Abstract
The coupling between cell-cycle exit and onset of differentiation is a common feature throughout the developing nervous system, but the mechanisms that link these processes are mostly unknown. Although the transcription factor Pax6 has been implicated in both proliferation and differentiation of multiple regions within the central nervous system (CNS), its contribution to the transition between these successive states remains elusive. To gain insight into the role of Pax6 during the transition from proliferating progenitors to differentiating precursors, we investigated cell-cycle and transcriptomic changes occurring in Pax6 - retinal progenitor cells (RPCs). Our analyses revealed a unique cell-cycle phenotype of the Pax6-deficient RPCs, which included a reduced number of cells in the S phase, an increased number of cells exiting the cell cycle, and delayed differentiation kinetics of Pax6 - precursors. These alterations were accompanied by coexpression of factors that promote (Ccnd1, Ccnd2, Ccnd3) and inhibit (P27 kip1 and P27 kip2) the cell cycle. Further characterization of the changes in transcription profile of the Pax6-deficient RPCs revealed abrogated expression of multiple factors which are known to be involved in regulating proliferation of RPCs, including the transcription factors Vsx2, Nr2e1, Plagl1 and Hedgehog signaling. These findings provide novel insight into the molecular mechanism mediating the pleiotropic activity of Pax6 in RPCs. The results further suggest that rather than conveying a linear effect on RPCs, such as promoting their proliferation and inhibiting their differentiation, Pax6 regulates multiple transcriptional networks that function simultaneously, thereby conferring the capacity to proliferate, assume multiple cell fates and execute the differentiation program into retinal lineages.
Introduction
During retinal development in vertebrates, a single pool of rapidly proliferating multipotent retinal progenitors cells (RPCs) gives rise to six different types of neurons and the Muller glia. Differentiation of all retinal cell types occurs in an evolutionarily conserved order and begins after terminal exit from the cell cycle [1-3]. Neurogenesis and progenitor proliferation occur simultaneously, thus at any given developmental stage some RPCs exit the cell cycle and differentiate while others continue to divide. The rate of proliferation and the fraction of RPCs that exit the cell cycle determine the size of the remaining progenitor pool as well as the number of each type of neuron generated. Thus, tight regulation of cell-cycle progression and exit is required to ensure the timely generation and correct amount of the various retinal cell types.
One of the greatest challenges in studying the regulation of RPC proliferation is the close coupling and interdependence of cell-cycle exit, cell-fate specification and neuronal differentiation. Thus, factors promoting neurogenesis or altering the cell fate acquired by ensuing postmitotic precursors may also directly regulate cell-cycle components or affect the timing of cell-cycle exit. For example, this dual activity was reported for the proneural gene Neurog2, which was found to promote neuronal differentiation in the spinal cord and at the same time suppress the expression of cell-cycle genes such as Ccnd1 [4]. Similarly, Ccnd1, which promotes cell-cycle progression, was recently found to play a role in cell-fate acquisition and in establishing the correct proportion of the various retinal cell types [5].
The paired and homeodomain-containing transcription factor (TF) Pax6 has been one of the most extensively investigated factors with respect to neural differentiation and progenitor proliferation. Its function has been studied in various animal models, in different areas of the CNS and at different stages of development. The roles of Pax6 in retinogenesis have been investigated by systemic and Cre-mediated somatic mutations [6-9]. Deletion of Pax6 from the peripheral optic cup (OC) revealed that the roles of Pax6 within RPCs depend on their state of maturation: RPCs located in the distal OC are late to differentiate and require Pax6 for inhibition of Crx and completion of neurogenesis. In contrast, RPCs located centrally, within the differentiation zone, require Pax6 to retain their multipotency and differentiate into most retinal cell types, excluding subtypes of GABAergic amacrine cells [6,8].
In addition to its role in neuronal differentiation, Pax6 is required for normal proliferation in the developing retina as well as in other regions of the CNS, including the telencephalon (reviewed in 10), diencephalon [11] and spinal cord [12]. In the optic vesicle (OV), as well as in the retina, Pax6 is required to maintain high levels of proliferation as Pax6 - progenitors display a significant reduction in bromodeoxy-uridine (BrdU) incorporation [6-8]. Nevertheless, it remains unclear whether these proliferative changes are due to a discrete role of Pax6 in regulating RPC proliferation or are an indirect outcome of the altered cell fates observed in the various knockout models.
The aim of this study was to distinguish between Pax6’s role in cell-cycle dynamics and its activity in cell specification, and to elucidate the transcriptional network mediating its involvement in RPC proliferation. We therefore characterized the dynamics of the cell cycle and cell differentiation following conditional mutation of Pax6 in RPCs, as well as the global changes in gene expression in the Pax6-mutant cells. These analyses revealed that the processes of cell-cycle exit, cell-fate specification and neuronal differentiation, which normally occur in close tandem succession, are temporally separated in Pax6-deficient RPCs, thus demonstrating a role for Pax6 in both regulating progenitor proliferation and coupling cell-cycle exit and neuronal differentiation. Furthermore, our findings reveal that rather than conveying a positive linear effect on RPC proliferation, Pax6 intersects with multiple retinal programs, some of which promote RPC proliferation, while others promote cell-cycle exit and differentiation.
Materials and Methods
Mouse Lines
The Pax6 flox allele contains loxPs flanking the initiator ATG and exons 4–6 encoding the paired domain [13]. The α-Cre-transgenic line contains the Pax6 P0 promoter and the peripheral retina enhancer (termed α) followed by Cre which was cloned 5′ of IRES-intron-gfp-pA [6]. All animal work was conducted according to national and international guidelines and all efforts were made to minimize suffering. The protocol was approved by the Tel Aviv University institutional animal care and use committee (IACUC permit: M08092).
Immunofluorescence and In Situ Hybridization (ISH)
Immunofluorescence analysis was performed as described previously [8]. The primary antibodies used are shown in Table S3. Secondary antibodies were conjugated to Alexa488, Alex594 (1:1,000, Invitrogen) or Aminomethylcoumarin Acetate (AMCA) (1:100, Jackson Immuno Research Laboratories).
ISH analysis was performed on frozen sections. Hybridization was conducted overnight at 65°C and performed as described previously [8]. Probes used for ISH were: FoxN4 (king gift of Xiang Ming [14]), Nr2e1 (amplified from cDNA using tcctgaacggcagactctcg and cgaggttgcctgacctacgg cloned into pGEM-T-easy, Promega), Plagl1 (king gift of Carol Schuurmans [15]), and Gli1, Gli2, Gli3 (king gift of Alexandra Joyer).
Slides were viewed with an Olympus BX61 fluorescent microscope or laser-scanning confocal microscope CLSM 410 (Zeiss) and images were analyzed using the image analysis system 'AnalySIS'.
Analysis of Cell-Cycle Exit
To label proliferating cells, 5-bromo-2-deoxyuridine (BrdU, 140 µg/g body weight, Sigma) was injected intraperitoneally (IP) into pregnant females at the desired developmental stage. To assay for cell-cycle exit, BrdU was injected 24 h before embryo harvesting. Tissues were then processed for embedding in paraffin and sectioned. Sections were stained for BrdU incorporation and either proliferating cell nuclear antigen (PCNA) or the proliferating cell marker Ki67 using immunofluorescence. No less than three central sections from three different eyes were counted for both BrdU+ and BrdU + Marker+. Cell-cycle exit index was calculated by dividing the total number of BrdU + Marker+ cells scored in all sections of the same eye by the total number of BrdU+ cells scored in all sections of the same eye. All of the values for a single developmental stage and genotype were averaged and standard deviation (SD) was calculated. Values obtained for control and Pax6-mutant animals were compared using either Student’s t-test or Mann–Whitney test to determine statistical significance.
