Abstract
Asthma is an idiopathic disease characterized by episodic inflammation and reversible airway obstruction triggered by exposure to environmental agents. Because this disease is heterogeneous in onset, exacerbations, inflammatory states, and response to therapy, there is intense interest in developing personalized approaches to its management. Of focus in this review, the recognition that a component of the pathophysiology of asthma is mediated by inflammation has implications for understanding its etiology and individualizing its therapy. Despite understanding how Th2 polarization mediates asthma exacerbations by aeroallergen exposure, we do not yet fully understand how RNA virus infections produce asthmatic exacerbations. This review will summarize the explosion of information that has revealed how patterns produced by RNA virus infection trigger the innate immune response (IIR) in sentinel airway cells. When the IIR is triggered, these cells elaborate inflammatory cytokines and protective mucosal interferons whose actions activate long-lived adaptive immunity and limit organismal replication. Recent work has shown the multifaceted way that dysregulation of the IIR is linked to viral-induced exacerbation, steroid insensitivity, and T helper polarization of adaptive immunity. New developments in quantitative proteomics now enable accurate identification of subgroups of individuals that demonstrate activation of IIR (“innate endotype”). Potential applications to clinical research are proposed. Together, these developments open realistic prospects for how identification of the IIR endotype may inform asthma therapy in the future.
Keywords: Asthma, Multivariate adaptive regression splines, Innate immune response (IIR), Respiratory syncytial virus, Interferon (IFN), Nuclear factor-κB (NFκB), Interferon response factor (IRF), Pattern recognition receptors, Toll-like receptors (TLRs), Retinoic acid like helicases (RLH), Personalized medicine, Endotypes, Innate immune response endotypes
Introduction
Asthma is an idiopathic disease whose lifetime prevalence is as high as 10% of the general population [1]. In asthmatics, acute episodes of inflammation that produce obstructive symptoms (wheezing, chest tightness, shortness of breath, mucous production) are the primary reasons for emergency room visits and reductions in the quality of life [2]. It is well known that a predominant group of asthmatics have aeroallergen sensitivity and established atopy. In these individuals, episodic deterioration can be induced by aero-allergen exposure, triggering IgE-mediated release of mast cell degranulation products [1].
Another mechanism for acute decompensation is mediated by RNA virus infection, with viruses from the picornaviridae (rhinovirus) or paramyxoviridae (respiratory syncytial virus, humanmetapneumovirus) families. Respiratory virus infection is a major cause of acute exacerbations of obstructive symptoms in patients with well-controlled asthma, accounting for up to ¾ of emergency visits in otherwise well-controlled asthmatics [2–5]. These infections stimulate a distinct inflammatory pathway known as the innate immune response (IIR; [6]). The IIR is a rapid, but non-pathogen-specific, intracellular inflammatory response produced by detection of a pathogen signature in sentinel airway cells. These cells include lining airway epithelial cells, macrophages, and specialized antigen presenting cells (dendritic cells). Cells with the IIR activated undergo an induced phenotypic change, elaborating inflammatory cytokines and protective mucosal interferons. These factors limit the spread of the organism, and regulate the nature and magnitude of the adaptive immune response.
Recent studies have shown that adult asthmatics have increased susceptibility to rhinovirus (RV) infections, leading to increased lower respiratory tract inflammation and hyperreactivity. These studies suggest that RV fails to induce type I IFNs and therefore replicates more efficiently. Together, these findings provide compelling evidence that dysregulation of the IIR is closely linked with viral-induced asthma exacerbations and warrants further clinical investigation.
The advent of the post-human genome era opens the door for applications of personalized medicine to this complex disease. Personalized medicine relies on the measurement of molecular profiles that identify distinct pathological subtypes (endotypes) of disease. Molecular profiling provides a level of information that is not otherwise apparent by conventional clinical diagnostic approaches. Understanding these profiles leads to application of individualized, tailored therapies directed at the underlying pathophysiology. To this end, two advances are needed: (1) objective definition pathophysiological subtypes, and (2) development of therapies specifically tailored to these underlying pathophysiologies. In previous issues of this journal, some preliminary strategies for molecular classification of asthma phenotypes have been presented [7]. Here, I will extend this consideration to focus on the identification of disease process-relevant endotype associated with innate inflammation.
