The Drs2 flippase increases membrane curvature and anionic phospholipid composition of the membrane by flipping phosphatidylserine, which is critical for vesicular transport between the trans-Golgi network and early endosomes.
Abstract
Vesicle-mediated protein transport between organelles of the secretory and endocytic pathways is strongly influenced by the composition and organization of membrane lipids. In budding yeast, protein transport between the trans-Golgi network (TGN) and early endosome (EE) requires Drs2, a phospholipid translocase in the type IV P-type ATPase family. However, downstream effectors of Drs2 and specific phospholipid substrate requirements for protein transport in this pathway are unknown. Here, we show that the Arf GTPase-activating protein (ArfGAP) Gcs1 is a Drs2 effector that requires a variant of the ArfGAP lipid packing sensor (+ALPS) motif for localization to TGN/EE membranes. Drs2 increases membrane curvature and anionic phospholipid composition of the cytosolic leaflet, both of which are sensed by the +ALPS motif. Using mutant forms of Drs2 and the related protein Dnf1, which alter their ability to recognize phosphatidylserine, we show that translocation of this substrate to the cytosolic leaflet is essential for +ALPS binding and vesicular transport between the EE and the TGN.
Introduction
The membranous organelles of eukaryotic cells have unique lipid compositions that greatly influence their function (Andersen and Koeppe, 2007; Phillips et al., 2009; Alexander et al., 2011; Iwamoto and Oiki, 2013). Component lipids not only provide the physical barrier role of the membrane, but also interact with membrane proteins to control their localization and activity. A variety of integral membrane proteins are known to be acutely sensitive to their lipid surroundings, and many peripheral membrane proteins rely on specific phospholipid headgroup interactions for membrane association. Moreover, the phospholipid composition of each leaflet of a membrane bilayer can be strikingly different, providing two distinct surfaces for protein interactions. The cytosolic leaflet of the plasma membrane, for example, is enriched in phosphatidylserine (PS) and phosphatidylethanolamine (PE), whereas these phospholipids are nearly absent in the extracellular leaflet (van Meer et al., 2008; Leventis and Grinstein, 2010). This asymmetric membrane organization is produced by the action of type IV P-type ATPases (P4-ATPases), which flip PS and PE across the membrane to the cytosolic leaflet (van Meer et al., 2008; Leventis and Grinstein, 2010).
The P4-ATPases responsible for generating membrane asymmetry also have crucial functions in vesicular transport pathways emanating from Golgi and endosomal membranes (Sebastian et al., 2012). The precise role P4-ATPases play in vesicle formation is unclear, although it is thought that the unidirectional transport of phospholipid from the luminal leaflet to the cytosolic leaflet bends the membrane, which contributes to the membrane curvature needed to bud small-diameter vesicles or tubular carriers. It is also possible that enrichment of specific phospholipid molecules in the cytosolic leaflet controls the localization and/or function of vesicle budding machinery recruited from the cytosol (Graham, 2004). The best characterized P4-ATPases for their functions in establishing membrane asymmetry and vesicular transport are Drs2 and Dnf1 from budding yeast. Drs2 localizes to the TGN, where it flips PS and PE from the luminal leaflet to the cytosolic leaflet (Natarajan et al., 2004). Drs2 flippase activity, along with Arf-GTP, is also required to bud transport vesicles coated with clathrin and the clathrin adaptor AP-1 (adaptor protein 1) from the TGN (Liu et al., 2008). Drs2 is incorporated into these AP-1 vesicles and delivered to an early endosome (EE), where its flippase activity is required to form retrograde vesicles that deliver Drs2 and an exocytic SNARE, Snc1, back to the TGN (Hua et al., 2002; Furuta et al., 2007; Liu et al., 2008). Dnf1 localizes to the TGN, EEs, and plasma membrane (Hua et al., 2002; Takagi et al., 2012), and preferentially flips phosphatidylcholine (PC) and PE (Pomorski et al., 2003; Baldridge and Graham, 2012; Hachiro et al., 2013). Although Dnf1 contributes to vesicular transport pathways exiting the Golgi, Dnf1 cannot replace Drs2 function in forming AP-1/clathrin-coated vesicles or the Snc1 retrograde vesicles (Liu et al., 2008). Whether the strict requirement for Drs2 in the AP-1 and Snc1 pathways is caused by a unique substrate, localization, or set of protein interactions is unknown. In addition, downstream effectors of Drs2 flippase activity have not been identified.
Here, we show that the Arf GTPase-activating protein (ArfGAP) Gcs1 is a downstream effector of Drs2 PS flippase activity at TGN/EE. Gcs1 contains an amphipathic, α-helical ArfGAP lipid packing sensor (ALPS) motif that is an exquisite sensor of membrane curvature, although the function of this ALPS motif in vivo has not been characterized (Bigay et al., 2005). We find that this ALPS motif is necessary but not sufficient for recruitment of Gcs1 from the cytosol to TGN/EE membranes. However, a slightly larger Gcs1 fragment containing a critical basic residue along with the ALPS motif was sufficient for membrane association. This new variant of the ALPS motif, which we refer to as a +ALPS, senses both membrane curvature and negative charge imparted to the cytosolic leaflet by Drs2 flippase activity. We have recently mapped residues that define substrate specificity of Drs2 and Dnf1. A point mutation in Dnf1 that allows it to recognize and flip PS (Dnf1-N550S; Baldridge and Graham, 2013; Baldridge et al., 2013) also allows Dnf1 to replace the function of Drs2 in recruiting Gcs1 to TGN/EE membranes. Remarkably, Dnf1-N550S effectively replaces the function of Drs2 in protein transport between the TGN and EE, demonstrating a crucial role for PS translocation in these trafficking pathways.
Results
Gcs1 endosomal localization is dependent on the +ALPS motif and Drs2
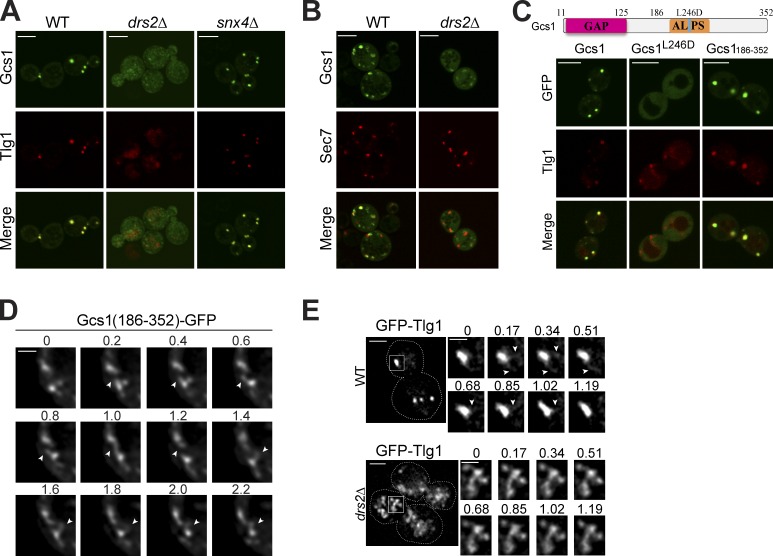
Gcs1 is an ArfGAP in budding yeast that localizes to cytoplasmic punctae, which colocalize with the TGN/EE marker mCherry-Tlg1 and the TGN marker Sec7-DsRed (Fig. 1, A and B; Robinson et al., 2006). In addition, Gcs1 and Drs2 have been functionally linked through genetic interaction studies and both contribute to protein transport from EEs to the TGN (Hua et al., 2002; Sakane et al., 2006). Therefore, we tested if Drs2 influenced the peripheral membrane association of Gcs1 with the EE and TGN. Indeed, Gcs1-GFP was significantly dispersed throughout the cytoplasm of drs2Δ cells, and the punctae that remained did not colocalize with Tlg1 or Sec7 (Fig. 1, A and B). As previously reported, the compact, punctate morphology of the EE was also perturbed and Tlg1 localized to enlarged structures or the vacuole (Fig. S1; Furuta et al., 2007). Similar to drs2Δ, the snx4Δ mutant also has a strong defect in Snc1 recycling from the EE to the TGN (Hettema et al., 2003). No vesicle coat protein is known to act in the Snc1 recycling pathway, and Snx4, with its PX-BAR domain, could be another potential driver of membrane curvature in this pathway. But unlike drs2Δ, Gcs1 localized normally to the TGN/EE in the snx4Δ strain, and TGN/EE morphology was not perturbed (Fig. 1 A).
