropipette tip. The wound closure, as an index of cell migration, was photographed and measured at the indicated times from the same area. Results were expressed as percentage of wound closure normalized to the initial width.
Colony formation assay in soft-agar
Cells were suspended in 0.4% top agar (Novel agar, Sigma-Aldrich) over a bottom layer of 0.8% base agar. After incubating for 5 wks, colonies were visualized by staining with p-iodonitrotetrazolium violet (Sigma-Aldrich, 1mg/ml in 50% ethanol). Macroscopic photographs of colonies were taken and analyzed using ImageJ. Microscopic representative photographs are represented.
Xenograft tumor model in nude mice
Cells (1×107) were subcutaneously injected into the back of 5-week old female BALB/c nude mice (n = 15) (Taconic Farms, Cranbury, NJ). The tumor size was measured twice per week, and tumor volume was estimated using the following formula: tumor volume (mm3) = (longer diameter) × (shorter diameter)2 × 0.5. When the diameter reached 10 mm, the mice were sacrificed. All animal experiments were conducted in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by Lifespan Animal Welfare Committee of Rhode Island Hospital, Providence, RI.
Harvested tumor samples fixed in 10% buffered formalin and embedded in paraffin were stained with hematoxylin and eosin, and subjected to immunohistochemistry. RNA and cell lysates were extracted from homogenized tumor samples, and used in quantitative real-time PCR and Western blot analysis for evaluating expression of mRNA and protein, respectively.
Statistical analysis
Data are expressed as means ± SD or SE. Differences between groups were assessed by the χ2-test, Fisher’s exact test or the Mann-Whitney U test. A P value < 0.05 was considered statistically significant. Data analyses were performed using StatView (version 5.0; SAS Institute Inc., Cary, NC).
Results
TCF-4C and D isoforms exhibit differential expression in human HCC tissues
We examined expression levels that may reflect an exon 4-dependent regulation in human HCC. The relative mRNA levels of TCF-4C and D in 27 pairs of tumors and corresponding adjacent uninvolved liver tissue were measured by semi-quantitative RT-PCR with normalization to GAPDH, as previously described (7). Comparisons were also made to three normal liver specimens. As shown in Fig. 1B, TCF-4C expression was significantly upregulated in HCC compared to peritumor tissue and normal liver. Interestingly, no expression of TCF-4C was found in normal liver and only 7% (2 of 27) of peritumor expressed TCF-4C mRNA compared to tumor tissue (18 of 27). In contrast, the expression level of TCF-4D was not significantly different among normal, peritumor, and tumor tissue (Fig. 1C). These results were verified by analyses with normalization to other housekeeping genes including 18S rRNA and RPS9 (Fig. S1). These observations suggest that TCF-4C and D isoforms expression are modulated during HCC development due to alternative splicing of exon 4, so that TCF-4C may be more associated with generation of a HCC malignant phenotype.
TCF-4C expressing clones increase Wnt/β-catenin signaling activation
To develop a better understanding of the role of exon 4 in promoting a malignant phenotype in vitro, we established stable clones expressing TCF-4C or D isoform by transfection into OUMS-29, immortalized human fetal hepatocyte cell line (TCF-4C; OU-C13, OU-C22, and TCF-4D; OU-D13, OU-D21 clones respectively) or HAK-1A which is a well differentiated HCC cell line (TCF-4C; 1A-C12, 1A-C14 and TCF-4D; 1A-D8, 1A-D9 clones respectively) followed by selection with G418. Empty vector (EV) plasmid-transfected clones were also generated as controls (OU-EV4, 1A-EV2). To exclude individual clonal artifacts, two different stable clones of each TCF-4 isoform were used for analyses. Protein expression of each TCF-4 isoform was confirmed by Western blot analysis using anti-Myc-tag antibody (Fig. 2A). Whole cell lysates obtained from transient transfection of TCF-4C and D plasmid constructs were used as positive controls, which corresponded to previous reports (7). The subcellular localization of TCF-4 isoforms in HAK-1A clones was further examined by immunofluorescence and observed to be predominantly expressed in the nucleus (Fig. S2).
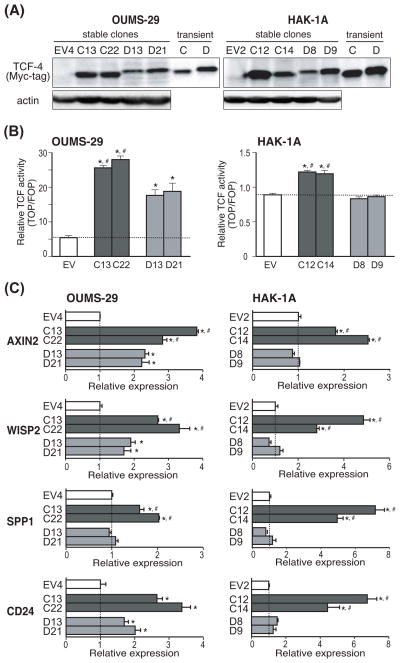
Fig. 2. Activation of the Wnt/β-catenin signaling pathway in TCF-4C and D expressing stable clones.
