Abstract
Peanut allergy (PA) is a common and serious food allergy and its prevalence has increased in the past decade. Although there is strong evidence of inheritance, the genetic causes of this disease are not well understood. Previously, a large-scale genome-wide association study described an association between human leukocyte antigen (HLA)-DQB1 and asthma; the aim of this study was to evaluate the association between HLA-DQB1 and PA. Genotypic and allelic profiles were established for 311 Caucasian members of a well-described Canadian group of children with PA and 226 Caucasian controls. Firth's logistic regression analyses showed associations between HLA-DQB1 alleles and PA for DQB1*02 (P=1.1 × 10−8, odds ratio (OR)=0.09 (CI=0.03–0.23)) and DQB1*06:03P alleles (P=2.1 × 10−2, OR=2.82 (CI=1.48–5.45)). This study of HLA in PA demonstrates specific association between two allelic groups of the HLA-DQB1 gene (DQB1*02 and DQB1*06:03P) and PA, highlighting its possible role in the development of this disease.
Keywords: HLA-DQB1, peanut allergy, Caucasian, association, food allergy
Introduction
Peanut allergy (PA) is a common food allergy that is associated with substantial morbidity and mortality. It generally presents before 2 years of age and resolves in about 20% of children.1 Its prevalence has increased over the past decade in the United States, although it may have recently stabilized in Canada and the United Kingdom at about 1.7%.1, 2 Peanut hypersensitivity is the most common cause of fatal anaphylactic reactions to food in North America and the United Kingdom.1 Although several environmental factors, such as the time of first exposure, have been postulated to have an important role in the development of PA, there is strong evidence for inheritance from twin and family studies.1 However, the genetic cause of this disease remains elusive.
In a large-scale genome-wide association study, the human leukocyte antigen (HLA)-DQB1 locus was found to be associated with asthma.3 Genes of the HLA family, which are part of the major histocompatibility complex (MHC) locus, are involved in antigen presentation to T cells. As asthma and food allergy are known to be clinically correlated,4 a few studies have examined MHC class II genes (HLA-DR, -DQ and -DP families) in the context of PA5, 6, 7 but only one study has shown a link between these genes and hypersensitivity to peanut.7 To better understand the genetic component of PA, we compared the HLA-DQB1 allele and genotype frequencies in children with PA and children without PA and demonstrated an association between the HLA-DQB1 gene and this disease.
Materials and methods
Sample
This study included 590 children with PA, recruited from a well-described Canadian PA case group.8 Children were considered to be allergic to peanut if either of the following criteria were fulfilled: (1) a convincing clinical history of an allergic reaction to peanut and a positive skin prick test (SPT) to peanut or a peanut-specific IgE level ≥ 0.35 kU/l (87% of cases) or (2) no previous exposure to peanut or an uncertain history of an allergic reaction to peanut and either a positive SPT to peanut and a peanut-specific IgE level ≥15 kU/l or a positive food challenge to peanut (13% of cases). Only those who self-reported Caucasian ethnicity were included in the study. A total of 332 subjects were selected as controls from two Canadian studies: the Canadian Asthma Primary Prevention Study (CAPPS)9 and the Study of Asthma Genes and the Environment (SAGE).10 Those individuals were required to (1) have a negative SPT to peanut, (2) have no history of atopy or asthma, (3) be of Caucasian ethnicity. See Table 1 for a more complete description of the subjects. The project was approved by the local ethics committee and all subjects gave written informed consent.
Table 1. Phenotypic description of the subjects.
| Peanut allergy cases (n=311)a,b | |||
|---|---|---|---|
| |
Controls (n=226) a |
Convincing history (87%) |
No exposure/uncertain history (13%) |
| Sex ratio, M:Fc | 1:1.4 | 1:0.6 | |
| Age, mean (SD)d | 4 (4) | 11 (4) | |
| SPT to peanut, mm (SD) | 0 | 9.9 (5.1) | 11.1 (5.0) |
| SPT to peanut, range | 0 | 3–31 | 3–20 |
| IgE, kU/l (SD) | ND | 46.2 (39.9) | 63.7 (36.1) |
| IgE, range | ND | 0.35–100 | 5.20–100 |
| Asthma, n (%)e | 0 | 205 (66) | |
Abbreviations: ND, not done; SPT, skin prick test.
