Abstract
Purpose
To use genetically engineered mouse models (GEMMs) and orthotopic syngeneic murine transplants (OSTs) to develop gene-expression based predictors of response to anti-cancer drugs in human tumors. These mouse models offer advantages including precise genetics and an intact microenvironment/immune system.
Experimental Design
We examined the efficacy of four chemotherapeutic or targeted anti-cancer drugs, alone and in combination, using mouse models representing three distinct breast cancer subtypes: Basal-like (C3(1)-T-antigen GEMM), Luminal B (MMTV-Neu GEMM), and Claudin-low (T11/TP53−/− OST). We expression-profiled tumors to develop signatures that corresponded to treatment and response, then tested their predictive potential using human patient data.
Results
Although a single agent exhibited exceptional efficacy (i.e. lapatinib in the Neu-driven model), generally single-agent activity was modest, while some combination therapies were more active and life-prolonging. Through analysis of RNA expression in this large set of chemotherapy-treated murine tumors, we identified a pair of gene expression signatures that predicted pathological complete response to neoadjuvant anthracycline/taxane therapy in human patients with breast cancer.
Conclusions
These results show that murine-derived gene signatures can predict response even after accounting for common clinical variables and other predictive genomic signatures, suggesting that mice can be used to identify new biomarkers for human cancer patients.
Introduction
Gene expression profiling has identified five molecular subtypes of breast cancer (Luminal A, Luminal B, Basal-like, HER2-Enriched, Claudin-low) and a normal-like group, which show significant differences in epidemiologic associations and clinical features including survival(1-3). Mounting evidence suggests that these subtypes vary in their responsiveness to chemotherapeutics(2, 4-6) and to biologically targeted agents(7-9). Methods for selecting the optimal chemotherapeutic agent for each breast tumor subtype have yet to be determined. Instead, chemotherapy choices for breast cancer patients have been mainly empiric and based upon large clinical trials using unselected patient populations, and population-based benefits. The Basal-like subtype of breast tumor, of which the majority are also “triple-negative” breast cancers, is particularly challenging due to its lack of validated biological targets (i.e. ER-, PR- and HER2 normal)(10, 11). Other breast cancer subtypes with poor prognosis also exist including the Luminal B subtype(2, 5) and the recently discovered Claudin-low subtype, which exhibits high numbers of tumor initiating cells(12).
Genetically Engineered Mouse Models (GEMMs) have proven valuable for validating the causal role of oncogenes and tumor suppressor genes in cancer(13), but their use in efficacy testing is less mature, with most studies being low-throughput efforts examining model-specific compounds in small numbers of tumor-bearing mice (<50)(14). Recently, academic and industry researchers have begun simultaneous efficacy testing at medium throughput, employing larger numbers of compounds (5-50) in larger numbers of GEMMs (100-1000)(15, 16). In particular, these efforts have attempted to mirror and inform ongoing human clinical trials, by testing novel therapeutics in faithful murine models as “co-clinical trials”(17). While this approach has been promising, we believe an additional untapped power of medium-throughput GEMM testing is the ability to use murine models to identify biomarkers of response for human cancer patients.
Previously, we performed RNA expression profiling of 13 distinct GEMMs of breast cancer(12, 18) and compared these signatures to human expression subtypes using an across-species expression analysis. These analyses identified murine models that faithfully represent multiple human breast tumor subtypes including Basal-like tumors (C3(1)-T-antigen)(19) and Luminal B tumors (MMTV-Neu)(20). No single Claudin-low GEMM was identified, but an orthotopic, transplantable syngeneic tumor from a BALB/c TP53−/− mouse was found to exhibit a stable Claudin-low expression phenotype(12). In this work, we used these credentialed murine tumor models and determined their sensitivities to a variety of chemotherapeutic and biologically targeted agents in routine clinical use. This analysis identified a heterogeneity of responses to certain cytotoxics in the Basal-like model. We exploited this existence of sensitive and resistant tumors from GEMMs to develop genomic signatures of chemotherapy response, which we tested in a large, clinically annotated human cohort of breast cancer patients.
Methods
Genetically Engineered Mouse Models
All work was done under protocols approved by the UNC Institutional Animal Care and Use Committee (IACUC). GEMMs of strain FVB/n carrying a transgene for Tg(MMTVneu)202Mul/J(MMTV-Neu)(20) and C3(1)SV40 T-antigen (C3(1)-T-antigen or C3-TAg)(21) were bred in-house and observed until the onset of a mammary tumor ∼0.5 cm in any dimension. Tumors derived from BALB/c TP53−/− orthotropic mammary gland transplant line (T11) were passaged in BALB/c wild-type mice by subcutaneous injection of one half million cells resuspended in matrigel into the flank as previously described(22). Mice were randomized into treatment groups and monitored with tumor growth measurements. Tumor volumes were measured by caliper in two dimensions and/or by ultrasound (Vevo 770 ultrasound imaging system (Visualsonics Inc.)). Chemotherapy was started at time zero and repeated weekly for a total of three injections over a twenty-one day period. The mice were further assessed for long-term survival as follows: if after a one week break from treatment a tumor increased in volume more than 1mm in any dimension, then an additional three cycles of therapy were initiated. This continued until either the mouse developed a tumor burden sufficient to warrant euthanasia (2 cm in any dimension or 3 tumors present) or until weight loss totaling 20% of the initial starting body mass was observed or because of any other severe health problems. Orally administered biological inhibitors were given continuously with no dose interruption. In the case of a chemotherapeutic plus an oral inhibitor, the chemotherapy agent was dosed once weekly for 21 days and stopped until progression, while the small molecule inhibitors were dosed continuously.
Compounds
Compounds were obtained from commercial sources: Carboplatin (Hospira, Inc), cyclophosphamide (Hospira, Inc), doxorubicin (Bedford Laboratories), paclitaxel (Ivax Pharmaceuticals, Inc), erlotinib (Genentech, Inc) and lapatinib (GlaxoSmithKline). Oral biological inhibitors (erlotinib and lapatinib) were milled into chow by Research Diets, Inc. while carboplatin and paclitaxel were delivered via intraperitoneal injection.
