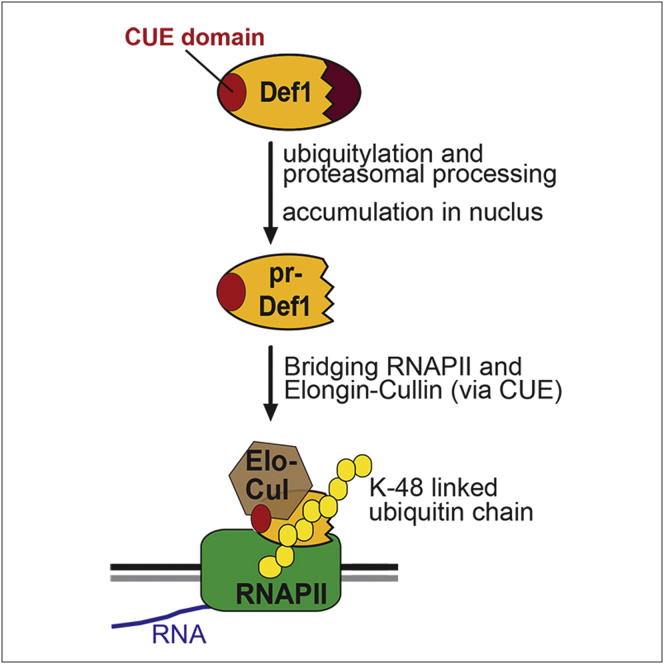
Summary
DNA damage triggers polyubiquitylation and degradation of the largest subunit of RNA polymerase II (RNAPII), a “mechanism of last resort” employed during transcription stress. In yeast, this process is dependent on Def1 through a previously unresolved mechanism. Here, we report that Def1 becomes activated through ubiquitylation- and proteasome-dependent processing. Def1 processing results in the removal of a domain promoting cytoplasmic localization, resulting in nuclear accumulation of the clipped protein. Nuclear Def1 then binds RNAPII, utilizing a ubiquitin-binding domain to recruit the Elongin-Cullin E3 ligase complex via a ubiquitin-homology domain in the Ela1 protein. This facilitates polyubiquitylation of Rpb1, triggering its proteasome-mediated degradation. Together, these results outline the multistep mechanism of Rpb1 polyubiquitylation triggered by transcription stress and uncover the key role played by Def1 as a facilitator of Elongin-Cullin ubiquitin ligase function.
Graphical Abstract
Highlights
-
•
Def1 protein is proteolytically processed in response to transcription stress
-
•
Processing is ubiquitin and proteasome dependent
-
•
Processed Def1 (pr-Def1) accumulates in the nucleus, where it binds RNAPII
-
•
Using its CUE domain, pr-Def1 then recruits Elongin-Cullin via Ela1’s UbH domain
DNA damage induces proteasome-dependent proteolytic processing of Def1, releasing an activated protein fragment, which highlights how irreversible proteasome-mediated cleavage of a protein can act as a posttranslational modification in this signaling pathway.
Introduction
Substantial research effort is presently focused on understanding the cellular processes maintaining genome integrity and allowing faithful replication after DNA damage (Branzei and Foiani, 2010). Although such processes are of utmost importance for the long-term survival and fitness of cells and organisms, the key immediate response of cells suffering genotoxic insult is arguably to maintain gene expression. Indeed, without continued transcription, cells cannot proceed through the cell cycle, and even nondividing cells will perish. Like DNA replication, transcription is severely affected by DNA damage, with various DNA lesions resulting in RNAPII stalling, pausing, arrest, and/or backtracking (hereafter collectively referred to as transcription stress). It is therefore not surprising that cells have evolved a number of mechanisms to ensure that transcription can rapidly resume upon DNA damage (Svejstrup, 2010). One important mechanism is transcription-coupled nucleotide excision repair (TC-NER), which removes transcription-blocking lesions so that RNAPII can continue (Gaillard and Aguilera, 2013). In budding yeast, TC-NER is dependent on Rad26, the homolog of human Cockayne syndrome B (van Gool et al., 1994). Intriguingly, Rad26 interacts with another protein, Def1 (Woudstra et al., 2002). The phenotypes of cells lacking DEF1 indicate a role for this factor in the DNA damage response, but Def1 is not involved in repair. Instead, it is required for a “mechanism of last resort.” During this alternative process, the largest subunit of RNAPII, Rpb1, becomes ubiquitylated and degraded, which results in disassembly of the large RNAPII complex and allows the lesion to be dealt with by other means (Wilson et al., 2013). Although it was originally identified as a response to DNA damage (Bregman et al., 1996; Beaudenon et al., 1999), it is now known that Rpb1 ubiquitylation and degradation occurs under a number of conditions that result in transcription stress (Hobson et al., 2012; Somesh et al., 2005; Sigurdsson et al., 2010). Obviously, Rpb1 ubiquitylation must be tightly regulated to specifically target the small subset of elongating polymerases that cannot otherwise be salvaged, as any unnecessary Rpb1 degradation will severely affect general gene expression and cell survival. Results obtained over the last decade have provided insight into the mechanisms by which Rpb1 is ubiquitylated and degraded (reviewed in Wilson et al., 2013), but although it is required for Rpb1 ubiquitylation, the precise role of Def1 has remained elusive.
Degradation of Rpb1 occurs by the addition of lysine 48-linked polyubiquitin chains, disassembly of the chromatin-associated RNAPII elongation complex, and proteasomal degradation (Wilson et al., 2013). Notably, ubiquitylation of Rpb1 is a two-step process, involving distinct ubiquitin ligases (E3s) (Harreman et al., 2009). Briefly, stalled RNAPII in budding yeast is targeted by a HECT domain E3, Rsp5 (Beaudenon et al., 1999), which cooperates with Uba1 (E1, ubiquitin-activating enzyme) and Ubc5 (E2, ubiquitin-conjugating enzyme) to add a single ubiquitin moiety, probably at more than one site on Rpb1 (Somesh et al., 2007; Harreman et al., 2009). A second E3 ligase, a complex containing the Elc1, Ela1, Cul3, and Rbx1 proteins (“Elongin-Cullin complex”), then takes over and adds lysine 48-linked ubiquitin chains to the premonoubiquitylated Rpb1 (Harreman et al., 2009; Ribar et al., 2006, 2007). Following polyubiquitylation, a ubiquitin-specific ATPase, Cdc48, then delivers Rpb1 from the RNAPII elongation complex to the proteasome (Verma et al., 2011). The mechanism of Rpb1 ubiquitylation is highly conserved, with the process in mammals being catalyzed by NEDD4 and the Elongin ABC-Cullin 5 complex, homologs of the budding yeast E3 proteins (Huibregtse et al., 1997; Anindya et al., 2007; Yasukawa et al., 2008; Harreman et al., 2009).
As mentioned above, polyubiquitylation and degradation of Rpb1 also requires the Def1 protein, both in vivo (Woudstra et al., 2002) and in vitro (Reid and Svejstrup, 2004). Def1 is an unusual protein, consisting largely of domains of low complexity, with a predicted N-terminal CUE (ubiquitin-binding) domain as the only notable feature (Ponting, 2002) (Figure 1A). In this study, we show that ubiquitylation and degradation of Rpb1 encompasses an unusually wide variety of ubiquitin-related mechanisms centered on the Def1 protein. Together, these mechanisms facilitate nuclear accumulation of a proteasome-processed version of Def1, which then acts as a bridging factor between RNAPII and the Elongin-Cullin complex, triggering Rpb1 polyubiquitylation.
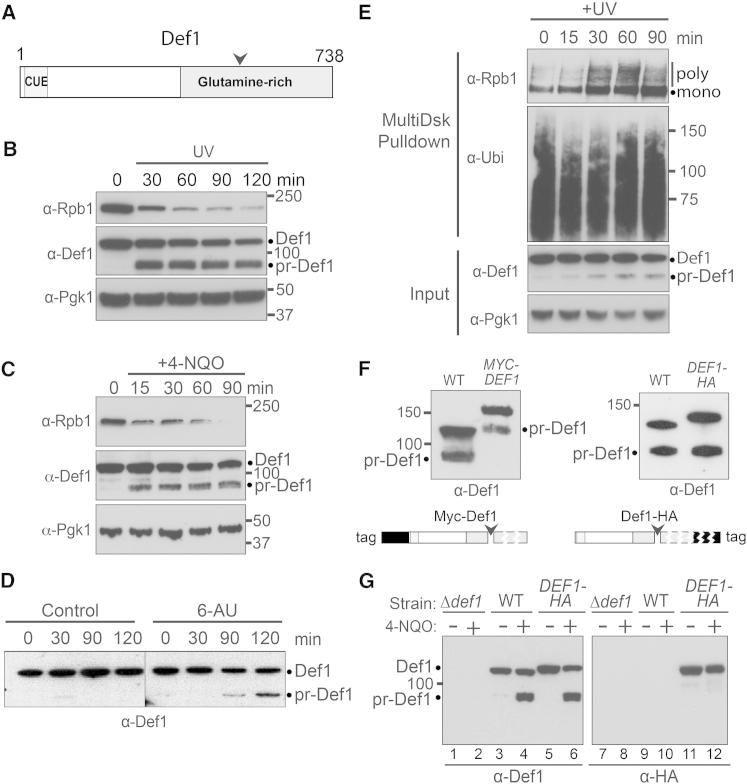
Figure 1.
Def1 Is Processed in Response to Transcription Stress
(A) Schematic representation of Def1 indicating the CUE domain, area of processing (arrow), and glutamine rich region.
(B) Western blot of cell extracts at the indicated times after UV irradiation, using antibodies against Rpb1, Def1, and Pgk1 (loading control).
(C) As in (B), but after incubation with 4-nitroquinoline 1-oxide (4-NQO).
(D) Def1 processing induced by 6-azauracil (6-AU). Superfluous lanes between lanes 4 and 5 were removed.
(E) As in (B), but ubiquitylated proteins isolated using MultiDsk pull-down (Wilson et al., 2012).
(F) Western blot probed using anti-Def1 antibody, showing WT, or N-terminally 9xMyc-tagged Def1 (left), or WT, or C-terminally 6xHA-tagged Def1 (right), after incubation with 4-NQO for 1 hr.
(G) Western blots of identical samples from Δdef1, WT, or DEF1-3xHA, probed with anti-Def1 antibodies (left panel), or anti-HA antibodies (right).
Results
Def1 Is Processed in Response to DNA Damage and Other Transcription Stress
Under conditions that generate transcription-impeding DNA damage, such as UV irradiation and treatment with 4-nitroquinoline 1-oxide (4-NQO), we noticed the appearance of a faster migrating protein, hereafter called processed Def1 (pr-Def1), which specifically cross-reacted with a Def1 antibody (Figures 1B, 1C, and 1G, left, and Figure S1A available online). The faster migrating Def1 form was also observed upon treatment with the transcription elongation inhibitor, 6-azauracil (6-AU) (Figure 1D), suggesting that it is generated in response to transcription stress, rather than as a damage response per se. pr-Def1 formation correlated with polyubiquitylation (Figure 1E) and degradation (Figures 1B and 1C) of Rpb1.
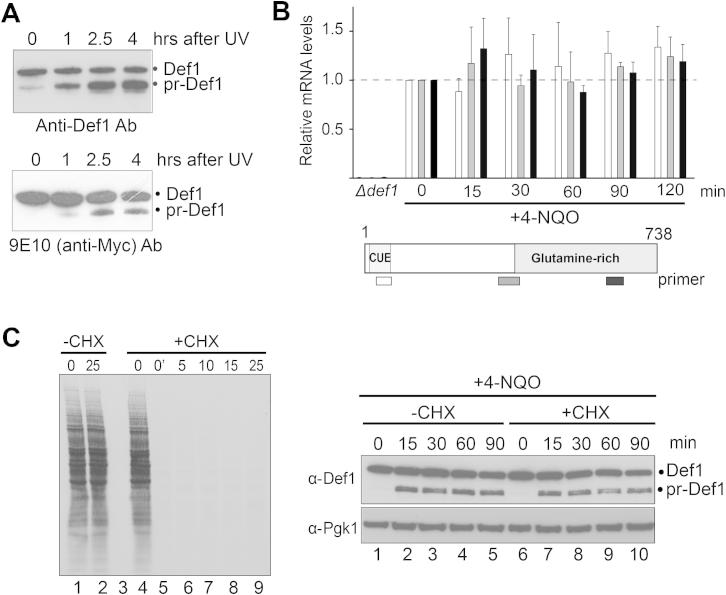
Figure S1.
pr-Def1 Is Not Formed from Alternate Transcription or Translation, Related to Figure 1
(A) pr-Def1 may in some cases appear to be generated without equivalent disappearance of full-length Def1. This is due to the anti-Def1 antibody exhibiting an unusual and pronounced preference for pr-Def1 over the full-length protein: only a small fraction of Def1 is actually processed, making it difficult to appreciate its depletion. To illustrate this point, the same samples from cells expressing N-terminally Myc-tagged Def1 were subjected to western blotting and probed with either polyclonal anti-Def1 antibodies (upper panel), or anti-Myc antibodies (lower panel). Note the strong preference for pr-Def1 exhibited by the anti-Def1 antibody (upper), which gives the false impression that pr-Def1 may not have been generated from full-length protein, which is only slightly depleted at the same time-points. However, weak, concomitant depletion of full-length Def1 can be appreciated by the use of anti-Myc antibody (lower). Because introducing N-terminal tags in DEF1 in all of the many strains used in this study would be impractical, Def1 processing was detected with the polyclonal Def1 antibody unless otherwise stated.