Expression Microarray
The control and mutant Pax6 - RPCs were isolated by fluorescent-activated cell sorting (FACS) from Pax6 +/+;α-Cre or Pax6 loxP/loxP;α-Cre E12 (embryonic day 12) eyes. Three pools of 1.8 x 106 cells each were collected for the analyses. RNA was isolated using TRIZOL reagent. Hybridization was to Affymetrix Mouse 430.2 gene-expression arrays according to the manufacturer’s protocol. For microarray data processing, we used remote analysis computation for gene expression data (RACE [16]) front-end for the Bioconductor [17] package of the R language utilizing the RMA (Robust Multichip Average; [18]) algorithm. Results were filtered by fold-change (1.5), p-value (0.05), and FDR (false discovery rate, 10%). The results were further verified using SAM (Significance Analysis of Microarrays) software [19]. The expression data were submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under series accession no. GSE45143.
Results
Delayed Neuronal Differentiation in the Pax6-Deficient Retina
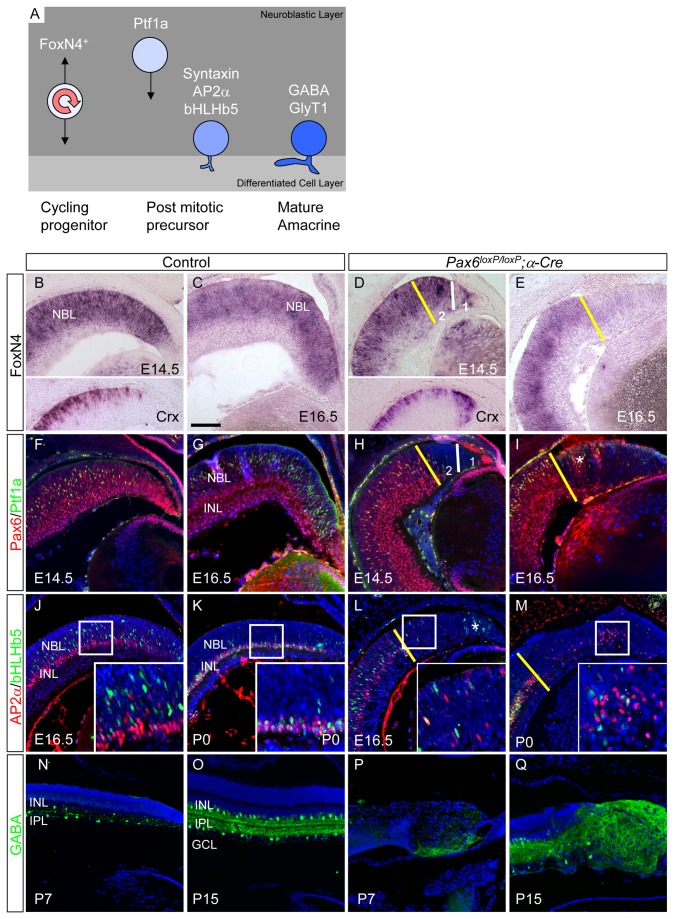
Previous studies of systemic and conditional Pax6 deletions have shown both reduced proliferation and limited differentiation of the Pax6-deficient RPCs exclusively to the GABAergic (producing γ-aminobutyric acid) amacrine lineage [6-8]. To distinguish the proliferative roles of Pax6 from its effects on cell-fate acquisition, we characterized the dynamics of differentiation into amacrine interneurons in Pax6 conditional knockout (Pax6 loxP/loxP;α-Cre) and control mice. We determined the temporal and spatial expression patterns of genes that are known to participate in the regulation of amacrine cell differentiation based on gene-expression studies of mutant mice (Figure 1A). The TF FoxN4 is an upstream regulator of amacrine and horizontal lineages [14]. FoxN4 is expressed by proliferating RPCs located in the neuroblastic layer (NBL) of the embryonic retina (E14.5, E16.5, Figure 1B,C and [14]). In the Pax6 loxP/loxP;α-Cre OC (Figure 1D,E), we defined distal and proximal Pax6 - populations by detection of Crx and Pax6 using indirect immunofluorescence (IIF) analysis on adjacent sections (Crx – inset in Figure 1B,D; Pax6 – Figure 1H,I). We found that in the Pax6-mutant region, FoxN4 is initially retained in Pax6 - Crx - neurogenic progenitors (Figure 1D area 2) and only shows a decrease in distally located Pax6 - Crx + cells (Figure 1D area 1). However, at E16.5, FoxN4 expression was lost from virtually all Pax6 - cells (Figure 1E).
Figure 1. Delayed differentiation of amacrine precursors in the Pax6 loxP/loxP ;α-Cre retina.
(A) A scheme of the stages of amacrine interneuron differentiation. Amacrine cells evolve from FoxN4-expressing RPCs. In the postmitotic amacrine precursors, FoxN4 is reduced and Ptf1a expression is initiated. The Ptf1a-positive precursors migrate to the prospective INL, lose Ptf1a expression and initiate expression of TFs involved in the differentiation of amacrine subtypes (e.g. Ap2α and bHLHb5). The final differentiation of amacrine cells occurs a few days after birth with accumulation of neurotransmitters and transporters (e.g. GABA, glycine transporter GlyT1). Expression of amacrine specification and differentiation markers in control (B,C,F,G,J,K,N,O) and Pax6loxP/loxP;α-Cre OC (D,E,H,I,L,M,P,Q). Expression of FoxN4 (B–E, the inset in B and D is Crx on adjacent section) as detected using ISH. Indirect immunofluorescence (IIF) was employed to detect the expression of Ptf1a and Pax6 (green and red, respectively, F–I; adjacent sections to B-E respectively), bHLHb5 and Ap2α (green and red, respectively, J–M) and GABA (green, N–Q) during various stages of development as indicated. Pax6 (red in F–I and not shown) and Crx (inset in B,D and not shown) expression determined by IIF or ISH was used to identify the Pax6-deficient area (yellow line in D,E,H,I,L,M) and to delineate the neurogenic and nonneurogenic RPC populations in the Pax6 loxP/loxP;α-Cre retina (numbered 1 and 2 and separated by a dotted white line in D and H). Abbreviations: GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; NBL, neuroblastic layer. Scale bar in C is 100 µm.
As FoxN4 is downregulated once cells exit the cell cycle [14], the reduced number of FoxN4-expressing cells may reflect accumulation of postmitotic amacrine precursors. We therefore examined the expression of TFs known to be expressed in these precursors and to be required for generation of amacrine subtypes. Ptf1a is transiently expressed in postmitotic amacrine and horizontal precursors located in the NBL (Figure 1F,G, green and [20,21]). Once Ptf1a expression is extinguished, amacrine precursors start to express Bhlhb5 and AP2a (Figure 1J,K, green and red respectively). These are first detected in migrating precursors in the NBL (E16.5, Figure 1J and [22,23]) and are later restricted to the prospective inner nuclear layer (INL, P0, Figure 1K).
Surprisingly, although Pax6-deficient RPCs expressed FoxN4 at E14, correlating with their competence to differentiate to amacrine cells, the dynamics of amacrine differentiation was perturbed in the Pax6 - retina based on the reduced number of cells expressing Ptf1a at E14.5 and E16.5 in both Pax6 - Crx - and Pax6 - Crx + areas of the Pax6 loxP/loxP;α-Cre retina (Figure 1H,I). Reduced Ptf1a expression was further accompanied by a similar reduction in the expression of other amacrine precursor markers such as bHLHb5, Ap2α, Ap2β, VC1.1, BarHL2 and syntaxin (E16.5, Figure 1L,M and Figure S1). However, despite reduced expression of amacrine precursor genes at embryonic stages, by P0 expression of some of the amacrine TFs, such as Ap2α (Figure 1M, red), was detected in an increasing number of Pax6 - cells, while expression of other markers, such as bHLHb5, remained low (Figure 1M green). Taken together, these results demonstrate that while Pax6 is not required to convey amacrine competence in RPCs, it is needed for the correct timing of expression and full repertoire of TFs involved in differentiation of the amacrine subtypes.