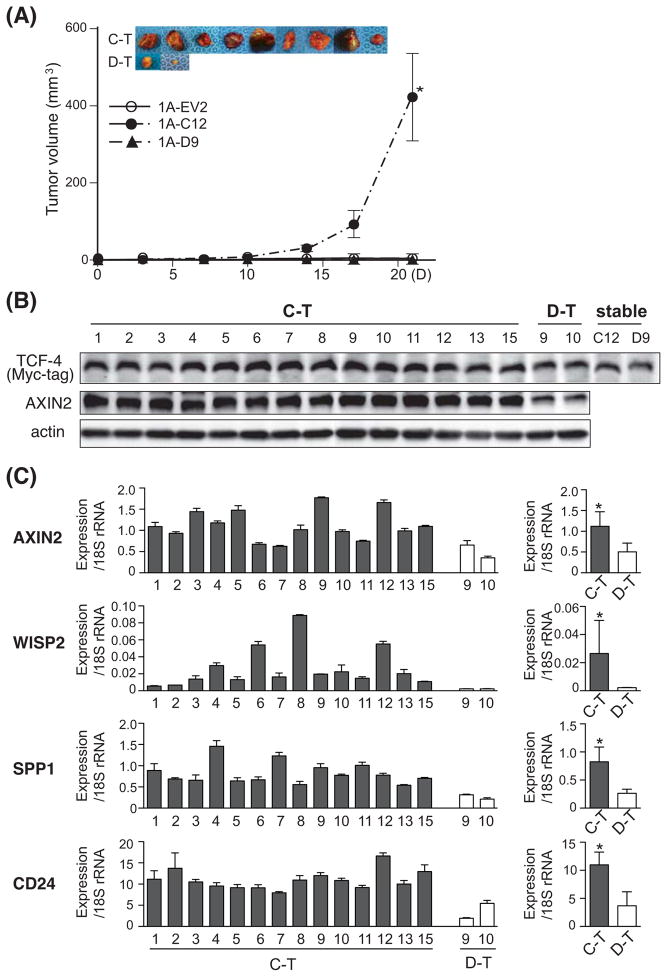
(A) Protein expression of TCF-4C and D isoforms in stable clones derived from OUMS-29 and HAK-1A parental cell lines. Expression was confirmed by Western blot analysis using antibodies against Myc-tag and actin. Transiently transfected cell lysates were used as positive controls for TCF-4C and TCF-4D. (B) TCF transcriptional activity in stable clones overexpressing TCF-4C and D isoforms. The transcriptional activity was calculated based on luciferase and β-galactosidase activity. The dotted lines represent the transcriptional activity level of EV control. The results are expressed as mean ± SD. *, P < 0.05 and #, P < 0.05 compared to EV and TCF-4D clones, respectively. (C) Expression levels of Wnt-responsive target genes including AXIN2, WISP2, SPP1, and CD24 in the stable clones. The expression levels were measured by quantitative real-time PCR and the values were normalized to 18S rRNA. Relative expression depicts the ratio of each gene expression to that of EV control. The dotted lines represent level of EV control. The results are expressed as mean ± SD. *, P < 0.05 and #, P < 0.05 compared to EV and TCF-4D clones, respectively.
Next, to evaluate activation of Wnt/β-catenin signaling pathway induced by constitutive expression of the TCF-4C or D isoform, TCF transcriptional activity was measured by a TOP/FOP reporter assay (Fig. 2B). In this regard, stable clones expressing TCF-4C exhibited significantly higher transcriptional activity than TCF-4D and the EV control, consistent with previous reports (7). Transcriptional activity exhibited by HAK-1A-derived TCF-4D clones was similar to the EV control, which was similar to a previous report using the Huh7 HCC cell line (7). To further confirm activation of Wnt/β-catenin signaling, we performed quantitative real-time PCR to measure expression levels of known Wnt-responsive target genes; AXIN2 (14), WNT1 inducible signaling pathway protein 2 (WISP2) (15), secreted phosphoprotein 1 (SPP1) (16), and CD24 (17) (Fig. 2C). In the stable clones derived from both parental cell lines, AXIN2, WISP2, and SPP1 were significantly upregulated in TCF-4C expressing clones compared to TCF-4D and EV clones. In HAK-1A-derived clones, upregulation of CD24 was evident in TCF-4C compared to TCF-4D and the EV control clones. In general, levels of target genes in TCF-4C and D clones were comparable to the observed difference in magnitude of TCF transcriptional activity. Taken together, these results demonstrate that TCF-4C expressing cells exhibit higher activation of the Wnt/β-catenin signaling cascade compared to TCF-4D clones.