Figure 1.
Drs2 flippase activity and the ALPS motif are required for Gcs1 TGN/EE localization. (A) Gcs1-GFP localization in wild-type (WT), drs2Δ, and snx4Δ cells relative to mCherry-Tlg1. (B) Gcs1-GFP localization in WT and drs2Δ cells relative to Sec7-DsRed. (C) GFP-tagged Gcs1, Gcs1L246D, and Gcs1(186–352) expressed in WT cells were imaged relative to mCherry-Tlg1. (D) Time-lapse images of Gcs1(186–352)-GFP decorating small diameter tubules (arrows) extending from TGN/EE. Time is indicated in seconds. (E) Membrane dynamics of TGN/EE in WT and drs2Δ cells. GFP-Tlg1 was imaged for several seconds (Videos 1 and 2), and time-lapse series for the boxed TGN/EE membranes are shown. Time is indicated in seconds. Bars: (A–C) 5 µm; (D) 1 µm; (E, main panels) 2 µm; (E, time lapse) 1 µm.
To test whether localization of Gcs1 is influenced by its ALPS motif, we examined the localization of a Gcs1 L246D mutant, in which a conserved leucine within the ALPS motif was substituted for an aspartic acid to disrupt the amphipathic motif (Fig. 1 C). Mutation of the analogous leucine in the ArfGAP1 ALPS motif abrogates interaction with curved membranes (Bigay et al., 2005; Mesmin et al., 2007). In wild-type cells, Gcs1-GFP resided on TGN/EE membranes as it colocalized with mCherry-Tlg1. In contrast, Gcs1-GFP harboring the L246D point mutation was diffusely localized in the cytoplasm. In addition, the C-terminal half of Gcs1 (Gcs1186–352) containing the ALPS motif was sufficient to target GFP to TGN/EE membranes (Fig. 1 C). The membranes decorated with Gcs1-GFP were highly dynamic organelles that rapidly extended small diameter tubular protrusions (Fig. 1 D). Thus, Gcs1 binds highly curved membranes in vivo in a Drs2- and ALPS-dependent manner.
To assess the influence of Drs2 on membrane dynamics, we acquired time-lapse movies of GFP-Tlg1 with high spatial and temporal resolution in wild-type and drs2Δ cells (Fig. 1 E and Videos 1 and 2). Again, the wild-type cells displayed compact TGN/EEs that extended small-diameter tubules that rapidly fragmented and disappeared. In contrast, the drs2Δ membranes marked with GFP-Tlg1 were significantly larger membrane networks that were less active in forming tubules than wild-type TGN/EEs (Fig. 1 E). Tubules could be observed extending from these drs2Δ membranes (Fig. 1 E, arrowheads), although these tubules typically had a larger diameter and were slow to fragment. By electron microscopy, the endosomes in Drs2-deficient cells are substantially enlarged tubular or cup-shaped structures with 200–250-nm diameters (Furuta et al., 2007), whereas the EE in wild-type cells is a tubular-vesicular membrane with diameters closer to 100 nm (Mulholland et al., 1999; Prescianotto-Baschong and Riezman, 2002).
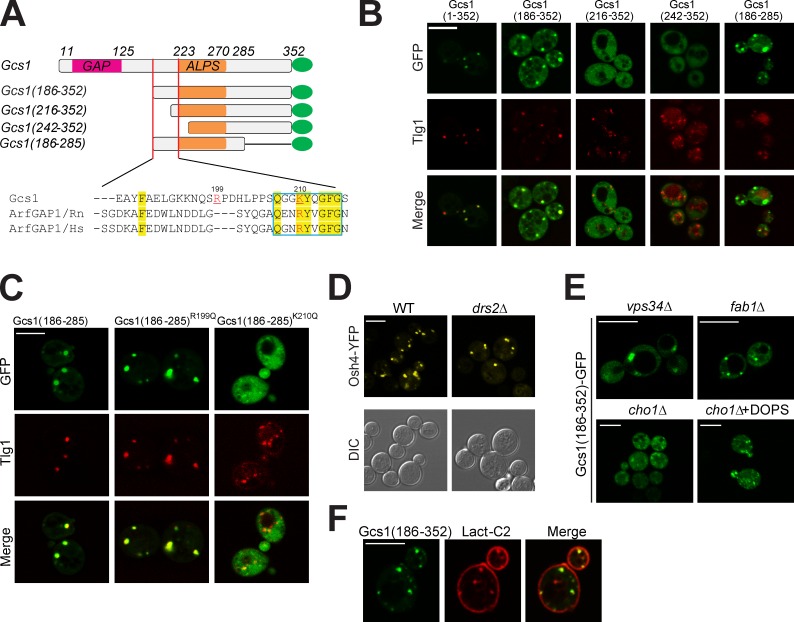
Many yeast organelles have highly curved membranes and yet Gcs1 is recruited preferentially to TGN/EE membranes, implying the presence of additional localization determinants in Gcs1. Therefore, we mapped the minimal sequence required for membrane association and found that residues 186–285 in Gcs1 were sufficient (Fig. 2, A and B). In addition to the ALPS motif (residues 223–270), the sequence from 186 to 216 was also critical for TGN/EE localization. The Drs2 requirement suggested that anionic phospholipids contributed to membrane recruitment of Gcs1 and so we targeted two basic amino acids in the region 186–216. Indeed, the K210Q mutation, within a small motif well conserved in mammalian ArfGAPs (Fig. 2 A), markedly perturbed membrane association of Gcs1(186–285)-GFP, whereas the R199Q mutation had no effect (Fig. 2 C). We refer to this new ALPS variant as a +ALPS motif to highlight the importance of a positively charged residue upstream of ALPS for membrane association. Osh4 contains an ALPS motif and localizes to the TGN (Drin et al., 2007). However, deletion of Drs2 has no influence on Osh4 membrane association, implying a specific relationship between Drs2 and the +ALPS motif in Gcs1 (Fig. 2 D).
Figure 2.
Gcs1 TGN/EE localization requires its +ALPS motif. (A) Diagram of truncated Gcs1-tagged GFP. The positive charged resides Lysine at position 210 and Arginine at position 199 are highlighted in red. Conserved sequences between Gcs1 and mammalian ArfGAP1 are highlighted in yellow. (B) Besides the ALPS motif, a conserved region between the GAP domain and ALPS motif (residues 186–223) is required for Gcs1 TGN/EE localization. Truncated Gcs1-tagged GFP expressed in wild-type (WT) cells and imaged relative to mCherry-Tlg1. (C) Mutation of K210 in Gcs1 disrupts TGN/EE localization. GFP-tagged Gcs1(186–285), Gcs1(186–285)R199Q, and Gcs1(186–285)K210Q expressed in WT cells and imaged relative to mCherry-Tlg1. (D) Osh4-YFP localization in WT and drs2Δ cells. (E) Gcs1(186–352)-GFP localization in mutants deficient for endosomal anionic lipids including PI3P (vps34Δ), PI(3,5)P2 (fab1Δ), and PS (cho1Δ). An aliquot of the cho1Δ cells was incubated with 1 µM DOPS/0.001% NP-40 mixed micelles for 1 h (cho1Δ+DOPS). (F) Gcs1(186–352)-GFP colocalized to the punctae with the PS biosensor mRFP-Lact-C2. Bars, 5 µm.
Phosphatidylinositol 3-phosphate (PI3P) is enriched on endosomes and plays an important role in recruiting several peripheral membrane proteins (Burd and Emr, 1998). Disruption of VPS34, encoding the sole phosphatidylinositol 3-kinase in yeast, caused enlargement of the endosomes but did not substantially perturb Gcs1(186–352)-GFP membrane association (Fig. 2 E). Neither did disruption of FAB1, the PI3P 5-kinase, indicating that endosome targeting of Gcs1 did not require PI3P or phosphatidylinositol-3, 5-bisphosphate (PI3,5P2). In contrast, a cho1Δ strain, deficient for PS synthase, displayed a primarily cytoplasmic distribution of Gcs1(186–352)-GFP. Addition of exogenous dioleoylphosphatidylserine (DOPS) to cho1Δ reversed the defect and enhanced Gcs(186–352)-GFP TGN/EE association (Fig. 2 E). PS is enriched in the cytosolic leaflet of the plasma membrane as shown by the localization of mRFP-Lact-C2, a PS biosensor (Yeung et al., 2008). mRFP-Lact-C2 also localized to cytoplasmic punctae in a small percentage of cells, which were typically decorated with GFP-Gcs1(186–352). However, GFP-Gcs1(186–352) did not associate with the PS-rich and relatively flat plasma membrane surface (Fig. 2 F).
