Abstract
Preclinical and clinical studies showed that autologous transplantation of epidermis derived from genetically modified epithelial stem cells (EpSCs) leads to long-term correction of inherited skin adhesion defects. These studies were based on potentially genotoxic retroviral vectors. We developed an alternative gene transfer strategy aimed at targeting a “safe harbor” locus, the adeno-associated virus integration site 1 (AAVS1), by zinc-finger nuclease (ZFN)-induced homologous recombination (HR). Delivery of AAVS1-specific ZFNs and a GFP-expressing HR cassette by integration-defective lentiviral (LV) vectors (IDLVs) or adenoviral (Ad) vectors resulted in targeted gene addition with an efficiency of >20% in a human keratinocyte cell line, >10% in immortalized keratinocytes, and <1% in primary keratinocytes. Deep sequencing of the AAVS1 locus showed that ZFN-induced double-strand breaks are mostly repaired by nonhomologous end joining (NHEJ) in primary cells, indicating that poor induction of the HR-dependent DNA repair pathway may be a significant limitation for targeted gene integration. Skin equivalents derived from unselected keratinocyte cultures coinfected with a GFP-IDLV and a ZFN-Ad vector were grafted onto immunodeficient mice. GFP-positive clones were observed in all grafts up to 18 weeks post-transplantation. By histological and molecular analysis, we were able to demonstrate highly efficient targeting of the AAVS1 locus in human repopulating EpSCs.
Introduction
Gene replacement therapy for human monogenic diseases has shown its therapeutic efficacy in a number of seminal clinical studies.1,2,3,4,5,6,7,8 However, the risks related to insertional mutagenesis showed the limits of the current integrating gene transfer technology.9,10,11,12 Epidermolysis bullosa (EB) is a family of severe skin adhesion genetic defects characterized, in the nonlethal forms, by disfiguring blistering, recurrent infections, visual impairment, and an increased risk of skin cancer.13,14,15 There is no cure for EB, and current therapies are palliative, aimed at treating infections and trauma and maintaining an acceptable quality of life. Junctional EB is due to autosomal recessive mutations in any of the three genes (LAMA2, LAMB3, and LAMG2) encoding the laminin 332 heterotrimer. In 2006, we reported the successful correction of the skin adhesion defect in a patient affected by junctional EB by transplantation of cultured skin derived from autologous epithelial stem cells (EpSCs) genetically corrected by a Moloney murine leukemia virus-derived retroviral vector carrying the LAMB3 cDNA under the control of the viral long-terminal repeat.16 While the study provided clinical validation for the therapeutic potential of this approach, safety concerns were raised by the use of a γ-retroviral vector to deliver and express the LAMB3 gene. We, therefore, developed and tested a potentially safer, human immunodeficiency virus-derived lentiviral (LV) vector in which the LAMB3 cDNA is under the control of a keratinocyte-specific enhancer/promoter, and demonstrated its efficacy in a preclinical model.17 LV vectors, however, do not overcome all the problems associated to uncontrolled integration in the human genome,9 and in particular, the post-transcriptional deregulation of target endogenous genes by aberrant splicing.9,18,19 Moreover, they are unsuitable for delivering large genes, such as COL7A1, responsible for the dystrophic forms of EB.20,21 Inserting LAMB3 or COL7A1 expression cassettes at a precise and predetermined location in the genome would overcome the problems and limitations associated with the current integrating vectors, and increase both the safety and efficacy of EpSC-mediated gene therapy.
To this end, we designed a gene-targeting approach aimed at site-specific insertion of a gene into a putative “safe harbor” location, the adeno-associated virus integration site 1 (AAVS1) locus on chromosome 19, in the genome of human keratinocytes. The strategy is based on the use of AAVS1-specific zinc-finger nucleases (ZFNs) to induce a targeted double-strand break and stimulate a specialized form of homologous recombination (HR) known as homology-directed DNA repair. Simultaneous provision of a suitably designed donor DNA cassette, in which the gene of interest is flanked by sequences homologous to the target site, results in the site-specific addition of the corrective DNA to the targeted site.22,23,24,25 The AAVS1 locus allows for robust transgene expression without perturbation of the neighboring gene expression.26,27,28 ZFNs can be delivered via integration-defective LV vectors (IDLVs),29 AAVs,30 or adenoviral (Ad) vectors,26 which do not persist in actively replicating cells. In this study, we provide proof of principle that ZFN-mediated, targeted gene addition can be achieved in human keratinocytes and in long-term repopulating EpSCs in a validated preclinical model of xenotransplantation of human skin equivalents on immunodeficient mice.
Results
Targeted gene integration at high efficiency in a human keratinocyte cell line
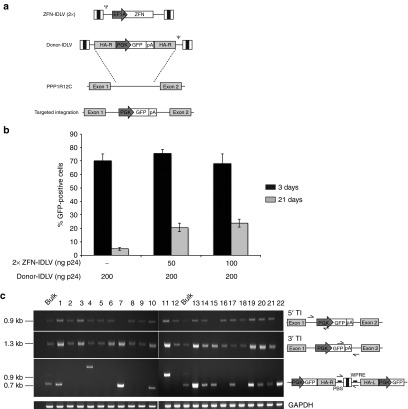
To investigate the feasibility of a ZFN-mediated approach to achieve site-specific integration in human keratinocytes, we used IDLVs for delivering the ZFNs and an AAVS1-specific HR DNA donor template, as previously described.29 Two IDLVs were used to deliver a pair of ZFNs designed against intron 1 of the PPP1R12C gene (the AAVS1 locus), each carrying a ZFN monomer driven by the eukaryotic elongation factor 1α promoter (ZFN-IDLVs). A third IDLV carried the donor template, a GFP gene driven by the phosphoglycerate kinase promoter and flanked by two 800-bp long AAVS1 homology arms (donor-IDLV) (Figure 1a).
Figure 1.
Targeted gene addition into the adeno-associated virus integration site 1 (AAVS1) locus in human HaCaT keratinocytes. (a) Schematic representation of two IDLVs-ZFN, each expressing one ZFN monomer, and the donor IDLV vector; endogenous AAVS1 locus showing the ZFNs target site; and targeted integration (TI) of the GFP cassette into AAVS1 locus. (b) HaCaT cells infected with the indicated doses (ng p24) of two ZFNs-expressing IDLVs and donor-IDLV. GFP expression was evaluated by flow cytometry 3 and 21 days post-transduction. Data are representative of three independent experiments (mean ± SEM; n = 3). (c) PCR analyses on genomic DNA from HaCaT clones derived from the bulk population infected at highest dose to determine TI of the GFP expression cassette into the AAVS1 target locus. Two couple of primers specific for the 5′ and 3′ integration junctions, amplifying 0.9- and 1.3-kb band, respectively, are indicated by black arrows. The middle panel shows concatemers-specific PCR products. The expected bands correspond to 0.7 or 0.9 kb, depending on the presence of one or two long-terminal repeats. The bottom gel shows control PCR amplification targeting the GAPDH gene. Bulk: HaCaT infected bulk population. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IDLV, integration-defective lentiviral vector; PBS, primer binding site; PGK, phosphoglycerate kinase; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element; ZFN, zinc-finger nuclease.