Endotyping
Advances in high dimensional, “omics”, technologies, now allows the routine measurement of large numbers of proteins, genes, or metabolites (analytes) in human health and disease. This approach has the promise to provide precise information about pathophysiologies in a disease that is not otherwise possible using conventional physiological or clinical assessments. Previous efforts at phenotypic classifications of asthma have used age of onset, exacerbating factors (aspirin sensitivity, exercise-induced, occupational), cellular type of inflammation (eosinophilic vs. noneosinophilic), pattern of severity, or lack of response to conventional therapy. In the absence of biochemical marker measurement, the multiple underlying pathological processes that contribute to the clinical syndrome of asthma, such as allergy, inflammation, airway remodeling, smooth muscle hypertrophy, neurogenic dysregulation, etc., are not considered [8]. Precise quantification of these processes would enable pathophysiological-focused treatments and refine clinical management.
Inflammation in asthma
It is widely accepted that asthma is driven by a specific type of T helper (Th)-2 lymphocyte inflammation, mediated by production of IL-4, -5, -9, and -13 cytokines resulting in pulmonary eosinophil activation, B cell IgE production, and recruitment of mast cells [1]. The Th2 hypothesis has provided an important conceptual and therapeutic framework for management of asthma. However, this hypothesis does not completely account for many aspects of asthma, including airway remodeling, smooth muscle hypertrophy, lack of response to T cell-directed therapies, and acute exacerbations produced by RNA virus infections [8].
Recent applications of bipartite network analysis as a visualization tool, to examine naturally occurring groups of cytokine measured in bronchoalveolar lavage samples obtained from patients with moderate and severe asthma, have revealed the presence of distinct inflammatory subtypes [7, 9]. This analysis has indicated that, although a subgroup have characteristic Th2 cytokine grouping (IL4 and eotaxin), another predominant subset was enriched in MIP-1α/β, MCP-1, MIG, and others [7, 9]. These cytokines are signatures of the activated IIR, suggesting that distinct patterns of adaptive and innate inflammatory subtypes are found in asthmatics. This seminal finding has important implications for the pathophysiology of asthma, as well as for its management.
The Pulmonary Innate Immune Response (IIR)
Alveolar macrophages, dendritic cells (DCs), and epithelial cells represent the primary sentinel cells of the naïve airways, patrolling for the presence of non-self antigens in the airway. Non-self antigens, so-called pathogen-associated molecular patterns (dsRNA, lipoteichoic acids, mannans, flagellins) are detected by germline-encoded pattern recognition receptors (PRRs). The two major PRR classes in the airways are the membrane-resident Toll-like receptors (TLRs) and the cytoplasmic Retinoic Acid inducible Gene (RIG)-I-like RNA helicases (RLHs; [10]).
Currently, we understand the role of these two PRRs in RNA virus infection as being quite distinct. Gene knockout studies in mice and in vitro experiments in human airway epithelial cells indicate that the RLHs play the major role in signaling in response to RSV infections [11].
RIG-I and the melanoma differentiation-associated gene-5 (Mda5) are two major members of the RLH family, and are essential PRRs for cytoplasmic double-stranded (ds) RNA [12, 13]. RIG-I and Mda5 recognize different types of viral and dsRNAs, with RIG-I responding to most ssRNA viruses and short viral RNAs, whereas Mda5 responds to picornavirus RNA and longer dsRNAs. Activation of RLHs is triggered by binding cytoplasmic RNA, triggering a non-destructive ubiquitylation, promoting formation of an activated signaling complex on the surface of mitochondria [14].
