Abstract
The nuclear envelope (NE) is connected to the different types of cytoskeletal elements by linker of nucleoskeleton and cytoskeleton (LINC) complexes. LINC complexes exist from yeast to humans, and have preserved their general architecture throughout evolution. They are composed of SUN and KASH domain proteins of the inner and the outer nuclear membrane, respectively. These SUN–KASH bridges are used for the transmission of forces across the NE and support diverse biological processes. Here, we review the function of SUN and KASH domain proteins in various unicellular and multicellular species. Specifically, we discuss their influence on nuclear morphology and cytoskeletal organization. Further, emphasis is given on the role of LINC complexes in nuclear anchorage and migration as well as in genome organization.
Keywords: LINC complex, SUN domain, KASH, Nesprins, Nuclear migration, Nuclear envelope
Introduction
Linker of nucleoskeleton and cytoskeleton (LINC) complexes are built from members of two conserved protein families, SUN and KASH domain proteins. SUN domain proteins are integral to the inner nuclear membrane (INM), whereas KASH domain proteins reside in the outer nuclear membrane (ONM). SUN and KASH proteins directly bind each other in the perinuclear space (PNS), thereby forming a bridge across the nuclear envelope (NE). On the nucleoplasmic face of the NE, SUN domain proteins engage into interactions with other INM proteins, chromatin, and, in metazoans, the nuclear lamina. In the cytoplasm, KASH domain proteins associate with all major cytoskeletal elements, i.e., actin, intermediate filaments, and microtubules. Based on their bridging molecular architecture, LINC complexes serve as handles for the cytoskeleton on the nucleus, as well as for force transduction between nucleo- and cytoplasm. Consistently, functions of LINC complexes involve mechanical action and crosstalk between both sides of the NE (Burke and Roux 2009; Razafsky and Hodzic 2009; Starr and Fridolfsson 2010).
SUN domain proteins are type II membrane proteins. They comprise an N-terminal, nucleo/cytoplasmic portion and one transmembrane segment, followed by a coiled-coil region and the SUN domain in the C-terminal, luminal part of the protein. On the sequence level, the SUN domain is highly conserved, while other regions of SUN proteins are rather diverse (Hiraoka and Dernburg 2009; Razafsky and Hodzic 2009).
KASH domain proteins are tail-anchored membrane proteins. They are composed of an N-terminal, cytoplasmic part, and the C-terminal KASH domain, which includes both the membrane anchor and a luminal peptide of typically 20-30 amino acids. The cytoplasmic portions of KASH domain proteins differ in size, structure, and function. The KASH domain, in contrast, is conserved both in the transmembrane segment and in the luminal peptide (Razafsky and Hodzic 2009; Starr and Fischer 2005). Importantly, a motif of two or three prolines followed by one variable amino acid (PPPX) features the very C terminus of typical metazoan KASH peptides, and hydrophobic residues are conserved further upstream in the peptide sequence. Both elements are essential for interaction between KASH and SUN domains, as well as for SUN-mediated localization of KASH to the ONM (Sosa et al. 2012; Stewart-Hutchinson et al. 2008). In contrast to metazoan KASH domains, ONM constituents of LINC complexes in yeast contain atypical luminal peptides with low sequence conservation (Razafsky and Hodzic 2009; Starr and Fischer 2005). Based on their preserved biological role, the term KASH domain protein will here be used for both metazoan and yeast ONM components.
The hallmark of LINC complexes is the interaction of SUN domains and KASH peptides in the PNS. In crystal structures of mammalian LINC, this centerpiece of the complex is hexameric, composed of three protomers of each SUN and KASH. The three SUN domains form a globular assembly, which is organized by a preceding trimeric coiled-coil. This arrangement of SUN domains is essential for KASH binding, and each SUN trimer harbors three binding sites for KASH peptides at the interfaces of neighboring SUN domains (Sosa et al. 2012).
The repertoire of LINC components is evolutionary diverse. One SUN domain protein has been identified in both budding and fission yeast, two are found in Caenorhabditis elegans and Drosophila melanogaster, and mammals posses at least five family members. Mammalian SUN1 and SUN2 are expressed in most tissues and organs, whereas SUN3 to 5 are specific to the male germline (Hiraoka and Dernburg 2009; Razafsky and Hodzic 2009).
KASH domain proteins have diverged even further than their INM counterparts. Two KASH proteins have been identified in both budding and fission yeast. Three family members are found in C. elegans, and two in D. melanogaster. In vertebrates, six KASH domain proteins have been described: Nesprin-1 to Nesprin-4, KASH5, and LRMP (Mellad et al. 2010; Morimoto et al. 2012; Lindeman and Pelegri 2012; Noegel and Neumann 2011). While Nesprin-1, Nesprin-2, and Nesprin-3 are expressed ubiquitously, Nesprin-4 is specific to epithelial cells and KASH5 is only expressed in meiosis (Morimoto et al. 2012; Mellad et al. 2010). LRMP has originally been described as lymphoid-restricted protein in mammals and its zebrafish homolog has recently been characterized in the zygote and the early embryo (Behrens et al. 1994; Lindeman and Pelegri 2012). Mammalian Nesprin-1 and Nesprin-2 exist in various splice isoforms, including giant variants of up to 800 MDa as well as many smaller ones. Most Nesprins reside in the ONM and form part of classical LINC complexes that connect the nucleus to the cytoskeleton. Some isoforms have also been described at the INM, and soluble Nesprin variants lacking the KASH domain were found in the nucleo- and cytoplasm (Mellad et al. 2010; Warren et al. 2005; Noegel and Neumann 2011).
Little is known about specificities in SUN–KASH association. With some exceptions, SUN and KASH domain proteins seem to bind each other promiscuously. Some LINC constituents are, however, tissue- or cell-type-specific and their expression patterns hence determine the composition of complexes. Plasticity and regulation of LINC complexes have so far remained unexplored (Burke and Roux 2009; Razafsky and Hodzic 2009; Starr and Fridolfsson 2010).
In their position as NE bridges and linkers of nucleo- and cytoskeleton, SUN–KASH assemblies form integral elements of cellular architecture. Here, we summarize the influence of LINC complexes on nuclear and cytoskeletal structure, and review their functions in nuclear positioning and chromatin organization.
LINC complexes determine nuclear morphology and membrane structure
Size and shape of the cell nucleus are largely determined by its structural constituents—the lamina, NE proteins, and chromatin in the first place (Walters et al. 2012). Aberrant nuclear morphologies are a hallmark of laminopathic diseases, indicating that the maintenance of nuclear structure may be critical for function (Worman et al. 2010). As integral NE components, SUN and KASH domain proteins are thought to organize the nuclear membrane system and the nucleus as a whole (Fig. 1). Size and shape of both animal and plant nuclei are influenced by LINC complexes (Table 1) (Luke et al. 2008; Lu et al. 2012; Zhou et al. 2012; Zhang et al. 2007a). In cultured mammalian cells, an interplay between different KASH domain proteins has been implicated in nuclear size control. Giant isoforms of Nesprin-1 and Nesprin-2 were shown to bind to Nesprin-3, and disruption of Nesprin interchain associations led to larger nuclei (Taranum et al. 2012; Lu et al. 2012). Furthermore, LINC complexes are required to maintain the even spacing of the nuclear membranes. LINC disruption in Hela cells caused irregular expansions of the PNS and bulges of the ONM (Crisp et al. 2006). In fact, structural analyses suggest that SUN–KASH assemblies would be well suited to determine the width of the NE (Sosa et al. 2012; Rothballer et al. 2013).
Fig. 1.