(B) Total RNA was isolated, reverse transcribed, and relative DEF1 mRNA levels were measured by qPCR, in the relevant areas of the transcript, before and after DNA damage. Quantification was normalized to β-actin mRNA levels in each sample, then against untreated conditions (time0). Primers hybridized to the start (white), middle (gray), and end (black) of the DEF1 mRNA. Error bars correspond to the standard deviation across three biological replicates. None of the mRNA areas, in particular the region encoding Def1’s C terminus, are significantly up- or downregulated in response to DNA damage and Def1 processing.
(C) Def1 processing occurs posttranslationally. (left) Autoradiogram of SDS-PAGE gel, showing cellular proteins, radioactively labeled in vivo. Protein synthesis was inhibited by the addition of 25 μg/ml cycloheximide (CHX) to midlog cells at time0, and new protein synthesis measured by incorporation of radioactive label in proteins (10 min S35-methionine pulses initiated at the times indicated after cycloheximide inhibition). Time-point 0’ was transferred to grow in radioactive methionine immediately after the addition of cycloheximide. Note that cycloheximide completely, and immediately, inhibits new protein synthesis. (Right) Western blot showing that Def1 processing occurs even in the presence of effective cycloheximide inhibition of new protein synthesis (compare lanes 7-10 with 2-5). Cycloheximide was added 5 minutes prior to the addition of 4-NQO.
Three distinct processes might theoretically explain the generation of pr-Def1. First, it might be caused by changes in the production of DEF1 mRNA (e.g., alternative start- or stop-site usage or alternative splicing). Second, it might be caused by changes in mRNA translation. Third, it might occur at a posttranslational stage, via partial proteolysis. No significant changes in the production of DEF1 mRNA were detectable in response to DNA damage (Figure S1B), and pr-Def1 was still generated in response to DNA damage in cells that no longer produced new Def1 protein (Figure S1C). This indicates that Def1 processing is a posttranslational event, a conclusion that was further supported by all subsequent experiments.
To investigate whether it is the N or C terminus of Def1 that is missing from pr-Def1, yeast strains expressing tagged versions of Def1 were generated. When N-terminally tagged Def1 was detected using the polyclonal anti-Def1 antibody, the processed version was larger than wild-type (WT) pr-Def1 (Figure 1F, left), whereas no mobility shift of pr-Def1 was observed when a HA-tag was positioned at the (degraded) C terminus (Figure 1F, right). Indeed, Def1, but not pr-Def1, could be detected with anti-HA antibody (Figure 1G, compare lanes 6 and 12). Together, these experiments demonstrate that pr-Def1 lacks the C-terminal region of full-length Def1. This short fragment has persistently evaded detection, suggesting that it is degraded during processing, or immediately thereafter.
Processing of Def1 Produces an Activated Form
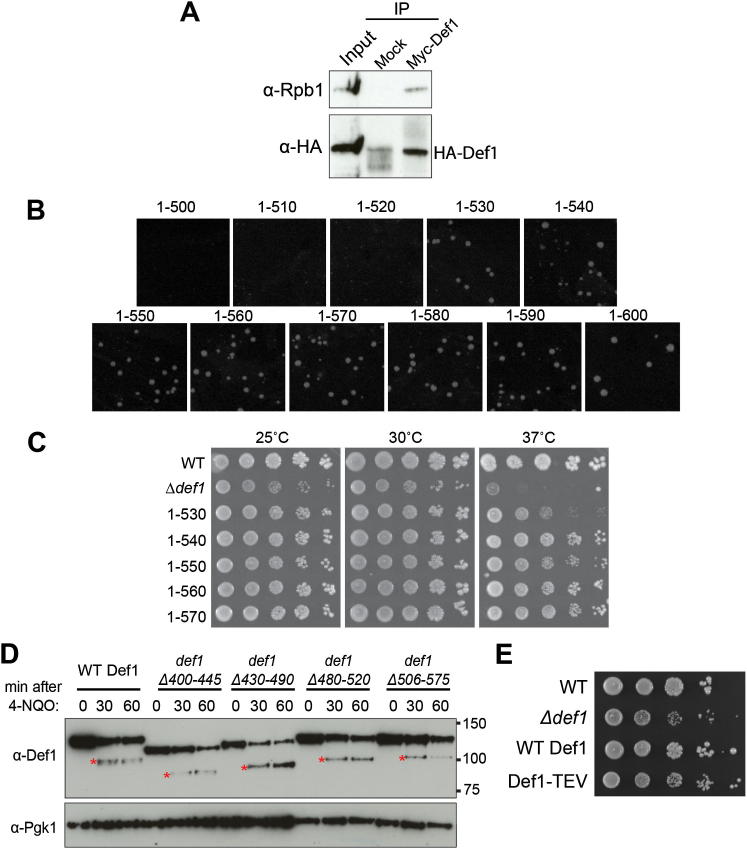
If pr-Def1 plays a causative role in the response to transcription stress, it might be expected that artificial generation of pr-Def1-like protein species would result in Def1 activation and possibly lead to Rpb1 ubiquitylation and degradation. To address this possibility, the endogenous DEF1 gene was shortened by genomic recombination. Small C-terminal deletions (generating Def11–600 and Def11–700) had little or no effect on cell growth, whereas large deletions (generating Def11–100 to Def11–300) resulted in a slow-growth phenotype, similar to that of a complete DEF1 deletion (Figure 2A). Strikingly, haploid strains expressing Def11–400 and Def11–500 (that would theoretically encode protein fragments of a size similar to that of pr-Def1) could not be generated despite repeated attempts, but a diploid strain expressing both WT Def1 and Def11–500 was viable. However, upon sporulation and tetrad dissection of this diploid into haploid spores, only two of four spores were viable; these contained the WT version of DEF1 (Figure 2B), indicating that the Def11–500 fragment is toxic. Def1 self-associates (Figure S2A) and might be either a dimer or a multimeric protein. This could explain why the expression of genetically truncated versions of Def1 is not lethal when a WT copy of DEF1 is also present. In apparent agreement with this idea, other proteolytically processed proteins have been reported to be dimeric (Rape and Jentsch, 2002).
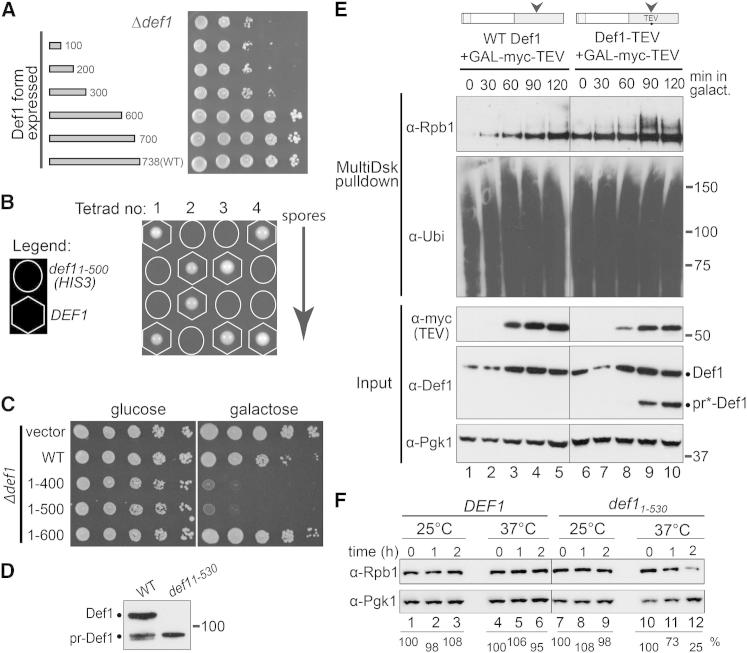
Figure 2.
Processing of Def1 Produces an Activated Form
(A) Dilution growth series of cells expressing Def1 fragments.
(B) A ratio of 2:2 segregation of spores from four different tetrads from a heterozygous DEF1/def11–500::HIS3 diploid, with spore genotype indicated. Viable HIS3 colonies were never observed.
(C) Dilution series of Δdef1 yeast cells (with GAL-driven plasmid indicated on left), grown on glucose or galactose. Overexpression of WT DEF1 from a 2 μm plasmid is slightly detrimental to Δdef1 cells.
(D) Western blot showing Def1 from 4-NQO-treated WT cells, and def11–530, respectively.
(E) Western blot of extract from cells expressing Myc-tagged TEV protease, as well as WT Def1, or Def1 containing a TEV protease cleavage site. Blots of ubiquitylated proteins shown in two upper panels, with extract blots shown below. Lanes 1 and 2 were underloaded, giving the false impression that growth in galactose results in higher Rpb1 monoubiquitylation. Superfluous lanes between lanes 5 and 6 were removed.
(F) Western blots of extracts from WT and def11–530 cells grown at permissive (25°C) or restrictive temperature (37°C). Rpb1 signal was quantitated relative to the Pgk1 control, and the values at time 0 set to 100; other values are expressed relative to that. Superfluous lanes between lanes 6 and 7 were removed.
See also Figure S2.
Figure S2.
Mapping the Site of Def1 Processing, Related to Figure 2
(A) Western blot showing Def1-HA and Rpb1 retained on immobilized Myc-Def1 or mock beads after immuno-precipitation in extracts derived from cells expressing both Myc-Def1 and Def1-HA.
(B) Yeast minimal media plates showing viability of transformed cells. Cells were transformed with equal amounts of PCR product for genomic truncation of Def1 at 10 amino acid intervals between 500-600 amino acids, as indicated. This simple procedure for comparing viability after transformation of similar amounts of slightly different DNA constructs is highly reliable and reproducible when performed on the same transformation-competent, yeast culture.
(C) Dilution series of yeast cells of the genotype indicated on the left, grown at 25°C, 30°C and 37°C.
(D) Def1 internal deletions result in the processing site shifting to another position. Western blots of extracts from WT Def1 and the indicated Def1 internal deletion strains, before and after treatment with 4-NQO for 30 and 60 min. The exact site of processing is not fixed, as the size of pr-Def1 changes (red asterisks). The mobility of the full-length Def1 proteins vary more than would be expected based on the difference in amino acids number between them. This is because glutamine-rich regions - deleted in some, but not others - contribute disproportionally to SDS-PAGE mobility.
(E) Def1-TEV does not exhibit growth defect. Δdef1 cells are slow-growing (second line of cell dilution), but not when expressing WT, or Def1-TEV (lower lines). Def1-TEV can also still be normally processed in response to transcription stress (data not shown).
Expression of Def11–400 and Def11–500 proteins from an inducible promoter also inhibited cell growth, whereas expression of WT Def1 and Def11–600 had little or no effect (Figure 2C, galactose). Together, these data indicate that expression of genetically generated, shorter forms of Def1 that mimic pr-Def1 is strongly detrimental to the cell.
Repeated attempts to map the site of damage-induced Def1 processing by mass spectrometric analysis proved unsuccessful. As an alternative approach to better define processing, we therefore used genetic recombination to generate C-terminal DEF1 deletions in the equivalent of ten amino acid intervals between Def1 residues 500 and 600. DEF1 genes expressing proteins between 500 and 520 amino acids in length were lethal, whereas those generating Def1 proteins of 530 amino acids or more were viable (Figure S2B). Interestingly, a strain expressing Def11–530 (def1–530) was temperature-sensitive (Figure S2C). This version of the Def1 protein migrated slightly slower than pr-Def1 (Figure 2D), suggesting that the natural processing site lies somewhere in the region prior to amino acid 530. We attempted to disrupt Def1 processing by making internal deletions in this region, but these merely resulted in shifting the site to another position (Figure S2D). This suggests that Def1 processing does not occur at a specific, short amino acid motif, as has also been found for other processed proteins (Piwko and Jentsch, 2006).