The TLRs are a family of 12 related cell surface-localized PRRs that bind diverse pathogen-associated molecular patterns, including lipopolysaccharide (LPS), lipoteichoic acids, mannans, flagellins, dsRNA, and DNA oligonucleotides[15]. The primary TLR binding to dsRNA is TLR3, a receptor primarily localized in endosomal compartment in epithelial cells, dendritic cells, and macrophages. This localization is highly regulated by the action of interferons. Our studies have shown that, after viral infection, TLR3 expression is upregulated and redistributes to the cell surface [11]. As a result, previous activation of the IIR will elicit distinct patterns of inflammatory responses. This is clinically important phenomenon, and explains why prior exposure to LPS or RNA viruses alters subsequent inflammatory responses [16] [17]. Upon binding to dsRNA, both TLR3 and RLHs form activated signaling complexes that converge on the cytoplasmic NF-κB and interferon response factor (IRF)-3 transcription factors. These proteins translocate into the cell nucleus and trigger gene expression. NF-κB triggers acute inflammation and cellular apoptosis [18, 19], whereas the actions of IRF3 triggers type I IFN expression [20, 21].
Linkage of the IIR to adaptive immunity
Current work suggests that the epithelial cell plays a major role in initiating and sustaining pulmonary IIR. High density microarrays have shown that epithelial cells are a major source of CC, CXC, and C-classes of chemokines in response to viral infections [22, 23]. The epithelial IIR triggers acute inflammation through the coordinated and distinct actions of the diverse actions cytokines produced. CXC chemokines stimulate the recruitment of specific classes of leukocytes via integrin–adressin interactions to invade the interstitial spaces and lumen of the airway [24]. These activated leukocytes then migrate down chemotactic gradients to the site of infection and are stimulated to induce phagocytosis and antigen uptake.
Another important cell mediating airway inflammation is the dendritic cell (DC), a highly specialized cell type that coordinates the development of adaptive immunity. Through CCL20–MIP-1α chemokine signaling, DCs are recruited into the lung in response to IIR activation. In combination with microenvironmental cues, distinct classes of DCs play an important role in shaping the adaptive immunity by determining polarization of CD4 T helper (TH) lymphocytes [25]. One important microenvironmental signal of special relevance to asthma is the epithelial-derived thymic stromal lymphopoietin, an IIR-inducible cytokine-like molecule that enhances DC maturation and promotes TH differentiation [26]. Depending on microenvironmental cofactors and cell type interactions, TH lymphocytes differentiate into phenotypically distinct subsets, known as TH-1 and -2), or the more recently identified TH17 subsets; these types are distinguished by immune regulatory function and cytokine secretion patterns [1, 27]. TH1 cells produce IFN-γ and mediate cellular immunity, whereas TH2 cells produce interleukin (IL)-4, -5, and -13 that mediate humoral immunity, IgE production, and allergic responses. TH17 cells express ILs-17 and -6, and, although their role in asthma is not fully understood, TH17 cells have been associated with chronic inflammation [27]. It is currently thought that TH polarization events alter the underlying pathophysiology of the lung. In this way, IIR induces both acute inflammation, TH polarization, and sensitization in the airway.
Dysregulation of the IIR in asthma
Respiratory tract infections with either viruses from the Picornaviridae (rhinovirus) or Paramyxoviridae (respiratory syncytial virus, humanmetapneumovirus) families is well-established as a cause of acute exacerbations of obstructive symptoms in patients with well-controlled asthma. In fact, RNA virus infections account for up to ¾ of emergency visits [2, 28, 29]. The interactions of these viruses with host epithelial cells have been intensively studied, and it is well established that both of these viruses are potent activators of the IIR, inducing both NF-κB and IRF3 activation and downstream cytokine expression [30, 31]. Mouse studies have shown that inhibition of the NF-κB arm of the IIR prevents acute disease induced by RSV infection, suggesting that an important component of airway inflammation is the host inflammatory response [32].
Interestingly, adult asthmatics have increased susceptibility to RV infections, leading to increased lower respiratory tract inflammation and hyperreactivity. Two studies have shown that sentinel respiratory cells replicate rhinovirus more efficiently and show deficient induction of type I IFNs, IFNs-β and -λ , with impaired ability to induce apoptosis [29, 33]. This defect in type I IFN production was highly correlated with severity of rhinovirus-induced asthma exacerbation and virus load in experimentally infected human volunteers, suggesting that asthmatics have an acquired defect in type I IFN production in sentinel cells of the airway. The mechanisms underlying this defect in the IIR will require further investigation.
