Abstract
The epithelial-mesenchymal transition (EMT) imparts metastatic competence on otherwise non-metastatic cancer cells through decreased inter-cellular adhesions, increased migratory capacity, stem cell properties and anoikis and chemotherapy resistance. In this study, we profiled changes in microRNA expression during EMT in conjunction with changes in DNA methylation at microRNA promoters to discover essential mediators of EMT-imparted stemness properties. MicroRNA-203 (miR-203) expression is repressed following EMT induced by multiple different stimuli and in established claudin-low cell lines as well as the CD44hi/CD24lo stem cell-enriched fraction. Expression of miR-203 in mesenchymal cells compromises migratory and invasive capacity in vitro, and tumor initiation and metastasis in vivo. Unexpectedly, miR-203 expression affects the sphere-forming capacity of neighboring cells by indirectly enhancing expression of DKK1, a secreted inhibitor of Wnt signaling and stemness resulting in suppression of β-catenin protein levels. Our data suggest that restoring miR-203 expression levels may inhibit metastasis and combat deregulated Wnt signaling.
The epithelial-mesenchymal transition (EMT) is a complex series of profound morphological changes that culminates in the loss of epithelial characteristics and the acquisition of a mesenchymal motile phenotype. In the context of cancer, EMT facilitates the dissemination of cancer cells and endows them with properties essential for metastasis including stemness, invasiveness, and the ability to survive in the circulation and seed a secondary site1,2,3,4. The reversal of EMT–known as mesenchymal-epithelial transition (MET)–is also an integral part of the metastatic cascade, in particular the last steps: colonization and establishment of macroscopic tumors at distant sites5,6. Due to their dynamic nature and reversibility, epigenetic alterations are proposed to facilitate EMT, MET and metastasis7. Indeed a number of epigenetic regulators are known to functionally regulate genes important for an EMT8. For instance, HDAC3, which deacetylates H3K4, acts preferentially upon EMT marker genes downstream of a hypoxia-induced EMT9. The removal of this repressive mark (H3K4ac) is balanced by methylation of this residue leading to activation of EMT marker genes. Additionally, a TGF-β induced EMT leads to increased expression of the histone demethylase LSD1 (KDM1), which plays a role in E-Cadherin (CDH1) repression through interaction with the EMT-promoting, transcription factor Snail10.
In addition to histone modification, DNA methylation of key genes facilitates an EMT11. Hypermethylation of the CDH1 promoter is associated with cancer progression in multiple tumor types12,13. Several microRNAs (miRNAs) are known to play important roles in regulating EMT and cancer stem cell (CSC) properties during cancer progression14,15. Moreover, epigenetic regulation also impacts microRNAs that regulate gene networks involved in EMT. In particular, the miR-200 family of miRNAs, which target the EMT-inducing transcription factors Zeb1 and Zeb216,17,18, is repressed by DNA methylation and histone modifications following an EMT and during the early stages of carcinogenesis19,20,21. In prostate cancer, the survival-associated miR-23b is also repressed by DNA methylation which relieves its repression of the proto-oncogene Src kinase22.
To discover novel methylation-regulated mediators of EMT and stem cell properties, we performed analyses to identify microRNAs which are regulated by DNA methylation and which regulate EMT-derived stemness properties. We found that the promoter of microRNA-203 (miR-203)–a known regulator of skin cell differentiation23,24–is methylated significantly in cells that have undergone EMT due to Twist expression and that its downregulation facilitates the gain of EMT/stemness properties. Thus, activating miR-203–either epigenetically or by other means–may inhibit invasion and metastasis.
Results
MiR-203 expression is downregulated via promoter methylation
To examine epigenetic regulation and in particular, the role of DNA methylation in regulating the expression of microRNAs during an EMT, we analyzed global changes in DNA methylation by genome-wide digital restriction enzyme (DREAM) assay using immortalized human mammary epithelial cells (HMLE)25 and HMLE cells induced to undergo EMT through overexpression of the transcription factor Twist (HMLE-Twist)26. In parallel, we also conducted a microRNA microarray using the same cells and found multiple microRNAs up- and down-regulated consistently in the EMT-induced, HMLE-Snail, -Twist, -TGFbeta, and -Gsc cells (Fig. 1a).
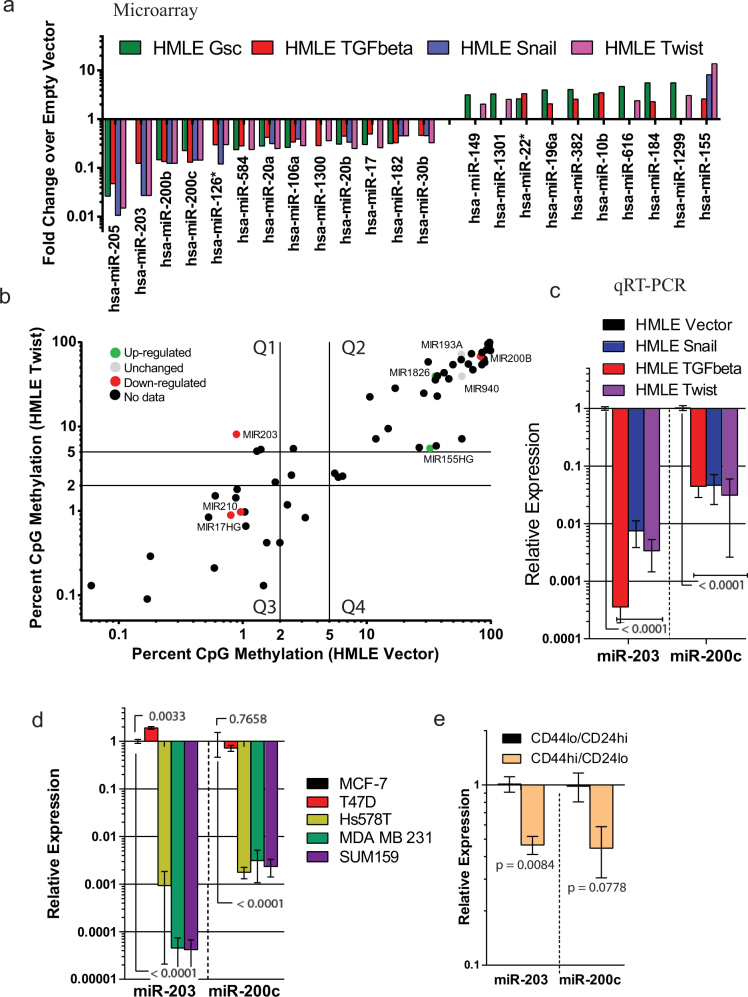
Figure 1. Low miR-203 expression is associated with EMT, claudin-low breast cancer and stem-like cells and the promoter of miR-203 is methylated in HMLE-Twist.
