Abstract
Purpose
We have previously characterized a tumor stroma expression signature in a subset of breast tumors that correlates with better clinical outcome. The purpose of this study is to determine whether this stromal signature, termed the ‘DTF fibroblast’ signature, is specific to breast cancer or is a common stromal response found in different types of cancer.
Experimental Designs
The DTF fibroblast signature was applied to gene expression profiles from five ovarian, five lung, two colon and three prostate cancer expression microarray datasets. Additionally, two different tissue microarrays of 204 ovarian tumors and 140 colon tumors were examined for the expression of previously characterized protein markers of DTF fibroblast signature. The DTF fibroblast stromal response was then correlated with clinicopathologic features.
Results
The DTF fibroblast signature is robustly present in ovarian, lung, and colon carcinomas. Both expression microarray data and immunohistochemistry show that the subset of ovarian tumors with strong DTF fibroblast signature expression has statistically significant worse survival outcomes. No reproducible survival differences were found in either the lung or the colon cancers. The prostate cancers failed to demonstrate a DTF fibroblast signature. Multivariant analysis showed that DTF fibroblast signature was significantly more prognostic than the proliferation status in ovarian carcinomas.
Conclusion
Our results suggest that the DTF fibroblast signature is a common tumor stroma signature in different types of cancer including ovarian, lung and colon carcinomas. Our findings provide further insight into the DTF fibroblast stromal responses across different types of carcinomas and their potential as prognostic and therapeutic targets.
Background
Tumor stroma plays an important role in cancer development and progression. Our previous studies have shown that gene signatures derived from desmoid-type fibromatosis (DTF), a soft tissue tumor composed of homogenous fibroblasts, can be used as a surrogate to recapitulate the expression features of some tumor stroma. We use gene expression signatures of soft tissue tumors as surrogates for expression signatures of non-neoplastic stromal cell types in the tumor microenvironment. Similar to lymphomas where many tumors retain markers specific for a particular lymphoid cell type, some types of soft tissue tumors can also be regarded as a clonal outgrowth of a particular connective tissue cell type. In multiple studies, we have found that the DTF fibroblast signature, when applied to breast cancers, identifies a subset of breast cancers with favorable clinical outcomes (1, 2).
In previous work, analysis of the stromal expression patterns of synchronous breast cancers and a comparison of matched primary and metastatic tumors have suggested that the DTF fibroblast response is host-specific (3) and that the genesis of the response originates within the stromal compartment and not the malignant epithelium (4). If the DTF fibroblast response is derived from the stromal cells, it is quite possible that the signature occurs in other carcinomas aside from breast cancer.
To determine whether the DTF fibroblast signature is specific to breast cancer or more widely present in different types of cancer, we performed a survey of common cancers using gene expression profiling datasets of lung, colon, prostate and ovarian tumors. These carcinomas have been extensively expression profiled with publicly available datasets and are well clinically annotated.
Using publicly available datasets, we examined the DTF fibroblast gene signature in a total of 1127 ovarian tumors in five datasets (5–9), three datasets of 279 prostate tumors (10–12), three datasets of 573 colon tumors (13–15), and five datasets of 519 lung tumors (16–20). Tissue microarrays of ovarian and colon tumors were also constructed to offer an additional platform for examining the abundance of DTF fibroblast core proteins, SPARC and CSPG2, and their prognostic values in these malignant carcinomas.
Material and Methods
Cancer Data Sets
We searched publicly-available databases to find carcinoma datasets containing not only gene expression profiles but also clinical annotations with at least one of the following records: overall survival (OS), disease-free survival (DFS), and disease-specific survival (DSS). Datasets without the information were excluded in our analysis. Following this inclusion exclusion criteria, a total of five ovarian datasets were identified (TCGA data (5), GSE9891 (6), GSE26712 (7), GSE31245 (8), GSE17260 (9)), containing gene expression data of 1127 patient tumors and the clinical follow-up in 1105 cases of them. The ovarian tumors profiled in these datasets were all pre-treatment samples except in 18 out of 285 tumors of GSE9891 were from patients who had neoadjuvant, platinum based chemotherapy. These tumors were acquired from the primary debulking surgery of patients. Three colon cancer datasets were identified (GSE14333 (13), GSE17538 (14), GSE5851 (15)), containing gene expression data on 573 patient tumors and the clinical follow-up in 538 cases of them. Five lung cancer datasets were identified (caArray-beer-00153 (16), GSE4573 (17), GSE10245 (18), GSE10445 (19), GSE11969 (20)), containing gene expression data on 519 patient tumors and the clinical follow-up in 492 cases of them. Three prostate cancer datasets (GSE1431 (10), GSE3933 (11), GSE25136 (12)) were identified, including 279 prostate tumors.
Data Analysis
Raw expression data were log2 normalized with RMA (21). Unsupervised hierarchical clustering was performed using Cluster3.0 with correlation (uncentered) and average linkage clustering. Treeview and Java Treeview were used to view the resulting heatmap and dendrograms.