The observation that TCF-4C (lacking exon 4) expressing cells exhibited increased activation of Wnt/β-catenin signaling compared to TCF-4D (bearing exon 4) raised the question if exon 4 may affect levels of co-factor proteins association within the β-catenin/TCF-4 transcriptional complex. The TCF proteins mediate Wnt signals in the nucleus as bipartite factors that act as either transcriptional activators or repressors according to their association with other proteins. Transcriptional activation is driven, in part, by β-catenin and the co-activator CREB-binding protein (CBP), whereas repression is mediated by co-repressors such as transducin-like enhancer of split (TLE) (18–22). Therefore, these co-factor proteins levels in the β-catenin/TCF-4 transcriptional complex can positively or negatively regulate activation of the Wnt/β-catenin signaling pathway. In this regard, we hypothesized that exon 4 could alter amounts of co-factor proteins in β-catenin/TCF-4 transcriptional complex, resulting in differential Wnt/β-catenin signaling activation. In this context, levels of co-factor proteins present in the complex of OUMS-29-derived clones (OU-EV4, OU-C22, OU-D21) and HAK-1A-derived clones (1A-EV2, 1A-C12, 1A-D9) were examined. The β-catenin/TCF-4 transcriptional complex was prepared from nuclear extracts by immunoprecipitation with an anti-c-Myc antibody. The amount of co-factor proteins in the complex was quantified after normalization of band intensity as detected by an anti-Myc-tag antibody. As shown in Fig. S3, in stable clones derived from both cell lines, levels of β-catenin in the complex was significantly increased in TCF-4C as compared to TCF-4D expressing clones. In the HAK-1A-derived clones, the amount of co-activator CBP protein was also significantly increased in TCF-4C compared to TCF-4D clones. In contrast, the level of co-repressor TLE was significantly decreased in TCF-4C compared to TCF-4D clones in both stable transfected parental cell lines. These findings suggest that exon 4 alters the protein levels of these co-factors in the β-catenin/TCF-4 transcriptional complex and may influence or modulate activation of Wnt/β-catenin signaling.
TCF-4C clones exhibit increased cell proliferation and resistance to chemotherapeutic agents
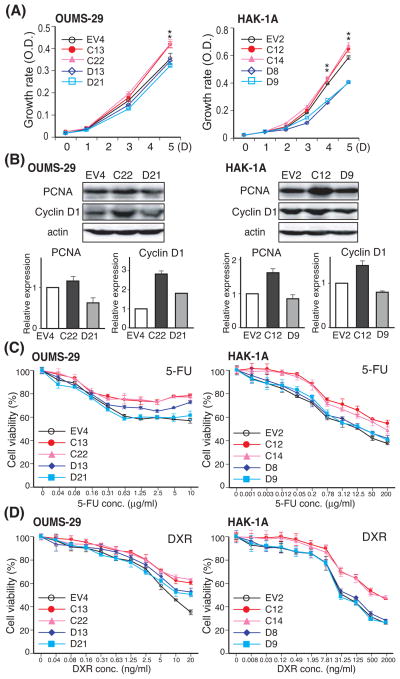
To evaluate the functions of TCF-4C and D isoforms during hepatic oncogenesis, we examined cell proliferation as measured by a MTS assay and evaluated cell cycle progression by flow cytometric analysis. As shown in Fig. 3A, TCF-4C expressing clones derived from both OUMS-29 and HAK-1A cell lines showed enhanced cell proliferation compared to the corresponding TCF-4D expressing clones. Cell proliferation rate of OUMS-29-derived TCF-4D clones (OU-D13, D21) was similar to control (OU-EV4), whereas HAK-1A-derived TCF-4D clones (1A-D8, D9) exhibited slower cell growth compared to control (1A-EV2). Clones were synchronized in the G0/G1 phase by serum starvation (0.5% FBS) for 72 hrs, followed by supplying with medium containing 10% FBS for analysis of cell cycle progression. The proportion of cells in G0/G1 phase and S phase at the end of the starvation (0 hr) was comparable between the clones. Nevertheless, 24 hrs after the starvation, a significant increase in proportion of cells in the S phase of the cell cycle in TCF-4C was observed compared to TCF-4D and EV clones in both stable cell lines (Table 1 and Fig. S4). This finding suggests that cell cycle progression was more rapid in TCF-4C expressing clones, which is compatible with the cell proliferation results. Cell proliferation rate and cell cycle progression were also assessed by the expression of proliferative cell nuclear antigen (PCNA) and Cyclin D1, a nuclear protein required for cell cycle progression and a well described Wnt-responsive target gene (23), respectively. In an agreement with the above results, expression levels of PCNA as well as Cyclin D1 were increased in TCF-4C compared to TCF-4D clones (Fig. 3B).
Fig. 3. Effect of TCF-4C and D isoform expression on cell proliferation, cell cycle progression, and resistance to chemotherapeutic reagents including 5-FU and DXR.
(A) Cell proliferation rate of the stable clones derived from OUMS-29 (left panel) and HAK-1A (right panel). The growth rate was measured by MTS assay over 5 days and are expressed as mean ± SD. *, P < 0.05 compared to EV and TCF-4D clones. (B) Protein expression of PCNA and Cyclin D1 in stable clones derived from OUMS-29 (left panel) and HAK-1A (right panel) were confirmed by Western blot analysis using antibodies against PCNA, Cyclin D1, and actin as a loading control. Expression level was determined by densitometry and normalized to actin. Relative expression depicts as the fold changes compared to EV control (lower panel). The results are shown as mean ± SD. (C, D) Stable clones derived from OUMS-29 (left panel) and HAK-1A (right panel) were treated with 5-FU (C) or DXR (D) for 72 hrs. The cell viability of stable clones was determined by MTT assay. Each value is expressed mean ± SD.
Table 1.
