Abstract
Background
The treatment efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors like erlotinib has not met expectations for glioblastoma therapy, even for EGFR-overexpressing tumors. We determined possible mechanisms of therapy resistance using the unique BS153 glioblastoma cell line, which has retained amplification of the egfr gene and expression of EGFR variant (v)III.
Methods
Functional effects of erlotinib, gefitinib, and cetuximab on BS153 proliferation, migration, and EGFR-dependent signal transduction were systematically compared in vitro. The tumor-initiating capacity of parental and treatment-resistant BS153 was studied in Naval Medical Research Institute/Foxn1nu mice. Potential mediators of resistance were knocked down using small interfering (si)RNA.
Results
Erlotinib and gefitinib inhibited proliferation and migration of BS153 in a dose-dependent manner, whereas cetuximab had no effect. BS153 developed resistance to erlotinib (BS153resE) but not to gefitinib. Resistance was associated with strong upregulation of EGFRvIII and subsequent activation of the phosphatidylinositol-3-OH kinase (PI3K) pathway in BS153resE and an increased expression of the regulatory 110-kDa delta subunit of PI3K (p110δ). Knockdown of EGFRvIII in BS153resE largely restored sensitivity to erlotinib. Targeting PI3K pharmacologically caused a significant decrease in cell viability, and specifically targeting p110δ by siRNA partially restored erlotinib sensitivity in BS153resE. In vivo, BS153 formed highly invasive tumors with an unusual growth pattern, displaying numerous satellites distant from the initial injection site. Erlotinib resistance led to delayed onset of tumor growth as well as prolonged overall survival of mice without changing tumor morphology.
Conclusions
EGFRvIII can mediate resistance to erlotinib in EGFR-amplified glioblastoma via an increase in PI3Kp110δ. Interfering with PI3Kp110δ can restore sensitivity toward the tyrosine kinase inhibitor.
Keywords: BS153, EGFR, glioma, invasion, PI3K, resistance
Glioblastoma multiforme (GBM) is characterized by a variety of genomic rearrangements and mutations that underlie resistance to conventional chemo- and radiotherapy as well as to novel experimental therapeutic strategies. One common alteration is an amplification of the epidermal growth factor receptor (EGFR) gene,1 which is present in 40%–60% of all GBM. Amplification is often associated with expression of the oncogenic variant III of the receptor (EGFRvIII), which lacks exons 2–7 and is constitutively active.2,3 Amplification-dependent overexpression of full-length EGFR and EGFRvIII at the protein level contributes to the proliferative and migratory/invasive phenotype of malignant GBM.4,5 Therefore, EGFR is considered an attractive therapeutic target.
Targeting of EGFR has been attempted by various approaches, including tyrosine kinase inhibition by small molecules such as erlotinib (Tarceva) or gefitinib (Iressa) and antibodies, including cetuximab (Erbitux). However, clinical trials have been disappointing so far, as they have not proven superior to standard treatment.6–8 Potential reasons for treatment failure are multifold and include insufficient tissue penetration of the tyrosine kinase inhibitor (TKI), insufficient target inhibition, and compensatory activation of EGFR-independent signaling pathways by tumor cells, as well as cellular heterogeneity of EGFR-amplified GBM, as not all cells carry genetic aberrations of the egfr gene.9–11
A major obstacle to identifying the exact mechanisms underlying resistance to EGFR-directed therapies has been the sparsity of preclinical models that faithfully recapitulate the in vivo situation. Cells from EGFR-amplified tumors usually lose amplification rapidly in vitro, rendering them unsuitable for research.12 Therefore, approaches have been used based on forced overexpression of wild-type EGFR (wtEGFR) or EGFRvIII in a non-amplified background, such as the U87MG cell line and subsequent blockade of the artificially overexpressed proteins.13 Alternatively, freshly resected patient material can be directly xenografted into immunocompromised rodents, a method that maintains amplification present in the original tumor12,14 but is highly laborious and difficult to standardize. The only relatively well-known glioma-derived, adherent EGFR-amplified cell line is SKMG-3.15 However, this cell line shows only moderate egfr amplification (∼8-fold), does not express EGFRvIII, and is not tumorigenic in nude mice. It is thus lacking important features of a representative GBM research model.16
In the current study, we used a GBM-derived cell line, BS153, which has only recently come to broader scientific attention.9,17 BS153, originally described by Jones et al,18 is highly amplified for the egfr gene (∼50-fold), expresses EGFRvIII, and grows as a monolayer in the presence of serum. Furthermore, we demonstrate BS153 to be tumorigenic in the brains of nude mice. We systematically compared the effects of erlotinib, gefitinib, and cetuximab on EGFR-induced proliferation, signaling, migration, and tumorigenicity, to identify possible mechanisms of resistance to these agents in a bona fide egfr-amplified background. BS153 cells developed resistance to erlotinib but not to gefitinib, while cetuximab failed to show any inhibition of proliferation or migration. Resistance was accompanied by a strong increase of EGFRvIII protein and upregulation of PI3K, in particular p110δ. Knockdown of PI3Kp110δ inhibited proliferation of erlotinib-resistant cells, suggesting that targeting downstream effectors of EGFRvIII signaling can circumvent EGFRvIII-dependent erlotinib resistance.
Materials and Methods
Chemicals and Antibodies
Erlotinib, gefitinib, and PX-866 were purchased from LC Laboratories. EGF came from PeproTech. The antibody recognizing the intracellular domain of EGFR used for western blot and immunohistochemistry came from Santa Cruz (clone 1005). The mouse monoclonal antibody recognizing wtEGFR (EGFR.1) used for fluorescence activated cell sorting analysis was obtained from Pierce. The EGFRvIII-specific monoclonal antibody (clone L8A4) was provided by D. Bigner.3 Antibodies recognizing EGFR phosphorylated tyrosine (Tyr)1068, phosphorylated phosphatase and tensin homolog (PTEN)/PTEN, phosphorylated PI3K, p110α, p110β, phosphorylated extracellular signal-regulated kinase (ERK)/ERK, and phosphorylated Akt/Akt came from Cell Signaling Technology. The tubulin antibody came from Merck Millipore. The p110δ antibody came from Abcam. The antibody recognizing Ki-67 (MIB-1) came from Dako. Cetuximab was a kind gift from ImClone Systems.
Cell Lines and Cell Culture
The cell line BS153 was generated by Jones et al18 from a primary glioblastoma. The cells grew as an adherent monolayer cell culture in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, and 1 mM sodium pyruvate (all from Life Technologies). BS153resE cells were grown in the presence of erlotinib (10 µg/mL; ∼25 µM) for at least 20 passages (>6 months). For phosphorylation analysis, cells were serum starved for 4 h and preincubated with inhibitors for 10min prior to the addition of mitogens as described previously.19
EGFR-specific PCR and Small Interfering RNAs
RNA was extracted using the NucleoSpin RNA XS Kit (Macherey & Nagel). cDNA was amplified with Superscript II (Life Technologies) and used for semiquantitative PCR to detect expression of wtEGFR and EGFRvIII as described previously.16 For quantitative gene expression analysis of all EGFR transcripts and wtEGFR-specific transcripts, validated TaqMan Gene Expression Assays were purchased (Hs01076088_m1 and Hs01076078_m1, respectively, Life Technologies). Primer/probe detecting EGFRvIII has been described by Rae et al.20 Reactions were performed in a 7500 Fast Real-Time PCR System (Life Technologies). Relative amounts of target mRNAs were normalized to RPL–13A as internal control. Expression values were calculated according to the delta-delta cycle threshold method. The EGFRvIII-specific small interfering (si)RNA has been described by Yamoutpour et al.21 The p110δ-specific siRNA was described by Luk et al.22 Cells were transfected using Lipofectamine RNAiMax (Life Technologies) as described previously.19
EGFR-specific Fluorescence In situ Hybridization
Fluorescence in situ hybridization (FISH) analysis for the egfr gene and centromere of chromosome 7 for glioma cells and xenograft tumors was performed as described previously16 using a probe derived from Homo sapiens PAC clone RP5–1091E12 (GenBank accession no. AC006977) labeled with Spectrum Orange–deoxyuridine triphosphate (Abbott Molecular), a centromere 7 probe (Spectrum Green), and mounted with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories).