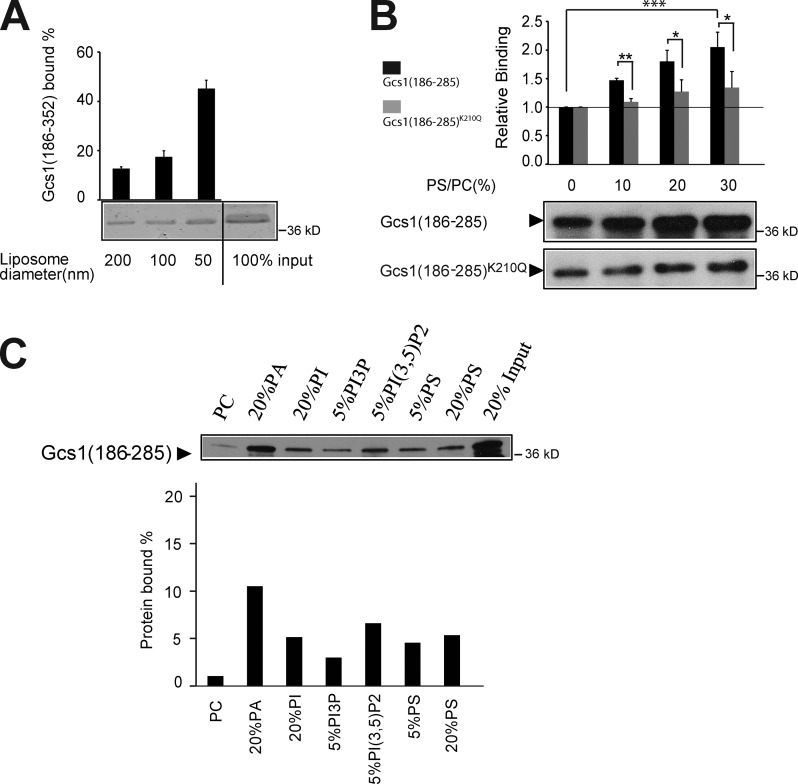
The ALPS motif shows a striking preference for interaction with highly curved membrane surfaces (Bigay et al., 2005; Drin et al., 2007). To determine if the +ALPS motif retained this binding specificity, we tested binding of Gcs1(186–352) to liposomes of 50–200 nm in diameter composed of Golgi-mix lipids (50% dioleoylphosphatidylcholine [DOPC], 19% dioleoylphosphatidylethanolamine [DOPE], 5% DOPS, 10% liver phosphatidylinositol (PI), and 16% cholesterol; Bigay et al., 2005; Fig. 3 A). The preferential binding to 50-nm liposome indicates that the +ALPS motif retains the same curvature-sensing property as the ALPS motif of ArfGAP1 (Bigay et al., 2005). We also tested the influence of electrostatic interactions on +ALPS motif binding to membranes. +ALPS bound weakly to 200-nm PC liposomes, but binding was enhanced in a PS dose-dependent manner, whereas the K210Q mutation blunted this effect (Fig. 3 B). The +ALPS binding to 100-nm PC liposomes was also enhanced by other anionic lipids, such as phosphatidic acid (PA), PI, and PI3,5P2 (Fig. 3 C). PS is likely the most abundant anionic phospholipid in the cytosolic leaflet of TGN/EE membranes and therefore has the greatest influence on +ALPS recruitment in vivo.
Figure 3.
The +ALPS motif binds highly curved membranes with anionic phospholipid. (A) GST-tagged Gcs1(186–352) (0.5 mM) was incubated with Golgi-mix liposomes (0.5-mM lipids) of decreasing size produced by extrusion through filter pores of the size indicated. After centrifugation in a density gradient to float liposomes, the top fractions were collected and analyzed by SDS-PAGE and Coomassie staining. Gcs1(186–352) in the top fraction from a liposome-free control was subtracted from the band intensities shown for quantitation. Error bars show variation observed between three independent experiments using different batches of extruded liposomes. (B) GST-tagged Gcs1(186–285) and Gcs1(186–285)K210Q were incubated with 200 nm DOPC liposomes with increasing percentages of DOPS. After centrifugation, the top fractions were collected and quantified by Western blotting using anti-GST antibody. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate mean + SD. (C) Binding of GST-Gcs1(186–285) to 100 nm DOPC liposomes with or without the indicated anionic phospholipids. Data are expressed as the percentage of input protein. The data shown are from a single representative experiment out of two repeats.
Drs2 flippase activity is acutely required for +ALPS recruitment
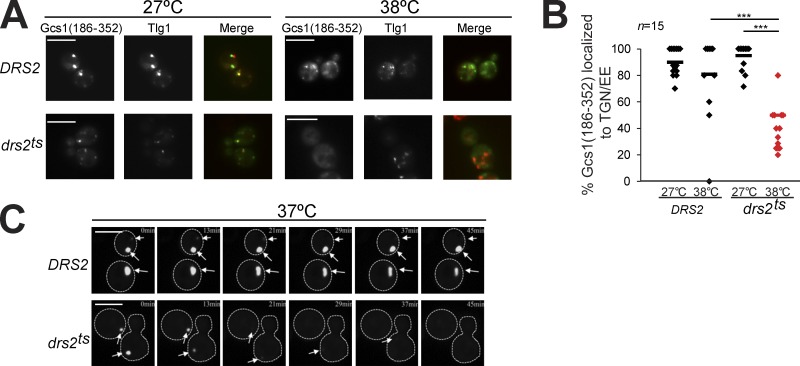
To determine if Drs2 flippase activity is acutely required for membrane recruitment of the +ALPS motif, we expressed Gcs1(186–352)-GFP in strains harboring either a wild-type copy of DRS2 or a temperature-sensitive drs2ts allele. At 27°C, Drs2ts is functional and Gcs1(186–352)-GFP localized normally to the TGN/EE with Tlg1. However, after shifting drs2ts cells to 38°C for 1 h, which inactivates flippase activity (Natarajan et al., 2004), Gcs1(186–352)-GFP membrane association was lost and the endosome became enlarged. In contrast, Gcs1(186–352)-GFP localized normally to TGN/EE membranes in an isogenic DRS2 strain that was shifted to 38°C (Fig. 4, A and B). We also monitored Gcs1-GFP localization in real time after shifting drs2ts or DRS2 cells to 37°C, and found that Gcs1-GFP dissociated from membranes as drs2ts cells warmed from room temperature (at 0 min) to ∼37°C over a 45-min period (Fig. 4 C). A GFP-tagged form of Drs2ts is stable after temperature shift and so the loss of Gcs1 membrane association upon Drs2 inactivation is unlikely to be caused by disrupting a Gcs1–Drs2 physical association. Consistently, we failed to detect the presence of Gcs1 in a Drs2 immunoprecipitate, whereas Cdc50, a known binding partner for Drs2 (Saito et al., 2004), showed a strong interaction (Fig. S2).
Figure 4.
Drs2 flippase activity is acutely required for Gcs1 endosomal localization. (A) Gcs1(186–352)-GFP localization was monitored in drs2Δ cells carrying wild-type DRS2 or a drs2 temperature-sensitive allele (drs2ts) grown at 27°C or shifted to 38°C for 1 h. Bars, 5 µm. (B) The percentage of mCherry-Tlg1 punctae showing colocalization with Gcs1(186–352)-GFP from 15 cells of each strain was quantified. ***, P < 0.001. (C) Time-lapse images of Gcs1-GFP expressed on 2μ plasmids, which exist in multiple copies per cell, in wild-type and drs2ts as cells warmed from room temperature to ∼37°C on a heated stage. Arrows indicate the positions of TGN/EE structures marked with Gcs1-GFP. Bar, 5 µm.
