Summary
Fluorescent calcium sensors are widely used to image neural activity. Using structure-based mutagenesis and neuron-based screening, we developed a family of ultra-sensitive protein calcium sensors (GCaMP6) that outperformed other sensors in cultured neurons and in zebrafish, flies, and mice in vivo. In layer 2/3 pyramidal neurons of the mouse visual cortex, GCaMP6 reliably detected single action potentials in neuronal somata and orientation-tuned synaptic calcium transients in individual dendritic spines. The orientation tuning of structurally persistent spines was largely stable over timescales of weeks. Orientation tuning averaged across spine populations predicted the tuning of their parent cell. Although the somata of GABAergic neurons showed little orientation tuning, their dendrites included highly tuned dendritic segments (5 - 40 micrometers long). GCaMP6 sensors thus provide new windows into the organization and dynamics of neural circuits over multiple spatial and temporal scales.
Introduction
Neural activity causes rapid changes in intracellular free calcium1-4. Calcium imaging experiments have relied on this principle to track the activity of neuronal populations5,6 and to probe excitation of small neurons and neuronal microcompartments2,7-10. Genetically-encoded protein sensors can be targeted to specific cell types2,9,11,12 for non-invasive imaging of identified neurons and neuronal compartments8,13-15 over chronic timescales6.
Calcium indicator proteins include the single fluorophore sensor GCaMP11,16,17 and several families of Förster resonance energy transfer based sensors18-22. However, none of these protein-based indicators have yet surpassed the sensitivity and speed of commonly used synthetic calcium indicators (e.g., Oregon Green Bapta-1-AM, OGB1-AM). Therefore, depending on the experimental goals, investigators choose between sensitive synthetic indicators delivered by invasive chemical or physical methods, or less sensitive protein sensors delivered with genetic methods.
Multiple rounds of structure-guided design have made GCaMPs the most widely used protein calcium sensors11,16,17. But past efforts in optimizing GCaMPs and other indicators of neuronal function were limited by the throughput of quantitative and physiologically relevant assays. Because neurons have unusually fast calcium dynamics and low peak calcium accumulations4, sensors designed to probe neuronal function are best tested in neurons11,13,23,24, rather than in non-neuronal systems, most of which show much slower and larger calcium changes19. We thus screened GCaMP variants produced by mutagenesis in neurons, and subsequently validated lead sensors in several in vivo systems.
GCaMP protein engineering
GCaMP17 and its progeny11,16 consist of circularly permuted green fluorescent protein (cpGFP)25, the calcium-binding protein calmodulin (CaM), and CaM-interacting M13 peptide26 (Fig. 1a). The CaM/M13 complex is in proximity to the chromophore inside the cpGFP β barrel27. Calcium-dependent conformational changes in CaM/M13, including modulation of solvent access and the pKa of the chromophore, cause increased brightness with calcium binding. Despite extensive structure-guided optimization11,16, GCaMP and other protein sensors still suffer from low sensitivity and slow kinetics.
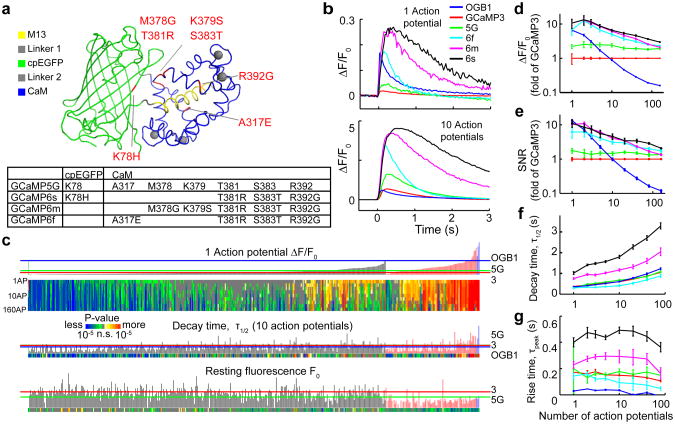
Figure 1. GCaMP mutagenesis and screening in dissociated neurons.
a, GCaMP structure27,64 and mutations in different GCaMP variants relative to GCaMP5G.
b, Responses averaged across multiple neurons and wells for GCaMP3, 5G, 6f, 6m, 6s, and OGB1-AM. Top, fluorescence changes in response to 1 action potential. Bottom, 10 action potentials.
c, Screening results, 447 GCaMPs. Top, fluorescence change in response to 1 action potential (vertical bars, ΔF/F0; green bar, OGB1-AM, left; black bars, single GCaMP mutations; red bars, combinatorial mutations; blue, GCaMP6 indicators) and significance values for different action potential stimuli (color plot). Middle, half decay time after 10 action potentials. Bottom, resting fluorescence, F0 normalized to nuclear mCherry fluorescence. Red line, GCaMP3 level; green line, GCaMP5G level; blue line, OGB1-AM level.
d-g, Comparison of GCaMP sensors and OGB1-AM (blue) as a function of stimulus strength (colors as in b). d, response amplitude; e, SNR; f, half decay time; g, time to peak (after stimulus offset). Error bars correspond to s.e.m (n=300, 16, 8, 11, 13, 11 wells for GCaMP3, GCaMP5G, OGB1-AM, 6f, 6m, 6s, respectively).
We produced numerous additional GCaMP variants and tested them in automated neuronal assays (Fig. 1). With the aim of improving sensitivity, we focused mutagenesis on the interface between cpGFP and CaM at 16 amino acid positions, some mutagenized to near completion (Fig. 1a, Supplementary Table 5)16. Mutations were made at 18 additional sites, notably at the M13/CaM interface which can affect calcium affinity28 (A317) and in CaM (R392)16 (Fig. 1a).
Dissociated rat hippocampal neurons in 24-well plates were transduced with GCaMP variants (one per well), together with nuclear mCherry29, using lentivirus-mediated gene transfer. Electrodes triggered trains of action potentials in all neurons within each well (Methods). Time-lapse images (35 Hz) of ∼800 μm fields of view containing 10-100 neurons were acquired, while delivering a series of action potential trains (Fig. 1c). Fluorescence changes extracted from single neurons were used to compare the sensitivity, dynamic range, and kinetics of individual GCaMP variants and OGB1-AM (Fig. 1d). We monitored the resting brightness of the sensor by measuring green fluorescence relative to red mCherry fluorescence.
The assay revealed 43/348 point mutants with improved sensitivity compared to GCaMP3 (ΔF/F0 in response to one action potential; p < 0.01; Wilcoxon rank sum test) (Fig. 1c; Supplementary Fig. 1). In addition, one mutation at the M13/CaM interface (A317E) accelerated kinetics (4-fold), but also reduced response amplitude (2-fold), compared to parent GCaMP variants (Supplementary Fig. 2). In a second round of mutagenesis we combined beneficial mutations, selected based on improved response amplitudes (1-3 action potentials) and/or kinetics, without compromising baseline fluorescence or maximal response (160 action potentials) (94 variants, up to 8 beneficial point mutations; Fig. 1a, c, Supplementary Table 5). In some cases the beneficial effects were additive (Supplementary Fig. 2). A317E consistently accelerated the kinetics compared to the parent sensors. In total, we screened 447 GCaMP variants (Supplementary Table 5).