Western Blotting
Western blotting was performed as described previously.23 Proteins were extracted with 1% Triton in phosphate buffered saline and complete protease inhibitor (Roche) in the presence of 2 mM sodium orthovanadate. Antibodies at a final concentration of 1–2 µg/mL were incubated overnight at 4°C. Bound antibody was detected using species-specific peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) and visualized using enhanced chemiluminescence or SuperSignal West Femto substrate (Pierce). Scans of x-ray films were saved as .tif files and quantified by densitometry using ImageJ software (version 1.44p).
Flow Cytometry
Cells were analyzed by flow cytometry as described previously.19 In brief, cells were scraped off cell culture dishes in ice cold phosphate buffered saline containing 0.01% NaN3 and incubated with either EGFR.1 or an isotype-matched control antibody (immunoglobulin [Ig]G2b; 3 µg/mL). For detection of EGFRvIII, cells were incubated with the EGFRvIII antibody3 or an isotype-matched control antibody (IgG1; 3 µg/mL). Bound primary antibody was detected using a secondary phycoerythrin-conjugated anti-mouse IgG (Jackson). Fluorescence was measured on a PAS Particle Analysing System (Partec).
Proliferation Assay
Proliferation was assessed in octuplicates as described previously.16 In brief, 2.5 × 103 cells/well were seeded in a black 96-well plate with a transparent bottom (Nunc). On days 1 and 3, cells were supplied with fresh medium containing 2% FCS and inhibitors. On days 3 and 6, cells were analyzed for their ATP content using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions. Luminescence was recorded using a SpectraFluor Plus luminometer (Tecan).
Migration Analysis
Glioma cell migration in response to EGF was analyzed using a modified Boyden chamber chemotaxis assay as described.23 Cells were preincubated with inhibitors for 10min prior to analysis. After 5 h of incubation at 37°C/5% CO2, migrated cells were stained as described and counted in 10 high-power fields using a 40× objective with a calibrated ocular grid.
Soft Agar Colony Formation Assay
The assay was performed as described previously.24 Cells were treated with erlotinib (10 µg/mL) in full medium. Fresh full medium with or without erlotinib was added every second day. After incubation for 7 d, cells were stained with 5% crystal violet for 90min at 37°C and analyzed by bright field microscopy.
In vivo Tumorigenicity
All animal experiments were approved by the local authority in Hamburg (Behörde für Soziales, Familie, Gesundheit und Verbraucherschutz) and were conducted according to the institution's guidelines for animal husbandry. Cells were washed twice (DMEM without additives). Anesthetized 8-week-old Naval Medical Research Institute (NMRI)/Foxn1nu mice (Harlan) were each injected with 1.5 × 105 cells/4 µL of DMEM without supplements into the right caudate/putamen as described previously.23 Mice were sacrificed when they either lost more than 10% body weight or developed neurological symptoms. Following resection, brains were fixed in 10% formalin and embedded in paraffin.
Immunohistochemistry
Paraffin-embedded tissue was analyzed as described previously.23 Four-micrometer sections were stained with hematoxylin and eosin (H&E) for assessing tumor burden. Antigens were recovered by heat-induced antigen retrieval (total EGFR, EGFRvIII, MIB-1).
Methods of Data Analysis
Statistical survival comparisons were carried out with the MedCalc program (Kaplan–Meier, log-rank test). Comparisons of cell proliferation and migration were carried out using the unpaired t-test for normally distributed samples. Differences were considered statistically significant at P < .05 (*) and as highly significant at P < .005 (**). Error bars represent SDs of a minimum of quadruplicates.
Results
Erlotinib and Gefitinib Inhibit Proliferation and Migration of EGFR-amplified BS153 Cells In vitro
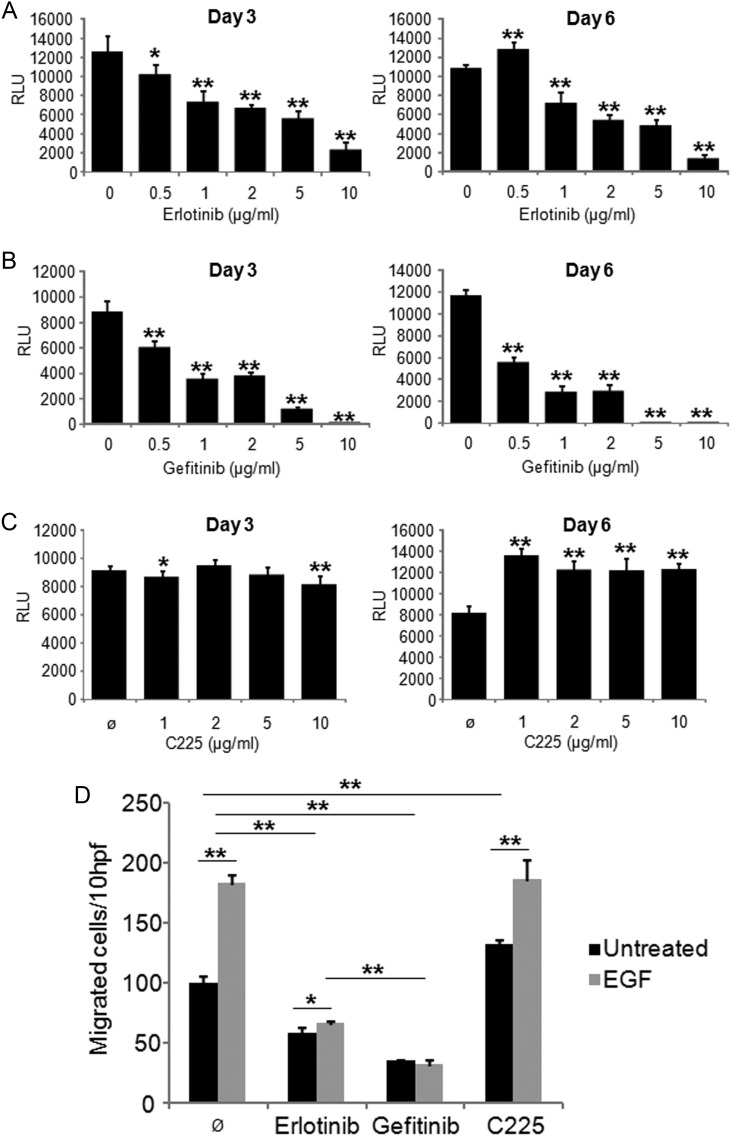
To systematically compare the effect of erlotinib, gefitinib, and cetuximab on GBM-derived, natively egfr-amplified cells in vitro, we first treated BS153 glioma cells with increasing concentrations of the drugs and assessed proliferation. As expected, erlotinib (Fig. 1A) and gefitinib (Fig. 1B) were able to reduce proliferation of BS153 significantly in a concentration-dependent manner within 72 h of treatment, in either the absence or presence of EGF (20 nM; Supplementary Fig. S1). For gefitinib, lower concentrations sufficed to inhibit proliferation (>50% reduction after 72 h at 1 µg/mL, P < .005, t-test; Fig. 1B) compared with erlotinib (>50% reduction after 72 h at 5 µg/mL, P < .005; Fig. 1A). After 6 days, gefitinib had killed most cells at concentrations of 5 or 10 µg/mL (<2% viable cells detectable by trypan blue staining; data not shown), while erlotinib-treated cells still proliferated even at 10 µg/mL (∼25 µM). Notably, BS153 reproducibly showed increased proliferation at 0.5 µg/mL erlotinib when compared with untreated cells (day 6, P < .005; Fig. 1A), suggesting that adaptive mechanisms allowed BS153 proliferation even in the presence of erlotinib, which was not observed in gefitinib-treated cells. Strikingly, cetuximab had almost no influence on proliferation after 72 h but consistently stimulated proliferation after 6 days at all concentrations tested (Fig. 1C).