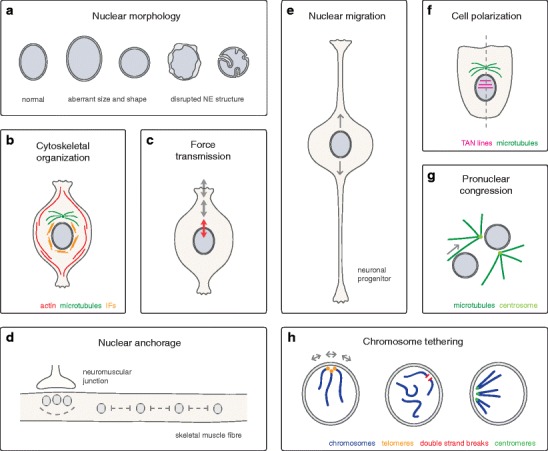
Cellular functions of LINC complexes. a Nuclear morphology. LINC complexes are required to maintain nuclear size and shape in mammals (Lu et al. 2012; Luke et al. 2008) and in A. thaliana (Zhou et al. 2012), as well as structure and integrity of the mammalian NE (Crisp et al. 2006; Zhang et al. 2007a). b Cytoskeletal organization. LINC complexes influence structure and distribution of perinuclear actin and intermediate filaments (IFs) in vertebrate cells (Khatau et al. 2009; Lombardi et al. 2011; Schneider et al. 2011; Morgan et al. 2011; Postel et al. 2011; Chambliss et al. 2013), and tether centrosomes to the NE in various metazoans (Schneider et al. 2011; Zhang et al. 2009; Malone et al. 2003; Morgan et al. 2011; Roux et al. 2009; Patterson et al. 2004). c Force transmission. LINC complexes transmit forces across the NE and affect mechanical properties of cultured mammalian cells (Lombardi et al. 2011; Anno et al. 2012; Stewart-Hutchinson et al. 2008). d Nuclear anchorage. LINC complexes mediate anchorage and positioning of nuclei in syncytial systems of various metazoans (Zhang et al. 2010; Zhang et al. 2007b; Grady et al. 2005; Lei et al. 2009; Starr and Han 2002; Malone et al. 1999; Elhanany-Tamir et al. 2012). In mammalian skeletal muscle (shown here), LINC complexes are required for the even spacing of extrasynaptic nuclei throughout myotubes, as well as for the clustering of synaptic nuclei beneath the neuromuscular junction (Zhang et al. 2010; Zhang et al. 2007b; Grady et al. 2005; Lei et al. 2009). e Nuclear migration. LINC complexes function in nuclear migration during various metazoan developmental events (Yu et al. 2011; Zhang et al. 2009; Malone et al. 1999; McGee et al. 2006; Starr et al. 2001; Meyerzon et al. 2009a; Fridolfsson et al. 2010; Mosley-Bishop et al. 1999; Fischer-Vize and Mosley 1994; Patterson et al. 2004; Kracklauer et al. 2007). In neural progenitors of the mammalian neocortex and retina (shown here), LINC complex-mediated nuclear migration processes are essential for proliferation and differentiation (Yu et al. 2011; Zhang et al. 2009). f Cell polarization. LINC complexes are required for nuclear positioning and orientation of the nuclear–centrosomal axis during fibroblast polarization (Lombardi et al. 2011; Luxton et al. 2010). LINC complexes form transmembrane actin-associated nuclear (TAN) lines to couple the nucleus to retrograde actin flow (Luxton et al. 2010). g Pronuclear congression. LINC complexes function in congression of male and female pronuclei in the fertilized zygote of C. elegans and vertebrates. Dedicated KASH domain proteins connect pronuclei to microtubule asters and the centrosome to allow their migration towards each other (Malone et al. 2003; Lindeman and Pelegri 2012). h Chromosome tethering. LINC complex-mediated tethering of chromosomes to the NE plays a role in various biological processes. In S. cerevisiae, tethering of telomeres and DNA double strand breaks has been implicated in silencing, stabilization, and repair (Bupp et al. 2007; Schober et al. 2009; Oza et al. 2009). In S. pombe, tethering of centromeres is important for mitotic chromosome segregation (Hou et al. 2012). Meiotic chromosomes are anchored to the NE via LINC complexes in both yeast and metazoans with impact on homolog pairing and recombination (Conrad et al. 2008; Conrad et al. 2007; Kosaka et al. 2008; Wanat et al. 2008; Chikashige et al. 2006; Shimanuki et al. 1997; Penkner et al. 2007; Penkner et al. 2009; Sato et al. 2009; Ding et al. 2007; Morimoto et al. 2012)
Table 1.
Functions of LINC complexes
| Cytoplasmic factors | KASH domain proteins | SUN domain proteins | Nucleoplasmic factors | Functions | References | |
|---|---|---|---|---|---|---|
| Vertebrates | Actin | Nesprin-1 giant | SUN1/2 | nd | Nuclear anchorage in muscle fibers | (Grady et al. 2005; Zhang et al. 2010; Zhang et al. 2007b; Lei et al. 2009) |
| Dynein, kinesin-1 | Nesprin-1/2 | SUN1/2 | nd | Nucleus-centrosome attachment and nuclear migration in neural progenitors | (Zhang et al. 2009) | |
| Dynein, kinesin-1 | Nesprin-2 | SUN1/2 | nd | Nuclear migration in photoreceptor progenitors | (Yu et al. 2011; Razafsky et al. 2012) | |
| Actin | Nesprin-2 giant | SUN2 | Lamin A/C | Formation of TAN lines, polarization of cultured cells | (Folker et al. 2011; Luxton et al. 2010; Luke et al. 2008) | |
| Kinesin-1 | Nesprin-2 | SUN1/2 | Lamin A/C | Cytoskeletal organization, nucleus-centrosome attachment, polarization of cultured cells | (Schneider et al. 2011; Lombardi et al. 2011; Rashmi et al. 2011) | |
| nd | Nesprin-2 | SUN1/2 | nd | Cell proliferation and differentiation during wound healing in vivo | (Rashmi et al. 2011) | |
| nd | Nesprin-1, other Nesprins | SUN1/2 | nd | Mechanics and force transduction in cultured cells | (Stewart-Hutchinson et al. 2008; Lombardi et al. 2011; Anno et al. 2012) | |
| Plectin | Nesprin-3α | SUN1/2 | Lamin A/C | Cytoskeletal organization, nucleus-centrosome attachment, polarization of cultured cells | (Ketema et al. 2007; Morgan et al. 2011; Wilhelmsen et al. 2005; Postel et al. 2011) | |
| nd | Nesprin-1/2/3 | SUN1/2 | nd | Organization of NE morphology in cultured cells | (Crisp et al. 2006) | |
| nd | Nesprin-1/2 giant and Nesprin-3 | SUN1/2 | nd | Nuclear size control in cultured cells | (Lu et al. 2012) | |
| nd | nd | SUN1/2 | DNAPK | DNA damage response in primary cells | (Lei et al. 2012) | |
| Kinesin-1 | Nesprin-4 | SUN1 | nd | Nuclear positioning in outer hair cells of the cochlea | (Roux et al. 2009; Horn et al. 2013) | |
| Dynein | KASH 5 | SUN1 | nd | Tethering of telomeres to the NE in meiosis | (Morimoto et al. 2012; Ding et al. 2007; Boateng et al. 2013) | |
| nd | LRMP | nd | nd | Pronucleus-centrosome attachment and pronuclear congression in the zygote | (Lindeman and Pelegri 2012) | |
| nd | Nesprin-3 | SUN1η | nd | Sperm development | (Gob et al. 2010) | |
| nd | Nesprin-1 | SUN3 | nd | Sperm development | (Gob et al. 2010) | |
| C. elegans | Actin | ANC-1 | UNC-84 | nd | Nuclear anchorage in syncytia | (Starr and Han 2002; Malone et al. 1999) |
| Dynein, kinesin-1 | UNC-83 | UNC-84 | nd | Nuclear migration in various cell types | (Starr et al. 2001; Fridolfsson et al. 2010; McGee et al. 2006; Meyerzon et al. 2009a; Malone et al. 1999) | |
| Dynein, ZYG-12A | ZYG-12 | SUN-1 | nd | Pronucleus-centrosome attachment and pronuclear congression in the zygote | (Malone et al. 1999; Minn et al. 2009) | |
| Dynein | ZYG-12 | SUN-1 | nd | Cytoskeletal organization and nuclear positioning in the gonad | (Zhou et al. 2009) | |
| Dynein | ZYG-12 | SUN-1 | HIM-8/ZIM1-3 | Tethering of chromosomes to the NE in meiosis | (Labella et al. 2011; Penkner et al. 2007; Penkner et al. 2009; Sato et al. 2009) | |
| nd | KDP-1 | SUN-1/ UNC-84 | nd | Cell cycle regulation in the germline and the embryo | (McGee et al. 2009) | |
| nd | nd | SUN-1 | nd | Apoptosis in the embryo | (Tzur et al. 2006) | |