Determination of DTF fibroblast-core gene-like (DTF Positive) case clusters
Expression values of the DTF fibroblast core genes were extracted and tumors were clustered using the program Cluster 3.0. Tumors were clustered based on similar level of expression of the DTF fibroblast core gene signature. We were interested in tumors showing highly coordinated expression of this group of genes and chose the correlation value of 0.6 among tumors with upregulated expression of these genes to identify the clustered nodes of tumors from different gene expression datasets with similarly upregulated DTF fibroblast gene expression patterns, similar to the correlations examined in our prior studies on tumor stroma in gene expression profiling datasets (1,2,4).
Analysis of Clinical-pathological Variables
Clinical annotations were collected from GEO or as indicated in the original publication for each dataset. Kaplan-Meier survival curves, Log-rank (Mantel-Cox) Test, Gehan-Breslow-Wilcoxon Test and Hazard Ratio calculations were performed with Graphpad Prism 5. Multivariant analysis, Pearson’s Chi-square test, Wilcox test and t test were performed with R.
Case material
For immunohistochemistry studies we used 204 high-grade serous ovarian carcinomas and 140 colon carcinomas distributed over two different TMAs. The TMAs were constructed using 0.6mm cores with a manual tissue arrayer from Beecher Instrument, Silver Spring, MD. For each specimen, a pathologist reviewed the H&E slides. A block with viable tumor was chosen. Scarred, necrotic, and hypocellular areas were avoided. A 0.6-mm tissue core was taken from these areas in each of the blocks and inserted into the tissue microarray block. We did not take duplicate cores. Survival information was available for all of the cases with a mean follow up of 5.4 years (0.4–16.5 years) for high-grade serous ovarian carcinomas, and 3.5 years (0.08–11.9 years) for colon cancer patients.
Immunohistochemistry
Slides were cut at 4μm, deparaffinized in xylene and hydrated in a graded series of alcohol. For immunohistochemistry, the primary antibodies used were SPARC (osteonectin) (mouse monoclonal antibody, 1:1000, Invitrogen, Catalog No: 33-5500, Clone ON1-1, Camarillo, California, USA), and CSPG2 (versican) (mouse monoclonal, 1:150, Santa Cruz Biotechnology Incorporated, Catalog No:47777, Clone 5C1, Santa Cruz, California, USA). Stains were interpreted as negative when the staining was less than 30% of the spindle stromal cells and positive when the staining was greater than 30%. A tumor was DTF positive when it had dual positivity of SPARC and CSPG2. The immunohistochemistry results were scored by a pathologist (IE) in a semi-quantitative manner, based on prior experience with these antibodies [1–4]. In our experience, manual scoring of robustly staining markers such as the ones used in this study perform as reliably as automated fully quantitative techniques (22), and we believe the findings from manual scoring are more generalizable as this is the practice most commonly used in the clinical setting.
Statistical analysis of tumors on the tissue microarrays
The Kaplan-Meier method was used to estimate survival distributions. Disease specific survival (DSS) was calculated from the time of diagnosis until the death from the specific disease. Overall survival (OS) was defined as the time interval from the diagnosis to death. Recurrence free survival (RFS) was calculated from the time of diagnosis to the date of recurrence. Multivariate Cox Regression analysis was used to study the relationship between survival and different covariates.
Use of the proliferation signature to examine the proliferation status of microarray-profiled tumors
The proliferation gene signature previously developed (23) was used to examine the proliferation status of breast and ovarian tumors with differential DTF fibroblast stromal responses. Expression values of the DTF fibroblast core genes and the proliferation genes were extracted and tumors were clustered using the program Cluster 3.0. The clustered patterns of tumors were examined in both breast and ovarian cancer datasets to determine the relative associations between the proliferation status and the DTF fibroblast stromal responses.
Results
DTF fibroblast signature in ovarian, lung, colon and prostate carcinomas
To examine the DTF fibroblast signature across different types of carcinomas, we selected publicly available gene expression datasets of common carcinomas which included data from at least 250 samples (total) on high quality arrays and clinical annotation. Datasets of study ovarian, lung, colon and prostate cancers met these criteria.