Fig. 1.
Response to EGFR-directed therapy in EGFR-amplified BS153. Erlotinib (A), gefitinib (B), and C225 (cetuximab; C) were applied to BS153 in different concentrations. Proliferation was assessed after 3 and 6 days. Erlotinib and gefitinib significantly inhibited BS153 growth in a dose-dependent manner (*P < .05, **P < .005, t-test), while C225 had no effect. Similar results were obtained when migration toward EGF (20 nM) was analyzed (D; RLU, relative luminescence units; hpf, high power fields); values are means ± SD from octuplicate determinations. One of 3 independent experiments is shown.
A key pathological characteristic of glioblastomas is their invasive phenotype, making complete surgical resection impossible. Therefore, an effective drug to treat GBM should ideally target migration as well as proliferation. To assess the impact of erlotinib, gefitinib, and cetuximab on the migratory capacity of BS153, we performed modified Boyden chamber assays16 (Fig. 1D). Erlotinib and gefitinib (10 µg/mL) inhibited migration significantly in the absence and presence of EGF, with gefitinib being more potent than erlotinib at equimolar concentrations (65% ± 1 vs 43% ± 5, respectively, without EGF; P < .005). Cetuximab, in contrast, induced migration at concentrations from 0.5 to 10 µg/mL (30% ± 5 at 10 µg/mL, P < .005; Fig. 1D and data not shown). Stimulation of BS153 with EGF in the presence of cetuximab yielded similar results as did stimulation of BS153 with EGF alone. These findings indicate that cetuximab has a partially agonistic effect and does not sufficiently block EGF binding to prevent the activation of EGFR.
Differing Signaling in BS153 in Response to EGFR Inhibition by Erlotinib, Gefitinib, and Cetuximab
We next examined key signaling components of EGF-induced signal transduction in the absence or presence of erlotinib, gefitinib, and cetuximab by western blot (Fig. 2). Erlotinib as well as gefitinib were able to abolish EGFR autophosphorylation and EGF-induced activation at Tyr1068 (Fig. 2A and quantification in Fig. 2B) and Tyr1173 (not shown) for wtEGFR at concentrations ranging from 0.5 µg/mL (∼1.25 µM; data not shown) to 10 µg/mL (∼25 µM; Fig. 2A), but we detected little influence on EGFRvIII autophosphorylation. Phosphorylation of Akt, a mediator of PI3K signaling, was nearly absent in the presence of gefitinib as a consequence of EGFR inhibition, whereas erlotinib caused only a small reduction of Akt activity. Conversely, erlotinib inhibited Ras-dependent signal transduction via ERK to a greater extent than gefitinib did. No suppression of EGFR phosphorylation was detectable when the cells were treated with cetuximab. Paradoxically, an increase in EGFR-phosphorylation and subsequent downstream activation of Akt in response to cetuximab was observed, while ERK remained relatively unaffected. In the presence of EGF, ERK was phosphorylated, again indicating insufficient inhibition of EGF binding to EGFR by cetuximab and/or a partially agonistic effect of cetuximab on BS153. Similar signaling patterns were observed for transforming growth factor–α, another important EGFR ligand in the brain (data not shown).
Fig. 2.
Western blot analysis of EGFR downstream signaling. BS153 cells were serum starved and stimulated with EGF (20 nM) in the presence or absence of inhibitors (10 µg/mL) or left untreated (A). Serum starvation led to downregulation of wtEGFR phosphorylation (P-), while EGFRvIII phosphorylation (Tyr1068) remained unaffected. EGFR inhibition with erlotinib and gefitinib affected Akt phosphorylation as well as ERK phosphorylation, in the presence or absence of EGF. Cetuximab induced EGFR and Akt phosphorylation in the absence of EGF. Densitometric quantification of phosphorylation events is shown in (B). One of 3 independent experiments is shown.
Long-term Exposure of BS153 to Erlotinib Leads to a Resistant Phenotype, Whereas Exposure to Gefitinib Does Not
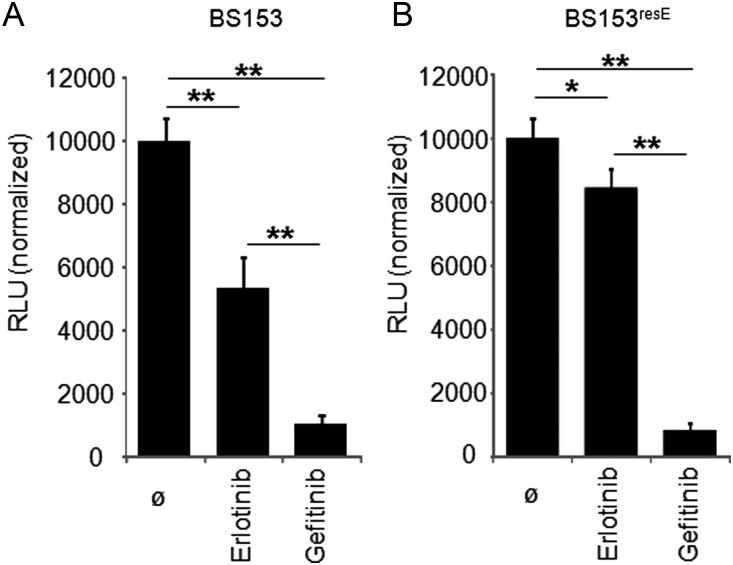
The aforementioned data indicate that both TKIs inhibit cell growth significantly, but gefitinib has a more pronounced effect than erlotinib, which is associated with considerably stronger inhibition of Akt activation by gefitinib. To investigate whether growth inhibition by both TKIs was a temporary or a permanent effect, we continuously treated BS153 cells with increasing concentrations of either inhibitor up to 10 µg/mL (∼25 µM). After a transient decrease in cell number, continuous incubation of BS153 with erlotinib at concentrations as high as 10 µg/mL produced an erlotinib-tolerant cell line (BS153resE; Fig. 3) that survived drug treatment and expanded persistently in the presence of the TKI, with a calculated doubling rate of 44 h versus a doubling rate of 48 h for the parental BS153. In contrast, we could not establish a gefitinib-resistant cell line using any concentrations tested, even as low as 0.5 µg/mL. Interestingly, BS153resE was highly sensitive to gefitinib (Fig. 3A), which eventually killed the erlotinib-resistant cells after prolonged exposure. Notably, when erlotinib was withdrawn from BS153resE, a slight increase in proliferations was detected, probably due to regained wtEGFR activity in the absence of the inhibitor. Since gefitinib has been effective in non–small cell lung cancers with activating mutations in the EGFR tyrosine kinase domain, we sequenced exon 18–21 of the EGFR gene as well as full-length EGFRvIII-cDNA in resistant and parental BS153, but no mutations were found (data not shown). These findings indicate that there are different inhibitory mechanisms for erlotinib and gefitinib. This is further supported by the finding that SW620 colorectal cancer cells, which lack wtEGFR as well as EGFRvIII,25 are insensitive to erlotinib up to 10 µg/mL, whereas gefitinib inhibits proliferation of SW620 cells at 5 µg/mL after 3 days of treatment (Supplementary Fig. S2), suggesting off-target effects of gefitinib at higher concentrations.
