Abstract
In an isolated population, individuals are likely to share large genetic regions inherited from common ancestors. Identity by descent (IBD) can be inferred from SNP genotypes, which is useful in a number of applications, including identifying genetic variants influencing complex disease risk, and planning efficient cohort-sequencing strategies. We present ANCHAP – a method for detecting IBD in isolated populations. We compare accuracy of the method against other long-range and local phasing methods, using parent–offspring trios. In our experiments, we show that ANCHAP performs similarly as the other long-range method, but requires an order-of-magnitude less computational resources. A local phasing model is able to achieve similar sensitivity, but only at the cost of higher false discovery rates. In some regions of the genome, the studied individuals share haplotypes particularly often, which hints at the history of the populations studied. We demonstrate the method using SNP genotypes from three isolated island populations, as well as in a cohort of unrelated individuals. In samples from three isolated populations of around 1000 individual each, an average individual shares a haplotype at a genetic locus with 9–12 other individuals, compared with only 1 individual within the non-isolated population. We describe an application of ANCHAP to optimally choose samples in resequencing studies. We find that with sample sizes of 1000 individuals from an isolated population genotyped using a dense SNP array, and with 20% of these individuals sequenced, 65% of sequences of the unsequenced subjects can be partially inferred.
Keywords: IBD, isolates, resequencing
Introduction
In isolated populations, most individuals share relatively recent common ancestors. If more than one individual inherited the same ancient haplotype in a region, we call them haplotype sharers. Segments of their chromosomes are identical by descent – their haplotypes ‘descend from a common ancestor without either of them experiencing a recombination'.1 Although SNP arrays and next-generation sequencing do not reveal gametic phase, haplotype sharers can be identified using computational methods. The applications of inferred regions of identity by descent (IBD) include optimization of resequencing studies and mapping genetic effects on complex traits.2, 3
In the first application, when the SNP genotypes are available and next-generation sequencing is planned, haplotype sharing between the individuals can save resources. With sharing inferred from SNP genotypes, it is possible to choose a minimally redundant subset of individuals to be sequenced, and then to impute sequence data into other subjects with SNP genotype data. Imputations that rely on IBD are now recognized to increase the power of sequencing studies in population isolates.4
In the second application, shared haplotypes may make it possible to detect the effects of genes in which functional variants that are rare in the general population have drifted to high frequency in the isolated population. Furthermore, the reduced allelic heterogeneity in an isolate provides an opportunity to detect associations with these otherwise rare variants. However, conventional GWAS studies may fail to detect associations with rare variants, as these may not be in linkage disequilibrium (LD) with SNPs on genotyping arrays that have been optimized to tag common variants.5, 6 The uncovered ancestral haplotypes can be in stronger association with rare functional variants and hence improve the power of association tests. The most ambitious attempts to map effects of shared haplotypes reconstruct descent trees, but this approach has been found computationally infeasible.7
Genomic phase can be revealed by long-range phasing methods that exploit regions of IBD between related individuals, or by local phasing methods that rely on patterns of LD.8 The resulting haplotypes from either of the models make it possible to revise the IBD regions in the former case, or find them in the latter case. The first rule-based algorithm for long-range phasing was described by Kong et al,9, 10 and is similar to the method presented later by Hickey et al.11 Both of these methods detect IBD sharing only in pre-specified genetic regions, identical for all pairs of compared diplotypes, whereas in reality boundaries of IBD regions can occur anywhere across the genome. Systematic long-range phasing (SLRP)12 is a fully probabilistic model for phasing and IBD detection in isolated populations, yet the inference algorithm used by SLRP requires significant computational resources. FastIBD is an example of a local phasing method for cohorts of unrelated individuals that was adapted for inference of IBD sharing.13 The method constructs a model of haplotypes, and even though it was designed for outbred populations and may struggle to capture long-range haplotypes present in an isolated populations, multiple resampling of haplotypes may make fastIBD also suitable for more closely related individuals. Long-range methods explicitly capture all shared haplotypes, whereas the local methods build more parsimonious models, which, however, may carry enough information for the detection of IBD sharing also in population isolates.
We describe a new long-range algorithm for the detection of identical by descent haplotypes in isolated populations named ANCHAP. Our method is designed to detect borders of regions of IBD precisely, at minimal computation time and with state-of-art sensitivity and false discovery rate. We compare ANCHAP with other long-range methods and a local method, and demonstrate an application of the identified IBD regions for optimization of sequencing studies.