We transduced a human keratinocyte cell line (HaCaT) with the donor-IDLV either alone or together with increasing doses of the two ZFN-IDLVs. Under all conditions tested, we achieved up to 70% transduction, as measured 3 days after infection by cytofluorimetric analysis of GFP expression (Figure 1b). Twenty-one days postinfection, the percentage of GFP+ cells decreased in all samples, falling to <5% in control cells treated with the donor-IDLV alone (4.83 ± 1.09, n = 3) (Figure 1b). By contrast, in cells transduced with the ZFN-IDLVs and the donor-IDLV, we observed up to 25% (23.9 ± 3.0, n = 3) of the cells retaining GFP expression at both doses of ZFN-IDLV vector tested (Figure 1b). To confirm that the GFP expression resulted from ZFN-mediated, targeted gene addition at the AAVS1 locus, we performed PCR analyses on randomly isolated, GFP-expressing individual cell clones obtained by limiting dilution of a bulk population infected with the donor-IDLV and the ZFN-IDLVs. PCR analyses on 5′ and 3′ junctions between the transgene expression cassette and the chromosomal AAVS1 locus gave PCR products consistent with those expected for precise homology-directed integration events in 19 out of 22 HaCaT clones (86%) (Figure 1c). In two clones (#7 and #15), we were able to amplify only the 3′ junctions, while just one clone (#22) scored negative for both the 5′- and 3′-specific PCR products, suggesting a bona fide off-target integration event. PCR analysis with primers annealing to the primer binding site and the Woodchuck hepatitis virus post-transcriptional regulatory element in the viral backbone was also performed to detect the presence of head-to-tail concatemers of the integration cassette. Twelve out 22 clones (54%) harbored at least two copies of the GFP cassette in the target locus (Figure 1c). Sequencing of the band of unpredicted size in clone #4 revealed an aberrant duplication of the Woodchuck hepatitis virus post-transcriptional regulatory element and a short fragment of the viral backbone (data not shown).
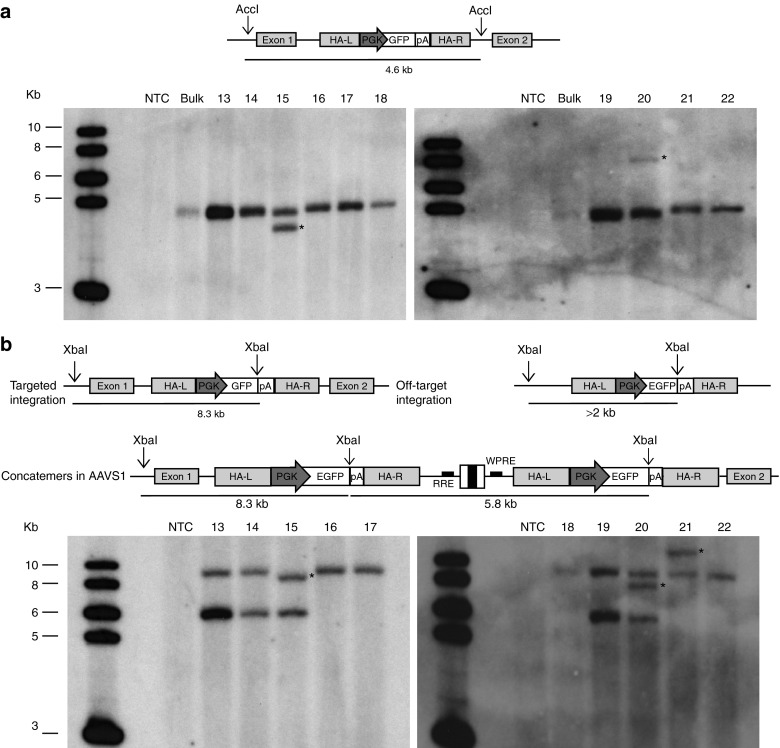
To confirm targeting of the AAVS1 site, genomic DNA was also analyzed by Southern blotting. Hybridization with a GFP-specific probe showed the expected 4.6-kb band in all clones analyzed (Figure 2a). An extra band was observed in 4/22 clones (18%), most likely the result of additional off-target integration events (Figure 2a and Supplementary Figure S1). Further Southern analysis revealed that 90% of the clones had the GFP cassette integrated as either a monomer (65%) or a dimer (35%) in the AAVS1 site. In two clones (#20 and #21 in Figure 2b), harboring three and two copies of the integration cassette, respectively, the GFP probe hybridized to a >2-kb extra band indicating an off-target integration of one copy of the GFP cassette (indicated by a star in Figure 2b). Clone #15 revealed a shorter ~8.1-kb band which, together with the results of the previous Southern blot (Figure 2a) and the PCR analysis (Figure 1b), is consistent with a minor truncation (Figure 2b). One clone (#7 in Supplementary Figure S2) showed a band of higher than expected size, suggesting an off-target integration or most probably a rearrangement at the 5′ junction of the integrated cassette within the AAVS1 locus, as previously detected by PCR analysis (Figure 1c).
Figure 2.
Molecular characterization of the zinc-finger nuclease-mediated integration events in HaCaT cell clones. (a) Southern blot analysis of genomic DNA extracted from HaCaT cell clones, digested with AccI flanking the AAVS1 homology arms, and hybridized to a GFP probe. The expected band, corresponding to 4.6 kb, indicates the presence of a GFP expression cassette correctly integrated into the target locus. Clones showing rearrangements or off-target integrations are highlighted by black asterisks. (b) Southern blot analysis of genomic DNA from the same cell clones digested with XbaI, single cutter in the donor cassette, and hybridized to a GFP probe. A band corresponding to 8.3 kb indicates integration of one copy of the GFP expression cassette into the AAVS1 target locus. A band higher than 2 kb indicates random integration of one copy of the GFP cassette (black asterisks). Finally, a 5.8-kb band indicates the presence of integrated head-to-tail vector concatemers. The first lane on the left-hand side of each Southern blot corresponds to the molecular weight marker. AAVS1, adeno-associated virus integration site 1; NTC, nontreated cells; PGK, phosphoglycerate kinase; RRE, rev-responsive element; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element.
Together, these data demonstrate that IDLVs can drive ZFN-mediated, site-specific gene addition at high frequency and efficiency in a human keratinocyte cell line.
Optimization of ZFN and donor DNA delivery in human keratinocytes
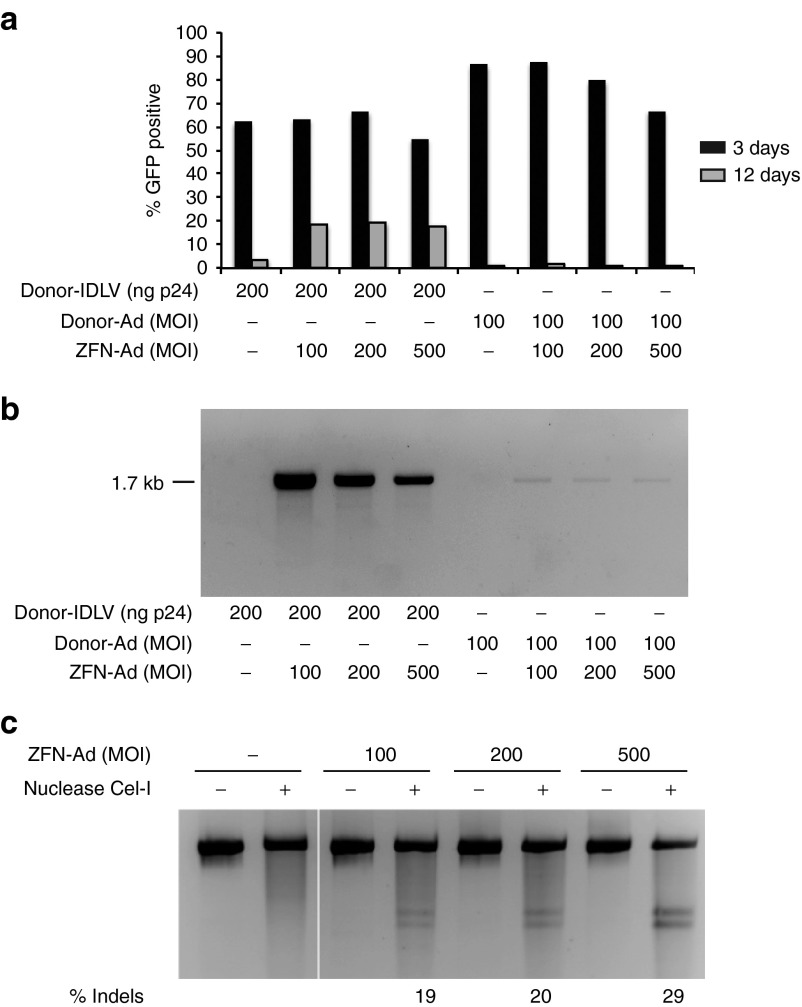