Based on screening in cultured neurons (Fig. 1), we chose three ultrasensitive GCaMP6 sensors (GCaMP6s, 6m, 6f; for slow, medium and fast kinetics, respectively) for characterization in vivo. These sensors vary in kinetics, with the more sensitive sensors having slower kinetics. Compared to GCaMP5G, the GCaMP6 sensors have similar baseline brightness and a 1.1-1.6 fold increase in dynamic range (ΔF/F0 at 160 action potentials). For small numbers of action potentials the most sensitive sensor, GCaMP6s, produced 7-fold larger signals (>10-fold larger than GCaMP3, Fig. 1b-e; Supplementary Table 1). Underlying this sensitivity gain are multiple factors (Supplementary Table 2). Compared to GCaMP5G, GCaMP6s exhibited 3-fold higher apparent affinity for calcium and 1.3-fold higher saturated fluorescence, with similar baseline fluorescence. Calcium-saturated GCaMP6s is 27% brighter than enhanced GFP (EGFP), its parent fluorescent protein. The fastest sensor, GCaMP6f, had 2-fold faster rise time and 1.7-fold faster decay time than GCaMP5G (Fig. 1f, g) (Supplementary Table 1). GCaMP6f is the fastest genetically-encoded calcium indicator for cytoplasmic free calcium in neurons, with sensitivity comparable to OGB1-AM (Fig. 1d-g). GCaMP6 indicators were more sensitive and/or faster than other GCaMP variants in presynaptic boutons of the Drosophila larval neuromuscular junction24 (Supplementary Fig. 3; Supplementary Table 4), in projection neurons of the Drosophila adult antennal lobe during odor presentation16 (Supplementary Fig. 4), and in neuropil and somata of the zebrafish tectum in vivo16 (Supplementary Fig. 5).
Imaging neuronal populations in the mouse visual cortex
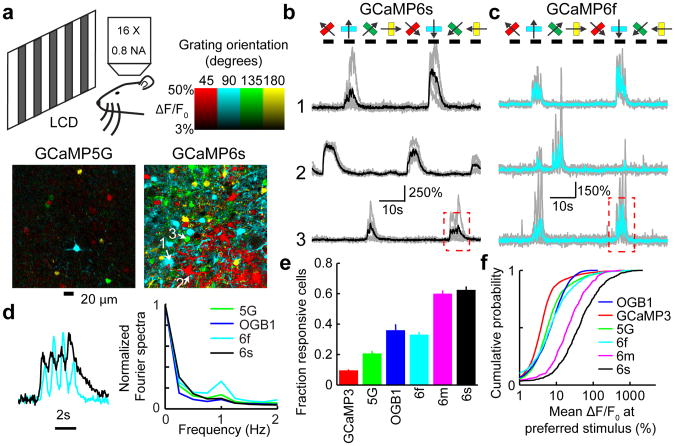
We next tested GCaMP6 in layer (L)2/3 pyramidal neurons in the mouse visual cortex V1 in vivo (Fig. 2a). The majority of V1 neurons can be driven to fire action potentials in response to drifting gratings30. V1 was infected with adeno-associated virus (AAV) expressing GCaMP variants (AAV-hsyn1-GCaMP variant)11. Three weeks after AAV infection, the vast majority of L2/3 neurons showed fluorescence mainly in the neuronal cytoplasm (Supplementary Fig. 6). Sensory stimuli consisted of moving gratings presented in eight directions to the contralateral eye12,16. Two-photon imaging revealed visual stimulus-evoked fluorescence transients in subsets of neurons (Fig. 2a-c). These responses were stable across trials (Supplementary Fig. 8) and tuned to stimulus orientation (Fig. 2a, b and Supplementary Fig. 9). Orientation tuning was similar for GCaMP5G, GCaMP6f, GCaMP6m, and bulk-loaded OGB1-AM5 (Supplementary Fig. 9). Fluorescence transients were faster with GCaMP6f compared to other sensors and faithfully tracked dynamic sensory stimuli (Fig. 2d).
Figure 2. GCaMP6 performance in the mouse visual cortex.
a, Top, schematic of the experiment. Bottom, field of view showing neurons color-coded according to their preferred orientation (hue) and response amplitude (brightness) for GCaMP5G (left) and GCaMP6s (right).
b, Example traces from three neurons expressing GCaMP6s. Single sweeps (grey) and averages of 5 sweeps (black) are overlaid. Directions of grating motion (8 directions) are shown above traces (arrows).
c, Example traces from three neurons expressing GCaMP6f. Single sweeps (grey) and averages of 5 sweeps (cyan) are overlaid.
d, Top, high magnification view of fluorescence changes corresponding to the red boxes in b (black) and c (cyan), normalized to the peak of the response. Bottom, Fourier spectra normalized to the response amplitude at 0 Hz for neurons driven with 1 Hz drifting gratings, transduced with GCaMP5G, OGB1-AM, 6f, 6s.
e, The fraction of cells scored as responding to visual stimulation when loaded with different calcium indicators. Error bars correspond to s.e.m. (n=70, 39, 23, 38, 21, 34 FOVs for GCaMP3, 5G, OGB1-AM, 6f, 6m, 6s, respectively). GCaMP3, 5G, and OGB1-AM data are from ref 16.
f, The distribution of fluorescence changes across cells at the preferred orientation.
GCaMP6 performance was compared to other sensors in several ways. The fraction of responding neurons detected with GCaMP6s was 3-fold higher than for GCaMP5G (5-fold higher than GCaMP3) (Fig. 2e). Notably, the fractions of active neurons detected with GCaMP6s and GCaMP6m were also significantly higher than for OGB1-AM (Fig. 2e, f, p<0.01, Wilcoxon rank sum test). GCaMP6 sensors thus reveal neuronal dynamics that were previously undetectable with protein sensors.
We imaged GCaMP6s-expressing neurons through a chronic imaging window6,31 over several weeks in V1 (Supplementary Fig. 6c)18. Sensory responses and orientation tuning of neurons were largely stable (Supplementary Fig. 6c, e). Responses of a small fraction of neurons were lost over time, which was balanced by other neurons that started responding. The orientation preference of the neurons responding in all imaging sessions (∼56% of cells that responded in the first session) was remarkably stable (Supplementary Fig. 6d). Over months of expression a small fraction of highly expressing neurons acquired nuclear fluorescence; these neurons eventually also developed aberrant responses11 (Supplementary Fig. 7). These experiments indicate that expression of GCaMP6s over 1-2 months does not obviously perturb the function of cortical circuits.
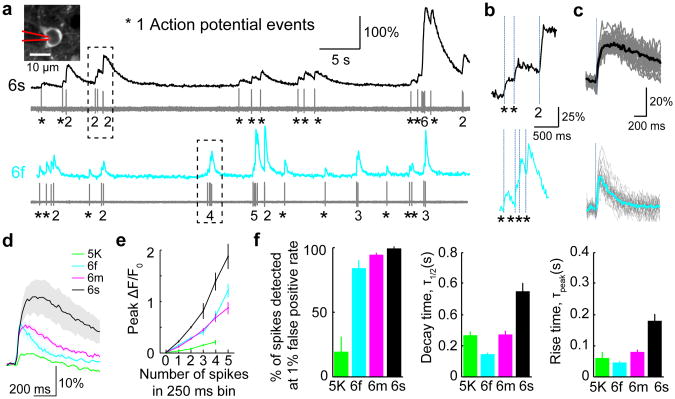
We directly compared cellular fluorescence changes and spiking using loose-seal, cell-attached recordings. The contrast of the visual stimulus was adjusted online to maintain a moderate spike rate. GCaMP6s produced large fluorescence transients even in response to single action potentials (> 6 times larger than for GCaMP5K, Fig. 3 and Supplementary Video 1), yielding high detection rates for single spikes (99 ± 0.2%; at 1% false-positive rate, n=9 cells, 250 spikes). GCaMP6f and GCaMP6m showed slightly lower spike detection efficiencies, but with faster kinetics (Fig. 3, Supplementary Table 3). Individual spikes within a burst resulted in step-wise fluorescence increases (Fig. 3b), which were resolvable if they were separated by an interval on the order of the rise time of the sensor or more (100-150 ms, GCaMP6s; 75-100 ms, GCaMP6m; 50-75 ms, GCaMP6f; Fig. 3f, Supplementary Fig. 10, Supplementary Table 3). These data show that GCaMP6s can detect activity with near 100% action potential detection in pyramidal cells.