Materials and methods
Cohorts under study
In our study of ancestral haplotypes, we analyzed four European cohorts, three of which (Orkney Complex Disease Study (ORCADES), CROATIA-VIS, CROATIA-KORCULA) are from isolated island populations and one from a mainland population (Study of Colorectal Cancer in Scotland (SOCCS)). The ORCADES is a family-based, cross-sectional study in the isolated Scottish archipelago of Orkney.14 Genetic diversity in this population is decreased compared with mainland Scotland, consistent with the high levels of endogamy throughout history. Orkney has been inhabited for over 5000 years, but the original population was almost completely replaced by Norse Vikings about 800–900 CE. From about 1300 to 1600 CE, there was an influx of mainland Scots.15 For this analysis, we used data from 749 participants aged 18–100 years from 10 islands; however, for the purposes of evaluation of methods we removed parents from the genotyped parent–offspring pairs, which reduced the cohort size to 597 individuals. Genotyping in the study was done using the Illumina HumanHap300 array (San Diego, CA, USA). The CROATIA-VIS study is a family-based, cross-sectional study in the villages of Komiza and Vis on the isolated island of Vis, which included 1056 examinees aged 18–93 years.16 The CROATIA-VIS study genotyping used the Illumina Hap300v1 SNP chip (San Diego, CA, USA). The CROATIA-KORCULA study is a family-based, cross-sectional study in the villages of Lumbarda, Zrnovo and Racisce on the isolated island of Korcula in Croatia.17 The study included 965 examinees aged 18–95 years. The CROATIA-KORCULA study genotyping used the Illumina Hap370CNV SNP chip (San Diego, CA, USA). The SOCCS study is a case–control study of prospectively collected colorectal cancer cases from all Scottish hospitals, and matched controls. One thousand participants in each group in the first phase of the study were genotyped with the Illumina HumanHap300 array. The participants for the control group were matched by age, sex and region to cases according to a nearly complete population-based register, and then selected at random. We analyzed the genotypes from the control group so as to obtain a sample representative of the Scottish population as a whole. Details of data pre-processing are given in the Supplementary Materials.
Recent IBD
Haplotypes that are identical by descent originate ‘from a common ancestor without either of them experiencing a recombination'.1 Unless a recent mutation occurred, the alleles on IBD haplotypes should be identical, also at untyped loci. For two individuals sharing a haplotype inherited from a common ancestor, the lengths of the shared regions are exponentially distributed with a mean equal to (2n)−1 morgans, where n is number of generations back to most recent common ancestor (MRCA).18 However, the distribution of segment lengths has a variance of (2n)−2 morgans; hence, the correspondence of segment length with time to common ancestor is only approximate. In an isolated population such as the Orkney population, founded by the Viking settlement about 50 generations ago, and where population size has been constrained over many generations, the time to MRCA is either of the order of 1000 generations ago, during the early settlement of Europe, or less than 50 generations ago. To be able to use IBD sharing to infer sharing of rare variants, taking into account mutation rates,19, 20 we restrict the definition of IBD sharing to sharing via a recent common ancestor. In practice, we can only do this by setting a minimum length for the shared region. For this study we set the cut-off value at 2 cM, equal to the expected length of sharing given a time to MRCA of 25 generations. In addition, the cut-off value at 2 cM has been suggested in literature as a threshold above which accurate detection from contemporary genotyping arrays can be obtained.18
Algorithm of ANCHAP
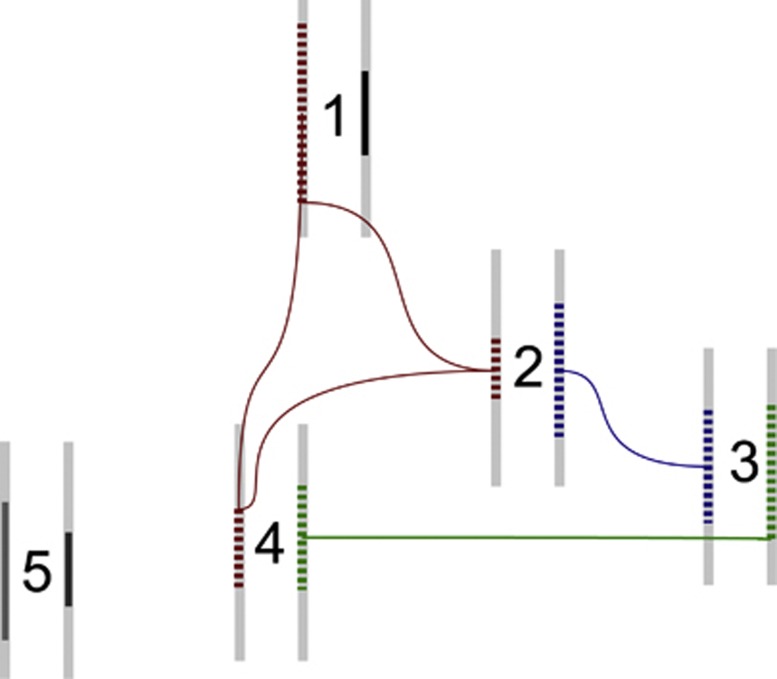
The objective of ANCHAP is to infer recent IBD from the SNP data with maximum sensitivity and specificity. The algorithm should declare IBD only where the haplotype was co-inherited from a recent common ancestor, and find all of such regions (see Figure 1).