Proteomics methods for sampling the IIR
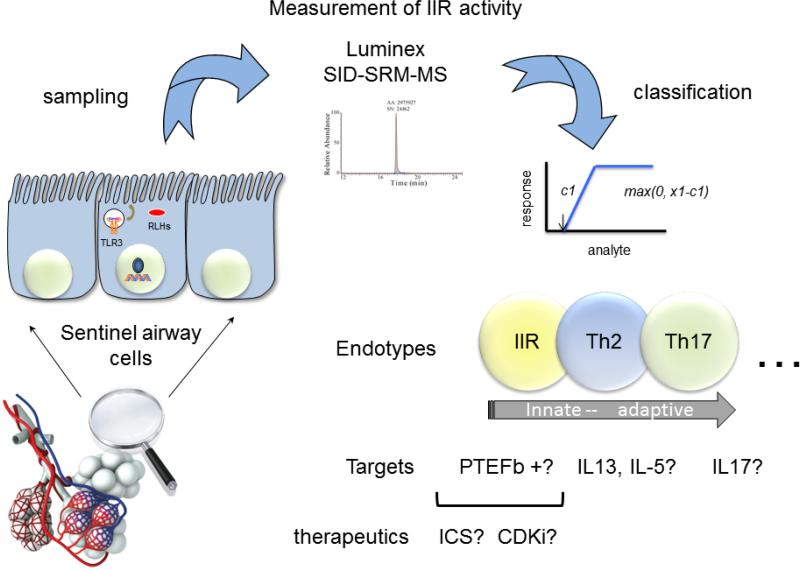
The ability to quantify the status of the activated IIR will open new opportunities for the treatment and management of asthma (Fig. 1). A significant advance in the ability to monitor multiple cytokines has been the development multiplex bead-based immunoassays with the Luminex technology. This technology uses panels of colored microspheres, with each color conjugated with a unique capture antibody. The amount of cytokine in a sample that binds to each bead type can be tracked and quantified. For each cytokine, a standard curve is generated by using recombinant proteins to estimate protein concentration in the unknown sample. Ten to 20 distinct cytokines can be measured, and in our hands, these assays have a sensitivity comparable to enzyme-linked immunosorbent assay measurements, with a detection limit of 10–30 ng/L, low interassay variation and a large dynamic range [34, 35].
Figure 1.
Personalized medicine for innate endotypes in asthma. A schematic view of a sampling strategy, inflammatory measurement, and potential targets for developing actionable individual interventions in asthma. CDKi cyclin-dependent kinase inhibitor, ICS inhaled corticosteroids, SID stable-isotope dilution, SRM-MS selective reaction monitoring mass spectrometry
Although the multiplex assays are simple and highly quantitative, the measure of cytokine concentration only indicates that the IIR has been activated at some time in the past. For the purposes of clinical investigation and manipulation of the IIR, a more direct method for detecting the activation status of the IIR is needed.
Selected reaction monitoring (SRM)-mass spectrometry (MS)
Work at the UTMB NHLBI Proteomic Center for Airway Inflammation has resulted in development of mass spectrometry-based techniques for the quantification of the IIR, known as selected reaction monitoring-mass spectrometry (SRM-MS; [36–38]). In an SRM-MS assay, the mass spectrometer monitors the sample for the presence of unique signature proteotypic peptides unique to the protein. When that peptide is detected, the triple quadrupole mass spectrometer fragments the peptide into specific fragment ions. SRM assays do not require the generation of high affinity antibodies, and yet have a lower limit of detection to pg/ml protein concentrations.