Only subjects with genotypic data were included in the analyses.
Peanut allergy cases can be classified in two groups, the first one (called convincing history in the table) is characterized by a convincing clinical history of an allergic reaction to peanut and a positive SPT to peanut or a peanut-specific IgE level ≥0.35 kU/l, and the second one (no exposure/uncertain history) is characterized by no previous exposure to peanut or an uncertain history of an allergic reaction to peanut and either a positive SPT to peanut and a peanut-specific IgE level ≥15 kU/l or a positive food challenge to peanut.
Sex available for 223 controls and 311 peanut allergy cases.
Age available for 124 controls and 310 peanut allergy cases.
Asthma status available for 226 controls and 310 peanut allergy cases.
Genotyping
Genomic DNA was extracted from saliva for the PA group (Oragene•DNA saliva kit, DNA Genotek Inc., Kanata, ON, Canada) and from blood for control samples (Blood and Cell Culture Midi Kit, QIAGEN Inc., Toronto, ON, Canada) and were stored at −80 °C. Sequencing of exons two and three was performed using the AlleleSEQR HLA-Sequence-Based Typing Packs by Abbott Molecular Diagnostics (Wiesbaden, Germany) at the genomic and proteomic laboratory at the Université du Québec à Chicoutimi. DNA was run on an ABI PRISM 3100 Genetic Analyzer (ABI, Foster City, CA, USA) at the ECOGENE-21 laboratory at the Centre hospitalier affilié universitaire régional (CAUR) de Chicoutimi. Genotyping was done for 48 samples per batch. Sequences were analyzed with Assign software (Conexio Genomics, version 3.5.1; Fremantle, WA, Australia). Genotypes were identified according to the ImMunoGeneTics (IMGT)/HLA database version 3.5.0 (July 2011) (EMBL-EBI, Hinxton, UK).11, 12 Calls were made at the peptide-binding domain (PBD) resolution level.13 In HLA nomenclature, the first set of two digits of an allele refers to the type and the second set of two digits refers to the subtype. P groups represent alleles that share the same PBD and are identified by the addition of the letter ‘P' at the end of the allele (eg, HLA-DQB1*03:01P or DQB1*05:01P).14 Sequencing of the 922 DNA samples resulted in 807 sequences of good quality (88%) and unambiguous genotype calls (without multiple possibilities of allele call at the type level) for 63% (311/491) of the PA cases and 72% (226/316) of the controls (see Supplementary Table S1 for the complete list of primary genotypes).
Statistical analyses
All analyses were done using the R statistical program (version 2.14.1, http://www.r-project.org/). The following quality control checks and measures were performed: (1) genotyping call rates were evaluated and confirmed to be in line with standards for HLA genotyping; (2) type ambiguities and alleles with mismatches (a sequence that does not match any known allele) were removed and subtype ambiguities were resolved using P group classification;13 (3) alleles not included in any P groups and rare P groups of a same type (<20 carriers) were grouped together in order to have stable odds ratio (OR) estimates and higher statistical power (indicated by allele names with only one set of two digits, such as DQB1*02);13 (4) only samples of Caucasian descent were used (Caucasian ethnicity was confirmed by genome-wide association studies in the controls from CAPPS and SAGE studies). Logistic regression analyses were performed to examine differences in frequencies of alleles or frequent genotypes (≥20 carriers) between PA samples and controls. For each analysis, samples were classified as non-carrier or carrier (heterozygous or homozygous) of the allele or non-carrier or carrier of the genotype. We considered the possibility of confounding by asthma status and sex (see Table 1 and Supplementary Table S2 in Supplementary Material for phenotypic differences between genotypes) and performed analyses adjusted for sex and asthma status. This was particularly important as HLA-DQB1 is an asthma susceptibility gene3 and there is a high prevalence of asthma in the PA cases (2/3 have asthma) and no asthma in the controls; we considered the possibility that any association observed may be due to the presence of asthma. An alternative hypothesis is that HLA-DQB1 is a susceptibility gene for both asthma and PA (ie, pleiotropy). In addition, the controls did not have PA (as indicated by a negative SPT), reducing the misclassification probability for controls,10 thereby increasing our statistical power. Owing to small sample sizes and correlation between asthma status and the other variables, a quasi-complete separation of the data resulted. To account for this complexity, we used Firth's logistic regression using the Heinze's logistf package in R without prior specification of weights.15 Finally, a Bonferroni correction for 12 independent tests (12 alleles tested) was applied.