Figure 1.
Haplotype sharing structure within a population isolate. Genotyped individuals are identified by numbers 1 to 5. Each individual has two haplotypes, represented by thick bars. Red, blue and green dotted lines represent IBD of two haplotypes in a genomic region. The dark gray-shaded haplotypes are unique in the sample, and they are not shared between the sampled subjects.
The algorithm consists of three stages:
Stage I. First scan for IBD sharing from comparisons of multilocus genotypes of all pairs of individuals (Algorithm 1 in Supplementary Materials).
Stage II. Splitting haplotype sharers by alignment and phasing. Individuals carrying parts of the individual's maternal haplotype are distinguished from ones that carry the paternal haplotype (Algorithm 2 in Supplementary Materials).
Stage III. Second scan for haplotype sharing: a more sensitive and specific scan for IBD sharing, by pairwise comparisons of partially uncovered haplotypes (Algorithm 3 in Supplementary Materials).
At Stage I, ANCHAP detects IBD sharing between pairs of unphased diplotypes, using a variable-length sliding window to screen for long regions with no opposing homozygotes between each pair of individuals. Where there are no opposing homozygotes over a long region, a haplotype is likely to be shared IBD. To account for uncertain sharing near the boundaries of a region with no opposing homozygotes, a number of markers at the margins are trimmed and are not included into the shared region. The parameters of the method are as follows: the IBD threshold – the minimum genetic length in centimorgans of a region without opposing homozygotes between a pair of diplotypes, and the number of markers to be trimmed.
In a given region, to reconstruct a phase, the proband's haplotype sharers can be split into two groups by alignment at Stage II. If sharers of the proband's haplotypes on both the gametes are present, they will form two groups. If sharers of only one gamete are present, or if the proband is homozygous by descent, they will form one group. When sharers of each proband's haplotypes are identified, phasing becomes possible. We can recover the proband's haplotypes at each locus where at least one of the haplotype sharers is homozygous, but information about all of the homozygotes among the sharers eliminates phasing errors. As a haplotype is shared, the haplotype sharer's allele at a homozygous locus must be the same as the allele on the proband's haplotype.9 As the method distinguishes groups of individuals sharing each of the proband's haplotypes in a region without recourse to pedigree, the actual paternal or maternal origin of these proband's haplotypes is not known, and in any case is irrelevant for phasing.
At Stage III of ANCHAP, we make use of the phase information obtained from the IBD regions that were detected earlier. When partially complete haplotypes have been inferred, a second scan for IBD sharing is undertaken, exploiting the additional phase information gained. A pair of completely known haplotypes can mismatch at any phased locus, whereas in a pair of unphased genotypes only loci homozygous for both individuals are indicative of sharing or lack thereof. Therefore, partially complete haplotypes carry more information for IBD detection, and thus the second scan can be more accurate. The idea for this second scan for haplotype sharing was inspired by the hidden Markov model described by Genovese et al.21 To detect IBD from comparisons of nearly complete haplotypes, we can use a threshold shorter than the one that delineates IBD sharing from IBS in diplotype–diplotype comparisons, and we no longer need to trim the borders of the shared regions. Recent IBD is declared in a region that spans a sufficient genetic distance, and when number of phased markers in the region exceeds the threshold of minimum phase information.