Considering the delayed onset of expression of amacrine precursor genes, we next monitored the accumulation of GABA (Figure 1N-Q), an inhibitory neurotransmitter which is detected in most Pax6-deficient amacrine interneurons in the Pax6 loxP/loxP ; α-Cre retina (Figure 1Q and [6]). Normally, GABA is first detected in the mouse retina at around birth [24]. However, in Pax6 - amacrines, GABA accumulation was delayed and typically appeared only around P7 (Figure 1P,Q). Nevertheless, by P15, all Pax6 - cells expressed GABA (Figure 1Q). Thus, despite reduced proliferation of Pax6 - RPCs, the differentiation of Pax6-mutant cells to the GABAergic amacrine lineages is delayed, revealing a role for Pax6 in the timing of neuronal differentiation.
Pax6 Regulates the Timing of Cell-Cycle Exit of RPCs
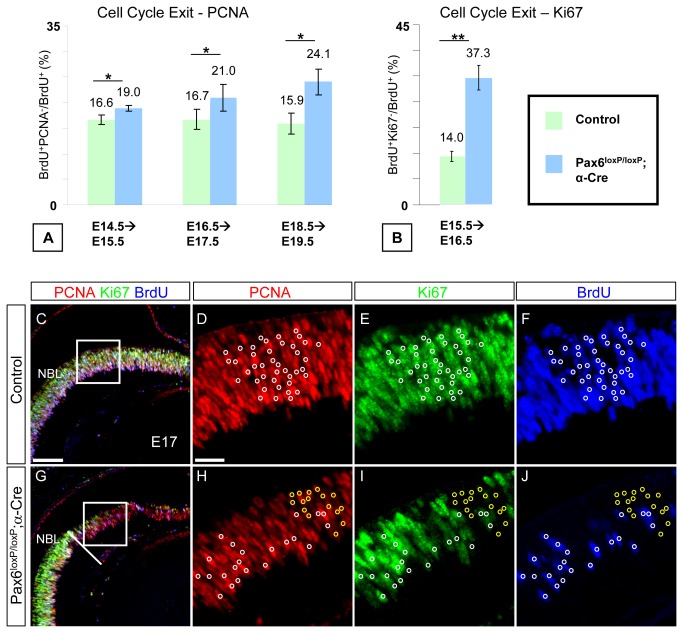
The reduced proliferation previously documented in the Pax6-mutant retina [8] and the delayed differentiation based on reduced expression of amacrine differentiation genes (Ptf1a, Figure 1) detected in the Pax6 - OC implicate altered cell-cycle kinetics. To investigate the cell-cycle kinetics of Pax6 - RPCs, we compared the number of cells exiting the cell cycle in control and Pax6 loxP/loxP;α-Cre distal retinas. Cycling cells were labeled with a pulse of BrdU 24 h prior to sacrifice followed by detection of PCNA and BrdU by double IIF [5]. Total BrdU-positive cells as well as PCNA-negative BrdU-positive (PCNA-BrdU+) cells were counted and postmitotic cell index was calculated as PCNA-BrdU + /BrdU+ (Total) in control and Pax6 loxP/loxP;α-Cre retinas (Figure 2, Figure S2). This index reports the total number of postmitotic RPCs generated during a 24-h period following BrdU administration.
Figure 2. Neurogenic Pax6- RPCs in the Pax6 loxP/loxP ;α-Cre retina display increased cell-cycle exit and sustained expression of cell-cycle factors.
A single pulse of BrdU was administered at E14.5, E15.5, E16.5 and E18.5, 24 h prior to sacrifice as indicated (A, B). Pax6 expression was detected on adjacent sections to identify the recombination area in the Pax6loxP/loxP;α-Cre OC. (A) The percentage of BrdU + PCNA-/BrdU+ Total was determined for the Pax6loxP/loxP control (green bars) and Pax6 loxP/loxP ;α-Cre (blue bars) distal retina. Pax6 - RPCs show increased cell-cycle exit at all stages tested (19% (SD=0.6%), 21% (SD=2.7%) and 24.1% (SD=2.6%) compared to 16.6% (SD=0.93%), 16.7% (SD=2%) and 15.9% (SD=2%) in control at E14.5, E16.5 and E18.5, respectively; p≤0.05, n≥3). (B) The number of BrdU+ and BrdU + Ki67- cells was quantified and the ratio BrdU + Ki67-/BrdU+ Total was used to compare cell-cycle exit rate in control (green bar) and Pax6 loxP/loxP ;α-Cre (blue bar) distal retina at E15.5. Pax6 - RPCs show increased cell-cycle exit (37.3% (SD=3.6%) in Pax6 - compared to 14% (SD=1.5%) in control; p<0.01, n=6 for both genotypes). Triple immunofluorescence for PCNA, Ki67 and BrdU (red, green and blue, respectively, in C–J) in control (C–F) and Pax6 loxP/loxP ;α-Cre (G–J) distal retina showing mitotic PCNA + Ki67 + BrdU+ in both control and mutant (white circles in D–F,H–J) and abnormal PCNA + Ki67-BrdU- cells detected only in Pax6 loxP/loxP ;α-Cre (yellow circles in H–J) OC. Scale bars in C,D are 100 and 25 µm, respectively.
In the control retina, cell-cycle exit index was 16.6 ± 0.93%, 16.7 ± 2% and 15.9 ± 2% at E14.5, E16.5 and E18.5, respectively (green bars in Figure 2A). A significant (P ≤ 0.05) increase in the cell-cycle exit index was documented in the Pax6 loxP/loxP;α-Cre OC at all time points tested: 19 ± 0.6%, 21 ± 2.7% and 24.1 ± 2.6% at E14.5E15.5, E16.5E17.5, E18.5E19.5/P0, respectively (blue bars in Figure 2A). Thus, despite the obvious delay in differentiation of Pax6 - RPCs and in agreement with reduced proliferation, there is a significant increase in cell-cycle exit of the Pax6-deficient RPCs as compared to controls and this increase becomes more prominent during the course of retinogenesis (1.14-, 1.25- and 1.51-fold at E14.5, E16.5 and E18.5, respectively).
In an attempt to reconcile the seemingly contrasting findings of increased cell-cycle exit but delayed differentiation, we tested control and Pax6 loxP/loxP;α-Cre retinas for expression of additional proliferating-cell markers. Ki67 has been previously shown to be expressed in proliferating cells and is downregulated in quiescent and differentiated cells [25-27]. In the retina, Ki67 expression fully overlapped with PCNA expression in proliferating cells (Figure 2C-E and [28]); however, postmitotic cells were previously reported to extinguish Ki67 prior to downregulating PCNA expression [29]. Surprisingly, we found progressive accumulation of PCNA + Ki67- cells in the distal retina of Pax6 loxP/loxP ; α-Cre mice. These were initially detected at E14.5 and gradually increased, until by P0 most PCNA+ cells were Ki67- (Figure 2G–I and Figure S2J-L). To determine the proliferative potential of the abnormal PCNA + Ki67- progenitors, we tested their ability to incorporate BrdU during a 1.5-h pulse of BrdU administered at E17.5. As expected, in the control embryos, all PCNA+ cells were also Ki67+ and some of these were also BrdU+ (white circles in Figure 2D–F). In the Pax6 - retina, BrdU incorporation was detected only in Ki67+ cells (white circles in Figure 2H–J) but BrdU was not incorporated into the PCNA + Ki67- population (yellow circles in Figure 2H–J). To compare accumulation of normal (PCNA-Ki67-) and aberrant (PCNA + Ki67-) postmitotic cells, the cell-cycle index was recalculated using Ki67 instead of PCNA. This analysis, conducted from E15.5 to E16.5, revealed a 2.67-fold increase in the number of cells that exited the cell cycle in Pax6 loxP/loxP;α-Cre mice, as 37.3% (SD=3.6%) of the proliferating cells that incorporated BrdU at E15.5 were Ki67 negative by E16.5 compared to only14% (SD=1.5%) in the control (P≤0.001) (Figure 2B). Consistent with reduction in the number of Ki67-expressing cells, we observed a reduced number of cells expressing cyclin B1 (Ccnb1), which in the NBL is located in phase G2/M cells (Figure S2M,N [28]).