(a) A microarray was performed to gauge changes in microRNA expression due to EMT induction in HMLE cells by Snail, Twist, Goosecoid or TGF-β1 overexpression. MicroRNAs with greater than 2-fold change in any at least two of the indicated cell lines are plotted. (b) DNA methylation levels at CpG islands were determined by DREAM analysis of HMLE and HMLE-Twist cells. MiRNAs for which microarray-derived expression data is available are color-coded to indicate the direction of their expression change after an EMT. (c/d) Relative levels of miR-200c and miR-203 were determined in cell lines induced to undergo EMT (c) and in established breast cancer cell lines (d) by qRT-PCR. (e) HMLE cells were FACS-sorted for CD44 and CD24. Relative levels of miR-200c and miR-203 were measured by qRT-PCR in the two subpopulations.
Among the differentially expressed miRNAs, in cells that had undergone an EMT (Fig. 1a), we found a 10-fold gain in DNA methylation at the promoter of microRNA-203 (miR-203) (Quadrant 1) which stood out among other microRNAs which had little to no change in DNA methylation (Quadrants 2 and 3) (Fig. 1b) compared to HMLE control cells (Fig. 1b). Additionally, among those microRNAs which acquired DNA methylation in HMLE Twist cells compared to HMLE control cells (Fig. S1a), only miR-203 expression changed in response to exposure to the DNA demethylating agent 5-azacytidine (5-azaC) (Fig. S1b). Based on these findings, we further tested the role of epigenetically regulated miR-203 in EMT and stem cell properties. Importantly, we also found that miR-203 is strongly downregulated in cells induced to undergo EMT by other stimuli including overexpression of Snail or TGF-β1 in addition to Twist (Fig. 1c). In addition, we examined a patient-derived panel of breast cancer cell lines for the expression of miR-203. We found that, as in Park et al.16, the expression of miR-203 is higher in more differentiated, epithelial-appearing, luminal cancer cell lines (MCF-7 and T47D) compared to the less differentiated and mesenchymal-appearing, EMT/CSC enriched claudin-low cancer cell lines (Hs578T, MDA-MB-231 and SUM159) (Fig. 1d). Finally, we analyzed the expression of miR-203 in the stem cell-enriched CD44hi/CD24lo fraction relative to the CD44lo/CD24hi fraction isolated from HMLE cells. Consistent with our previous results, we found that miR-203 expression is significantly lower in the mesenchymal, stem cell-enriched CD44hi/CD24lo subpopulation (Fig. 1e) compared to the CD44lo/CD24hi differentiated cell fraction. Collectively, these data indicate that the miR-203 promoter is highly methylated in Twist expressing mesenchymal cells and its expression is downregulated in cells induced to undergo EMT as well as in EMT/CSC enriched claudin-low tumors.
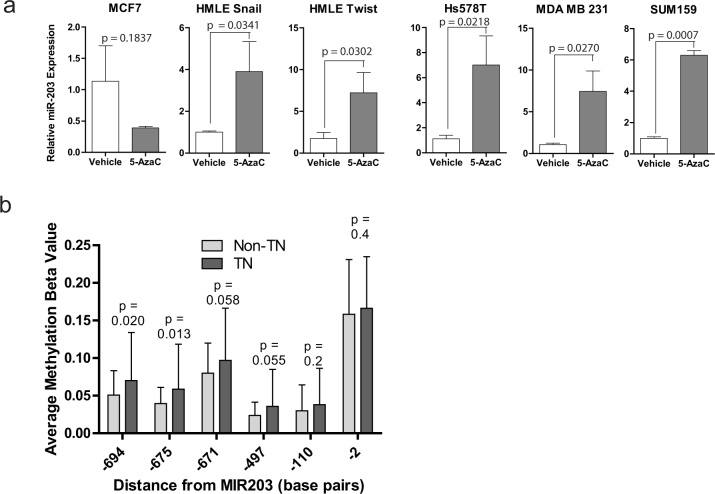
We next ascertained the functional relevance of DNA methylation in the promoter of miR-203 to its reduced expression. To test this, we treated HMLE-Snail and -Twist cells as well as a number of breast cancer cell lines either with an epithelial phenotype (MCF7), or with EMT/CSC properties including SUM159 and Hs578t, with the DNA methylation inhibitor 5-azaC. Strikingly, in all the cell lines, we found that miR-203 is re-expressed between four and seven fold excepting MCF7, which already expresses a high level of miR-203 (Fig. 2a). Analysis of expression of 5 other microRNAs revealed that they are not uniformly affected by 5-azaC treatment (Fig. S1b). To further extend our findings, we analyzed the methylation status of miR-203 in human tumors using The Cancer Genome Atlas (TCGA). Remarkably, we found that methylation at the miR-203 promoter is significantly higher in triple-negative breast cancers relative to the rest of the tumor subtypes (Fig. 2b). These findings suggest that DNA methylation is one of the major drivers for the suppression of miR-203 expression following the induction of EMT as well as in cancer cells with increased EMT/CSC properties including the triple negative breast cancers.