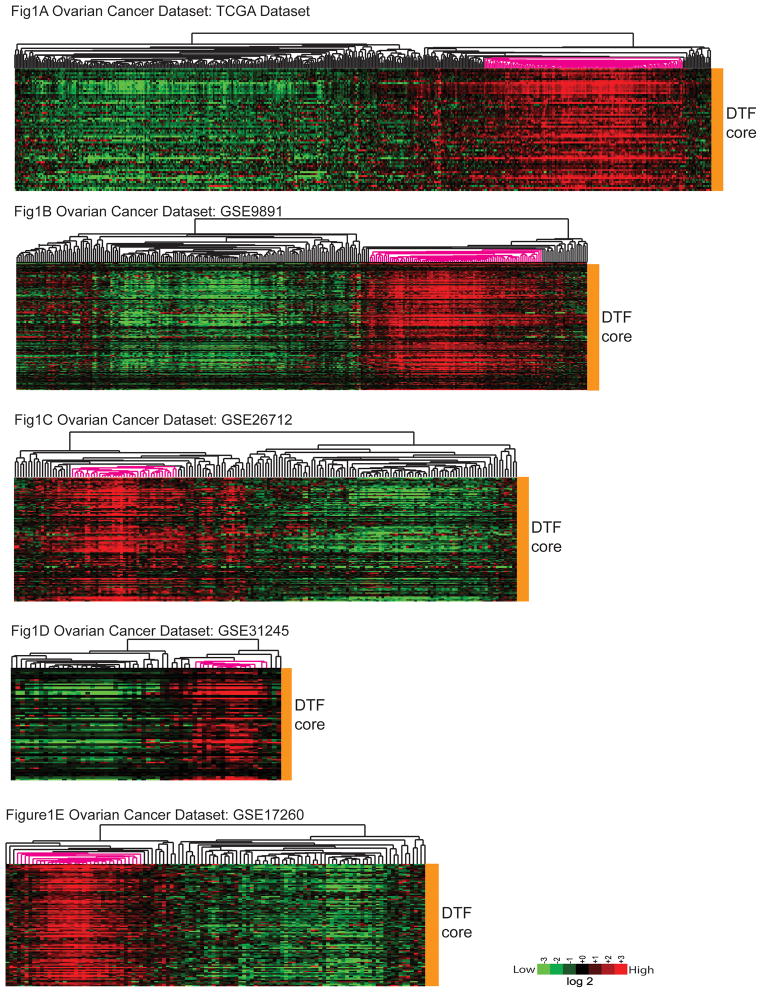
We examined each dataset for the presence of DTF fibroblast positive samples by clustering the tumors using the previously determined DTF fibroblast core gene list (2). We found a subset of the ovarian tumors with highly coordinated DTF fibroblast core gene expression in each of the five ovarian cancer datasets examined (Figure 1). This subset of ovarian tumors with robust DTF fibroblast response accounted for 28.1% of the total number of 1127 microarray-profiled ovarian tumors in the 5 datasets.
Figure 1.
The expression profiles of DTF fibroblast core gene signature in five ovarian cancer datasets, including TCGA dataset (Fig 1A), GSE9891 (Fig 1B), GSE26712 (Fig 1C), GSE31245 (Fig 1D) and GSE17260 (Fig 1E). The selected subgroup of ovarian tumors highlighted by pink in the clustered array trees are the ones with highly coordinated DTF fibroblast core gene expression thus categorized as DTF(+) ovarian tumors.
The pattern of highly coordinated DTF fibroblast core gene expression was also observed in each of the lung and colon cancer datasets. The 5 independent subset of lung tumors with strong DTF fibroblast response accounted for 21.6% of the total number of 519 lung tumors (Supp Fig 1). The DTF fibroblast pattern was also observed in each of the two colon cancer datasets and accounted for 28.5% of the total number of 493 colon tumors (Supp Fig 2). In prostate carcinoma, however, the fibroblast stromal response was not identified in any of three datasets (Supp Fig 3).
Clinical associations of the subset of tumors identified by DTF fibroblast signature
We found previously that the subset of breast tumors identified by DTF fibroblast signature with high DTF fibroblast stromal responses were correlated with a lower tumor grade, estrogen receptor expression and good outcome in breast cancer patients (1, 2). In this study, we looked for similar correlations in the ovarian carcinoma datasets.
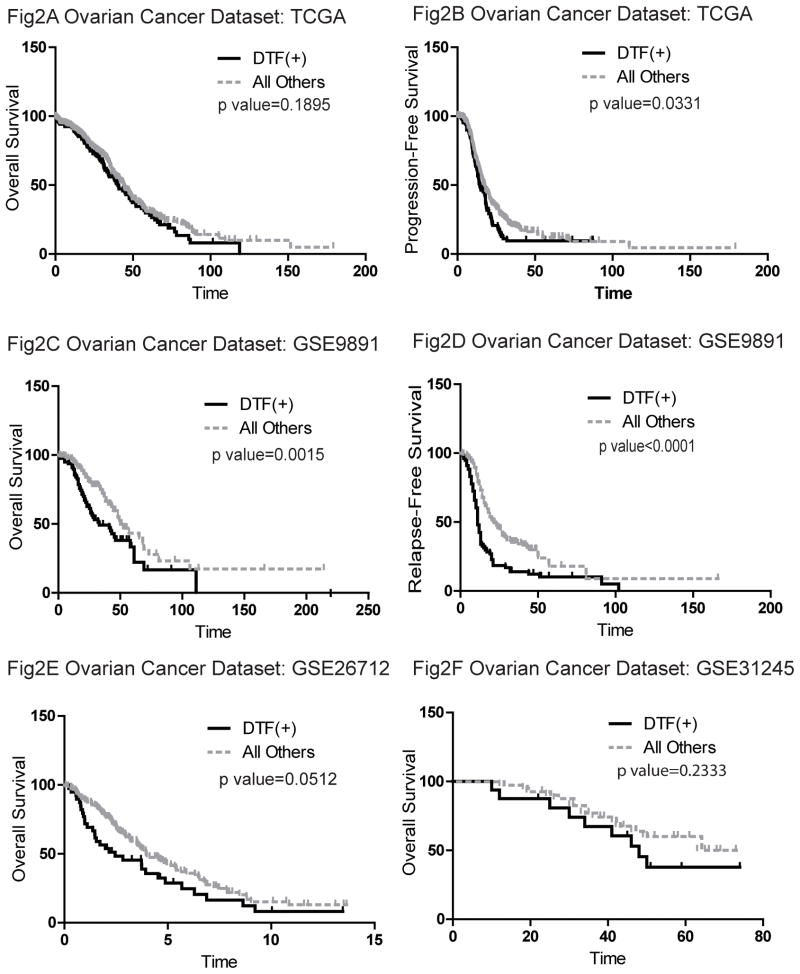
The DTF positive tumors in 4 of 5 ovarian datasets showed associations with worse overall survival and/or worse progression-free survival (TCGA, GSE9891, GSE26712 and GSE31245) (Figure 2). In one ovarian dataset we did not observe any survival association, GSE17260, though this was also the dataset with the fewest samples (110). Thus among the five ovarian datasets, out