| D. melanogaster | Actin, microtubules | MSP-300, Klarsicht | Klaroid | nd | Nuclear anchorage in muscle fibers | (Technau and Roth 2008; Rosenberg-Hasson et al. 1996; Volk 1992; Elhanany-Tamir et al. 2012) |
| Microtubules | Klarsicht | Klaroid | Lamin Dm(0) | Nucleus-centrosome attachment and nuclear migration during eye development | (Kracklauer et al. 2007; Mosley-Bishop et al. 1999; Patterson et al. 2004) | |
| Yuri Gagarin, dynein | nd | Spag-4 | nd | Nucleus-centrosome attachment during spermatogenesis | (Kracklauer et al. 2010) | |
| S. pombe | nd | Kms1, Kms2 | Sad1 | nd | SPB integrity, mitotic spindle formation | (Hagan and Yanagida 1995; Miki et al. 2004; Shimanuki et al. 1997) |
| nd | nd | Sad1 | Csi1 | Tethering of centromeres to the NE in interphase | (Hou et al. 2012) | |
| Dynein | Kms1 | Sad1 | Bqt1, Bqt2 | Tethering of telomeres to the NE in meiosis | (Chikashige et al. 2006; Shimanuki et al. 1997) | |
| S. cerevisiae | nd | Mps2 | Mps3 | nd | SPB integrity and duplication, mitotic spindle formation | (Jaspersen et al. 2002; Jaspersen et al. 2006; Munoz-Centeno et al. 1999; Winey et al. 1991; Friederichs et al. 2011) |
| nd | nd | Mps3 | Sir4 | Tethering of telomeres to the NE during vegetative growth | (Bupp et al. 2007) | |
| nd | nd | Mps3 | Sir4, Ebp2, Rrs1 | Clustering of tethered telomeres at the NE | (Horigome et al. 2011) | |
| nd | nd | Mps3 | Telomerase, Ku70/80 | Tethering of telomeres to the NE during vegetative growth | (Schober et al. 2009; Antoniacci et al. 2007) | |
| nd | nd | Mps3 | Telomerase, Ku70/Ku80 | Tethering of DNA double strand breaks to the NE | (Oza et al. 2009) | |
| nd | nd | Mps3 | TFIIIC | Tethering of extra TFIIIC sites to the NE | (Hiraga et al. 2012) | |
| nd | nd | Mps3 | Ctf7 | Sister chromatid cohesion | (Antoniacci et al. 2004) | |
| nd | nd | Mps3 | Replication factor C, Htz1 | Potentially sister chromatid cohesion or DNA repair | (Haas et al. 2012) | |
| Actin | Csm4 | Mps3 | Ndj1 | Tethering of telomeres to the NE in meiosis | (Conrad et al. 2008; Conrad et al. 2007; Kosaka et al. 2008; Wanat et al. 2008) | |
| A. thaliana | AtRanGAP1 | AtWIP1-3 | AtSUN1/2 | nd | Anchorage of AtRanGAP1 at the NE, maintenance of nuclear shape | (Zhou et al. 2012) |
The table lists characterized functions of LINC complexes in model organisms, the involved SUN and KASH domain proteins, as well as their cytoplasmic and nucleoplasmic interaction partners. For SUN and KASH domain proteins written in bold, the respective functions have directly been demonstrated; others have been implicated indirectly
nd not defined
LINC complexes organize the cytoskeleton
LINC complexes also act as cytoskeletal organizers (Fig. 1). In vertebrate cells, Nesprins determine the structure and distribution of perinuclear actin and intermediate filaments with impact on overall cellular shape and mechanics (Table 1) (Khatau et al. 2009; Lombardi et al. 2011; Schneider et al. 2011; Morgan et al. 2011; Postel et al. 2011; Chambliss et al. 2013). Giant isoforms of Nesprin-1 and Nesprin-2 directly bind to actin via N-terminal calponin-homology (CH) domains (Fig. 2) (Zhang et al. 2002; Zhen et al. 2002; Padmakumar et al. 2004). Intermediate filaments are connected to the nucleus by Nesprin-3 and the cytoskeletal crosslinker plectin (Fig. 2) (Ketema et al. 2007; Wilhelmsen et al. 2005). Consistent with their role in cytoskeletal organization, LINC components were found to influence organelle positioning, cell polarity, and ciliogenesis (Schneider et al. 2011; Hedgecock and Thomson 1982; Starr and Han 2002; Dawe et al. 2009; Elhanany-Tamir et al. 2012).
Fig. 2.

Nucleo-cytoskeletal interactions through LINC complexes. SUN-KASH pairs (dark and light blue) and their connections to the cytoskeleton in mammals (a), C. elegans (b), and D. melanogaster (c). Trimeric organization of SUN domain proteins (Sosa et al. 2012) has been omitted for clarity. Giant KASH domain proteins directly bind to actin (red) (Zhang et al. 2002; Zhang et al. 2001; Zhen et al. 2002; Padmakumar et al. 2004; Volk 1992; Starr and Han 2002). Intermediate filaments (orange) and microtubules (green) are linked via plectin (Ketema et al. 2007; Wilhelmsen et al. 2005) and motor proteins (Schneider et al. 2011; Yu et al. 2011; Zhang et al. 2009; Meyerzon et al. 2009a; Fridolfsson et al. 2010), respectively. C. elegans ZYG-12A directly associates with the centrosome (Malone et al. 2003). D. melanogaster Klarsicht colocalizes with microtubules, but their molecular connection has not been characterized (Fischer et al. 2004). Spag4 cooperates with the coiled-coil protein Yuri Gagarin and dynein, potentially via a KASH domain protein (Kracklauer et al. 2010). Question marks indicate that the specific protein or structure involved in the depicted complexes has not yet been defined
Importantly, metazoan SUN–KASH pairs mediate the attachment of the centrosome to the nucleus, which is thought to be essential during nuclear and cell migration (Table 1) (Schneider et al. 2011; Zhang et al. 2009; Malone et al. 2003; Morgan et al. 2011; Roux et al. 2009; Patterson et al. 2004). Similarly, yeast SUN and KASH domain proteins function at the spindle pole body (SPB; Table 1) (Jaspersen et al. 2002; Jaspersen et al. 2006; Winey et al. 1991; Hagan and Yanagida 1995). SPBs are multi-subunit protein complexes integrated into or closely associated with the NE. They serve as the main microtubule organizing centers (MTOC) in yeast cells. Duplication of the SPB occurs via formation and maturation of a cytoplasmic complex, which is subsequently inserted into the NE. In Saccharomyces cerevisiae, SPB insertion is coupled to its duplication, and mature complexes remain membrane-integrated throughout the cell cycle. Schizosaccharomyces pombe SPBs, in contrast, lie closely attached to the ONM during interphase and integrate into the NE only during mitosis to organize the intranuclear microtubule spindle (Ding et al. 1997; Jaspersen and Winey 2004).
SUN and KASH domain proteins are structural components of the SPB in S. cerevisiae (Munoz-Centeno et al. 1999; Jaspersen et al. 2002), and form part of the MTOC attachment site in the NE in S. pombe (Hagan and Yanagida 1995; King et al. 2008; Shimanuki et al. 1997; Miki et al. 2004). In both yeasts, they are essential for SPB integrity and duplication, and consequently for mitotic spindle formation and chromosome segregation (Jaspersen et al. 2002; Jaspersen et al. 2006; Winey et al. 1991; Hagan and Yanagida 1995). The S. cerevisiae SUN domain protein Mps3 has been implicated specifically in the insertion of SPBs into the NE (Friederichs et al. 2011). SPB insertion requires a local fusion between INM and ONM to generate an aqueous pore in the double membrane into which complexes are embedded (Jaspersen and Ghosh 2012). Mps3 was shown to influence the lipid composition of the NE, which was proposed to modulate biophysical membrane properties required for pore formation. Alternatively, Mps3 may function by recruiting membrane fusion factors to the site of SPB integration (Friederichs et al. 2011; Jaspersen and Ghosh 2012).