Comparison of methods
We compared ANCHAP against SLRP – a fully probabilistic method for long-range phasing, and against fastIBD – a local phasing method designed for populations of unrelated individuals. We evaluated their results genome-wide against recent IBD that can be reliably detected by comparison of haplotypes phased using parental genotypes. Among the individuals genotyped in ORCADES, there were 58 individuals with both parents genotyped, and on average 80% of heterozygous loci of such reference individuals were phased. We identified the regions of true recent IBD sharing between pairs of reference individuals where their haplotypes are identical for at least 2 cM. Each of the compared methods was used on genotype data from the 597 individuals in ORCADES, free from parent–offspring pairs. The results between the reference individuals were evaluated against the regions of true recent IBD. The total number of markers in true regions and in resulting regions is TP, in true regions but not in the resulting regions is FN, and not in true regions but in the resulting regions is FP. For each method, we quote sensitivity defined as the ratio TP/(TP+FN) and false discovery rate FP/(FP+TP).
Parameter tuning
All of the compared methods require setting different parameters. The methods were tuned according to their sensitivity and false discovery rate on a subset of the ORCADES data set from chromosome 2, using the reference individuals phased in parent–offspring trios. Other metrics, like inconsistencies between genotypes in supposed IBD regions are further described in Supplementary Materials.
We attempted to set the IBD threshold at Stage I of ANCHAP, such that the length of falsely assumed IBD regions is reduced while recovering as much of the true IBD regions as possible, and thus the phase recovery that uses the IBD segments is most accurate and maximally spread. The margin sizes were set by comparison of borders of IBD regions deduced from genotypes and the reference haplotypes. The setting of the alignment parameters at Stage II aimed at increasing the ratio of the IBD segments aligned into haplotypes and minimizing the inconsistencies between them, which indicate alignment errors. At Stage III, the minimum number of markers phased for both individuals in a putative IBD region was set using the reference haplotypes and the sensitivity and specificity values. The details of the experiments are shown in Supplementary Materials.
In addition, using the values of sensitivity and false discovery rate in data from chromosome 2, we adjusted the parameters of SLRP and fastIBD. SLRP required setting the expected length of IBD regions and expected regions of IBS, but not the IBD regions. The scale parameter in fastIBD controlled the parsimony of the haplotype model.
Optimization of resequencing studies in population isolates
The uncovered IBD sharing within a cohort can be used for efficient selection of individuals to resequence, with a view to using them as a reference for imputation. Selection of individuals for resequencing is based on maximizing representation of haplotypes and minimizing multiple resequencing of the same haplotypes. As the first individual for resequencing, we choose the one whose haplotypes have the most copies in the rest of the cohort. After excluding regions that have been covered by sharing with individuals already chosen, we repeat the procedure of selecting the individual with the most copies until a target level of coverage has been achieved (Algorithm 4 in Supplementary Materials).
Results
Phase propagation in ANCHAP
Table 1 shows the gain in sensitivity and the reduction in false discovery rate in the detection of recent IBD regions that are obtained at Stage III of our algorithm, as compared with Stage I. On chromosome 2, sensitivity of IBD detection between the 58 reference individuals per pair of individuals per marker grew from 0.75 to 0.81 in the second round. Detection of IBD for partially phased haplotypes in the second round helped to reduce the false discovery rate from 0.16 to 0.01.
Table 1. Part III of ANCHAP offers better accuracy in detecting regions of IBD than the first one.
| Method | ANCHAP Stage III | ANCHAP Stage I |
|---|---|---|
| Sensitivity | 0.81 | 0.75 |
| False discovery rate | 0.01 | 0.16 |
Abbreviations: IBD, identity by descent; ORCADES, Orkney Complex Disease Study.
Experiments with data from chromosome 2, and 597 ORCADES individuals with their genotyped parents removed. The identified regions of IBD were evaluated against phased haplotypes of 58 individuals who could be phased using the genotypes of their parents.
Comparison of ANCHAP against other methods
Table 2 compares different tuning settings of ANCHAP, SLRP and fastIBD. Using data from chromosome 2, we manipulated the parameters of SLRP and fastIBD, to match sensitivity and false discovery rate of ANCHAP. Notably, as the sensitivity of fastIBD grows to exceed ANCHAP's 0.81, false discovery rate of fastIBD reaches 0.024.