Results
After correction for multiple testing, a significant difference was observed for five HLA-DQB1 alleles in PA cases compared to controls: a higher frequency of the DQB1*06:03P (P=1.9 × 10−3, OR=2.59 (CI=1.56–4.44)) allele and a decreased frequency of the DQB1*02 (P=1.3 × 10−15, OR=0.12 (CI=0.07–0.21)), DQB1*03:02P (P=2.6 × 10−2, OR=0.52 (CI=0.34–0.79)), DQB1*05 (P=3.0 × 10−3, OR=0.21 (CI=0.08–0.50)) and DQB1*05:01P (P=9.3 × 10−5, OR=0.25 (CI=0.13–0.47)) alleles (see Table 2 and Figure 1). Adjusted analyses considering asthma status and sex continued to show a significantly higher frequency of the DQB1*06:03P (P=2.1 × 10−2, OR=2.82 (CI=1.48–5.45)) allele and decreased frequency of the DQB1*02 (P=1.1 × 10−8, OR=0.09 (CI=0.03–0.23)) allele.
Table 2. Significant associations for HLA-DQB1 alleles with peanut allergy performed with or without adjustment for asthma status and sex.
| Allele alone | Adjusted for asthma status and sexa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Allelesd |
Ctrl, n (%)b |
PA, n (%)b |
OR |
95% CI |
Xb |
P-value |
Pcc |
OR |
95% CI |
Xb |
P-value |
Pcc |
| DQB1*02 | 82 (18) | 25 (4) | 0.12 | 0.07–0.21 | 68.76 | 1.1 × 10−16 | 1.3 × 10−15 | 0.09 | 0.03–0.23 | 37.49 | 9.2 × 10−10 | 1.1 × 10−08 |
| DQB1*03:02P | 68 (15) | 62 (10) | 0.52 | 0.34–0.79 | 9.39 | 2.2 × 10−03 | 2.6 × 10−02 | 0.47 | 0.24–0.86 | 6.18 | 1.3 × 10−02 | NS |
| DQB1*05 | 20 (4) | 6 (1) | 0.21 | 0.08–0.50 | 13.43 | 2.5 × 10−04 | 3.0 × 10−03 | 0.26 | 0.05–0.84 | 5.24 | 2.2 × 10−02 | NS |
| DQB1*05:01P | 37 (8) | 16 (3) | 0.25 | 0.13–0.47 | 20.00 | 7.7 × 10−06 | 9.3 × 10−05 | 0.38 | 0.15–0.83 | 6.09 | 1.4 × 10−02 | NS |
| DQB1*06:03P | 23 (5) | 71 (11) | 2.59 | 1.56–4.44 | 14.23 | 1.6 × 10−04 | 1.9 × 10−03 | 2.82 | 1.48–5.45 | 9.80 | 1.7 × 10−03 | 2.1 × 10−02 |
Abbreviations: CI, confidence interval of 95% Ctrl, controls (subjects without allergy); NS, not significant; OR, odds ratio; PA, subjects with peanut allergy; Pc, corrected P-value; PBD, peptide-binding domain.
Analyses performed for 534 subjects: asthma status available for 310/311 subjects with peanut allergy (none of the control subjects have asthma) and sex available for 534/537 subjects.
Allelic frequency in control and peanut allergy cases shown in numbers of alleles (n) and percentages (%).
P-values with Bonferroni correction considering 12 independent comparisons (12 alleles).