Cell cycle distribution of OUMS-29- and HAK-1A-derived clones following synchronization.
| Time (hr) | Phase | OUMS-29
|
HAK-1A
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EV4 | C13 | C22 | D13 | D21 | EV2 | C12 | C14 | D8 | D9 | ||
| 0 | G0/G1 | 68.63 | 69.86 | 69.68 | 69.37 | 68.85 | 74.54 | 76.16 | 77.74 | 80.04 | 81.13 |
| S | 27.62 | 26.36 | 26.40 | 26.75 | 27.13 | 21.91 | 20.75 | 19.47 | 17.49 | 16.68 | |
| G2/M | 3.75 | 3.78 | 3.92 | 3.88 | 4.02 | 3.55 | 3.09 | 2.79 | 2.47 | 2.19 | |
|
| |||||||||||
| 24 | G0/G1 | 60.22 | 51.58 | 52.19 | 57.68 | 58.71 | 66.91 | 54.29 | 58.46 | 74.60 | 68.16 |
| S | 37.34 | 42.96 | 42.91 | 36.92 | 36.06 | 29.70 | 43.72 | 39.73 | 23.47 | 29.17 | |
| G2/M | 2.44 | 5.46 | 4.90 | 5.40 | 5.23 | 3.39 | 1.99 | 1.81 | 1.93 | 2.67 | |
Data are expressed as percentage. Raw data are available on Fig. S4.
We also examined resistance to chemotherapeutic agents such as 5-FU and DXR using a growth-inhibitory assay. As shown in Fig. 3C and D, in both stable cell lines, TCF-4C clones were more resistant to both drugs than TCF-4D clones; the resistance in TCF-4D clones was similar to the control EV clones.
TCF-4C expression accelerates cell migration and promotes colony formation in soft-agar
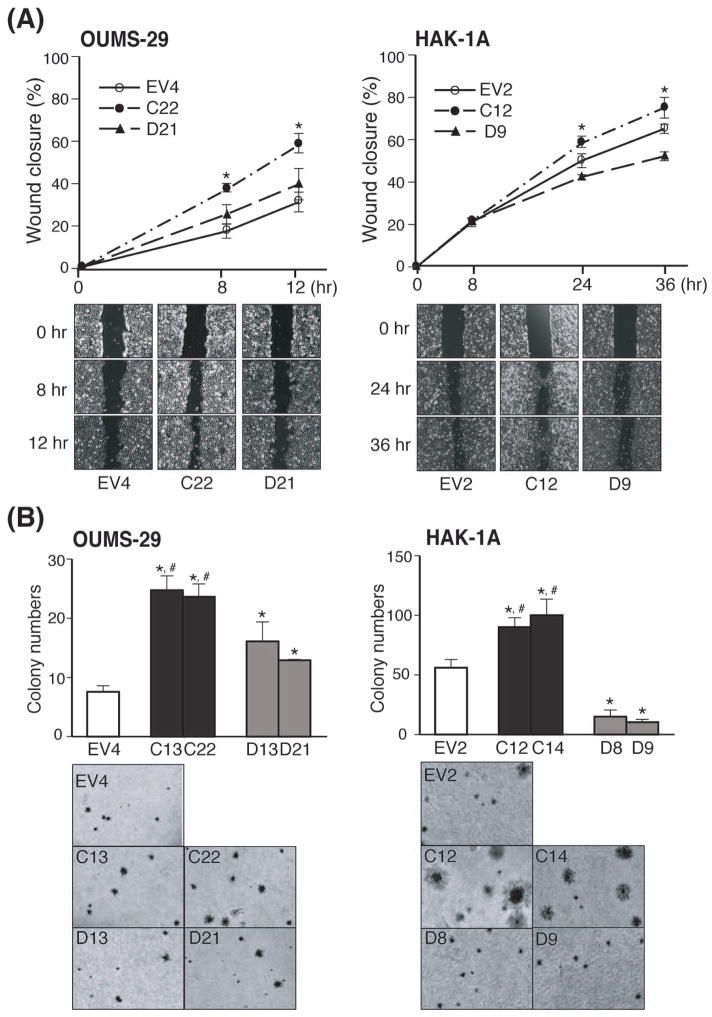
Cell migration was evaluated by a wound-healing assay. As shown in Fig. 4A, accelerated wound closure was observed for TCF-4C expressing clones derived from both parental cell lines (OU-C22, 1A-C12) compared to the corresponding TCF-4D clones (OU-D21, 1A-D9) as well as the controls (OU-EV4, 1A-EV2). The OUMS-29-derived TCF-4D clone (OU-D21) migrated faster than the control (OU-EV4), whereas the HAK-1A-derived TCF-4D clone (1A-D9) revealed a slower migration pattern compared to the control (1A-EV2).
Fig. 4. Effect of TCF-4C and D expression on cell migration and colony formation in soft-agar.
(A) Cell migratory capacity of the stable clones (OUMS-29: left panel, HAK-1A: right panel) assessed by wound healing assay. Upper panel: The graph represents percentage of wound closure and representative pictures at the indicated time are shown. The percentage of wound closure is expressed as mean ± SD. *, P < 0.05 compared to EV and TCF-4D clones. Lower panel: Representative wound-healing closure photographs were taken at the indicated time (40×). (B) Anchorage-independent cell growth of the stable clones (OUMS-29: left panel, HAK-1A: right panel) assessed by soft-agar colony formation assay. The bar graph (upper panel) indicates colony numbers and representative photographs (lower panel) are shown (40×). The colony numbers are expressed as mean ± SD. *, P < 0.05 and #, P < 0.05 compared to EV and TCF-4D clones, respectively.