LINC complexes transmit forces at the nuclear envelope
The cytoskeleton is the major determinant of the physical and mechanical properties of a cell, and mediates its responses to respective cues from the surrounding. LINC complexes connect the nucleus to the cytoskeleton, and modulate cellular mechanics and force transmission throughout the cell (Fig. 1; Table 1) (Lombardi et al. 2011; Anno et al. 2012; Stewart-Hutchinson et al. 2008). In cultured mammalian cells, LINC disruption was shown to affect the stiffness of the cytoplasm (Stewart-Hutchinson et al. 2008). Biophysical assays that directly measure force propagation in cells further revealed a role of SUN–KASH complexes in the mechanical coupling of nucleus and cytoskeleton. Forces were applied either at the cell surface or in the cytoplasm, e.g., by stretching of the growth substrate, and resulting changes in cellular and nuclear shape were analyzed. Deformation of the nucleus was strongly impaired upon LINC disruption indicating that the complexes transmit forces at the NE (Lombardi et al. 2011; Anno et al. 2012). Activation of mechanosensitive genes, however, was unaffected. LINC complexes thus seem to be critical for physical responses to mechanical cues but dispensable for mechanotransduction signaling (Lombardi et al. 2011).
LINC complexes anchor and move the nucleus
Many founding members of the SUN and KASH domain protein families have initially been identified in model organisms based on mutant phenotypes affecting nuclear anchorage or migration. The anchorage of nuclei in cells or syncytia mostly relies on the actin cytoskeleton. Nuclear migration events, in contrast, often involve microtubules, motor proteins, and centrosomes (Table 1). From yeast to humans, nuclear anchorage and migration are essential for reproduction, development, and differentiation (Burke and Roux 2009; Starr and Fridolfsson 2010; Razafsky et al. 2011).
Nuclear anchorage in syncytia
Nuclear anchorage is particularly important in syncytia, multinucleate cells developed through cell fusions (Daubenmire 1936) (Fig. 1). Anchorage is generally achieved by dedicated KASH domain proteins, which tether nuclei to the actin cytoskeleton (Fig. 2; Table 1). C. elegans ANC-1 (Starr and Han 2002), D. melanogaster MSP-300 (Volk 1992), and the giant isoforms of vertebrate Nesprin-1 and Nesprin-2 (Zhang et al. 2002; Zhang et al. 2001; Zhen et al. 2002; Padmakumar et al. 2004), share an enormous size of ∼800 kDa as well as their functional domain structure. They contain a tandem repeat of N-terminal actin-binding domains, an extended middle region built from repetitive elements, as well as the C-terminal KASH domain. While the middle regions of MSP-300 and Nesprins are composed of spectrin repeats, coiled-coils constitute the backbone of ANC-1 (Zhang et al. 2002; Zhang et al. 2001; Zhen et al. 2002; Padmakumar et al. 2004; Volk 1992; Starr and Han 2002).
Adult C. elegans are covered with several large syncytia containing more than a hundred nuclei in total (Sulston and Horvitz 1977). LINC complexes composed of ANC-1 and the SUN domain protein UNC-84 anchor syncytial nuclei to the actin cytoskeleton and aid to maintain their equal distribution. In ANC-1 or UNC-84 mutants, nuclei float freely in the cytoplasm or accumulate in clusters (Hedgecock and Thomson 1982; Starr and Han 2002; Malone et al. 1999).
Similarly, mammalian skeletal muscle is composed of syncytial myotubes. Nuclei are evenly spaced throughout myotubes with some specialized synaptic nuclei clustered beneath the neuromuscular junction (NMJ) (Bruusgaard et al. 2003). Nuclear anchorage and positioning in muscle cells relies primarily on Nesprin-1, in conjunction with SUN1 and SUN2, as deduced from knockout mice phenotypes (Zhang et al. 2010; Zhang et al. 2007b; Grady et al. 2005; Lei et al. 2009). The two SUN domain proteins seem to fulfill dedicated functions in muscle cells. SUN2 is expressed in both synaptic and extrasynaptic nuclei, whereas high levels of SUN1 could only be detected outside the synapse. Localization of both SUN2 and Nesprin-1 throughout the NE of synaptic, but not of extrasynaptic nuclei depended on integrity of the Lamin A network, suggesting that LINC complex defects may contribute to NMJ phenotypes observed in laminopathic diseases (Mejat et al. 2009).
Organization of nuclei in muscle fibers has recently also been characterized in D. melanogaster. A complex interplay between both membrane-bound and KASH-less isoforms of MSP-300, as well as the microtubule-associated KASH domain protein Klarsicht has been revealed (Elhanany-Tamir et al. 2012; Volk 2012). A function of MSP-300 in nuclear positioning in fly oocytes has also been suggested (Yu et al. 2006), but its role remains controversial (Xie and Fischer 2008; Technau and Roth 2008).
Nuclear migration in development
Nuclear migration events are critical at diverse steps of metazoan development (Fig. 1; Table 1) (Burke and Roux 2009; Starr and Fridolfsson 2010; Razafsky et al. 2011). In C. elegans, the SUN–KASH pair UNC-83/UNC-84 has been implicated in nuclear migration in a variety of tissues including epidermal precursors and P cells, which give rise to vulval cells and neurons. Mutants of either UNC protein have uncoordinated movement and egg-laying-defective phenotypes due to failure of P cell nuclear migration (Horvitz and Sulston 1980; Sulston and Horvitz 1981; Malone et al. 1999; McGee et al. 2006; Starr et al. 2001). UNC-83 interacts with kinesin-1 as well as with two distinct cytoplasmic dynein adaptor complexes (Fig. 2) (Meyerzon et al. 2009a; Fridolfsson et al. 2010). Based on mutant phenotypes, differential roles in nuclear migration have been ascribed to the motor proteins. Kinesin-mediated movement towards microtubule plus ends is thought to constitute the main driving force, whereas minus end-directed movement via dynein may overcome hindrances or modulate migration (Fridolfsson et al. 2010; Meyerzon et al. 2009a).
In D. melanogaster, nuclear migration is important for eye development (Tomlinson 1985). The KASH domain protein Klarsicht, in conjunction with the SUN domain protein Klaroid, have been implicated in this process (Mosley-Bishop et al. 1999; Fischer-Vize and Mosley 1994; Patterson et al. 2004; Kracklauer et al. 2007). Klarsicht was shown to colocalize with microtubules and is thought to mediate nuclear movements via motor proteins (Fig. 2) (Welte 2004; Welte et al. 1998; Mosley-Bishop et al. 1999; Patterson et al. 2004; Fischer et al. 2004).
Direct homologs of UNC-83 or Klarsicht are absent in mammals (Mellad et al. 2010; Razafsky and Hodzic 2009; Starr and Fischer 2005; Starr and Fridolfsson 2010). However, small Nesprin-1 and Nesprin-2 isoforms as well as Nesprin-4 seem to fulfill analogous functions (Fig. 2) (Roux et al. 2009; Yu et al. 2011; Zhang et al. 2009). Nesprin-1/-2, together with SUN1/2, function in nuclear migration during neuronal development (Yu et al. 2011; Zhang et al. 2009). The characteristic multi-layered structure of the neocortex and the retina are established by coordinated events of nuclear and cell migration (Baye and Link 2008; Schaar and McConnell 2005). Two fundamental processes are interkinetic nuclear migration (IKNM) and radial neuronal migration (RNM). IKNM in neural progenitor cells describes movements of the nucleus between the basal and the apical surface of the neuroepithelium in coordination with the cell cycle. The centrosome remains apical during IKNM. During G2, the nucleus moves towards centrosomes and mitosis occurs at the apical surface. In G1, the nucleus moves back to the basal side, where DNA replication takes place. Apical and basal nuclear movements were suggested to involve dynein and kinesin, respectively, but alternative models also exist (Kosodo 2012; Meyer et al. 2011; Spear and Erickson 2012). IKNM is essential for the asymmetric division of progenitor cells and for neuronal differentiation.