Fig. 3.
Chronic exposure of BS153 to erlotinib led to a resistant phenotype. BS153 was sensitive to erlotinib as well as to gefitinib (A; both 10 µg/mL), whereas BS153resE was relatively resistant to erlotinib (B; both 10 µg/mL) after 6 days of treatment. Proliferation increased when erlotinib was removed from BS153resE for 6 days. Values are means ± SD from octuplicate determinations. One of 3 independent experiments is shown. RLU, relative luminescence units.
Resistance to Erlotinib Is Associated With Enhanced Expression of EGFRvIII Protein and PI3K Activity
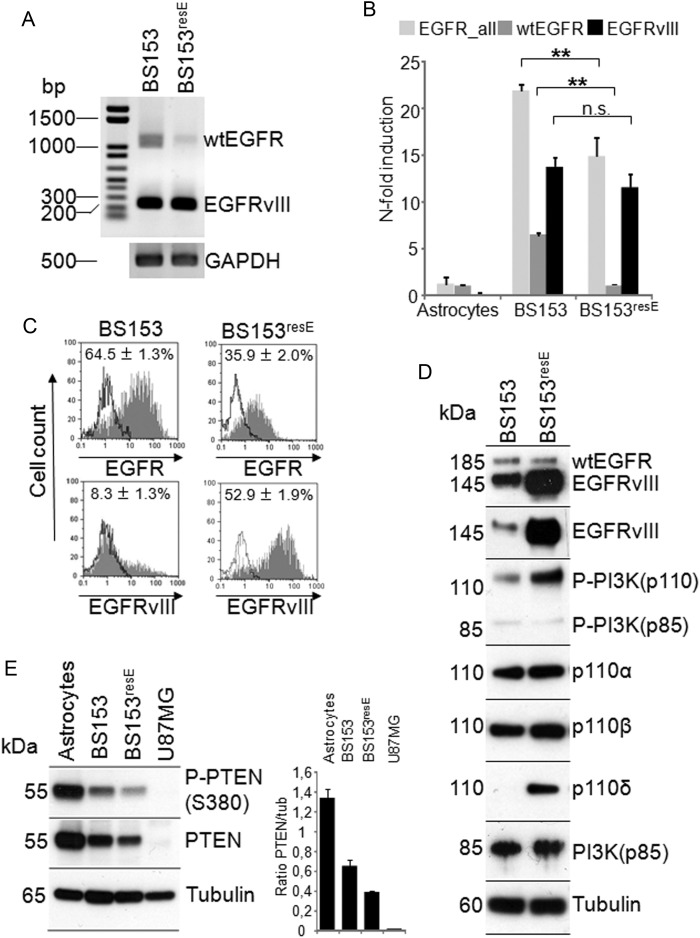
We next analyzed the underlying molecular changes mediating tolerance to high doses of erlotinib in BS153resE. Compared with parental BS153, BS153resE displayed an unaltered egfr copy number as determined by quantitative PCR (38.3 ± 6.1 for BS153 vs 39.5 ± 5.0 for BS153resE; Supplementary Fig. S3A) and by FISH analysis (45.9 ± 8.5 vs 41.4 ± 6.5, respectively; Supplementary Fig. S3B) and displayed no substantial alteration of cellular morphology (Supplementary Fig. S3C). However, we detected decreased mRNA levels for wtEGFR by semiquantitative and quantitative PCR, while the mRNA coding for EGFRvIII did not change significantly (Fig. 4A and B). At the protein level, EGFRvIII was expressed in only a subpopulation of wtEGFR-positive cells in parental BS153, as indicated by flow cytometry (Fig. 4C) and western blot of cells sorted by magnetic activated cell sorting (Supplementary Fig. S3D). The number of EGFRvIII-positive cells analyzed by flow cytometry increased from 8.3% ± 1.3 in BS153 to 52.9% ± 1.9 in BS153resE (6.6–fold; Fig. 4C), accompanied by an almost 2-fold decrease in wtEGFR-positive cells in BS153resE (Fig. 4C). Additionally, expression of EGFRvIII protein, as measured by mean fluorescence intensity (MFI), increased 3.7–fold from 19.8 ± 1.7 in BS153 to 72.2 ± 3.8 in BS153resE, while the MFI for wtEGFR decreased from 88.4 ± 1.4 in BS153 to 38.7 ± 3.5 in BS153resE (2.3-fold). Increased EGFRvIII protein expression in BS153resE could also be confirmed by western blot analysis (Fig. 4D). The discrepancy between unchanged mRNA levels for EGFRvIII and strongly increased cell surface expression may be due to either translational regulation or decreased turnover of the mutant receptor. Importantly, upregulation of EGFRvIII occurred only after prolonged exposure of BS153 to erlotinib (>12 passages) and not as an immediate response to acute erlotinib exposure (ie, 3–6 days; Supplementary Fig. S3E).
Fig. 4.
Comparison of BS153 and BS153resE at the mRNA and protein level. cDNA was analyzed by conventional PCR for the expression of EGFRvIII and wtEGFR mRNA (A) showing strong EGFRvIII expression in both cell lines and reduced wtEGFR expression in BS153resE. Quantitative real-time PCR confirmed reduced expression of wtEGFR mRNA in BS153resE compared with BS153 (**P < .005, t-test), while EGFRvIII mRNA remained unchanged. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Astrocyte cDNA was used as calibrator (B; values are means ± SD of quadruplicate determinations). Strongly enhanced expression of EGFRvIII protein and decreased expression of wtEGFR protein using variant-specific antibodies was detected in BS153resE by flow cytometry (C). Western blot analysis of overall EGFR levels using a pan–EGFR antibody as well as selective detection of EGFRvIII using a specific antibody confirmed increased expression of EGFRvIII at the protein level. The western blot for the different PI3K isoforms showed increased PI3Kp110 phosphorylation along with increased expression of PI3Kp110δ in BS153resE (D). PTEN protein levels were decreased, but PTEN was still phosphorylated in both cell lines compared with normal human astrocytes. U87MG served as a negative control (E).
As a possible consequence of deregulated EGFR signaling, phosphorylation of the catalytic p110 subunit of PI3K, a key mediator of EGFR signaling, was increased in BS153resE, as seen by western blot (Fig. 4D). Since for the p110 subunit, 4 different isoforms with different tissue distributions are known (designated α, β, γ, and δ), we performed isoform-specific western blots. Interestingly, we detected a strong increase of PI3Kp110δ protein and mRNA in BS153resE (Fig. 4D and Supplementary Fig. S3F). P110δ is usually not expressed in the brain but in the hematopoietic system26,27 and has only recently been linked to glioma cell migration and invasion.22 Increased expression of p110δ is also detected in only BS153resE and not in acutely treated BS153 (Supplementary Fig. S3E). Additionally, a slight but consistent decrease in PTEN protein, an important negative regulator of EGFR signaling, was detectable in BS153resE by western blot analysis (Fig. 4E). Taken together, BS153resE cells displayed increased expression of EGFRvIII in comparison with parental BS153, which was associated with higher activity of PI3K, in particular due to increased expression of p110δ.