of 1019 tumors with available clinical information, a total of 909 cases showed the trend that tumors with highly coordinated DTF fibroblast core gene expression were associated with worse survival outcomes while 851 of these tumors from 4 datasets had statistically significant associations. We found that ovarian cancer patients with tumors that had a robust DTF fibroblast stromal response also were correlated with higher tumor stage in TCGA dataset (Pearson’s Chi-square test: X-squared=8.2468, df=2, p-value=0.01619) and GSE9891 cohort (Pearson’s Chi-square test: X-squared=18.2699, df=3, p-value=0.0003869). However, there were no associations between high DTF stromal responses and the tumor grade.
Figure 2.
The results of the survival analysis of ovarian tumors showed that DTF(+) ovarian tumors are associated with worse overall survival (Fig 2A) and worse progression-free survival in TCGA dataset (Fig 2A and Fig 2B), GSE9891 (Fig 2C and Fig 2D), as well as GSE26712 (Fig 2E) and GSE31245 (Fig 2F).
In contrast to the ovarian carcinomas, we found no statistically significant clinical associations between the subset of DTF fibroblast positive lung tumors and survival either for individual datasets or the aggregate set of lung cancers (Supp Fig 4). In colon carcinomas, we found discordant results between datasets. The subset of DTF fibroblast positive colon tumors in GSE14333 dataset of 255 tumors were associated with better disease-free-survival (DFS) whereas the DTF fibroblast positive subset of the colorectal tumors in GSE17538 dataset of 238 tumors were significantly associated with worse overall survival (OS), worse relapse-free-survival (RFS), and worse disease-specific-survival (DSS) (Supp Fig 2). No clear differences between the datasets in terms of cohort characteristics could explain these different outcomes. There were no correlations of DTF status with tumor stage (Pearson’s Chi-square test, X-squared=4.5957, df=3, p-value=0.2039), tumor grade (Pearson’s Chi-square test, X-squared=0.1165, df=2, p-value=0.9434) and age (Wilcox test, p-value=0.0773) in GSE17538. There was also no correlation of DTF status with the Duke stage of tumor in GSE14333 (Pearson’s Chi-square test, X-squared=2.2041, df=3, p-value=0.5311). However, there was significant association between DTF status and age in GSE14333 (Wilcox test, p-value=0.01661).