Figure 2. Methylation of the MIR203 promoter region.
(a) Relative miR-203 expression was determined in the indicated cell lines treated with vehicle or with 10 μM 5-azaC for eight days. (b) Methylation data from TCGA within the miR-203 promoter demonstrate a significant difference at indicated sites between triple negative and non-triple negative tumors. The above experiments were performed in at least triplicate. Error bars represent the standard deviation of the mean.
Restoring miR-203 expression induces differentiation and suppresses mesenchymal and stem cell attributes
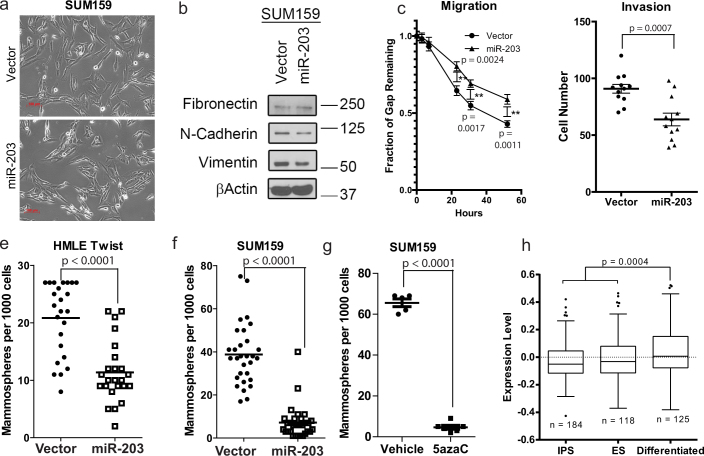
Since the EMT process is known to promote migration and invasion, we investigated the effect of restoring miR-203 expression on EMT properties. For this, we overexpressed miR-203 both in cells induced to undergo EMT (HMLE-Twist) as well as in a mesenchymal-appearing established breast cancer cell line (SUM159) using a retrovirus. In order to ensure that the overexpression of miR-203 did not affect either the cellular microRNA processing machinery or the levels of other processed microRNAs, we analyzed a spectrum of microRNAs by qRT-PCR and found that the overexpression of miR-203 did not alter either the expression of abundant or rare miRNAs (Fig. S2). Furthermore, overexpression of miR-203 did not impact cell growth in 2D cultures, in contrast to previous reports that miR-203 affects cell proliferation in other contexts27 (Fig. S3). Overexpression of miR-203 in SUM159 cells partially altered the mesenchymal morphology resulting in epithelial-looking cells (Fig. 3a) and reduced the expression of select mesenchymal markers such as N-cadherin and vimentin while not affecting fibronectin expression (Fig. 3b). To test the effect of miR-203 expression on migration and invasion, we performed in vitro wound closure and Boyden-chamber invasion assays. In concert with the observations of others that miR-203 expression is lost at the invasive front of certain cancers28, we found that miR-203 inhibited wound closure by 1.4-fold as well as the ability of cells to invade through a transmembrane by 1.4-fold relative to control vector-transduced counterparts (Fig. 3c,d).
Figure 3. Restoration of miR-203 expression abrogates certain EMT attributes including stem cell properties.
(A) retrovirus expressing miR-203 was transduced into SUM159 and HMLE-Twist cells. (a) The morphology of the SUM159 cells transduced with a control and with a miR-203 expressing vector are shown. (b) EMT-induced mesenchymal markers N-cadherin and vimentin are reduced due to miR-203 expression in SUM159 cells. (c) MiR-203 expressing SUM159 cells exhibit reduced wound-healing ability and migration. Migratory capacity was assessed by a wound healing assay. The distance between the two edges was measured at multiple positions and time points. (d) MiR-203 expressing SUM159 cells exhibit reduced invasiveness as determined by the number of cells invaded through a PET track-etched membrane. (e/f) Vector or miR-203 expressing HMLE-Twist (e) and SUM159 (f) cells were grown in mammosphere-forming conditions for 10 days and spheres with a diameter larger than 50 micrometers were counted. (g) Mammosphere formation was determined in SUM159 cells pre-treated with vehicle or 10 μM 5-azaC for eight days followed by 2 days in the absence of drug. (h) The gene expression signature of miR-203 expressing cells is compared to the gene expression signatures from differentiated, ES or iPS cells.
Since EMT and stem cell properties are interconnected, we also assessed the impact of miR-203 on CSC properties. For this, we performed a mammosphere assay, an in vitro surrogate assay of the self-renewal capabilities of mammary stem and progenitor cells29,30. Strikingly, we observed that the overexpression of miR-203 in HMLE-Twist cells as well as in SUM159 and Hs578t breast cancer cell lines reduced mammosphere formation by 2- to 6-fold (Fig. 3e/f, Fig. S4). In addition, overexpression of miR-203 doubled the proportion of differentiation-associated CD24-positive cells (Fig. S5) relative to vector-transduced SUM159 cells. Since we previously observed that the inhibition of DNA methylation using 5-azaC resulted in increased miR-203 expression, we also assessed whether restoration of miR-203 expression by 5-azaC treatment could recapitulate the impact of exogenous miR-203 overexpression on sphere formation. For this, we treated SUM159 cells with the DNA methylation blocker 5-azaC for 8 days followed by 48 hours in the absence of drug. As expected, drug-treated cells expressed higher level of miR-203 (Fig. 2a) and lost the ability to form spheres compared to vehicle treated cells (Fig. 3g). Additionally, we analyzed the gene expression signature of miR-203-overexpressing SUM159 cells and found that it associated more closely with gene expression profiles derived from differentiated as opposed to ES, or iPS cells (Fig. 3h).