Figure 3. Combined imaging and electrophysiology in the visual cortex.
a, Simultaneous fluorescence dynamics and spikes in a GCaMP6s (top) and a GCaMP6f (bottom) expressing neuron. The number of spikes for each burst is indicated below the trace (single spikes are indicated by asterisks). Left inset, a GCaMP6s expressing neuron with the recording pipette indicated schematically.
b, Zoomed-in view of bursts of action potentials. Top, GCaMP6s; bottom, GCaMP6f.
c, Fluorescence change in response to one action potential. Top, GCaMP6s; bottom, GCaMP6f.
d, Median fluorescence change in response to one action potential for different calcium indicators. Shading corresponds to s.e.m., n= 9 (GCaMP5K, data from ref 16), 11 (GCaMP6f), 10 (GCaMP6m), 9 (GCaMP6s) cells. GCaMP5K and GCaMP5G have similar properties16.
e, Peak fluorescence change as a function of number of action potentials in a 250 ms bin (5K: n=161, 65, 22, 4 events for 1, 2, 3, 4 action potentials; 6f: n=366, 120, 50, 15, 7 events for 1, 2, 3, 4, 5 action potentials; 6m: n=354, 105, 31, 11, 7 events for 1, 2, 3, 4, 5 action potential; 6s: n=250, 60, 20, 5, 4 events for 1, 2, 3, 4, 5 action potentials). Error bars correspond to s.e.m.
f, Comparison of GCaMP indicators. Left, fraction of isolated spikes detected at 1% false positive rate. Middle, half decay time. Right, rise time to peak. Error bars correspond to s.e.m.
Calcium transients in dendritic spines
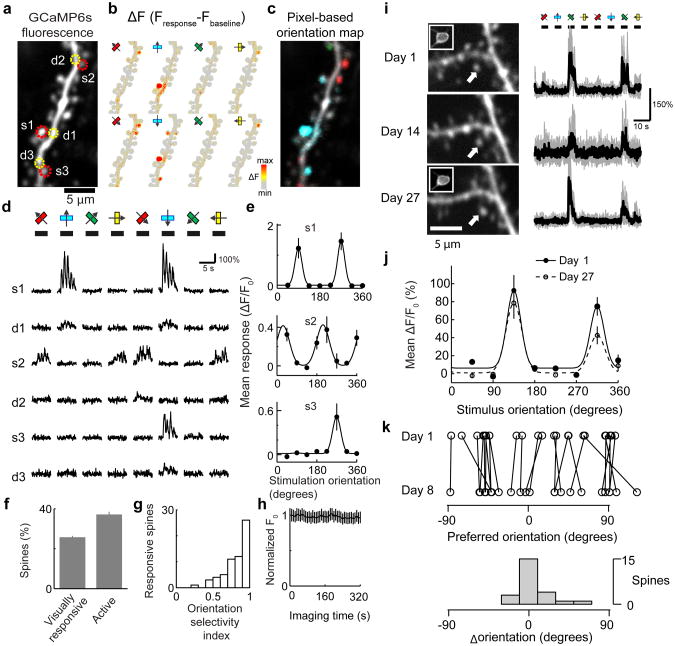
Pyramidal neuron output is shaped by thousands of excitatory synapses distributed across the dendritic arbor. Activation of single excitatory synapses causes calcium accumulations in individual dendritic spines, mediated by NMDA-Rs32,33, which can be imaged to measure the tuning of single synapses in vivo10,33. We used GCaMP6s to image synaptic calcium signals within dendritic spines over chronic timescales. In V1 with sparsely labeled L2/3 pyramidal neurons (Methods) small dendritic branches were imaged at high magnification during visual stimulation (Fig. 4a). We first focused on neurons that did not fire visually evoked action potentials (∼40% of neurons) to avoid calcium changes caused by action potentials back-propagating into dendrites7. Individual spines showed large fluorescence transients, often independent of their parent dendrites (Fig. 4b,d and Supplementary Video 2). Spine responses were orientation-tuned (Fig. 4b-e), as expected from the fact that most input to V1 neurons originates from (presumably orientation-tuned) V1 neurons34. Neighboring spines were often tuned with different preferred orientations. The correlation between orientation tuning and distance between spines was weak (R=0.08; p>0.05). Overall, 27% of spines (62/228, 15 dendrites, 4 mice) were visually responsive, and most were orientation-tuned (Fig. 4f, g). Furthermore, close to 40% of spines were active at some time during the imaging session (Fig. 4f). Bleaching of GCaMP6 was negligible over 40 imaging trials (320 s of continuous imaging, Fig. 4h).
Figure 4. Imaging activity in dendritic spines in the visual cortex.
a, Image of an L2/3 dendritic branch expressing GCaMP6s. Regions of interest (ROIs) are indicated as dashed circles (red, spines; yellow, dendrites).
b, Map of fluorescence change (ΔF=Fresponse-Fbaseline) in response to drifting gratings of 8 different orientations.
c, Pixel-based map of orientation preference.
d, Responses of dendritic spines (s1-s3) and neighboring dendritic shafts (d1-d3) to drifting gratings with different orientations (corresponding to ROIs indicated in a).
e, Orientation tuning of individual spines (s1, s2, s3). Error bars correspond to s.e.m. (n=5 trials).
f, Fraction of spines that show detectable calcium transients (active) and respond to visual stimulation (responsive) (see Methods for definitions) (228 spines; 15 dendrites; 4 mice).
g, Distribution of the orientation selectivity index across visually responsive spines (62 spines).
h, Baseline fluorescence across individual dendritic spines over 320 seconds of continuous imaging (228 spines; 15 dendrites; 4 mice; error bars reflect s.e.m. across spines).
i, Left, the same GCaMP6s labeled spine imaged over weeks. Right, fluorescence responses to oriented drifting gratings. Insets, parent soma of imaged spines.
j, Orientation selectivity of single spines measured over time (same as i).
k, Top, preferred orientation for spines that responded in two imaging sessions separated by one week. Opposing stimulus directions are considered as equivalent in this analysis. Bottom, the distribution of ΔOri (difference in preferred orientation between two sessions).
We imaged the same dendritic segments over imaging sessions separated by weeks. Most spines persisted over this timescale, although other spines appeared and disappeared31. We analyzed the visual responses of persistent spines. The percentage of spines that responded to visual stimulation was stable over a week (37/139 on day 1; 34/139 on day 8). Spines that responded during one imaging session were also likely to respond a week later (65%). Furthermore, visually responsive spines mostly retained their orientation tuning (Fig. 4i-k).