Anchorage-independent cell growth was also assessed by a soft-agar colony formation assay. The number of colonies was increased in TCF-4C compared to TCF-4D expressing clones (Fig. 4B). In addition, TCF-4C clones derived from HAK-1A HCC cells were associated with formation of larger colonies compared to those derived from TCF-4D expressing clones (Fig. 4B, right lower panel). Thus, TCF-4D clones derived from HAK-1A HCC cells exhibited decreased colony formation compared to EV control as well as reduced cell proliferation and migration.
TCF-4C isoform expression confers a tumorigenic phenotype to HCC cells in a xenograft model
The tumorigenic potential of TCF-4C, TCF-4D, and EV control clones was assessed in a nude mouse xenograft model. Parental HAK-1A cells do not form tumors in nude mice, and therefore, are suitable for assessment of malignant transformation due to overexpression of TCF-4 isoform (12). OUMS-29-derived clones (OU-EV4, OU-C22, OU-D21) did not produce visible tumors 4 wks after subcutaneous injection. On the other hand, the TCF-4C clone (1A-C12) derived from the parental HAK-1A HCC cell line was highly tumorigenic. Interestingly, 93% (14 of 15) of mice injected with the TCF-4C clone (1A-C12) developed tumors, whereas the TCF-4D clone (1A-D9) produced 20% (3 of 15) tumors. Although the TCF-4D clone generated tumors, they grew significantly slower than TCF-4C clone-derived tumors and representative photographs of the excised tumors are shown (Fig. 5A). The control EV clone (1A-EV2) did not form tumors, similarly to the parental cells (12).
Fig. 5. Tumorigenic potential of the stable TCF-4C and D expressing clones in a nude mouse xenograft model.
(A) In vivo tumor growth curve and representative photographs of excised tumors. A total of 1×107 cells of HAK-1A clones (1A-EV2, 1A-C12, and 1A-D9) were injected subcutaneously. The representative photographs of the tumors derived from 1A-C12 (C-T) and 1A-D9 (D-T) are shown. The tumor volume (mm3) is estimated using the following formula: tumor volume (mm3) = (longer diameter) × (shorter diameter)2 × 0.5. The results are expressed as means ± SE. *, P < 0.05 compared to 1A-EV and 1A-D9. (B) Protein expression of TCF-4C and D, and AXIN2 in the excised tumors evaluated by Western blot analysis using antibodies against Myc-tag and AXIN2. Proteins extracted from stable clones (1A-C12, 1A-D9) were used as positive controls and actin was a loading control. (C) Expression level of Wnt-responsive target genes including AXIN2, WISP2, SPP1, and CD24 in the excised tumors. Left panel shows expression level of each sample, and the average of expression is depicted on right panel. The expression levels were measured by quantitative real-time PCR and the values were normalized to 18S rRNA. The results are shown as mean ± SD. *, P < 0.05 compared to D-T.
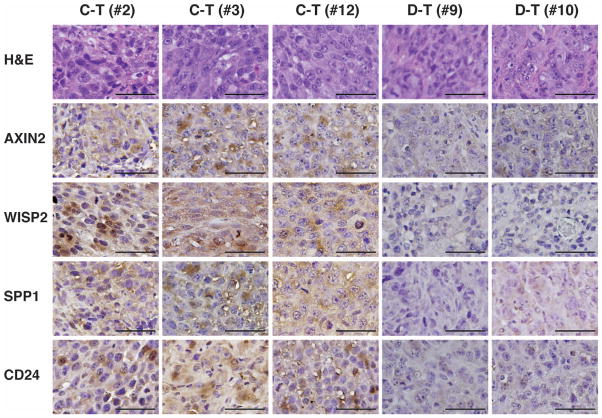
Western blot analysis was performed to confirm protein expression of TCF-4C and D in the xenograft tumors. The Myc-tag antibody detected exogenous TCF-4C and D expression in the excised tumor samples; in addition, AXIN2, one of the known Wnt-responsive genes, showed increased protein expression in TCF-4C clone-derived tumors (Fig. 5B). Expression levels of Wnt-responsive target genes were also measured by quantitative real-time PCR analysis. Consistent with the above results from cell lines, AXIN2, WISP2, SPP1, and CD24 were significantly upregulated in TCF-4C clone-derived tumors compared to TCF-4D clone-derived tumors (Fig. 5C). We also assessed protein expression of these genes by immunohistochemical staining. The histological features of the tumor tissues as well as cellular morphology did not differ among tumors induced by either TCF-4C or D clone using hematoxylin and eosin staining (Fig. 6, upper panel). However, there was increased expression of AXIN2, WISP2, SPP1, and CD24 in TCF-4C clone-derived tumors compared to TCF-4D clone-derived tumors (Fig. 6).
Fig. 6. Immunohistochemical staining for Wnt-responsive target genes in TCF-4C and D clones-derived tumors.
The tumors derived from 1A-C12 (C–T) and 1A-D9 (D–T) were stained with hematoxylin and eosin (H&E) and immunohistochemically using antibodies against AXIN2, WISP2, SPP1, and CD24. Photographs show stained sections in representative samples (C–T: #2, #3, #12, and D–T: #9, #10) (400×). Bar = 100 μm.