SRM assays offer several attractive features. First, because only preselected precursor–product ion transitions are monitored in SRM mode, the noise level is significantly reduced, and thereby SRM assays decrease the lower detection limit for peptides by up to 100-fold in comparison to a conventional full scan MS/MS analysis. Second, the two filtering stages in SRM result in near-absolute structural specificity for the target protein. Third, SRM-MS assay is compatible with stable-isotope dilution (SID) for direct quantification of target proteins in a complex mixture [36]. Our evaluation of these assays indicates that SRM-MS assays are highly sensitive and specific for components of the IIR, enabling quantification of the direct activation state of the IIR in mucosal samples. This exciting new technology affords direct, mechanistic read out of the IIR and its component signaling arms.
Interpretation and prediction using high dimensional datasets: molecular classification
Simply measuring multiple panels of cytokines or biomarkers is of limited value in predicting disease phenotypes. An active area of investigation is how to relate multidimensional measurements to clinically meaningful subtypes. This approach is referred to as molecular classification, and relies on machine-learning tools. Although much work has been done to develop molecular classification methods based on gene expression data, our experience using protein patterns requires that a distinct approach is needed [39].
A characteristic of high-dimensional datasets is that only some of the measurements are informative and many are not. Classification approaches using unprocessed features leads to the “curse of dimensionality”, where the presence of many features often leads to poorer, rather than better, classifier performance. A key step in developing robust and generalizable predictive models, therefore, is to reduce the dimensionality using a technique called feature reduction. To accomplish this, we have employed a widely used permutation-based approach to identifying differentially expressed features in high dimensional datasets using adjustment for false discovery rate [40].
The next major step is to determine how to combine groups of proteins into meaningful models that predict a clinical phenotype. In a systematic comparison of a variety of modeling techniques, we have determined that multivariate regression splines (MARS) is a robust nonparametric modeling approach that outperforms other approaches including linear regression modeling, classification trees [41]. Some of the advantages of MARS are that it can model predictor variables of many data types, continuous or categorical, and it can tolerate large numbers of input predictor variables. Importantly, MARS does not make any underlying assumptions about the distribution of the predictor variables of interest. This characteristic is extremely important in high-dimensional modeling because many of the cytokine and protein expression values are not normally distributed. The basic concept behind spline models is to model using potentially discrete linear or nonlinear functions of any analyte over differing intervals. The classifier combines these piecewise curves into a predictive model in an approach similar to multivariate regression modeling. As with the other modeling techniques, MARS uses cross-validation to avoid over-fitting. The optimal end result is a classification model based on single variables and interaction terms which will optimally determine predictive outcome. To this end, we have demonstrated this nonparameteric MARS modeling approach yields highly accurate classifiers of inflammatory endotypes in asthma, outperforming all other machine-learning approaches tested [34].
Personalized medicine in asthma
It is widely appreciated that asthmatics show wide variations in therapeutic response to β adrenergic agonists, glucocorticoids and leukotriene inhibitors [42]. Here, the identification of biomarkers that predict a successful therapeutic response will have revolutionary impact on the field by allowing more rapid response with reduction in side effects. A schematic view of personalized medicine approach using inflammatory subtypes is shown in Fig. 1.
One proof-of-principle for personalized medicine in asthma was provided by the prospective, randomized, double-blind, placebo-controlled trial of the humanized IgG4 monoclonal IL-13 neutralizing antibody (Lebrikizumab). This study included 219 subjects with severe asthma, exemplified by reduction in FEV1 (the average FEV1 was 65 % predicted) that showed reversibility with bronchodilator treatment. In the study design, pre-treatment measurements of Th2 inflammation (serum IgE and osteopontin) were obtained. In airway epithelial cells, osteopontin is induced by the Th2 cytokine, IL13, and thereby represents a biomarker for IL-13 action. This study reported that the response of the overall population was minimal, yet a highly responsive subgroup was identified by measurement of pre-treatment Th2 activity [43]. This subgroup showed a rapid and sustained increase in prebronchodilator FEV1 measurements after 12 weeks compared to the low osteopontin subgroup. These exciting studies suggest that highly responsive individuals can potentially be identified by measurements of biomarkers of the inflammatory endotype.