Comparison of DTF fibroblast features in breast and ovarian carcinomas
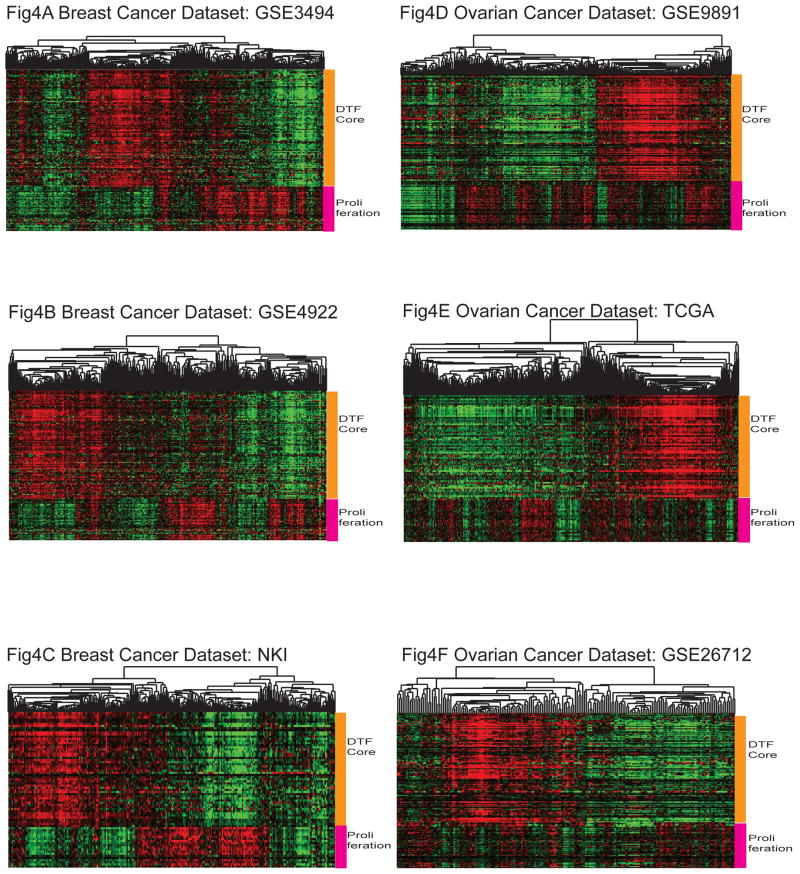
Our current data showing worse outcome in ovarian cancer when the DTF fibroblast signature is present contrasts with a previously found correlation with good outcome in breast cancer. To address the difference between these two carcinomas, we examined the correlation between the proliferation status of breast and ovarian tumors and the DTF fibroblast stromal responses. Across a wide variety of cancers, proliferation is a dominant feature of many signatures that predict outcome. To identify tumors with a high and low proliferation index in the context of gene expression profiling data, we used a proliferation gene signature to stratify cases by proliferation index. There has been several proliferation signatures discovered in gene expression profiling studies published by different groups which have significant overlap in genes and biology (23–27). We used the proliferation gene signature developed by Dai et al.(23) which demonstrated reproducible and significantly worse survival associations of breast tumors with high proliferation.
In breast cancer datasets (GSE3494, GSE4922 and NKI), the DTF fibroblast stromal response was negatively correlated with the proliferation signature (Figure 4). Breast tumors with a strong DTF fibroblast stromal response had low expression of the proliferation genes compared to non-DTF tumors (6.9% vs 19.7% in GSE3494, 7.1% vs 22.8% in GSE4922 and 5.3% vs 30% in the NKI, all p-values < 0.01), which was consistent with the finding that a positive DTF fibroblast stromal response selected for better survival. However, in the ovarian cancer datasets, tumors with high DTF fibroblast stroma expression consisted of a mixture of tumors with high and low proliferation signatures (Figure 4) (17% vs 13.6% in GSE9891, 7.2% vs 12.3% in TCGA, and 15.4% vs 6.2% in GSE 26712). Multivariate analysis found that proliferation status was not as strong as the DTF fibroblast signature in predicting the survival of patients with ovarian carcinomas (Table 1).
Figure 4.
The expression profiles of DTF fibroblast core gene signature and the proliferation signature in breast cancer datasets of GSE3494 (Fig 4A), GSE4922 (Fig 4B) and NKI dataset (Fig 4C) as well as ovarian cancer datasets of GSE9891 (Fig 4D), TCGA dataset (Fig 4E) and GSE26712 (Fig 4F).
Table 1.
Multivariate Analysis for Ovarian Tumor Group Status versus Clinical Risk Factors including Proliferation, Grade, Age and Survival
| Dataset | Tissue Type | Analysis Group | Survival Data | Statistical Significance | Hazard Ratio | 95% CI Lower | 95% CI Upper |
|---|---|---|---|---|---|---|---|
| GSE9891 | Ovarian | Tothill | OS | 0.05481 | 1.9 | 0.9869 | 3.657 |
| DTF | OS | 0.89902 | 1.037 | 0.5928 | 1.814 | ||
| Proliferation | OS | 0.71203 | 1.106 | 0.6487 | 1.885 | ||
| Stage | OS | 0.00372** | 1.774 | 1.2043 | 2.612 | ||
| Grade | OS | 0.74833 | 1.059 | 0.7472 | 1.5 | ||
| Age | OS | 0.00758** | 1.029 | 1.0075 | 1.05 | ||
| GSE9891 | Ovarian | Tothill | RFS | 0.066 | 1.6546 | 0.9673 | 2.83 |
| DTF | RFS | 0.284 | 1.2744 | 0.8179 | 1.986 | ||