Treatments
The drug-specific approach to determine schedule and dose is described in Supplemental Table 5. A minimum tumor volume of ∼0.5cm in size was required for randomization into a treatment group (including a control group). Combination treatments were given at the same doses as the individual treatments. Chemotherapy was started at time zero and repeated weekly for over a 14-day (T11/TP53−/−) or 21-day (C3(1)-T-antigen and MMTV-Neu) period.
Pharmacokinetic (PK) Studies
PK studies were performed after administration of paclitaxel (Supplemental Figure 1), erlotinib, and lapatinib (data not shown). For paclitaxel, seventeen transgenic FVB/n mice bearing the MMTV-Neu transgene were administered a single intraperitoneal dose of paclitaxel at 10 mg/kg. Plasma and tumor samples (3 mice used at each time point; 2 mice used for the 48 hour time point) were collected at 0.083, 1, 4, 8, 24, and 48 hours after administration and flash frozen in liquid nitrogen. The samples were analyzed via liquid chromatography/tandem mass spectrometry (LC-MS/MS) as described previously(23). The concentration versus time profiles of paclitaxel in plasma and tumor c plasma following IP administration were 2.1 μg/mL ± 1.5 and 6.3 μg/mL·h respectively. The mean ± SD of paclitaxel Cmax and AUC0-∞ in tumor following IP administration were 3.7 μg/g ± 2.1 and 42.4 μg/g·h respectively.
Response Criteria
Tumor volume was calculated from two-dimensional measurements as (Volume = [(width)2 × length]/2). The percent change in volume at 21 days was used to quantify response, except in the case of the T11/TP53−/− model where its faster growth rate required a 14-day treatment response assessment. Twenty-one day response was chosen as our primary response endpoint based on the fact that most of the untreated animals do not survive much longer than 21 days when starting with a tumor of >0.5 cm. Survival was measured from the first day of drug treatment.
Microarray Analysis
DNA microarray analyses of murine tumors was performed as described in Herschkowitz et al.(12). We used using Agilent 4 × 44,000 feature mouse DNA microarrays and a common reference strategy. For hierarchical clustering analyses, the genes/rows were median centered and clustering of arrays was performed using Cluster v3.0(24) with correlation centered genes and arrays, and centroid-linkage. Array cluster viewing and display was performed using JavaTreeview v1.1.4(25).
Statistical Analyses
(A) Identification of significant differential genes in response to treatments
We performed two unpaired two-class SAM(26) analyses to identify genes that showed differential expressions as following: (i) between carboplatin/paclitaxel treated C3(1)-T-antigen tumors that responded versus those that did not and (ii) between carboplatin/paclitaxel treated C3(1)-T-antigen tumors versus those untreated. The primary SAM analysis to identify tumor response related genes included three responding tumors (shrinkage >20%) versus nine non-responding tumors (growth >20%). The secondary SAM analysis to identify treatment up-regulated or down-regulated genes included seven untreated tumors versus the twelve treated tumors. Two gene lists were obtained with a FDR of 1%: 348 genes (428 probes) showing significantly high expression in the untreated samples (called UNTREATED) and 61 genes (74 probes) showing significantly high expression in the samples from responders (called RESP-HIGH);the identified genes are listed in Supplemental Table 2. Using the Mouse Genome Database(27), these lists were converted to orthologous human genes. In order to refine the list of these candidate genes relevant to human tumors, a hierarchical clustering analysis of these orthologous human gene lists was performed using the 337 tumor samples from Prat et al.(1). From these clusters, we chose a dendrogram node based on the criteria that it would include the largest number of highly expressed genes and have a node correlation of >0.4. Supplemental Figure 2B illustrates the gene set called UNTREATED-HUM that includes 30 unique genes. Supplemental Figure 2D illustrates the gene set called RESP-HUM that includes 12 unique genes.
In the UNC337 human tumors sets, these two gene lists showed “homogeneous” expression patterns, and thus we decided that taking the mean of the genes within each list/dendrogram node was the most appropriate method to assign the signature score for each tumor sample. In brief, an UNTREATED-HUM score was assigned to each test sample by taking the mean of the 26 genes in the list. A RESP-HUM score was assigned to each test sample by taking the mean of the 12 genes in the list. Since we also aimed to compare the performance of these two signatures as well as including published genomic signatures, we standardized the signature scores with a standard deviation equivalent to 1 to bring all the signature scores to the same scale. We applied this same methodology to two independent data sets of neoadjuvant human tumors described below.
(B) Association of the identified signatures with tumor response for neoadjuvant anthracycline/taxane containing chemotherapy regimens
The performance of UNTREATED-HUM and RESP-HUM signatures to predict pathological complete response (pCR) was first tested on 462 patients with HER2 normal tumors in MDACC data set (Hatzis et al.(28), GEO # GSE25066) and validated on 81 patients with HER2 normal tumors in JSE data set (Miyake et al.(29) GEO #GSE32646). Patients on both data sets were treated with neoadjuvant anthracycline-taxane containing regimens. Univariable logistic regression analysis was used to assess the odds ratio and significance of the two signatures to predict pCR. Multivariable logistic regression analysis was used to determine the adjusted odds ratio and significance taking into account for the standard clinical variables measured at baseline and other published genomic signatures as appropriate. The area under the curve (AUC) value was calculated from the Receiver Operating Characteristics analysis of the univariable and multivariable logistic model respectively. The published genomic signatures included the PAM50 intrinsic subtypes(2), Claudin-low predictor(1), and 11-gene proliferation signature(9); we also included signatures developed by Hatzis and colleagues (including Hatzis Sensitivity to endocrine therapy (SET) index, Hatzis signature chemo sensitive RCB-I predict, and Hatzis signature chemo resistance (RCB-III predict)) that were available for the data set(28). Finally, survival outcome data after neoadjuvant treatment was available for the Hatzis et al. data set and Kaplan Meier analysis and log-rank test were used to determine the differential survival estimates of the two signatures to distant relapse free survival.