To investigate whether Def1 clipping in itself can trigger RNAPII ubiquitylation even in the absence of transcription stress, we now constructed a yeast strain expressing Def1 with a tobacco etch virus (TEV) protease cleavage site inserted between amino acids 522 and 523, around the predicted region of normal processing. Such modification had no obvious effect on DEF1 function (Figure S2E). Upon induction of GAL-regulated Myc-TEV protease (Figure 2E, third panel from bottom), a fraction of Def1 was indeed cleaved, but only in the strain with the inserted TEV site (pr∗-Def1; Figure 2E, second panel from bottom). Importantly, a fraction of Rpb1 reproducibly became polyubiquitylated concurrently with the emergence of pr∗-Def1 (Figure 2E, top, compare lanes 9 and 10 with 4 and 5, respectively). These results indicate that Def1 processing can in itself trigger detectable Rpb1 polyubiquitylation, independently of DNA damage. Not surprisingly, the effect of TEV-induced cleavage was less pronounced than after DNA damage and did not result in appreciable degradation of the Rpb1 protein, presumably because TEV-mediated Def1 nicking does not induce other pathways that are activated by transcription stress.
Next, the temperature-sensitive def11–530 strain was used to further investigate the effect of Def1 processing. We surmised that because def11–530 is slow growing at 37°C, the restrictive temperature likely activates this version of Def1. Overall protein levels and that of the Pgk1 control protein remained largely unchanged at 37°C. In contrast, Rpb1 protein levels decreased with time in def11–530, but not WT cells (Figure 2F, compare lanes 11 and 12 with 5 and 6), again suggesting that the processed version of Def1 triggers RNAPII ubiquitylation and degradation.
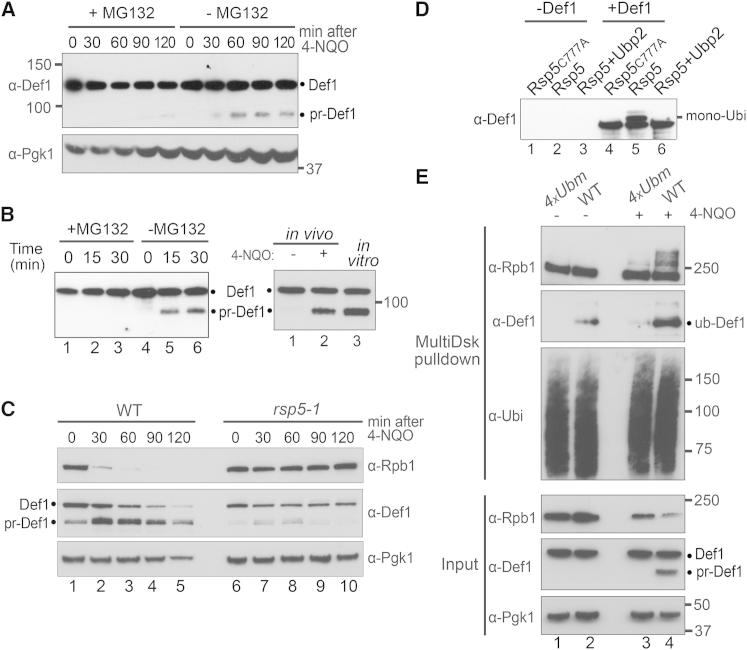
Def1 Is Processed in a Ubiquitin- and Proteasome-Dependent Manner
Ubiquitin- and proteasome-dependent protein processing, whereby a part of a protein is degraded to release a biologically active fragment, has been described for other proteins (reviewed in Rape and Jentsch, 2002). In agreement with a similar role for the proteasome in the processing of Def1, treatment of yeast cells that are sensitized to proteasome inhibitor (Collins et al., 2010) with MG132 inhibited the formation of pr-Def1 (Figure 3A).
Figure 3.
Proteasome- and Ubiquitin-Dependent Def1 Processing
(A) Western blot of extracts from cells treated with (or without) MG132 prior to incubation with 4-NQO for the indicated times.
(B) Left: Def1 processing reconstituted using pure Def1 and 26S proteasome (see Figure S3A), and its sensitivity to MG132. Right: western blot comparing pr-Def1 with Def1 processed by pure proteasome.
(C) Western blot of extract from WT and rsp5-1 cells grown at 37°C for 2 hr before addition of 4-NQO for the indicated times. Lanes 4 and 5 are underloaded, giving the false impression that Def1 disappears at those time points. Normal processing was observed in rsp5-1 at the permissive temperature (data not shown).
(D) Western blots of Def1 ubiquitylation, reconstituted using highly purified proteins (Def1, ubiquitin, Uba1, Ubc5, Rsp5, and Ubp2; see Figure S3A).
(E) Western blots of ubiquitylated proteins (upper three panels) and extracts (input; lower three panels) from UV-irradiated WT and Def1 ubiquitylation site mutant (4xUbm) cells, respectively.
See also Figure S3.
In order to test whether the proteasome can directly process Def1, we reconstituted proteasomal processing in vitro using Def1 and 26S proteasome purified to virtual homogeneity (Figure S3A). Strikingly, in this simple assay correct proteasome-dependent Def1 processing was observed, with full-length Def1 processed to the shortened version (Figure 3B, lanes 5 and 6). As expected, this was greatly reduced in the presence of proteasome inhibitor (lanes 2 and 3). Significantly, pr-Def1 was not proteolyzed further: smaller proteolysis products were not observed, and the overall Def1 protein amount remained constant. Moreover, the size of processed Def1 generated by purified proteasome in vitro was similar to that of pr-Def1 generated by transcription stress in vivo (Figure 3B, right panel, compare lanes 2 and 3).
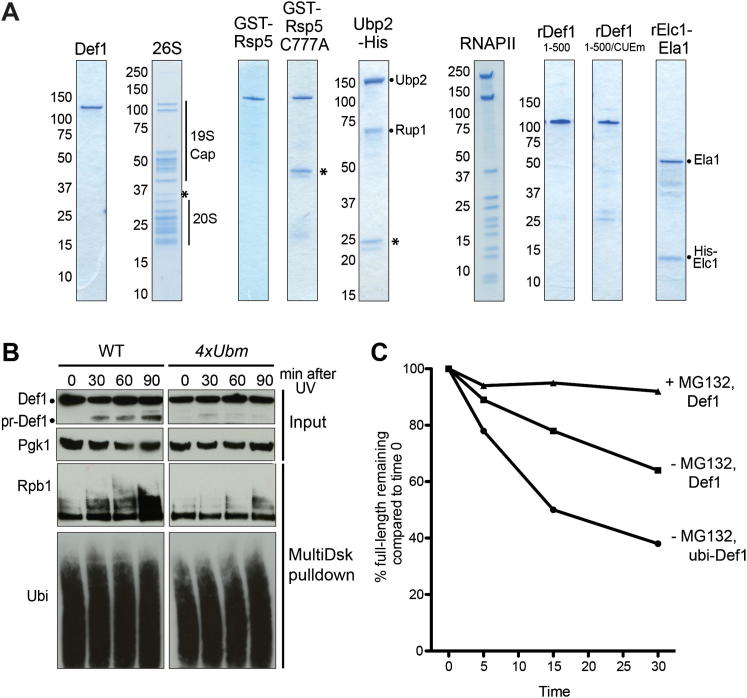
Figure S3.
Ubiquitylation Stimulates Def1 Processing, Related to Figures 3, 5, and 6
(A) Purified proteins used throughout this study. Asterisks indicate minor contaminants.
(B) Time course of damage response in WT and Def1 ubiquitylation site mutant (4xUbm). Note that some residual Def1 truncation and Rpb1 poly-ubiquitylation can still be observed in the mutant.
(C) Proteolysis of full-length, ubiquitylated Def1 (ubi-Def1), or un-modified Def1 (Def1) over time in response to incubation with proteasome in the absence (-) or presence (+) of MG132. Def1 was ubiquitylated in vitro prior to the assay (as in Figure 3D). Western blot quantification of the disappearance of the full-length band, normalized to 100% at starting time 0.
Not all proteasome-mediated protein degradation requires ubiquitylation of the target protein (Jariel-Encontre et al., 2008). However, in other reported cases of proteasomal protein processing, ubiquitylation of the target protein has been implicated, at least in vivo (Palombella et al., 1994; Hoppe et al., 2000; Pan et al., 2006). Moreover, monoubiquitylation of NF-κB p105 is sufficient for its proteasomal processing (Kravtsova-Ivantsiv et al., 2009), and we recently found that Def1 becomes monoubiquitylated in response to DNA damage (Wilson et al., 2012). The ubiquitin ligase Rsp5 binds and monoubiquitylates Rpb1 (Huibregtse et al., 1997; Harreman et al., 2009). Furthermore, this E3 has been implicated in proteasomal processing of other yeast proteins (Hoppe et al., 2000). We therefore investigated whether Rsp5 might be involved in Def1 ubiquitylation and proteasomal processing. Indeed, in a strain carrying a temperature-sensitive rsp5 allele, Def1 failed to be efficiently processed in response to 4-NQO at the restrictive temperature (Figure 3C, middle panel on right), whereas Def1 processing and accompanying Rpb1 degradation was observed in the WT control strain (Figure 3C, left). The remaining proteasomal processing of Def1 observed in this experiment might be explained by residual Rsp5 activity at the restrictive temperature, or by ubiquitylation merely playing a stimulatory role in Def1 processing.
To ascertain whether Rsp5 can directly ubiquitylate Def1, we reconstituted the process with highly purified proteins. Def1 ubiquitylation was indeed observed upon incubation with Uba1 (E1), Ubc5 (E2), and ubiquitin, but only when active Rsp5 was included (Figure 3D, lane 5). Ubiquitylation was not observed when the active site of Rsp5 was mutated (lane 4), or in the presence of ubiquitin protease Ubp2 (lane 6), which associates with Rsp5 in vivo (Kee et al., 2005).
Using Def1 that had been ubiquitylated in vitro, two pairs of adjacent, potential ubiquitylation sites were now identified by mass spectrometry. Simultaneous mutation of these four sites (K281R, K288R, K328R, K329R; creating 4xUbm) resulted in strongly reduced Def1 ubiquitylation and perturbed Rpb1 polyubiquitylation, (Figure 3E, upper panels, compare lanes 3 and 4) and also affected Def1 processing and Rpb1 degradation (Figure 3E, lower panels), in response to DNA damage. Interestingly, 4xUbm did not always completely abrogate Def1 processing (see Figure S3B). This suggests that other potential ubiquitylation sites still remain available, or that proteasomal processing of Def1 is only accelerated by ubiquitylation, but not absolutely dependent on it, in apparent agreement with the results obtained in rsp5-1 (Figure 3C). Furthermore, ubiquitylation of Def1 was not absolutely required for proteasomal processing in the reconstituted in vitro system, although it could accelerate the process (Figure S3C).
Taken together, these results strongly indicate that processing of Def1 involves Rsp5-mediated ubiquitylation and partial degradation by the proteasome.