How orientation-tuned neurons connect to other orientation-tuned neurons remains controversial10,35. Some measurements suggest that individual neurons sample heterogeneous input, from neurons with diverse orientation-tuning10. Models indicate that specific connectivity is not required for orientation tuning36. However, other measurements emphasize that neurons prefer to make synapses with neurons sharing similar orientation tuning35, but orientation tuning at the level of subthreshold membrane potential is weak37,38. We thus compared the orientation tuning of populations of individual spines and the output of their parent neuron. We identified neurons with tuned output (OSI, 0.91 ± 0.04, n=5) (Fig. 5a) and measured the orientation tuning of large numbers of dendritic spines per neuron (average, 201 spines; range, 120-298 spines) (Fig. 5b-d). Contributions to the signal from back-propagating action potentials were removed using a computational subtraction procedure (Methods; Supplementary Fig. 11). For individual neurons, the orientation tuning averaged across all spines was biased towards the orientation tuning of the parent neurons (Fig. 5e, f) (p < 0.01, Wilcoxon rank sum test), although the modulation depth for spines was smaller (p < 0.01, Wilcoxon rank sum test). A similar trend was also apparent in the distributions of preferred orientations across spines (Fig. 5g, h). Our results show that spine fluorescence transients, which are not necessarily good predictors of the strength of excitatory synaptic input39, averaged across a neuron predict the orientation tuning of the cell's output.
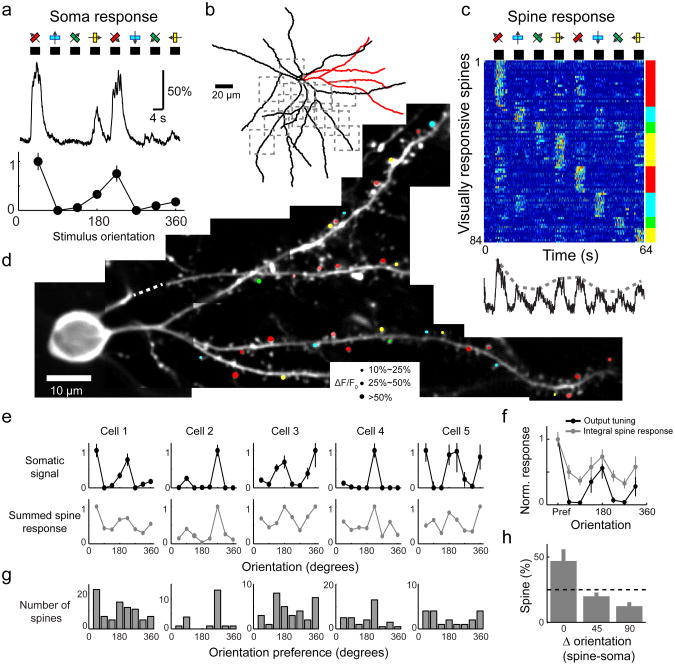
Figure 5. The orientation preference of populations of dendritic spines predicts the orientation preference of their parent neuron.
a, Somatic fluorescence responses of a GCaMP6s-expressing layer 2/3 pyramidal neuron (depth, 120 μm) to oriented drifting gratings (Top) and the corresponding tuning curve (Bottom, normalized).
b, Reconstruction of the dendritic arbor (red dendrites, dendrites shown in d; dashed squares, additional imaged regions).
c, Top, fluorescence responses of visually responsive spines (84/298) sorted by their preferred orientation (averaged over 5 trials). Each row shows one spine normalized to its peak. Bottom, summed ΔF/F0 across all spines (without normalization).
d, Locations of orientation selective spines on a subset of imaged dendrites (corresponding to red dendrites in b). The size of the circle corresponds to the averaged ΔF/F0 at the preferred stimulus, the color indicates the preferred orientation, and the saturation of the color encodes the orientation selectivity index (OSI =1, saturated color; OSI=0, white).
e, Top, tuning curve of somatic ΔF/F. Bottom, summed spine ΔF/F. Cell 1 corresponds to panels a-d.
f, Averaged output tuning (black) and integral spine response (gray) across the 5 neurons (same cells as in e). The turning curves were aligned to the preferred orientation of the output response (0 degree). The average was normalized.
g, The distribution of preferred orientation of dendritic spines (5 cells; number of spines sampled: 298,166,137,278,116).
h, Fraction of visually responsive spines preferring orientations 0, 45 or 90 degree away from the postsynaptic cell's preferred orientation. Opposing stimulus directions are considered as equivalent in this analysis. Error bars correspond to s.e.m.
Calcium transients in GABAergic dendrites
Consistent with previous results40,41, somatic fluorescence changes in GABAergic neurons were broadly tuned with respect to stimulus orientation (Fig. 6a, b; Supplementary Fig. 12). However, their dendrites showed pronounced orientation-tuned domains (Fig. 6c, d). Individual dendritic branches often had multiple domains with distinct preferred orientations, consistent with the diverse preferences of inputs to GABAergic neurons42,43. The underlying fluorescence responses were modulated at the temporal frequency of the drifting grating (1 Hz) (Fig. 6e), a characteristic feature of V1 excitatory neurons in the superficial layers of V130, suggesting that the responses reflect local excitatory synaptic input. Tuned dendritic domains were seen in parvalbumin-positive (Fig. 6) and somatostatin-positive (data not shown) interneurons.
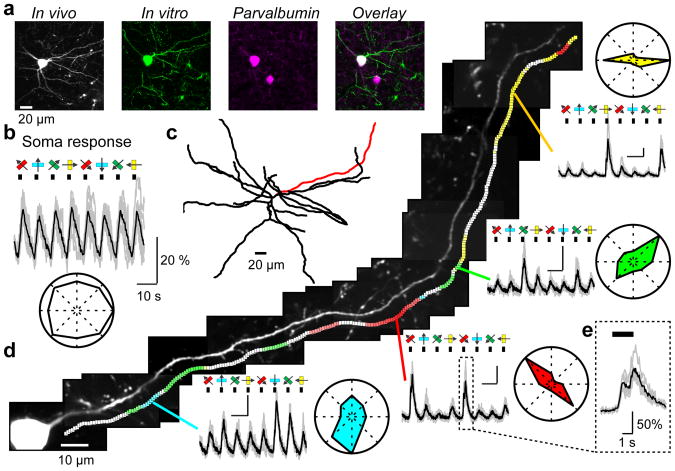
Figure 6. Orientation-tuned domains in dendrites of GABAergic interneurons.
a, A GCaMP6s-expressing interneuron (soma depth, 250 μm), identified post hoc as a parvalbumin-positive interneuron.
b, Somatic fluorescence changes to oriented drifting grating (same cell as in a). Bottom, polar plot.
c, Reconstruction of the dendritic arbor based on GCaMP6s fluorescence.
d, Left, a dendrite of the cell (red in c) was imaged along its entire length. Colored squares indicate dendritic sites showing significant orientation tuning (p < 0.01, ANOVA across 8 stimulus directions). The color of each square indicates the local preferred orientation, and the saturation of the color encodes the orientation selectivity index (OSI =1, saturated color; OSI=0, white). Right, example dendritic fluorescence changes and the corresponding polar plots for four locations with distinct orientation preferences. Scale bars: 10s; 50% ΔF/F.
e, Zoomed-in view of the dendritic calcium signal corresponding to the box in d. The signal shows modulation at the frequency of the drifting grating (1 Hz).