Results
Sensitivity of GEMMs to chemotherapeutic agents
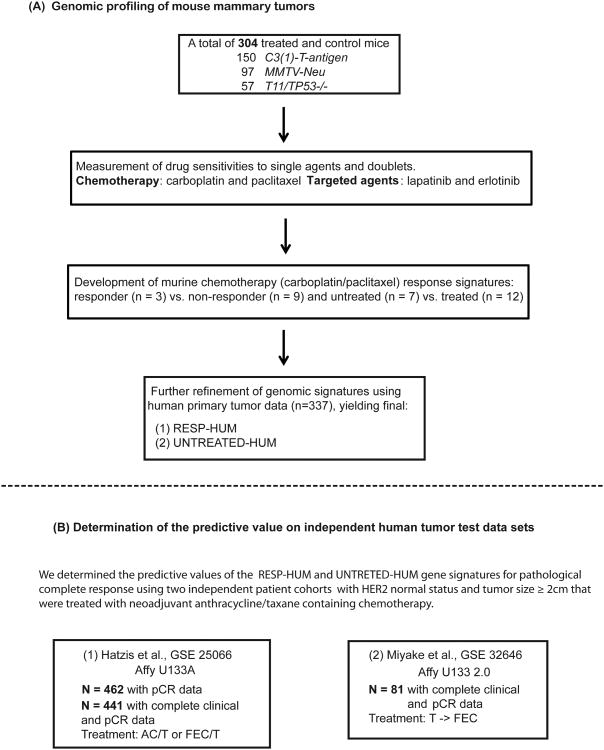
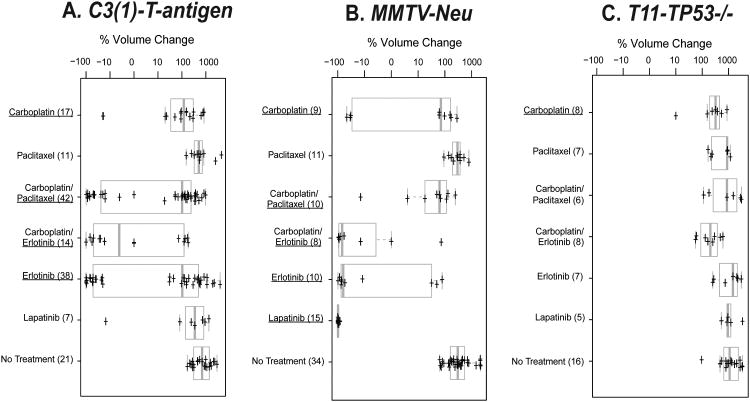
Our ultimate goal was to use GEMMs to develop predictors of therapeutic response for humans. Details of the work flow are outlined in the study design Figure 1. As a first step, we tested three different mammary cancer GEMMs with multiple therapeutics to find a GEMM, and a drug regimen, which gave a range of responses; from this GEMM, we then profiled sensitive and resistant tumors in order to identify a signature associated with response. We first therefore, determined the sensitivity of three distinct GEMMs/OSTs models of human breast cancer subtypes versus two cytotoxic chemotherapeutics and two small molecule kinase inhibitors. The models used were C3(1)-T-antigen, MMTV-Neu, and T11/TP53−/−, with these models chosen based on their similarity in gene expression to Basal-like, Luminal B and Claudin-low human tumor subtypes respectively(12, 18). Tumor volume changes at 21 days (or 14 days in the T11/TP53−/− model), and long-term survival were the primary endpoints. Response at 21 days (or 14 days for T11/TP53−/−) was measured for 304 treated and control mice (150 C3(1)-T-antigen, 97 MMTV-Neu, 57 T11/TP53−/−) with the percent volume change of each model's non-treated controls (i.e. growth rate) shown in Figure 2(bottom rows). Although there was overlap in the average growth rates of tumors from each GEMM, the untreated T11/TP53−/− tumors grew significantly faster than their MMTV-Neu counterparts (p<0.01, Student's t-test), with the C3(1)-T-antigen model exhibiting an intermediate growth rate (Figure 2).
Figure 1. Study design overview.
(A). Drug treatment and genomic profiling of mouse mammary tumors for the development of chemotherapy response signatures. (B). Testing of genomic signatures on two human tumor neoadjuvant treatment data test data sets.
Figure 2. Short-term treatment responses for three mouse models of mammary cancer.
Box and whisker plots are shown as measures of tumor responsiveness. In each case, 2-3 cycles of therapy was administered for all chemotherapeutics (1 dose/week), while in the case of erlotinib and lapatinib, the drug was continuously administered via the chow. Tumor size was measured at baseline and at weekly intervals thereafter. The change in tumor volume over a 21-day treatment period is plotted for A) C3(1)-T-antigen model, B) MMTV-Neu model, and C) T11/TP53−/− model; note that the T11/TP53−/− model is based upon a 14-day treatment period due to its faster growth rate. Drugs that elicited a statistically significant response as assessed by a t-test when compared versus its matched untreated controls are identified by being underlined. The number of animals in each treatment group is indicated in parentheses.
With the growth kinetics of these models established, we next tested two chemotherapeutics that are widely used to treat many solid epithelial human cancers, namely paclitaxel and carboplatin. Although the standard of care for most breast cancer patients is doxorubicin/cyclophosphamide with or without a taxane (i.e. AC-T)(30), platinum agents (carboplatin/cisplatin) are also gaining in use(31), and thus are relevant to breast cancers, especially triple-negative breast cancers (TNBC). As a single agent, carboplatin elicited a modest but significant responses in all three models, while paclitaxel alone elicited no response; however, systemic and tumor drug delivery was confirmed for paclitaxel (Supplemental Figure 1).