Table 2. Parameter tuning of ANCHAP, SLRP and fastIBD.
| Method | Parameters and values | Sensitivity | False discovery rate |
|---|---|---|---|
| ANCHAP | IBD threshold Stage I: 3 cM | 0.81 | 0.010 |
| IBD threshold Stage III: 2 cM | |||
| Overlap threshold: 10 markers | |||
| Mismatch tolerance: 2% | |||
| Minimum phase information: 100 markers | |||
| SLRP | Default | 0.76 | 0.008 |
| ExpectedIBS: 1 cM | |||
| ExpectedIBD: 10 cM | |||
| SLRP | Empirical | 0.77 | 0.011 |
| ExpectedIBS: 0.42 cM | |||
| ExpectedIBD: 9.17 cM | |||
| fastIBD | Scale: 1 | 0.27 | 0.000 |
| fastIBD | Scale: 2.8 | 0.80 | 0.021 |
| fastIBD | Scale: 2.9 | 0.81 | 0.024 |
| fastIBD | Scale: 3 | 0.83 | 0.024 |
| fastIBD | Scale: 4 | 0.87 | 0.044 |
Abbreviations: IBD, identity by descent; SLRP, systematic long-range phasing; ORCADES, Orkney Complex Disease Study.
Experiments with data from chromosome 2, and 597 ORCADES individuals with their genotyped parents removed. The identified regions of IBD were evaluated against phased haplotypes of 58 individuals who could be phased using the genotypes of their parents. Highlighted rows indicate parameters used in genome-wide analysis.
Table 3 shows the accuracy of IBD detection of ANCHAP against the other methods and their running times. Genome-wide, the methods achieved similar sensitivity of IBD: from 0.75 for SLRP, through 0.78 for ANCHAP and to 0.82 for fastIBD. Long-range methods, ANCHAP and SLRP resulted in similar false discovery rates of 0.009 and 0.007, respectively, whereas for fastIBD it is 0.025. Genome-wide inference of IBD with the SLRP model took much longer than for the other methods: the analysis with SLRP took 207 h, whereas ANCHAP handled the same task in 20 h and fastIBD in 12 h.
Table 3. Comparison of accuracies of methods for IBD detection.
| Method | ANCHAP | SLRP | FastIBD |
|---|---|---|---|
| Sensitivity | 0.78 | 0.75 | 0.82 |
| False discovery rate | 0.009 | 0.007 | 0.025 |
| Runtime (h) | 20 | 207 | 12 |
Abbreviations: IBD, identity by descent; SLRP, systematic long-range phasing; ORCADES, Orkney Complex Disease Study.
ANCHAP is compared with SLRP – a probabilistic method for phasing in isolated populations, and with fastIBD – a method designed for general populations. This genome-wide comparison was run on the subset of 597 individuals from ORCADES, such that their parents were not included. Regions of IBD were also evaluated using parent–offspring trios. Experiments were run on a computer with a 2.0 GHz and 16 GB of RAM.
Sharing in different cohorts and across the genome
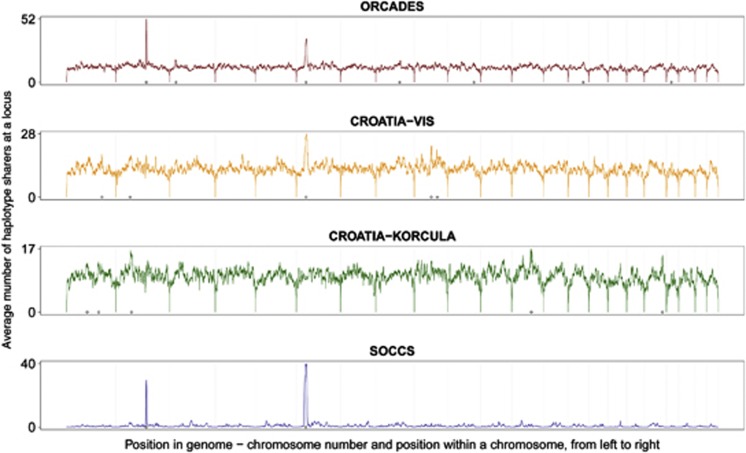
The average number of haplotype sharers per locus varied from 9.4 in CROATIA-KORCULA, through 12.3 in ORCADES and to 12.6 in CROATIA-VIS. In SOCCS, which consists of genotypes of nominally unrelated individuals, there were only 0.9 sharers per locus on average.
The frequency of haplotype sharing varies not only between the cohorts but also across the genome. Figure 2 shows average counts of the haplotype sharers in different locations across the genome. Drops at the telomeres can be consistently observed, as well as the peaks on chromosomes 2, 6, 8 and 9. In SOCCS, particularly notable are the peaks on chromosomes 2 and 6, which also occur in ORCADES and CROATIA-VIS but not in CROATIA-KORCULA.