Preliminary experiments showed that unlike HaCaT cells, human primary keratinocytes grown on a standard feeder layer of irradiated NIH3T3-J2 cells are infected at a relatively low efficiency (<15%) by VSV-G-pseudotyped IDLVs (Supplementary Figure S3). To improve gene delivery to keratinocytes, we cloned the same GFP donor cassette in a first-generation Ad vector equipped with a chimeric fiber consisting in the tail of serotype 5 and the shaft and knob motifs of serotype 50 (Ad5/50). Cells were infected with either the donor-IDLV (300 ng of p24) or the donor-Ad vector at a multiplicity of infection (MOI) varying from 200 to 1,000, and GFP expression measured 48 hours after infection by flow cytometry. Transduction efficiency was at least 2.5-fold higher in cells transduced with the donor-Ad vector (35–50%, depending on the MOI; Supplementary Figure S3). To determine the optimal combination of donor and ZFN vector, we infected HaCaT cells with the either the donor-Ad or the donor-IDLV together with increasing doses of an Ad5/35 vector, belonging to the Adenovirus subgroup B as the Ad5/50 vector, carrying the two AAVS1-specific ZFNs driven by a cytomegalovirus promoter and linked by a 2A self-cleaving peptide sequence (ZFN-Ad, a kind gift of Sangamo Biosciences, Richmond, CA).26 Twelve days after transduction, the culture coinfected with ZFN-Ad and the donor-IDLV contained 20% GFP+ cells compared with just 1% GFP+ cells observed in the culture coinfected with the ZFN-Ad (MOI: 100–500) and the donor-Ad at MOI 100 (Figure 3a). Increasing the donor-Ad MOI over 500 resulted in some cell toxicity without any gain in the frequency of stable integration. Nonquantitative PCR analysis of the expected 3′ junction between the GFP cassette and the chromosomal AAVS1 locus confirmed that HR-mediated targeted integration occurred with both types of donor vector, although at markedly higher efficiency with the donor-IDLV (Figure 3b). The extent of ZFN cleavage at the AAVS1 locus repaired by nonhomologous end joining (NHEJ) (% indels) was evaluated by Surveyor nuclease (Cel-I) assay on genomic DNA from infected cells. We observed increasing % indels in cells infected with the ZFN-Ad vector, reaching a maximum of 29% at MOI 500 (Figure 3c), suggesting that NHEJ repair is favored when the donor cassette is carried by an Ad vector, while HR is used at a much higher frequency in the presence of an IDLV donor template.
Figure 3.
ZFN-Ad delivery in HaCaT cells. (a) HaCaT cells were infected with the indicated doses of ZFN-Ad (MOI) and donor-IDLV (ng p24) or Ad donor (MOI) and analyzed by fluorescence-activated cell sorting for GFP expression 3 and 12 days after transduction. (b) AAVS1-targeted PCR on genomic DNA isolated from HaCaT bulk populations 12 days after infection using primers specific for the 3′ integration junction amplifying a 1.7-kb band. (c) Cel-I assay on HaCaT cells infected with ZFN-Ad (MOI: 100, 200, and 500). The PCR product spanning the ZFN target site was loaded before and after nuclease treatment. Frequencies of gene disruption are indicated as % indels. Ad, adenoviral; IDLV, integration-defective lentiviral vector; MOI, multiplicity of infection; ZFN, zinc-finger nuclease.
Targeted gene integration in immortalized human EB keratinocytes
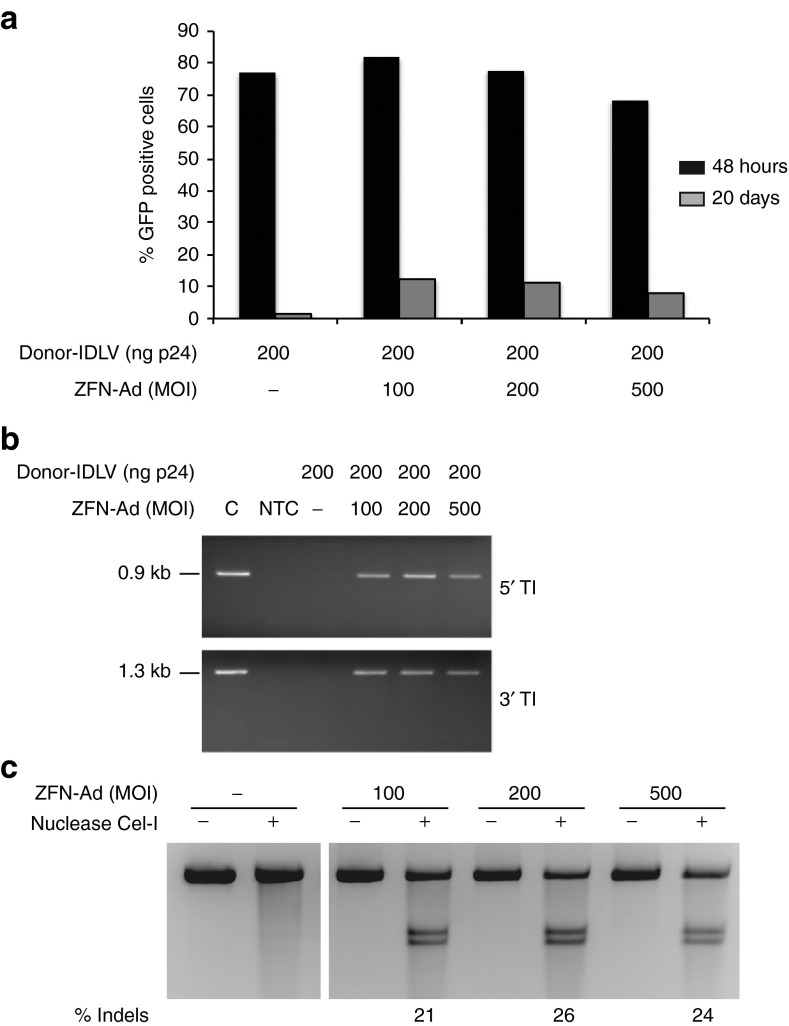
The efficiency of the donor-IDLV + ZFN-Ad combination in targeting gene integration was then tested in SV40-immortalized primary keratinocytes derived from a patient affected by generalized atrophic benign EB (GABEB).31 Unlike HaCaT cells, immortalized GABEB keratinocytes showed an almost normal karyotype by cytogenetic analysis: >50% of the scored nuclei were diploid with minor abnormalities on chromosome 3, 6, 8, 11, 13, 20, and X (data not shown). No abnormality was observed in the number and structure of chromosome 19, hosting the AAVS1 locus. Immortalized GABEB keratinocytes were then transduced with the donor-IDLV and increasing doses of the ZFN-Ad. We achieved >65% transduction efficiency of the donor-IDLV 48 hours postinfection under all conditions tested, as analyzed by flow cytometry of GFP expression (Figure 4a). Twenty days after infection, we observed up to 12.3% GFP+ cells in keratinocytes infected with donor-IDLV + ZFN-Ad, compared with 1.3% of GFP+ cells in control cultures infected with the donor-IDLV alone (Figure 4a). HR-mediated targeted gene addition was confirmed by PCR assays specific for the expected 5′ and 3′ junctions between the GFP cassette and the AAVS1 locus (Figure 4b). The Cel-I assay showed 21–26% indels at the ZFN target site in the AAVS1 locus at all three doses of ZFN-Ad vector (MOI: 100, 200, and 500) (Figure 4c), indicating an almost equal efficiency in HR- versus NHEJ-mediated DNA repair in the presence of an IDLV donor template in immortalized keratinocytes. Increasing the MOI of ZFN-Ad >200 did not result in higher cleavage activity but, if anything, in a modest decrease of both % indels and % GFP+ cells (Figure 4).
Figure 4.