pr-Def1 Accumulates in the Nucleus
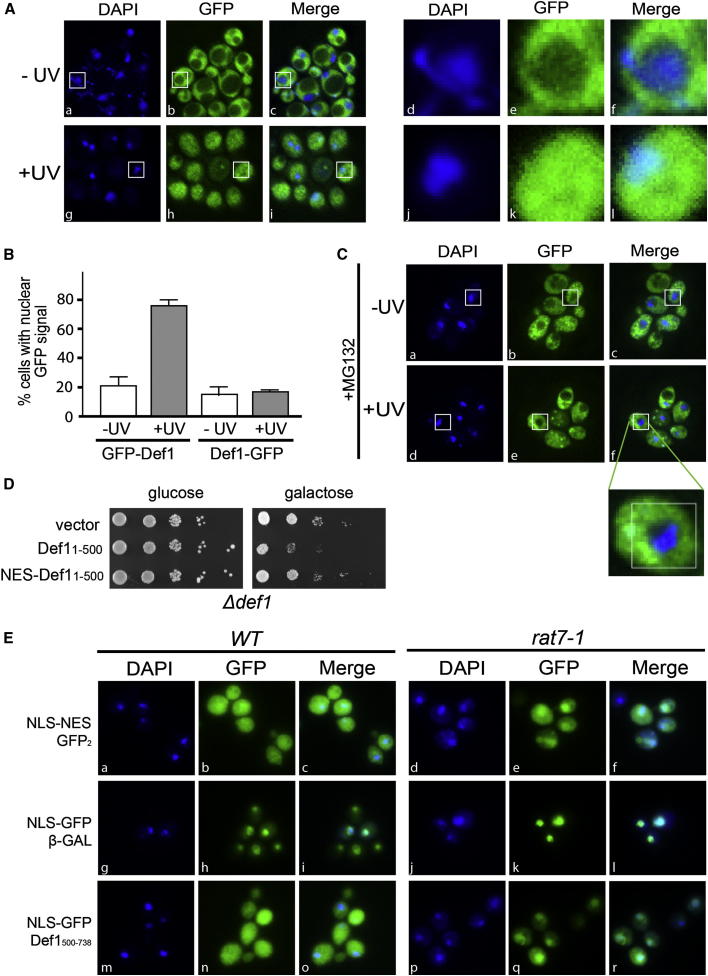
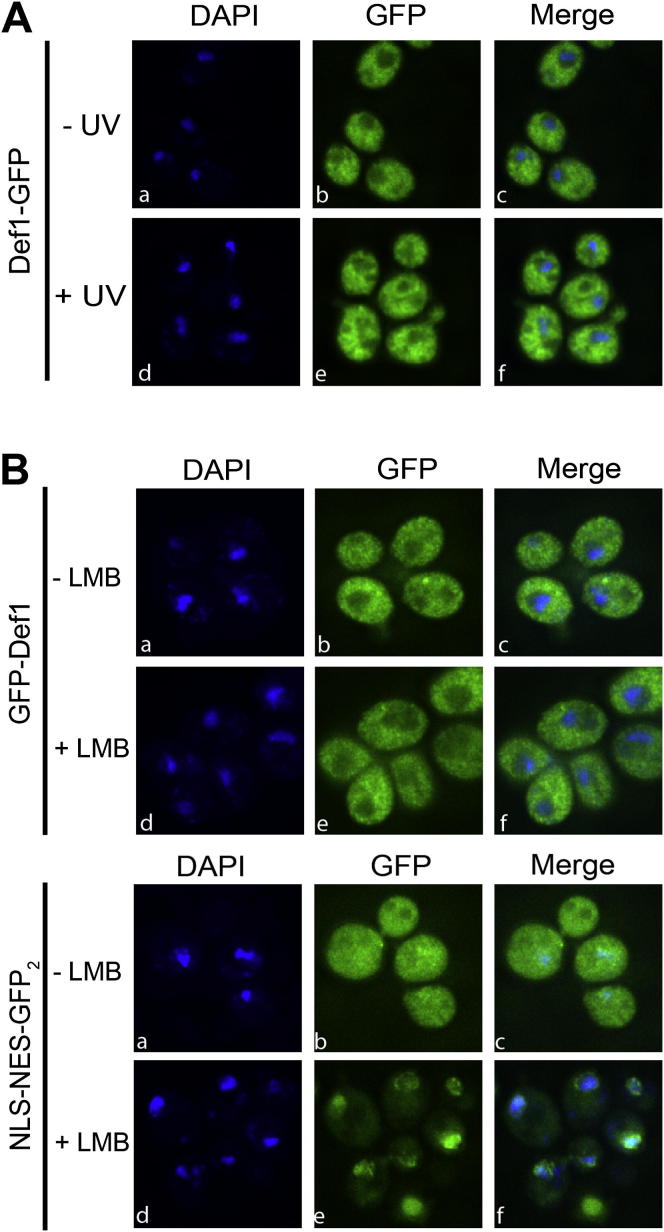
The experiments above indicate that proteasomal processing activates Def1, but how pr-Def1 promotes polyubiquitylation of Rpb1 remained unclear. We began to characterize this connection by examining the cellular localization of Def1. Previous studies indicated that Def1 is cytoplasmic (Huh et al., 2003; Tkach et al., 2012), despite being required for Rpb1 polyubiquitylation in chromatin (Woudstra et al., 2002; Verma et al., 2011). However, the earlier localization studies relied on a C-terminal GFP-tag on Def1, which would not visualize pr-Def1. By using an N-terminal GFP-tag on Def1 instead, we ensured that both forms of Def1 could be detected. Without UV treatment, GFP-Def1 was cytoplasmic, as expected (Figure 4A, panels c and f). After treating yeast cells with UV light, however, the GFP signal spread across the entire cell, including the DAPI-stained nucleus (Figure 4A, panels i and l). In contrast, C-terminally tagged Def1-GFP did not show relocalization after treatment with UV (Figures 4B and S4A).
Figure 4.
pr-Def1 Accumulates in the Nucleus
(A) Subcellular localization of N-terminally GFP-tagged Def1 after UV irradiation. Nucleus is marked by DAPI staining. Left: a typical field of cells. Right: enlargement of the yeast cell indicated by white box in panel on left. The number and size of vacuoles differed from experiment to experiment, but it had no influence on the nuclear accumulation observed.
(B) Quantification of damage-induced, nuclear accumulation of N-terminally tagged GFP-Def1 and C-terminally tagged Def1-GFP, respectively. Error bars indicate SD of three biological replicates, counting 200–300 cells for each condition.
(C) Localization of GFP-Def1 in cells incubated with MG132 for 1 hr prior to UV irradiation. Image below shows enlargement of yeast cell indicated by white box.
(D) Dilution series of Δdef1 cells (carrying the GAL-driven CEN plasmid indicated on the left) grown on glucose or galactose.
(E) Localization of GFP in fixed cells expressing NLS-NES-GFP2 (panels a–f), NLS-GFP-β-galactosidase (panels g–l), or NLS-GFP-Def1500–738 (panels m–r), in WT cells (left), or rat7-1 cells (right).
See also Figure S4.
Figure S4.
pr-Def1 Changes Subcellular Localization Independently of Crm1, Related to Figure 4
(A) In contrast to N-terminally tagged Def1 (Figure 4A, panel i), nuclear accumulation after UV irradiation is not observed when the Def1 tag is at the C terminus (panel f in A). This figure complements the data in Figure 4B.
(B) Leptomycin B (LMB) inhibition of Crm1-dependent nuclear export (in the hypersensitive crm1 strain MNY8 [Neville and Rosbash, 1999]) has no effect on the cytosolic sub-cellular localization of GFP-Def1 (upper panels), but results in nuclear accumulation of a control protein whose cytoplasmic localization depends on a classical NES (lower panels).
Def1 was also GFP-tagged in the MG132-sensitized strain (Collins et al., 2010). When these cells were treated with proteasome inhibitor to block pr-Def1 formation, Def1 remained in the cytosol even after DNA damage (compare Figure 4C, panel f, with 4A, panel i), further indicating that only pr-Def1 accumulates in the nucleus.
It was now investigated whether altering steady-state subcellular localization might suppress the previously described toxicity of GAL-expressed Def11-500. Indeed, even though Def11–500 was overexpressed, appending a prototypical leucine-rich nuclear export signal (NES) (Gerace, 1995) to its N terminus reproducibly decreased toxicity (Figure 4D), suggesting that the detrimental effect of Def11–500 expression is at least partly due to nuclear localization.
We next examined whether the C-terminal region of Def1 might harbor a domain, which directly or indirectly leads to cytoplasmic localization by either inhibiting nuclear import or promoting nuclear export (Figure 4E). When a GFP control protein containing both a nuclear localization signal (NLS) and an NES was expressed, the fusion-protein was detected in the whole cell (Figure 4E, panels a–c) (Taura et al., 1998). In contrast, if the GFP construct contained an NLS, but lacked the NES and instead carried a C-terminal control domain (β-galactosidase), it was exclusively nuclear (Figure 4E, panels g–i) (Lee et al., 1996). Importantly, when this control domain was replaced with the Def1 C terminus (amino acids 500–738), the resulting GFP fusion protein was found in both the nucleus and the cytoplasm (Figure 4E, panels m–o), showing that the C terminus of Def1 promotes cytoplasmic localization.
Although the best-characterized mechanism of nuclear export occurs via NESs recognized by the Crm1 receptor (Hutten and Kehlenbach, 2007), 13 additional members of the karyopherin family of nuclear transport receptors are found in Saccharomyces cerevisiae (Ström and Weis, 2001). For the vast majority of these receptors, the motif targeting the cargo for transport has not been defined. However, export from the nucleus is typically dependent on components of the nuclear pore, such as Rat7 (Nup159) (Brykailo et al., 2007 and references therein). The Def1 C-terminal domain does not contain a classical NES motif, and as expected, Def1 indeed remained cytoplasmic when Crm1 was inhibited (Figure S4B). Importantly, however, the C-terminal domain of Def1 failed to bring about cytoplasmic localization and thus accumulated in the nucleus in the nuclear pore mutant rat7-1 (Figure 4E, panels p–r), opening the possibility that this Def1 domain works by promoting export from the nucleus rather than by inhibiting nuclear import.
Together, these data indicate that Def1 is normally efficiently exported from the nucleus so that it is mainly cytoplasmic at steady state. Upon DNA damage or other transcriptional stress, however, a region directing cytoplasmic localization is removed by proteolysis, allowing pr-Def1 to accumulate in the nucleus, facilitating Rpb1 ubiquitylation/degradation.
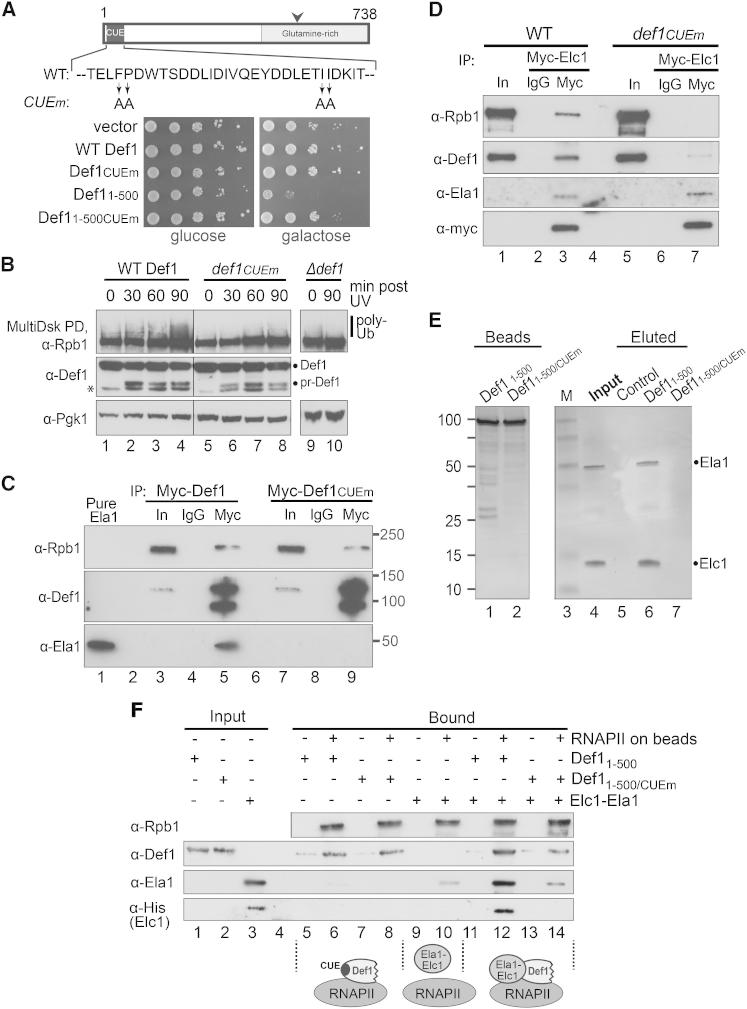
The CUE Domain Is Critical for Def1 Function
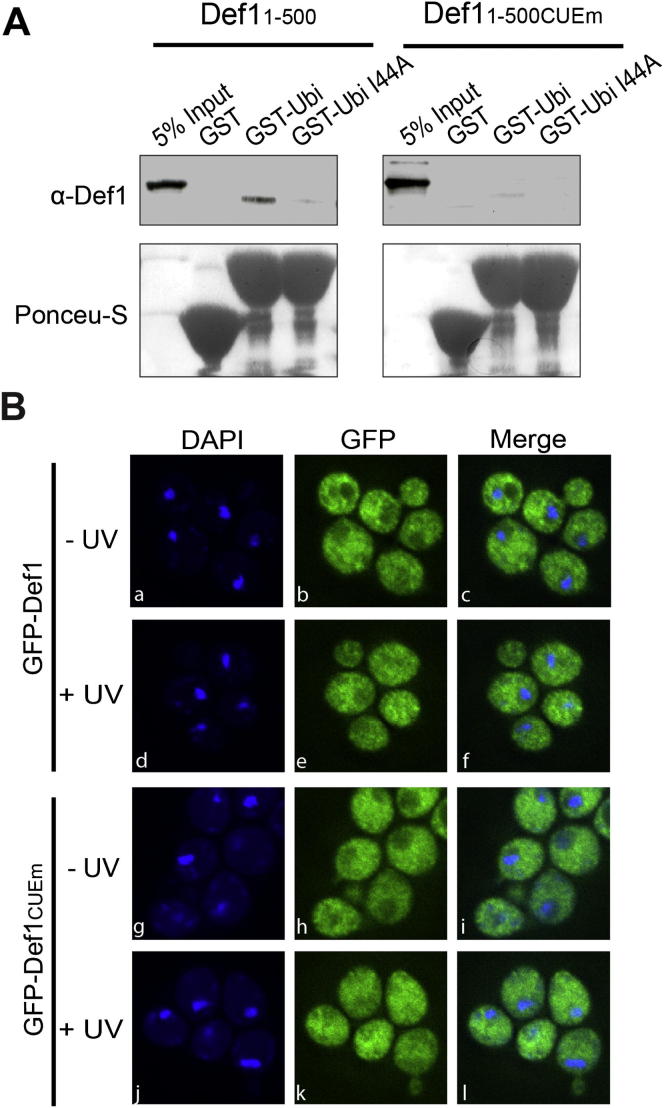
The data above indicate that transcription stress induces proteasomal Def1 processing, leading to pr-Def1 accumulation in the nucleus. However, the role of Def1 in Rpb1 ubiquitylation inside the nucleus remained to be established. We began by examining the role of the N-terminal CUE domain (Ponting, 2002), predicted to bind ubiquitin (Shih et al., 2003). Indeed, Def1’s CUE domain bound to ubiquitin in vitro and when four key CUE residues were mutated (Shih et al., 2003) (Figure 5A, schematic) it reduced such binding (Figure S5A; see also Figure 6D). Strikingly, although expression of Def11–500 was highly toxic, CUE domain mutation effectively suppressed this effect (Figure 5A). CUE mutation also abrogated DNA damage-induced Rpb1 polyubiquitylation (Figure 5B, upper panel; compare lanes 4 and 8). This was not due to a lack of Def1 activation: CUE mutation had no effect on Def1 processing (Figure 5B, middle panel) or on its accumulation in the nucleus (Figure S5B).