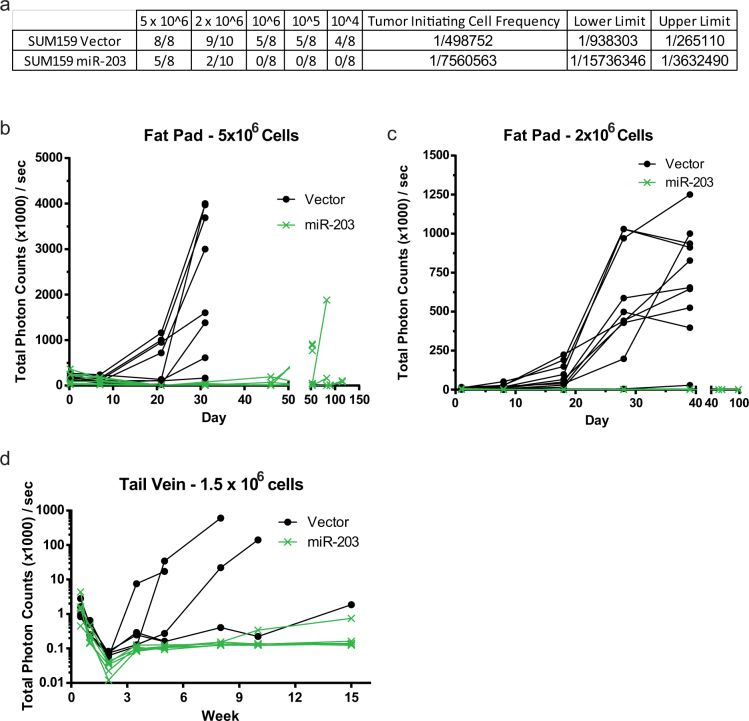
Both tumor formation and dissemination of cancer cells depend on EMT and CSC properties31. Therefore, a crucial test for the suppressive effect of miR-203 expression on stem cell properties is its ability to inhibit tumor formation in limiting dilution tumor initiation assays. Indeed, we found that as few as 104 SUM159 control cells were sufficient to form tumors within 60 days, whereas 2 × 106 SUM159 cells expressing miR-203 were necessary to form tumors within the same timeframe (Fig. 4a). Furthermore, 5 × 106 vector-transduced SUM159 cells grew palpable tumors within 20 days, while the same number of miR-203 expressing SUM159 cells took more than 60 days to form a palpable tumor (Fig. 4b), despite equivalent levels of luciferase activity observed at the time of inoculation of these cells into the mammary fat-pad (Fig. S6). Injection of 2 × 106 cells yielded similar results whereupon vector-transduced cells formed tumors between 30 and 40 days and miR-203 expressing SUM159 cells failed to form tumors up to 100 days after injection (Fig. 4c). Through extreme limiting dilution analysis32, we found that the tumor initiating cell frequency for the vector-transduced cells is 1/498,752 whereas the frequency for the miR-203 transduced cells is 1/7,560,563 ( = 44.2, P < .0001) (Fig. 4a). These findings show that stable expression of miR-203 dramatically decreases tumor formation and latency of the tumorigenic breast cancer cell line SUM159.
= 44.2, P < .0001) (Fig. 4a). These findings show that stable expression of miR-203 dramatically decreases tumor formation and latency of the tumorigenic breast cancer cell line SUM159.
Figure 4. Restoration of miR-203 expression blocks tumor formation and experimental metastasis in mice.
(a) Indicated numbers of luciferase-labeled, control and miR-203 SUM159 cells were injected into the mammary fat pads of NOD/Scid mice. Tumors larger than 0.5 cm, formed within 70 days, were counted. Limiting dilution assays were performed on a total of 42 cultures (injection sites) in each of the two populations, SUM159 Vector and SUM159 miR-203. The estimate of the tumor initiating cell frequency in the Vector group is 1/498752 with a 95% confidence interval 1/938303 to 1/265110. The estimate of tumor initiating cell frequency in the miR-203 group is 1/7560563 with a 95% confidence interval 1/15736346 to 1/3632490. The estimated cell frequencies between the Vector and miR-203 populations are significantly different ( = 44.2, P < .0001). (b/c) Timecourse of luciferase activity following fat pad injection of 5 × 106 (b) or 2 × 106 (c) control and miR-203 expressing SUM159 cells. (d) SUM159 vector and miR-203 expressing luciferase-labeled cells were injected into the tail vein of NOD/SCID mice and clearance from the lung and metastasis formation was monitored by luciferase activity.
= 44.2, P < .0001). (b/c) Timecourse of luciferase activity following fat pad injection of 5 × 106 (b) or 2 × 106 (c) control and miR-203 expressing SUM159 cells. (d) SUM159 vector and miR-203 expressing luciferase-labeled cells were injected into the tail vein of NOD/SCID mice and clearance from the lung and metastasis formation was monitored by luciferase activity.
Since EMT is known to facilitate and regulate metastasis, we also evaluated the effect of miR-203 expression on experimental metastasis. For this, we injected 1.5 × 106 luciferase-labeled SUM159 cells expressing either miR-203 or the control vector into mice via the tail vein. We found that mice injected with the vector-transduced cells developed metastases within 4–5 weeks, at which time the mice were sacrificed. Conversely, mice injected with miR-203 expressing cells failed to develop metastases in the lung even after 15 weeks (Fig. 4d). Consistent with this, a recent report indicated that metastasis-prone Lewis lung carcinoma cells express low levels of miR-20333. Collectively, these data demonstrate that expression of miR-203 promotes differentiation and lends strong support for the potential use of miR-203 as a therapeutic agent to promote differentiation.