RNM specifies the migration of neurons from the neuroepithelium into outer layers of the cortex (Lambert de Rouvroit and Goffinet 2001). The centrosome is constantly positioned behind the leading edge of the migrating cell, whereas the nucleus follows in a saltatory mode. Nuclear movement during RNM depends on dynein (Bellion et al. 2005; Tsai et al. 2007; Schaar and McConnell 2005). RNM critically contributes to formation of the striated structure of neocortex and retina (Lambert de Rouvroit and Goffinet 2001).
LINC complexes composed of Nesprin-1/-2 and SUN1/2 function in both IKNM and RNM. Double knockout of either both KASH or both SUN domain proteins in mice causes reduced brain size, severe cortical malformation, and lethality. Mice show depletion of neural progenitor pools and detachment of the centrosome from the nucleus in migrating neurons (Zhang et al. 2009). Similar phenotypes are observed in the retina upon disruption of Nesprin-2 or SUN1 and SUN2 (Yu et al. 2011). Importantly, Nesprin-2 was found to colocalize and interact with both dynein and kinesin-1 during cortical and retinal development (Fig. 2), strongly supporting a direct role of LINC complexes in neuronal nuclear migration (Yu et al. 2011; Zhang et al. 2009).
Nesprin-4 also binds to kinesin-1 (Fig. 2). Nesprin-4 is specific to epithelial cells, and its heterologous expression in Hela cells induces microtubule plus end-directed movement of nuclei into the cell periphery (Roux et al. 2009). Mouse knockout studies revealed that Nesprin-4, in collaboration with SUN1, is essential for hearing. In absence of either LINC component, the outer hair cells of the cochlea exhibited nuclear positioning defects and degeneration eventually leading to deafness (Horn et al. 2013).
Nuclear movement during cell polarization
Major reorganization of cells occurs during polarization as for instance in fibroblasts that prepare for cell migration (Fig. 1). Rearrangements include positioning of the nucleus and orientation of the nuclear–centrosomal axis. Specifically, the nucleus moves away from the leading edge towards the rear of the cell, while the centrosome stays in place. Nuclear movement is driven by retrograde actin flow, and depends on actin-associated LINC complexes built of Nesprin-2 giant and SUN2 (Fig. 2) (Gomes et al. 2005; Lombardi et al. 2011; Luxton et al. 2010; Luxton and Gundersen 2011). Interestingly, during fibroblast polarization in wounded cell monolayers, LINC components organize into linear arrays in the NE, termed transmembrane actin-associated nuclear (TAN) lines. TAN lines align with actin cables perpendicular to the leading edge, and are thought to couple the nucleus to retrograde actin flow (Luxton et al. 2010; Luxton et al. 2011).
Pronuclear congression in the zygote
A particular type of nuclear migration takes place in fertilized oocytes, where the male and female pronuclei congress to unite their haploid genomes and initiate the first mitotic division (Fig. 1). In many species, the centrosome is closely associated with the male pronucleus, and the female pronucleus migrates towards it along astral microtubules (Reinsch and Gonczy 1998; Schatten 1994). In the C. elegans zygote, nucleus-centrosome attachment and pronuclear migration are mediated by the KASH domain protein ZYG-12 and the SUN domain protein SUN-1 (Wood et al. 1980; Malone et al. 2003). ZYG-12 contains an N-terminal Hook domain, which anchors organelles to microtubules, and interacts with dynein (Walenta et al. 2001; Malone et al. 2003). KASH domain-containing isoforms of ZYG-12 localize to the ONM, whereas KASH-less forms are found at the centrosome (Fig. 2) (Malone et al. 2003). Pronucleus–centrosome association is thought to involve two steps. Membrane-bound ZYG-12 first captures microtubule asters via dynein and pulls them towards the nucleus (Malone et al. 2003; Meyerzon et al. 2009b). Attachment is then completed by direct interaction between ZYG-12 isoforms at the centrosome and the ONM (Malone et al. 2003).
Vertebrates do not posses a direct ZYG-12 homolog. Another potential KASH domain protein, LRMP, however, seems to fulfill the same principal task as recently characterized in zebrafish (Lindeman and Pelegri 2012). In absence of LRMP, the centrosome detaches from the male pronucleus, and male and female pronuclei fail to congress. LRMP localizes at the NE in the zygote and early embryo, and concentrates in microtubule-associated membrane regions, suggesting a direct function in NE–microtubule attachment. The existence of LRMP-containing LINC complexes and participating SUN domain proteins still await their identification.
LINC complexes organize the genome
Although the center stage for nuclear anchorage and migration seems to be placed on the cytoplasmic face of the NE, functions of LINC complexes in the nuclear interior are not less intricate. From yeast to humans, SUN and KASH domain proteins connect chromatin to the NE (Fig. 1). Diverse physical links between SUN domain proteins and chromosomes have been revealed in recent years, and many of them could be implicated in chromatin organization and function (Table 1).
Chromosome tethering to the nuclear envelope in vegetative and somatic cells
The spatial organization of the genome within the nucleus is not random. In general, the nuclear periphery is considered as repressive environment associated with heterochromatin and inactive genes. Lamins and INM proteins have been implicated in chromatin condensation and silencing (Mekhail and Moazed 2010; Towbin et al. 2009; Van de Vosse et al. 2011). The physical tethering of chromosomes to the NE is best understood in yeast. In S. cerevisiae, telomeres are sequestered at the nuclear periphery during interphase, which aids to suppress telomere transcription and subtelomeric recombination (Gartenberg 2009; Mekhail and Moazed 2010). Different pathways of NE–telomere tethering act in parallel, two of which converge at Mps3 as membrane anchor (Bupp et al. 2007; Schober et al. 2009). The first mechanism centers around the Sir complex of chromatin silencing factors. Sir4 interacts with Mps3, and is assisted by several other proteins to mediate the anchorage and clustering of telomeres at the NE (Bupp et al. 2007; Horigome et al. 2011). The second mechanism requires the telomere replication machinery, including the telomerase catalytic core, as well as the Ku70/Ku80 DNA-binding subunits (Schober et al. 2009). Linkage to the NE is thought to involve the interaction between Mps3 and the telomerase subunit Est1 (Antoniacci et al. 2007).
Interestingly, DNA double strand breaks (DSBs) seem to be handled by yeast cells similarly to telomeres. Persistent DSBs are shuttled to the nuclear periphery, where they are retained by Mps3 in cooperation with Ku70/Ku80 (Oza et al. 2009). Sequestration at the NE is thought to separate DSBs from bulk chromatin to allow for their processing along several possible routes. DSBs may be stabilized by the de novo addition of telomeres, they may be repaired by homologous recombination, or they may be passed on to NPCs to meet nuclear basket-associated DNA repair pathways (Gartenberg 2009; Oza and Peterson 2010). The spectrum of Mps3 interactions with chromatin regulators is constantly increasing with connections to insulator elements, cohesion factors, and histone variants uncovered lately (Antoniacci et al. 2004; Haas et al. 2012; Hiraga et al. 2012; Gardner et al. 2011).
Another link between a SUN domain protein and chromosomes has been described in S. pombe (Hou et al. 2012). During interphase, centromeres localize at the NE near the SPB, which is thought to assist their capturing by spindle microtubules upon mitotic entry. Centromere clustering is mediated by Sad1 and the nucleoplasmic adaptor Csi1, and disruption of this anchor was shown to cause defects in chromosome segregation and mitotic progression (Hou et al. 2012).
Connections between vertebrate LINC complexes and chromatin remain vague. Recently, SUN1 and SUN2 have been implicated in the DNA damage response (DDR) (Lei et al. 2012). SUN1/2 double knockout fibroblasts exhibited excessive DNA damage, genomic instability, and impaired activation of the DDR pathway. SUN1 and SUN2 were found to interact with DNA-dependent protein kinase, including the Ku70/Ku80 subunits, involved in DDR and DNA repair. Although no physical NE–chromatin tether could be revealed by this study, the principal role of SUN domain proteins in the handling of DNA damage may well be conserved from yeast to mammals.