Figure 5.
The CUE Domain of Def1 Is Essential for Rpb1 Polyubiquitylation
(A) Top: schematic of Def1 showing four key mutated residues of the CUE domain. Bottom: dilution series of Δdef1 cells (carrying the GAL-driven CEN plasmid indicated on the left), grown on glucose or galactose.
(B) Western blot of extracts from WT, def1CUEm, or Δdef1 cells, UV-irradiated at time 0. Ubiquitylated Rpb1 isolated by MultiDsk pull-down. Asterisk denotes an additional band occasionally observed with the anti-Def1 antibody. Superfluous lanes between lanes 4 and 5 were removed.
(C) Western blot of Myc-Def1 and Myc-Def1CUEm, immunoprecipitated from extracts prepared 1 hr after UV irradiation. Recombinant Ela1 is loaded as an antibody specificity control (lane 1).
(D) Western blot of Myc-Elc1 immunoprecipitated from chromatin-enriched extracts from WT or Def1CUEm cells, prepared 1 hr after UV irradiation.
(E) Binding of recombinant Ela1-Elc1 heterodimer to immobilized, recombinant Def11–500 or Def11–500/CUEm (see also Figure S3A). Proteins were separated by SDS-PAGE and stained with silver.
(F) Western blot analyzing binding of recombinant Ela1-Elc1 heterodimer to immobilized RNAPII (or empty control beads), in the absence or presence of recombinant Def11-500, or Def11-500/CUEm. Schematics of results are shown below the relevant lanes.
See also Figure S5.
Figure S5.
The CUE Domain of Def1 Can Bind to Ubiquitin and Does Not Affect Subcellular Localization, Related to Figure 5
(A) Binding of purified, recombinant Def11-500, or Def11-500/CUEm to immobilized GST, GST-ubiquitin or GST-ubiquitinI44A. 5% of input, and bound proteins were analyzed by SDS-PAGE, followed by anti-Def1 immunoblotting. Ponceau-S is shown below as a loading control for immobilized proteins. See also Figure 6D.
(B) The Def1CUEm protein still accumulates in the nucleus after DNA damage (compare panels k-l with e-f). Localization of N-terminally GFP-Def1 or GFP-Def1CUEm in untreated and UV-treated cells is shown. DNA was counterstained with DAPI.
Figure 6.
The CUE Domain of Def1 Binds Ela1-Elc1
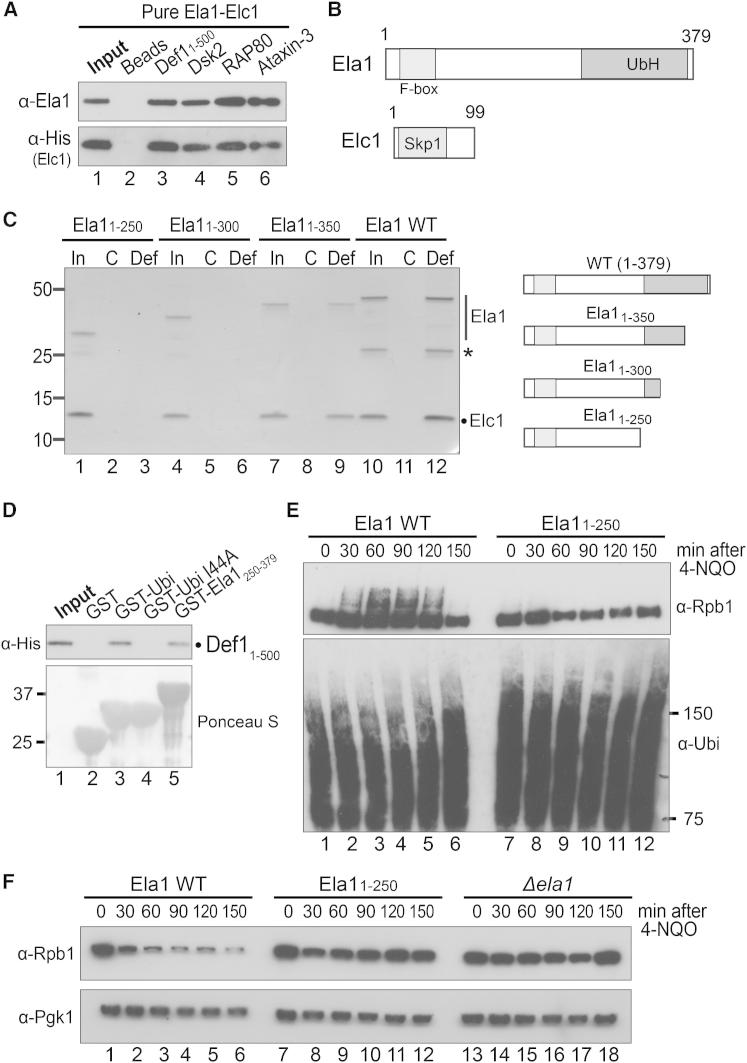
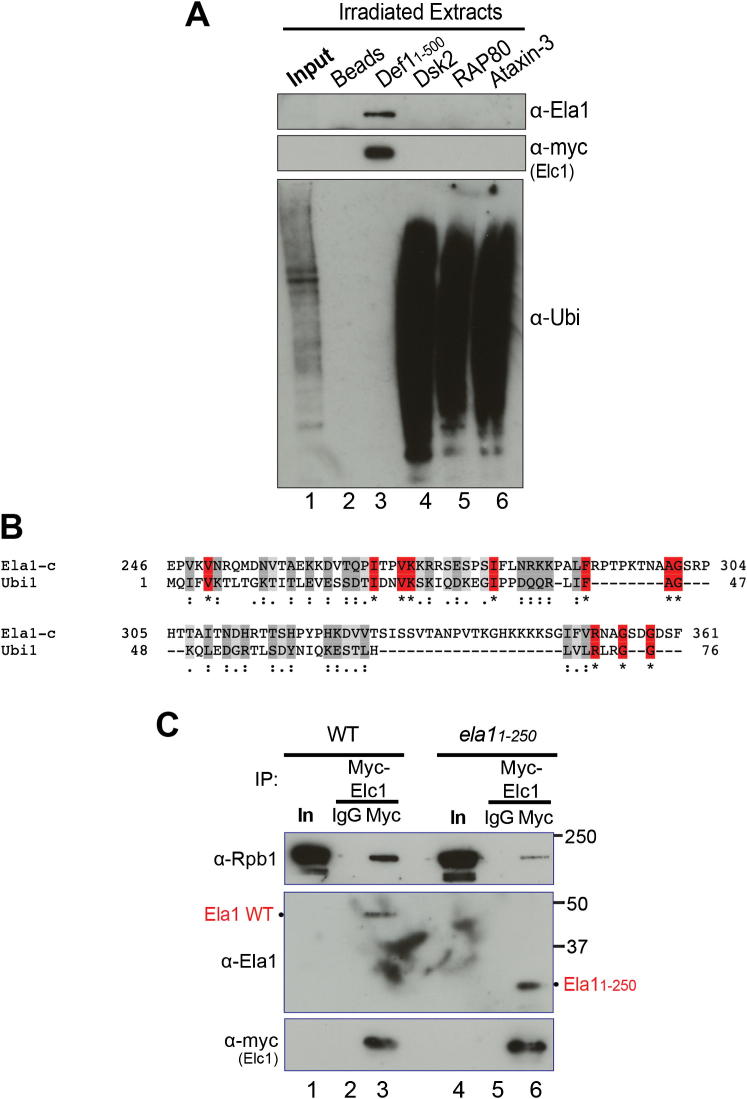
(A) Western blot to detect binding of recombinant Ela1-Elc1 heterodimer to immobilized, recombinant Def11–500 and Dsk2, or the ubiquitin-binding domains from Rap80 and Ataxin-3. See also Figures S3A and S6A.
(B) Schematic of Ela1 and Elc1, indicating relevant domains.
(C) Binding of recombinant Ela1-Elc1 heterodimers (Ela1 form schematically depicted on the right) to immobilized, recombinant Def11–500 (Def), or control beads (C). Protein gel stained with silver. Asterisk indicates a contaminating protein from the purification of full-length Ela1-Elc1 complex.
(D) Western blot showing binding of purified, recombinant Def11–500 to immobilized GST-fusion proteins, including GST-Ela1250–379. Ponceau S stain of membrane shown as a loading control for GST-fusion proteins.
(E) MultiDsk pull-down, comparing damage-induced Rpb1 ubiquitylation in WT Ela1 (Ela1 WT) and cells lacking Ela1’s UbH (ela11–250).
(F) As in (E), but comparing total Rpb1 levels after DNA damage.
Together, these results indicate that the ubiquitin-binding CUE domain is critical to the nuclear function of pr-Def1.
pr-Def1 Associates with Ela1-Elc1 Complex via Its CUE Domain
Rpb1 is monoubiquitylated by Rsp5 in a Def1-independent manner and then polyubiquitylated by the Elongin-Cullin complex in a Def1-dependent fashion (Woudstra et al., 2002; Harreman et al., 2009). Consequently, the data presented above might be explained if Def1 helps mediate the interaction of monoubiquitylated polymerase with the Elongin-Cullin complex. In this model, Def1 would use its CUE domain to bind monoubiquitylated RNAPII, the Elongin-Cullin complex, or both.
We first performed coimmunoprecipitation experiments using extracts from UV-irradiated yeast cells. RNAPII and the Ela1 subunit of the Elongin-Cullin complex both coimmunoprecipitated with Myc-Def1 (Figure 5C, lane 5). Somewhat unexpectedly, however, mutation of the Def1 CUE domain had little or no effect on the interaction with RNAPII, but resulted in a reproducible decrease in the interaction with Ela1 (compare lanes 5 and 9).
To further analyze the intriguing interaction between Def1 and Elongin-Cullin complex, the same factors were now immunoprecipitated from a strain expressing a Myc-tagged version of the Elc1 protein. Myc-Elc1 coprecipitated Def1, but only if Def1 had a functional CUE domain (Figure 5D, compare lanes 3 and 7). Notably, Myc-Elc1 also failed to coprecipitate RNAPII unless Def1 had a functional CUE domain, indicating that Def1 is required to bridge the interaction of Elongin-Cullin with RNAPII.
Ela1 and Elc1 form a heterodimer, akin to the F-box/Skp1 substrate adaptor of other Cullin ligases (Koth et al., 2000), which we now produced in Escherichia coli. To test the interaction of this dimer with Def1, pure recombinant Def11–500, mimicking the active pr-Def1 fragment, was immobilized on beads. A similar amount of the corresponding CUE mutant was also immobilized (Figure 5E). Remarkably, although the recombinant, purified Ela1-Elc1 complex was not ubiquitylated, it associated with immobilized Def11–500, but not when the CUE domain was mutated (Figure 5E, compare lanes 6 and 7).