Next we tested the commonly used chemotherapy doublet of carboplatin/paclitaxel (CT). A varied response profile was seen for the CT combination where the combination demonstrated no activity in the T11/TP53−/− model, and only modest activity in the MMTV-Neu model. Importantly, in the C3(1)-T-antigen model, a clear bimodal response was observed to the CT combination: ∼2/3 of the tumors showed little response and ∼1/3 showed near complete regression (Figure 2A). This finding is in accord with the observation that human Basal-like tumors exhibit a ∼30-40% complete pathological response rate (pCR) to taxane containing neoadjuvant regimens, while the other 60-70% show residual disease and a worse overall survival (1, 5, 10).
Sensitivity to targeted agents
Two classes of biologically targeted agents are used in patients with breast cancer: agents blocking estrogen and progesterone receptor (ER/PR) signaling (e.g. tamoxifen or aromatase inhibitors) and drugs targeting HER2 (e.g. trastuzumab and lapatinib). Given that none of our GEMMs were ER+ or PR+(12), we chose to focus on the HER2/EGFR family of kinases by using the small molecule inhibitor lapatinib (which targets HER2/ERBB2 primarily(27)), and the EGFR inhibitor erlotinib(32). In the MMTV-Neu model, erlotinib and lapatinib were both highly effective, with lapatinib causing near 100% regression in all MMTV-Neu tumors. Conversely, neither erlotinib nor lapatinib were effective at reducing the growth rate of the T11/TP53−/− tumors. Lapatinib was similarly ineffective in the C3(1)-T-antigen tumors, but as was the case for the CT doublet, erlotinib showed potent activity in a subset (∼40%) of treated mice. These data show that HER2/EGFR inhibitors exhibit potent activity in the Neu/ERBB2/HER2-driven model as expected, and provide further evidence for at least two subtypes of C3(1)-T-antigen tumors with regard to therapeutic sensitivity.
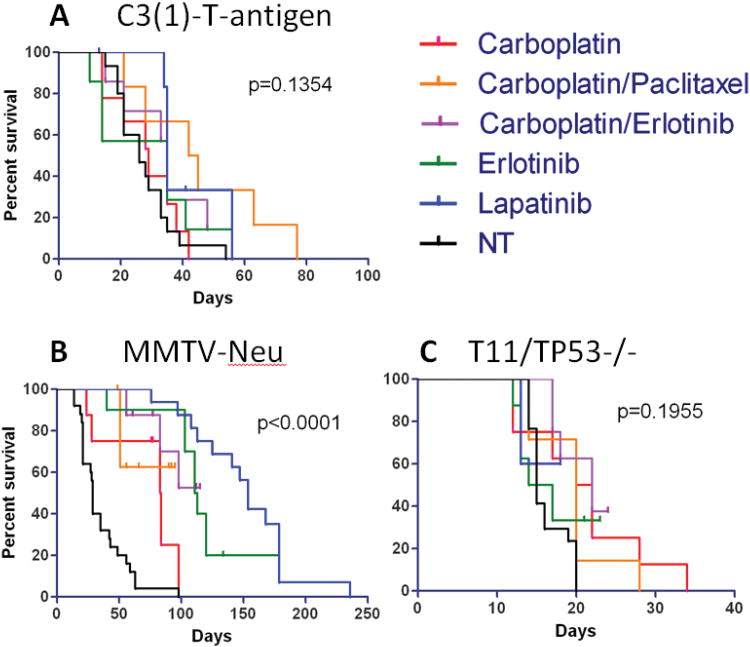
We also assessed the effects of anti-cancer therapies on the overall survival of tumor-bearing mice. Baseline survival for the MMTV-Neu (29 days) and C3(1)-T-antigen models (33 days) was similar in the absence of therapy, while the T11/TP53−/− animals showed significantly shorter median survival (15 days) (Figure 3). In the MMTV-Neu model, single-agent lapatinib (and to some extent erlotinib) greatly extended lifespan from a median of 29 days to 154 days (Figure 3B). Conversely, no single or combination regimen was able to extend survival in the C3(1)-T-antigen or T11/TP53−/− models.
Figure 3. Long term survival results for three mouse models of mammary cancer.
Kaplan-Meier analyses for overall survival of tumor bearing mice was performed. A) C3(1)-T-antigen, B) MMTV-Neu, and C) T11/TP53−/− results for chemotherapeutic treatments, targeted agents, and combinations. A log-rank test was performed to determine significance of all treatment groups and is shown.
Development of murine chemotherapy response signatures
A heterogeneous response to CT was seen in the C3(1)-T-antigen tumors that ranged from progressive disease to complete response (Figure 2A). We sought to explore these findings and develop a genomic predictor of this response using this GEMM by performing RNA expression profiling of treated vs. untreated tumors. For these experiments, we treated C3(1)-T-antigen tumors with carboplatin/paclitaxel for two or three cycles and measured response (n=12), and then harvested the tumor for molecular analysis. In addition, an independent set of seven untreated tumors was used as the non-treated controls (Supplemental Table 1).
Significance Analysis of Microarray (SAM)(26) was used to derive two sets of differentially expressed genes by (A) comparing those mice that responded to treatment (n=3) versus those that did not (n=9), and by (B) comparing the untreated (n=7) versus treated tumors (n=12)(Supplemental Tables 1 and 2). When testing untreated versus treated tumors at a FDR of 1%, this analysis identified 428 probes corresponding to 348 mouse genes that were more highly expressed in untreated tumors (called UNTREATED gene list, Supplemental Table 2A); a Gene Ontology analysis of the UNTREATED list identified multiple significant terms including “cellular macromolecule metabolic process”, “nucleic acid metabolic process”, “regulation of macromolecule biosynthetic process”, “chromosome organization”, “DNA metabolic process” and “cell cycle”. We applied a modules/signatures analysis to the untreated versus treated tumors where we examined if 302 previously defined expression signatures(33) varied with treatment (Supplemental Table 3). This modules/signatures analysis showed that multiple signatures of fibroblasts/extracellular matrix, and signatures of the Claudin-low phenotype(1, 18) were more highly expressed after treatment, with this last result recapitulating findings observed in post-chemotherapy treated human tumors(34). Multiple signatures decreased after treatment including one of proliferation and one of HER1-RAS-pathway activation. These data show that CT treatment induced expression of genes associated with Claudin-low/mesenchymal phenotype, and reduced cellular proliferation.