BS153resE Exhibits Delayed Spheroid Growth In Vitro and Retarded Tumor Growth In Vivo
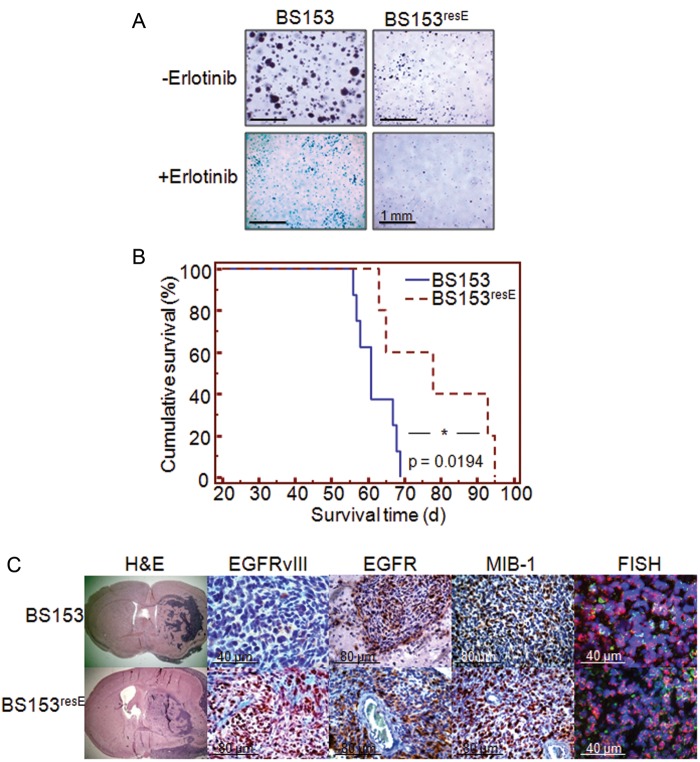
To determine the tumor-initiating capacity of BS153resE, we first analyzed the anchorage-independent growth of BS153 and BS153resE in a colony formation assay (Fig. 5A).24,28 BS153 readily formed spheroids in soft agar after 48 h of incubation, while erlotinib-treated BS153 started to form spheroids as late as 7 days after seeding. Unexpectedly, BS153resE spheroid formation was compromised in the presence and in the absence of erlotinib, indicating that erlotinib induced permanent changes in BS153resE, resulting in a less aggressive phenotype.
Fig. 5.
Tumor-initiating capacity of BS153 cells. Anchorage-independent growth of BS153 in soft agar showed delayed sphere formation either for cells acutely treated with erlotinib or for chronically exposed BS153resE cells in the absence or presence of erlotinib after 7 days of incubation (A). When implanted into nude mice, BS153resE (n = 5) gave rise to symptomatic tumors significantly later than parental BS153 (B; n = 7, Kaplan–Meier, P = .0194, log-rank test). Histological analysis identified unusual morphology and an invasive phenotype for tumors from both cell lines (C, H&E), while strong expression of EGFRvIII as observed in vitro remained low in BS153 and high in BS153resE (C, EGFRvIII). Total EGFR expression (wtEGFR + EGFRvIII) was high in tumors from both cell lines (C, EGFR), while EGFRvIII was overexpressed in BS153resE tumors (C, EGFRvIII). BS153- and BS153resE-derived tumors were highly proliferative, as visualized by staining with MIB-1, and preserved their gene amplification (C, FISH).
To further validate these findings, BS153 and BS153resE were implanted intracranially into nude mice. Animals carrying BS153 xenografts developed tumor-related symptoms significantly earlier than did animals carrying BS153resE (61 d vs 78 d, P = .0194, log-rank test; Fig. 5B). Additionally, animals with BS153 tumors sacrificed 4 weeks after cell implantation already displayed clear tumor formation (Supplementary Fig. S3), whereas animals implanted with BS153resE showed no pathology at this point (not shown), confirming the in vitro observations. Interestingly, BS153 grew as tumors with solid areas as well as prominent invasive features (Fig. 5C), recapitulating a major hallmark of GBM histopathology. Despite their delayed onset, tumors of BS153resE cells resembled BS153 tumors closely and exhibited identical histopathological features, while BS153resE exhibited no difference in proliferation relative to tumors from BS153 as indicated by MIB–1 staining (Fig. 5C). Importantly, the molecular characteristics observed for BS153 and BS153resE cells in vitro were preserved in vivo. At the genomic level, amplification of the egfr gene was preserved, as FISH analysis of paraffin-embedded tissue showed (Fig. 5C). At the protein level, high expression of total EGFR was confirmed for BS153 and BS153resE by immunohistochemistry. In particular, the upregulation of EGFRvIII detected in vitro in BS153resE was maintained in BS153resE xenograft tumors, whereas only a few cells expressed EGFRvIII in BS153-derived tumors.
EGFRvIII Is Essential for Resistance to Erlotinib, and Resistant Cells Are Sensitive to Inhibition of Downstream Mediators of EGFR Signaling
To substantiate the role of EGFRvIII for the continued proliferation of BS153resE in the presence of erlotinib, we performed siRNA knockdown in BS153 and BS153resE (Supplementary Fig. S5).21 Knockdown of EGFRvIII in the absence of erlotinib reduced proliferation of BS153resE by 25% (P < .005; Fig. 6A). More importantly, EGFRvIII knockdown resensitized BS153resE to erlotinib and reduced proliferation by almost 50% relative to control cells (P < .005). These data indicate a central role for EGFRvIII in BS153resE proliferation, as well as for their tolerance for high doses of erlotinib. Notably, knockdown of EGFRvIII in parental BS153, although significantly inhibiting proliferation (P < .05), did not increase sensitivity to erlotinib upon acute treatment with the TKI.
Fig. 6.
A central role for EGFRvIII and PI3Kp110δ in erlotinib resistance. Transient knockdown of EGFRvIII by specific siRNA led to a reduced proliferation of BS153resE and restored sensitivity to erlotinib (A; **P < .005, *P = .05, t-test); RLU, relative luminescence units. At the protein level, EGFRvIII knockdown reduced downstream phosphorylation of PI3Kp110 and phosphorylation of Akt and ERK (B). Pharmacological interference with overall PI3K activity using PX-866 significantly reduced proliferation at concentrations above 1 µM for both cell lines after 6 d of incubation. Below 1 µM, only BS153resE was significantly affected (C, grey asterisks). PX-866 inhibited proliferation of BS153resE stronger than proliferation of BS153 at all concentrations tested. Specifically targeting p110δ by siRNA-mediated knockdown significantly decreased proliferation of BS153resE. Additionally, BS153resE was resensitized to treatment with erlotinib (D); values are means ± SD of octuplicate determinations. One of 3 independent experiments is shown.