Figure 2.
Density of surrogate parents across the genome in the four cohorts (3 cM threshold, 2 cM in the second round). The horizontal axis shows index of a SNP, and not its physical or genetic location. Supplementary Materials give the locations of the peaks highlighted on x axes.
Optimization of resequencing studies
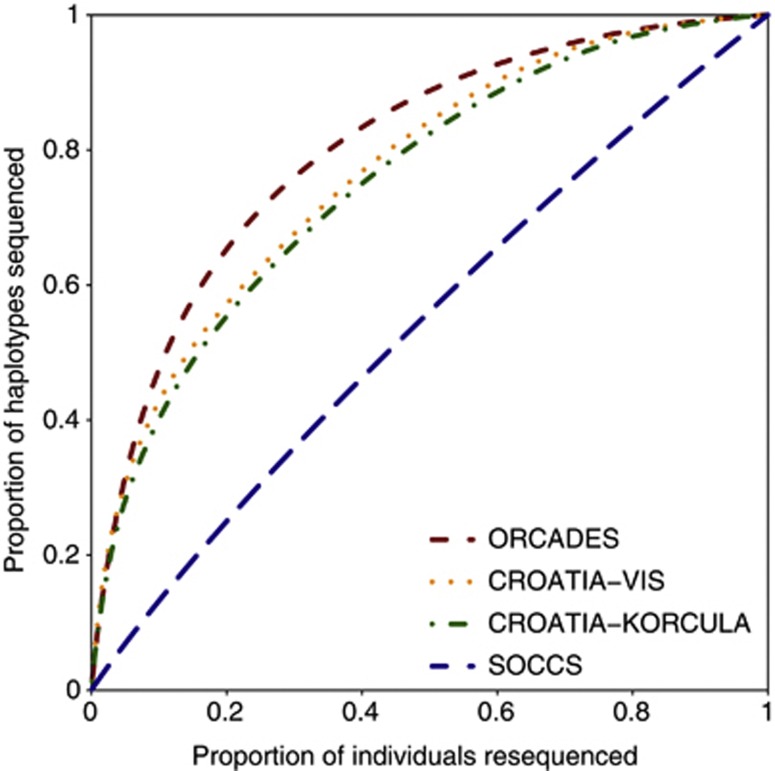
The uncovered sharing in ORCADES was used to choose an optimal subset of 200 individuals to be sequenced. Figure 3 shows that if such an optimal 20% of ORCADES individuals are sequenced, 65% of haplotypes in the cohort would be genotyped either directly or through an IBD copy. For CROATIA-VIS and CROATIA-KORCULA, the corresponding coverage would be 57% and 55%, respectively, whereas for SOCCS it would be 25%.
Figure 3.
Coverage of surrogate parents for different resequencing scenarios in the four cohorts.
Discussion
Comparison with other methods for IBD detection
Design of the algorithms affects the performance of the methods for IBD inference. SLRP is a model-based probabilistic approach for simultaneous IBD detection and phasing. It can simultaneously handle genotyping errors and phase uncertainty, yet this comes at the price of high computational demand. The loopy belief propagation that SLRP uses for inference may not find the optimal solution and is not guaranteed converge. ANCHAP does not explicitly model genotyping errors; phasing and IBD detection are separate steps, yet our tests detects recent IBD similarly well. When a genotyping error gives rise to a pair of opposing homozygotes in a region of IBD sharing, the region of sharing detected by ANCHAP may be shorter or missed altogether. However, in ORCADES we encountered on average only 1 opposing homozygote per 10 000 markers in genotypes of parent–offspring pairs; hence, genotyping errors will not prevent most of the sharing regions from being detected. FastIBD is a method for IBD detection designed for general, not necessarily isolated populations.13 It builds a model of haplotypes, which can capture only short-range allele correlations. This deficiency is then ameliorated by sampling multiple haplotypes for each individual, and checking overlap of such samples between pairs of individuals. FastIBD turned out to be more sensitive than ANCHAP or SLRP, but also returned more false discoveries. A possible explanation for why fastIBD yields more false discoveries is that haplotype resampling of short blocks may occasionally yield matches between individuals by chance.