A major advance in personalized medicine will be the identification of selectively druggable targets that modify the activity of the innate immune response in asthmatics who manifest pathway activation (Fig. 1). In this regard, there have been significant advances in the understanding how the innate pathway is activated. At the biochemical level, recent work has shown that the rapid activation of inflammatory genes in the innate pathway is mediated by the action of a nuclear complex termed the positive transcriptional elongation factor (PTEF-b). Transcriptional elongation allows cytokine genes to rapidly respond to the activated innate pathway independent of extensive chromatin remodeling or promoter assembly [44]. The core activity of PTEF-b is a cyclin-dependent kinase [44–46]. Interestingly, inducible transcriptional elongation is blocked by the actions of glucocorticoids; this is one of several mechanisms by which glucocorticoids reduce inflammation [47]. Whether individuals with the innate endotype will be more responsive to glucocorticoid treatment will require systematic study. Additionally, selective cyclin-dependent kinase inhibitors are under clinical development for the treatment of chronic lymphocytic leukemia [48], and may represent a class of well-tolerated compounds that could be repurposed for treatment of acute asthma exacerbations associated with RNA viral infections.
Potential clinical applications
Accurate (and reliable measurement) of the IIR is the first step in developing a personalized approach to modulating the innate endotype for therapeutic purposes. One potential way that these new technologies could be applied to monitoring the IIR would be to sample epithelial brushings of asthmatics using SID-SRM, or to measure products of the IIR in airway secretions (or BAL) using multiplex immunoassays. This information can be used in clinical research, where the activity of the IIR can be related to outcomes or to stratify response to various therapies, similar to the example of the anti-IL13 mAb discussed above. Additionally, these tools can be used to investigate the dysregulation of the IIR in asthmatics with viral infections and to evaluate the impact of immunomodulation on the pulmonary response to RNA viral infections.
Conclusions
Because of its central role in mediating the effect of RNA virus on acute decompensation and shaping the adaptive immune response to produce allergic sensitization/atopy, much attention is being placed on investigating the pathophysiological role of the IIR in asthma. The recent development of quantitative proteomics methods for measurement of the IIR in complex samples will stimulate clinical investigation, both to determine the influence of the IIR on the natural history of the disease, and to understand the response of the IIR to conventional therapies.
Acknowledgments
Work in the author's laboratory is supported by NIAID PO1 AI062885, NCATS UL1TR000071, NIAID Clinical Proteomics Center HHSN272200800048C, NHLBI Proteomics Center for Airway Inflammation NIH-NHLBI-HHSN268201000037C, and NIEHS P30 ES006676.
Footnotes
Conflict of Interest
Allan R. Brasier declares that he has no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
* of importance
** of major importance
- 1.Busse WW, Lemanske RF. Asthma. New England Journal of Medicine. 2001;344:350–362. doi: 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]
- 2.Johnston NW, Sears MR. Asthma exacerbations · 1: Epidemiology. Thorax. 2006;61:722–728. doi: 10.1136/thx.2005.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardin PG, Johnston SL, Pattemore PK. Viruses as precipitants of asthma symptoms. II. Physiology and mechanisms. Clinical and Experimental Allergy. 1992;22:809–822. doi: 10.1111/j.1365-2222.1992.tb02825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corne JM, Holgate ST. Mechanisms of virus induced exacerbations of asthma. Thorax. 1997;52:380–389. doi: 10.1136/thx.52.4.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gern JE, Busse WW. Relationship of viral infections to wheezing illnesses and asthma. Nature Reviews Immunology. 2002;2:132–138. doi: 10.1038/nri725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brasier AR, Calhoun WJ. Proteomic Insights Into Inflammatory Airway Diseases. Current Proteomics. 2011;8:84–96. [Google Scholar]