When the cohort of treated tumors was subdivided into responders versus non-responders at a FDR of 1%, a list of 74 differentially expressed probes corresponding to 61 mouse genes was obtained (Supplemental Table 2B). These genes were more highly expressed in the mice that responded to treatment and the list was named RESP-HIGH. A gene ontology analysis of the RESP-HIGH list revealed the presence of no significant GO terms after Bonferroni or Benjamini corrections. We also applied the 302 signatures analysis above on the responder versus non-responder sample set, and only a small number of proliferation signatures were more highly expressed in non-responders.
Human testing of the murine chemotherapy response signatures
Next, the murine 348 gene UNTREATED and 74 gene RESP-HIGH lists were converted into human lists using gene orthology, and both lists were then further refined using hierarchical cluster analyses of 337 human breast tumors from Prat et al.(1) (Supplemental Figure2). This mouse-to-human filtering was necessary because a homogenous gene list from a cell line, or murine experiment, when applied to human primary tumors, will typically fragment into multiple signatures/modules when using in vivo human data(35). We observed this type of gene list heterogeneity here, and thus, from these cluster analyses we chose a single dendrogram node that contained the highest homogenously expressed gene set observed within this human primary tumor data set, and for each gene list separately. This gave a set of 30 genes from the UNTREATED list that we call UNTREATED-HUM, and 12 genes from the RESP-HIGH list that we call RESP-HUM (Supplemental Figure 2 B and D); it should be noted that we did not test all possible dendrogram nodes, but instead limited our analyses to a single node from each cluster analysis. These two refined gene lists were also analyzed for GO terms with the UNTREATED-HUM list enriched for the terms ‘cell cycle’, ‘M phase’, ‘nuclear division’ and ‘mitosis’, and we also noted that 12/30 entries were ATP-binding proteins. The RESP-HIGH was not enriched for any GO term.
We next tested both humanized gene lists for their ability to predict distant relapse-free survival (DRFS), and most importantly, pathological complete response (pCR) using a completely independent set of human breast cancer patients treated with neoadjuvant chemotherapy. For both clinical endpoints, we used the Hatzis et al. data set (See Figure 1), which is a combined data set of patients who were treated with a taxane and anthracycline-containing neoadjuvant chemotherapy regimen(28). We first stratified patients into low-medium-high (tertiles) groups based upon their rank-ordered mean expression values for the RESP-HUM and UNTREATED-HUM signature and then tested these stratifications for their ability to predict DRFS. These analyses showed that the RESP-HUM (p < 0.001) and UNTREATED-HUM (p = 0.003) signatures were able to predict DRFS, as was pCR vs. not, intrinsic subtype, and an 11-gene proliferation signature (Supplemental Figure 3). In multivariable analyses, however, neither of these murine signatures added prognostic information beyond that conveyed by the PAM50 11-gene proliferation signature(9) (data not shown).
We then tested the humanized gene lists for their ability to predict pathological complete response (pCR), which is the most relevant endpoint for these chemotherapy response-based signatures. Within this patient set, 462 patients had pathological response data; 91 patients achieved a pCR and 371 did not (20% overall pCR rate). The pCR rates varied according to intrinsic subtype as follows: Basal-like (n=129, 40% pCR), Claudin-low (n=70, 23% pCR), HER2-Enriched (n=27, 19% pCR), Luminal A (n=140, 3% pCR), Luminal B (n=68, 16% pCR), and Normal-like (n=28 total, 14% pCR). To determine the possible significance of our two response signatures on this test set of human patients, the mean expression values for each gene list was calculated and the distribution of values between pCR patients versus not pCR patients determined. As shown in Table 1, when all 473 patients were considered, the UNTREATED-HUM signature was significantly correlated with pCR (p<0.001)and the RESP-HUM signature was trending toward significance (p=0.051). As we further stratified patients into the five, and even six intrinsic subtypes the UNTREATED-HUM signature continued to maintain significance. Interestingly, the RESP-HUM signature predicted pCR more strongly in the Normal-like and Claudin-low subtypes while the UNTREATED-HUM signature better tracked response within the Basal-like subtype (Table 1). Lastly, the triple-negative breast cancer distinction is a highly clinically relevant group because these patients are not candidates for the current targeted therapies in the breast clinic(10, 11); within this group, the UNTREATED-HUM signature was also a significant predictor (p=0.003).
Table 1. Pathological Complete Response (pCR) rates across different patient subsets for the RESP-HUM and UNTREATED-HUM signatures.
The P-value and Area-Under-the-Curve (AUC) columns indicate whether the RESP-HUM or UNTREATED-HUM signature (as a continuous variable from low to high expression) was associated with response (italics) within that patient set/subset.