The analysis of downstream signaling events in response to EGFRvIII knockdown revealed a decrease in PI3K, Akt, and ERK phosphorylation in BS153resE (Fig. 6B) without changes in overall protein levels. We therefore hypothesized that interfering with PI3K signaling should effectively target BS153resE. We treated BS153 as well as BS153resE with PX–866, a Wortmannin-derived PI3Kp110 inhibitor directed against all 4 isoforms of p110 that is currently being tested in a phase II clinical trial for the treatment of recurrent GBM.29 PX-866 inhibited BS153resE proliferation significantly more strongly than BS153 at all concentrations tested (P < .005, Fig. 6C). Additionally, BS153 responded to PX-866 only at concentrations above 1 µM, pointing to a pivotal role for PI3K activity in BS153resE. Since BS153resE displayed a strong upregulation of p110δ (Fig. 4F), we tested whether specifically interfering with this p110 isoform using siRNA would lead to results similar to those obtained by interfering with all p110 isoforms (Supplementary Fig. S6).22 Both BS153 and BS153resE proliferated significantly more slowly when transfected with p110δ siRNA, yet the effect was more pronounced in BS153resE (Fig. 6D). Knockdown of p110δ combined with erlotinib treatment of BS153 did not show cumulative effects on proliferation compared with erlotinib treatment alone. However, p110δ knockdown resensitized BS153resE to erlotinib in a fashion similar to EGFRvIII knockdown, indicating the crucial role of p110δ in mediating EGFRvIII-dependent erlotinib resistance in BS153resE.
Discussion
The significance of EGFR amplification and overexpression for glioblastoma biology and the relevance for choice of treatment is still unclear despite enormous research efforts. Recently, we were able to demonstrate that the effects of natural EGFR overexpression differ considerably from those described for engineered overexpression,16 indicating that results obtained from artificial model systems might not be representative of original tumors. Since controversy exists regarding the efficacy of EGFR-directed therapies in the context of EGFR amplification, we used BS153, a GBM-derived, egfr-amplified, EGFRvIII-positive cell line, to systematically compare the functional effects of erlotinib, gefitinib, and cetuximab.
We found that erlotinib and gefitinib inhibited phosphorylation of wtEGFR and thereby proliferation and migration of BS153, with gefitinib being more potent than erlotinib. This was accompanied by effective inhibition of Akt phosphorylation by gefitinib, while Akt inhibition by erlotinib was less efficient. In cultured lung cancer cell lines, sensitivity to gefitinib has been correlated with dependence on Akt signaling.30 Our data suggest that the PI3K/Akt pathway is a more potent driver of proliferation and migration in BS153 than is the Ras/ERK pathway. This may also explain the stronger effect of gefitinib on BS153, which significantly inhibited Akt phosphorylation, whereas erlotinib predominantly reduced signaling via ERK. Importantly, only little inhibition of EGFRvIII phosphorylation by both TKIs occurred in BS153. We recently obtained similar results in glioma stemlike cells that retained EGFR amplification and EGFRvIII expression present in the original tumor.16 In those cells, erlotinib and gefitinib also inhibited wtEGFR phosphorylation but had no effect on EGFRvIII phosphorylation. These findings are in contrast to observations of U87MG cells engineered to overexpress EGFRvIII, in which EGFR TKIs were shown to effectively inhibit EGFRvIII phosphorylation.31 These discrepancies are probably due to the use of different cell line models that either express EGFRvIII endogenously or artificially overexpress the mutant receptor.13 Impaired inhibition of EGFRvIII in BS153 may partially explain why downstream signaling events were incompletely blocked by erlotinib and gefitinib.
Interestingly, cetuximab had no inhibitory effect in our in vitro analysis and even stimulated phosphorylation of wtEGFR and EGFRvIII, followed by stimulation of proliferation and migration of BS153 via Akt. Agonistic effects of cetuximab were described previously in H292 non–small cell lung cancer cells, in which cetuximab also induced phosphorylation of wtEGFR.32 Furthermore, Akt downstream signaling has been shown to persist despite cetuximab treatment in egfr-amplified SKMG-3 glioma cells.33 Cetuximab was also reported to bind to EGFRvIII in transfected U373 glioma cells, causing enhanced phosphorylation of the mutant receptor.34 Importantly, the effect of cetuximab in vivo differs from its effect in vitro. Previously, we and others have shown that cetuximab effectively inhibits proliferation of EGFR-amplified, EGFRvIII-expressing xenograft tumors derived from freshly resected patient material, while it failed to inhibit nonamplified xenografts.35–37 The tumor microenvironment in vivo may play an important role in mediating the antitumor effects of cetuximab. For example, antibody-dependent cellular cytotoxicity has been suggested as a possible mechanism of cetuximab function rather than direct inhibition of EGFR, even in NMRI/Foxn1nu mice.38,39
BS153 displayed resistance to erlotinib but not to gefitinib; that is cells survived treatment with the drug and expanded persistently even in the presence of 25 µM of the TKI, possibly due to insufficient inhibition of Akt signaling. Moreover, erlotinib-resistant cells were still susceptible to treatment with gefitinib, showing that erlotinib and gefitinib have substantially different downstream effects. In lung cancer, gefitinib has been approved as a first-line monotherapy for patients harboring activating mutations in the tyrosine kinase domain of the EGFR.40 These mutations were shown to increase the affinity of gefitinib to the ATP-binding pocket of the EGFR.41 In contrast to what is observed in lung cancer, mutations in the tyrosine kinase domain of EGFR are virtually absent in GBM.42 Our findings demonstrate that in EGFR-amplified, EGFRvIII-expressing glioma cells, erlotinib and gefitinib have different molecular mechanisms, although they share a similar chemical backbone and both compete for the ATP-binding site in the tyrosine kinase domain of EGFR.43 Additionally, off-target effects of gefitinib may in part explain the differential effect on BS153 and the effect of the TKI on EGFR-negative SW620 colorectal cancer cells, since gefitinib has been reported to also inhibit other kinases—Lyn, RICK, BLK, and JNK2—with a half-maximal inhibitory concentration comparable to that of EGFR.44
The relevance of EGFRvIII for the response to erlotinib in glioma is disputed. In patients with recurrent glioma treated with erlotinib or gefitinib, EGFRvIII expression, if coexpressed with PTEN, was described as a positive predictor for response to tyrosine kinase inhibition.45 In contrast, no association between EGFRvIII, PTEN, and response to erlotinib was found in a large, randomized phase II trial analyzing recurrent GBM.7 A phase I trial described a positive response to erlotinib to be associated with EGFR overexpression and amplification combined with low levels of phosphorylated Akt in glioma patients, but none of the responders expressed EGFRvIII.46 Our data show that EGFRvIII-expressing BS153 initially responds to erlotinib. Still, a strong upregulation of EGFRvIII is a major mechanism of resistance, since its knockdown resensitized resistant cells to erlotinib in our study. This could be explained by the selective depletion of cells expressing only wtEGFR, which are thus reliant on ligand-induced receptor activation and can be efficiently blocked by TKIs. This is supported by our finding that other glioma cell lines, which express only wtEGFR endogenously but not EGFRvIII, such as U87MG and G55, do not survive continued treatment with erlotinib (data not shown). Interestingly, studies by Sampson et al47 showed that vaccination against EGFRvIII selectively eradicated EGFRvIII-positive cells. This suggests selective vulnerability of individual tumor cells, based on their EGFR status, which is known to be intratumorally heterogeneous; that is, only a subpopulation of cells express EGFRvIII and are amplified for egfr.16