We mapped the visual response along 104 interneuron dendrites (total imaged length 6.3 mm) with regions of interest placed every micrometer along the dendrite (Fig. 6d). Visually-evoked GCaMP6s responses were detected in 5.7 mm (90 %) of dendritic length, with 1.79 mm (28%) showing significant orientation-tuning (p < 0.01, ANOVA across 8 conditions). The majority of imaged dendrites (33/54; longer than 50 μm) had multiple domains preferring different orientations. The OSI (0.44 ± 0.15; mean ± s.d.) of tuned dendritic segments was higher than for interneuron somata (p < 0.005, Supplementary Fig. 12), but lower than for typical pyramidal neurons (p < 0.001, Supplementary Fig. 9). Back-propagation44 of broadly tuned somatic action potentials probably lowers the OSI of the dendritic calcium signals. Consistent with this, both the dendritic OSI and the percentage of orientation selective sites increased with distance from the soma (Supplementary Fig. 13a).
The sizes of individual domains (12 ± 7 μm, mean ± s.d. FWHM, n=107 domains) were considerably larger than the spacing between excitatory synapses on interneuron dendrites (<1 μm)45 (Supplementary Fig. 13b, c). Nearby domains with distinct preferred orientations often overlapped (Supplementary Fig. 13b). The large size of these domains might reflect spatially clustered input with shared orientation preference. Alternatively, individual domains might be dominated by a few sparse, randomly distributed strong inputs46, amplified by local postsynaptic mechanisms47
Conclusions
We developed a new class of genetically-encoded indicators of neuronal function with greatly improved properties. GCaMP6 indicators cross important performance thresholds. They have higher sensitivity than commonly used synthetic calcium dyes (e.g., OGB1) and detect individual action potentials with high reliability at reasonable microscope magnifications. These indicators can be used to image large groups of neurons as well as tiny synaptic compartments over multiple imaging sessions separated by months. It is likely that these sensors will find widespread applications for diverse problems in brain research and calcium signaling. Future engineering efforts could focus on red fluorescent calcium indicator proteins48,49, which promise imaging considerably deeper in scattering tissue50.
Methods
All experiments were conducted according to National Institutes of Health guidelines for animal research and were approved by the Janelia Farm Research Campus Institutional Animal Care and Use Committee and Institutional Biosafety Committee.
Neuronal culture screen
GCaMP variants were made in a modified SIV-based lentiviral construct, pGP-syn-GCaMP-nls-mCherry-WPRE, derived from pCL20cSLFR MSCV-GFP51. The prolentiviral vector included a 476-bp human synapsin promoter, GCaMP, a nuclear localization sequence fused to mCherry, and the woodchuck hepatitis post-transcriptional regulatory element. Site-directed mutagenesis was conducted by PCR and mutated regions were incorporated into the lentiviral constructs by gene assembly52.
Hippocampi were dissected and dissociated in papain. Cells were plated at a density of 225,000 viable cells/well in 24-well glass-bottom plates (Mattek, #1.5 glass coverslips), pre-coated with Matrigel (BD Biosciences). Cells were cultured in growth medium (28 mM glucose, 2.4 mM sodium bicarbonate, 100 μg/mL transferrin, B-27 supplement (1X, Invitrogen), 500 μM L-glutamine, 50 units/mL penicillin, 50 mg/mL streptomycin, 5% fetal bovine serum in MEM).
Lentiviral particles were made in a biosafety level 2 laboratory by transfecting a prolentiviral construct and packaging and coat pseudotyping DNA constructs (pCAG-SIVgprre, pCAG4-RTR-SIV, pCMV-VSV-G)51,53 into HEK293T/17 cells (ATCC) in 10-cm plates. After 72 h, supernatant was collected (6 mL) and filtered. Neuronal cultures were infected at 3 days in vitro. Each well of a 24-well plate was incubated overnight with 0.5 mL of lentivirus in conditioned growth medium. The growth medium was supplemented with 4 μM AraC to inhibit glial proliferation. In some experiments, OGB1-AM was loaded into cells by incubating neurons in 1 mL of 2 μM OGB1-AM (Invitrogen) for 30 min and rinsing 3 times with imaging buffer (145 mM NaCl, 2.5 mM KCl, 10 mM glucose, 10 mM HEPES pH 7.4, 2 mM CaCl2, 1 mM MgCl2).
Neurons were stimulated in imaging buffer containing a drug cocktail to inhibit synaptic receptors (10 μM CNQX, 10 μM (R)-CPP, 10 μM gabazine, 1 mM (S)-MCPG, Tocris). Under these conditions, intracellular calcium increases are presumably caused by the opening of voltage sensitive calcium channels.
Action potentials (APs) (83 Hz) were evoked by field stimulation with a Grass Technologies S48 stimulation unit and a custom-built 24-well cap stimulator with pairs of parallel platinum wires. The microscope was an Olympus IX81 with a 10× (0.4 NA) air objective lens and EMCCD camera (Andor 897, 512 × 512 pixels, 35 frames/s), Cairn OptoLED illumination system, and GFP (Excitation: 450-490 nm; Dichroic: 495 nm long-pass; Emission: 500-550 nm) and TxRed (Excitation: 540-580 nm; Dichroic: 585 nm long-pass; Emission: 593-668 nm) filter sets. The field of view was 800 μm × 800 μm. Images were background subtracted (mean of 5% lowest pixel values). Responses were quantified for each cell as change in fluorescence divided by baseline fluorescence measured one second prior to stimulation. Signal-to-noise ratio (SNR) was quantified as peak ΔF/F0 response over the standard deviation of the signal during a one second period prior to stimulation.
Control experiments varying stimulation voltage, frequency, and pulse width insured suprathreshold stimulation of neurons. Voltage imaging using the ArchWT-GFP archaerhodopsin-based voltage sensor54 confirmed that individual pulses (1 ms, 40 V, 83 Hz) reliably triggered single APs. The imaging and stimulation system was controlled by custom scripts written in MetaMorph software (version 7.7.5, Molecular Devices) and Ephus software55 (ephus.org). Detailed neuronal culture screening methods will be described elsewhere (T.J.W., T.W.C., E.R.S., R.A.K., V.J., L.L.L., K.S., and D.S.K., manuscript in preparation).
Labeling V1 neurons
Constructs used to produce AAV included pGP-AAV-syn-GCaMP-WPRE and the Cre recombinase-activated construct pGP-AAV-syn-flex-GCaMP-WPRE. Virus was injected slowly (30 nL in 5 minutes) at a depth of 250 μm into the primary visual cortex (two sites, 2.5 and 2.9 mm lateral from the lambda suture). For population imaging and electrophysiology (Fig 2-3), AAV2/1-syn-GCaMP-WPRE virus (titer: ∼1011 -1012 genomes/mL) was injected into the visual cortex of C57BL/6J mice (1.5-2 months old)6. For dendritic imaging (Fig 4, 5 and 6a-f), sparse labeling was achieved by injecting a mixture of diluted AAV2/1-syn-Cre particles (titer: ∼1012 genomes/mL, diluted 8000-20,000 fold in PBS) and high titer, Cre-dependent GCaMP6s virus (∼8×1011 genomes/mL). This produces strong GCaMP6 expression in a small subset of neurons (∼3-5 cells in a 250 μm × 250 μm × 250 μm volume), defined by Cre expression56. Both pyramidal (Fig. 4-5) and GABAergic (Fig. 6) neurons were labeled using this approach, but they could be distinguished based on the presence or absence of dendritic spines. Post hoc immunolabeling further identified the imaged cells. For specific labeling of parvalbumin interneurons (Fig. 6g and Supplementary Fig. 12), Cre-dependent GCaMP6s AAV was injected into the visual cortex of PV-IRES-Cre mice57. Individual somata (Supplementary Fig. 12) and dendritic segments could be recognized (Fig. 6 g, h, total length of imaged dendrite: 2.86 mm), but the high labeling density made it difficult to track individual dendrites over long distances.