| RESP-HUM | UNTREATED-HUM | |||||||
|---|---|---|---|---|---|---|---|---|
| pCR | RD | P-value | AUC | Odds ratio | P-value | AUC | Odds ratio | |
| All patients | 91(19.7%) | 371(80.3%) | 0.051 | 0.586 | 0.788(0.61-0.99) | <0.001 | 0.752 | 2.72(2.08-3.63) |
| ER-negative only | 62(33.3%) | 124(66.7%) | 0.821 | <0.001 | 0.683 | 2.09(1.44-3.11) | ||
| ER positive only | 29(10.5%) | 246(89.5%) | 0.405 | <0.001 | 0.747 | 2.64(1.67-4.31) | ||
| PAM50 (5 intrinsic subtypes) | ||||||||
| Basal-like | 62(35.8%) | 111(64.2%) | 0.707 | 0.001 | 0.649 | 2.05(1.33-3.24) | ||
| HER2-enriched | 5(17.9%) | 23(82.1%) | 0.430 | 0.076 | ||||
| Luminal A | 4(2.8%) | 141(97.2%) | 0.208 | 0.172 | ||||
| Luminal B | 12(16.7%) | 60(83.3%) | 0.638 | 0.079 | ||||
| Normal-like | 8(18.2%) | 36(81.8%) | 0.009 | 0.837 | 4.47(1.69-16.9) | 0.274 | ||
| PAM50 + Claudin-low (6 intrinsic subtypes) | ||||||||
| Basal-like | 51(39.5%) | 78(60.5%) | 0.356 | 0.001 | 0.680 | 2.33(1.44-3.93) | ||
| Claudin-low | 16(22.9%) | 54(77.1%) | 0.054 | 0.660 | 1.55(1-2.47) | 0.474 | ||
| HER2-enriched | 5(18.5%) | 22(81.5%) | 0.426 | 0.086 | ||||
| Luminal A | 4(2.9%) | 136(97.1%) | 0.223 | 0.169 | ||||
| Luminal B | 11(16.2%) | 57(83.8%) | 0.976 | 0.272 | ||||
| Normal-like | 4(14.3%) | 24(85.7%) | 0.052 | 0.137 | ||||
| Triple-Negative only | 56(33.5%) | 111(66.5%) | 0.651 | 0.003 | 0.651 | 1.8(1.24-2.68) | ||
To more rigorously test the predictive significance of these new expression signatures, multivariable analysis using logistic regression was performed that included the common clinical variables, the intrinsic subtypes, the RESP-HUM and UNTREATED-HUM signatures, and three predictive genomic signatures identified by Haztis et al. (Table 2). For these analyses, we used the subset of patients that had pCR/response data, survival data, and who were treated with an anthracycline and taxane chemotherapy regimen (n=441). As shown in Table 2, multiple biomarkers were predictive in univariate analyses, but only the UNTREATED-HUM, Basal-like, Normal-like, and one of the Haztis et al. chemotherapy predictor signatures (i.e. RCB-III/resistance) were found significant in both the univariate and multivariate tests. To further assess the strength of the predictive powers of these genomic signatures, each was used to calculate an Area Under the Curve (AUC) for pCR, both alone (univariate AUC) and in the multivariate model (Table 2). The UNTREAT-HUM signature provided a good univariate AUC, and the multivariate model provided improvement with a high AUC (0.879). When the three Hatzis et al. signatures were removed from the multivariate analysis, most of the variables that were significant in the initial MVA remained significant, and the overall model continued to show a high AUC (0.82) (data not shown). Lastly, an additional test data set of anthracycline and taxane treated human patients was tested, which represents 81 patients treated neoadjuvantly from Japan(29); similar predictive results were seen for the UNTREAT-HUM signature, which was again a significant predictor in both the univariate and multivariate analyses (Supplemental Table 4). These data show that the UNTREATED-HUM signature (and possibly the RESP-HUM) provided predictive information for pCR beyond 1) the commonly used clinical variables, 2) breast cancer subtype, and 3) other genomic signatures derived from one of the data sets tested here.
Table 2. Univariate and Multivariate Analysis for pCR using clinical and genomic features including the RESP-HUM and UNTREATED-HUM signatures on the Hatzis et al. data set.
Univariate and Multivariate analyses were performed using all Hatzis et al. patients who received anthracycline and taxane chemotherapy only, and who had overall survival data (n=441).
| # pts* | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| p-value | odds ratio | AUC | p-value | odds ratio | AUC | ||
|
| |||||||
| UNTREATED-HUM | 441 | <0.001 | 2.57 (1.96-3.45) | 0.740 | 0.013 | 2.3 (1.21-4.52) | 0.879 |
|
| |||||||
| RESP-HUM | 441 | 0.073 | 0.796 (0.618-1.02) | 0.583 | 0.058 | 1.45 (0.99-2.15) | |
|
| |||||||
| PAM50 proliferation | 441 | <0.001 | 2.57 (1.9-3.56) | 0.730 | 0.917 | 0.96 (0.443-2.11) | |
|
| |||||||
| ER | |||||||
| Negative | 175(40%) | 1 | 0.562 | 1 | |||
| Positive | 266(60%) | <0.001 | 0.234 (0.139-0.385) | 0.992 | 0.99 (0.391-2.52) | ||
|
| |||||||
| PR | |||||||
| Negative | 227(51%) | 1 | 0.467 | ||||
| Positive | 214(49%) | <0.001 | 0.30 (0.176-0.51) | 0.683 | 0.83 (0.363-1.97) | ||
|
| |||||||
| Clinical T Stage | |||||||
| 1 | 27(6%) | 1 | 0.571 | 1 | |||
| 2 | 226(51%) | 0.364 | 0.652 (0.269-1.75) | 0.902 | 0.92 (0.261-3.39) | ||
| 3 | 126(29%) | 0.746 | 0.854 (0.34-2.36) | 0.844 | 0.87 (0.236-3.38) | ||
| 4 | 62(14%) | 0.054 | 0.306 (0.0885-1.02) | 0.195 | 0.35 (0.071-1.69) | ||
|
| |||||||
| Clinical Grade | |||||||
| 1 | 28(6%) | 1 | 0.481 | 1 | |||
| 2 | 170(39%) | 0.498 | 2.05 (0.38-38.1) | 0.967 | 1.05 (0.13-23.7) | ||
| 3 | 243(55%) | 0.019 | 11.1 (2.3-201) | 0.574 | 2.02 (0.235-46.6) | ||
|
| |||||||
| PAM50 | |||||||
| LumA | 141(32%) | 1 | 0.633 | 1 | |||
| Basal | 167(38%) | <0.001 | 18.2 (7.21-61.5) | 0.026 | 5.76 (1.3-29.4) | ||
| Her2 | 24(5%) | 0.010 | 6.85 (1.51-31.1) | 0.161 | 3.55 (0.59-21.7) | ||
| LumB | 67(15%) | 0.003 | 6.01 (1.92-22.6) | 0.400 | 1.92 (0.438-9.49) | ||
| Normal | 42(10%) | 0.001 | 8.06 (2.39-31.7) | 0.002 | 10 (2.34-47.6) | ||
|
| |||||||
| Hatzis signature SET index | |||||||
| 1 | 386(88%) | 1 | 0.136 | 1 | |||
| 2 | 36(8%) | 0.091 | 0.353 (0.0835-1.02) | 0.729 | 1.34 (0.223-6.49) | ||
| 3 | 19(4%) | 0.302 | 0.457 (0.0715-1.64) | 0.953 | 0.94 (0.105-6.17) | ||
|
| |||||||
| Hatzis signature chemo sensitive (RCB-I predict) | |||||||
| 1 | 296(67%) | 1 | 0.553 | 1 | |||
| 2 | 145(33%) | <0.001 | 2.97 (1.82-4.84) | 0.127 | 1.76 (0.855-3.67) | ||
|
| |||||||
| Hatzis signature chemo resistance (RCB-III predict or 3 year survival) | |||||||
| 1 | 197(45%) | 1 | 0.603 | 1 | |||
| 2 | 244(55%) | <0.001 | 0.089 (0.0449-0.166) | <0.001 | 0.129 (0.0565-0.277) | ||
The number of patients with clinical ER status, PR status, T stage, grade and pCR status available.