It is conceivable that depletion of wtEGFR-expressing cells also accounts for the delay in sphere formation and tumor initiation observed for BS153resE. WtEGFR signaling sustains a stemlike phenotype, which is thought to be a prerequisite for tumor initiation.16,48,49 EGFRvIII by itself does not increase tumor-initiating capacity and has been described as enhancing tumor growth of U87MG xenografts only when wtEGFR is overexpressed as well.13 EGFRvIII has rather been associated with tumor invasiveness, which is reflected in the unusual invasive growth pattern observed for BS153 and BS153resE in vivo.4,23,50
Targeting common downstream mediators of EGFR, like PI3K, perhaps the most important mediator of EGFRvIII signaling,51,52 might help improve EGFR-directed therapy. We found PI3K to be active even in the presence of erlotinib in BS153resE, causing persistent activation of Akt. The reduction of PTEN protein, an important negative regulator of Akt activity, additionally enforces PI3K/Akt pathway activity in BS153resE. This is in line with several in vitro studies that link a loss of PTEN to resistance against tyrosine kinase inhibition.53,54 Interfering with PI3K by applying inhibitors like PX-866 that target all isoforms of PI3K could bypass erlotinib resistance in BS153resE. However, a number of side effects would be expected in healthy tissue because PX-866 targets all PI3K isoforms.55 Therefore, the identification of p110δ as a major mediator of erlotinib resistance in BS153resE in our study is of particular importance because p110δ is usually not expressed in the healthy brain. P110δ can negatively control PTEN and further escalate PI3K/Akt signaling in a RhoA-dependent fashion and has only recently been described to mediate glioma cell migration and invasion.22,26,56 We now show for the first time that p110δ is upregulated in EGFR-amplified glioblastoma cells in response to pharmaceutical intervention with erlotinib treatment. Additionally, p110δ is crucial in mediating resistance to erlotinib downstream of EGFRvIII because its knockdown reduced proliferation and resensitized resistant BS153 to treatment with erlotinib. A specific inhibitor for p110δ, CAL-101, showed promising preclinical results in lymphoma, is currently being tested in a number of clinical trials, and might offer a new treatment approach for GBM, especially GBM with amplified egfr and expression of EGFRvIII.22,26,57
Supplementary Material
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (LA1300/4-1), Deutsche Krebshilfe (M.W., K.L.), Forschungsförderungsfonds der Medizinischen Fakultät Hamburg (T.M., A.S.), the Erich and Gertrud Roggenbuck-Stiftung Hamburg (M.W., K.L.), the Georg und Jürgen Rickertsen Stiftung, Hamburg (A.S., M.W.), and the Forschungs- und Wissenschaftsstiftung Hamburg (M.W., K.L.).
Supplementary Material
Acknowledgments
The authors thank Malgorzata Stoupiec from the Department of Tumor Biology, UKE Hamburg, for expert technical assistance and Dr. Darell Bigner, Duke University, for the EGFRvIII-specific antibody. The SW620 colorectal cancer cell line was a gift from Prof. Steven A. Johnsen, Department of Tumor Biology, UKE. The authors acknowledge Dr. Stéphane Marguet from the Centre for Molecular Neurobiology, Hamburg, for critically reading the manuscript.
Conflict of interest statement. None declared.
References
- 1.Libermann TA, Nusbaum HR, Razon N, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313(5998):144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 2.Wong AJ, Ruppert JM, Bigner SH, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89(7):2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphrey PA, Wong AJ, Vogelstein B, et al. Anti-synthetic peptide antibody reacting at the fusion junction of deletion-mutant epidermal growth factor receptors in human glioblastoma. Proc Natl Acad Sci U S A. 1990;87(11):4207–4211. doi: 10.1073/pnas.87.11.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lal A, Glazer CA, Martinson HM, et al. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 2002;62(12):3335–3339. [PubMed] [Google Scholar]
- 5.Lund-Johansen M, Bjerkvig R, Humphrey PA, Bigner SH, Bigner DD, Laerum OD. Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res. 1990;50(18):6039–6044. [PubMed] [Google Scholar]
- 6.Rich JN, Reardon DA, Peery T, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22(1):133–142. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 7.van den Bent MJ, Brandes AA, Rampling R, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27(8):1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, Diserens AC, Bady P, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib—a phase II trial. Mol Cancer Ther. 2011;10(6):1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- 10.Lassman AB, Rossi MR, Raizer JJ, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01–03 and 00–01. Clin Cancer Res. 2005;11(21):7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 11.Steinbach JP, Klumpp A, Wolburg H, Weller M. Inhibition of epidermal growth factor receptor signaling protects human malignant glioma cells from hypoxia-induced cell death. Cancer Res. 2004;64(5):1575–1578. doi: 10.1158/0008-5472.can-03-3775. [DOI] [PubMed] [Google Scholar]
- 12.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39(1):29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 13.Inda MM, Bonavia R, Mukasa A, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens T, Laabs Y, Gunther HS, et al. Inhibition of glioblastoma growth in a highly invasive nude mouse model can be achieved by targeting epidermal growth factor receptor but not vascular endothelial growth factor receptor-2. Clin Cancer Res. 2008;14(17):5447–5458. doi: 10.1158/1078-0432.CCR-08-0147. [DOI] [PubMed] [Google Scholar]
- 15.Filmus J, Pollak MN, Cairncross JG, Buick RN. Amplified, overexpressed and rearranged epidermal growth factor receptor gene in a human astrocytoma cell line. Biochem Biophys Res Commun. 1985;131(1):207–215. doi: 10.1016/0006-291x(85)91790-5. [DOI] [PubMed] [Google Scholar]
- 16.Schulte A, Gunther HS, Martens T, et al. Glioblastoma stem-like cell lines with either maintenance or loss of high-level EGFR amplification, generated via modulation of ligand concentration. Clin Cancer Res. 2012;18(7):1901–1913. doi: 10.1158/1078-0432.CCR-11-3084. [DOI] [PubMed] [Google Scholar]
- 17.Del Vecchio CA, Giacomini CP, Vogel H, et al. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene. 2013;32:2670–2681. doi: 10.1038/onc.2012.280. [DOI] [PubMed] [Google Scholar]
- 18.Jones G, Machado J, Jr, Merlo A. Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 2001;61(13):4978–4981. [PubMed] [Google Scholar]
- 19.Schulte A, Schulz B, Andrzejewski MG, et al. Sequential processing of the transmembrane chemokines CX3CL1 and CXCL16 by alpha- and gamma-secretases. Biochem Biophys Res Commun. 2007;358(1):233–240. doi: 10.1016/j.bbrc.2007.04.100. [DOI] [PubMed] [Google Scholar]
- 20.Rae JM, Scheys JO, Clark KM, Chadwick RB, Kiefer MC, Lippman ME. EGFR and EGFRvIII expression in primary breast cancer and cell lines. Breast Cancer Res Treat. 2004;87(1):87–95. doi: 10.1023/B:BREA.0000041585.26734.f9. [DOI] [PubMed] [Google Scholar]