Alleles as indicated in the ImMunoGeneTics (IMGT)/HLA database nomenclature version 3.5.0 (July 2011). Alleles with two sets of two digits and a ‘P' are P groups of alleles comprising HLA-DQB1 alleles that share the same PBD. Alleles with only one set of two digits are those that include a mix of rare alleles that share the same type because of their low frequency in the studied population (see Supplementary Table S4 in the Supplementary Materials for a list of those alleles).
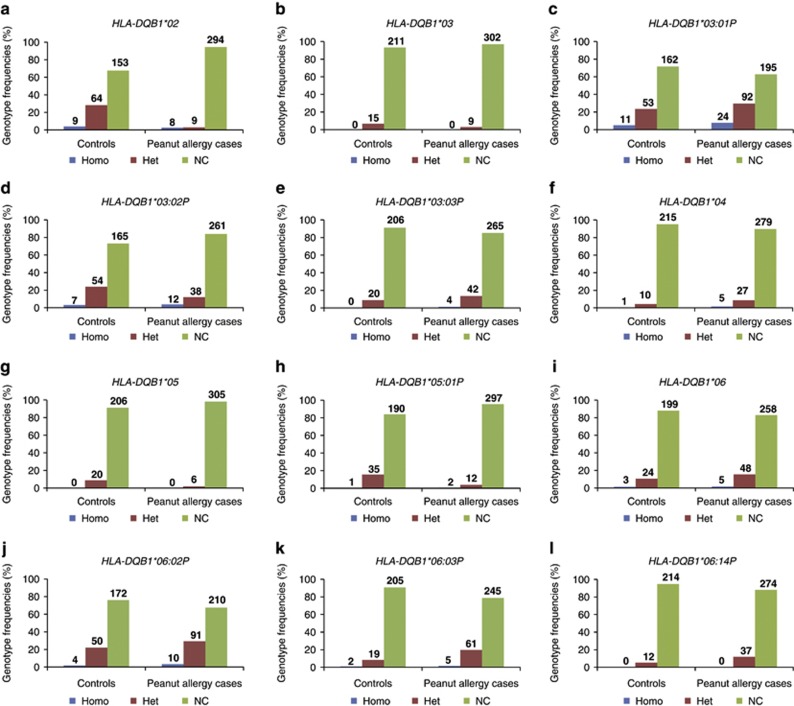
Figure 1.
Distribution of each HLA-DQB1 genotype for peanut allergic subjects and controls. Bar graphs represent the genotypic frequencies in percentages for peanut allergic subjects and controls for each studied allele of the HLA-DQB1 loci. Numbers of subjects for each genotype are also indicated. Results for HLA-DQB1*02 are shown in figure a, for HLA-DQB1*03 in b, for HLA-DQB1*03:01P in c, for HLA-DQB1*03:02P in d, for HLA-DQB1*03:03P in e, for HLA-DQB1*04 in f, for HLA-DQB1*05 in g, for HLA-DQB1*05:01P in h, for HLA-DQB1*06 in i, for HLA-DQB1*06:02P in j, for HLA-DQB1*06:03P in k, and for HLA-DQB1*06:14P in l. For example, Figure 1a shows a frequency of 4.0% among controls (representing 9 carriers) for the DQB1*02 homozygotes (DQB1*02+DBQ1*02) and a frequency of 2.6% (for 8 carriers) among PA cases, a frequency of 28.3% (for 64 carriers) of heterozygotes among controls for the DQB1*02 allele (DQB1*02+DQB1* any other allele) and 2.9% (for 9 carriers) among PA cases and finally a frequency of 67.7% (for 153 carriers) among controls for all other subjects that do not carry any DQB1*02 alleles and 94.5% (for 294 carriers) among PA cases. Alleles and genotypes as indicated in the ImMunoGeneTics (IMGT)/HLA database nomenclature version 3.5.0 (July 2011). Alleles with two sets of two digits and a ‘P' are P groups of alleles comprising HLA-DQB1 alleles that share the same PBD. Alleles with only one set of two digits are those that include a mix of rare alleles because of their low frequency in the studied population (see Supplementary Table S4 in the Supplementary Materials for a list of those alleles). Het, heterozygotes for the studied allele; Homo, homozygotes for the studied allele; NC, non-carrier of this allele.