- 7.Pillai RR, Divekar R, Brasier A, Bhavnani S, Calhoun WJ. Strategies for Molecular Classification of Asthma Using Bipartite Network Analysis of Cytokine Expression. Current allergy and asthma reports. 2012;12:388–395. doi: 10.1007/s11882-012-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. The Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 9.Bhavnani SK, Victor S, Calhoun WJ, Busse WW, Bleecker E, Castro M, Ju H, Pillai R, Oezguen N, Bellala G, et al. How cytokines co-occur across asthma patients: from bipartite network analysis to a molecular-based classification. J Biomed Inform. 2011;44(Suppl 1):S24–30. doi: 10.1016/j.jbi.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akira S, Uematsu S, Takeuchi O. Pathogen Recognition and Innate Immunity. Cell (Cambridge MA) 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. Retinoic Acid-Inducible Gene I Mediates Early Antiviral Response and Toll-Like Receptor 3 Expression in Respiratory Syncytial Virus-Infected Airway Epithelial Cells. The Journal of Virology. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature (London) 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 14.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 15.O'Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature Reviews Immunology. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez ML, Harris B, Lay JC, Bromberg PA, Diaz-Sanchez D, Devlin RB, Kleeberger SR, Alexis NE, Peden DB. Comparative airway inflammatory response of normal volunteers to ozone and lipopolysaccharide challenge. Inhalation toxicology. 2010;22:648–656. doi: 10.3109/08958371003610966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed CE, Milton DK. Endotoxin-stimulated innate immunity: A contributing factor for asthma. The Journal of allergy and clinical immunology. 2001;108:157–166. doi: 10.1067/mai.2001.116862. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annual Review of Immunology. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. 225-60. [DOI] [PubMed] [Google Scholar]
- 19.Brasier AR. The NF- k B Signaling Network: Insights from systems approaches. In: Brasier AR, Lemon SM, Garcia-Sastre A, editors. Cellular Signaling And Innate Immune Responses To RNA Virus Infections. American Society for Microbiology; 2008. pp. 119–135. [Google Scholar]
- 20.Hiscott J. Triggering the Innate Antiviral Response through IRF-3 Activation. Journal of Biological Chemistry. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 21.Pitha PM, Au WC, Lowther W, Juang YT, Schafer SL, Burysek L, Hiscott J, Moore PA. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie (Paris) 1998;80:651–658. doi: 10.1016/s0300-9084(99)80018-2. [Review] [50 refs] [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Luxon BA, Casola A, Garofalo RP, Jamaluddin M, Brasier AR. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J Virol. 2001;75:9044–9058. doi: 10.1128/JVI.75.19.9044-9058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian B, Brasier AR. Microarrays and Transcription Factor Networks. Landes Bioscience; Austin, TX: 2005. The Nuclear Factor- k B (NF- k B) Gene Regulatory Network. [Google Scholar]
- 24.Garrood T, Lee L, Pitzalis C. Molecular mechanisms of cell recruitment to inflammatory sites: general and tissue-specific pathways. Rheumatology. 2006;45:250–260. doi: 10.1093/rheumatology/kei207. [DOI] [PubMed] [Google Scholar]
- 25.Pulendran B, Smith JL, Caspary G, Brasel K, Pettit D, Maraskovsky E, Maliszewski CR. Distinct dendritic cell subsets differentially regulate the class of immune response in vivo. Proc Natl Acad Sci U S A. 1999;96:1036–1041. doi: 10.1073/pnas.96.3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, Gilliet M, Ho S, Antonenko S, Lauerma A, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–680. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- 27.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston SL. Innate Immunity in the Pathogenesis of Virus-induced Asthma Exacerbations. Proceedings of the American Thoracic Society. 2007;4:267–270. doi: 10.1513/pats.200701-030AW. [DOI] [PubMed] [Google Scholar]