Window surgery
After 2-4 weeks of expression, mice were anesthetized using isoflurane (3% for induction, 1.5-2% during surgery) and a circular craniotomy (2-3 mm diameter) was made above V1 (centered 2.7 mm lateral from the lambda suture). For acute experiments, the craniotomy was covered with agarose (1-1.3 %), and a round glass coverslip (Warner Instruments; 5mm diameter; #1 thickness) was cemented to the skull to reduce motion of the exposed brain. A custom titanium head post was fixed to the skull using black dental cement (Contemporary Ortho-Jet). For simultaneous imaging and cell-attached recording, the exposed brain was covered with ∼1 mm thick agarose (1.3%) without a coverslip. For chronic imaging experiments, the imaging window was constructed from two layers of microscope coverglass6. A larger piece (Fisher, #1 thickness) was attached to the bone and a smaller insert (#2 thickness) was fitted snugly into the craniotomy. Imaging experiments were started ∼1-2 weeks after chronic window implantation.
Visual stimuli
Moving grating stimuli were generated using the Psychophysics Toolbox58,59 in MATLAB. Each stimulus trial consisted of a 4 s blank period (uniform gray at mean luminance) followed by a 4 s drifting sinusoidal grating (0.05 cycles per degree, 1 Hz temporal frequency). Typically, 8 drifting directions were used (separated by 45 degrees) and 5 trials were recorded for each direction, giving a total of 40 stimulus trials per recording session (320 s recording time). The gratings were presented with an LCD monitor (30 × 40 cm), placed 25 cm in front of the center of the right eye of the mouse. The monitor subtended an angle of ±38° horizontally and -20° to +38° vertically around the eye of the mouse. For experiments with cell-attached recording (Fig. 3), pipette access required the use of a smaller LCD monitor (12 × 16 cm) placed ∼10 cm in front of the right eye. During simultaneous imaging and electrophysiology, the optimal grating stimulus was repeatedly played (duration 2 s, separated by a 4 s blank period), but the contrast of the stimulus grating was adjusted online to maintain moderate spike rates.
For analysis of dendritic spine populations (Fig. 5), the stimulus contrast was lowered to reduce action potential-related dendritic signals (average ΔF/F0 in dendritic shafts at the preferred orientations was 32%, 80%. 16%, 15%, 12% for cells 1 through 5, corresponding to stimulus contrast 10-40%, 5-20%, 10-40%, 20-40%, 20-40%). The orientation preference of dendritic shafts was identical to the soma, consistent with back-propagating action potentials (data not shown). Orientation tuning of GABAergic dendrites (Fig. 6) was mapped using the standard stimulus set at full contrast. The stimulus duration was 2 s; because of the slower decay of calcium transients41 we used a 6 s inter trial interval.
Imaging mouse V1 neurons and dendrites
Mice were placed on a warm blanket (37°C) and kept anesthetized with 0.5% isoflurane and sedated with chlorprothixene (20-40 μL at 0.33 mg/ml, i.m.)30. Imaging was performed using a custom-built two-photon microscope (designs available at research.janelia.org/Svoboda) equipped with a resonant galvo scanning module (Thorlabs), controlled by ScanImage (scanimage.org)60. The light source was a Mai Tai femtosecond pulsed laser (Spectra-Physics) running at 940 nm. The objective was a 16× water immersion lens (Nikon, 0.8 NA, 3 mm working distance). The power used was 35-50 mW for full field imaging (Fig. 2) and 20-40 mW for higher zoom imaging (Fig. 3-6).
Images were collected at 15 Hz (512 × 512 pixels, 250 μm × 250 μm; Fig. 2) or 60 Hz (256 × 256 pixels, 30 μm × 30 μm; Fig. 3), or 15 Hz (512 × 512 pixels, 30 μm × 30 μm; Fig. 4-5), or 15 Hz (512 × 512 pixels, 30 μm × 30 μm - 100 μm × 100 μm; Fig. 6). For dendritic imaging experiments (Fig. 4-6), fields of view were chosen so that extended dendritic segments were in one focal plane. At the end of each imaging session, z-stacks (1 μm step size) of the recorded cells were acquired. The coordinates of the imaged dendrites relative to the parent somata were recorded. The orientation, curvature, and the branching pattern of the dendrites together with the constellation of spines, helped to precisely identify the same field of view in long-term imaging experiments.
Electrophysiology
In vivo cell-attached recordings were performed using glass pipettes (∼5-7 MΩ) filled with solution containing the following (in mM): 125 NaCl, 5 KCl, 10 glucose, 10 HEPES, 2 CaCl2, 2 MgSO4, and 0.1 Alexa Fluor 594; pH 7.4). Signals were amplified using an AxoPatch 200B amplifier (Molecular Devices), filtered at 5 kHz, and digitized at 10 kHz. Spikes were recorded using current clamp mode. The frame trigger pulses of ScanImage 4.0 were also recorded and used offline to synchronize individual frames to electrophysiological recordings. After establishment of a low-resistance seal (15-50 MΩ), the orientation, spatial and temporal frequency of the stimuli was quickly optimized for individual neurons using recorded spikes. The optimal grating stimulus was repeated at a reduced contrast to maintain a moderate spiking rate.
Image analysis
Mechanical drift in the imaging plane was corrected using the TurboReg plug-in in ImageJ61. All remaining analyses were performed in MATLAB. Regions-of-interest (ROIs) corresponding to visually identifiable cell bodies were selected using a semi-automated algorithm (Supplementary Fig. 14). For GCaMP, ring-shaped ROIs were placed at the cytosolic regions of the cells (excluding the nucleus; GCaMP expression is typically restricted to the cytoplasm11). For OGB1-AM, circular ROIs covering the whole soma were used. For long-term GCaMP imaging, baseline fluorescence images of multiple sessions were inspected manually, and only the cells that could be clearly identified in all imaged sessions were included in the analysis. The fluorescence time course of each cell was measured by averaging all pixels within the ROI, with a correction for neuropil contamination41. The fluorescence signal of a cell body was estimated as Fcell_true(t)=Fcell_measured(t)-r*Fneuropil(t), with r=0.7. The neuropil signal Fneuropil(t) surrounding each cell was measured by averaging the signal of all pixels within a 20 μm region from the cell center (excluding all selected cells). Cell-attached recordings confirmed that neuropil-compensated fluorescence changes reflect action potentials in single neurons (Supplementary Fig. 15). To ensure robust neuropil subtraction, only cells that were at least 3% brighter than the surrounding neuropil were included. The neuropil correction was not applied for dendritic imaging experiments because sparse labeling provided negligibly low background. ΔF/F0 was calculated as (F-F0)/F0, where F0 is the baseline fluorescence signal averaged over a 2 second period immediately before the start of visual stimulation. Visual responses were measured for each trial as ΔF/F0, averaged over the stimulus period. Visually responsive neurons were defined as cells with significant stimulus-related fluorescence changes (ANOVA across blank and eight direction periods, p<0.01)5 with an average ΔF/F0 at preferred orientations greater than 6%.