Discussion
As new agents for breast cancers are developed, validated pre-clinical models for assessing these agents' activity alone and in combination with approved therapies are needed. In this study, we chose genomically credentialed GEMM representatives for three human breast tumor subtypes (Basal-like, Luminal B and Claudin-low) as our pre-clinical models. While using single representatives of different tumor subtypes does not allow for the identification of subtype-specific effects, we believe this approach does make future predictions of therapeutic efficacy more robust by including results from a biologically diverse group of tumor-bearing individuals.
For therapeutic efficacy, each GEMM was treated with identical regimens and for most drugs, variable responses were seen. Our findings show that the MMTV-Neu tumors were the most responsive in general, with multiple agents being able to achieve complete tumor regression, especially the HER2 targeted agent lapatinib. Next in sensitivities was the Basal-like C3(1)-T-antigen model, which was generally more resistant than the MMTV-Neu model, but in some cases complete responses were documented (CT and carboplatin/erlotinib); interestingly, a heterogeneity of responses was common in this GEMM (Figure 2A), suggesting that two or more sub-classes of tumors may be present. Importantly, a similar heterogeneous response pattern is seen within human Basal-like patients when treated with comparable agents where many patients achieve a pCR and have good overall survival, but the majority show residual disease and worse outcomes (Supplemental Figure 3C and see (5, 36)). Lastly, the Claudin-low T11/TP53−/− model was the most resistant with only small responses seen in this model.
We ultimately chose to focus our analysis on expression-signatures associated with chemotherapy treatment of one of our GEMMs and response for two main reasons. First, we reasoned transcripts highly expressed in sensitive murine tumors (i.e. the RESP-HIGH list) might also be highly expressed in sensitive human tumors; although this list was predictive in human tumors, it was not obvious from gene ontology analysis what molecular characteristics drive this biology, and this list was not significant when accounting for other variables (MVA p-value = 0.058). Second, in a tumor treated in vivo, we reasoned chemotherapy might deplete the most sensitive cells and their characteristic transcripts. Therefore, the collection of transcripts that were highly expressed in untreated cells and depleted with treatment (i.e. the UNTREATED list) similarly seemed rational for testing in humans. Specifically, an analysis showed the 26-gene UNTREAT-HUM signature (Supplemental Figure 3) was a significant predictor of response and may also provide mechanistic insight. This 26-gene list suggests that the cells actually undergoing DNA synthesis and mitosis (i.e. in S/G2/M-phase) are more sensitive to cytotoxic agents than cells in other parts of the cell cycle (G0 or G1), which is a concept dating back to the 1960's (reviewed in (37)). It is important to note that this list added independent information above and beyond strict assessments of proliferation (e.g. an 11-gene proliferation signature that contains Ki-67), suggesting this list may better capture specific features of the cell cycle (e.g. length of time spent in S/G2/M) associated with sensitivity to carboplatin/paclitaxel. The UNTREAT-HUM list is in fact a biologically rich list that contains at least two different sets of genes/proteins that physically form a multi-protein complex, namely SMC2 and SMC4, and MCM4 and MCM6. In addition, this list has two different E2F family members (E2F3 and E2F8), for which a poor prognostic signature has already been linked to E2F3(38). These data also suggest that no single gene/protein is likely to be a robust biomarker of chemosensitivity because a multitude of genes, each involved in different aspects of the cell cycle, were collectively identified as being predictive of response. These new expression signatures were derived from murine models that, despite their specific chemoresponses not being a mirror of their human counterparts (i.e. paclitaxel), added a significant predictive component to the multivariate model that at least equaled the ability of those tested signatures that were derived directly from this human tumor data set.
In terms of human biomarker advances, we made progress using the C3(1)-Tag GEMM. As shown in Tables 1 and 2, the UNTREATED-HUM signature was predictive of response to a multi-agent neoadjuvant chemotherapy regimen, not only across all HER2-normal human breast cancer patients but also within the clinically relevant triple-negative subset, as well as the more biologically relevant Basal-like subset. Interestingly, this UNTREATED-HUM signature was also able to predict pCR even when accounting for intrinsic subtype, the common clinical variables, and two other genomic signatures specifically designed to predict neoadjuvant response (Table 2). Although the murine treatment and human treatment involved the use of different chemotherapeutics, both species studies used paclitaxel and at least one DNA damaging agent (carboplatin in mice and doxorubicin/epirubicin in humans). Overall, a multivariate model that contained the UNTREAT-HUM, the intrinsic subtypes, and the common clinical variables showed an AUC of 0.82, which may be sufficiently predictive to be of value for routine clinical use.