Gcs1 is an effector of Drs2 PS flippase activity
Gcs1 participates in Golgi-ER and post-Golgi vesicle transport in yeast (Poon et al., 2001, 2006), but also facilitates Snc1,2 retrieval from the EE to the Golgi (Robinson et al., 2006). Snc1 is a SNARE that constantly cycles between the TGN, plasma membrane, EE, and back to the TGN. In wild-type cells, the rate-limiting step in this cycle is endocytosis from the plasma membrane, and so most GFP-Snc1 localizes to the plasma membrane at steady-state. In gcs1Δ cells expressing wild-type GCS1, most cells showed typical Snc1 localization to the plasma membrane and only 8% of the cells displayed GFP-Snc1 primarily in the cytoplasm. In contrast, 38% of gcs1Δ cells (vector), 23% of gcs1-L246D cells (mutation in the Gcs1 +ALPS motif), and 30% of gcs1-R54K cells deficient for Gcs1 GAP activity showed GFP-Snc1 primarily localized to cytoplasmic punctae (Fig. 5, A and B). Thus, the ALPS motif has a measurable influence on Gcs1 activity in the Snc1 recycling pathway.
Figure 5.
Gcs1 is a Drs2 flippase effector. (A and B) Both the GAP domain and ALPS motif are important for Gcs1 function in Snc1 recycling. gcs1Δ cells expressing GFP-Snc1 and an empty vector, wild-type Gcs1, Gcs1 GAP(R54K), or Gcs1 ALPS(L246D) were cultured to mid-log phase, and GFP-Snc1was imaged by fluorescence microscopy. More than 100 cells from three independent isolates of each strain were counted and categorized for GFP-Snc1 localization to the plasma membrane primarily (black bars) or intracellular punctae (gray bars). ***, P < 0.001. Error bars indicate mean + SD. (C) Schematic representation of the Gcs1 constructs used. The region (206–281) containing ALPS was replaced with either a FYVE domain from EEA1 or a PH domain from Osh1. (D) Localization of GFP-Snc1 in drs2Δ cells containing the constructs indicated. Bars, 5 µm.
To determine if providing an alternative mechanism for ArfGAP recruitment to membranes could bypass the function of Drs2 in protein transport, we replaced the Gcs1 ALPS domain with either a FYVE domain specific for PI3P binding (Burd and Emr, 1998) or a pleckstrin homology (PH) domain from Osh1 that binds specifically to phosphatidylinositol 4-phosphate (PI4P) at the TGN (Levine and Munro, 1998; Fig. 5 C). The Gcs1-ALPS::PH protein tagged to GFP colocalized nearly completely with the TGN marker Sec7-DsRed, whereas Gcs1-ALPS::FYVE partially overlapped with mCherry-Tlg1 (Fig. S3). Neither Gcs1-ALPS::FYVE nor Gcs1-ALPS::PH could bypass drs2Δ and restore GFP-Snc1 plasma membrane localization. Together, these data indicate that the Drs2- and ALPS-dependent recruitment of Gcs1 is functionally important for EE-to-TGN protein transport, but additional effectors of Drs2 must exist to explain the role of Drs2 in this pathway.
PS translocation is required for +ALPS membrane association
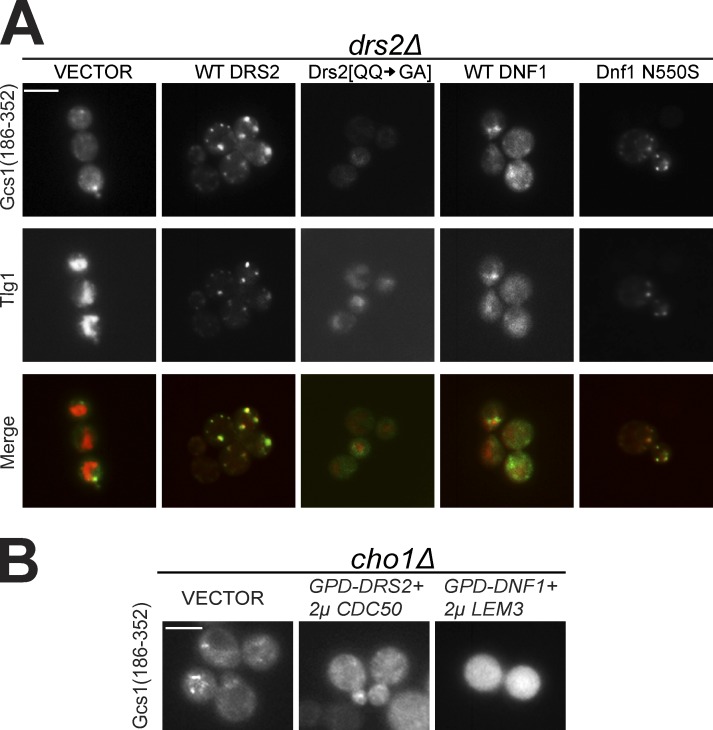
Thus far, our data imply an important role for PS translocation by Drs2 in Gcs1 membranes recruitment. To test this, we used a Drs2 mutation in the first transmembrane segment (Drs2[QQ → GA]) that specifically perturbs recognition of PS with no measurable effect on PE recognition (Baldridge and Graham, 2013). drs2Δ displayed cytoplasmic Gcs1(186–352)-GFP localization and enlarged endosome morphology, and these mutant phenotypes were complemented efficiently by a plasmid expressing wild-type Drs2. In contrast, these mutant phenotypes were not corrected by the Drs2[QQ → GA] mutant or empty vector control (Fig. 6 A). An extra copy of wild-type Dnf1 that flips PE and PC could not support recruitment of Gcs1(186–352)-GFP to membranes in drs2Δ cells, nor did it correct the TGN/EE morphology defect (Fig. 6 A). In contrast, Dnf1 N550S, which recognizes and flips PS, corrected both phenotypes as well as wild-type Drs2 for both Gcs1(186–352)-GFP and full-length Gcs1-GFP (Fig. 6 A and Fig. S4). Moreover, overexpression of Drs2 or Dnf1 failed to recruit Gcs1(186–352)-GFP to the TGN/EE membranes of a PS-deficient cho1Δ strain, implying that no other substrate of these flippases (e.g., PE or PC) can be used to recruit Gcs1 (Fig. 6 B). These data demonstrate that PS translocation by a P4-ATPase is essential for Gcs1 recruitment to TGN/EE membranes.
Figure 6.
Translocation of PS to the cytosolic leaflet by a flippase is required for Gcs1 TGN/EE localization. (A) Gcs1(186–352)-GFP localization to TGN/EE is restored in drs2Δ cells expressing wild-type (WT) Drs2, but not the PS-deficient Drs2 [QQ → GA] mutant. An extra copy of WT Dnf1 failed to suppress the Gcs1(186–352)-GFP localization defect, but the Dnf1 N550S mutant that can flip PS restored Gcs1(186–352)-GFP TGN/EE localization. (B) Neither DRS2 nor DNF1 overexpression could restore Gcs1(186–352)-GFP TGN/EE localization in PS-deficient cho1Δ cells. Bars, 5 µm.
PS translocation is required for protein transport between the EE and the TGN
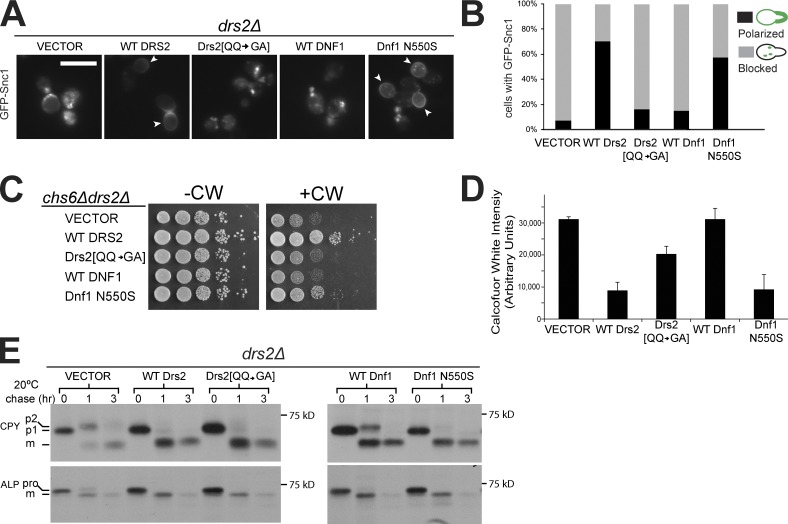
We then asked if the ability of Drs2 or Dnf1 to flip PS correlated with the ability to restore normal protein trafficking in a drs2Δ strain. Introduction of wild-type Drs2 restored plasma localization of GFP-Snc1 in most drs2Δ cells. In contrast, Drs2 [QQ → GA] was only slightly better than the empty vector at correcting this defect (Fig. 7, A and B). Although wild-type Dnf1 failed to suppress this drs2Δ defect, Dnf1 N550S restored GFP-Snc1 plasma membrane localization as well as wild-type Drs2 (Fig. 7, A and B).