Targeted integration in immortalized generalized atrophic benign epidermolysis bullosa (GABEB) keratinocytes. (a) Analysis of GFP expression 48 hours and 20 days after infection with donor-IDLV and ZFN-Ad. (b) PCR analysis of the 5′ and 3′ junction between the expression cassette and the adeno-associated virus integration site 1 locus. C, GFP+ HaCaT clone used as a positive control. (c) Cel-I assay performed on immortalized GABEB cells infected with MOI 100, 200, or 500 of ZFN-Ad vector, as described for HaCaT cells. % Indels: frequency of insertions–deletions. Ad, adenoviral; IDLV, integration-defective lentiviral vector; MOI, multiplicity of infection; NTC, nontreated cells; ZFN, zinc-finger nuclease.
Site-specific gene addition in human primary keratinocytes
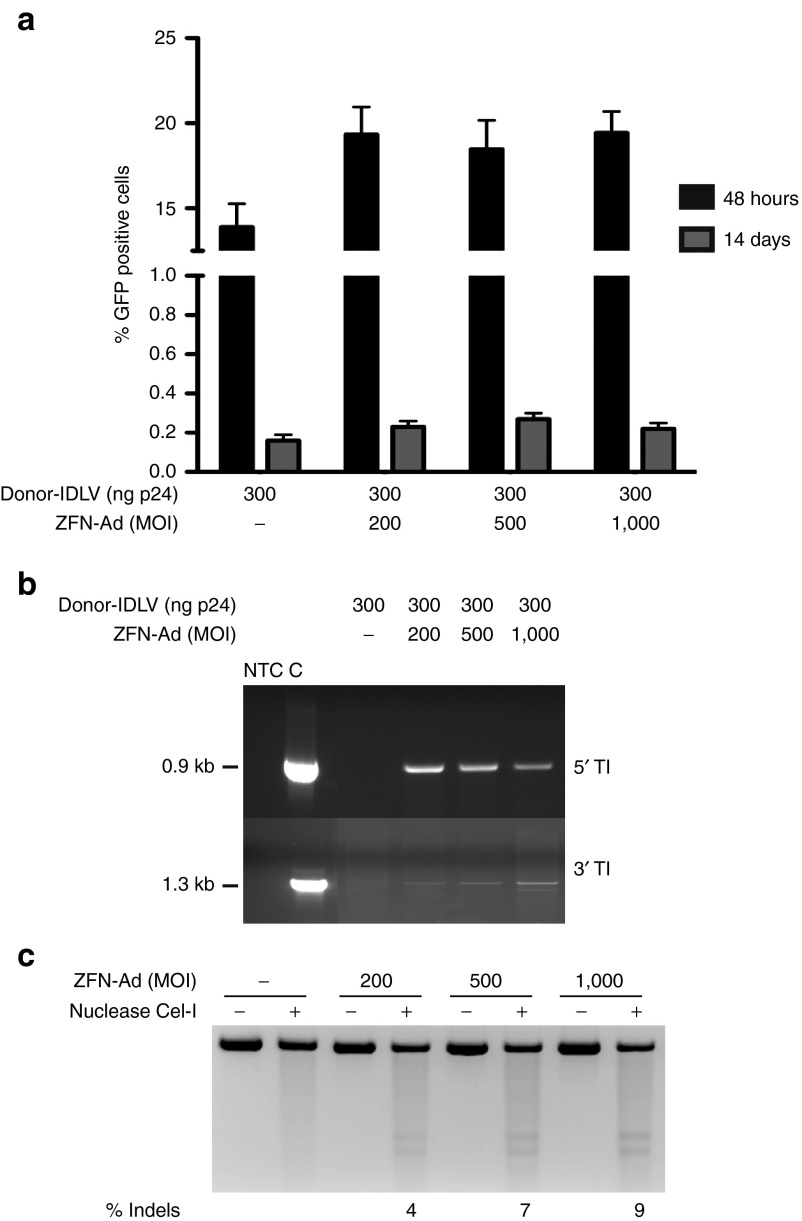
To assess the efficiency of the donor-IDLV + ZFN-Ad combination in targeting integration in primary cells, we infected early passage, foreskin-derived human keratinocytes with the donor-IDLV and increasing doses of the ZFN-Ad (MOI: 200–1,000). GFP expression analysis revealed a transduction efficiency of 14–20% 48 hours postinfection in all conditions, confirming the low transduction efficiency by IDLVs in primary keratinocytes. Fourteen days after infection, the number of GFP+ cells decreased to 0.3% in cultures infected with donor-IDLV + ZFN-Ad (0.27 ± 0.03, n = 3), and below 0.1% in cells infected with the donor-IDLV alone (Figure 5a). PCR analysis of the 5′ and 3′ junctions between the GFP cassette and the AAVS1 locus showed HR-mediated targeted integration in the bulk population of primary keratinocytes (Figure 5b), which was confirmed by sequencing both ends of the PCR products (data not shown). Cel-I assay carried out 48 hours postinfection revealed 4, 7, and 9% indels in the AAVS1 alleles in keratinocytes infected with the ZFN-Ad at MOIs of 200, 500, and 1,000, respectively (Figure 5c), indicating a 10- to 30-fold lower efficiency of HR- versus NHEJ-mediated repair in these cells (a summary of the relative efficiency of HR- and NHEJ-mediated gene repair in all tested cell types is reported in Supplementary Table S1).
Figure 5.
Targeted gene addition in adeno-associated virus integration site 1 (AAVS1) locus in human primary keratinocytes. (a) Human primary keratinocytes were infected with the indicated doses of ZFN-Ad (MOI: 200, 500, and 1000) and donor-IDLV (ng p24). The GFP expression was evaluated by fluorescence-activated cell sorting analysis 48 hours and 14 days after transduction. Data are representative of three independent experiments (mean ± SEM; n = 3). (b) PCR analyses on the 5′ and 3′ junction between the integrated cassette and the AAVS1 locus, as performed on HaCaT cells. C, HaCaT GFP+ clone as a positive control. (c) Cel-I assay on genomic DNA of HaCaT and immortalized generalized atrophic benign epidermolysis bullosa cells isolated 48 hours after infection with increasing doses (MOI) of ZFN-Ad. % Indels: frequency of insertion or deletions. Ad, adenoviral; IDLV, integration-defective lentiviral vector; MOI, multiplicity of infection; NTC, nontreated cells; ZFN, zinc-finger nuclease.
Deep sequencing of the AAVS1 target site in primary keratinocytes infected with the ZFN-Ad vector at MOI 500 allowed the identification of the types of indels introduced by NHEJ repair. More than 99% of the sequences retrieved from uninfected cells (7,538 out of 7,548) perfectly matched the AAVS1 sequence (UCSC hg19 release of the human genome). Alignment of the 14,195 sequences retrieved from cells infected with ZFN-Ad identified 1,214 sequences (8.5% of the reads, P < 2.2 × 10–16) carrying insertions and/or deletions within a 90-bp window (−45 to +45) around the 6-bp spacer of the ZFN target site. In particular, 624 sequences contained deletions of 1–45 bp, and predominantly of 1 bp (17.7% of sequences), 2 bp (6.7%), 4 bp (3.9%), and 10 bp (5.5%), while 559 sequences contained insertions ranging from 1 to 20 bp and predominantly (>93%) <4 bp. Twenty-nine sequences (2.4%) contained insertions and deletions of at least 1 bp. By restricting the analysis to a 24-bp window around the ZFN target site, we observed that 64% (776) of the 1,214 mutated sequences were clustered in the ZFN binding sites, with 424 sequences containing 1- to 25-bp deletions, 338 sequences containing 1- to 13-bp insertions, and 14 sequences containing insertions and deletions of at least 1 bp.