These results demonstrate a dramatic increase in the production of postmitotic cells in the Pax6 - retina and reveal, for the first time, that in the Pax6 loxP/loxP;α-Cre retina, there is accumulation of an abnormal population of postmitotic PCNA + Ki67- cells. These cells inhabit the NBL and seem to maintain neuroepithelial morphology until late stages of retinogenesis. Pax6 is therefore required to maintain progenitors in the cell cycle as well as for the downregulation of cell-cycle factors, such as PCNA and Ccnd1 (see below) in the postmitotic precursors.
High-Throughput Transcriptome Analysis of Pax6- RPCs Reveals Altered Expression of Genes Associated with Cell Proliferation
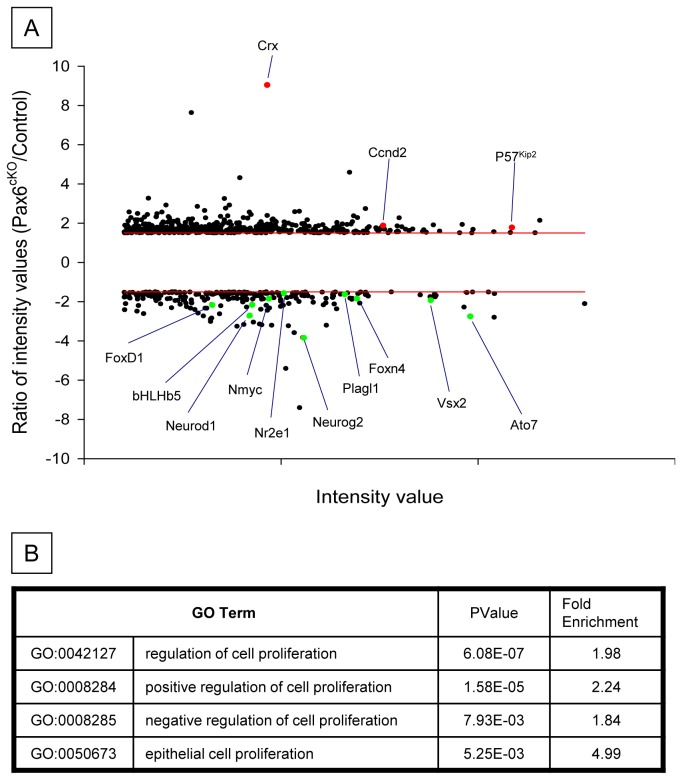
To identify the gene network operating downstream of Pax6 to regulate cell proliferation and fate, we determined the transcriptomic changes in Pax6-deficient RPCs using microarray analysis. The expression profile was examined in E12 retina when both the proliferative and cell-cycle-related phenotypes are first detected and prior to onset of cell differentiation. Pax6-deficient and control RPCs were collected by FACS from mutant (Pax6 loxP/loxP;α-Cre) and control (Pax6 +/+ α-Cre) E12.5 retinas. This was possible because GFP is expressed in conjunction with Cre from the α-Cre construct [6]. Three biological replicates were conducted for each of the two genotypes and the gene-expression profiles were compared using Affymetrix Gene Chip microarray (MOE430-2). The expression of 952 genes was found to be altered between control and mutant RPCs (fold-change >1.5, P<0.05, FDR<10%); of these, 316 were downregulated and 636 were upregulated (Figure 3A and Table S1). Expression of Pax6 was reduced threefold, as was the expression of the previously reported retinal Pax6 targets Atoh7, NeuroD1, Neurog2, FoxG1, FoxD1, delta-catenin (Ctnnd2) and Crx (Figure 3A and [6,8,30,31]). Altered expression of over 20 genes found to be differentially in the microarray was validated using either IF or in situ hybridization (Table S2). Furthermore, testing the list of altered genes for enriched gene ontology (GO) terms using DAVID Bioinformatics Resources [32,33] and clustering significantly enriched (P<0.05) terms into functional groups indicated that these genes have known roles in previously reported Pax6 functions such as eye development, cellular adhesion [34-36], neurogenesis and proliferation (Figure 3B and Figure S3). Therefore, the microarray data faithfully represented the molecular phenotype of the Pax6-mutant RPCs and reveals significant changes in expression of genes associated with cell proliferation.
Figure 3. High-throughput analysis of transcriptional alterations in Pax6- RPCs.
(A) Scatterplot representing the fold change in gene expression in Pax6 - versus control retinas (y axis) plotted against the intensity value in the control (x axis). Each spot corresponds to one gene; only genes whose expression changed at least 1.5-fold are shown. (B) Gene ontology (GO) analysis conducted using DAVID bioinformatics resource [32] for genes whose expression was altered in Pax6 - versus control retinas, showing enrichment in various proliferation- and cell-cycle-related GO terms.
Pax6- RPCs Overexpress Proteins Involved in both Cell-Cycle Withdrawal and Progression
The RPC decision to either differentiate or continue proliferating occurs during the G1 phase of the cell cycle. At this stage, proliferation can be promoted by the phosphorylation of Rb by a complex of D/E cyclins and cyclin-dependent kinases (CDKs). This phosphorylation is inhibited by CDK inhibitors (CKIs), which function to promote cell-cycle exit [5,37,38]. Altered expression of cell-cycle-related factors has been documented in the developing cortex and spinal cord of Pax6 mutants [39-41].
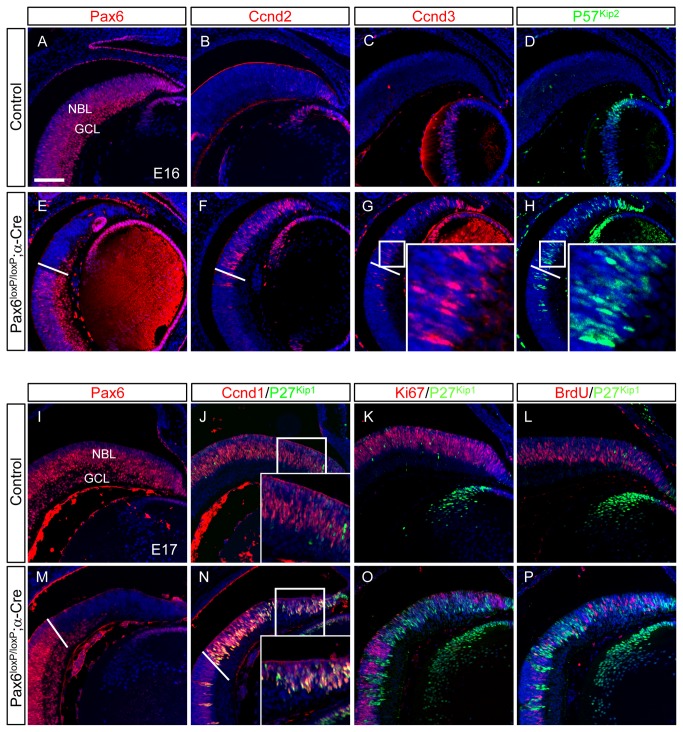
Our microarray analysis revealed upregulation of both CyclinD2 (Ccnd2) and the CKI P57 Kip2 in Pax6 loxP/loxP;α-Cre OC compared to controls (Table S1). We therefore monitored the expression of these and other cell-cycle factors in control and Pax6 loxP/loxP;α-Cre OCs during retinogenesis (Figure 4, Figure S4). In the control embryonic retina, Pax6 was detected in proliferating RPCs in the NBL as well as in differentiating neurons located in the inner nuclear layer (INL) and ganglion cell layer (GCL, Figure 4A,I). At these stages, Ccnd2, Ccnd3 and P57Kip2 were rarely detected in the RPCs (Figure 4B–D), whereas Ccnd1 and P27Kip1 were prominent in the NBL (Figure 4J [42] [43]). In contrast, we detected ectopic expression of Ccnd2, Ccnd3 and P57Kip2 (Figure 4F–H), as well as elevated expression of Ccnd1 and P27Kip1 (Figure 4N), within the Pax6-deficient RPCs, identified by monitoring loss of Pax6 protein on adjacent sections (Figure 4E,M). Moreover, while in the control retina Ccnd1+ and P27Kip1+ cells are mutually exclusive [43] (Figure 4J, inset), these factors were coexpressed in the Pax6 loxP/loxP;α-Cre retina (Figure 4N, inset). Despite the misexpression of Ccnd1, the Pax6-P27 Kip1+ Ccnd1+ mutant cells were negative for Ki67 and did not incorporate BrdU (Figure 4O,P), similar to P27 Kip1+ cells in the control retina (Figure 4K,L). Thus, aberrant expression of Ccnd1 in these cells is not sufficient to maintain them in a proliferative state.