Discussion
TCF transcriptional factor proteins are important in mediating the final transcriptional program in Wnt/β-catenin signaling and act as bi-functional proteins depending on their association with co-factors. Previous studies provided evidence that certain TCF-4 isoforms may function as either transcriptional activators or repressors, to produce distinct cellular phenotypes during hepatocarcinogenesis (7). Moreover, the different functional properties of TCF-4 isoforms generated by alternative splicing events may be regulated by expression of these conserved motifs in HCC tumors. Indeed, a specific TCF-4J isoform, which lacks a regulatory SxxSS motif, has robust tumor-initiating potential in HCC (7, 13). In this study, we explored a possible regulatory role of exon 4 in TCF-4 isoforms by comparing the functional properties of TCF-4C (lacking exon 4) and D (bearing exon 4) during hepatic oncogenesis. We demonstrated that the expression level of TCF-4C was significantly increased in human HCC tumors compared to corresponding peritumor and normal liver, whereas there was no difference of TCF-4D expression among all three tissues. In vitro experiments using stable clones overexpressing TCF-4C and D isoforms generated from two different parental human liver cell lines (OUMS-29, fetal liver; HAK-1A, well differentiated HCC) revealed that exogenous expression of TCF-4C induces characteristics of a malignant phenotype such as enhanced cell proliferation, cell migration, colony formation in soft agar plates and resistance to chemotherapeutic agents. Moreover, TCF-4C isoform expressing cells exhibited robust tumorigenic potential in vivo and formed large tumors in nude mice. Furthermore, we also observed that TCF-4C expression levels in human HCC tumor tissue correlate with large-size and poorly differentiated tumor characteristics [data not shown and (7)]. Stronger TCF-4 expression in the TCF-4C clones than the TCF-4D clones may raise a concern that the degree of malignancy in the clones is due to TCF-4 expression level regardless of the isoforms. However, in TCF-4D clones derived from both cell lines, malignant phenotypes such as proliferation and colony-formation capacity were not positively correlated with the expression level. In addition, HAK-1A-derived TCF-4D clones exhibited less malignant phenotypes than their EV control in spite of their higher TCF-4 expression level than the control, indicating that the malignant phenotypes in the stable clones are not due to TCF-4 expression difference. Taken together, these observations suggest that exon 4 of TCF-4 may play an important role in regulating the malignant characteristics of HCC.
Consistent with a previous report (7), ectopic expression of TCF-4C exhibited increased TCF transcriptional activity compared to TCF-4D; the relative activity of TCF-4C to TCF-4D was 1.3 and 1.4 fold in OUMS-29-derived clones and HAK-1A clones, respectively. Although difference of the transcriptional activity was modest, there was significant difference in expression levels of Wnt-responsive target genes such as WISP2, AXIN2, SPP1, and CD24 as well as Cyclin D1 between TCF-4C and D expressing clones (Figs. 2 and 3). In this regard, the TCF transcriptional activity may not directly represent the magnitude of Wnt-responsive target genes activation but may be considered to be one of the components necessary for Wnt/β-catenin signaling pathway activation. Consistent with previous reports that Wnt/β-catenin signaling activation is regulated by co-factor proteins associated with formation of a β-catenin/TCF-4 transcriptional complex (18–22), we found that the amount of co-factor proteins in the β-catenin/TCF-4 transcriptional complex between TCF-4C and D clones were different. This is probably important in the activation of Wnt/β-catenin signaling exhibited by the TCF-4C expressing clones.
It was observed that the HAK-1A-derived TCF-4C clone demonstrates an enhanced tumorigenic phenotype in a murine xenograft model and illustrates that aberrant activation of Wnt/β-catenin signaling is associated with carcinogenesis and promotes cancer progression in HCC (6, 8–10). Indeed, recent studies revealed that a pair of TCF-4 isoforms (TCF-4J and K) exhibits differential TCF transcriptional activity and gene expression profiles, and promoted a HCC malignant phenotype (7, 13). These observations raise the possibility that the enhanced tumorigenicity of the TCF-4C clones may be related to differential expression levels of Wnt-responsive target genes in different HCC cell lines. By evaluating Wnt-responsive target gene expression, we observed that activation of SPP1 is robust only in HAK-1A-derived TCF-4C clones (> 5-fold increased compared to TCF-4D and EV control), and is upregulated in in vivo TCF-4C clone-derived tumors compared to TCF-4D clone-derived tumors. SPP1, encoding osteopontin, is related to tumorigenicity and metastatic potential, which is known to be upregulated in various cancers and associated with a poor prognosis in HCC (24, 25). The level of CD24 was also increased 5–7 fold in HAK-1A-derived TCF-4C clones compared to EV control as well as the TCF-4D clones, and the upregulation was verified in in vivo tumors. In this regard, CD24 is associated with tumor growth and metastasis, and CD24-positive cells have cancer stem cell-like characteristics in HCC (26, 27). In addition, AXIN2 is known to serve as a negative regulator of canonical Wnt/β-catenin signaling in normal cells, but it has recently been reported that AXIN2 promotes colon carcinoma oncogenic activity by inducing a functional epithelial-mesenchymal transition program which drives metastatic activity (28). Thus, upregulation of AXIN2 in HAK-1A-derived TCF-4C clones may have a positive role in HCC pathogenesis. Collectively, these observations imply that activation of Wnt-responsive target genes induced by the TCF-4C isoform may be responsible, in part, for the significantly enhanced malignant phenotype exhibited by HAK-1A-derived TCF-4C clones.