Chromosome tethering to the nuclear envelope in meiosis
From yeast to humans, LINC complexes anchor chromosomes at the NE in the prophase of meiosis (Table 1). In most organisms, meiotic chromosomes are tethered via telomeres, which cluster at the NE giving rise to a typical bouquet configuration of chromosomes. NE tethering is often accompanied by dramatic movements of chromosomes within the nucleus or oscillation of the entire nucleus within the cell. Different cytoskeletal elements and meiosis-specific nuclear adaptor proteins cooperate with LINC complexes during these processes. Importantly, NE attachment and chromosome movements have been shown to assist homologous pairing and recombination and are required for faithful meiotic progression in both yeast and metazoans (Fridkin et al. 2009; Hiraoka and Dernburg 2009; Kracklauer et al. 2013).
In S. cerevisiae, meiotic bouquet formation is mediated by the SUN domain protein Mps3 and the meiosis-specific nuclear membrane protein Csm4, a potential atypical KASH domain protein (Conrad et al. 2008; Conrad et al. 2007; Kosaka et al. 2008; Wanat et al. 2008). They collaborate with the actin cytoskeleton (Trelles-Sticken et al. 2005; Scherthan et al. 2007; Koszul et al. 2008), and the meiosis-specific nuclear adaptor protein Ndj1, which connects Mps3 to telomeres (Conrad et al. 1997; Trelles-Sticken et al. 2000; Conrad et al. 2007). In meiotic prophase, Mps3 relocalizes from the SPB to telomere attachment sites at the NE consistent with establishment the molecular tether (Conrad et al. 2007). Subsequently, telomeres move along the NE concomitantly with deformation of the nuclear surface. Both processes are dependent on the actin cytoskeleton (Conrad et al. 2008; Scherthan et al. 2007; Trelles-Sticken et al. 2005; Koszul et al. 2008).
During meiotic prophase of S. pombe, telomeres are anchored to the NE by the SUN domain protein Sad1 and the KASH domain protein Kms1 (Chikashige et al. 2006; Chikashige et al. 2007; Shimanuki et al. 1997). The meiosis-specific adaptor proteins Bqt1 and Bqt2 connect Sad1 to telomeres (Chikashige et al. 2006), whereas microtubules and dynein act on the cytoplasmic side (Ding et al. 1998; Yamamoto et al. 1999; Miki et al. 2004; Goto et al. 2001; Miki et al. 2002). Similar to Mps3, Sad1 disperses from the SPB at the onset of meiosis and colocalizes with telomeres at the NE (Chikashige et al. 2006; Chikashige et al. 2007). Telomeres associate with the INM via a distinct mechanism during interphase (Chikashige et al. 2009), and are thought to be captured by Sad1 upon expression of meiotic adaptor proteins (Chikashige et al. 2006; Chikashige et al. 2007). Sad1 and telomeres subsequently refocus at a site close to the SPB. Led by this telomere attachment site, the nucleus then oscillates along the entire length of the cell, a process termed horsetail movement (Chikashige et al. 1994; Chikashige et al. 2006). In contrast to actin-dependent movements in S. cerevisiae, nuclear oscillation in S. pombe is mediated by dynein and microtubules (Ding et al. 1998; Yamamoto et al. 1999).
An exception from telomere-mediated NE attachment is found in C. elegans, where specific chromosomal regions, termed pairing centers, are involved (MacQueen et al. 2005; Phillips et al. 2005). Pairing centers are recognized by the meiosis-specific zinc-finger proteins HIM-8 and ZIM-1 to ZIM-3, which are required for the association of pairing centers with the NE (Phillips and Dernburg 2006; Phillips et al. 2005). LINC complexes composed of the SUN domain protein SUN-1 and the KASH domain protein ZYG-12 accumulate at chromosome attachment patches at the NE in meiotic prophase, where they function in chromosome tethering and homolog pairing (Penkner et al. 2007; Penkner et al. 2009; Sato et al. 2009). CHK2 and PLK-1/2 kinase activities are necessary for phosphorylation of the SUN-1 N terminus and for SUN-1 patch formation in meiotic prophase (Labella et al. 2011; Penkner et al. 2009). In the cytoplasm, ZYG-12 interacts with dynein (Fig. 2) (Malone et al. 2003), and dynein and microtubules have also been implicated in meiotic processes (Sato et al. 2009).
In mammals, the tethering of meiotic chromosomes to the NE requires SUN1 and KASH5 (Ding et al. 2007; Morimoto et al. 2012). Both SUN1 and SUN2 enrich at telomere attachment sites in meiotic prophase, but only SUN1 is essential for telomere tethering and homologous recombination in mice (Ding et al. 2007; Schmitt et al. 2007). KASH5 and dynein colocalize with SUN1 at telomere attachment sites, and are thought to form a NE bridge that links telomeres to microtubules and mediates meiotic chromosome movements (Morimoto et al. 2012). It will be interesting to see which nuclear adaptor proteins connect SUN1 to telomeres.
LINC components regulate signaling, cell division, and apoptosis
Besides their structural role in the NE, regulatory functions of LINC components have been described (Table 1). The C. elegans KASH domain protein KDP-1, in conjunction with SUN-1, regulates cell-cycle progression in the germline and in early embryos, and is essential for viability and development (McGee et al. 2009). SUN-1 also functions in apoptosis. Initiation of apoptosis involves the redistribution of the proapoptotic factor CED-4 from mitochondria to the NE (Chen et al. 2000). CED-4 redistribution depends on SUN-1, which binds to CED-4 in vitro and affects apoptosis in vivo (Tzur et al. 2006). Whether CED-4 enters the nucleus to contact SUN-1 directly, or is connected via an ONM KASH domain protein, is not resolved.
Mammalian Nesprin-2 has also been implicated in signal transduction and gene regulation. Nesprin-2 was shown to interact with α- and β-catenin through a conserved spectrin repeat region present in many isoforms (Neumann et al. 2010; Luke et al. 2008). Catenins are components of cell adhesions, and β-catenin additionally functions as a transcription factor of the Wnt pathway that localizes to the nucleus upon activation (Pandur et al. 2002). Nesprin-2 depletion affected nuclear levels of β-catenin, target gene expression, and cell proliferation in cultured cells (Neumann et al. 2010). In another study, a nucleoplasmic Nesprin-2 variant lacking the KASH domain was detected in complex with ERK1/2 (extracellular signal-regulated kinase 1 and 2) and PML (promyeolocytic leukemia protein). This isoform was required for association of ERK2 with PML nuclear bodies, and repressed ERK signaling and cell proliferation (Warren et al. 2010).
Concluding remarks
Their versatile molecular tasks make SUN and KASH domain proteins indispensable for cellular function. Not surprisingly so, disruption of LINC complexes severely affects viability, development, and reproduction (Kracklauer et al. 2013; Starr and Fridolfsson 2010). Mutations in LINC components have been associated with laminopathic diseases including Emery-Dreifuss muscular dystrophy caused by mutation of the LINC interaction partner Emerin or of Nesprin-1 or Nesprin-2 (Fridkin et al. 2009; Meinke et al. 2011; Mejat and Misteli 2011). An astonishing link between SUN1 and Hutchinson-Gilford progeria syndrome (HGPS) has recently been uncovered. SUN1 was found to accumulate in HGPS mouse models as well as in patient cells, and depletion of SUN1 alleviated progeric phenotypes, indicating that the SUN domain protein may play a causative role in HGPS (Chen et al. 2012). A future challenge will clearly be to dissect the relations between the molecular functions of LINC complexes in the NE and their impact on physiology and disease.
Acknowledgments
We thank the members of the Kutay lab for helpful discussions, and the Swiss National Science Foundation, and the European Research Council for funding.