Figure 4. Increased expression of cell-cycle factors in the Pax6 loxP/loxP ;α-Cre retina.
Expression of Pax6 (A,E,I,M) and of cell-cycle progression and withdrawal factors was monitored using immunofluorescence in control (A–D,I–L) and Pax6 loxP/loxP ;α-Cre (E–H,M–P) distal retina at E16 and E17, as indicated. Pax6 - area was delineated by staining for Pax6 protein (E,M) on adjacent sections (dotted line in E–H,M,N). Expression of Ccnd2 (B,F) Ccnd3 (C,G), P57Kip2 (D,H) and P27Kip1 (green in J–L,N–P) is upregulated in most of the Pax6 - RPCs of Pax6 loxP/loxP ;α-Cre retina. Ccnd1 and P27Kip1, which are expressed in different cells in the control (J) are coexpressed in Pax6 - RPCs (N). For the control and Pax6-deficient cells, P27Kip1-expressing cells are negative for Ki67 (red in K,O) and BrdU (red in L, P). Abbreviations: GCL, ganglion cell layer; NBL, neuroblastic layer. Scale bar in A is 100 µm.
Elevated expression of Ccnd2 and P57Kip2 proteins was also detected in a small number of cells at E12.5, consistent with the E12 transcriptomic data (not shown). Interestingly, the microarray data for Ccnd1 indicated reduced transcript levels in the distal Pax6 loxP/loxP;α-Cre retina compared to controls (fold change -1.5). Nevertheless, IIF for Ccnd1 protein indicated maintained and even elevated protein levels (E13.5, E15.5, Figure S4G,N; E17.5, Figure 4N). We therefore monitored Ccnd1 transcript and protein at E12.5, E13.5 and E15.5 (not shown and Figure S4). At these stages, the normal RPCs co-express Pax6 and Ccnd1 (Figure S4A,B,I,J), while the postmitotic Crx+ photoreceptor precursors do not express either Pax6 or Ccnd1 (Figure S4C,D,K,L and [8]). In contrast, in the Pax6-mutant RPCs, we detected a reduction in Ccnd1 transcript from E13.5 which was prominent by E15.5 (Figure S4F,N). Interestingly, although Ccnd1 transcript was gradually reduced, Ccnd1 protein was abnormally retained as it was detected in the Pax6-Crx+ cells at E13.5 (Figure S4G,H) and in the neurogenic population (Pax6-Crx- cells) at E15.5 and E17.5 (Figure S4O,P, Figure 4N). This finding suggests complex regulation of Ccnd1 by Pax6, as it is required for both normal levels of Ccnd1 transcription and regulation, and through posttranscriptional/translational mechanisms, of Ccnd1 protein levels.
Taken together, the Pax6 - retinal precursors display elevated expression of both cell-cycle-promoting (Ccnd1–3 and PCNA) and cell-cycle-inhibiting (P27 Kip1 and P57 Kip2) factors. These results indicate a major role for Pax6 in controlling the events leading to timely cell-cycle exit of RPCs.
TFs that Regulate RPC Proliferation and Cell Cycle Are Abnormally Expressed in Pax6- Retina
The above results indicate a key role for Pax6 in controlling cell-cycle parameters. We therefore focused our analysis on TFs whose expression was altered based on the microarray analysis and which have been previously shown to regulate RPC proliferation and thus could mediate Pax6’s role in the regulation of cell-cycle dynamics.
The homeodomain TF Vsx2 and the orphan nuclear receptor Nr2e1 (tailless, Tlx) have been previously reported to promote RPC proliferation [43-45]. Loss-of-function mutations in both genes result in increased expression of P27 Kip1, reduced expression of Ccnd1 and a hypoplastic retina [43-45]. Expression of these TFs was found to be reduced in the Pax6 loxP/loxP;α-Cre retina based on the microarray analysis (fold change -1.9 and -2.18 for Vsx2 and Nr2e1, respectively). We therefore examined the dynamics of their expression following Pax6 deletion in situ. Both Nr2e1 and Vsx2 expression was detected in most proliferating RPCs in the NBL of control retinas (Figure 5A–D); their expression was slightly reduced at E12.5 in Pax6 loxP/loxP;α-Cre retinas (Figure 5E,G; Pax6 mutant region identified by IIF on adjacent section) and it was virtually abolished by E14.5 in Pax6 loxP/loxP;α-Cre mutants (Figure 5F,H). A reduction in the expression of these neuronal progenitor genes could mediate the reduced proliferation of the Pax6-deficient RPCs.
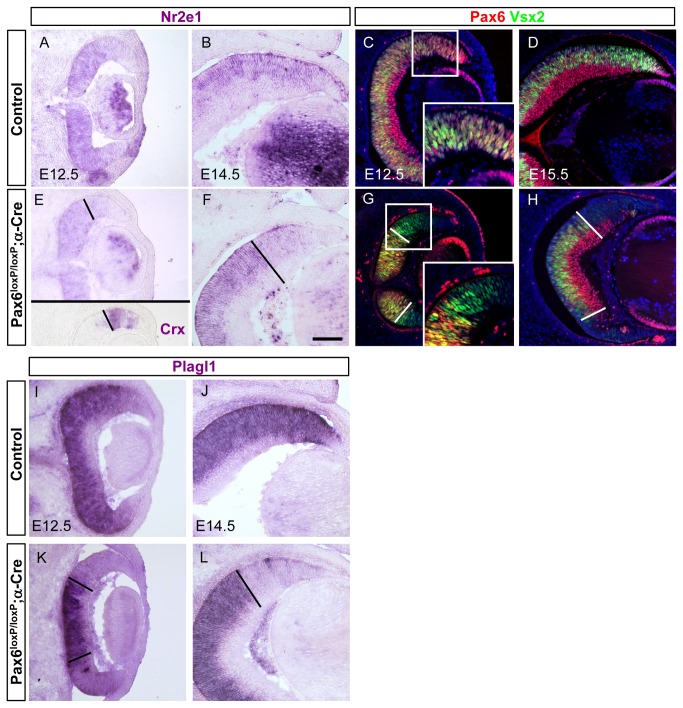
Figure 5. Abrogated expression of factors implicated in regulating RPC proliferation in Pax6 loxP/loxP ;α-Cre retina.
The expression pattern of different factors was monitored on sections from eyes of control (A-D,I,J) and Pax6 loxP/loxP ;α-Cre (E-H,K,L) mice. Nr2e1 (E12.5 A,E; E14.5 B,F) and Plagl1 (E12.5 I,K; E14.5 J,L) were detected using ISH while Vsx2 (green; E12.5 C,G; E15.5 D,H) Pax6 (red; E12.5 C,G; E15.5 D,H) were detected by IIF analysis. Pax6 (red in C,D,G,H and not shown) and Crx (inset in E and not shown) expression were used to delineate the different RPC populations in the Pax6loxP/loxP;α-Cre retina (marked with dotted line in E-H,K,L). Scale bar in F is 100 µm.