To further investigate whether Def1 bridges or stabilizes an interaction between RNAPII and the Elongin-Cullin complex, we used purified RNAPII, Def1, and recombinant Ela1-Elc1 complex in tripartite binding experiments (Figure 5F). In these experiments, RNAPII was immobilized on beads. Both Def11–500 and Def11–500/CUEm bound to such beads, but showed only background binding to the empty control beads (Figure 5F, compare lanes 6 and 8 with 5 and 7, respectively). In contrast, Ela1-Elc1 complex bound poorly to RNAPII (Figure 5F, compare lanes 9 and 10). However, when Def1 was included in the binding reaction, a strong association of Ela1-Elc1 complex with RNAPII was detected (Figure 5F, compare lanes 10 and 12). Importantly, the ability of Def1 to bridge the interaction between RNAPII and Ela1-Elc1 was dependent on the CUE domain: even though Def11–500/CUEm associated with RNAPII-containing beads as efficiently as its WT counterpart, it had little or no stimulating effect on Ela1-Elc1 binding (Figure 5F, compare lanes 12 and 14 with lane 10). Interestingly, addition of Ela1-Elc1 also clearly increased Def1-RNAPII interaction (Figure 5F, compare Def1 binding in lane 12 with that in 6, 8, and 14), thus contributing to a more stable RNAPII/Def1/Ela1-Elc1 ternary complex.
A Functionally Important Ubiquitin-Homology Domain in the Ela1 Protein
Recombinant Ela1-Elc1 complex interacts directly with Def1 in a CUE (ubiquitin-binding domain [UBD])-dependent manner, suggesting that, per definition, Ela1 or Elc1 must contain a domain that resembles ubiquitin, e.g., a ubiquitin homology domain (UbH). Indeed, Ela1-Elc1 complex was also capable of associating with other purified UBD proteins or domains, such as Dsk2, RAP80, and Ataxin-3 (Dikic et al., 2009) (Figure 6A, compare lanes 4–6 with lanes 2 and 3). Interestingly, however, only Def1 retrieved Ela1-Elc1 from crude yeast extracts, but did not bind ubiquitylated proteins. Conversely, Dsk2, Rap80, and Ataxin-3 bound ubiquitylated proteins, but did not bind Ela1-Elc1 under the same conditions (Figure S6A). This indicates that Def1’s CUE domain specifically interacts with Ela1-Elc1 through the putative UbH, but does not generally bind ubiquitylated proteins.
Figure S6.
The C Terminus of Ela1 Contains a UbH Domain, Specific to the CUE Domain of Def1, Related to Figure 6
(A) Western blot to detect binding to immobilized, recombinant Dsk2 and Def11-500, and ubiquitin-binding domains from Rap80 and Ataxin-3, in chromatin-enriched extracts from yeast cells expressing Myc-tagged Elc1. Membrane probed for Ela1 (anti-Ela1) and Elc1 (anti-myc) (upper two panels), as well as ubiquitylated proteins (anti-ubiquitin) (lower panel). Def1 pulls Ela1-Elc1 out of the extract, but does not bind significant amounts of ubiquitylated proteins, while Dsk2, Rap80, and Ataxin-3 pull unknown ubiquitylated proteins out of the extract, but do not bind significant amounts of Ela1-Elc1.
(B) Alignment of Ela1’s C-terminal region with yeast ubiquitin (alignment performed using ClustalW2) (Larkin et al., 2007).
(C) Western blot detecting binding of Myc-Elc1 to WT Ela1 (lanes 1-3) and Ela11-250 (lanes 4-6) in vivo. Note that while Elc1 binding to the shorter form of Ela1 is normal, binding to Rpb1 is appreciably decreased in ela11-250 cells.
Given that Elc1 is a very small, Skp1-like polypeptide (Figure 6B, lower), it seemed unlikely to contain a UbH. In contrast, Ela1 is significantly larger (upper), and the function of its C-terminal domain was unclear, prompting us to investigate this region. Constructs encoding different C-terminally truncated Ela1 forms (Figure 6C, schematic on right) were coexpressed with Elc1 in bacteria, and the resulting complexes purified and tested for their ability to bind Def1 immobilized on beads (Figure 6C, left). As expected from previous work (Koth et al., 2000), the different Ela1 forms were all capable of forming a complex with Elc1 (Figure 6C, lanes 1, 4, 7, and 10). However, Ela1-Elc1 complexes that only contained the first 250 or 300 amino acids of Ela1, respectively, were unable to bind Def1 beads (Figure 6C, compare lanes 3 and 6 with lanes 9 and 12). Moreover, the Ela1 C-terminal region was not only required, but also sufficient for association with purified Def11–500 (Figure 6D, compare lane 5 with lanes 2 and 3).
These data prompted us to perform an alignment of ubiquitin with the Ela1 C-terminal domain required for Def1 binding (Figure S6B), which further supported the finding that Ela1 contains a UbH. Ela11–250 also interacted with Elc1 inside cells (Figure S6C) but failed to support damage-induced ubiquitylation (Figure 6E) and degradation of Rpb1, similar to a strain lacking ELA1 altogether (Figure 6F, compare lanes 1–6 with 7–12 and 13–18, respectively), indicating that the UbH is important for cellular Ela1 function.
Together, these results indicate that Def1 bridges the interaction between RNAPII and the Elongin-Cullin ubiquitin ligase complex via an interaction between its CUE domain and a UbH in the Ela1 subunit of the Ela1-Elc1 adaptor complex.
Discussion
The process leading to disassembly of RNAPII elongation complexes in response to transcription stress is remarkable by encompassing so many disparate ubiquitin-based events. For example, Def1 is partially processed, but not entirely degraded, via a ubiquitin-directed, proteasome-dependent mechanism. Def1 also contains a ubiquitin-binding domain (UBD), but this domain does not have a ubiquitin-moiety as its primary target. Rather, it binds to a ubiquitin homology (UbH) domain in the Ela1 subunit of the yeast Elongin adaptor complex. This allows recruitment of the Elongin-Cullin E3 complex to RNAPII, which in turn results in Rpb1 becoming polyubiquitylated and degraded by the proteasome.
Proteasomal Processing and Activation of Def1
Several general principles of proteasome-mediated protein processing have emerged through the study of mammalian NF-κB proteins and their yeast homologs, Mga2 and Spt23 (Rape and Jentsch, 2002). For example, proteasomal processing typically involves ubiquitylation of the target protein, although not necessarily polyubiquitin chains: monoubiquitylation can be sufficient to elicit or speed up the reaction (Rape et al., 2001; Kravtsova-Ivantsiv et al., 2009). Similar results are reported here: proteasomal Def1 processing is greatly stimulated by, but does not absolutely require, monoubiquitylation. Remarkably, the reaction can even be reconstituted with purified proteins: incubation of highly purified Def1 with highly purified proteasome results in correct processing. This suggests that all signals for this intriguing process may be intrinsic to the factors involved, e.g., that it does not require another protease to perform the initial “protein nicking,” or accessory factors to help stop the proteasome from completely degrading the target. The fully defined Def1 processing system should prove helpful in further defining the underlying mechanisms.
At the cellular level, our data are consistent with a model, in which Def1 constantly shuttles in and out of the nucleus, but is predominantly found in the cytoplasm at steady state; this is dependent on the C-terminal region of the protein. Def1 processing removes this region, allowing nuclear accumulation. As far as we know, Def1 represents the first example of a protein in which nuclear accumulation occurs as a result of regulated, proteolytic removal of a domain that directs cytoplasmic transport. We note that studies on proteins such as Huntingtin and Ataxin-7 have indicated that polyglutamine domain expansion observed in disease-causing versions of these proteins affects subcellular localization (Nucifora et al., 2001; Chan et al., 2011). Further studies are needed to understand the details of Def1 subcellular transport, but it is an intriguing possibility that the extremely glutamine-rich regions in the C-terminal half of the protein also somehow regulate nucleocytoplasmic transport in the case of Def1.
Ubiquitylation and degradation of Rpb1 is used as a last-resort response to a variety of conditions, frequent or infrequent, that result in transcription stress (Bregman et al., 1996; Somesh et al., 2005; Sigurdsson et al., 2010; Hobson et al., 2012). Given that the C-terminal domain of Def1 promotes cytoplasmic localization, it is possible that Def1 normally shuttles in and out of the nucleus so that at least a small amount of Def1 is present in the nucleus at all times. In this scenario, the predominant cytoplasmic location of Def1 ensures that it is normally limiting for Rpb1 polyubiquitylation, helping to restrict Rpb1 degradation to a last resort solution to normal transcription elongation problems. Upon DNA damage or other genome-wide transcription stress, however, the protein is allowed to temporarily accumulate in the nucleus, so that RNAPII can be rapidly cleared and transcription restarted. Def1 ubiquitylation by Rsp5, as well as its processing by the proteasome, might occur in the nucleus, possibly even triggered at the stalled polymerase, but this remains to be determined.
Interestingly, Def1 is involved in a number of other, apparently unrelated cellular processes (Chen et al., 2005; Jordan et al., 2007; Suzuki et al., 2011), opening the possibility that other pathways might also rely on Def1 for their proper activation or inactivation in response to DNA damage and other stress. We note that the Def1 pathway has interesting parallels with the bacterial SOS response, as well as with metazoan apoptosis. In the bacterial SOS response, DNA damage results in the RecA-dependent autocatalytic clipping of the LexA repressor protein, resulting in upregulation of repair genes and allowing mutation-prone survival in the face of severe DNA damage (Schlacher and Goodman, 2007). In mammalian cells, severe DNA damage can elicit apoptosis by triggering the protease (caspase)-dependent cleavage and inactivation of ICAD/DFF45 to allow the nuclease CAD to enter the nucleus and fragment the DNA (Nagata, 2000). Proteolytic processing-dependent release of protein fragments, whose activity needs to be restricted to a severe cellular crisis situation, may thus have been utilized several times during evolution.
The Multiple Steps of Rpb1 Ubiquitylation and Degradation
Rpb1 ubiquitylation and degradation occurs via a complex, multistep mechanism (Wilson et al., 2013). In the initial step, Rpb1 is monoubiquitylated by Rsp5, before another E3, the Elongin-Cullin complex, takes over to perform polyubiquitylation (Figure 7, steps 1 and 4, respectively). Our data show how Def1 fits into this complex scheme: it is activated by Rsp5-mediated ubiquitylation, which allows its proteasomal processing and nuclear accumulation (step 2), so that Def1 can bind RNAPII and help recruit the Elongin-Cullin complex, promoting efficient polyubiquitylation (steps 3 and 4). Intriguingly, Def1 contains a CUE domain (UBD), shown here to be essential for this function. Interestingly, this domain is not used to bind the monoubiquitylated polymerase, but rather to recruit the Elongin-Cullin complex. Indeed, we found that the Ela1 subunit of the Elongin adaptor complex harbors a UbH. In humans, the related Elongin A complex comprises three subunits, Elongin A, B, and C (Aso et al., 1995). Budding yeast only has homologs of Elongin A (Ela1) and Elongin C (Elc1) (Koth et al., 2000). Remarkably, the small human Elongin B protein, which lacks a yeast homolog, contains a UbH (Garrett et al., 1995). Given that yeast Ela1 harbors a UbH in its C terminus (that is not conserved in mammalian Elongin A), it is an obvious possibility that sequences encoding this domain have become separated onto a separate gene, encoding Elongin B, during evolution. Our data provide evidence for an important role for this UbH in protein ubiquitylation. More specifically, we show that Elongin’s UbH plays a role in mediating target recognition, rather than, for example, mediating proteasome delivery as has previously been hypothesized (Welchman et al., 2005). We note that, as is true for all proteins containing a ubiquitin-binding domain or a ubiquitin-homology domain (Hofmann, 2009), binding specificity must occur through combination with other recognition domains and motifs in Def1 and the Elongin-Cullin complex.
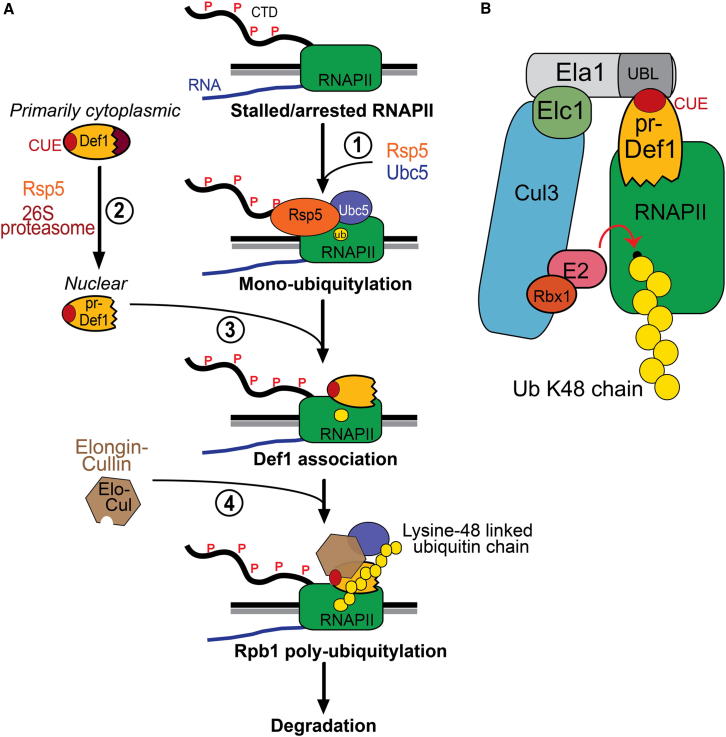
Figure 7.