References
- Anno T, Sakamoto N, Sato M. Role of nesprin-1 in nuclear deformation in endothelial cells under static and uniaxial stretching conditions. Biochem Biophys Res Commun. 2012;424(1):94–99. doi: 10.1016/j.bbrc.2012.06.073. [DOI] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Skibbens RV. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6(1):75–79. doi: 10.4161/cc.6.1.3647. [DOI] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Uetz P, Fields S, Skibbens RV. The spindle pole body assembly component mps3p/nep98p functions in sister chromatid cohesion. J Biol Chem. 2004;279(47):49542–49550. doi: 10.1074/jbc.M404324200. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TW, Jagadeesh J, Scherle P, Kearns G, Yewdell J, Staudt LM. Jaw1, A lymphoid-restricted membrane protein localized to the endoplasmic reticulum. J Immunol. 1994;153(2):682–690. [PubMed] [Google Scholar]
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25(24):5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD (2013) Homologous Pairing Preceding SPO11-Mediated Double-Strand Breaks in Mice. Dev Cell 24(2):196–205. doi:S1534-5807(12)00575-8 [DOI] [PMC free article] [PubMed]
- Bruusgaard JC, Liestol K, Ekmark M, Kollstad K, Gundersen K. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J Physiol. 2003;551(Pt 2):467–478. doi: 10.1113/jphysiol.2003.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179(5):845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17(5):587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Chambliss AB, Khatau SB, Erdenberger N, Robinson DK, Hodzic D, Longmore GD, Wirtz D. The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci Rep. 2013;3:1087. doi: 10.1038/srep01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang KT. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149(3):565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz HR. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287(5457):1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264(5156):270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma. 2007;116(6):497–505. doi: 10.1007/s00412-007-0114-8. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125(1):59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. Membrane proteins Bqt3 and −4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol. 2009;187(3):413–427. doi: 10.1083/jcb.200902122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276(5316):1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133(7):1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104(21):8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172(1):41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenmire RF. The use of the terms coenocyte and syncytium in biology. Science. 1936;84(2189):533. doi: 10.1126/science.84.2189.533. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Adams M, Wheway G, Szymanska K, Logan CV, Noegel AA, Gull K, Johnson CA. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J Cell Sci. 2009;122(Pt 15):2716–2726. doi: 10.1242/jcs.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8(8):1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12(6):863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol. 2012;198(5):833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Acosta S, Kenny A, Cater C, Robinson C, Hook J. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 2004;168(3):1385–1393. doi: 10.1534/genetics.104.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Vize JA, Mosley KL. Marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development. 1994;120(9):2609–2618. doi: 10.1242/dev.120.9.2609. [DOI] [PubMed] [Google Scholar]
- Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009;66(9):1518–1533. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338(2):237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 2011;7(11):e1002365. doi: 10.1371/journal.pgen.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JM, Smoyer CJ, Stensrud ES, Alexander R, Gogol M, Wiegraebe W, Jaspersen SL. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J Cell Biol. 2011;193(3):489–507. doi: 10.1083/jcb.201011017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR. Life on the edge: telomeres and persistent DNA breaks converge at the nuclear periphery. Genes Dev. 2009;23(9):1027–1031. doi: 10.1101/gad.1805309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gob E, Schmitt J, Benavente R, Alsheimer M (2010) Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS One 5(8):e12072. doi:10.1371/journal.pone.0012072 [DOI] [PMC free article] [PubMed]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121(3):451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Goto B, Okazaki K, Niwa O. Cytoplasmic microtubular system implicated in de novo formation of a Rabl-like orientation of chromosomes in fission yeast. J Cell Sci. 2001;114(Pt 13):2427–2435. doi: 10.1242/jcs.114.13.2427. [DOI] [PubMed] [Google Scholar]
- Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci U S A. 2005;102(12):4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas J, Lemoncelli A, Morozov C, Franke K, Dominder J, Antoniacci LM. Physical links between the nuclear envelope protein Mps3, three alternate replication factor C complexes, and a variant histone in Saccharomyces cerevisiae. DNA Cell Biol. 2012;31(6):917–924. doi: 10.1089/dna.2011.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129(4):1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Thomson JN. A gene required for nuclear and mitochondrial attachment in the nematode Caenorhabditis elegans. Cell. 1982;30(1):321–330. doi: 10.1016/0092-8674(82)90038-1. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Botsios S, Donze D, Donaldson AD. TFIIIC localizes budding yeast ETC. sites to the nuclear periphery. Mol Biol Cell. 2012;23(14):2741–2754. doi: 10.1091/mbc.E11-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17(5):598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Horigome C, Okada T, Shimazu K, Gasser SM, Mizuta K. Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. EMBO J. 2011;30(18):3799–3811. doi: 10.1038/emboj.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, Friedman LM, Roux KJ, Kozlov S, Jeang KT, Frydman M, Burke B, Stewart CL, Avraham KB (2013) The LINC complex is essential for hearing. J Clin Invest. 123 (2):740–750. doi:10.1172/JCI66911 [DOI] [PMC free article] [PubMed]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96(2):435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol. 2012;199(5):735–744. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Ghosh S (2012) Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes. Nucleus 3(3) [DOI] [PMC free article] [PubMed]
- Jaspersen SL, Giddings TH, Jr, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol. 2002;159(6):945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Martin AE, Glazko G, Giddings TH, Jr, Morgan G, Mushegian A, Winey M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol. 2006;174(5):665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci. 2007;120(Pt 19):3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106(45):19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134(3):427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka H, Shinohara M, Shinohara A. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 2008;4(9):e1000196. doi: 10.1371/journal.pgen.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y. Interkinetic nuclear migration: beyond a hallmark of neurogenesis. Cell Mol Life Sci. 2012;69(16):2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133(7):1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1(2):75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- Kracklauer MP, Link J, Alsheimer M. LINCing the nuclear envelope to gametogenesis. Curr Top Dev Biol. 2013;102:127–157. doi: 10.1016/B978-0-12-416024-8.00005-2. [DOI] [PubMed] [Google Scholar]
- Kracklauer MP, Wiora HM, Deery WJ, Chen X, Bolival B, Jr, Romanowicz D, Simonette RA, Fuller MT, Fischer JA, Beckingham KM. The Drosophila SUN protein Spag4 cooperates with the coiled-coil protein Yuri Gagarin to maintain association of the basal body and spermatid nucleus. J Cell Sci. 2010;123(Pt 16):2763–2772. doi: 10.1242/jcs.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S, Woglar A, Jantsch V, Zetka M. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell. 2011;21(5):948–958. doi: 10.1016/j.devcel.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM. Neuronal migration. Mech Dev. 2001;105(1–2):47–56. doi: 10.1016/S0925-4773(01)00396-3. [DOI] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009;106(25):10207–10212. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Zhu X, Xu R, Shao C, Xu T, Zhuang Y, Han M. Inner nuclear envelope proteins SUN1 and SUN2 play a prominent role in the DNA damage response. Curr Biol. 2012;22(17):1609–1615. doi: 10.1016/j.cub.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman RE, Pelegri F. Localized products of futile cycle/lrmp promote centrosome-nucleus attachment in the zebrafish zygote. Curr Biol. 2012;22(10):843–851. doi: 10.1016/j.cub.2012.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286(30):26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Schneider M, Neumann S, Jaeger VM, Taranum S, Munck M, Cartwright S, Richardson C, Carthew J, Noh K, Goldberg M, Noegel AA, Karakesisoglou I. Nesprin interchain associations control nuclear size. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, Padmakumar VC, Gloy J, Walz G, Noegel AA. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci. 2008;121(Pt 11):1887–1898. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329(5994):956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2(3):173–181. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23(5):579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123(6):1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126(14):3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115(7):825–836. doi: 10.1016/S0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17(4):1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J Cell Sci. 2009;122(Pt 16):2895–2905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P, Nguyen TD, Wehnert MS. The LINC complex and human disease. Biochem Soc Trans. 2011;39(6):1693–1697. doi: 10.1042/BST20110658. [DOI] [PubMed] [Google Scholar]