The maternally imprinted tumor suppressor TF Plagl1 (Zac1) is a zinc-finger protein that can act as both a transcriptional activator and repressor [46]. Loss of Plagl1 results in a hypercellular retina containing an additional INL composed of amacrine cells [15]. As previously described, Plagl1 was widely expressed throughout the NBL in control mice (Figure 5I,J and [15]) but its expression was lost from all Pax6 - RPCs (identified by IIF, not shown) in the distal retina of Pax6 loxP/loxP;α-Cre as early as E12.5 (Figure 5K,L).
These results reveal that Pax6 is required for the expression of TFs known to promote (Vsx2 and Nr2e1) and restrict (Plagl1) RPC proliferation and thus Pax6 loss in RPCs results in a dramatic alteration in cell-cycle dynamics (see discussion).
Figure S5A–D, I–L [48]). The microarray analysis showed reduced expression of Hes5 and Dll1 at E12. Indeed, expression of the Notch-pathway genes Notch1, Dll1 and Hes5 was reduced in the Pax6 - Crx + population of E13.5 Pax6 loxP/loxP;α-Cre retina (Figure S5E–H). However, at later stages of development, when the neurogenic population (Pax6 -;Crx -) is prevalent, the expression of Notch1 and Dll1 was similar to their expression levels in the control OC (E16.5, Figure S5N,O), although the expression of Hes5 was reduced in some, but not all of the Pax6-mutant OC, probably reflecting a reduction in RPCs (Figure S5P). These findings suggest that Pax6 may be required for the Notch pathway in the most distal OC (Pax6 - Crx +), but its activity is not essential for maintaining the expression of Notch-signaling components in the more central RPCs. Therefore, the reduced proliferation of the Pax6 -;Crx - neurogenic RPCs is probably not due to altered activity of Notch signaling.
HH signaling is required to maintain normal proliferation of RPCs, as loss of HH signaling results in precocious cell-cycle exit and depletion of the progenitor pool [51,52], whereas overactivation of the HH pathway results in increased proliferation and retinal hyperplasia [53]. HH signaling has also been shown to regulate the expression of various cell-cycle-related factors such as Ccnd1, P57 Kip2 and Mycn [51-54]. The microarray analysis conducted on E12 control and Pax6 loxP/loxP;α-Cre retinas yielded no differential expression of HH pathway-related factors, possibly because at this stage Shh components are not prominent in the peripheral OC in regions of α-Cre activity [55].
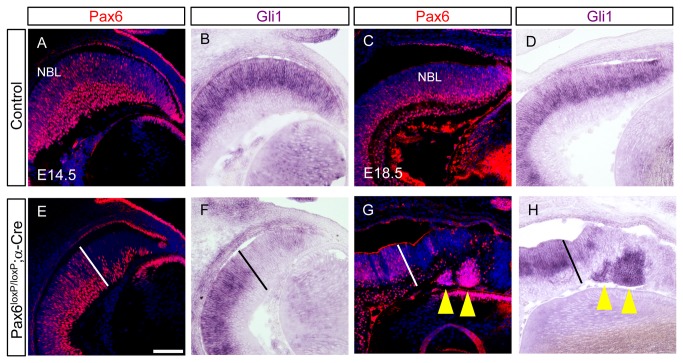
To test for possible alterations in HH signaling, we characterized Gli1 expression at various developmental stages, as Gli1 is both an effector and a target of the HH pathway (Figure 6B,D,F,H [56]). IIF analysis for Pax6 was performed on adjacent sections to determine the Pax6 mutant area (Figure 6A,C,E,G). As previously reported, extensive Gli1 expression was detected in the NBL of both E14.5, E1.5.5 and E18.5 control retinas (Figure 6B,D, Figure S6A). However, in the distal retina of Pax6 loxP/loxP ; α-Cre, Gli1 expression was undetectable throughout retinogenesis (E14.5, E18.5, Figure 6F,H, Figure S6D) suggesting HH-signaling arrest in Pax6 - RPCs. The expression of Gli2 and Gli3, which participate in HH signaling but are not HH-signaling targets [57], was unaltered in Pax6 loxP/loxP ; α-Cre compared to controls (Figure S6) and thus these mediators of the HH pathway are maintained despite the loss of Pax6. To further determine whether Pax6 is required cell autonomously for HH signaling, we examined Gli1 expression in RPCs that escaped Cre-mediated Pax6 deletion. This occurred because the α-Cre transgene is occasionally inactive in some RPCs resulting in patches of nonrecombinant cells surrounded by recombinant ones. In the distal OC of the Pax6 loxP/loxP ; α-Cre embryos, these patches of Pax6-expressing RPCs form rosette-like structures (Figure 6G,H, yellow arrowheads). These rosettes share the same environment as recombinant Pax6 - RPCs, yet extensive Gli1 expression was detected in the Pax6 + rosettes (Figure 6H, yellow arrowheads). These results suggest that Pax6 plays a cell-autonomous role within the RPCs for the normal activation of HH signaling.
Figure 6. Hedgehog signaling is disrupted in Pax6 loxP/loxP ;α-Cre retina.
Expression of Gli1 was detected using ISH (B,D,F,H) and Pax6 protein was detected by IIF on adjacent sections (A,C,E,G) for control (A-D) and Pax6 loxP/loxP ;α-Cre (E-H) OC. Gli1 expression was abrogated in all Pax6 - RPCs (F,H) compared to controls (B,D) at both E14.5 (B,F) and E18.5 (D,H). Pax6- area was determined by antibody labeling on adjacent sections and marked by dotted line (E-H). Scale bar in B is 100 µm.
Discussion
Pax6-Deficient RPCs Exhibit a Unique Cell-Cycle Phenotype
Previous studies have substantiated the requirement for Pax6 in establishing the neuronal progenitor pool during development of the vertebrate nervous system. Reduced proliferation was documented following knockdown of Pax6 in the embryonic chick retina and spinal cord [12,58]. Similarly, reduced BrdU incorporation was detected in the diencephalon and altered proliferation was observed in the dorsal telencephalon of Pax6-knockout mice [11,59,60]. Corresponding with alterations in the cell cycle, the expression of cell-cycle factors was noted in various Pax6 mutants, including increased expression of Ccnd1 in the Pax6-deficient cortex during early corticogenesis, and elevation of both Ccnd2 and P57 Kip2 during late stages of cortical development [40]. Increased levels of Ccnd1 were reported in zebrafish embryos following knockdown of Pax6 proteins [61].
In the current study, we observed that in Pax6-deficient retina there is a reduced number of cells in the S phase, accompanied by a significant and progressive increase in the number of cells exhibiting cell-cycle abnormalities. These were manifested in the elevated expression of cell-cycle-progression factors (Ccnd1, Ccnd2, Ccnd3) and cell-cycle-withdrawal factors (CKIs: P27 Kip1 and P57 Kip2, Figure 4, Figure 7). Despite their opposing functions, increased expression of both types of cell-cycle factors can contribute to delayed differentiation of the Pax6-deficient neuronal precursors. This is supported by the finding that overexpression of Ccnd1 in photoreceptor precursors delays their differentiation [62]. Beyond the CNS, Ccnd1 was further shown to inhibit adipocyte and myoblast differentiation [63-65]. In addition, while the increase in P27 Kip1 and P57 Kip2 promotes cell-cycle exit, it is now recognized that this increase is not sufficient to induce cell differentiation, which depends on additional cues [66-68]. Moreover, P27 Kip1 has been recently shown to contain CDK-independent functions that inhibit the differentiation of postmitotic cells. These oncogenic activities of P27 Kip1 were recognized in mice expressing a mutant isoform of P27 Kip1 which is unable to bind cyclin–CDK. In these mice, in contrast to P27 Kip1-knockout mice, ectopic proliferation and differentiation arrest were detected in the developing retina [69]. It is therefore likely that the elevation of both cell-cycle-promoting and inhibiting factors in the Pax6-deficient RPCs contributes to their delayed differentiation to retinal neurons.
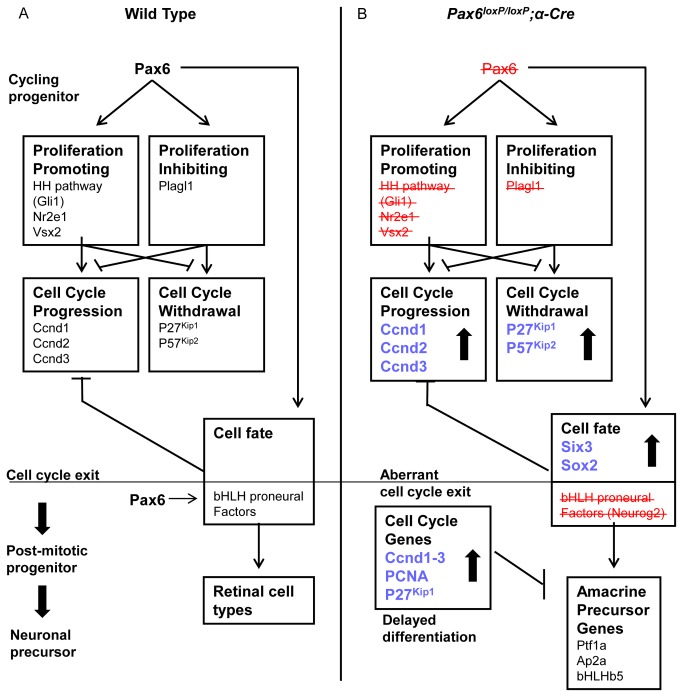
Figure 7. Scheme depicting Pax6 roles during the transition from proliferating retinal progenitor to differentiating retinal neuron.
(A) In normal cycling retinal progenitors, Pax6 regulates the balance between proliferation promoting (i.e. Nr2e1, Vsx2, Hedgehog (HH) signaling) and inhibiting factors (i.e. Plagl1). These in turn regulate the expression of genes which induce either progression of (i.e. Ccnd1–3) or withdrawal (P27 Kip1, P57 Kip2) from the cell cycle. It is also required for the expression of bHLH proneural factors (Neurog2, Atoh7, and Ascl1) presumed to inhibit cell-cycle factors as well as promote specific retinal lineages. (B) Pax6 loss from RPCs results in aberrant cell-cycle exit as Ccnd1–3, as well as P27 Kip1 and P57 Kip2, are elevated and several cell-fate determination factors show reduced (bHLH proneural factors) or increased (Six3, Sox2) expression. The combined outcome of these alterations is delayed differentiation of the Pax6-deficient cells to only one subclass of retinal interneurons.
Pax6 Regulates a Transcriptional Network Capable of both Promoting and Inhibiting the Transition of Proliferating RPCs into Postmitotic Precursors
A few studies have suggested the direct involvement of Pax6 in cell-cycle mechanisms by regulating cell-cycle-related factors such as P27 Kip1 [39] and interacting with Rb [70]. Other findings propose roles for Pax6 in cell division, including sister-chromatid separation [71], interkinetic nuclear movement and determination of the orientation of cell division [72,73]. . While these Pax6 functions may have some effect on the cell-cycle kinetics of Pax6 - RPCs, it is unlikely that they account for the profound alterations in the expression of multiple cell-cycle factors observed in the Pax6 loxP/loxP;α-Cre retina.
Through the characterization of cell-cycle and cell-differentiation dynamics together with transcriptional changes in Pax6 - RPCs, the current study reveals that Pax6 functions upstream of multiple factors, each of which is an important determinant in the regulation of RPC proliferation. We observed reduced expression of several factors that are required for proliferation of retinal progenitors during early stages of retinogenesis: NR2e1, Vsx2 and HH signaling (Figure 7). The loss of each of these components is expected to result in reduced proliferation, as observed in the Pax6-mutant retina.
Nr2e1 is an essential intrinsic regulator of neural stem cells during embryonic development and in adult neurogenesis [74,75]. Nr2e1 has been found to play a key role during multiple stages of eye development. Functional studies in frog embryos suggest that the Nr2e1 homolog Xtll is an eye-field gene and required for normal eye formation [76,77]. In murine embryos, Nr2e1 seems to be dispensable for specification of retinal cell types but consistent with its activities in the cortex, it is required for normal proliferation of RPCs through activation of Ccnd1 and inhibition of P27 Kip1 [44]. Similar to Nr2e1, Vsx2 is expressed in RPCs prior to the onset of retinal neurogenesis. It is initially required at the OV stage for ocular neuroectoderm patterning to the retinal pigmented epithelium (RPE) and retinal lineages, while at the OC stage it is primarily involved in promoting RPC proliferation [78,79]. Though Vsx2 expression is maintained during early stages in the optic rudiment of Pax6-knockout mice [30], its expression was lost at later stages of development in both the systemic and Pax6 loxP/loxP;α-cre OC (Figure 5 and [30]). From this we can conclude that Pax6 is required for Vsx2 maintenance in RPCs but not for initiating its expression in the OV. Vsx2-knockout retinas exhibit reduced expression of Ccnd1 and elevated levels of P27 Kip1, as well as cryptic coexpression of both Ccnd1 and P27 Kip1. The inhibition of P27 Kip1 by Vsx2 is thought to mediate its activity in RPCs, as proliferation is recovered in P27 Kip1;Vsx2-double-knockout mice [43]. The third positive regulator of proliferation that was lost in the Pax6-mutant RPCs is HH signaling, based on the loss of the target Gli1 [80]. HH is an established mitogen in the developing CNS, including the retina [51,52,81]. The deletion of Smoothened, an essential mediator of this signal-transduction pathway, from RPCs resulted in progenitor-pool depletion due to cell-cycle aberrations which included reduced expression of Ccnd1 and elevated expression of P27 Kip1 [51,52].
Considering the proliferation-promoting roles of Nr2e1, Vsx2 and HH in RPCs, it is expected that combined reduction of these factors following Pax6 loss will result in the reduced proliferation observed in the Pax6 loxP/loxP;α-Cre retina (Figure 7). Yet, in the Pax6-deficient RPCs, the reduced proliferation was accompanied by elevated expression of several cell-cycle-promoting factors: Ccnd1, Ccnd2 and Ccnd3. This elevated expression could be due to altered expression of factors which normally inhibit cell-cycle progression (Figure 7). Among these is the tumor suppressor Plagl1 [46]. Plagl1 is expressed from early stages of retinogenesis in RPCs, with higher levels in the distal OC and reduced levels in the central more mature RPCs [15]. Analysis of Plagl1-knockout mice revealed increased RPC proliferation and an increase in the number of Ccnd1-expressing cells [15]. In addition to loss of Plagl1, we detected increased expression of several progenitor factors in the Pax6-deficient RPCs such as Six3 and Sox2 (Figure 5, Figure S7). The increased expression of these progenitor genes combined with the loss of Plagl1 probably contributes to the unique cell-cycle phenotype observed in the Pax6-mutant retina.
The distinctive molecular phenotype of Pax6-deficient RPCs suggests that Pax6 simultaneously controls a number of genes which function during the transition of RPCs to differentiating precursors. Consistent with our findings, recent high-throughput analyses of Pax6 function in the developing cortex, conducted using chromatin immunoprecipitation and transcriptomic profiling of both Pax6 loss- and gain-of-function transgenic models, suggest a complex gene network mediating Pax6 activity in cell proliferation and differentiation during cortical development [41]. The analysis suggested that Pax6 can simultaneously regulate the expression of factors which promote cell proliferation and self-renewal (CDK4 and Hmga2 [82]) and cell-cycle inhibitors (tumor suppressor Pten [83]). Collectively, these observations suggest that Pax6 controls several key TFs and signaling pathways, some with opposing roles, and that the phenotype observed in Pax6 loxP/loxP;α-Cre retinas is due to the combined disruption of several pathways.