The orientation selectivity index (OSI) was calculated for visually responsive cells16,30. First, the preferred orientation (θpref) of the cell was determined as the stimulus that produced the strongest response. The orientation tuning curve was constructed by measuring the mean ΔF/F0, averaged over the stimulus period, for each orientation. We then fitted the tuning curve with the sum of two Gaussians centered on θpref and θpref + π, both with width σ (constrained to >15°), amplitudes A1 and A2, and a constant baseline B. The OSI was defined as OSI=(Rpref-Rortho)/(Rpref+Rortho), where Rpref and Rortho are the response amplitudes at the preferred (θpref) and the orthogonal orientation (θpref + π/2) respectively.
For simultaneous imaging and cell-attached recording, ring-shaped ROIs were placed over the cytosolic regions of the cells. Fluorescence transients at the soma were caused by action potentials, with little contribution from subthreshold activity62 (Supplementary Fig. 15). To quantify the efficiency for detecting single APs (Fig. 3), we identified single AP events with nearby APs at least 1 s away. Fluorescence traces consisting of 10 frames (0.17 s) before and 60 frames (1 s) after the ith 1 AP event were assembled in 70-dimensional vectors, fi. Segments of noisy traces, ni were taken from periods without APs. The average of all 1 AP traces was used as a template vector, ftemplate=Σifi/N. The vector was normalized after subtraction of the mean to create a unit vector f̂template. The projection of fi or ni along the direction of f̂template was calculated to obtain a scalar fi or ni, respectively. The AP detection threshold was defined as the 99th percentile of all ni values (i.e., 1% false positive), and the percentage of the fi values above the detection threshold was the AP detection efficiency.
For spine images (Fig. 4-5), circular ROIs were placed over individual dendritic spines to measure spine fluorescence and compute ΔF/F0_spine. To minimize contamination from back-propagating action potentials (BAPs), we either recorded from ‘silent cells’ (∼ 40 % of cells) showing few or no APs in response to a standard set of grating stimuli (Fig. 4), or used stimuli with reduced stimulus contrast (Fig. 5).
Occasional BAP related calcium signals that invaded the imaged spines were removed using a subtraction method, implemented in three steps (Supplementary Fig. 11). First, a region covering the entire parent dendritic shaft (∼30 micrometers of dendritic length; excluding all spines) was drawn for each recorded dendritic segment to estimate BAP related global dendritic signal, ΔF/F0_dendrite. Because of the much larger volume of the dendritic shaft compared to tuned dendritic spines (100-fold), spines above and below the focal plane were expected to contribute negligible signal to ΔF/F0_dendrite. This was verified using principle component analysis (data not shown). Plotting ΔF/F0_spine against ΔF/F0_dendrite reveals two components of spine signals, a BAP-related component and a spine-specific component. Second, the BAP-related component was removed from the spine signals by subtracting a scaled version of the dendritic shaft signal, ΔF/F0_spine_specific=ΔF/F0_spine - α ·ΔF/F0_dendrite. α was determined using robust regression (MATLAB function ‘robustfit.m’) of ΔF/F0_spine vs. ΔF/F0_dendrite (the slope of the fitted line in Supplementary Fig. 11b). Third, the visual responsiveness (ΔF/F0 > 10%) and the OSI of individual spines were calculated with the BAP signal removed. Active spines (Fig. 4f) were defined as spines showing at least three spine-specific (i.e., BAP independent) calcium events during the 5 minute imaging session, with an event defined as an episode of the calcium signal that crosses 3 s.d. of the baseline noise for at least three consecutive frames (∼50 ms).
We next confirmed the effectiveness of the BAP removal algorithm. First, BAP removed spine signals showed sharp orientation tuning (OSI = 0.84 ± 0.14, mean ± s.d., n=190 spines) comparable to V1 excitatory neurons (Supplementary Fig. 9, p>>0.05, Wilcoxon rank sum). This holds even for spines preferring the orthogonal orientation compared to the soma (OSI=0.82 ± 0.16, n=24 spines), indicating that the BAP contamination was cleanly removed. Second, the majority (79.7%) of visually responsive spines showed little trial-to-trial correlation with the dendritic shaft signal after BAP subtraction. 20.3% spines still displayed significant correlation with shaft responses compared to trial shuffled controls (p<0.01). This could reflect synchronously active pre-synaptic cells, or imperfect BAP action potential signal subtraction. Because we were unable to distinguish between these possibilities, these spines were excluded from further analysis. In two cells (cell 4 and cell 5; Fig. 5), we collected larger numbers of trials (15 trials per orientation), which made it possible to further exclude trials with detectable dendritic responses (ΔF/F0 > 6%). The result (i.e., the preferred orientation of the summed spine responses) was identical as with using the subtraction procedure alone.
For the analysis of GABAergic cells (Fig. 6), dendrites were traced using ‘Simple neurite tracer’ in ImageJ63. The program outputs a 1D sequence of coordinates traversing a dendrite and a 2D mask covering the traced dendrite, which were used to define ROIs along the dendrite (size, 1.5 μm of dendritic length; spacing, 1 μm). Visual responsiveness and orientation selectivity index were computed for individual ROIs. A region of dendrite is considered orientation selective if its response to least one stimulus orientation is significantly different from other orientations (p < 0.01, ANOVA across 8 conditions). Because synaptic and BAP signals were intermixed in the same dendritic compartment, no attempt was made to isolate synaptic signals from BAP-related components. We did not analyze the relationship between integrated synaptic signals and the output tuning in GABAergic cells.
Reagent distribution
DNA constructs and AAV particles with GCaMP6 variants were deposited for distribution at Addgene (www.addgene.org) and the University of Pennsylvania Vector Core (www.med.upenn.edu/gtp/vectorcore) and the Bloomington Drosophila Stock Center (flystocks.bio.indiana.edu), respectively.
Supplementary Material
Acknowledgments
We thank J. Akerboom and L. Tian for constructs and advice; J. Hasseman, M. Ramirez, G. Tsegaye for molecular cloning; B. Shields for neuronal culture; A. Hu for histology; B. Fosque, R. Behnam, K. Ritola for virus production; J. Macklin and R. Patel for spectroscopy; B. Coop and L. Ramasamy for multiwell electrode manufacturing; K. Smith for mouse viral transduction; K. Hibbard for fly husbandry; J. Yu, C. Niell, M. Stryker, J. Trachtenberg and A. Kerlin for advice on visual cortex experiments.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: V.J., R.A.K., L.L.L., and K.S. initiated the project. T.W.C., T.J.W, and D.S.K. conducted neuronal culture screening. T.W.C. performed mouse visual cortical experiments on pyramidal neurons. T.W.C. and A.B. performed experiments on inhibitory neurons. Y.S., T.J.W., and S.R.P. carried out fly larval NMJ studies. Y.S. and V.J. carried out adult fly antennal lobe imaging. S.L.R. and M.B.O. conducted zebrafish tectal imaging. E.R.S. performed protein assays. All authors analyzed data. T.W.C., R.A.K., M.B.O., V.J., L.L.L., K.S., and D.S.K. wrote the paper with comments from all authors.
Author Information: The authors have applied for a patent on materials and methods related to the GCaMP6 variants. S.L.R. is supported by a Fellowship from the Swiss National Science Foundation. M.B.O. is supported by a Marie Curie Career Integration Grant PCIG09-GA-2011-294049.