- 29.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, Kebadze T, Mallia P, Stanciu LA, Parker HL, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 30.Brasier AR, Tian B, Jamaluddin M, Kalita MK, Garofalo RP, Lu M. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J Virol. 2011;85:11752–11769. doi: 10.1128/JVI.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartlett NW, Slater L, Glanville N, Haas JJ, Caramori G, Casolari P, Clarke DL, Message SD, Aniscenko J, Kebadze T, et al. Defining critical roles for NF-kappaB p65 and type I interferon in innate immunity to rhinovirus. EMBO molecular medicine. 2012;4:1244–1260. doi: 10.1002/emmm.201201650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haeberle HA, Casola A, Gatalica Z, Petronella S, Dieterich HJ, Ernst PB, Brasier AR, Garofalo RP. IkappaB kinase is a critical regulator of chemokine expression and lung inflammation in respiratory syncytial virus infection. J Virol. 2004;78:2232–2241. doi: 10.1128/JVI.78.5.2232-2241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, Davies DE. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Brasier AR, Victor S, Ju H, Busse WW, Curran-Everett D, Bleecker E, Castro M, Chung KF, Gaston B, Israel E, et al. Predicting intermediate phenotypes in asthma using bronchoalveolar lavage-derived cytokines. Clin Transl Sci. 2010;3:147–157. doi: 10.1111/j.1752-8062.2010.00204.x. [This paper provides proof of principle that cytokine patterns in BAL of stable asthmatics provide information relative to dynamic airway physiology.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brasier AR, Victor S, Boetticher G, Ju H, Lee C, Bleecker ER, Castro M, Busse WW, Calhoun WJ. Molecular Phenotyping Of Severe Asthma Using Pattern Recognition Of Bronchoalveolar Lavage-Derived Cytokines. Journal Allergy and Clinical Immunology. 2008;121:30–37. doi: 10.1016/j.jaci.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Tian B, Edeh CB, Brasier AR. Quantitation of the dynamic profiles of the innate immune response using multiplex selected reaction monitoring-mass spectrometry. Molecular & cellular proteomics : MCP. 2013 doi: 10.1074/mcp.M112.023465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Brasier AR. Methods for Biomarker Verification and Assay Development. Current Proteomics. 2011;8:138–152. [Google Scholar]
- 38.Zhao Y, Brasier AR. Applications Of Selected Reaction Monitoring (SRM)-Mass Spectrometry (MS) For Quantitative Measurement Of Signaling Pathways. Methods. 2013 doi: 10.1016/j.ymeth.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt H, Ju H, Brasier AR. A structured approach to predictive modeling of a two-class problem using multidimensional data sets. Methods. 2013 doi: 10.1016/j.ymeth.2013.01.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman JH. Multivariate Adaptive Regression Splines. Annals of Statistics. 1991;19:1–67. [Google Scholar]
- 42.Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, Craig TJ, Dolovich M, Drazen JM, Fagan JK, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. Journal of Allergy and Clinical Immunology. 2002;109:410–418. doi: 10.1067/mai.2002.122635. [DOI] [PubMed] [Google Scholar]
- 43**.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, Harris JM, Scheerens H, Wu LC, Su Z, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–1098. doi: 10.1056/NEJMoa1106469. [This study demonstrates that stratification on the basis of Th2 bioactivity identifies subjects that respond to neutralizing antibody therapy. It is a demonstration of personalized medicine in asthma.] [DOI] [PubMed] [Google Scholar]
- 44.Brasier AR. Expanding Role of Cyclin Dependent Kinases in Cytokine Inducible Gene Expression. Cell Cycle. 2008;7:1–6. doi: 10.4161/cc.7.17.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian B, Zhao Y, Kalita M, Edeh C, Paessler S, Casola A, Teng M, Garofalo R, Brasier AR. CDK9-dependent transcriptional elongation in the innate ISG response to RSV infection in airway epithelial cells. Journal of Virology. 2013 doi: 10.1128/JVI.03399-12. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. RelA Ser276 Phosphorylation Is Required for Activation of a Subset of NF-{kappa}B-Dependent Genes by Recruiting Cyclin-Dependent Kinase 9/Cyclin T1 Complexes. Molecular and Cellular Biology. 2008;28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NF{kappa}B to effect promoter-specific transcriptional repression. Genes and Development. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro GI. Preclinical and Clinical Development of the Cyclin-Dependent Kinase Inhibitor Flavopiridol. Clinical Cancer Research. 2004;10:4270s–4275s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]