Figure 7.
Translocation of PS to the cytosolic leaflet by a flippase is required for protein transport between the EE and the TGN. (A) Dnf1 N550S and wild-type (WT) Drs2 restored normal Snc1 trafficking in drs2Δ cells, whereas Drs2 [QQ → GA] and WT Dnf1 failed. Arrowheads indicate Snc1 plasma membrane localization polarized to buds. Bar, 5 µm. (B) Three independent colonies of each strain were cultured to mid-log phase and Snc1 localization was quantified. (C) Dnf1 N550S but not Drs2 [QQ → GA] restored resistance of chs6Δdrs2Δ cells to CW. (D) Quantitation of CW fluorescent staining of strains used in C. Error bars indicate mean + SD. (E) The strains indicated were pulse-labeled with [35S]methionine for 10 min and chased for 3 h at 20°C. CPY or ALP were immunoprecipitated from cells collected at 0, 1, and 3 h of chase and subjected to SDS-PAGE. WT Drs2, Drs2 [QQ → GA], and Dnf1 N550S restored normal CPY and ALP transport kinetics, whereas WT Dnf1 partially suppressed the transport defects. pro, precursor; m, mature.
Drs2 is required with AP-1 and clathrin for trafficking chitin synthase III (Chs3) between the TGN and EE. In an exomer mutant (chs6Δ), Chs3 cannot be exported to the plasma membrane, and these cells are resistant to the chitin-binding compound calcofluor white (CW; Valdivia et al., 2002). A chs6Δdrs2Δ double knockout strain restores sensitivity to CW because the AP-1 pathway is disrupted, allowing Chs3 to move to the PM constitutively (Liu et al., 2008). A chs6Δdrs2Δ strain expressing wild-type Drs2 was resistant to CW but Drs2 [QQ → GA] failed to restore CW resistance, which indicates that AP-1–dependent trafficking requires PS translocation (Fig. 7 C). An additional copy of wild-type Dnf1 was incapable of suppressing the AP-1–dependent trafficking defect in chs6Δdrs2Δ cells. In contrast, Dnf1 N550S provided increased resistance to CW. We quantified CW binding to cell wall chitin of chs6Δdrs2Δ strains expressing the flippase variants, and P4-ATPases capable of flipping PS (wild-type Drs2, Dnf1-N550S) reduced chitin deposition, whereas Drs2 [QQ→GA] and wild-type Dnf1 were much less effective (Fig. 7 D). These data indicate that PS translocation to the cytosolic leaflet is essential to support Chs3 trafficking in the AP-1 pathway.
Drs2 functions redundantly with Dnf1 to deliver carboxypeptidase Y (CPY) from the TGN to late endosome, a pathway mediated by monomeric GGA clathrin adaptors (Hua et al., 2002). To test the requirement for PS translocation in this pathway, we evaluated the influence of P4-ATPase variants on CPY transport kinetics with a pulse-chase assay. The drs2Δ strain displayed a kinetic delay in CPY proteolytic processing (p2 precursor conversion to mature [m] form) at 20°C, and Drs2[QQ → GA] corrected the CPY trafficking defect as well as wild-type Drs2, which indicates that PS flip is not necessary to support this pathway (Fig. 7 E). Although an additional copy of wild-type Dnf1 partially corrected the CPY transport defect, Dnf1 N550S completely restored the CPY trafficking, indicating that PS translocation can enhance CPY trafficking even though it is not required (Fig. 7 E). These results are consistent with the prior observation that CPY transport kinetics are normal in a cho1Δ strain (Natarajan et al., 2004). We also examined the influence of PS translocation on AP-3 adaptor function in delivering alkaline phosphatase (ALP) from the Golgi to the vacuole, a pathway where Drs2 and Dnf1 also act redundantly (Hua et al., 2002). For ALP, the kinetics of precursor (pro) to mature (m) form proteolytic processing were also delayed in drs2Δ cells at 20°C (vector), but in this case each of the Drs2 and Dnf1 variants could correct the delay in AP-3–mediated transport to the vacuole (Fig. 7 E). Thus, PS translocation by a P4-ATPase is crucial for bidirectional protein transport between the Golgi and EE, but other flippase substrates can be used to support GGA and AP-3 trafficking routes.
Discussion
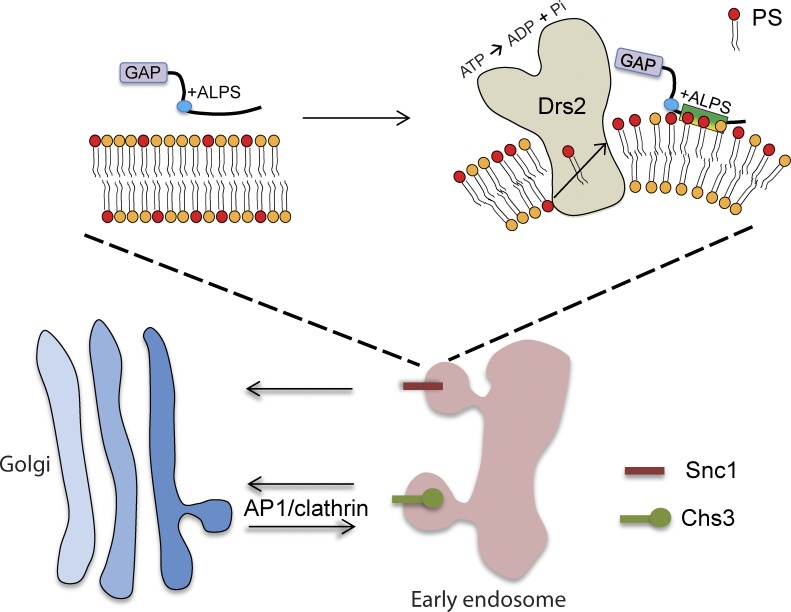
The vesicular transport machinery has a remarkable capacity for deforming biological membranes into tightly curved transport vesicles or tubular carriers with diameters of 50–100 nm (McMahon and Gallop, 2005; Graham and Kozlov, 2010). Although several distinct mechanisms for generating this curvature are possible, there is a limited set of tools available to address the contribution of each mechanism to specific transport pathways in living cells. For example, it is a matter of debate whether Bin/Amphiphysin/Rvs167 (BAR) domains induce membrane curvature in vivo or preferentially bind to membranes with the appropriate curvature (Galic et al., 2012). In this regard, the ALPS motif is particularly useful because it lacks the capacity to deform membranes and is a well-defined sensor of membrane curvature (Bigay and Antonny, 2012). The ALPS motif was first described in ArfGAP1, although no in vivo function had been reported for this motif (Bigay et al., 2005). Here, we show that a point mutation in the Gcs1 ALPS motif that disrupts association with curved membranes also disrupts TGN/EE localization and Gcs1 function in the EE-to-TGN protein transport pathway. In addition, our data indicate that the +ALPS variant in Gcs1 senses both curvature and negative charge imparted to the TGN/EE membrane by P4-ATPases with the ability to flip PS to the cytosolic leaflet (Fig. 8). Our data strongly imply that the Drs2- and +ALPS-dependent membrane recruitment of Gcs1 is functionally significant and that Gcs1 is an effector of Drs2 PS translocase activity. This PS flippase activity is essential to support vesicle-mediated protein transport between EE and the TGN, providing a strong correlation between the membrane-bending activity of Drs2 and vesicular transport.
Figure 8.
Model for the role Drs2 PS flippase activity plays in generating membrane curvature and negative charge in the cytosolic leaflet that is sensed by the +ALPS motif of Gcs1. PS flip is crucial for the AP-1–dependent trafficking of Chs3 between the TGN and EE, as well as the AP-1–independent transport of Snc1 from the EE to the TGN.
The PS specificity for +ALPS of Gcs1 recruitment to the TGN/EE in vivo is surprising when considering the promiscuous interaction of +ALPS with a variety of anionic phospholipids in liposome binding studies. However, dissociation of Gcs1 from TGN/EE membranes in PS-deficient cho1Δ cells may primarily result from loss of curvature rather than a dramatic change in anionic phospholipid composition. In these cells, Drs2 activity would be diminished by the lack of favored substrate, and the uncharged natural substrates of Dnf1 would not be as effective in recruiting the +ALPS motif. In addition, other effectors of PS in the TGN/EE cytosolic leaflet are almost certainly present, and these factors could help stabilize the Gcs1 membrane association, enhancing the specificity of its localization.