Although the malignant characteristics of the TCF-4C isoform were confirmed by in vitro experiments in both OUMS-29-derived and HAK-1A-derived clones, the robust tumorigenicity in the xenograft model was inducible only in the HAK-1A-derived TCF-4C, not in the OUMS-29-derived TCF-4C clone. Taking into consideration that Wnt/β-catenin signaling activation was found in both OUMS-29- and HAK-1A-derived TCF-4C clones, cellular transformation mediated by TCF-4C overexpression may not be sufficient to induce tumorigenicity in OUMS-29 cells derived from fetal hepatocytes. These results suggest that, in addition to Wnt/β-catenin signaling activation induced by TCF-4C overexpression, other factors may be required to induce tumorigenicity in non-transformed cells such as OUMS-29. This hypothesis is consistent with studies reporting that activation of Wnt/β-catenin signaling pathway alone is not sufficient to transform normal hepatocytes (29, 30). In this regard, comparison of gene signatures in hepatic cells between TCF-4C-inducible tumor formation as found in HAK-1A and the lack of tumor growth in OUMS-29 cells may be a useful model to determine other key factors necessary for hepatic transformation in non-transformed cells in addition to Wnt/β-catenin signaling activation.
The links between cancer and impaired alternative splicing have been well established and several reports demonstrated that cancer-specific splice variants and differential levels of spliced isoforms had an impact on carcinogenesis (31–35). Moreover, several mechanisms that regulate splicing events associated with tumor development and progression have been elucidated: 1) mutations which create or disrupt splice or regulatory site; 2) abnormal expression of splicing factors; 3) deregulated signaling pathway impacting activity of splicing machinery (31, 32, 35). Our results also imply a significant role of the TCF-4C isoform generated through the alternative splicing of exon 4 in HCC development. However, mechanisms related to the induction of the TCF-4C isoform have not been explored yet. Recently, Berasain et al. proposed different modes of alternative splicing events such as exon skipping or inclusion, selection of alternative splicing sites, intron retention, alternative polyadenylation, and using alternative promoters (35). In this regard, one of the possible mechanisms involved in the generation of TCF-4C isoform may be the skipping of exon 4 during hepatocarcinogenesis and further studies will be needed to clarify the mechanism, which may contribute to the development of new effective therapeutic drugs for HCC.
In summary, the present study demonstrated that TCF-4C isoform expression (lacking exon 4) is significantly associated with hepatocarcinogenesis. The presence (TCF-4D) or absence (TCF-4C) of exon 4 in TCF-4 isoform showed differential transforming activity both in vitro as well as in vivo, and suggests that exon 4 plays an important role in regulating the HCC malignant phenotype. This study provides new insights into the role of exon 4, one of several unique motifs found in TCF-4 isoforms and clarification of its functional properties may be useful for understanding the role of these TCF-4 isoforms during hepatocarcinogenesis.
Supplementary Material
Fig. S1. Verified expression profiles of TCF-4C and D in the human HCC samples.
Fig. S2. Nuclear localization of TCF-4C and D in HAK-1A-derived stable clones.
Fig. S3. Differential binding and accumulation of co-factor proteins associated with the β-catenin/TCF-4 transcriptional complex in the stable clones.
Fig. S4. Cell cycle distribution in the stable clones following synchronization.
Table S1. List of primers sequences used for semi-quantitative RT-PCR analysis.
Table S2. List of primers sequences used for quantitative real-time PCR analysis.
Acknowledgments
FINANCIAL SUPPORT
This work was supported, in part, by grants from National Institute of Health AA020587 (MK) and CA123544 (JRW).
We thank Dr. Namba and Kobayashi (Okayama University, Japan) and Dr. Yano (Kurume University, Japan) for OUMS-29 and HAK-1A cells, respectively. The authors are grateful to Rolf Carlson for the assistance of animal work.
Footnotes
Additional Supporting Information may be found in the online version of this article:
CONFLICTS OF INTERESTS
None
References
- 1.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. 2009;4:307–12. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Lee HC, Kim M, Wands JR. Wnt/Frizzled signaling in hepatocellular carcinoma. Front Biosci. 2006;11:1901–15. doi: 10.2741/1933. [DOI] [PubMed] [Google Scholar]
- 5.Nejak-Bowen KN, Monga SP. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M, Lee HC, Tsedensodnom O, et al. Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/beta-catenin signaling pathway in hepatocellular carcinoma cells. J Hepatol. 2008;48:780–91. doi: 10.1016/j.jhep.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsedensodnom O, Koga H, Rosenberg SA, et al. Identification of T-cell factor-4 isoforms that contribute to the malignant phenotype of hepatocellular carcinoma cells. Exp Cell Res. 2011;317:920–31. doi: 10.1016/j.yexcr.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 9.Merle P, de la Monte S, Kim M, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–22. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Nambotin SB, Wands JR, Kim M. Points of therapeutic intervention along the Wnt signaling pathway in hepatocellular carcinoma. Anticancer Agents Med Chem. 2011;11:549–59. doi: 10.2174/187152011796011019. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi N, Miyazaki M, Fukaya K, et al. Transplantation of highly differentiated immortalized human hepatocytes to treat acute liver failure. Transplantation. 2000;69:2027. doi: 10.1097/00007890-200001270-00002. [DOI] [PubMed] [Google Scholar]
- 12.Yano H, Iemura A, Fukuda K, et al. Establishment of two distinct human hepatocellular carcinoma cell lines from a single nodule showing clonal dedifferentiation of cancer cells. Hepatology. 1993;18:320–7. [PubMed] [Google Scholar]
- 13.Koga H, Tsedensodnom O, Tomimaru Y, et al. Loss of the SxxSS motif in a human T-cell factor-4 isoform confers hypoxia resistance to liver cancer: an oncogenic switch in Wnt signaling. PLoS ONE. 2012;7:e39981. doi: 10.1371/journal.pone.0039981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jho EH, Zhang T, Domon C, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–83. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longo KA, Kennell JA, Ochocinska MJ, et al. Wnt signaling protects 3T3-L1 preadipocytes from apoptosis through induction of insulin-like growth factors. J Biol Chem. 2002;277:38239–44. doi: 10.1074/jbc.M206402200. [DOI] [PubMed] [Google Scholar]
- 16.El-Tanani M, Fernig DG, Barraclough R, Green C, Rudland P. Differential modulation of transcriptional activity of estrogen receptors by direct protein-protein interactions with the T cell factor family of transcription factors. J Biol Chem. 2001;276:41675–82. doi: 10.1074/jbc.M103966200. [DOI] [PubMed] [Google Scholar]
- 17.Shulewitz M, Soloviev I, Wu T, et al. Repressor roles for TCF-4 and Sfrp1 in Wnt signaling in breast cancer. Oncogene. 2006;25:4361–9. doi: 10.1038/sj.onc.1209470. [DOI] [PubMed] [Google Scholar]
- 18.Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–54. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–9. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasperowicz M, Otto F. Mammalian Groucho homologs: redundancy or specificity? J Cell Biochem. 2005;95:670–87. doi: 10.1002/jcb.20476. [DOI] [PubMed] [Google Scholar]
- 21.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–71. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 22.Najdi R, Syed A, Arce L, et al. A Wnt kinase network alters nuclear localization of TCF-1 in colon cancer. Oncogene. 2009;28:4133–46. doi: 10.1038/onc.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shtutman M, Zhurinsky J, Simcha I, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–7. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Dong L, Lu B, et al. Down-regulation of osteopontin suppresses growth and metastasis of hepatocellular carcinoma via induction of apoptosis. Gastroenterology. 2008;135:956–68. doi: 10.1053/j.gastro.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 25.Brown LF, Papadopoulos-Sergiou A, Berse B, et al. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145:610–23. [PMC free article] [PubMed] [Google Scholar]
- 26.Yang XR, Xu Y, Yu B, et al. CD24 is a novel predictor for poor prognosis of hepatocellular carcinoma after surgery. Clin Cancer Res. 2009;15:5518–27. doi: 10.1158/1078-0432.CCR-09-0151. [DOI] [PubMed] [Google Scholar]
- 27.Lee TK, Castilho A, Cheung VC, et al. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Wu Z-Q, Brabletz T, Fearon E, et al. Canonical Wnt suppressor, Axin2, promotes colon carcinoma oncogenic activity. Proc Natl Acad Sci USA. 2012;109:11312–11317. doi: 10.1073/pnas.1203015109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longato L, de la Monte S, Kuzushita N, et al. Overexpression of insulin receptor substrate-1 and hepatitis Bx genes causes premalignant alterations in the liver. Hepatology. 2009;49:1935–43. doi: 10.1002/hep.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nambotin SB, Tomimaru Y, Merle P, Wands JR, Kim M. Functional consequences of WNT3/Frizzled 7 mediated signlaing in non-transformed hepatic cells. Oncogenesis. 2012;1:e31. doi: 10.1038/oncsis.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 32.Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Grosso AR, Martin S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettigrew CA, Brown MA. Pre-mRNA splicing aberrations and cancer. Front Biosci. 2008;13:1090–1105. doi: 10.2741/2747. [DOI] [PubMed] [Google Scholar]
- 35.Berasain C, Goni S, Castillo J, et al. Impairment of pre-mRNA splicing in liver disease: mechanisms and consequences. World J Gastroenterol. 2010;16:3091–3102. doi: 10.3748/wjg.v16.i25.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duval A, Rolland S, Tubacher E, et al. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Verified expression profiles of TCF-4C and D in the human HCC samples.
Fig. S2. Nuclear localization of TCF-4C and D in HAK-1A-derived stable clones.
Fig. S3. Differential binding and accumulation of co-factor proteins associated with the β-catenin/TCF-4 transcriptional complex in the stable clones.
Fig. S4. Cell cycle distribution in the stable clones following synchronization.
Table S1. List of primers sequences used for semi-quantitative RT-PCR analysis.
Table S2. List of primers sequences used for quantitative real-time PCR analysis.