| Proliferation | RFS | 0.827 | 0.9542 | 0.6272 | 1.452 | ||
| Stage | RFS | 0.00000382*** | 1.9462 | 1.4674 | 2.581 | ||
| Grade | RFS | 0.393 | 1.121 | 0.8625 | 1.457 | ||
| Age | RFS | 0.078 | 1.0144 | 0.9984 | 1.031 | ||
| TCGA | Ovarian | Tothill | OS | 0.165918 | 0.7029 | 0.4269 | 1.157 |
| DTF | OS | 0.117083 | 1.4147 | 0.9167 | 2.183 | ||
| Proliferation | OS | 0.996411 | 0.9991 | 0.6719 | 1.486 | ||
| Stage | OS | 0.008118** | 1.4474 | 1.1007 | 1.903 | ||
| Grade | OS | 0.13625 | 1.3227 | 0.9155 | 1.911 | ||
| Age | OS | 0.000564*** | 1.0206 | 1.0088 | 1.032 | ||
| TCGA | Ovarian | Tothill | PFS | 0.01411* | 0.5457 | 0.3364 | 0.8851 |
| DTF | PFS | 0.00177** | 1.9542 | 1.2839 | 2.9744 | ||
| Proliferation | PFS | 0.45604 | 0.864 | 0.5883 | 1.269 | ||
| Stage | PFS | 0.00707** | 1.4398 | 1.1044 | 1.8771 | ||
| Grade | PFS | 0.18877 | 1.26 | 0.8926 | 1.7787 | ||
| Age | PFS | 0.54974 | 1.0033 | 0.9925 | 1.0143 |
Localization of the DTF fibroblast signature expression in ovarian and colon tumor stroma
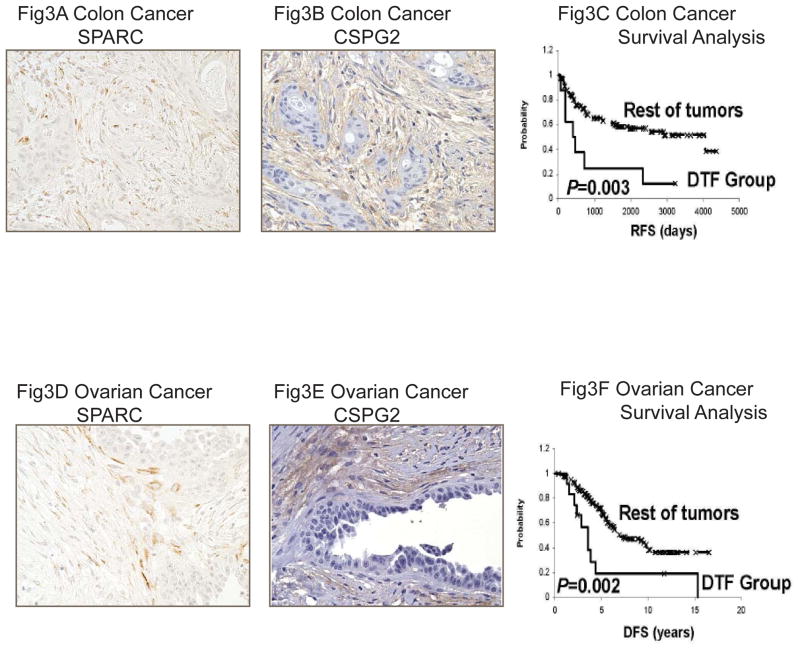
We have previously used a set of immunohistochemistry markers as surrogates for the DTF fibroblast gene expression signature, including SPARC and CSPG2, to localize the expression of the signature to the tumor stroma. We assayed the expressed of SPARC and CSPG2 on tissue microarrays of ovarian and colon cancer, which contained 204 high-grade serous ovarian carcinomas and 140 colon carcinomas respectively. These protein markers were expressed in the stromal compartment by cells with a fibroblast-like morphology (Figure 3). As found in the gene expression datasets, ovarian cancer patients with co-expression of DTF fibroblast markers (SPARC and CSPG2) had worse prognosis than those patients who did not co-express the DTF fibroblast markers in the tumor stroma (Figure 3). Multivariate analysis was conducted using clinical features of age, FICO stage, ECOG, grade and the DTF signature. We found that only DTF signature was prognostic (DSS, hazard ratio 2.4; 95% CI, 1.14 to 5.05; p=0.02) The results in the colon carcinoma array also demonstrated a worse outcome for DTF fibroblast positive cases, consistent with one, but not the other, of the colon cancer gene expression dataset results.
Figure 3.
Immunohistochemical stains positive of the DTF fibroblast core protein markers, SPARC and CSPG2, in colon cancers (Fig 3A and Fig 3B) and ovarian cancers (Fig 3D and Fig 3E). Patients with co-expressed SPARC and CSPG2 had worse survival in both colon cancers (Fig 3C) and ovarian cancers (Fig 3F).
Discussion
The importance of stroma in tumor development has been observed and recognized in a number of experimental models. Activated stroma comprised of inflammatory cells and fibroblasts found to be essential in supporting neoplasia proliferation and survival (28, 29). Irradiated stroma produces more mammary carcinomas compared with non-irradiated stroma in mouse models (30). In addition, higher expression of transforming growth factor-β1 (TGF-β1) in irradiated stroma fibroblasts enhances cancer progression in the mammary gland as well as in the pancreas (31).