Microarray analysis reveals novel putative miR-203 downstream targets
In order to investigate the molecular pathways downstream of miR-203 that potentially mediate its effects, we performed gene expression arrays using miR-203 expressing SUM159 cells relative to vector-transduced control counterparts. To identify the genes that mediate the effect of miR-203 downregulation during an EMT, we focused on genes which are highly downregulated in miR-203 expressing cells and at the same time, are upregulated in cells induced to undergo EMT in response to multiple different EMT-inducing stimuli, using the EMT core signature reported previously34. With these selection criteria, we found that overexpression of miR-203 reduces the expression of 112 genes by at least 1.5 fold and that only 8 out of these 112 genes are also upregulated following EMT in the EMT core signature (Fig. 5a). Next, we confirmed the negative effect of miR-203 on expression of these genes by quantitative RT-PCR. Among these eight genes, we confirmed downregulation of five genes (and only two to a significant degree) following miR-203 expression (Fig. 5b) and that three of these five downregulated genes contain interaction sites for miR-203 in their 3′UTR's by miRanda (Fig. 5c)35. Next, we knocked down the five miR-203 downregulated genes using shRNA in SUM159 cells and found that a decrease in three of these genes (NEBL, PPAP2B and TFPI) was able to significantly reduce sphere formation (Fig. 5d), phenocopying the effect of miR-203, while knockdown of the other genes did not affect sphere formation. Interestingly, expression of one of these genes (TFPI) is significantly higher in the mesenchymal-like, claudin-low breast cancer subtype. Nevertheless, re-expression of these genes was not sufficient to rescue the effect of miR-203 on sphere formation (data not shown) suggesting that the moderate suppression of multiple genes by miR-203 is responsible for the decreased stemness phenotype.
Figure 5. Individual targets of miR-203 are partially responsible for the effect of miR-203 on mammosphere formation.
(a) The list of genes downregulated by at least 1.5 fold due to miR-203 expression in SUM159 cells was compared to the list of genes upregulated by a set of five EMT inducers34. Eight genes were common to the two lists. (b) qRT-PCR was performed to confirm the downregulation of selected genes due to miR-203 expression in SUM159 cells. Only the presented genes showed any level of downregulation. Only NID1 and PPAP2B were downregulated to a statistically significant extent. (c) Three of these genes contain predicted miR-203 interaction sites according to miRanda35. (d) SUM159 cells with the indicated knockdown vectors were subjected to a mammosphere assay. (e) High expression of TFPI is significantly associated with the claudin-low and normal-like breast cancer subtype.
Paracrine effects of miR-203 via the secreted factor DKK1 affects β-catenin
We next tested whether the suppressive effect of miR-203 on sphere formation was intrinsic to the cell in which it is expressed or if expression of miR-203 could lead to suppressive effects on neighboring cells in a paracrine fashion. For this, we labeled control and miR-203 expressing cells with GFP and RFP respectively and mixed an equal number of these cells under normal cell culture conditions. Similar to culturing of these cells individually (Fig. S3), there was no significant difference in their representation up to 45 hours after plating (Fig. 6a). It is known that cells with stem cell properties are capable of surviving better in suspension culture relative to more differentiated cells36. To investigate whether miR-203 expressing cells undergo differentiation and are unable to evade anoikis and thus survive less well in suspension, we mixed GFP-labeled miR-203 cells and RFP-labeled control cells in a 1:1 ratio and cultured them in suspension using agar coated plates. Remarkably, the number of viable miR-203 expressing cells decreased to less than a third within 45 hours (Fig. 6b). In order to ensure that the effect that we observed is not due to the GFP or RFP, we reversed the color labels and observed similar results (data not shown). These findings indicate that, consistent with our earlier observation, overexpression of miR-203 induces differentiation and reduces stem cell properties.
Figure 6. DKK1 produced by miR-203 expressing cell suppresses the sphere-forming capacity of SUM159 cells through down-regulation of β-catenin.
(a) Control cells labeled with GFP and miR-203 expressing cells labeled with RFP were mixed at a 1:1 ratio under standard cell culture conditions, followed by periodic FACS analysis. (b) Control cells labeled with GFP and miR-203 expressing cells labeled with RFP were mixed at a 1:1 ratio under non-attachment cell culture conditions, followed by periodic FACS analysis. (c) 1000 control cells labeled with GFP and 1000 miR-203 expressing cells labeled with RFP were subjected to sphere formation. (d) 500 control cells labeled with GFP and 500 miR-203 expressing cells labeled with RFP were mixed and subjected to mammosphere-forming conditions. In these co-culture experiments, results were compared to expected values based on the sphere-forming efficiency of the corresponding cells grown independent of each other. (e) DKK1 expression was measured in SUM159 vector and SUM159 miR-203 cells by microarray. (f) Recombinant DKK1 reduces mammosphere formation by SUM159 cells. SUM159 cells were treated with vehicle or 200 ng/ml DKK1 for 48 hours followed by mammosphere growth in the presence of vehicle or DKK1. (g/h) The effect of miR-203 on β-catenin expression was determined by Western blot (g) and immunofluoresence (h).