It seems unlikely that the differences in sensitivity and false discovery rate between the methods would seriously affect the uses of detected IBD region in mapping complex traits or IBD-based imputations. Sensitivity approaching 100% would be desirable for downstream applications,8 but none of the methods achieves sensitivity of IBD detection of more than 81% for the ORCADES data. In case of ANCHAP, this probably results from inability to handle the incorrect assignments in Stages I and II, which trigger phasing errors, and IBD is no longer detected in Stage III. Ability to recover from sporadic phasing errors would certainly improve the sensitivity of IBD detection. For SLRP, incomplete IBD detection could be because of limitations of the inference algorithm, conservative approach to declaring IBD or low tolerance to inconsistencies between the IBD sharing relationship and, possibly, noisy data. In case of fastIBD, if for a pair of individuals sharing IBD there are few haplotypes that would explain the genotypes, the program may not sample the matching pair. In accordance with this observation, the highest sensitivity we could achieve was by runs of fastIBD with scale 4.0 repeated 10 times. Together with the sensitivity going up to 89%, the false discovery rate also grew to 7%.
The comparison of methods as well as parameter tuning are based on the presence of parent–offspring trios among the ORCADES samples. The borders of reference sharing regions as determined by the parent–offspring phasing are only as accurate as the SNP density allows. The reference regions may still have false endpoints, because a recombination may not be detectable from SNP alleles. However, the endpoints should not affect the results of the comparison, as they will be small compared with the regions themselves; long matching haplotypes that do not descent from a common ancestor are unlikely. In absence of the parent–offspring trios, several types of inconsistencies between the genotype data and IBD relationships recovered could indicate errors. For example, the multipoint genotypes of haplotype sharers, which are recognized to carry one of the proband's haplotypes, cannot have opposing homozygotes with respect to not just the proband but also each other.
Genetic maps and peaks of IBD
With a genetic map, we not only estimate minimal physical sizes of regions that could be IBD, but also try to account for extensive LD that existed in genotypes of isolate founders to avoid false detections of IBD. If haplotypes are very similar to each other, and we observe only unphased SNP genotypes, our method may declare IBD incorrectly even when two individuals do not share a recent common ancestor and their full sequences are not identical. To account for such regions of extended LD between isolate founders, inference of IBD requires longer sequences of SNPs without opposing homozygotes. ANCHAP's threshold for the length of segments identical by state required to declare THEM identical by descent, is expressed in centimorgans, referring to the HapMap genetic map. This map is based on modeling haplotype structure in outbred populations.22, 23 When the true recombination rate is low, or when there has been recent drift or selection, this will be reflected in the ‘recombination map', and using it will correct for extended LD. Such a correction is evident in the SOCCS data, where using the HapMap markedly reduces the size of the peaks for apparent IBD sharing on chromosomes 6 and 11 (Supplementary Figure 4).
The remaining peaks of IBD sharing observed in Figure 2 could result from either increased sensitivity to IBD sharing, increased IBD false discovery rate, or selection pressure. A possible explanation for these peaks may be that the HapMap genetic map too does not appropriately adjust for extended LD in the European and possibly the Scottish populations. Excess sharing in SOCCS, a cohort composed mainly of unrelated individuals from across Scotland, concentrates around the two peaks on chromosome 2 and 6 (exact locations in Supplementary Materials). Furthermore, these peaks also occur in ORCADES and partially in the Croatian cohorts. The peaks are less visible in CROATIA-KORCULA, where a different SNP array was used. In Supplementary Figure 5c, we show there is no marked variation of marker density with respect to physical or genetic maps at the peak regions. Alternatively, if the peaks are not the result of an inappropriate adjustment for the background LD, they could indicate a drift or selection in the Scottish populations and the isolates.24 A list of genes present within these peaks, including the HLA genes, is presented in Supplementary Materials.