To assess cell toxicity associated to ZFN-Ad infection in human primary keratinocytes, we measured the induction of apoptosis 48 hours after infection by flow cytometry after Annexin V-PE staining. In both uninfected and ZFN-Ad–infected primary keratinocytes, the percentage of apoptotic cells was ~1.5%, independently from the MOI (200–1,000). A slightly higher level of apoptotic cells (2.3–3.5%) was observed in the control, GFP-Ad–infected cells. To look at the effect of IDLV + ZFN-Ad combined infection on proliferation and colony forming efficiency (CFE) of clonogenic keratinocyte progenitors, we infected keratinocyte cultures with the donor-IDLV and the ZFN-Ad vector at MOI 200 and 500, and determined the number of cells and CFE at different time points. The cell proliferation assay showed a slightly reduced number of cells 72 hours postinfection (T1) but not at later time points (Supplementary Figure S4). A CFE assay was carried out at passage T0 (uninfected cells) up to T5 after infection, corresponding to 32 days of culture. No significant difference was observed at any time point between control uninfected and infected keratinocytes (Supplementary Figure S5). These results indicate that coinfection with donor-IDLV + ZFN-Ad does not affect proliferation and clonogenic efficiency of human keratinocytes in vitro, and has no significant toxic effect on keratinocyte progenitors.
ZFN-mediated targeted gene addition in human repopulating keratinocyte stem cells
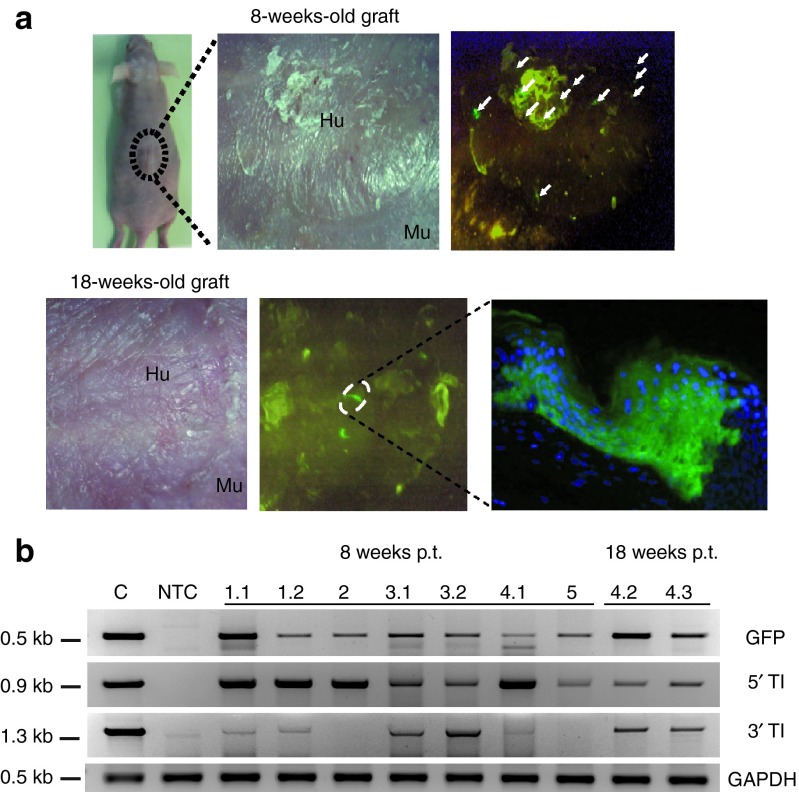
To determine the efficiency of targeted gene addition in keratinocyte stem cells, we used an in vivo assay measuring the long-term engrafting potential of genetically modified EpSCs upon xenotransplantation on immunodeficient (nu/nu) mice. In this assay, skin equivalents are grown ex vivo by seeding keratinocytes on a dermal equivalent composed of a fibroblast-containing fibrin matrix and grafted on the dorsal region of immunodeficient mice, as previously described.32,33 Human primary keratinocytes were infected with donor-IDLV + ZFN-Ad (MOI: 500) as described above. One week after infection, the cultures contained 3–8% of GFP+ cells, and were plated on fibrin matrix to prepare 10 transplantable human skin equivalents to be grafted on the back of 10 nu/nu immunodeficient mice. Eight out of 10 mice that received the genetically modified skin equivalents were successfully grafted, an expected efficiency for this type of assay, indicating that the combined vector infection did not affect the in vivo regeneration capacity of EpSCs. The grafts were analyzed macroscopically, histologically, and molecularly 8 and 18 weeks after transplantation. Epifluorescent illumination of the transplanted zone allowed in vivo monitoring of the genetically modified graft up to 18 weeks after grafting. Five out of eight successfully grafted mice showed GFP+ spots 8 weeks after transplantation, indicating that the GFP transgene cassette had been stably integrated into transplantable clonogenic cells. Seven GFP+ skin biopsies (1.1, 1.2, 2, 3.1, 3.2, 4.1, and 5) were obtained from five independent grafts representative of five mice (#1–5) 8 weeks post-transplantation, and two more GFP+ spots (4.2 and 4.3) were obtained from the same graft (mouse #4) 18 weeks post-transplantation. The skin biopsies were sectioned and analyzed for GFP expression by fluorescence microscopy. In all cases, we observed columns of GFP+ cells extending from the basal cell layer to the surface of the graft, indicating engraftment of basal layer stem/progenitor cells expressing the transgene throughout terminal differentiation (Figure 6a). Molecular analysis performed on genomic DNA extracted from all nine biopsies showed the presence of the GFP transgene by PCR analysis (Figure 6b). Seven out of nine biopsies (1.1,1.2, 3.1, 3.2, 4.1, 4.2, and 4.3) showed the PCR products expected for HR-mediated 5′ and 3′ AAVS1-transgene junctions (Figure 6b). The correct, site-specific genomic insertion was confirmed by DNA sequencing in at least one, randomly selected amplified junction (Supplementary Figure S6a). The remaining two biopsies (2 and 5) resulted positive only for the 5′ AAVS1-transgene junctions (Figure 6b). To validate the positive signal of those junctions, we sequenced the PCR products. The sequences demonstrated a genuine genome-expression cassette junction (Supplementary Figure S6b,c).
Figure 6.
Transplantation of engineered human primary keratinocyte onto immunodeficient mice. (a) Representation of one mouse engrafted with human keratinocytes transduced with zinc-finger nuclease-Ad and integration-defective lentiviral vector donor and examined 8 and 18 weeks post-transplant (p.t.), and GFP expression monitored in vivo under an ultraviolet lamp is shown. Fluorescence analysis of GFP-expressing cells was performed in histological cryosections from single GFP-positive spots of regenerated epidermis. (b) PCR analyses on nine biopsies (1.1, 1.2, 2, 3.1, 3.2, 4.1, 4.2, 4.3, and 5) harvested from five treated mice (1–5) 8 and 18 weeks after grafting. GFP-specific PCR shows the correct 0.5-kb amplified fragment. 5′ TI and 3′ TI indicate the 5′ and 3′ GFP cassette-genome junctions. At bottom, control PCR amplification targeting the GAPDH gene is shown. The size of the expected bands is indicated on the left. C, HaCaT clone carrying a single copy of GFP expression cassette integrated into adeno-associated virus integration site 1 target locus. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Hu, human skin; Mu, murine skin; NTC, nontreated cells; TI, targeted integration.
Taken together, these data indicate that transduction of human keratinocyte cultures with the ZFN-Ad and an AAVS1-IDLV vector induces targeted gene addition, at low frequency but with remarkable precision, in EpSCs able to maintain a skin graft for up to 18 weeks.