Similar to our earlier observation, the RFP-labeled miR-203 expressing cells formed significantly fewer spheres relative to vector-transduced cells (Fig. 6c). Moreover, when we mixed RFP-labeled miR-203 expressing cells (500 cells) with GFP-labeled vector-transduced cells (500 cells), we observed entire spheres made of either RFP+ or GFP+ cells even though both GFP and RFP labeled cells were mixed together for this assay. This is due to the presence of methylcellulose. However, relative to control vector-transduced SUM159 cells cultured alone, we observed at least a 1.4 fold reduction in the number of spheres formed by these control cells when miR-203 expressing cells were added to the culture (Fig. 6d). Notably, reversing the color labeling did not change the outcome (data not shown). These results suggest that the miR-203 expressing cells reduce the sphere forming ability of control cells in co-culture conditions, potentially in a paracrine manner. Analysis of our microarray data revealed that expression of the Wnt antagonist DKK137,38 was significantly higher in miR-203 expressing SUM159 cells (Fig. 6e). To test whether DKK1 could inhibit sphere formation, we treated parental SUM159 cells with recombinant DKK1 and found that the sphere formation decreased by nearly two fold (Fig. 6f). As DKK1 inhibits Wnt signaling by preventing Frizzled-Wnt-LRP6 complex formation leading to decreased β-catenin stability39, we next examined the effect of miR-203 expression on β-catenin expression by Western blot and immunofluoresence. Indeed we found that β-catenin expression was decreased in SUM159 cells expressing miR-203 compared to vector-transduced cells (Fig 6g/h). Together, these results indicate that miR-203's paracrine effect on stemness properties is mediated, at least in part, through inhibition of the Wnt signaling pathway.
Discussion
The role of EMT in cancer progression is becoming increasingly well understood40. In addition to cellular detachment and increased migratory capacity, EMT has also been correlated with the acquisition of stemness properties that contribute to metastatic competence4,41. However, molecules that function downstream of multiple EMT pathways and that contribute specifically to a gain in stemness properties have not yet been uncovered. A Snail- or Twist- induced EMT leads to profound changes in gene expression and cellular behavior26,34 including the repression of the miR-200 family of microRNAs, which are linked to both the EMT and stemness properties42,43. However, we observed that miR-203, which is repressed to an even greater degree than miR-200 family members, is linked directly to the EMT-generated stemness properties.
Prior studies have linked the loss of miR-203 expression to stem cell properties and EMT in several contexts27,28,44,45,46. However, our study is the first to show: 1) miR-203 silencing via DNA methylation in breast cancer cell lines 2) the ability of miR-203 to markedly reduce tumor initiation in breast cancer cell lines and 3) the ability of miR-203 to upregulate DKK1 and downregulate β-catenin expression and thereby affect the stemness properties of neighboring cells. While miR-203 has been shown to reduce the expression of a number of targets in other contexts (most notably p63, Bmi1 and Snai2)23,42,44, the expression of these genes was not affected by expression of miR-203 in HMLE Twist or SUM159 cells. MicroRNA target selection depends on the profile of expressed mRNAs in a particular cell. In SUM159 breast cancer cells miR-203 affects a unique gene set leading to upregulation of the Wnt inhibitor DKK1, seemingly independently of its effects on other established targets.
Herein, we describe how suppression of miR-203, downstream of an EMT, is essential for stemness properties, yet largely independent of other mesenchymal attributes. This is significant as recent work has demonstrated that EMT reversal is important for the final step in the metastatic cascade, namely the progression of micrometastases to macrometastatic nodules5,47. Therefore, therapies focused on a complete reversal of EMT may inadvertently promote the growth of nascent micrometastases. Instead of complete EMT reversal, expression of miR-203 in mesenchymal cells affects only select properties. Collectively, the repression of miR-203 in EMT/CSC-enriched cell lines as well as triple-negative breast tumors and the ability to restore its expression following treatment with 5-azaC in HMLE-Snail and -Twist cells–despite the continued expression of Snail or Twist–suggest that miR-203 expression is downregulated epigenetically by methylation of its promoter. This could lead to the use of miR-203, or epigenetic therapy to relieve miR-203 repression, as a selective inhibitor of stemness properties in cancer treatment.
Methods
MicroRNA microarray, data processing, and statistical methods
Quantification of microRNA expression by microarray was performed as in48. Briefly, 5 μg of RNA from each cell type was labeled with biotin and hybridization was carried out on a miRNA-chip (ArrayExpress accession number A-MEXP-25849), which contained 238 probes for mature miRNA and 143 probes for precursor miRNAs. Hybridization signals were detected by biotin binding of a Streptavidin-Alexa647 conjugate (one-color signal) using a GenePix 4000B scanner (Axon Instruments). Images were quantified using the GenePix Pro 6.0 (Axon Instruments). Raw data were analyzed in BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam (Version: 3.6.1,May 2008; National Cancer Institute) http://linus.nci.nih.gov/BRB-ArrayTools.html. Probes were normalized over the entire array. A two-sample T-test (with random variance model) was used to analyze each comparison.
The ELDA webtool was used to perform extreme limiting dilution analyses. Unlike the standard least squares regression approach, the maximum likelihood (ML) method underlying ELDA does not ignore 0% or 100% positive cultures as often appear in limiting dilution assays. ELDA computes a maximum likelihood estimate and 95% confidence interval for the active frequency of tumor initiating cells in defined cell populations and performs a diagnostic test on the assumed Poisson single-hit model.
Cell culture and drug treatments
Immortalized human mammary epithelial cells (HMLE), including cells expressing the empty vector (pWZL), Snail, Twist, Goosecoid or TGF-β1 were maintained as in4. Established human breast cancer cell lines were cultured in cell specific media as in 5-azacytidine (5-azaC; Sigma) was used at a concentration of 10 μM, dissolved as per manufacturer's instructions.
DNA methylation analysis
DREAM was performed as reported previously50. Briefly, genomic DNA was sequentially digested with a pair of enzymes recognizing the same restriction site (CCCGGG) containing a CpG dinucleotide. The first enzyme, SmaI, cuts only at unmethylated CpG and leaves blunt ends. The second enzyme, XmaI, is not blocked by methylation and leaves a short 5′ overhang. The enzymes thus create methylation-specific signatures at the ends of digested DNA fragments. These are deciphered by next generation sequencing using the Illumina Gene Analyzer II and Hiseq2000 platforms. Methylation levels for each sequenced restriction site are calculated based on the numbers of DNA molecules with the methylated or unmethylated signatures. Overall, we acquired around 36 million sequence tags per sample that were mapped to unique CpG sites in the human genome using hg18 version. Details of the DREAM method were previously reported in50.