We have previously demonstrated that gene signatures generated from soft tissue tumors such as desmoid-type fibromatosis and solitary fibrous tumors, highly homogeneous neoplasms with different fibroblastic phenotypes, can identify differential stromal responses in breast tumors with different associated survival outcomes (1). In this study, we sought to extend our findings in cancer stroma gene expression signatures by surveying some of the most common carcinomas for which numerous well-annotated datasets were available. We looked for the presence of the DTF fibroblast gene signature in ovarian, lung, colon, and prostate cancer datasets. We discovered the DTF fibroblast gene signature was present in a subset of ovarian, lung, and colon tumors. However, only significant and reproducible outcome differences were identified in ovarian tumors.
No stromal gene expression signature was identified in prostate carcinoma. This is interesting because the prostatic stroma is quite different from the stroma of the other organs, in that it has a pronounced myofibroblastic phenotype. One possible explanation for the lack of the DTF fibroblast signature is that the myofibroblastic nature predisposes the tumor microenvironment to a different stromal signature that is not covered by our fibroblast-derived expression profiles.
Tothill et al. (6) found a “poor prognosis subtype” identified by k-means clustering of six subtypes of ovarian tumors. This subtype had a reactive stromal gene expression signature, which was composed of upregulated expression of 289 stroma genes. 45 of these 289 genes were overlapping with the DTF fibroblast core gene signature of 63 genes. Although the two different gene signatures containing different gene numbers were originally generated by different methods and groups of researchers, both of them proved that stromal responses in ovarian carcinoma were associated with worse survival outcomes. These results provided solid evidence for the critical role of cancer stroma in the prognosis of ovarian cancer patients. An RNAi screening of the common stromal genes proposed by these two groups for their functions and effects in ovarian cancer is worth pursuing. Other work on ovarian cancer stroma has studied morphologic differences between cancers. Labiche et al. (32), focusing on proportional representation of stroma to malignant epithelium, have shown that the stroma compartment influences ovarian cancer invasiveness and histological differentiation. They used an automated image analysis software approach to quantify the stroma proportion, and found that higher stroma content (≥50%) is associated with poor prognosis. From our study, we found that the subgroup of microarray-profiled tumors showing strong DTF fibroblast stromal signature were associated with worse survival of patients. However, a collection of ovarian tumors to be simultaneously examined for their stroma abundance and genetically-profiled for differential stromal gene expression will be required to establish the direct relationship between the signature score and stroma abundance, which remains to be an interesting study in the future.
While we found that the DTF fibroblast stromal signature is evident in both breast and ovarian cancers, the presence of this signature was associated with different outcomes for these two types of tumors: positive breast cancers had a good outcome whereas positive ovarian cancers had a poor outcome. These results could partly be explained by differences in proliferation. Breast tumors with high DTF fibroblast stromal response had a low proliferation gene expression signature. In ovarian carcinoma, however, there was no significant correlation between DTF stromal response and proliferation. In survival multivariate analysis, only the DTF fibroblast signature, but not the proliferation signature, has the prognostic value in ovarian cancer. These differences suggest that cancer cell proliferation and stromal responses might be closely related in breast tumors. In addition to the closely-related role of proliferation and stromal responses in breast carcinoma but not ovarian carcinoma, the contrasting associations between signature score and survival in breast and ovarian cancer may also come from differences of tissue specificity, the stage and grade of the ovarian and breast tumor samples profiled, and the potentially different cancer-stroma interaction in these two types of cancers. Further analyses to investigate chromosomal changes and methylation status on breast and ovarian tumors of differential stromal responses will help identify potential regulating factors, in addition to the fundamental differences brought by tissue specificity.
We have demonstrated that the DTF fibroblast signature is present in multiple cancer types, including breast (1, 2), ovarian, lung and colon carcinomas, which suggested a recurring relationship between cancer cells and the surrounding stroma. These findings suggest that the DTF fibroblast signature is a common stromal response to cancer. It is interesting to consider that this signature is found in only a subset of the carcinomas from any given organ system. It is unclear why the same stromal response is not present in every tumor, rather than just subset. An a priori hypothesis might be that different tumors form different stromal signatures based on the tumor intrinsic biology. But this does not seem to be the case from our earlier synchronous breast cancer study (3) where clinically distinct cancers from the same patient tended to have the same DTF fibroblast response despite having other pathologic differences. Further work to define the genesis of the tumor stromal response is needed. Besides the DTF fibroblast stromal signature, several other stromal signatures discovered in breast cancer (33, 34) may also be present in other types of malignant carcinomas. It would be interesting to further examine and compare these stromal signatures and their responses in a variety of cancers to develop a more complete understanding of the stromal patterns in cancer.
Supplementary Material
Translational relevance.
This study examines the stromal responses in ovarian, lung, colon and prostate carcinomas with the DTF fibroblast gene signature, which was previously found to stratify breast tumors and identify breast cancer patients with better clinical outcomes. The stromal responses defined by this fibroblast gene signature can stratify tumors in ovarian, lung, and colon tumors and identify ovarian cancer patients with worse clinical outcomes. Additionally, the DTF fibroblast stromal response is found to be more prognostic than the proliferation status associating survival outcome in ovarian cancer patients. The broader survey provided here offers characterized genetic information in stromal responses of different types of tumors from a large number of cancer patients, which can lead to better understanding of individual tumors and aid in the design of targeted therapy in the future.