- 21.Yamoutpour F, Bodempudi V, Park SE, et al. Gene silencing for epidermal growth factor receptor variant III induces cell-specific cytotoxicity. Mol Cancer Ther. 2008;7(11):3586–3597. doi: 10.1158/1535-7163.MCT-08-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luk SK, Piekorz RP, Nurnberg B, Tony To SS. The catalytic phosphoinositol 3-kinase isoform p110delta is required for glioma cell migration and invasion. Eur J Cancer. 2012;48(1):149–157. doi: 10.1016/j.ejca.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Schulte A, Gunther HS, Phillips HS, et al. A distinct subset of glioma cell lines with stem cell-like properties reflects the transcriptional phenotype of glioblastomas and overexpresses CXCR4 as therapeutic target. Glia. 2011;59(4):590–602. doi: 10.1002/glia.21127. [DOI] [PubMed] [Google Scholar]
- 24.Lamszus K, Jin L, Fuchs A, et al. Scatter factor stimulates tumor growth and tumor angiogenesis in human breast cancers in the mammary fat pads of nude mice. Lab Invest. 1997;76(3):339–353. [PubMed] [Google Scholar]
- 25.Yang JL, Qu XJ, Russell PJ, Goldstein D. Interferon-alpha promotes the anti-proliferative effect of erlotinib (OSI-774) on human colon cancer cell lines. Cancer Lett. 2005;225(1):61–74. doi: 10.1016/j.canlet.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 26.Herman SE, Gordon AL, Wagner AJ, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078–2088. doi: 10.1182/blood-2010-02-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herman SE, Lapalombella R, Gordon AL, et al. The role of phosphatidylinositol 3-kinase-delta in the immunomodulatory effects of lenalidomide in chronic lymphocytic leukemia. Blood. 2011;117(16):4323–4327. doi: 10.1182/blood-2010-11-315705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Courtenay VD, Mills J. An in vitro colony assay for human tumours grown in immune-suppressed mice and treated in vivo with cytotoxic agents. Br J Cancer. 1978;37(2):261–268. doi: 10.1038/bjc.1978.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitz MW, MacNeil MV, Macdonald DR, et al. Phase II study of PX-866 in recurrent glioblastoma. J Clin Oncol. 2012 doi: 10.1093/neuonc/nou365. ASCO Annual Meeting Proceedings (Post-Meeting Edition). 2012;30(15_suppl):2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono M, Hirata A, Kometani T, et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3(4):465–472. [PubMed] [Google Scholar]
- 31.Learn CA, Hartzell TL, Wikstrand CJ, et al. Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Clin Cancer Res. 2004;10(9):3216–3224. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida T, Okamoto I, Okabe T, et al. Matuzumab and cetuximab activate the epidermal growth factor receptor but fail to trigger downstream signaling by Akt or Erk. Int J Cancer. 2008;122(7):1530–1538. doi: 10.1002/ijc.23253. [DOI] [PubMed] [Google Scholar]
- 33.Hasselbalch B, Lassen U, Poulsen HS, Stockhausen MT. Cetuximab insufficiently inhibits glioma cell growth due to persistent EGFR downstream signaling. Cancer Invest. 2010;28(8):775–787. doi: 10.3109/07357907.2010.483506. [DOI] [PubMed] [Google Scholar]
- 34.Jutten B, Dubois L, Li Y, et al. Binding of cetuximab to the EGFRvIII deletion mutant and its biological consequences in malignant glioma cells. Radiother Oncol. 2009;92(3):393–398. doi: 10.1016/j.radonc.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 35.Johns TG, Perera RM, Vernes SC, et al. The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res. 2007;13(6):1911–1925. doi: 10.1158/1078-0432.CCR-06-1453. [DOI] [PubMed] [Google Scholar]
- 36.Martens T, Schmidt NO, Eckerich C, et al. A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo. Clin Cancer Res. 2006;12(20 Pt 1):6144–6152. doi: 10.1158/1078-0432.CCR-05-1418. [DOI] [PubMed] [Google Scholar]
- 37.Lamszus K, Brockmann MA, Eckerich C, et al. Inhibition of glioblastoma angiogenesis and invasion by combined treatments directed against vascular endothelial growth factor receptor-2, epidermal growth factor receptor, and vascular endothelial-cadherin. Clin Cancer Res. 2005;11(13):4934–4940. doi: 10.1158/1078-0432.CCR-04-2270. [DOI] [PubMed] [Google Scholar]
- 38.Fukai J, Nishio K, Itakura T, Koizumi F. Antitumor activity of cetuximab against malignant glioma cells overexpressing EGFR deletion mutant variant III. Cancer Sci. 2008;99(10):2062–2069. doi: 10.1111/j.1349-7006.2008.00945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roda JM, Joshi T, Butchar JP, et al. The activation of natural killer cell effector functions by cetuximab-coated, epidermal growth factor receptor positive tumor cells is enhanced by cytokines. Clin Cancer Res. 2007;13(21):6419–6428. doi: 10.1158/1078-0432.CCR-07-0865. [DOI] [PubMed] [Google Scholar]
- 40.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26(15):2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 41.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 42.Marie Y, Carpentier AF, Omuro AM, et al. EGFR tyrosine kinase domain mutations in human gliomas. Neurology. 2005;64(8):1444–1445. doi: 10.1212/01.WNL.0000158654.07080.B0. [DOI] [PubMed] [Google Scholar]
- 43.Siegel-Lakhai WS, Beijnen JH, Schellens JH. Current knowledge and future directions of the selective epidermal growth factor receptor inhibitors erlotinib (Tarceva) and gefitinib (Iressa) Oncologist. 2005;10(8):579–589. doi: 10.1634/theoncologist.10-8-579. [DOI] [PubMed] [Google Scholar]
- 44.Brehmer D, Greff Z, Godl K, et al. Cellular targets of gefitinib. Cancer Res. 2005;65(2):379–382. [PubMed] [Google Scholar]
- 45.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 46.Haas-Kogan DA, Prados MD, Tihan T, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97(12):880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 47.Sampson JH, Aldape KD, Archer GE, et al. Greater chemotherapy-induced lymphopenia enhances tumor-specific immune responses that eliminate EGFRvIII-expressing tumor cells in patients with glioblastoma. Neuro Oncol. 2011;13(3):324–333. doi: 10.1093/neuonc/noq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J, Jung J, Lee SJ, Lee JS, Park MJ. Cancer stem-like cells persist in established cell lines through autocrine activation of EGFR signaling. Oncol Lett. 2012;3(3):607–612. doi: 10.3892/ol.2011.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzoleni S, Politi LS, Pala M, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2011;70(19):7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 50.Okada Y, Hurwitz EE, Esposito JM, Brower MA, Nutt CL, Louis DN. Selection pressures of TP53 mutation and microenvironmental location influence epidermal growth factor receptor gene amplification in human glioblastomas. Cancer Res. 2003;63(2):413–416. [PubMed] [Google Scholar]
- 51.Choe G, Horvath S, Cloughesy TF, et al. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63(11):2742–2746. [PubMed] [Google Scholar]
- 52.Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HK. Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Oncogene. 2004;23(26):4594–4602. doi: 10.1038/sj.onc.1207602. [DOI] [PubMed] [Google Scholar]
- 53.Bianco R, Shin I, Ritter CA, et al. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene. 2003;22(18):2812–2822. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- 54.Sos ML, Koker M, Weir BA, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69(8):3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koul D, Shen R, Kim YW, et al. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12(6):559–569. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papakonstanti EA, Ridley AJ, Vanhaesebroeck B. The p110delta isoform of PI 3-kinase negatively controls RhoA and PTEN. EMBO. 2007;26(13):3050–3061. doi: 10.1038/sj.emboj.7601763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meadows SA, Vega F, Kashishian A, et al. PI3Kdelta inhibitor, GS-1101 (CAL-101), attenuates pathway signaling, induces apoptosis, and overcomes signals from the microenvironment in cellular models of Hodgkin lymphoma. Blood. 2012;119(8):1897–1900. doi: 10.1182/blood-2011-10-386763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.