Discussion
Integration of a therapeutic transgene at a precise location into the human genome is a major goal in the development of new gene transfer technology. A number of gene therapy applications, such as transplantation of genetically modified somatic stem cells, require permanent expression of the therapeutic gene, and thus stable genomic integration of the transgene expression cassette. Retroviral vectors have been the only clinically applicable integrating technology for almost 20 years. It is becoming obvious, however, that the uncontrolled insertion of a transgene expression cassette mediated by γ-retroviral or LV vectors may severely interfere with genetic and epigenetic mechanisms of gene regulation at both transcriptional and post-transcriptional levels, and lead to harmful genotoxic consequences in patients.34 There are several possible solutions to this problem, which range from improving the current retroviral vector design to developing entirely new technology. An attractive possibility is the design of new vectors aimed at driving transgene addition into “safe” genomic loci in the human genome. The preferred integration site of AAV on human chromosome 19, the AAVS1 locus, represents one such putative site. Originally, attempts to target AAVS1 employed the AAV Rep replicase-integrase to drive site-specific integration.35,36,37 More recently, ZFN technology has been used to mediate HR at this site.26,38
In this study, we have investigated the feasibility of a ZFN-mediated approach to achieve AAVS1-specific integration of a GFP reporter gene in the genome of human keratinocytes and EpSCs with in vivo repopulating activity. We have described two delivery platforms. The first is entirely based on IDLVs, two of which carry ZFNs designed to introduce a double-strand break in the first intron of the PPP1R12C gene (the AAVS1 locus), and a third carrying a GFP expression cassette flanked by AAVS1 homology arms. The second platform is based on a first-generation Ad vector expressing both AAVS1-specific ZFNs, and the donor cassette again provided by an IDLV vector. Under both conditions, we achieved >20% stable integration of the donor GFP cassette in the human keratinocyte cell line HaCaT, and >10% in SV40-immortalized keratinocytes from a GABEB patient. Molecular analyses on isolated HaCaT clones showed that almost 90% of the integration events were mediated by homology-directed repair. Unfortunately, our attempts to increase the frequency of HR-mediated integration by delivering also the donor integration cassette by an Ad vector failed, as the Ad DNA genome apparently provided a much less efficient HR template compared with the IDLV carrying the same donor cassette. Both Ad and LV vector genomes have a mostly linear, double-stranded conformation in cell nuclei, and it is, therefore, difficult to understand why an IDLV would be superior to an Ad genome as an HR template. One possibility is that a LV preintegration complex is capable of actively recruiting the HR-mediated DNA repair complex as part of its integration process, may be during the strand invasion step, thus facilitating HR-mediated integration of an LV genome associated to a defective human immunodeficiency virus integrase. The inefficiency of the Ad genome to serve as a donor template is unfortunate: large-capacity Ad genomes would be ideal for delivering large payloads to keratinocytes, such as a COL7A1 expression cassette necessary for gene therapy of recessive dystrophic EB. Additional studies are, therefore, required to optimize the conformation of the template and the cofactors involved in mediating HR from an Ad-encoded donor template.
Encouraged by the results obtained in cell lines, we investigated the feasibility of using the ZFN-mediated approach to insert a gene expression cassette in primary keratinocytes, keratinocyte progenitors, and EpSCs. Targeted integration occurred in vitro in <1% of transduced, unselected keratinocytes, a much lower efficiency compared with HaCaT cells or SV40-immortalized keratinocytes, even considering the overall lower transient transduction efficiency obtained with the ZFN-Ad + donor-IDLV combination in primary cells. Comparing the efficiency of ZFN-mediated double-stranded DNA cleavage, analyzed by both a gel mobility assay and deep sequencing of the AAVS1 locus, we observed that in HaCaT cells, the majority of the cleaved DNA is repaired by HR with the incorporation of the donor cassette, whereas in primary keratinocytes, HR occurs with a frequency 30-fold lower than NHEJ, apparently the preferred mechanism for repairing ZFN-induced double-stranded breaks. These data suggest that the major limitation of ZFN-mediated targeted integration in primary cells is not a reduced cleavage activity of the ZFNs but rather a failure to activate the HR pathway to repair DNA. Interestingly, the efficiency of homology-directed DNA repair in SV40-immortalized keratinocytes is somewhat intermediate between HaCaT and primary cells with almost 50% of the double-stranded break repaired by HR-mediated mechanisms. Apparently, immortalization somehow activates the HR pathway may be as a consequence of the altered cell cycle progression, thus resulting in significant level of ZFN-mediated targeted integration (>10%).
Despite the overall poor efficiency obtained in primary keratinocytes, we decided to test the efficiency of targeted gene delivery in EpSCs, defined by their ability to maintain stable human skin xenografts on immunodeficient mice for more than 8 weeks. EpSCs are relatively abundant in a primary keratinocyte culture, and can be cloned, tested for site-specific integration, selected, and expanded before transplantation, as previously demonstrated with retrovirus-transduced human keratinocytes.33 To demonstrate HR-mediated gene addition in EpSCs, we transplanted skin equivalents derived from keratinocyte cultures cotransduced with donor-IDLV and ZFN-Ad on nu/nu mice, without selecting for GFP-positive cells. In situ analysis of the transplanted skin and histological and molecular analysis confirmed that HR-mediated integration occurred in long-term repopulating, bona fide stem cells. Although the in vivo assay cannot be considered quantitative, it clearly demonstrated that the efficiency of targeted integration is similar in keratinocytes in vitro and clonogenic EpSCs in vivo, and that the major limitation of the ZFN-mediated approach is the lack of efficient engagement of the HR-mediated DNA repair pathway. Previous studies clearly showed similar difficulties in targeting the CCR5 or the AAVS1 locus in human hematopoietic stem/progenitor cells compared with established hematopoietic cell lines,26,29 suggesting that human stem cells might be generally difficult to engineer by ZFN-based technology unless strategies are found to induce HR-mediated DNA repair more efficiently. Nonetheless, our study shows that gene targeting in transplantable somatic stem cells is feasible and lead to stable, site-specific gene addition in their progeny. These results open some hopes for the development of a “gene correction” approach to gene therapy for skin adhesion defects, where rare corrected stem cells can be isolated by cloning and used to derive corrected skin implants.
Materials and Methods
Vectors and ZFNs. The IDLV containing the donor GFP cassette and the AAVS1-specific ZFNs have been described previously.28 The Ad vector containing the AAVS1-specific ZFN expression cassettes was previously described.26,39 The Ad donor vector was obtained by cloning the phosphoglycerate kinase. GFP expression cassette flanked by sequences homologous for the AAVS1 locus into a pShuttle vector (Stratagene, La Jolla, CA). Given the higher capacity of the Ad vector compared with that of the LV vector, the homologous regions were extended in the former by including nucleotide from 670 through 2,183 of the AAVS1 locus for the arm upstream of the expression cassette and from nucleotide 2,190 through 3,386 for the arm downstream of the expression cassette. The shuttle vector was then recombined in BJ5183 competent cells with an AdEasy-based backbone encoding chimeric 5/50 fibers, resulting in the assembly of molecular clones containing the first-generation Ad5/50 genome. Next, these clones were treated with PacI enzyme and subsequently transfected by using ExGen 500 (Fisher Scientific, The Netherlands) into Ad serotype 5 early region 1 (E1)-complementing PER.C6 cells. The resulting rescued vector particles were propagated in PER.C6 cells, after which they were purified through CsCl density gradient ultracentrifugation. The viral vector titer was determined via 50% tissue culture infectious dose assay in E1-complementing 911 cells.
Cell culture. Human HaCaT cells were maintained in Dulbecco's modified Eagle's medium (Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (Lonza). Clones were obtained by limiting dilution, plating the cells in 96-well plates at a concentration of 0.3 cells/well. Keratinocytes from homozygous GABEB patients immortalized with SV40 (kindly provided by Johann W. Bauer) were cultivated in Epilife medium supplemented with human keratinocyte growth supplement (Invitrogen, Milan, Italy). Epilife is a serum-free keratinocyte culture medium with a low calcium (0.06 mmol/l) concentration supplemented with human keratinocyte growth supplement which results in a final concentration of 0.2% (vol/vol) bovine pituitary extract, 5 µg/ml bovine insulin, 0.18 µg/ml hydrocortisone, 5 µg/ml bovine transferrin, and 0.2 ng/ml human epidermal growth factor. Swiss mouse 3T3-J2 cells (a kind gift from Yan Barrandon, EPFL, Lausanne, Switzerland) were grown in Dulbecco's modified Eagle's medium supplemented with 10% donor bovine serum (GIBCO, Milan, Italy), 50 IU/ml penicillin–streptomycin, and 4 mmol/l glutamine. Primary human keratinocytes from foreskin biopsy were plated onto lethally irradiated 3T3-J2 cells and cultured in keratinocyte growth cFAD medium, a Dulbecco's modified Eagle's medium and Ham's F12 media mixture (3:1) containing fetal bovine serum (10%), penicillin–streptomycin (1%), glutamine (2%), insulin (5 µg/ml), adenine (0.18 mmol/l), hydrocortisone (0.4 µg/ml), cholera toxin (0.1 nmol/l), and triiodothyronine (2 nmol/l). After 3 days, epidermal growth factor at a concentration of 10 ng/ml was added to the culture. Keratinocytes were trypsinized at subconfluency and replated onto a new feeder layer.