To determine if specific allelic combinations were associated with PA, we performed the analyses with the more frequent genotypes (genotypes with >20 carriers). However, only three genotypes met the frequency criteria (DQB1*03:01P+DQB1*03:01P, DQB1*03:01P+DQB1*06:02P and DQB1*03:01P+DQB1*06:03P) and none remained significant after correction for multiple testing and adjustment for asthma and sex (see Supplementary Table S3 in Supplementary Materials for significant results before correction).
Discussion
This is the largest genetic study of HLA and PA in children, involving 311 PA cases, which is more than triple the number in previous studies.5, 6, 7 An association has been previously demonstrated between DQB1 allele and PA by Howell et al.7 The present results do not replicate the increase of the DQB1*04 allele in PA cases as previously published;7 however, we showed an increase of the DQB1*06:03P allele and a decrease of the DQB1*02, DQB1*03:02P, DQB1*05 and DQB1*05:01P alleles in PA subjects compared with controls. The association of the DQB1*06:03P and DQB1*02 alleles with PA remained significant after adjusting for asthma status and sex.
As HLA loci are expressed co-dominantly, analyses of genotypic frequencies for the most frequent genotypes were conducted to determine if specific allelic combinations may be associated with PA. However, there were very few genotypes with sufficient carriers to be included in the analyses and hence, more subjects are required to examine allelic combinations.
This study is limited by the relatively high number of ambiguous genotypes at the allele type level; this can be explained by the high failure rate previously observed in the HLA-DQB1 region in a recent linkage-disequilibrium study on the HLA regions.16 Owing to the high polymorphic rate of this locus16 and to increase the power of the study, analyses were performed at the P group resolution, rarer alleles and rarer P groups were grouped together and logistic regression adapted to small samples and to complete separation of variables (Firth's logistic regression) was performed.13, 15 Although these grouping methods increase statistical power, they decrease the diversity of the genetic information as groups of rare alleles can include risk-conferring and protective alleles together and may lead to the failure to identify some possible associations. As HLA loci are highly variable and are influenced by ethnicity, making population stratification a concern, our study is limited to individuals of Caucasian descent. Caucasian descent was validated using principal components analyses for control subjects, but not for PA cases because of the absence of genome-wide association data on the cases; hence, we were unable to include principal components to control for any population differences between the cases and the controls.
In this study, the HLA-DQB1 gene has been selected based on its association with asthma in a previous large-scale genome-wide association study.3 However, linkage disequilibrium has been documented between the HLA-DQB1 and the HLA-DRB1 genes in diseases such as multiple sclerosis and HLA-DRB1 has been previously associated with PA and IgE levels.3, 17 A genetic association test with HLA-DRB1 to refine the present finding could provide insight, and should be included in future research.
The results of this study suggest that DQB1*06:03P allele may be a risk factor for PA and support the role of DQB1*02 allele as a protective factor. As PA is a complex trait involving multiple environmental and genetic factors, more research is needed to understand the possible interactions between HLA-DQB1 and other genes and environmental factors involved in PA.