The requirement of PS translocation in protein transport
PS is known to stimulate the Arf-GTP–dependent assembly of AP-1 and clathrin onto liposomes (Zhu et al., 1999). Consistently, we found that PS translocation by Drs2 is required for AP-1/clathrin function in yeast. However, it is striking that Arf, AP-1, and clathrin appear to be recruited normally to the TGN of drs2Δ cells, even though AP-1/clathrin function in protein transport appears to be abolished (Liu et al., 2008). Both Arf-GTP and clathrin are thought to drive membrane curvature during vesicle budding, and yet the highly tubular Golgi cisternae and endosomes collapse into enlarged cup-shaped structures, as viewed by electron microscopy, when Drs2 is inactivated (Chen et al., 1999; Gall et al., 2002; Saito et al., 2004). These observations strongly argue that penetration of the Arf1-GTP amphipathic helix into the TGN membrane, along with the association of AP-1 and clathrin, is insufficient to drive curvature in the membrane in the absence of PS translocation by Drs2. We suggest that the requirement of Drs2 in the AP-1 pathway is primarily to induce membrane curvature that can be molded into vesicles by clathrin. There also appears to be an undefined PS-specific effector acting in the AP-1 pathway because GGA and AP-3 function, while still requiring flippase activity, is much less dependent on PS translocation.
PS is also known to have an essential function in protein transport from recycling endosomes to the TGN in mammalian cells (Uchida et al., 2011). In this case, PS concentrated on the cytosolic leaflet is important for recruiting evectin-2, a PS-binding protein, to the endosomal membrane. Depletion of evectin-2 or overexpression of Lact-C2 (to mask PS) specifically perturbed the recycling endosome–to–TGN retrograde pathway (Uchida et al., 2011). There is no obvious homologue to evectin-2 in yeast, although our data indicate that PS flip is required in an analogous endosome to the TGN pathway, and perhaps other PS effectors, in addition to Gcs1, act in this pathway. No vesicle coat protein is known to act in the EE-to-TGN transport pathway. The only other potential driver of curvature in this pathway is the PX-BAR domain of the Snx4 sorting nexin (Hettema et al., 2003). However, Gcs1-GFP is recruited normally to the endosome of snx4Δ cells even though trafficking of Snc1-GFP is perturbed (Hettema et al., 2003). None of the 14 human P4-ATPases have yet been implicated in endosome-to-TGN protein transport, although mutations in the Drs2 orthologue ATP8A2 cause intellectual disability in humans and neurodegeneration in mice (Zhu et al., 2012; Onat et al., 2013). Similar pathologies are found in human patients with an AP-1 subunit mutation or mice deficient for a GARP subunit involved in endosome-to-TGN transport (Schmitt-John et al., 2005; Tarpey et al., 2006). In addition, the Caenorhabditis elegans Drs2 orthologue Tat-1 localizes to recycling endosomes and is required for its tubular morphology and protein trafficking through this organelle (Ruaud et al., 2009; Chen et al., 2010). Thus, PS translocation by P4-ATPases appears to be a highly conserved mechanism for inducing membrane deformation to support vesicular transport.
Materials and methods
Yeast strains and plasmid construction
Strains used in this study are listed in Table S1. Standard media and techniques for growing and transforming yeast were used (Sherman, 1991; Gietz and Schiestl, 2007). Plasmid constructions were performed using standard molecular manipulation. Point mutations were introduced by site-directed mutagenesis using the QuikChange II kit (Agilent Technologies). p416-mCherry-Tlg1 was provided by D. Katzmann (Mayo Clinic, Rochester, MN). A complete list of plasmids is provided in Table S2.
Cell imaging
To visualize GFP- or mCherry-tagged proteins, cells were grown to early-to-mid-logarithmic phase, harvested, and resuspended in imaging buffer (10 mM Na2PHO4, 156 mM NaCl, 2 mM KH2PO4, and 2% glucose). Cells were mounted on glass slides and observed immediately at room temperature. Micrographs were acquired using a TCS-SP5 scanning confocal microscope equipped with a 63× objective lens (Leica) and a camera using AF Lite software (both from Leica) or using an microscope (Axioplan; Carl Zeiss) equipped with 63× objective lens with a charge-coupled device (CCD) camera using MetaMorph 4.5 software (Molecular Devices). Overlay images were taken using Merge Channels functions of ImageJ software.
Time-lapse live cell microscopy
Cells were grown to mid-log phase, concentrated, and mounted on a microscope slide in synthetic media. Time-lapse movies of Gcs1(186–352) in drs2ts and wild-type DRS2 cells were acquired using a microscope (TE2000; Nikon) with a spinning disk confocal head equipped with 63× oil immersion lens and a cooled CCD camera (Princeton Instruments). Temperature was controlled with a temperature controller. Images were acquired by means of commercially available software (IPLab Spectrum, version 3.1; Scan Analytics). Images were further assembled using ImageJ.
Time-lapse movies of TGN/EE membrane were acquired using a DeltaVision Elite workstation (Applied Precision) based on an inverted microscope (IX-70; Olympus) equipped with a 100×, 1.4 NA oil immersion lens. Videos were captured (1 frame/ 200 ms) at room temperature (24°C) with a 12-bit charge-coupled device camera (CoolSNAP HQ; Photometrics) and deconvolved using an iterative-constrained algorithm and the measured point spread function (softWoRx 5.5 software; Applied Precision).
Growth and induction of protein expression cultures
BL21-CodenPlus@(DE3)-RIL (Agilent Technologies) Escherichia coli cells containing plasmids encoding each fusion protein were grown in 1,000 ml of Luria-Bertani media containing 100 mg/ml ampicillin in a 2-liter flask shaken at 240 rpm at 37°C to an OD600 of 0.5. For the GST-Gcs1(186–352) fusion proteins, expression was induced with 1 mM IPTG and cell growth was continued for 3 h at 30°C. Expression of the GST-Gcs1(186–285), GST-Gcs1(186–285)R199Q, and GST-Gcs1(186–285)K210Q fusion proteins was induced with 1 mM IPTG, and cell growth was continued at 240 rpm at 30°C for 1 h. Cells were harvested by centrifugation at 5,000 g for 10 min and stored at −80°C.
Purification of GST fusion proteins
Cells were lysed with GST lysis buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, pH 8.0, 0.5% Nondidet P-40, 2 µg/µl aprotinin, 0.7 µg/ml pepstatin, and 25 µg/ml PMSF) for 30 min on ice and centrifuged at 12,000 g for 10 min after sonication cycles (sonicate 30 s, rest 10 s) 10 times. The supernatant was incubated with glutathione-agarose beads (Sigma-Aldrich) for 2 h at 4°C. The beads were washed in a column with 10 bed volumes of washing buffer (20 mM Tris-HCl and 200 mM NaCl, pH 8.0), then eluted in 1-ml fractions with GST elution buffer (50 mM Tris-HCl and 20 mM reduced glutathione, pH 9.5). The protein was equilibrated to neutral buffer (20 mM Tris-HCl and 100 mM NaCl, pH 6.8) using dialysis, and the detergent was completely removed using Bio-Beads SM-2 (Bio-Rad Laboratories). Protein concentration and purity was measured by standard gel electrophoresis and comparison to BSA standards. Protein samples were immediately flash frozen before being stored at −80°C.
Liposome preparation
Lipids in chloroform were purchased from Avanti Polar Lipids, Inc. Liposomes used in flotation and binding assays were produced by the extrusion as described previously (Bigay et al., 2005). In brief, a dried film prepared by evaporation of a mixture of the indicated lipids in chloroform was resuspended in 20 mM Tris-HCl and 100 mM NaCl, pH 6.8. After five steps of freezing in dry ice and thawing, the liposome suspension was extruded through 200-nm or 100-nm polycarbonate filters using a hand extruder (Avanti Polar Lipids, Inc.) at a final lipid concentration of 1 mM. For preparation of 50-nm liposomes, the liposome suspension sequentially extruded through 0.2-, 0.1-, and 0.05-µm (pore size) polycarbonate filters. The liposome size was confirmed by negative stain EM. Liposomes were stored at room temperature and used at the same day after preparation. The composition of Golgi-mix liposomes was 50% DOPC, 19% DOPE, 5% DOPS, 10% liver phosphatidylinositol, 16% cholesterol, and 0.2% NBD-PS (mol%).