A CFE assay on treated and untreated keratinocytes cultured for 4–5 weeks was performed as follows. At each passage, 5 × 102–4 × 103 keratinocytes (untreated, donor-IDLV, or donor-IDLV/ZFN-Ad–treated cells) were plated in a 100-mm culture dish onto lethally irradiated 3T3-J2 cells and cultured in keratinocyte growth cFAD medium; epidermal growth factor (10 ng/ml) was added in the first medium change. Cultures were maintained at 37 °C, 5 % CO2 for 12 days and fed three times weekly. After that time, culture medium was discarded and stained with Rhodamine B solution at 2% (formaldehyde 37% diluted 1:10 and Rhodamine B) for 24 hours; then Rhodamine B was discarded and washed with water. The CFE was calculated as follows: % CFE = (grown colonies/seeded cells) × 100.
Among the colonies, we counted specifically the abortive colonies and we calculated the % abortive colonies as follows: % abortive colonies = (number of abortive colonies/number of grown colonies) × 100.
Transduction of keratinocytes. HaCaT cells (1 × 105) and immortalized GABEB cells (2.5 × 105) were seeded in 6-well plates and infected by spinoculation (one round at 1,800 rpm for 45 minutes) with 200 ng p24 of donor IDLV and/or 50 and 100 ng p24 of each IDLV-ZFN. Ad transduction was performed in serum-free medium containing the viral particles at MOIs 100, 200, or 500 in the presence of polybrene (8 µg/ml). After 1 hour at 37 °C, the transduction mixture was replaced with regular medium. In the samples treated with the donor-IDLV/ZFN-Ad platform, infection with Ad vector always followed infection with LV vector. Human primary keratinocytes were seeded onto 3T3-J2 feeder cells and infected after 16 hours under the same conditions used for the cell lines. Infected cells were grown for at least 14 days to avoid scoring GFP expression from unintegrated vector. Transduction efficiency was evaluated at different time points by flow cytometry for GFP expression (FACS Canto II; Becton Dickinson/Pharmingen, Milan, Italy). For apoptosis analysis, Annexin V (Becton Dickinson) was added to 1 × 105 cells at 48 hours after transduction and analyzed by fluorescence-activated cell sorting according to the manufacturer's instruction.
Targeted integration analysis. Genomic DNA was isolated with either Blood & Cell Culture DNA Mini or Micro Kit (Qiagen, Milan, Italy). Briefly, 100 and 500 ng of genomic DNA from HaCaT cells and primary keratinocytes, respectively, was subjected to PCR (PCR GoTaq; Promega, Milan, Italy) using primers TI5 and TI3 and the following conditions: 95 °C for 5 minutes, then 30 cycles of 95 °C for 30 seconds, 61 °C for 45 seconds, 72 °C for 30 seconds, followed by 72 °C for 7 minutes. When using the Ad5/50 donor vector, targeted integration was assessed only at the 3′ junction with a primer specific for a region in AAVS1 outside the larger homology sequence present in the vector (TI3Ad primer). To detect the presence of integrated concatemers, 100 ng genomic DNA was PCR amplified using specific primers and the following cycling conditions: 95 °C for 5 minutes, then 30 cycles of 95 °C for 30 seconds, 61 °C for 45 seconds, 72 °C for 30 seconds, followed by 72 °C for 7 minutes. PCR amplicons were resolved on 1% agarose gel and visualized by ethidium bromide staining. All primers are listed in the Supplementary Table S2. For Southern blot analyses, 10 µg of genomic DNA was digested overnight with AccI or XbaI, run on a 0.8% agarose gel, transferred to a nylon membrane (Hybond-XL; Amersham, Milan, Italy), and probed with 2 × 107 cpm 32P-labeled GFP probe.
Cel-I (Surveyor nuclease) assay. Genomic DNA was extracted with the Blood & Cell Culture DNA Mini Kit (QIAGEN) according to manufacturer's instructions. Frequency of gene modification by NHEJ was evaluated by Surveyor endonuclease assay (Cel-I; Transgenomics, Glasgow, UK) according to manufacturer's instructions. Briefly, the genomic DNA purified from ZFN-Ad–infected keratinocytes was used as a template to amplify a short region of the AAVS1 site flanking the ZFN target site using the specific primers Cel-1F and Cel-1R (Supplementary Table S2) and the following conditions (Taq High Fidelity; Invitrogen): 95 °C for 5 minutes, then 40 cycles of 95 °C for 30 seconds, 61 °C for 30 seconds, and 68 °C for 30 seconds. As control, genomic DNA purified from uninfected human keratinocytes was subjected to PCR amplification using the same conditions. The PCR products were then heated, allowed to reanneal followed by a treatment with the nucleotide mismatch-sensitive Surveyor nuclease40 in order to detect insertions and deletions caused by NHEJ. The percentage of indels was calculated by densitometric analysis comparing between digested with undigested amplicons.
454 Deep sequencing. Genomic DNA was isolated from ZFN-Ad–infected and uninfected keratinocytes using the QIAamp DNA Micro Kit (Qiagen). A 500 ng genomic DNA was then PCR amplified using Platinum Taq High Fidelity (Invitrogen) using primers HS (Supplementary Table S2) joint to 454 adaptor sequences (forward 5′-CTATGCGCCTTGCCAGCCCGCTCAG-3′, reverse 5′-CGTATCGCCTCCCTCGCGCCATCAG-3′) and four nucleotides. DNA barcodes to discriminate between untreated (barcode: TAGC) and treated (barcode: CGTA) cells. The cycling conditions were 95 °C for 5 minutes, then 20 cycles of 95 °C for 30 seconds, 55 °C for 3 seconds, 68 °C for 30 seconds; 1/50 of the product was then subjected to a second round of amplification using the same conditions. Following PCR amplification, the PCR product was analyzed on a 1% agarose gel, extracted and then purified using QIAquick Gel extraction kit (QIAGEN). DNA samples were then pooled at a molar ratio of 1:3 favoring the infected cells and run on a Roche/454 GS FLX using titanium chemistries by GATC Biotech AG Next Gen Lab (Konstanz, Germany). Approximately, 25,000 reads were obtained for both amplicons. Sequences derived from ZFN-Ad–infected and uninfected cells were separated thanks to their different barcode. Any reads containing ambiguous base calls or without a perfect match to barcode and primer were discarded. The reference sequence was assembled using reads from both ZFN-Ad–infected and uninfected cells assembled and the software Newbler (version 2.6). The resulting contigs were searched against the reference sequence of Hg19 with BLASTN to verify the percentage of identity between reference target and contigs. Contigs that matched with the best similarity were further used as reference to map reads from ZFN-Ad–infected and uninfected cells using gsMapper (version 2.6). Deviations from the AAVS1 consensus sequence were quantified using ad-hoc R scripts. All deviations (deletions and insertions) from AAVS1 consensus sequence were determined considering 45 and 12 bp up- or downstream from the ZFN binding site, respectively.