Acknowledgments
Grant Support:
NIH (R01 CA129927) and California Breast Cancer Research Program (15NB-0156)
References
- 1.West RB, Nuyten DS, Subramanian S, Nielsen TO, Corless CL, Rubin BP, et al. Determination of stromal signatures in breast carcinoma. PLoS biology. 2005 Jun;3(6):e187. doi: 10.1371/journal.pbio.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck AH, Espinosa I, Gilks CB, van de Rijn M, West RB. The fibromatosis signature defines a robust stromal response in breast carcinoma. Laboratory investigation; a journal of technical methods and pathology. 2008 Jun;88(6):591–601. doi: 10.1038/labinvest.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JM, Beck AH, Pate LL, Witten D, Zhu SX, Montgomery KD, et al. Endogenous versus tumor-specific host response to breast carcinoma: a study of stromal response in synchronous breast primaries and biopsy site changes. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011 Feb 1;17(3):437–46. doi: 10.1158/1078-0432.CCR-10-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster JA, Beck AH, Sharma M, Espinosa I, Weigelt B, Schreuder M, et al. Variations in stromal signatures in breast and colorectal cancer metastases. The Journal of pathology. 2010 Oct;222(2):158–65. doi: 10.1002/path.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 30;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008 Aug 15;14(16):5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 7.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer research. 2008 Jul 1;68(13):5478–86. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spentzos D, Levine DA, Kolia S, Otu H, Boyd J, Libermann TA, et al. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005 Nov 1;23(31):7911–8. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 9.Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PloS one. 2010;5(3):e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart RO, Wachsman W, Berry CC, Wang-Rodriguez J, Wasserman L, Klacansky I, et al. In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004 Jan 13;101(2):615–20. doi: 10.1073/pnas.2536479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004 Jan 20;101(3):811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Goodison S. Optimizing molecular signatures for predicting prostate cancer recurrence. The Prostate. 2009 Jul 1;69(10):1119–27. doi: 10.1002/pros.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009 Dec 15;15(24):7642–51. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010 Mar;138(3):958–68. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007 Aug 1;25(22):3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 16.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature medicine. 2002 Aug;8(8):816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 17.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer research. 2006 Aug 1;66(15):7466–72. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 18.Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung cancer. 2009 Jan;63(1):32–8. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Broet P, Camilleri-Broet S, Zhang S, Alifano M, Bangarusamy D, Battistella M, et al. Prediction of clinical outcome in multiple lung cancer cohorts by integrative genomics: implications for chemotherapy selection. Cancer research. 2009 Feb 1;69(3):1055–62. doi: 10.1158/0008-5472.CAN-08-1116. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006 Apr 10;24(11):1679–88. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003 Apr;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Turbin DA, Leung S, Cheang MC, Kennecke HA, Montgomery KD, McKinney S, et al. Automated quantitative analysis of estrogen receptor expression in breast carcinoma does not differ from expert pathologist scoring: a tissue microarray study of 3,484 cases. Breast cancer research and treatment. 2008 Aug;110(3):417–26. doi: 10.1007/s10549-007-9736-z. [DOI] [PubMed] [Google Scholar]
- 23.Dai H, van’t Veer L, Lamb J, He YD, Mao M, Fine BM, et al. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer research. 2005 May 15;65(10):4059–66. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- 24.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proceedings of the National Academy of Sciences of the United States of America. 1999 Aug 3;96(16):9212–7. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000 Aug 17;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 26.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001 Sep 11;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002 Jan 31;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 28.Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiological reviews. 1996 Jan;76(1):69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 29.Tlsty TD, Hein PW. Know thy neighbor: stromal cells can contribute oncogenic signals. Current opinion in genetics & development. 2001 Feb;11(1):54–9. doi: 10.1016/s0959-437x(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 30.Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer research. 2000 Mar 1;60(5):1254–60. [PubMed] [Google Scholar]
- 31.Ohuchida K, Mizumoto K, Murakami M, Qian LW, Sato N, Nagai E, et al. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor-stromal interactions. Cancer research. 2004 May 1;64(9):3215–22. doi: 10.1158/0008-5472.can-03-2464. [DOI] [PubMed] [Google Scholar]
- 32.Labiche A, Heutte N, Herlin P, Chasle J, Gauduchon P, Elie N. Stromal compartment as a survival prognostic factor in advanced ovarian carcinoma. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2010 Jan;20(1):28–33. doi: 10.1111/IGC.0b013e3181bda1cb. [DOI] [PubMed] [Google Scholar]
- 33.Bergamaschi A, Tagliabue E, Sorlie T, Naume B, Triulzi T, Orlandi R, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. The Journal of pathology. 2008 Feb;214(3):357–67. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
- 34.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nature medicine. 2008 May;14(5):518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.