Liposome binding assay
0.5-µM proteins and 0.5–1-µM liposomes were incubated in flotation buffer (20 mM Tris-HCl, 100 mM NaCl, pH 6.8, and 1 mM DTT) at room temperature for 5 min in a total volume of 150 µl. The suspension was adjusted to 30% sucrose by adding and mixing 100 µl of a 75% wt/vol sucrose solution. The resulting high-sucrose suspension was overlaid with 200 µl of flotation buffer containing 25% wt/vol sucrose and 50 µl of flotation buffer without any sucrose. The sample was centrifuged at 55,000 rpm (240,000 g) in a swinging bucket rotor (TLS 55; Beckman Coulter) for 1 h. The top layer (50 µl) was collected and analyzed by Western blotting using the anti-GST antibody (1:4,000) or Coomassie blue staining. Bands were quantified using the Odyssey Image System (LI-COR Biosciences).
Immunoprecipitation and Western blotting
Immunoprecipitation was performed as described previously (Saito et al., 2004) with the following modifications. Cells in 400 ml of minimal medium were harvested when grown to 0.5 OD600/ml at 30°C. The pellet was washed twice with ice-cold water and resuspended in 1 ml of lysis buffer (10 mM Tris-HCl, pH 7.5, 0.3 M sorbitol, 0.1 M NaCl, and 5 mM MgCl2) containing protease inhibitors (1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 2 mM benzamidine, and 1 mM PMSF) per 200 OD600. Cells were lysed by using acid-washed glass beads (vortex 10 times; each time, vortex 30 s and rest 30 s). The supernatant taken by centrifuging at 1,000 g for 5 min was then centrifuged at 100,000 g for 1 h at 4°C using a rotor (TLS55; Beckman Coulter). The pellets were taken as the total membranes. Membrane pellets were solubilized in 0.8 ml of immunoprecipitation buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% CHAPS) containing protease inhibitors. Then pellets were discarded after the centrifugation at 20,000 g for 10 min at 4°C. The supernatants were separated into two equal parts: one of them was incubated with Drs2 antiserum (0.5 µl per OD600) overnight. We then incubated these samples with 20 µl of protein G–Sepharose 4 Fast Flow (GE Healthcare) for 1 h at 4°C. The protein G–Sepharose beads were then pelleted and washed three times with immunoprecipitation buffer without detergents. The beads were finally heated with SDS-urea sample buffer at 65°C for 5 min. The immunoprecipitates were separated by SDS-PAGE after the Western blot. Membranes were then incubated with anti-HA antibody (1:4,000) at room temperature for 1 h. After three washes with TBS with Tween 20 (TBST), the membrane was incubated in anti–mouse IgG-HRP (1:2,000 in TBST) for 2 h at room temperature. After three washes in TBST, membranes were visualized by enhanced chemiluminescence (PerkinElmer). Protein bands were detected using scientific imaging film (Kodak).
Pulse-chase labeling and immunoprecipitation of CPY
This procedure was performed as described previously (Graham, 2001). We subcultured starting at 0.15 OD600/ml and allowed cells to grow to mid-log phase. The cells were pelleted and resuspended at 5 OD600/ml, then we labeled 1-OD600 cells with 25 µCi EXPRESS35S (PerkinElmer) at 20°C for 10 min. The chase was performed with 1 mM methionine and 0.1 mM cysteine for the indicated times. To stop the reaction, we added one-fourth volume of 50% TCA. The reactions were incubated on ice for 15 min, then centrifuged at top speed for 10 min. The supernatant was aspirated and the remaining pellets were resuspended in 2 volumes of ice-cold acetone, and recentrifuged for 10 min. The acetone wash was repeated, and the pellets were dried to completion in a SpeedVac system (Thermo Fischer Scientific). We resuspended the pellets in 100 µl SDS/urea buffer (50 mM Tris-Cl, pH 7.5, 1 mM EDTA, 1% SDS, and 6 M urea) and incubated at room temperature for 15 min. Glass beads were added to ∼70% volume, vortexed for 1 min, heated to 85°C for 4 min, and vortexed for 15 s. We added 900 µl of detergent IP buffer (50 mM Tris Cl, pH 7.5, 0.1 mM EDTA, 150 mM NaCl, and 0.5% Tween-20), the reactions were pelleted at top speed for 15 min, and ∼850 µl of supernatant was transferred to a new tube. To immunoprecipitate protein from these samples, we added 75 µl protein A–Sepharose CL-4B and 1–2 µl of antibody per OD600 cell. The reactions were incubated overnight with end-over-end rotation at 4°C. The samples were centrifuged at 3,000 g for 5 min, and the pellets were then resuspended in detergent/urea buffer (100 mM Tris Cl, pH 7.5, 200 mM NaCl, 0.5% Tween-20, and 2 M urea) and repelleted. The supernatants were cleared with a second batch of protein A–Sepharose and kept for a second immunoprecipitation. The pellets were resuspended a second time in detergent/urea buffer, repelleted, resuspended in detergent IP buffer, repelleted, and dried in a SpeedVac system. The dried protein A–Sepharose was resuspended in Laemmli sample buffer with fresh 5% β-mercaptoethanol (vol/vol) and heated to 80°C for 5 min before separation by SDS-PAGE.
CW staining
Overnight cultures were subcultured to 0.15 OD600/ml and allowed to grow to mid-log phase. 1,000 µl of cells were pelleted, resuspended in water, repelleted, and resuspended in 100 µl of a 0.05% CW solution (wt/vol; BD) for 10 min. The cells were washed (pelleted and resuspended) twice with water and finally suspended in imaging buffer (10 mM Tris-HCl, pH 7.4, and 2% glucose [wt/vol]). For quantification, mid-log phase cells were stained at 0.1 OD600/ml in 1 mg/ml CW in 10 mM Tris, pH 7.5, for 10 min at room temperature. The cells were pelleted, washed once with water, and transferred to a 96-well plate at a final concentration of 0.01 OD600/ml, and the fluorescence signal intensity was measured using excitation at 360/40 and emission at 460/40 with a multimode plate reader (Synergy HT; BioTek).
Statistical analysis
Results are expressed as the mean + SD from the specified number of experiments, as indicated in the figure legends. Student’s t test was used to analyze statistical significance.
Online supplemental material
Fig. S1 displays the mCherry-Tlg1 localization in wild-type and drs2Δ cells. Fig. S2 shows no physical interaction between Drs2 and Gcs1 by coimmunoprecipitation. Fig. S3 displays Gcs1-ALPS::FYVE and Gcs1-ALPS::PH localization in wild-type yeast cells. Fig. S4 shows wild-type Gcs1 localization affected by Drs2 mutants and Dnf1 mutants. Table S1 lists the yeast stains used in this study. Table S2 lists the plasmids used in this study. Video 1 shows GFP-Tlg1 dynamics in wild-type cells. Video 2 shows GFP-Tlg1 dynamics in drs2Δ cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305094/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201305094.dv.
Supplementary Material
Acknowledgments
We thank Pak Poon, Gerald Johnston, Susan Wente, David Katzmann, Sergio Grinstein, Chris Beh, and David Cortez for yeast strains and plasmids. We thank Dr. Xiaodong Zhu for help with the confocal imaging taken from Vanderbilt Medical Center Core Facility. We also thank our laboratory colleagues for comments on this manuscript.
These studies were supported by National Institutes of Health grant R01GM062367 (to T.R. Graham).