Model for Def1 Function
(A) Model for Def1-dependent polyubiquitylation of Rpb1. (1) Rsp5 monoubiquitylates Rpb1. (2) Concurrently, Rsp5 ubiquitylates Def1, resulting in proteasomal processing and nuclear accumulation. (3) Nuclear pr-Def1 binds RNAPII, (4) recruiting Elongin-Cullin complex to carry out Rpb1 polyubiquitylation. See text for details.
(B) Schematic depicting the bridging function of Def1. Ela1-Elc1 complex may also have weak Def1-independent interactions with RNAPII, as suggested by the findings of Figure 5F.
The Elongin complex is akin to the Skp1/F-box proteins of other Cullin-ligases (Koth et al., 2000). These proteins are substrate-specific adaptors, which mediate correct target recognition. Ela1/Elc1 complex is unusual in that this adaptor does not actually recognize its target protein, RNAPII, but rather the bridging factor Def1, which then connects it to the target (Figure 7B). The APC/C complex, a Cullin-like ligase, employs distinct substrate-specificity factors (e.g., Cdc20 and Cdh1), but these are integral components of the complex in specific phases of the cell cycle (Bassermann et al., 2013). In contrast, Def1, to the best of our knowledge, represents the first example of a discrete bridging factor being required for target recognition by a Cullin E3 ligase. Whether substrate-specific bridging factors are required for the diverse functions of Elongin-Cullin complex(es), and for Cullin-type E3 ligases in general, is an intriguing possibility that deserves further investigation.
Experimental Procedures
Yeast Strains and Plasmids
Tables S1 and S2 describe S. cerevisiae strains and plasmids. Standard techniques were employed for their construction.
In Vivo Techniques
Whole-cell extracts were prepared by alkaline extraction (Kushnirov, 2000) or via bead-beating, with ubiquitylated proteins being isolated using MultiDsk resin (Wilson et al., 2012). A list of antibodies used in this study can be found in Table S3.
In Vitro Assays
Def1 ubiquitylation assays were performed essentially as described for RNAPII ubiquitylation (Somesh et al., 2005). Binding experiments were performed largely as described (Anindya et al., 2010). Reconstituted proteasome assays were performed with purified yeast proteasome (10 nM) and Def1 (1.5 μM) in reactions with and without MG132 (10 μM).
Fluorescence microscopy was carried out on live cells or in formaldehyde-fixed cells (Figures 4E and S4B). Deltavision microscopy was used to visualize yeast using an X100 UplanSApo 1.40 NA oil objective lens on an Olympus inverted microscope (IX71).
For further details, please see the Extended Experimental Procedures.
Extended Experimental Procedures.
Yeast Strains and Plasmids
Saccharomyces cerevisiae strains used in this study were grown and manipulated using standard techniques (Sherman, 1991). All strains, except GAC202, DEF1-GFP, RPB1-3HA and PRE1-FLAG-6xHIS, are in the W303 background.
Def1 genomic truncations were created by homologous recombination of PCR products (5′- truncation sequence - HA tag - STOP codon – HIS3 selection marker - 3′ truncation sequence) into haploid or diploid yeast, and selection on minimal media plates. Primer sequences are available upon request. The truncations were confirmed by genomic PCR and western blot analysis. Heterozygous diploids were sporulated and tetrads dissected by standard methods.
Strains JSY1191 and 1193-1199, and 1208-1211 were created by inserting, via recombination, different versions of DEF1 into the DEF1 genomic locus of the Δdef1::URA strain (JSY568). Strains in which the URA3 marker had been replaced by the relevant version of DEF1 were selected on 5-Fluroorotic acid. Correct integration was checked by PCR analysis and western blotting. JSY1190 serves as a control for experiments with these strains. In JSY1190, the WT DEF1 gene was integrated into Δdef1::URA (recreating a WT DEF1 locus), as described above.
Def1 was N- and C-terminally tagged as described previously (Harreman et al., 2009). Strain JSY1201 was created by inserting GFP at the N terminus of Def1 via recombination, without altering the DEF1 promoter. The PCR product (5′- homology to upstream DEF1 promoter – URA3 selection marker – DEF1 promoter – GFP in frame with beginning of Def1 coding sequence), created by two-step PCR, contained a URA3 resistance cassette 1 kb upstream of the GFP tag. Correct intergrants were checked via genomic PCR and western blotting. Strains JSY1203, 1204, 1215 and 1216 were created by homologous recombination of PCR products (5′- Ela1 truncation sequence - HA tag - STOP codon – HIS3 selection marker - 3′ Ela1 noncoding sequence) into haploid WT, or Myc-His-TEV-Elc1, yeast strains. The truncations were confirmed by genomic PCR and expression by RT-PCR and by coimmunoprecipitation with Elc1. Further details and primer sequences are available on request.
Def1 mutations and N-terminal tags were introduced into a yeast DEF1-expression plasmid (pRS414-DEF1), containing the endogenous DEF1 promoter, ORF, and terminator regions, using standard PCR methods. For galactose-inducible overexpression of DEF1, the appropriate DNA sequence was amplified by PCR from these plasmids and cloned into the pYC2 vector (Invitrogen). Site-directed mutagenesis was carried out using the QuikChange II XL kit (Stratagene) according to the manufacturer’s instructions. The presence of point mutations was confirmed by sequencing.
The sequence coding for Def1 amino acids 1-500 was codon-optimized and synthesized by Genscript. The coding region was sub-cloned into pGex-6P1 (GE Healthcare). Mutation in the CUE domain was performed by PCR using mutant primers. Sequences can be provided upon request.
The coding regions of Elc1 and Ela1, complete with polycistronic spacer sections (Tan, 2001) were codon-optimized and synthesized by Genscript and sub-cloned into vector pST44 (Tan, 2001). C-terminal Ela1 deletions were created by amplifying the corresponding regions from codon-optimized Elc1-Ela1, and sub-cloning into vector pST44.
Sequences expressing the TEV protease were sub-cloned from construct 118 in Uhlmann et al. (2000) into pRS425. The TEV recognition site was incorporated between amino acid residues 522 and 523 of Def1, inserting the sequence Glu-Asn-Leu-Tyr-Phe-Gln (using the native residue 523 of Def1 as the final part of the TEV recognition consensus site), by site-directed mutagenesis using the QuikChange II XL kit (Stratagene) according to the manufacturer’s instructions.
To create the NLS-GFP-DEF1500-738 plasmid, the DEF1 region coding for amino acids 500-738 was first cloned into a pRS423-based plasmid (pBOW3), containing the ADH1 promoter and terminator separated by multiple cloning sites. Sequences encoding NLS-GFP were amplified from pYM28 (eGFP; EUROSCARF) (SV40 nuclear localization signal (NLS) encoded in primers) and cloned in frame with DEF1500-738. The construct has 4 amino acid spacers between each feature. The final construct was confirmed by DNA sequencing.
Further details and sequences are available on request.
Quantitative Real-Time PCR of DEF1 mRNA Transcripts
Total RNA was isolated from zymolase-treated yeast cells, using a QIAGEN RNeasy kit, following the manufactures instructions. Total mRNA was reverse transcribed, using oligo-dT primers. qPCR reactions were performed on this material, using iQ Custom SYBR Green SuperMix (BioRad), with primers used at a final concentration of 0.2 μM. Δdef1 cells were used as a negative control for mRNA levels. Standard thermal cycling and melting-curve conditions were used in a CFX-96 Real-Time System (BioRad). Quantification was performed in Excel (Microsoft), normalized to β-actin mRNA. Primer sequences are available upon request.
Yeast Growth and Treatment
Damage-induced Rpb1 degradation in vivo was investigated using early logarithmic cells, as previously described (Woudstra et al., 2002). Cells were irradiated with 300 J/m2 UV-light throughout, but similar effects were observed at lower doses. 6-AU sensitivity was tested with 250 μg/ml 6-AU (5 mg/ml stock solution in water), added directly to the medium. 4-NQO was used at a final concentration of 8 μg/ml (10 mg/ml stock solution in acetone). Proteasome inhibitor, MG132 (50 μM final), was added to the medium one hour prior to treatments, where indicated. The translation inhibitor cycloheximide was used at 25 μg/ml (25 mg/ml stock in DMSO). Leptomycin B (30 min incubation) was used to inhibit Crm1-mediated export in MNY8-derived strains at a final concentration of 100 ng/ml (stock 100 μg/ml in ethanol). In experiments using drugs, incubation with an equivalent amount of the relevant solvent was used as a control. Some Def1 processing is observed in response to a variety of stress and growth conditions, including elevated temperature and in cells at high density (>3x107/ml).
For visualization of pr-Def1 formation in vivo, whole cell extracts (WCE) were always prepared by alkaline (denaturing) extraction (Kushnirov, 2000). Some pr-Def1 forms in native extracts (even in the absence of DNA damage), and it is extremely rapidly degraded in native extracts. pr-Def1 is even somewhat unstable in SDS-loading buffer when prepared by alkaline extraction, making it necessary to visualize it by western blotting immediately after preparation. 8WG16 was used to visualize Rpb1 degradation after DNA damage.
For the TEV induction experiment, cells were grown overnight in SC medium containing 2% glucose, washed twice, resuspended in SC containing 2% raffinose, and incubated for 3 hr at 30°C. Galactose was then added to a final concentration of 2% and time points taken as indicated.
For 35S-methionine labeling of proteins, cells were grown to early logarithmic phase, and transferred to medium lacking methionine for 30 min. Cycloheximide (25 μg/ml) was added at the indicated time-points, before 35S-chase was performed. 100 nCi of 35S-labeled methionine (Perkin Elmer) was added to the media for 10 min to label any protein synthesized in this time period radioactively. 1x107 cells were harvested and washed extensively in media containing cycloheximide to remove unincorporated methionine, and cell extracts prepared by alkaline (denaturing) extraction (Kushnirov, 2000).
Protein Purification and Detection
Ubiquitin, Uba1, RAP80, and Ataxin-3 were purchased from Boston Biochem. RNAPII (Rpb1-3HA) and GST-Rsp5 were purified as described previously (Somesh et al., 2005). His-Ubc5, Def11-500-His, and Def11-500/CUEm-His were purified by Ni-affinity chromatography using standard techniques. Def1 proteins were further purified by GST purification, using PreScission protease (GE healthcare) for elution. A final Ni-affinity purification step was used prior to dialysis in A-buffer (100 mM HEPES, 5% glycerol, 2 mM DTT). Ubp2 was purified from JSY1175 as described (Greenwood et al., 2009), and copurifying FLAG-Rsp5 was removed by incubation with M2-agarose (Sigma-Aldrich). Proteasome was purified from a PRE1-FLAG-HIS strain as described (Verma et al., 2000). Full-length yeast Def1 was purified as a Mono-Q side fraction from a RNAPII purification (Somesh et al., 2005). Dsk2 was purified as described (Anindya et al., 2007) and eluted using Factor Xa cleavage, prior to dialysis in A-buffer. His-Ela1 (including Ela1 C-terminal deletions, Ela11-150, Ela11-250, Ela11-300, and Ela11-350) and Elc1 were expressed in E.coli, using 0.5 mM IPTG at 25°C for 4 hr. Clarified extract was subject to standard Ni-affinity purification. The eluate was subject to MonoQ and MonoS chromatography (both columns from GE healthcare). The resulting Ela1-Elc1 complexes were dialyzed into A-buffer. Ela1-Elc1 complexes in which the Ela1 C terminus was deleted expressed and behaved like WT, although they eluted at a lower salt concentration on the MonoS column.
Proteins were subjected to SDS-PAGE using 4%–12% or 3%–8% BioRad Criterion precast gels. Pure proteins were detected by InstantBlue staining (Expedeon) or SilverQuest silver staining kit (Invitrogen). For detection of Def1 by western blotting, a purified rabbit polyclonal anti-Def1 antibody raised against the C-terminal 350 amino acids of the protein was used. As outlined in Figure S1A, this antibody strongly preferentially recognizes pr-Def1 over full-length Def1. Western blots were performed following standard techniques using the following antibodies: secondary antibody; anti-rabbit HRP (GE Healthcare) 1:10,000; anti-mouse HRP (GE Healthcare) 1:10,000; anti-mouse TrueBlot HRP (eBioscience) 1:1000; anti-rabbit TrueBlot HRP (eBioscience) 1:1000.