Antibodies, immunoblotting, immunofluoresence and flow cytometry
For immunoblotting, proteins were extracted by lysing cells in ice-cold radio-immunoprecipitation (RIPA) buffer containing protease and phosphatase inhibitors (Roche). Protein was quantified using the Bradford Assay (BioRad). Cell lysates (50 μg) were resolved using SDS-PAGE and transferred to PVDF membranes. Membranes were probed with primary antibodies against fibronectin (BD Biosciences), vimentin (Neomarkers) and β-actin (Abcam). Immunofluoresence was performed as described in4 with anti-β-catenin (BD Biosciences). Fluorescence activated cell sorting for CD44 and CD24 cell surface marker expression was performed as described in4 using the Flow Cytometry and Cellular Imaging Facility at MD Anderson Cancer Center.
microRNA and mRNA quantitation and statistical methods
RT-PCR for microRNAs was performed using miR-specific primers from Applied Biosystems. Quantitative PCR was performed on an Applied Biosystems Viia7 using SYBR for mRNAs and Taqman for microRNAs. All experiments were performed with at least three biological replicates with at least three technical replicates each and the results are reported as the mean of the biological replicates plus or minus the standard error of the mean. In all cases *** denotes p < 0.001, ** denotes p < 0.01, and * denotes p < 0.05.
Microarray
For changes in mRNA expression we harvested RNA using Trizol (Ambion). Reverse transcription, hybridization to the Human Genome U133 Plus 2.0 microarray (Affymetrix) and image processing were performed by SeqWright Inc, Houston, TX. Data from this microarray is available at GSE50697.
Growth, migration and invasion assays
Cell growth rates were determined by counting cells after the indicated number of days in culture. For migration cells were serum-starved overnight and scratch wounds were created using a sterile pipette tip on the cell monolayer. The distance between the two edges at multiple points was quantified using Photoshop CS4 (Adobe) at the indicated timepoints. For invasion, the number of cells invaded through a PET track-etched membrane towards serum-containing media was quantified by staining with methylene blue.
Sphere and non-adherent culture
Mammospheres were grown as in29,30. Spheres larger than 50 micrometers were counted. For measuring cell survival under non-adherent conditions, plates were coated with 1% agarose and cells were plated in their own media. The proportion of surviving RFP+ and GFP+ cells was determined by FACS. In co-culture experiments, we used the number of spheres formed by each cell line on their own to calculate the expected number of spheres from co-culture of the labeled cells.
Animal studies
NOD/SCID mice were purchased from the Jackson Laboratory (Maine, USA). All mouse procedures were approved by the Animal Care and Use Committees of MD Anderson Cancer Center under protocol #10-10-08531 and performed in accordance with institutional policies. For xenograft tumor-initiation studies, luciferase-labeled cells suspended in DMEM were mixed 1:1 with matrigel and then injected into the inguinal or thoracic fat pads of NOD/SCID mice. For experimental metastasis studies, luciferase-labeled cells were suspended in PBS for tail vein injection. Mice were assessed periodically for tumor growth via the intraperitoneal injection of D-Luciferin (Caliper LifeSciences) at 150 mg/kg in PBS and bioluminescent imaging using the IVIS imaging system 200 series (Xenogen Corporation). All mice were monitored for tumor growth and lung metastasis. Once mammary gland tumors reached 1.5 cm in diameter, mice were euthanized and their organs harvested and fixed using Bouin's fixative.
Associations with differentiated cell gene expression and triple negative breast cancer
To evaluate the methylation of the miR-203 promoter in breast cancer, we acquired the methylation data (Illumina Human Methylation 450 array) for 672 breast tumor samples in TCGA by downloading the beta values (a measure of the percent methylation) processed by the Broad Firehose pipeline. We distinguished the tumors into ones that are triple-negative and others using the clinical annotations provided by the TCGA. Then, using annotations from Illumina, we identified the probes targeting the region upstream of the miR-203 transcription start site. We compared the methylation levels of the triple negative and non-triple negative tumors using a two-sided t-Test with unequal variance.
To score the association of miR-203 with stem cells, we generated a gene expression signature that consists of the genes with at least a 2-fold change in gene expression and are correlated with the expression of miR-203. Using a SAM analysis51 we found 66 genes that are positively correlated with miR-203, and 65 negatively correlated genes. Then, we collected data from the Gene Expression Omnibus 25 Affymetrix gene expression data sets that contained both differentiated cells and either embryonic stem (ES) or induced pluripotent stem (iPS) cells. We preprocessed each data set with RMA and normalized each gene to mean 0 and variance 1. Then, to derive the miR-203 score, we averaged the positively correlated genes with the inverse of the negatively correlated genes. We compared the miR-203 scores of the differentiated cells and stem cells using a t-Test with unequal variance.
Author Contributions
J.H.T. carried out the molecular genetics and cellular studies, performed statistical analysis, designed the study, and drafted the manuscript. E.L. and J.T.C. carried out the bioinformatics and performed statistical analysis. M.N., S.R. and G.A.C. performed the microRNA microarray. P.P.R., V.V., R.U. and S.G. performed cell experiments. G.G.M. and J.J.I. performed the DREAM analysis. J.I.H. and J.M.R. performed the claudin-low association. S.A.M. designed the study and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Figures
Acknowledgments
We acknowledge the entire Mani Lab for invaluable discussion and advice. Also, we appreciate the assistance of Dr. Rudy Guerra of Rice University for additional statistical analysis. This work was supported by DOD Postdoctoral Fellowship BC101115 (JHT). JTC is supported by NIH R00LM009837 and grant R1006 from the Cancer Prevention and Research Institute of Texas. Flow cytometry, and animal imaging were in part funded by the Cancer Center Support Grant from the National Cancer Institute (CA16672). This work was supported in part by an MD Anderson Research Trust Fellow Award, funded by the George and Barbara Bush Endowment for Innovative Cancer Research (SAM). GM was also funded by the following non-for-profit organizations: Fondation Nadine Midy, Foundation Nelia et Amadeo Barletta (F.N.A.B.) and the Association pour l'Aide à'la Recherche et l'Enseignement en Cancérologie (A.A.R.E.C.).