Resequencing optimization
The inferred shared haplotypes in an isolated population can be exploited to increase the efficiency of a sequencing study given a fixed budget. One possible strategy is to identify an optimal subset of individuals for resequencing at high coverage so as to obtain an accurate sequence data, then to impute these sequences into the other cohort members with whom they share the IBD. For the selection of individuals, our algorithm favors individuals who share the largest identical by descent regions with individuals who were not chosen for resequencing. The imputation of sequences of not chosen individuals could be most effective when for each haplotype throughout the genome there were few sequenced sharers. We have examined the strategy based on resequencing an optimal 20% of individuals from ORCADES, which would reduce the cost fivefold, with 65% of the unsequenced diploid genomes sharing with the sequenced individuals. Had we chosen the individuals randomly or based on kinship coefficients, the IBD coverage of unsequenced haplotypes would have been 61% or 62%, respectively (details of comparison in Supplementary Materials). A recent study in another population isolate also confirmed the merit of IBD-based optimization procedures.25
A number of factors determine the accuracy of imputations based on IBD. First, only correctly detected IBD would result in correct imputations. Second, a variant could only be correctly imputed if it is older than the most common ancestor from whom the haplotype was co-inherited. As long-range methods detect IBD from recent common ancestors, they should allow for more accurate imputation of more recent variants. Finally, the ease of sequence imputations with the IBD sharing information depends on whether the sequences can be phased, and whether it is possible to overlay the array and sequence haplotypes. There is work underway both on algorithmic and laboratory methods for phasing of sequences.8 If the sequences are not phased, the IBD sharing can still be exploited for imputation, but this would result in more uncertainty about imputed alleles.
Acknowledgments
This work was supported by the Medical Research Council grant G0800604 to PM, and by MRC's scholarship to DG. The ORCADES study was supported by a Royal Society fellowship for JFW, by the Chief Scientist Office of the Scottish Government Health Directorates, the EU Framework Programme 6 EUROSPAN award and by the facilities of the Wellcome Trust Clinical Research Facility, Edinburgh. The Croatian studies were supported by MRC and the Croatian Ministry of Science, Education and Sport. The SOCCS study was funded by Cancer Research UK Programme grant funder number C348/A12076.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Powell JE, Visscher PM, Goddard ME. Reconciling the analysis of IBD and IBS in complex trait studies. Nat Rev Genet. 2010;11:800–805. doi: 10.1038/nrg2865. [DOI] [PubMed] [Google Scholar]
- Gusev A, Kenny EE, Lowe JK, et al. Dash: a method for identical-by-descent haplotype mapping uncovers association with recent variation. Am J Hum Genet. 2011;88:706–717. doi: 10.1016/j.ajhg.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Thompson EA. Detecting rare variant associations by identity-by-descent mapping in case-control studies. Genetics. 2012;190:1521–1531. doi: 10.1534/genetics.111.136937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E. Next-generation association studies for complex traits. Nat Genet. 2011;43:287–288. doi: 10.1038/ng0411-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality. Curr Opin Biotechnol. 1998;9:578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- Johnson G, Esposito L, Barratt BJ, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Morris AP, Whittaker JC, Balding DJ. Fine-scale mapping of disease loci via shattered coalescent modeling of genealogies. Am J Hum Genet. 2002;70:686–707. doi: 10.1086/339271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Masson G, Frigge ML, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey JM, Kinghorn P, Tier B, et al. A combined long-range phasing and long haplotype imputation method to impute phase for snp genotypes. Genet Sel Evol. 2011;43:12. doi: 10.1186/1297-9686-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin K, Campbell H, Wright AF, Wilson JF, Durbin R. Identity-by-descent-based phasing and imputation in founder populations using graphical models. Genet Epidemiol. 2011;8:853–860. doi: 10.1002/gepi.20635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet. 2011;88:173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R, et al. Runs of homozygosity in european populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JF, Weiss DA, Richards M, et al. Genetic evidence for different male and female roles during cultural transitions in the British isles. Proc Natl Acad Sci USA. 2001;98:5078–5083. doi: 10.1073/pnas.071036898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart V, Biloglav Z, Hayward C, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–4876. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- Polasek O, Marusic A, Rotim K, et al. Genome-wide association study of anthropometric traits in Korcula Island, Croatia. Croat Med J. 2009;50:7–16. doi: 10.3325/cmj.2009.50.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. High-resolution detection of identity by descent in unrelated individuals. Am J Hum Genet. 2010;86:526–539. doi: 10.1016/j.ajhg.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. Mutation patterns in the human genome: more variable than expected. PLoS Biol. 2009;7:e1000028. doi: 10.1371/journal.pbio.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Leibon G, Pollak MR, Rockmore DN. Improved IBD detection using incomplete haplotype information. BMC Genet. 2010;11:58. doi: 10.1186/1471-2156-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A, Moltke I, Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics. 2010;186:295–308. doi: 10.1534/genetics.110.113977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Kenny EE, Lowe JK, et al. Low-pass genome-wide sequencing and variant inference using identity-by-descent in an isolated human population. Genetics. 2012;190:679–689. doi: 10.1534/genetics.111.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.