Footnotes
Abbreviations used in this paper:
- ALP
- alkaline phosphatase
- ALPS
- Arf GTPase-activating protein lipid packing sensor
- AP-1
- adaptor protein 1
- ArfGAP
- Arf GTPase-activating protein
- Chs3
- chitin synthase III
- CPY
- carboxypeptidase Y
- CW
- calcofluor white
- DOPC
- dioleoylphosphatidylcholine
- DOPS
- dioleoylphosphatidylserine
- EE
- early endosome
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PH
- pleckstrin homology
- PI3P
- phosphatidylinositol 3-phosphate
- PS
- phosphatidylserine
References
- Alexander R.T., Jaumouillé V., Yeung T., Furuya W., Peltekova I., Boucher A., Zasloff M., Orlowski J., Grinstein S. 2011. Membrane surface charge dictates the structure and function of the epithelial Na+/H+ exchanger. EMBO J. 30:679–691 10.1038/emboj.2010.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen O.S., Koeppe R.E., II 2007. Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 36:107–130 10.1146/annurev.biophys.36.040306.132643 [DOI] [PubMed] [Google Scholar]
- Baldridge R.D., Graham T.R. 2012. Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. USA. 109:E290–E298 10.1073/pnas.1115725109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge R.D., Graham T.R. 2013. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl. Acad. Sci. USA. 110:E358–E367 10.1073/pnas.1216948110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge R.D., Xu P., Graham T.R. 2013. Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 288:19516–19527 10.1074/jbc.M113.476911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J., Antonny B. 2012. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 23:886–895 10.1016/j.devcel.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Bigay J., Casella J.F., Drin G., Mesmin B., Antonny B. 2005. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 24:2244–2253 10.1038/sj.emboj.7600714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G., Emr S.D. 1998. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 2:157–162 10.1016/S1097-2765(00)80125-2 [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Ingram M.F., Rosal P.H., Graham T.R. 1999. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147:1223–1236 10.1083/jcb.147.6.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Jiang Y., Zeng S., Yan J., Li X., Zhang Y., Zou W., Wang X. 2010. Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet. 6:e1001235 10.1371/journal.pgen.1001235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. 2007. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell. 18:295–312 10.1091/mbc.E06-05-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic M., Jeong S., Tsai F.C., Joubert L.M., Wu Y.I., Hahn K.M., Cui Y., Meyer T. 2012. External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat. Cell Biol. 14:874–881 10.1038/ncb2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall W.E., Geething N.C., Hua Z., Ingram M.F., Liu K., Chen S.I., Graham T.R. 2002. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 12:1623–1627 10.1016/S0960-9822(02)01148-X [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2:31–34 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- Graham T.R. 2001. Metabolic labeling and immunoprecipitation of yeast proteins. Curr. Protoc. Cell Biol. Chapter 7:6. [DOI] [PubMed] [Google Scholar]
- Graham T.R. 2004. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 14:670–677 10.1016/j.tcb.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Graham T.R., Kozlov M.M. 2010. Interplay of proteins and lipids in generating membrane curvature. Curr. Opin. Cell Biol. 22:430–436 10.1016/j.ceb.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiro T., Yamamoto T., Nakano K., Tanaka K. 2013. Phospholipid flippases Lem3p-Dnf1p and Lem3p-Dnf2p are involved in the sorting of the tryptophan permease Tat2p in yeast. J. Biol. Chem. 288:3594–3608 10.1074/jbc.M112.416263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Lewis M.J., Black M.W., Pelham H.R. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22:548–557 10.1093/emboj/cdg062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Fatheddin P., Graham T.R. 2002. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell. 13:3162–3177 10.1091/mbc.E02-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Oiki S. 2013. Amphipathic antenna of an inward rectifier K+ channel responds to changes in the inner membrane leaflet. Proc. Natl. Acad. Sci. USA. 110:749–754 10.1073/pnas.1217323110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis P.A., Grinstein S. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39:407–427 10.1146/annurev.biophys.093008.131234 [DOI] [PubMed] [Google Scholar]
- Levine T.P., Munro S. 1998. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8:729–739 10.1016/S0960-9822(98)70296-9 [DOI] [PubMed] [Google Scholar]
- Liu K., Surendhran K., Nothwehr S.F., Graham T.R. 2008. P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol. Biol. Cell. 19:3526–3535 10.1091/mbc.E08-01-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Gallop J.L. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- Mesmin B., Drin G., Levi S., Rawet M., Cassel D., Bigay J., Antonny B. 2007. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 46:1779–1790 10.1021/bi062288w [DOI] [PubMed] [Google Scholar]
- Mulholland J., Konopka J., Singer-Kruger B., Zerial M., Botstein D. 1999. Visualization of receptor-mediated endocytosis in yeast. Mol. Biol. Cell. 10:799–817 10.1091/mbc.10.3.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P., Wang J., Hua Z., Graham T.R. 2004. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. USA. 101:10614–10619 10.1073/pnas.0404146101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onat O.E., Gulsuner S., Bilguvar K., Nazli Basak A., Topaloglu H., Tan M., Tan U., Gunel M., Ozcelik T. 2013. Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur. J. Hum. Genet. 21:281–285 10.1038/ejhg.2012.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R., Ursell T., Wiggins P., Sens P. 2009. Emerging roles for lipids in shaping membrane-protein function. Nature. 459:379–385 10.1038/nature08147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski T., Lombardi R., Riezman H., Devaux P.F., van Meer G., Holthuis J.C. 2003. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 14:1240–1254 10.1091/mbc.E02-08-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon P.P., Wang X., Rotman M., Huber I., Cukierman E., Cassel D., Singer R.A., Johnston G.C. 1996. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA. 93:10074–10077 10.1073/pnas.93.19.10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon P.P., Nothwehr S.F., Singer R.A., Johnston G.C. 2001. The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J. Cell Biol. 155:1239–1250 10.1083/jcb.200108075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescianotto-Baschong C., Riezman H. 2002. Ordering of compartments in the yeast endocytic pathway. Traffic. 3:37–49 10.1034/j.1600-0854.2002.30106.x [DOI] [PubMed] [Google Scholar]
- Robinson M., Poon P.P., Schindler C., Murray L.E., Kama R., Gabriely G., Singer R.A., Spang A., Johnston G.C., Gerst J.E. 2006. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell. 17:1845–1858 10.1091/mbc.E05-09-0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud A.F., Nilsson L., Richard F., Larsen M.K., Bessereau J.L., Tuck S. 2009. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 10:88–100 10.1111/j.1600-0854.2008.00844.x [DOI] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. 2004. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:3418–3432 10.1091/mbc.E03-11-0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane H., Yamamoto T., Tanaka K. 2006. The functional relationship between the Cdc50p-Drs2p putative aminophospholipid translocase and the Arf GAP Gcs1p in vesicle formation in the retrieval pathway from yeast early endosomes to the TGN. Cell Struct. Funct. 31:87–108 10.1247/csf.06021 [DOI] [PubMed] [Google Scholar]
- Schmitt-John T., Drepper C., Mussmann A., Hahn P., Kuhlmann M., Thiel C., Hafner M., Lengeling A., Heimann P., Jones J.M., et al. 2005. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat. Genet. 37:1213–1215 10.1038/ng1661 [DOI] [PubMed] [Google Scholar]
- Sebastian T.T., Baldridge R.D., Xu P., Graham T.R. 2012. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta. 1821:1068–1077 10.1016/j.bbalip.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 10.1016/0076-6879(91)94004-V [DOI] [PubMed] [Google Scholar]
- Takagi K., Iwamoto K., Kobayashi S., Horiuchi H., Fukuda R., Ohta A. 2012. Involvement of Golgi-associated retrograde protein complex in the recycling of the putative Dnf aminophospholipid flippases in yeast. Biochem. Biophys. Res. Commun. 417:490–494 10.1016/j.bbrc.2011.11.147 [DOI] [PubMed] [Google Scholar]
- Tarpey P.S., Stevens C., Teague J., Edkins S., O’Meara S., Avis T., Barthorpe S., Buck G., Butler A., Cole J., et al. 2006. Mutations in the gene encoding the Sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am. J. Hum. Genet. 79:1119–1124 10.1086/510137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y., Hasegawa J., Chinnapen D., Inoue T., Okazaki S., Kato R., Wakatsuki S., Misaki R., Koike M., Uchiyama Y., et al. 2011. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA. 108:15846–15851 10.1073/pnas.1109101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R.H., Baggott D., Chuang J.S., Schekman R.W. 2002. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2:283–294 10.1016/S1534-5807(02)00127-2 [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T., Gilbert G.E., Shi J., Silvius J., Kapus A., Grinstein S. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 319:210–213 10.1126/science.1152066 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Drake M.T., Kornfeld S. 1999. ADP-ribosylation factor 1 dependent clathrin-coat assembly on synthetic liposomes. Proc. Natl. Acad. Sci. USA. 96:5013–5018 10.1073/pnas.96.9.5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Libby R.T., de Vries W.N., Smith R.S., Wright D.L., Bronson R.T., Seburn K.L., John S.W. 2012. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8:e1002853 10.1371/journal.pgen.1002853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.