References
- Kalluri R. & Weinberg R. A. The basics of epithelial-mesenchymal transition. J Clin Invest 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K. & Weinberg R. A. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9, 265–273 (2009). [DOI] [PubMed] [Google Scholar]
- Thiery J. P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2, 442–454 (2002). [DOI] [PubMed] [Google Scholar]
- Mani S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. H., Donaher J. L., Murphy D. A., Chau S. & Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn D. M. et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 4, e7181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J. R., Gabrielson E., Fujii H., Baylin S. B. & Herman J. G. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem 275, 2727–2732 (2000). [DOI] [PubMed] [Google Scholar]
- Wu C. Y., Tsai Y. P., Wu M. Z., Teng S. C. & Wu K. J. Epigenetic reprogramming and post-transcriptional regulation during the epithelial-mesenchymal transition. Trends Genet 28, 454–463 (2012). [DOI] [PubMed] [Google Scholar]
- Wu M. Z. et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell 43, 811–822 (2011). [DOI] [PubMed] [Google Scholar]
- McDonald O. G., Wu H., Timp W., Doi A. & Feinberg A. P. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol 18, 867–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. & Shang Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp Cell Res 319, 160–169 (2013). [DOI] [PubMed] [Google Scholar]
- Kanai Y., Ushijima S., Tsuda H., Sakamoto M. & Hirohashi S. Aberrant DNA methylation precedes loss of heterozygosity on chromosome 16 in chronic hepatitis and liver cirrhosis. Cancer Lett 148, 73–80 (2000). [DOI] [PubMed] [Google Scholar]
- Nass S. J. et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res 60, 4346–4348 (2000). [PubMed] [Google Scholar]
- Bullock M. D., Sayan A. E., Packham G. K. & Mirnezami A. H. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell 104, 3–12 (2012). [DOI] [PubMed] [Google Scholar]
- Nicoloso M. S., Spizzo R., Shimizu M., Rossi S. & Calin G. A. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer 9, 293–302 (2009). [DOI] [PubMed] [Google Scholar]
- Park S. M., Gaur A. B., Lengyel E. & Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22, 894–907 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10, 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- Bracken C. P. et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68, 7846–7854 (2008). [DOI] [PubMed] [Google Scholar]
- Vrba L. et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One 5, e8697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves R. et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes 3, 219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez C. S. et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res 71, 3087–3097 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S. et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res 72, 6435–6446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Poy M. N., Stoffel M. & Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452, 225–229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. J. et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development 140, 1882–1891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B. et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 15, 50–65 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004). [DOI] [PubMed] [Google Scholar]
- Wang C., Zheng X., Shen C. & Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res 31, 58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kempen L. C. et al. Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch 461, 441–448 (2012). [DOI] [PubMed] [Google Scholar]
- Dontu G. et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17, 1253–1270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J. & Clarke M. F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. & Weinberg R. A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- Hu Y. & Smyth G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- Rask L., Fregil M., Hogdall E., Mitchelmore C. & Eriksen J. Development of a metastatic fluorescent Lewis Lung carcinoma mouse model: Identification of mRNAs and microRNAs involved in tumor invasion. Gene 517, 72–81 (2013). [DOI] [PubMed] [Google Scholar]
- Taube J. H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A 107, 15449–15454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D. S. & Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 36, D149–153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberio R. et al. Keratinocytes enriched for stem cells are protected from anoikis via an integrin signaling pathway in a Bcl-2 dependent manner. FEBS Lett 524, 139–144 (2002). [DOI] [PubMed] [Google Scholar]
- Kuhnert F. et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A 101, 266–271 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A., Liu G., Goldin L., Harris V. & Aaronson S. A. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell 6, 497–506 (2004). [DOI] [PubMed] [Google Scholar]
- Li Y., Lu W., King T. D., Liu C. C., Bijur G. N. & Bu G. Dkk1 stabilizes Wnt co-receptor LRP6: implication for Wnt ligand-induced LRP6 down-regulation. PLoS One 5, e11014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B. & Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13, 97–110 (2013). [DOI] [PubMed] [Google Scholar]
- Morel A. P., Lievre M., Thomas C., Hinkal G., Ansieau S. & Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 3, e2888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U. et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11, 1487–1495 (2009). [DOI] [PubMed] [Google Scholar]
- Shimono Y. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y. et al. MiR-182 and miR-203 induce mesenchymal to epithelial transition and self-sufficiency of growth signals via repressing SNAI2 in prostate cells. Int J Cancer 133, 544–555 (2013). [DOI] [PubMed] [Google Scholar]
- DeCastro A. J. et al. MiR203 mediates subversion of stem cell properties during mammary epithelial differentiation via repression of DeltaNP63alpha and promotes mesenchymal-to-epithelial transition. Cell Death Dis 4, e514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Park S. I., McCauley L. K. & Wang C. Y. Signaling between Transforming Growth Factor beta (TGF-beta) and Transcription Factor SNAI2 Represses Expression of MicroRNA miR-203 to Promote Epithelial-Mesenchymal Transition and Tumor Metastasis. J Biol Chem 288, 10241–10253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M. et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med 17, 1101–1108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A. et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 41, 365–370 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Calin G. A. & Croce C. M. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol 427, 193–213 (2007). [DOI] [PubMed] [Google Scholar]
- Challen G. A. et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44, 23–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R. & Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures