Abstract

The pH-regulated M2 proton channel from the influenza A virus has a His-tetrad in its transmembrane (TM) domain that is essential for proton conduction. At neutral pH, the tetrad has been observed in two distinct configurations, the “His-box” and “dimer-of-dimers”, with similar backbone structures suggesting competing models for proton conduction. Here, we propose that both conformations can play a role. In support of this hypothesis, we used molecular dynamics simulations based on density functional theory to simulate the M2-TM domain and force-field-based simulations to estimate the relevant free-energy barriers. Both configurations are stable on accessible simulation time scales, and transitions between them occur faster than the millisecond time scale of proton conduction. Moreover, the deprotonation energy is too high for spontaneous conduction, consistent with their occurrence in the low-current regime. Our computations support a multiconfiguration model with different population levels, thereby connecting experimental data obtained under different conditions.
The diffusion of protons (H+) through water is a ubiquitous phenomenon with unique characteristics among those involving atomic cations. Few examples exist of controlled proton diffusion near neutral pH; one of them comes from biology, the M2 protein of the influenza A virus, which is a tetrameric channel embedded in the viral membrane, conducting protons from the exterior of a viral particle via its lumen.1,2 The conduction is modulated by the presence of a lower pH level at the exterior; after entering a host cell, a viral particle lowers its pH to match the cellular level and to release its viral genetic material and trigger replication. Therefore, significant effort has been devoted to understand the mechanism of proton conduction through a lipid membrane by the M2 channel.3−5
Extensive experimental6−14 and computational15−21 work has been devoted to study the structure of the tetrameric M2 channel protein, especially various M2-TM constructs. Not surprisingly, the structure of its backbone is sensitive to pH, and an allosteric mechanism was hypothesized to model pH regulation.8,10,20 Central to this mechanism are the histidine amino acids, evolutionarily required for pH-regulated conduction of protons and other cations.22−24 When histidine is present in the amino acid sequence of M2 at position 37 (His37), the conduction rate is highly nonlinear with the proton concentration, suggesting that the two mechanisms of gating and conduction are intimately connected.22,23 To understand both mechanisms, knowledge of the structural ensemble of the His37 tetrad is paramount.
One of the first atomic-resolution experiments specifically aimed at the His37 tetrad measured the magnetic chemical shielding of their nitrogen atoms by solid-state nuclear magnetic resonance (SSNMR) spectroscopy, at a temperature of 277 K and pH levels in the range between 8.5 and 5.0. The magnitude of the measured shieldings suggested a dimer-of-dimers arrangement for the imidazole groups,6 in which each of the charged His37 forms a strong hydrogen bond (HB) with an adjacent neutral His37. On the basis of this hypothesis, the structure of the His37 tetrad was further refined by additional SSNMR measurements on the protein backbone and by quantum mechanical calculations (PDB entry: 2L0J).14 In apparent contrast, a high-resolution X-ray structure obtained at 100 K and pH 6.5 (PDB entry: 3LBW; 1.65 Å) exhibited a tightly packed His-box,10 with the imidazoles forming HBs with adjacent water molecules rather than with each other.6,14
Extensive experimental work emerged to support either the dimer-of-dimers25−27 or the His-box4,12,28−30 configuration, suggesting that the two are mutually exclusive. However, another possibility is that the two configurations coexist at neutral pH but that their respective populations are finely tuned by experimental conditions such as pH, temperature, and the composition of the viral membrane mimic. In this work, we explore the latter hypothesis using molecular dynamics (MD) protocols that span multiple time scales.
We first studied the stability of the two alternative configurations, which are putatively in the doubly protonated (+2 charge) state at neutral pH conditions (Figure 1a and b),6,28 by simulating these tetrameric M2-TM proteins embedded in fully hydrated phospholipid bilayers. To accurately model the structure of the proton-binding chemical groups, we used a quantum mechanical/molecular mechanical (QM/MM) methodology, explicitly calculating the electronic structure of His37, the adjacent Trp41, plus neighboring water molecules. To do so, we made use of density functional theory (DFT) with the Becke exchange31 and Lee–Yang–Parr correlation32 functionals, including semiempirical dispersion corrections.33 To model the equilibrium structure of the protein and the distribution of water in the lumen, we embedded the high-resolution X-ray structure10 in a hydrated lipid bilayer with the His37 and Trp41 constrained in configuration and the rest of the system simulated with the CHARMM and TIP3P force fields.34−36 We equilibrated each system for 40 ns and extracted two uncorrelated snapshots as the starting points for subsequent QM/MM MD simulations; each of the four were accumulated for ∼25 ps (100 ps in total).
Figure 1.
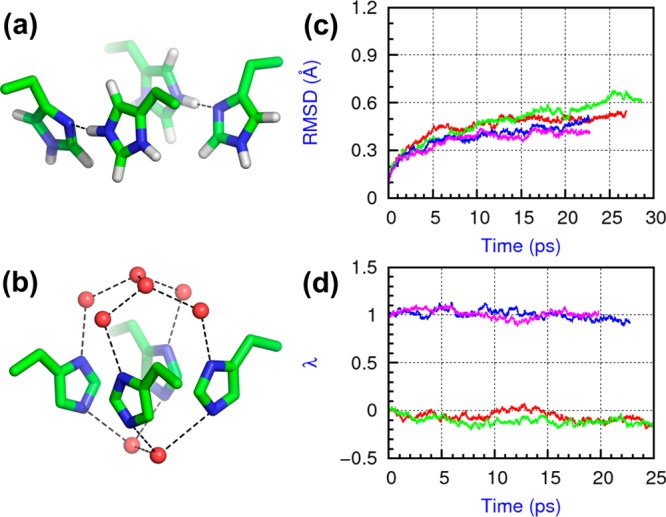
Two His37 configurations in the M2 protein at the 2+ state and their stabilities. (a) The dimer-of-dimers configuration (PDB entry: 2L0J); (b) the His-box configuration (PDB entry: 3LBW); (c) RMSDs of the backbone Cα atoms in the simulations; (d) principal component of the His37 tetrad configurations as a function of time. In both (c) and (d), two parallel simulations of the dimer-of-dimers model are colored in red and green, and two parallel simulations of the His-box model are in blue and pink.
Within the time scale explored, both models appear to represent locally stable or metastable minima on the free-energy surface of the system. The root-mean-square deviations (RMSDs) of the backbone Cα atoms from the starting snapshot quickly saturate to a value of 0.6 Å (Figure 1c), indicating that the protein backbone had been fully relaxed during the 40 ns pre-equilibration steps (Figure S1, Supporting Information). To detect possible transitions of the His37 tetrad between the two configurations, we calculated a principal component, λ, along the vector connecting the two configurations (see the Supporting Information): λ = 0 represents the dimer-of-dimers configuration and λ = 1 the His-box. During all simulations, λ-values remained within 0.1 from their initial values (Figure 1d), showing the stability of both configurations. We then monitored the specific structural motifs of them.
On the basis of chemical shift measurements, in the dimer-of-dimers model, each dimer was proposed to form a low-barrier hydrogen bond (LBHB),6 with the shared proton freely moving between the donor and the acceptor.37 Our simulations confirm the ∼2.75 Å distance between the donor and acceptor N atoms and explicitly show multiple proton “jumps” between the two (Figures 2 and S2, Supporting Information). Although the proton was treated as a classical particle, a high rate of jumps indicates a rather delocalized wave function.38 We did not observe a strong preference for either the Nδ position (π tautomer) or the Nε position (τ tautomer); the relative occupancies were ∼40% and ∼60%, respectively. This suggests that the structure of the histidine tetrad tunes the pKa’s of the two sites to be approximately equal, providing optimal conditions to form LBHBs. Furthermore, the LBHBs appear explicitly to be robust structural motifs; even though one LBHB pair temporarily broke in one of the simulations, it quickly re-formed in ∼2 ps (Figure S2c, Supporting Information).
Figure 2.

Correlation between the HB distance and proton population in two independent simulations of the dimer-of-dimers model. The inset graph shows the jump frequency of the proton within a dimer in the dimer-of-dimers model by monitoring the hydrogen-bonding-involved Nδ−H (red) and Nε–H (green) distances.
In the His-box configuration, despite the tight packing, there are no direct HBs between the four His37; instead, each forms HBs with the adjacent clusters of water molecules,10 and the overall His37–water HB network is well-conserved in our simulations. The distribution of water molecules fits well to the electron density determined in the X-ray structure (Figure S3, Supporting Information).10 A few HB distances changed with respect to the X-ray structure; the two outermost waters of the “entry cluster”, separated by ∼2.6 Å in the X-ray structure,10 featured an average O···O distance of ∼2.8 Å during our simulations. The Nδ1···O distance between all four His37 and water is ∼2.8 Å in the X-ray structure; in our simulations, one charged His37 reduced its HB distance to ∼2.7 Å, while the remaining three increased to 3.0–3.1 Å. HB distances with the two waters at the Nε2 positions did not differ from the X-ray structure beyond their observed fluctuations. However, the magnitude of such fluctuations is larger than that of the six waters at the Nδ1 positions, a fact consistent with the 15N–1H dipolar couplings measured by SSNMR.12
The Trp41 side chains, neighboring His37, constitute a structural motif for both configurations, as shown by the high-resolution structures10,14 and suggested by Raman spectroscopy.39 At pH < 6, when 3 or 4 of the His37 are thought to be charged, a cation−π interaction has been suggested between each charged His37 and the indole of the neighboring Trp41.39 For the 2+ charge state studied here, such cation−π interactions are obviously weaker due to the delocalization of charge.10,14,21 Nevertheless, they are preserved during our simulations; in the dimer-of-dimers, the distance and the angle between the His37 N–H and the indole plane remain at 3.15 Å and 72°, respectively; in the His-box, the two are 3.15 Å and 45°. These cation−π interactions may play a role in stabilizing the rotameric state of Trp41, which is the tp rotamer (χ1 = 180°, χ2 = −90°) in the dimer-of-dimers and the tm rotamer (χ1 = 180°, χ2 = 90°) in the His-box. If additional protons bind to the His37, cation−π interactions are likely to become prominent in both models, reaching the threshold for being detected by Raman spectroscopy.39
Because the presence of water through the channel is the prerequisite for proton conduction,40 we monitored during all simulations the distribution of water and the size of the channel’s pore. In both configurations, two constriction regions could be identified along the pore (Figure 3). One region, formed by His37 and Trp41, is responsible for gating and conduction,22,23,41 and the other region, formed by Val27, is at the outer end of the channel and is shown in previous simulations to partially hinder the water flow through the channel.4,10,17,20 While these two regions are narrow in both configurations, the dimer-of-dimers configuration features a wider hydrophobic portion (radius = 1.6 Å) and a narrower hydrophilic portion (0.9 Å); the His-box has, instead, radii of 1.0 and 1.3 Å, respectively. This alternation is reminiscent of the allosteric mechanism of proton conduction proposed for the M2 channel,8,10,20 where pH regulates the dynamical equilibrium between two states, labeled “openout–closedin” and “closedout–openin”.42
Figure 3.
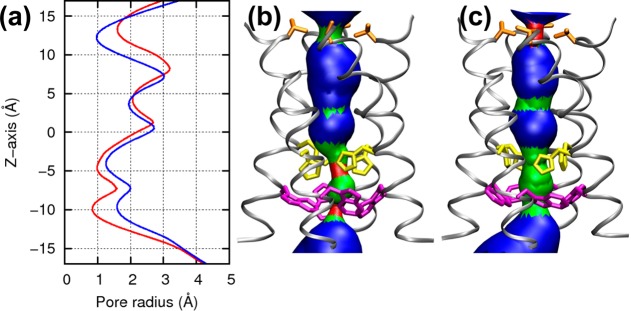
Pore size of the two configurations. (a) Averaged radius of the pore in the dimer-of-dimers (red line) and the His-box (blue line) models. The pore dimension of the dimer-of-dimers model (b) and the His-box model (c) were generated by the HOLE program. The backbone is shown with a silver ribbon, and side chains of Val27, His37 and Trp41 are shown in orange, yellow, and magenta, respectively, in stick mode.
Having failed to observe spontaneous loss of protons in the previous simulations, we then calculated explicitly, for both configurations, the energy differences for deprotonation. To this end, we computed the difference between the final structure of each QM/MM MD simulation and a structure where one proton was removed from the Nε2 position of one charged His37 and bound to the adjacent water molecule to form hydronium (H3O+). We then optimized this structure (Figure S4, Supporting Information) for both configurations. The energy difference in the dimer-of-dimers reaches ∼40 kcal/mol, which compares well with the estimated LBHB formation energy of ∼31 kcal/mol in the gas phase;43 no stable HBs with water are formed in the product. In the His-box configuration, the calculated energy difference between the two is ∼12 kcal/mol, which is high enough to prevent fast spontaneous release of protons. The relatively high energies can be most likely attributed to the presence of only two acceptor water molecules; when one of them is transformed into an H3O+, the lack of a complete solvation shell destabilizes the product. These results indicate that the proton release, bringing the His37 tetrad from a 2+ to a 1+ charge state, is a kinetically unfavorable event, if not thermodynamically. In the SSNMR experiment that first proposed the dimer-of-dimers model, the second pKa constant of the His37 tetrad was estimated at 8.2, two units higher than the third pKa of 6.3.6 In another SSNMR experiment using a different membrane mimic, consistent instead with the His-box model,28 these were estimated as 6.8 and 4.9, showing the same difference of two units. Therefore, the higher affinity of the protein for the second proton than that for the third does not appear to depend on the membrane environment, and the doubly protonated M2 channel can be considered to be in its resting, nonconducting state.6 Finally, the calculated energy differences are further evidence of the ability by M2 to stabilize excess charge within a lipid membrane by delocalizing among the proton-binding groups.10,14,19,21
Having established that both the dimer-of-dimers and the His-box are stable configurations of the system, we finally attempted to estimate the time scale of conversion between the two. A direct computation of a kinetic constant is currently not possible because the large collections of states associated with the protein backbone and the water distribution are likely to be greatly undersampled by subnanosecond, and possibly even submicrosecond, MD simulations. This task is further complicated by a predictably small free-energy difference between the two configurations because both were observed experimentally under different conditions. Therefore, we performed a simulation with enhanced sampling to evaluate the order of magnitude of the energies involved, using the classical CHARMM force field34 and a metadynamics biasing potential44 acting on the two RMSD functions of the His37 and Trp41 tetrads, respectively. On the basis of these calculations, we estimate that the highest energy required to transition between the dimer-of-dimers and the His-box configurations is on the order of 10 kcal/mol (Figure 4). Due to the classical force field description of the system, this number does not account for the LBHBs characteristic of the dimer-of-dimers and may be overestimated. Thus, we did not observe significant energy barriers between the two configurations; this is consistent with a putative transition pathway composed of individual ring flips rather than one concerted motion. On the basis of this assumption, we predict that the time scale of conversion between the dimer-of-dimers and the His-box is larger than or equal to the time of a ring flip of a single His37, measured by SSNMR to be approximately microseconds.12 Due to the enhanced sampling protocol, we were able to access many configurations (Figures 4 and S6, Supporting Information), which we analyzed in terms of rotameric states of the His37 tetrad.
Figure 4.

2D free-energy surface of the transformation between the dimer-of-dimers and the His-box models. The reference positions for both RMSD variables are the His-box configurations. Due to thermal motions, states with zero RMSD values have very low populations.
The distribution of the χ1 and χ2 angles of His37 is shown in Figures S5 and S6 (Supporting Information). In both the charged and the neutral His37, most of the values of χ1 are around 180°, the value observed in all experimental structures.7,8,10,11,14 The values of χ2 are more widely distributed; in our simulation, by effect of the biasing potential, χ2 spans reversibly all accessible values throughout the entire 360° period. In particular, values characteristic of the dimer-of-dimers as well as the His-box configurations are observed. In addition, we also observed values characteristic of the low-pH structures,8 which are typically associated with an expanded protein backbone; however, the latter was held restrained during our simulations (RMSD < 1.5 Å). This suggests that the His37 tetrad may sample both the neutral and low-pH configurations, with minor structural changes in the protein backbone and a relatively low free-energy cost. For example, we show in Figure S6 (Supporting Information) a “hybrid” configuration, where two His37 form a dimer connected by a HB, while the other two are in a His-box-like geometry. We therefore conclude that the two configurations can experience fast exchange and that the membrane environment ultimately dictates the precise balance between their relative populations.