Acknowledgments
We thank all the individuals involved in this study for their participation and the funding organizations for their support. This study was supported by the AllerGen NCE (the Allergy, Genes and Environment Network of Centers of Excellence). VT Vaillancourt is an AllerGen NCE trainee and is supported by the Corporation de recherche et d'action sur les maladies héréditaires (CORAMH) and the Campagne de développement de l'Université du Québec à Chicoutimi. Y Asai is supported by a Canadian Institutes for Health Research (CIHR) Research fellowship, an AllerGen CAIDATI training award and the Canadian Dermatology Foundation (CDF). M Ben-Shoshan is an Emerging Clinician-Scientist of the AllerGen NCE. A Kozyrskyj is the WCHRI Research Chair in Maternal-Child Health and the Environment. D Daley has received grant support from CIHR and is a Michael Smith Foundation for Health Research Career Scholar and she holds a Tier II Canadian Research Chair (Genetics of Common Complex Diseases). A Clarke is a National Research Scholar of the Fonds de recherche du Québec–santé (FRQS). P Hull is supported by the CDF and the University of Saskatchewan's Department of Medicine Research Fund. C Laprise holds the Canada Research Chair on Genetic Determinants in Asthma and is the director of the Inflammation and Remodeling Strategic Group of the Respiratory Health Network of the FRQS. A Kozyrskyj, A Becker, D Daley, A Sandford and C Laprise are AllerGen NCE members. The Canadian studies (SAGE and CAPPS studies) were supported by the AllerGen NCE. The McGill PA case group is supported by the Foundations of the McGill University Health Center and the Montreal Children's Hospital as well as by grants from the Canadian Allergy, Asthma and Immunology Foundation and the AllerGen NCE.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Hourihane JO.Peanut allergy Pediatr Clin North Am 201158445–458.xi. [DOI] [PubMed] [Google Scholar]
- Ben-Shoshan M, Harrington DW, Soller L, et al. A population-based study on peanut, tree nut, fish, shellfish, and sesame allergy prevalence in Canada. J Allergy Clin Immunol. 2010;125:1327–1335. doi: 10.1016/j.jaci.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AB, Yousef E, Hossain J. Association between peanut allergy and asthma morbidity. J Pediatr. 2010;156:781. e771. doi: 10.1016/j.jpeds.2009.11.080. [DOI] [PubMed] [Google Scholar]
- Shreffler WG, Charlop-Powers Z, Sicherer SH. Lack of association of HLA class II alleles with peanut allergy. Ann Allergy Asthma Immunol. 2006;96:865–869. doi: 10.1016/S1081-1206(10)61351-8. [DOI] [PubMed] [Google Scholar]
- Dreskin SC, Tripputi MT, Aubrey MT, et al. Peanut-allergic subjects and their peanut-tolerant siblings have large differences in peanut-specific IgG that are independent of HLA class II. Clin Immunol. 2010;137:366–373. doi: 10.1016/j.clim.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell WM, Turner SJ, Hourihane JO, Dean TP, Warner JO. HLA class II DRB1, DQB1 and DPB1 genotypic associations with peanut allergy: evidence from a family-based and case-control study. Clin Exp Allergy. 1998;28:156–162. doi: 10.1046/j.1365-2222.1998.00224.x. [DOI] [PubMed] [Google Scholar]
- Nguyen-Luu NU, Ben-Shoshan M, Alizadehfar R, et al. Inadvertent exposures in children with peanut allergy. Pediatr Allergy Immunol. 2012;23:133–139. doi: 10.1111/j.1399-3038.2011.01235.x. [DOI] [PubMed] [Google Scholar]
- Chan-Yeung M, Manfreda J, Dimich-Ward H, Ferguson A, Watson W, Becker A. A randomized controlled study on the effectiveness of a multifaceted intervention program in the primary prevention of asthma in high-risk infants. Arch Pediatr Adolesc Med. 2000;154:657–663. doi: 10.1001/archpedi.154.7.657. [DOI] [PubMed] [Google Scholar]
- Liem JJ, Huq S, Kozyrskyj AL, Becker AB. Should younger siblings of peanut-allergic children be assessed by an allergist before being fed peanut. Allergy Asthma Clin Immunol. 2008;4:144–149. doi: 10.1186/1710-1492-4-4-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA Database. Nucleic Acids Research. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA - a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- Hollenbach JA, Mack SJ, Gourraud PA, et al. A community standard for immunogenomic data reporting and analysis: proposal for a STrengthening the REporting of Immunogenomic Studies statement. Tissue Antigens. 2011;78:333–344. doi: 10.1111/j.1399-0039.2011.01777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HLA nomenclature: nomenclature for factors of the HLA system; 2012; Accessed on September 2012. . http://hla.alleles.org/alleles/p_groups.html .
- Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21:2409–2419. doi: 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
- Miretti MM, Walsh EC, Ke X, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Ebers GC. Multiple sclerosis: major histocompatibility complexity and antigen presentation. Genome Med. 2009;1:105. doi: 10.1186/gm105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.