In order to visualize ubiquitylated Rpb1 or Def1, the total ubiquitylated protein pool was isolated from WCE via multiDsk chromatography (Wilson et al., 2012), followed by western blotting using anti-Rpb1 antibody (4H8), anti-Def1 antibody, or anti-ubiquitin antibody, P4D1 (Enzo). Myc-Def1 immuno-precipitation was performed in D-buffer (150 mM Tris-Acetate pH 7.4, 100 mM potassium acetate, 1 mM EDTA, 0.1% Triton X-100, 10% glycerol, 1x Protease Inhibitor mix [284 ng/ml leupeptin, 1.37 ug/ml pepstatin A, 170 ug/ml phenylmethylsulfonyl fluoride and 330 ug/ml benzamindine]) from WCEs prepared from cells UV-irradiated 1 hr prior to harvesting. For Myc-Elc1 immuno-precipitation, cell extracts were prepared from similarly irradiated cells by mechanical grinding in liquid nitrogen (Kong and Svejstrup, 2002). The crude extract was subjected to centrifugation (10 min, 4000 g), and a chromatin-enriched fraction prepared by re-suspending the pellet in D-buffer with 4 mM MgCl2 and subjecting it to sonication and benzonase treatment (20 min at 25°C with 2 units of benzonase/ml of extract). The resulting extract was subjected to centrifugation (10 min, 4000 g) and the supernatant, now enriched for chromatin proteins, used for immuno-precipitation (IP). IP was done using 9E10 and 9E11 (anti-Myc) antibody, or mouse IgG control antibody (Sigma-Aldrich), chemically crosslinked to Protein A/G Agarose beads (Lane and Harlow, 1988), and the IPs washed extensively before being directly re-suspended in SDS-loading buffer and subjected to western blotting.
Def1 Experiments In Vitro
Def1 Binding Experiments
Def11-500, Def11-500/CUEm and Dsk2 were immobilized on Affigel-15 (BioRad) as per manufacturers instructions, at 2 mg/ml of resin. Empty Affigel-15 beads - with crosslinker blocked by ethyl-ethanolamine - were used as control beads. RNAPII-HA was immobilized on anti-HA affinity matrix (Roche). A 10-fold molar excess of the relevant pure, recombinant protein(s) was added to the reactions as indicated, in B-buffer (20 mM Tris pH 7.5, 200 mM NaCl, 0.05% Triton X-100, 1x Protease Inhibitors, 15% glycerol, 75 μg/ml BSA). After a 2 hr incubation, beads were washed extensively, re-suspended in SDS-PAGE loading buffer, and analyzed by western blotting. Experiments to test Def1 binding to ubiquitin with purified proteins were performed essentially as described for Cockayne syndrome B protein (Anindya et al., 2010). For the ubiquitin-binding assay in crude extracts (Figure S6A) extracts were prepared from chromatin as described above, and incubated with the indicated beads with a crosslinked UBD-containing protein (immobilized at 2 mg/ml of resin), for 2 hr. Beads were washed extensively before directly re-suspending in 1.5x SDS loading buffer and subjected to SDS-PAGE.
Def1 Ubiquitylation
Ubiquitylation assays were performed essentially as described (Somesh et al., 2005, 2007). Briefly, Def1 (1.5 μM) was incubated with Uba1 (90 nM), His-Ubc5 (100 nM), Rsp5 or Rsp5 C777A (4.5 nM), Ubp2-His (300 nM), and ubiquitin (100 μM) in ubiquitylation buffer (25 mM Tris pH 7.5, 125 mM NaCl, 2 mM MgCl2, 1 mM DTT, 3.75 mM ATP) at 30°C for 90 min, as indicated. The reaction was stopped by the addition of SDS loading buffer. Sites of ubiquitylation in Def1 protein were uncovered by mass spec analysis, as previously described for RNAPII (Somesh et al., 2007). The identified ubiquitylated peptides contained two lysine residues each; which of these was ubiquitylated could not be determined. As a result, both K281 and K288 from one peptide, and K328 and K329R from the other were mutated.
Def1 Processing In Vitro
In vitro proteasome assays were performed in P-buffer (25 mM Tris pH 7.5, 100 mM NaCl, 10% glycerol, 2 mM ATP, 5 mM MgCl2, 1 mM DTT and 0.25 mg/ml [4.5 μM] BSA) at 30°C or 4°C. Purified yeast proteasome (10 nM) was added to reactions containing 1.5 μM Def1, with and without MG132 (typically 10 μM). The reaction was stopped by the addition of SDS-PAGE loading buffer.
Fluorescence Microscopy
Cells were grown to logarithmic phase, and treated with MG132, leptomycin B or UV-irradiated, where relevant. Cells were spun at 600 g before incubation in Vectashield (Vector labs) containing DAPI. In Figures 4E and S4B, cells were fixed for 30 min using 3.75% formaldehyde (final concentration). Deltavision microscopy, with an X100 UplanSApo 1.40 NA oil objective lens on an Olympus inverted microscope (IX71) was used to visualize enhanced GFP (eGFP)-tagged Def1. Images were acquired and deconvolved using SoftwoRX software (Applied Precision), using 5 iterations with low noise reduction.
Acknowledgments
This work was supported by grants from Cancer Research UK and European Research Council (to J.Q.S.). We thank Fermentation Services and the Protein Analysis and Proteomics Laboratory at the London Research Institute (LRI) for their expert assistance. We thank William Tansey, Raymond Deshaies, Pamela Silver, Anita Corbett, and Frank Uhlmann for providing reagents. We thank Peter Verrijzer, Helle Ulrich, Kay Hofmann, Nico Dantuma, Anita Corbett, James Omichinski, and members of the Svejstrup laboratory for comments on the manuscript.
Published: August 29, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Extended Experimental Procedures, six figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.07.028.
Supplemental Information
References
- Anindya R., Aygün O., Svejstrup J.Q. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Anindya R., Mari P.O., Kristensen U., Kool H., Giglia-Mari G., Mullenders L.H., Fousteri M., Vermeulen W., Egly J.M., Svejstrup J.Q. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell. 2010;38:637–648. doi: 10.1016/j.molcel.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T., Lane W.S., Conaway J.W., Conaway R.C. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Bassermann F., Eichner R., Pagano M. The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbamcr.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudenon S.L., Huacani M.R., Wang G., McDonnell D.P., Huibregtse J.M. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Bregman D.B., Halaban R., van Gool A.J., Henning K.A., Friedberg E.C., Warren S.L. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykailo M.A., McLane L.M., Fridovich-Keil J., Corbett A.H. Analysis of a predicted nuclear localization signal: implications for the intracellular localization and function of the Saccharomyces cerevisiae RNA-binding protein Scp160. Nucleic Acids Res. 2007;35:6862–6869. doi: 10.1093/nar/gkm776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.M., Tsoi H., Wu C.C., Wong C.H., Cheng T.C., Li H.Y., Lau K.F., Shaw P.C., Perrimon N., Chan H.Y. Expanded polyglutamine domain possesses nuclear export activity which modulates subcellular localization and toxicity of polyQ disease protein via exportin-1. Hum. Mol. Genet. 2011;20:1738–1750. doi: 10.1093/hmg/ddr049. [DOI] [PubMed] [Google Scholar]
- Chen Y.B., Yang C.P., Li R.X., Zeng R., Zhou J.Q. Def1p is involved in telomere maintenance in budding yeast. J. Biol. Chem. 2005;280:24784–24791. doi: 10.1074/jbc.M413562200. [DOI] [PubMed] [Google Scholar]
- Collins G.A., Gomez T.A., Deshaies R.J., Tansey W.P. Combined chemical and genetic approach to inhibit proteolysis by the proteasome. Yeast. 2010;27:965–974. doi: 10.1002/yea.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., Walters K.J. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H., Aguilera A. Transcription coupled repair at the interface between transcription elongation and mRNP biogenesis. Biochim. Biophys. Acta. 2013;1829:141–150. doi: 10.1016/j.bbagrm.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Garrett K.P., Aso T., Bradsher J.N., Foundling S.I., Lane W.S., Conaway R.C., Conaway J.W. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc. Natl. Acad. Sci. USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- Harreman M., Taschner M., Sigurdsson S., Anindya R., Reid J., Somesh B., Kong S.E., Banks C.A., Conaway R.C., Conaway J.W., Svejstrup J.Q. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl. Acad. Sci. USA. 2009;106:20705–20710. doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson D.J., Wei W., Steinmetz L.M., Svejstrup J.Q. RNA polymerase II collision interrupts convergent transcription. Mol. Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst.) 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H.D., Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M., Yang J.C., Beaudenon S.L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Kehlenbach R.H. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Jariel-Encontre I., Bossis G., Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Jordan P.W., Klein F., Leach D.R. Novel roles for selected genes in meiotic DNA processing. PLoS Genet. 2007;3:e222. doi: 10.1371/journal.pgen.0030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., Lyon N., Huibregtse J.M. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koth C.M., Botuyan M.V., Moreland R.J., Jansma D.B., Conaway J.W., Conaway R.C., Chazin W.J., Friesen J.D., Arrowsmith C.H., Edwards A.M. Elongin from Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:11174–11180. doi: 10.1074/jbc.275.15.11174. [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol. Cell. 2009;33:496–504. doi: 10.1016/j.molcel.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Henry M., Silver P.A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Nagata S. Apoptotic DNA fragmentation. Exp. Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- Nucifora F.C., Jr., Sasaki M., Peters M.F., Huang H., Cooper J.K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V.L. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando O.J., Goldberg A.L., Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Pan Y., Bai C.B., Joyner A.L., Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko W., Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat. Struct. Mol. Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M., Jentsch S. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 2002;4:E113–E116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Reid J., Svejstrup J.Q. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J. Biol. Chem. 2004;279:29875–29878. doi: 10.1074/jbc.C400185200. [DOI] [PubMed] [Google Scholar]
- Ribar B., Prakash L., Prakash S. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3999–4005. doi: 10.1128/MCB.00293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar B., Prakash L., Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol. Cell. Biol. 2007;27:3211–3216. doi: 10.1128/MCB.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Goodman M.F. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- Shih S.C., Prag G., Francis S.A., Sutanto M.A., Hurley J.H., Hicke L. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 2003;22:1273–1281. doi: 10.1093/emboj/cdg140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S., Dirac-Svejstrup A.B., Svejstrup J.Q. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh B.P., Reid J., Liu W.F., Søgaard T.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Somesh B.P., Sigurdsson S., Saeki H., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007;129:57–68. doi: 10.1016/j.cell.2007.01.046. [DOI] [PubMed] [Google Scholar]
- Ström A.C., Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Yokoyama A., Tsuji T., Ikeshima E., Nakashima K., Ikushima S., Kobayashi C., Yoshida S. Identification and characterization of genes involved in glutathione production in yeast. J. Biosci. Bioeng. 2011;112:107–113. doi: 10.1016/j.jbiosc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem. Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Taura T., Krebber H., Silver P.A. A member of the Ran-binding protein family, yrb2p, is involved in nuclear protein export. Proc. Natl. Acad. Sci. USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool A.J., Verhage R., Swagemakers S.M., van de Putte P., Brouwer J., Troelstra C., Bootsma D., Hoeijmakers J.H. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Oania R., Fang R., Smith G.T., Deshaies R.J. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wilson M.D., Saponaro M., Leidl M.A., Svejstrup J.Q. MultiDsk: a ubiquitin-specific affinity resin. PLoS ONE. 2012;7:e46398. doi: 10.1371/journal.pone.0046398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.D., Harreman M., Svejstrup J.Q. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim. Biophys. Acta. 2013;1829:151–157. doi: 10.1016/j.bbagrm.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Woudstra E.C., Gilbert C., Fellows J., Jansen L., Brouwer J., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- Yasukawa T., Kamura T., Kitajima S., Conaway R.C., Conaway J.W., Aso T. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 2008;27:3256–3266. doi: 10.1038/emboj.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Gorsch L.C., Dockendorff T.C., Cole C.N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood C., Selth L.A., Dirac-Svejstrup A.B., Svejstrup J.Q. An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J. Biol. Chem. 2009;284:141–149. doi: 10.1074/jbc.M805312200. [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kong S.E., Svejstrup J.Q. Incision of a 1,3-intrastrand d(GpTpG)-cisplatin adduct by nucleotide excision repair proteins from yeast. DNA Repair (Amst.) 2002;1:731–741. doi: 10.1016/s1568-7864(02)00080-0. [DOI] [PubMed] [Google Scholar]
- Lane D., Harlow E. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. Antibodies: A Laboratory Manual. [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Neville M., Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.L., Ulrich H.D. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Guthrie C., Fink G.R., editors. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc.; San Diego: 1991. pp. 3–20. [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R.J. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.