References
- 1.Baker PF, Hodgkin AL, Ridgway EB. Depolarization and calcium entry in squid giant axons. J Physiol. 1971;218:709–755. doi: 10.1113/jphysiol.1971.sp009641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerr R, et al. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26:583–594. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]
- 3.Tank DW, Sugimori M, Connor JA, Llinas RR. Spatially resolved calcium dynamics of mammalian Purkinje cells in cerebellar slice. Science. 1988;242:773–777. doi: 10.1126/science.2847315. [DOI] [PubMed] [Google Scholar]
- 4.Sabatini BL, Oertner TG, Svoboda K. The life cycle of Ca(2+) ions in dendritic spines. Neuron. 2002;33:439–452. doi: 10.1016/s0896-6273(02)00573-1. [DOI] [PubMed] [Google Scholar]
- 5.Ohki K, Chung S, Ch'ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 6.Huber D, et al. Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature. 2012;484:473–478. doi: 10.1038/nature11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- 8.Petreanu L, et al. Activity in motor-sensory projections reveals distributed coding in somatosensation. Nature. 2012;489:299–303. doi: 10.1038/nature11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Stereotyped odor-evoked activity in the mushroom body of Drosophila revealed by green fluorescent protein-based Ca2+ imaging. J Neurosci. 2004;24:6507–6514. doi: 10.1523/JNEUROSCI.3727-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia H, Rochefort NL, Chen X, Konnerth A. Dendritic organization of sensory input to cortical neurons in vivo. Nature. 2010;464:1307–1312. doi: 10.1038/nature08947. [DOI] [PubMed] [Google Scholar]
- 11.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zariwala HA, et al. A Cre-Dependent GCaMP3 Reporter Mouse for Neuronal Imaging In Vivo. J Neurosci. 2012;32:3131–3141. doi: 10.1523/JNEUROSCI.4469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao T, O'Connor DH, Scheuss V, Nakai J, Svoboda K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PloS One. 2008;3:e1796. doi: 10.1371/journal.pone.0001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreosti E, Odermatt B, Dorostkar MM, Lagnado L. A genetically encoded reporter of synaptic activity in vivo. Nat Methods. 2009;6:883–889. doi: 10.1038/nmeth.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerrero G, et al. Heterogeneity in synaptic transmission along a Drosophila larval motor axon. Nat Neurosci. 2005;8:1188–1196. doi: 10.1038/nn1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akerboom J, et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 18.Mank M, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods. 2008 doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 20.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer AE, et al. Ca2+ Indicators Based on Computationally Redesigned Calmodulin-Peptide Pairs. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Horikawa K, et al. Spontaneous network activity visualized by ultrasensitive Ca(2+) indicators, yellow Cameleon-Nano. Nat Methods. 2010;7:729–732. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- 23.Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiff DF, et al. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 27.Akerboom J, et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J Biol Chem. 2009;284:6455–6464. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayley PM, Findlay WA, Martin SR. Target recognition by calmodulin: dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5:1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004 doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 30.Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trachtenberg JT, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 32.Mainen ZF, Malinow R, Svoboda K. Synaptic calcium transients in single spines indicate that NMDA receptors are not saturated. Nature. 1999;399:151–155. doi: 10.1038/20187. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Leischner U, Rochefort NL, Nelken I, Konnerth A. Functional mapping of single spines in cortical neurons in vivo. Nature. 2011;475:501–505. doi: 10.1038/nature10193. [DOI] [PubMed] [Google Scholar]
- 34.Binzegger T, Douglas RJ, Martin KA. A quantitative map of the circuit of cat primary visual cortex. J Neurosci. 2004;24:8441–8453. doi: 10.1523/JNEUROSCI.1400-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko H, et al. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansel D, van Vreeswijk C. The mechanism of orientation selectivity in primary visual cortex without a functional map. J Neurosci. 2012;32:4049–4064. doi: 10.1523/JNEUROSCI.6284-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu BH, et al. Broad inhibition sharpens orientation selectivity by expanding input dynamic range in mouse simple cells. Neuron. 2011;71:542–554. doi: 10.1016/j.neuron.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan AY, Brown BD, Scholl B, Mohanty D, Priebe NJ. Orientation selectivity of synaptic input to neurons in mouse and cat primary visual cortex. J Neurosci. 2011;31:12339–12350. doi: 10.1523/JNEUROSCI.2039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobczyk A, Scheuss V, Svoboda K. NMDA receptor subunit-dependent [Ca2+] signaling in individual hippocampal dendritic spines. J Neurosci. 2005;25:6037–6046. doi: 10.1523/JNEUROSCI.1221-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bock DD, et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofer SB, et al. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat Neurosci. 2011;14:1045–1052. doi: 10.1038/nn.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldberg JH, Tamas G, Yuste R. Ca2+ imaging of mouse neocortical interneurone dendrites: Ia-type K+ channels control action potential backpropagation. J Physiol. 2003;551:49–65. doi: 10.1113/jphysiol.2003.042580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gulyas AI, Megias M, Emri Z, Freund TF. Total number and ratio of excitatory and inhibitory synapses converging onto single interneurons of different types in the CA1 area of the rat hippocampus. J Neurosci. 1999;19:10082–10097. doi: 10.1523/JNEUROSCI.19-22-10082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldberg JH, Tamas G, Aronov D, Yuste R. Calcium microdomains in aspiny dendrites. Neuron. 2003;40:807–821. doi: 10.1016/s0896-6273(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 47.Katona G, et al. Roller Coaster Scanning reveals spontaneous triggering of dendritic spikes in CA1 interneurons. Proc Natl Acad Sci USA. 2011;108:2148–2153. doi: 10.1073/pnas.1009270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y, et al. An expanded palette of genetically encoded Ca(2) indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobat D, et al. Deep tissue multiphoton microscopy using longer wavelength excitation. Opt Express. 2009;17:13354–13364. doi: 10.1364/oe.17.013354. [DOI] [PubMed] [Google Scholar]
- 51.Hanawa H, et al. Efficient gene transfer into rhesus repopulating hematopoietic stem cells using a simian immunodeficiency virus-based lentiviral vector system. Blood. 2004;103:4062–4069. doi: 10.1182/blood-2004-01-0045. [DOI] [PubMed] [Google Scholar]
- 52.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 53.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suter BA, et al. Ephus: multipurpose data acquisition software for neuroscience experiments. Front Neural Circuits. 2010;4:100. doi: 10.3389/fncir.2010.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu NL, et al. Nonlinear dendritic integration of sensory and motor input during an active sensing task. Nature. 2012;492:247–251. doi: 10.1038/nature11601. [DOI] [PubMed] [Google Scholar]
- 57.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brainard DH. The Psychophysics Toolbox. Spatial vision. 1997;10:433–436. [PubMed] [Google Scholar]
- 59.Pelli DG. The VideoToolbox software for visual psychophysics: transforming numbers into movies. Spat Vis. 1997;10:437–442. [PubMed] [Google Scholar]
- 60.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser-scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thevenaz P, Ruttimann UE, Unser M. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 62.Bandyopadhyay S, Shamma SA, Kanold PO. Dichotomy of functional organization in the mouse auditory cortex. Nat Neurosci. 2010;13:361–368. doi: 10.1038/nn.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Longair MH, Baker DA, Armstrong JD. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- 64.Wang Q, Shui B, Kotlikoff MI, Sondermann H. Structural basis for calcium sensing by GCaMP2. Structure. 2008;16:1817–1827. doi: 10.1016/j.str.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.