Bioengineered skin preparation and grafting to immunodeficient mice. The bioengineered human skin equivalent is based on the use of a fibrin matrix containing live fibroblasts as a dermal component.32 Briefly, 3 ml of fibrinogen (from swine plasma cryoprecipitates) were added to 12 ml of Dulbecco's modified Eagle's medium with 10% fetal calf serum containing 5 × 105 dermal fibroblasts and 500 IU of bovine aprotinin (Trasylol; Bayer, West Haven, CT). Immediately afterwards, 1 ml of 0.025 mmol/l CaCl2 (Sigma, St Louis, MO) containing 11 IU of bovine thrombin (Sigma) were added. Finally, the mixture was distributed on a 6-well culture plate (Transwell; Costar, Cambridge, MA) and allowed to clot at 37 °C for 2 hours. Human keratinocytes, including those genetically modified (5 × 105 per well in 3 ml of keratinocyte medium), were then seeded on top of the dermal equivalents and allow to grow to confluence. Graft recipient mice were aseptically cleansed and grafted with bioengineered skin equivalents as previously described.17,41 Briefly, full-thickness 35-mm circular wounds were created on the dorsum of the 6-week old female nu/nu mice (Janvier Labs, Saint Berthevin, France). Bioengineered skin equivalents were manually detached from the wells and placed orthotopically on the wound and covered with freeze-thawed (three cycles) devitalized mouse skin. Regenerated human skin became visible, upon mouse devitalized skin spontaneously detached, 3–4 weeks after grafting. Mice were housed for the duration of the experiment at the CIEMAT Laboratory Animals Facility (Spanish registration number: 28079-21 A) in pathogen-free conditions using individually ventilated type II cages (25 air changes per hour) and 10 kGy-irradiated soft wood pellets as bedding.
In vivo skin analysis and sampling. Surviving individual mice with successful engraftment (n = 8) were analyzed at different time points postgrafting. The presence of GFP-positive epidermal spots within engrafted human skins was determined with a stereomicroscope equipped with a GFP illumination source and filters. GFP fluorescent spots were carefully excised either as biopsies (taken with a 2-mm skin biopsy punch) on anesthetized mice or after sacrifice. Samples were either fixed in 4% buffered paraformaldehyde (pH 7.4) for 40 minutes at 4 °C, equilibrated in 30% sucrose in primer binding site overnight for GFP analysis on frozen sections or snap frozen for molecular analysis.
SUPPLEMENTARY MATERIAL Figure S1. Southern blot analysis of HaCaT cell clones digested with AccI. Figure S2. Southern blot analysis of HaCaT cell clones digested with XbaI. Figure S3.Transduction efficiency of IDLV and Ad vector in human primary keratinocytes. Figure S4.Cell proliferation assay on keratinocytes untreated and infected with IDLV donor alone or together with the ZFN-Ad vector. Figure S5.Colony forming efficiency assay on keratinocytes untreated and infected with IDLV donor alone or together with the ZFN-Ad vector. Figure S6.Sequence of 5′ and 3′ TI junctions in mouse biopsies. Table S1.Comparison between cleavage efficiency (% indels) and percentage of GFP integration in the cell types tested. Table S2.List of primers used for targeted integration analyses.
Acknowledgments
We thank Michael C Holmes and Philip D Gregory for the gift of the Ad vector expressing the AAVS1-specific ZFN pairs, and for critically reading the manuscript. We also thank Blanca Duarte for her help in the xenotransplantation experiments. This work was supported by grants from the European Commission (FP7, PERSIST), the European Research Council (ERC-2010-AdG, GT-SKIN), and the Italian Ministry of University and Research (FIRB 2008). F.L. is supported by grants PI11/0125 from Spanish ISCIII and grant P2010BMD-2359 from Comunidad de Madrid. M.D.R. is supported by grants SAF2010-16976 from MICINN and P2010BMD-2420 from Comunidad de Madrid. The authors declared no conflict of interest.
Supplementary Material
References
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Díez IA, Dewey RA, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN Jr, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Christiano AM, Uitto J. Molecular complexity of the cutaneous basement membrane zone. Revelations from the paradigms of epidermolysis bullosa. Exp Dermatol. 1996;5:1–11. doi: 10.1111/j.1600-0625.1996.tb00086.x. [DOI] [PubMed] [Google Scholar]
- Borradori L, Sonnenberg A. Structure and function of hemidesmosomes: more than simple adhesion complexes. J Invest Dermatol. 1999;112:411–418. doi: 10.1046/j.1523-1747.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- Epstein EH Jr. The genetics of human skin diseases. Curr Opin Genet Dev. 1996;6:295–300. doi: 10.1016/s0959-437x(96)80005-0. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- Di Nunzio F, Maruggi G, Ferrari S, Di Iorio E, Poletti V, Garcia M, et al. Correction of laminin-5 deficiency in human epidermal stem cells by transcriptionally targeted lentiviral vectors. Mol Ther. 2008;16:1977–1985. doi: 10.1038/mt.2008.204. [DOI] [PubMed] [Google Scholar]
- Moiani A, Paleari Y, Sartori D, Mezzadra R, Miccio A, Cattoglio C, et al. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J Clin Invest. 2012;122:1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Marinkovich MP, Veis A, Cai X, Rao CN, O'Toole EA, et al. Interactions of the amino-terminal noncollagenous (NC1) domain of type VII collagen with extracellular matrix components. A potential role in epidermal-dermal adherence in human skin. J Biol Chem. 1997;272:14516–14522. doi: 10.1074/jbc.272.23.14516. [DOI] [PubMed] [Google Scholar]
- Bruckner-Tuderman L, Höpfner B, Hammami-Hauasli N. Biology of anchoring fibrils: lessons from dystrophic epidermolysis bullosa. Matrix Biol. 1999;18:43–54. doi: 10.1016/s0945-053x(98)00007-9. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Kandavelou K, Mani M, Durai S, Chandrasegaran S. “Magic” scissors for genome surgery. Nat Biotechnol. 2005;23:686–687. doi: 10.1038/nbt0605-686. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y, et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc Natl Acad Sci USA. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Moehle EA, Rock JM, Rock JM, Lee YL, Lee YL, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradori L, Chavanas S, Schaapveld RQ, Gagnoux-Palacios L, Calafat J, Meneguzzi G, et al. Role of the bullous pemphigoid antigen 180 (BP180) in the assembly of hemidesmosomes and cell adhesion–reexpression of BP180 in generalized atrophic benign epidermolysis bullosa keratinocytes. Exp Cell Res. 1998;239:463–476. doi: 10.1006/excr.1997.3923. [DOI] [PubMed] [Google Scholar]
- Del Rio M, Larcher F, Serrano F, Meana A, Muñoz M, Garcia M, et al. A preclinical model for the analysis of genetically modified human skin in vivo. Hum Gene Ther. 2002;13:959–968. doi: 10.1089/10430340252939069. [DOI] [PubMed] [Google Scholar]
- Larcher F, Dellambra E, Rico L, Bondanza S, Murillas R, Cattoglio C, et al. Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy. Mol Ther. 2007;15:1670–1676. doi: 10.1038/sj.mt.6300238. [DOI] [PubMed] [Google Scholar]
- Cavazza A, Moiani A, Mavilio F. Mechanisms of retroviral integration and mutagenesis. Hum Gene Ther. 2013;24:119–131. doi: 10.1089/hum.2012.203. [DOI] [PubMed] [Google Scholar]
- Recchia A, Parks RJ, Lamartina S, Toniatti C, Pieroni L, Palombo F, et al. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc Natl Acad Sci USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves MA, van Nierop GP, Tijssen MR, Lefesvre P, Knaän-Shanzer S, van der Velde I, et al. Transfer of the full-length dystrophin-coding sequence into muscle cells by a dual high-capacity hybrid viral vector with site-specific integration ability. J Virol. 2005;79:3146–3162. doi: 10.1128/JVI.79.5.3146-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A, Perani L, Sartori D, Olgiati C, Mavilio F. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol Ther. 2004;10:660–670. doi: 10.1016/j.ymthe.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Llames SG, Del Rio M, Larcher F, García E, García M, Escamez MJ, et al. Human plasma as a dermal scaffold for the generation of a completely autologous bioengineered skin. Transplantation. 2004;77:350–355. doi: 10.1097/01.TP.0000112381.80964.85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.