We were surprised to find that the results from mice treated with single agent paclitaxel did not mimic the effectiveness of this drug in human breast cancer patients. Delivery of higher therapeutic doses of paclitaxel to the mice (i.e. doses closer to those received by human patients)may have proven more efficacious; however, our chosen formulation of paclitaxel contained chremaphor and ethanol in amounts that precluded higher dosing. Another caveat to our studies is that these two GEMM-derived signatures were both predictive and prognostic; however, it must be noted that it is often difficult, if not impossible, to disentangle these two features. For example, both ER and HER2 in breast cancer are prognostic (they predict outcomes in the absence of therapy) and they are predictive (ER predicts hormone therapy benefit and HER2 predicts trastuzumab benefit) and thus, our new signatures are showing dual properties similar to those seen for the existing breast cancer biomarkers. Much additional validation work is needed before these two murine-derived signatures could be used to guide patient treatment. However, this study has laid the groundwork of a general strategy for evaluating new drugs, combinations, and schedules using GEMMs and has shown it is possible to use mice as a tool to identify a biomarker that may be of predictive value for human cancer patients.
Supplementary Material
Statement of Translational Relevance.
The identification of new predictive biomarkers is a difficult task which often requires treatment of human patients with experimental drugs. Genetically Engineered Mouse Models (GEMM) hold the promise of providing a preclinical arena for this early drug testing and possible biomarker discovery. A signature of chemotherapy response was identified through the use of credentialed GEMM of mammary cancer. This signature was then shown to predict neoadjuvant response in human breast cancer patients. If validated in additional studies, this signature may show clinical value for selecting patients who will benefit from neoadjuvant anthracycline/taxane regimens, and shows the value of drug testing in mice as a means for identifying new biomarkers.
Acknowledgments
This work was supported by funds from the NCI Breast SPORE program (P50-CA58223-09A1), by RO1-CA138255 and RO1-CA148761, by the Breast Cancer Research Foundation and by a generous gift to the UNC Mouse Phase I Unit.
Footnotes
Disclosure of Potential Conflicts of Interest: C.M Perou serves as a board member and has an ownership interest in University Genomics and Bioclassifier LLC. C.M.P and M.C.U. Cheang are listed as inventors on pending patents on the PAM50 intellectual property.
References
- 1.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker JS, Mullins M, Cheang MC, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hugh J, Hanson J, Cheang MC, et al. Breast Cancer Subtypes and Response to Docetaxel in Node-Positive Breast Cancer: Use of an Immunohistochemical Definition in the BCIRG 001 Trial. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 6.Martin M, Romero A, Cheang MC, et al. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1461-y. [DOI] [PubMed] [Google Scholar]
- 7.Gluck S, Ross JS, Royce M, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/− trastuzumab. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1412-7. [DOI] [PubMed] [Google Scholar]
- 8.Dunbier AK, Anderson H, Ghazoui Z, et al. Association between breast cancer subtypes and response to neoadjuvant anastrozole. Steroids. 2011 doi: 10.1016/j.steroids.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–32. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 11.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular Characterization of Basal-Like and Non-Basal-Like Triple-Negative Breast Cancers. Oncologist. 2013 doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herschkowitz JI, Zhao W, Zhang M, et al. Breast Cancer Special Feature: Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–44. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 14.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–54. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 15.Roberts PJ, Usary J, Darr D, et al. Combined PI3K/mTOR and MEK Inhibition Provides Broad Anti-Tumor Activity in Faithful Murine Cancer Models. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nardella C, Lunardi A, Patnaik A, Cantley LC, Pandolfi PP. The APL paradigm and the “co-clinical trial” project. Cancer discovery. 2011;1:108–16. doi: 10.1158/2159-8290.CD-11-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci U S A. 1994;91:11236–40. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green JE, Shibata MA, Yoshidome K, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19:1020–7. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 22.Jerry DJ, Kittrell FS, Kuperwasser C, et al. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene. 2000;19:1052–8. doi: 10.1038/sj.onc.1203270. [DOI] [PubMed] [Google Scholar]
- 23.Hou W, Watters JW, McLeod HL. Simple and rapid docetaxel assay in plasma by protein precipitation and high-performance liquid chromatography-tandem mass spectrometry. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2004;804:263–7. doi: 10.1016/j.jchromb.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spector NL, Xia W, Burris H, 3rd, et al. Study of the biologic effects of lapatinib, a reversible inhibitor of ErbB1 and ErbB2 tyrosine kinases, on tumor growth and survival pathways in patients with advanced malignancies. J Clin Oncol. 2005;23:2502–12. doi: 10.1200/JCO.2005.12.157. [DOI] [PubMed] [Google Scholar]
- 28.Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–81. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyake T, Nakayama T, Naoi Y, et al. GSTP1 expression predicts poor pathological complete response to neoadjuvant chemotherapy in ER-negative breast cancer. Cancer science. 2012;103:913–20. doi: 10.1111/j.1349-7006.2012.02231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 Update of Recommendations for the Use of Tumor Markers in Breast Cancer. Journal of Clinical Oncology. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 31.Tutt AN, Lord CJ, McCabe N, et al. Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol. 2005;70:139–48. doi: 10.1101/sqb.2005.70.012. [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo M, Siu LL, Nemunaitis J, et al. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19:3267–79. doi: 10.1200/JCO.2001.19.13.3267. [DOI] [PubMed] [Google Scholar]
- 33.Fan C, Prat A, Parker JS, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoadley KA, Weigman VJ, Fan C, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 37.Norton L. Implications of kinetic heterogeneity in clinical oncology. Seminars in oncology. 1985;12:231–49. [PubMed] [Google Scholar]
- 38.Huang E, Ishida S, Pittman J, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–30. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.