- Mejat A, Decostre V, Li J, Renou L, Kesari A, Hantai D, Stewart CL, Xiao X, Hoffman E, Bonne G, Misteli T. Lamin A/C-mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J Cell Biol. 2009;184(1):31–44. doi: 10.1083/jcb.200811035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejat A, Misteli T. LINC complexes in health and disease. Nucleus. 2011;1(1):40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11(5):317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellad JA, Warren DT, Shanahan CM. Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol. 2010;23(1):47–54. doi: 10.1016/j.ceb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. Interkinetic nuclear migration is a broadly conserved feature of cell division in pseudostratified epithelia. Curr Biol. 2011;21(6):485–491. doi: 10.1016/j.cub.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136(16):2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Gao Z, Liu J, Wu JC, Malone CJ, Starr DA. Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol. 2009;327(2):433–446. doi: 10.1016/j.ydbio.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics. 2004;270(6):449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell. 2002;13(3):930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JT, Pfeiffer ER, Thirkill TL, Kumar P, Peng G, Fridolfsson HN, Douglas GC, Starr DA, Barakat AI. Nesprin-3 regulates endothelial cell morphology, perinuclear cytoskeletal architecture, and flow-induced polarization. Mol Biol Cell. 2011;22(22):4324–4334. doi: 10.1091/mbc.E11-04-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol. 2012;198(2):165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9(21):1211–1220. doi: 10.1016/S0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- Munoz-Centeno MC, McBratney S, Monterrosa A, Byers B, Mann C, Winey M. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol Biol Cell. 1999;10(7):2393–2406. doi: 10.1091/mbc.10.7.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Schneider M, Daugherty RL, Gottardi CJ, Eming SA, Beijer A, Noegel AA, Karakesisoglou I. Nesprin-2 interacts with {alpha}-catenin and regulates Wnt signaling at the nuclear envelope. J Biol Chem. 2010;285(45):34932–34938. doi: 10.1074/jbc.M110.119651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel AA, Neumann S. The role of nesprins as multifunctional organizers in the nucleus and the cytoskeleton. Biochem Soc Trans. 2011;39(6):1725–1728. doi: 10.1042/BST20110668. [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23(8):912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Peterson CL. Opening the DNA repair toolbox: localization of DNA double strand breaks to the nuclear periphery. Cell Cycle. 2010;9(1):43–49. doi: 10.4161/cc.9.1.10317. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295(2):330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Pandur P, Maurus D, Kuhl M. Increasingly complex: new players enter the Wnt signaling network. Bioessays. 2002;24(10):881–884. doi: 10.1002/bies.10164. [DOI] [PubMed] [Google Scholar]
- Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15(2):600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007;12(6):873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y, Jantsch V. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139(5):920–933. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006;11(6):817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123(6):1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel R, Ketema M, Kuikman I, de Pereda JM, Sonnenberg A. Nesprin-3 augments peripheral nuclear localization of intermediate filaments in zebrafish. J Cell Sci. 2011;124(Pt 5):755–764. doi: 10.1242/jcs.081174. [DOI] [PubMed] [Google Scholar]
- Rashmi RN, Eckes B, Glockner G, Groth M, Neumann S, Gloy J, Sellin L, Walz G, Schneider M, Karakesisoglou I, Eichinger L, Noegel AA (2011) The nuclear envelope protein Nesprin-2 has roles in cell proliferation and differentiation during wound healing. Nucleus 3(2):172–186. doi:10.4161/nucl.19090 [DOI] [PMC free article] [PubMed]
- Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186(4):461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Zang S, Hodzic D. UnLINCing the nuclear envelope: towards an understanding of the physiological significance of nuclear positioning. Biochem Soc Trans. 2011;39(6):1790–1794. doi: 10.1042/BST20110660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Blecher N, Markov A, Stewart-Hutchinson PJ, Hodzic D (2012) LINC complexes mediate the positioning of cone photoreceptor nuclei in mouse retina. PLoS One 7(10):e47180. doi:10.1371/journal.pone.0047180 [DOI] [PMC free article] [PubMed]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111(Pt 16):2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Rothballer A, Schwartz TU, Kutay U (2013) LINCing complex functions at the nuclear envelope: what the molecular architecture of the LINC complex can reveal about its function. Nucleus 4(1) [DOI] [PMC free article] [PubMed]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106(7):2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139(5):907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102(38):13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165(2):299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104(43):16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci U S A. 2007;104(18):7426–7431. doi: 10.1073/pnas.0609198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Lu W, Neumann S, Brachner A, Gotzmann J, Noegel AA, Karakesisoglou I. Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell Mol Life Sci. 2011;68(9):1593–1610. doi: 10.1007/s00018-010-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23(8):928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254(3):238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149(5):1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Interkinetic nuclear migration: a mysterious process in search of a function. Dev Growth Differ. 2012;54(3):306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27(11):1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298(5592):406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128(24):5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314(8):1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol. 1981;82(1):41–55. doi: 10.1016/0012-1606(81)90427-9. [DOI] [PubMed] [Google Scholar]
- Taranum S, Sur I, Muller R, Lu W, Rashmi RN, Munck M, Neumann S, Karakesisoglou I, Noegel AA. Cytoskeletal interactions at the nuclear envelope mediated by nesprins. Int J Cell Biol. 2012;2012:736524. doi: 10.1155/2012/736524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau M, Roth S. The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis. Fly (Austin) 2008;2(2):82–91. doi: 10.4161/fly.6288. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morphol. 1985;89:313–331. [PubMed] [Google Scholar]
- Towbin BD, Meister P, Gasser SM. The nuclear envelope—a scaffold for silencing? Curr Opin Genet Dev. 2009;19(2):180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol. 2005;170(2):213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E, Dresser ME, Scherthan H. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J Cell Biol. 2000;151(1):95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10(8):970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tzur YB, Margalit A, Melamed-Book N, Gruenbaum Y. Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc Natl Acad Sci U S A. 2006;103(36):13397–13402. doi: 10.1073/pnas.0604224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Wozniak RW, Aitchison JD. Role of the nuclear envelope in genome organization and gene expression. Wiley Interdiscip Rev Syst Biol Med. 2011;3(2):147–166. doi: 10.1002/wsbm.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development. 1992;116(3):721–730. doi: 10.1242/dev.116.3.721. [DOI] [PubMed] [Google Scholar]
- Volk T (2012) Positioning nuclei within the cytoplasm of striated muscle fiber: cooperation between microtubules and KASH proteins. Nucleus 4(1) [DOI] [PMC free article] [PubMed]
- Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152(5):923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AD, Bommakanti A, Cohen-Fix O. Shaping the nucleus: factors and forces. J Cell Biochem. 2012;113(9):2813–2821. doi: 10.1002/jcb.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008;4(9):e1000188. doi: 10.1371/journal.pgen.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DT, Tajsic T, Mellad JA, Searles R, Zhang Q, Shanahan CM. Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. J Biol Chem. 2010;285(2):1311–1320. doi: 10.1074/jbc.M109.032557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DT, Zhang Q, Weissberg PL, Shanahan CM. Nesprins: intracellular scaffolds that maintain cell architecture and coordinate cell function? Expert Rev Mol Med. 2005;7(11):1–15. doi: 10.1017/S1462399405009294. [DOI] [PubMed] [Google Scholar]
- Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14(13):R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92(4):547–557. doi: 10.1016/S0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171(5):799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114(4):745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB, Hecht R, Carr S, Vanderslice R, Wolf N, Hirsh D. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. Dev Biol. 1980;74(2):446–469. doi: 10.1016/0012-1606(80)90445-5. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2(2):a000760. doi: 10.1101/cshperspect.a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Fischer JA. On the roles of the Drosophila KASH domain proteins Msp-300 and Klarsicht. Fly (Austin) 2008;2(2):74–81. doi: 10.4161/fly.6108. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145(6):1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20(6):1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Starr DA, Wu X, Parkhurst SM, Zhuang Y, Xu T, Xu R, Han M. The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev Biol. 2006;289(2):336–345. doi: 10.1016/j.ydbio.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Zhang J, Felder A, Liu Y, Guo LT, Lange S, Dalton ND, Gu Y, Peterson KL, Mizisin AP, Shelton GD, Lieber RL, Chen J. Nesprin 1 is critical for nuclear positioning and anchorage. Hum Mol Genet. 2010;19(2):329–341. doi: 10.1093/hmg/ddp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, Wheeler MA, Ellis JA, Skepper JN, Vorgerd M, Schlotter-Weigel B, Weissberg PL, Roberts RG, Wehnert M, Shanahan CM. Nesprin-1 and −2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16(23):2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80(5):473–481. doi: 10.1006/geno.2002.6859. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114(Pt 24):4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64(2):173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134(5):901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115(Pt 15):3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol. 2012;196(2):203–211. doi: 10.1083/jcb.201108098. [DOI] [PMC free article] [PubMed] [Google Scholar]