In summary, we investigated the stability of two competing models for the His37 tetrad at neutral pH, the His-box and the dimer-of-dimers. Both were found to be either stable or metastable within the subnanosecond time scale by QM/MM MD simulations, and the unique characteristics of them were well-reproduced. The release of a proton to a water molecule in the direction of proton flow is energetically unfavorable in both models, consistent with the low current at neutral pH conditions. The energy cost associated with converting between the two configurations is comparable to that of individual ring flips. On the basis of the above data, we propose a multiconfiguration model, in which the dimer-of-dimers and the His-box configurations are populated with different probabilities and where they are both able to stabilize two protons within the viral membrane. By this mechanism, the precise relationship between proton flow and pH may be preserved against changes in the membrane environment. This conclusion sets the grounds for a quantitative description of the mechanism of proton conduction.
Acknowledgments
This work was supported by the National Institute of Health through Grant U01-AI-74571, the Commonwealth of Pennsylvania Health Research Formula Fund. Supercomputing time was awarded by the National Science Foundation (XSEDE allocation TG-MCA93S020) and by the Temple High-Performance GPU/CPU System that was purchased in part with funds from NSF Grant MRI-R2 0958854.
Supporting Information Available
Additional computational details, including the ONIOM protocol, classical and QM/MM MD simulations and optimization, and enhanced sampling calculations, and figures on the hydrogen bond distance, the water density, and the side chain dihedral angles. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
¶ H.D. and G.F. contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Pinto L. H.; Lamb R. A. The M2 Proton Channels of Influenza A and B Viruses. J. Biol. Chem. 2006, 281, 8997–9000. [DOI] [PubMed] [Google Scholar]
- Cross T. A.; Dong H.; Sharma M.; Busath D. D.; Zhou H.-X. M2 Protein from Influenza A: From Multiple Structures to Biophysical and Functional Insights. Curr. Opin. Virol. 2012, 2, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Antiviral Agents Active against Influenza A Viruses. Nat. Rev. Drug Discovery 2006, 5, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Ma C.; Fiorin G.; Carnevale V.; Wang T.; Hu F.; Lamb R. A.; Pinto L. H.; Hong M.; Klein M. L.; DeGrado W. F. Molecular Dynamics Simulation Directed Rational Design of Inhibitors Targeting Drug-Resistant Mutants of Influenza A Virus M2. J. Am. Chem. Soc. 2011, 133, 12834–12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Wu Y.; Ma C.; Fiorin G.; Pinto L. H.; Lamb R. A.; Klein M. L.; DeGrado W. F. Structure and Inhibition of the Drug-Resistant S31N Mutant of the M2 Ion Channel of Influenza A Virus. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.; Fu R.; Nishimura K.; Zhang L.; Zhou H.-X.; Busath D. D.; Vijayvergiya V.; Cross T. A. Histidines, Heart of the Hydrogen Ion Channel from Influenza A Virus: Toward an Understanding of Conductance and Proton Selectivity. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 6865–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell J. R.; Chou J. J. Structure and Mechanism of the M2 Proton Channel of Influenza A Virus. Nature 2008, 451, 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer A. L.; Acharya R.; Salom D.; Levine A. S.; Di Costanzo L.; Soto C. S.; Tereshko V.; Nanda V.; Stayrook S.; DeGrado W. F. Structural Basis for the Function and Inhibition of an Influenza Virus Proton Channel. Nature 2008, 451, 596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Luo W.; Hu F.; Hong M. Structure and Function of the Influenza A M2 Proton Channel. Biochemistry 2009, 48, 7356–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya R.; Carnevale V.; Fiorin G.; Levine B. G.; Polishchuk A. L.; Balannik V.; Samish I.; Lamb R. A.; Pinto L. H.; DeGrado W. F.; Klein M. L. Structure and Mechanism of Proton Transport through the Transmembrane Tetrameric M2 Protein Bundle of the Influenza A Virus. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Schmidt-Rohr K.; Wang J.; Soto C. S.; DeGrado W. F.; Hong M. Structure of the Amantadine Binding Site of Influenza M2 Proton Channels in Lipid Bilayers. Nature 2010, 463, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F. H.; Luo W. B.; Hong M. Mechanisms of Proton Conduction and Gating in Influenza M2 Proton Channels from Solid-State NMR. Science 2010, 330, 505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielak R. M.; Chou J. J. Solution NMR Structure of the V27A Drug Resistant Mutant of Influenza A M2 Channel. Biochem. Biophys. Res. Commun. 2010, 401, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M.; Yi M. G.; Dong H.; Qin H. J.; Peterson E.; Busath D. D.; Zhou H.-X.; Cross T. A. Insight into the Mechanism of the Influenza A Proton Channel from a Structure in a Lipid Bilayer. Science 2010, 330, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. J.; Voth G. A. A Computational Study of the Closed and Open States of the Influenza A M2 Proton Channel. Biophys. J. 2005, 89, 2402–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Wu Y.; Voth G. A. Proton Transport Behavior through the Influenza A M2 Channel: Insights from Molecular Simulation. Biophys. J. 2007, 93, 3470–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M.; Cross T. A.; Zhou H.-X. A Secondary Gate As a Mechanism for Inhibition of the M2 Proton Channel by Amantadine. J. Phys. Chem. B 2008, 112, 7977–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M. G.; Cross T. A.; Zhou H.-X. Conformational Heterogeneity of the M2 Proton Channel and a Structural Model for Channel Activation. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 13311–13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale V.; Fiorin G.; Levine B. G.; DeGrado W. F.; Klein M. L. Multiple Proton Confinement in the M2 Channel from the Influenza A Virus. J. Phys. Chem. C 2010, 114, 20856–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E.; Dal Peraro M.; DeVane R.; Vemparala S.; DeGrado W. F.; Klein M. L. Molecular Dynamics Calculations Suggest a Conduction Mechanism for the M2 Proton Channel from Influenza A Cirus. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.; Yi M.; Cross T. A.; Zhou H.-X. Ab Initio Calculations and Validation of the pH-Dependent Structures of the His37–Trp41 Quartet, The Heart of Acid Activation and Proton Conductance in the M2 Protein of Influenza A Virus. Chem. Sci. 2013, 4, 2776–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould J. A.; Drury J. E.; Frings S. M.; Kaupp U. B.; Pekosz A.; Lamb R. A.; Pinto L. H. Permeation and Activation of the M2 Ion Channel of Influenza A Virus. J. Biol. Chem. 2000, 275, 31038–31050. [DOI] [PubMed] [Google Scholar]
- Mould J. A.; Li H. C.; Dudlak C. S.; Lear J. D.; Pekosz A.; Lamb R. A.; Pinto L. H. Mechanism for Proton Conduction of the M2 Ion Channel of Influenza A Virus. J. Biol. Chem. 2000, 275, 8592–8599. [DOI] [PubMed] [Google Scholar]
- Venkataraman P.; Lamb R. A.; Pinto L. H. Chemical Rescue of Histidine Selectivity Filter Mutants of the M2 Ion Channel of Influenza A Virus. J. Biol. Chem. 2005, 280, 21463–21472. [DOI] [PubMed] [Google Scholar]
- Andreas L. B.; Eddy M. T.; Pielak R. M.; Chou J.; Griffin R. G. Magic Angle Spinning NMR Investigation of Influenza A M2(18–60): Support for an Allosteric Mechanism of Inhibition. J. Am. Chem. Soc. 2010, 132, 10958–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can T. V.; Sharma M.; Hung I.; Gor’kov P. L.; Brey W. W.; Cross T. A. Magic Angle Spinning and Oriented Sample Solid-State NMR Structural Restraints Combine for Influenza A M2 Protein Functional Insights. J. Am. Chem. Soc. 2012, 134, 9022–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y. M.; Qin H. J.; Fu R. Q.; Sharma M.; Can T. V.; Hung I.; Luca S.; Gor’kov P. L.; Brey W. W.; Cross T. A. M2 Proton Channel Structural Validation from Full-Length Protein Samples in Synthetic Bilayers and E. coli Membranes. Angew. Chem., Int. Ed. 2012, 51, 8383–8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F.; Schmidt-Rohr K.; Hong M. NMR Detection of pH-Dependent Histidine–Water Proton Exchange Reveals the Conduction Mechanism of a Transmembrane Proton Channel. J. Am. Chem. Soc. 2012, 134, 3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. H.; Hong M. Protonation, Tautomerization, and Rotameric Structure of Histidine: A Comprehensive Study by Magic-Angle-Spinning Solid-State NMR. J. Am. Chem. Soc. 2011, 133, 1534–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M.; Fritzsching K. J.; Williams J. K. Hydrogen-Bonding Partner of the Proton-Conducting Histidine in the Influenza M2 Proton Channel Revealed from 1H Chemical Shifts. J. Am. Chem. Soc. 2012, 134, 14753–14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke A. D. Density-Functional Exchange-Energy Approximation with Correct Asymptotic-Behavior. Phys. Rev. A 1988, 38, 3098–3100. [DOI] [PubMed] [Google Scholar]
- Lee C. T.; Yang W. T.; Parr R. G. Development of the Colle–Salvetti Correlation-Energy Formula into a Functional of the Electron-Density. Phy. Rev. B 1988, 37, 785–789. [DOI] [PubMed] [Google Scholar]
- Grimme S. Semiempirical Hybrid Density Functional with Perturbative Second-Order Correlation. J. Chem. Phys. 2006, 124, 034108. [DOI] [PubMed] [Google Scholar]
- MacKerell A. D. Jr.; Banavali N.; Foloppe N. Development and Current Status of the CHARMM Force Field for Nucleic Acids. Biopolymers 2000, 56, 257–265. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- Klauda J. B.; Venable R. M.; Freites J. A.; O’Connor J. W.; Tobias D. J.; Mondragon-Ramirez C.; Vorobyov I.; MacKerell A. D.; Pastor R. W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W.; Kreevoy M. M. Low-Barrier Hydrogen-Bonds and Enzymatic Catalysis. Science 1994, 264, 1887–1890. [DOI] [PubMed] [Google Scholar]
- Marx D.; Tuckerman M. E.; Hutter J.; Parrinello M. The Nature of the Hydrated Excess Proton in Water. Nature 1999, 397, 601–604. [Google Scholar]
- Okada A.; Miura T.; Takeuchi H. Protonation of Histidine and Histidine–Tryptophan Interaction in the Activation of the M2 Ion Channel from Influenza A Virus. Biochemistry 2001, 40, 6053–6060. [DOI] [PubMed] [Google Scholar]
- Balannik V.; Carnevale V.; Fiorin G.; Levine B. G.; Lamb R. A.; Klein M. L.; DeGrado W. F.; Pinto L. H. Functional Studies and Modeling of Pore-Lining Residue Mutants of the Influenza A Virus M2 Ion Channel. Biochemistry 2010, 49, 696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. J.; Zaitseva F.; Lamb R. A.; Pinto L. H. The Gate of the Influenza Virus M-2 Proton Channel Is Formed by a Single Tryptophan Residue. J. Biol. Chem. 2002, 277, 39880–39886. [DOI] [PubMed] [Google Scholar]
- Polishchuk A. L.; Lear J. D.; Ma C. L.; Lamb R. A.; Pinto L. H.; DeGrado W. F. A pH-Dependent Conformational Ensemble Mediates Proton Transport through the Influenza A/M2 Protein. Biochemistry 2010, 49, 10061–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W. Low-Barrier Hydrogen-Bonds and Low Fractionation Factor Bases in Enzymatic-Reactions. Biochemistry 1992, 31, 317–319. [DOI] [PubMed] [Google Scholar]
- Laio A.; Parrinello M. Escaping Free-Energy Minima. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 12562–12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