The seemingly opposing activities of Pax6 in RPCs may reflect dosage- and context-dependent activity of this TF in the heterogeneous population of progenitors. Indeed, the levels of Pax6 vary according to cell-cycle stage; cells in the G1 and S phase express low levels of Pax6, whereas cells in G2 and M may display either low levels, high levels or no Pax6 expression at all [58]. An additional level of complexity in deciphering the mechanism of Pax6 activity is that this TF also regulates the expression of differentiation factors, particularly that of bHLH proneural factors Ato7 [84,85], Neurog2 [86] and Ascl1 [6-8]. These differentiation-promoting TFs are thought to directly inhibit cell-cycle-promoting factors as was demonstrated for Neurog2 [4]), and thus their combined loss may contribute not only to the limited differentiation potential of Pax6-deficient RPCs but also to the persistent expression of Ccnd1 in the Pax6-deficient precursors. Finally, in addition to regulating multiple downstream TFs, it is now established that Pax6 controls the expression of microRNAs [87]. In future studies, microRNAs should be considered important mediators of Pax6 activity in posttranscriptionally controlling the expression levels of multiple genes simultaneously.
Pax6 Regulates Multiple Factors that Affect Retinal Cell-Fate Specification
In addition to reduced proliferation, the Pax6-deficient RPCs are limited in their differentiation potential as they only give rise to subclasses of GABAergic amacrine, while other retinal lineages fail to differentiate [6]. This limited multipotency was previously attributed to abrogated expression of the proneural bHLH proteins [6,8]. In the present study, we recognized additional perturbation in the Pax6-mutant OC that might account for the eventual acquisition of the amacrine fate (Figure S7). These changes included elevated expression of the amacrine-promoting factors Sox2 (Figure S7A,D) and Six3 (Figure S7B,E), as well as maintained expression of NeuroD1 in the NBL (Figure S7C,F, green). Ectopic expression of Sox2 was shown to induce amacrine cells in the mouse retina [88] and similarly, overexpression of Six3 together with Neurod1 promoted amacrine cell genesis [89]. Furthermore, we detected reduced expression of Plagl1 (Figure 5I-L). In addition to its role in regulating RPC proliferation, Plagl1 functions in the regulation of amacrine cell number by activating a feedback mechanism by which amacrine cells can inhibit the formation of additional amacrines from RPCs [15]. Thus, loss of Plagl1 in the Pax6 loxP/loxP;α-Cre retina might contribute to the formation of excessive numbers of amacrine cells. Collectively, these findings suggest that Pax6 loss results in a unique transcription profile in the retinal precursors, which promotes the generation of amacrine interneurons. Interestingly, although these Pax6-mutant amacrine cells express GABA, their molecular phenotype was distinct from that of normal amacrine cells. In the Pax6-mutant retina, Sox2 protein was detected in many of the amacrine cells but only a small subset of these coexpressed Isl1 and only a few of these coexpressed choline acetyltransferase (ChAT; Figure S8E–H and [88]). In contrast, in the normal retina, the expression of Sox2 in amacrine cells is restricted to Isl1- and ChAT-expressing cells (Figure S8A–D). These findings suggest that while Pax6 is dispensable for the generation of GABAergic interneurons, it is required for normal differentiation of the amacrine cell types.
Future studies, employing Cre lines which are expressed in late progenitors and in postmitotic precursors, are required to determine the roles of Pax6 in the generation of the late-born amacrine cell types (glycinergic and non-GABAergic-non-glycinergic [90,91]) as well as in the differentiation of the postmitotic amacrine precursors.
Supporting Information
Reduced expression of amacrine precursor markers in the Pax6loxP/loxP ;α-Cre retina. Expression of amacrine specification and differentiation markers in control (A–D) and Pax6 loxP/loxP ;α-Cre (E–H) OC. IIF was employed for the detection of Pax6 and VC1.1 (E15.5, green and red, respectively, in A,E) Ptf1a (E15.5, green in B,F), syntaxin and Ap2β (E16.5, red and green, respectively, C, G). BarHL2 (E16.5, D,H) was detected using ISH. The recombinant area in the Pax6loxP/loxP;α-Cre retina (marked with dotted line in E–H) was determined by monitoring Pax6 expression by IIF on an adjacent section (E,G adjacent to F,H respectively). Abbreviations: GCL, ganglion cell layer; NBL, neuroblastic layer. Scale bar in A is 100 μm for A,B,E,F. Scale bar in C is 100 µm for C,D ,G,H.
(TIF)
Expression of PCNA and Ki67 does not overlap in a subset of Pax6- RPCs. A single pulse of BrdU was administered 24 h prior to sacrifice at E18.5 (A,B) or E15.5 (C,D). Sections of control (Pax6loxP/loxP;A,C) and Pax6 loxP/loxP ;α-Cre (B,D) optic cup were double-stained by IIF with antibodies against BrdU (green in A–D) and either PCNA (red in A, B) or Ki67 (red in C,D). Pax6 expression was detected on adjacent sections and used to identify the recombination area in the Pax6loxP/loxP;α-Cre OC (dotted line in B,D,I–L, N). Coexpression of PCNA and Ki67 (red and green, respectively, in E–L) determined by IIF in control (E–H) and Pax6 loxP/loxP ;α-Cre (I–L) retinas at E12.5 (E,I), E14.5 (F,J), E16,5 (G,K) and P0 (H,L). CyclinB1 expression detected by IIF at E13.5 in control and Pax6loxP/loxP;α-Cre OC (M,N). Abbreviation: NBL, neuroblastic layer. Scale bar in A is 100 µm.
(TIF)
Gene ontology (GO) analysis of genes altered in Pax6loxP/loxP ;α-Cre compared to control RPCs. Histogram depicting average significance of significantly enriched (p<0.05) GO terms as calculated using DAVID Bioinformatics Resources [32,33] clustered into functional and previously reported Pax6 functions.
(TIF)
Aberrant expression of cell-cycle factors in the Pax6 mutant OC. IIF analysis for detection of Pax6 (A,E,I,M) ISH for detection of Ccnd1 transcript (B,F,J,N), IIF for Ccnd1 and Crx (red and green, respectively, C,D,G,H,K,L,O,P) in control (A–D, I–L) and Pax6 loxP/loxP ;α-Cre (E–H, M–P) distal retina. Scale bar in A is 100 µm.
(TIF)
Characterization of components of the Notch signaling pathway during retinogenesis in control and Pax6loxP/loxP ;α-Cre embryos. Expression of Notch-pathway components at E13.5 (A-H) and E16.5 (I-P) in control (A-D, I-L) and Pax6 loxP/loxP ;α-Cre (E-H, M-P) retinas. Pax6 - area was delineated by staining for Pax6 protein on the same or adjacent sections (red in D-H,I,M; dotted line in E–H,M-P). Expression of Notch1 (B,F,J,N), Dll1 (C,G,K,O) and Hes5 (D,H,L,P) detected using fluorescent (A-H) or regular (I-P) ISH. Scale bar in A is 100 µm.
(TIF)
Expression of Gli1 but not of Gli2 or Gli3 is decreased in the Pax6loxP/loxP ;α-Cre retina. Expression of Gli1 (A,D) Gli2 (B,E) and Gli3 (C,F) in Pax6 loxP/loxP control (A–C) and Pax6 loxP/loxP ;α-Cre (D–F) optic cups as detected by ISH at E15.5. Scale bar in A is 100 µm.
(TIF)
Altered expression of amacrine-differentiation-promoting and inhibiting factors in Pax6loxP/loxP ;α-Cre RPCs. Control (A–C) and Pax6 loxP/loxP ;α-Cre (D–F) embryonic retina labeled by IIF for Pax6 (E15, red, A,D,C,F) Sox2 (E15, green, A,D) and by ISH for detection of Six3 (E16, B, E), NeuroD1 (E15, green, C,F). Scale bar in A is 100 µm.
(TIF)
Pax6- amacrines display an abnormal molecular phenotype. Control (A–D) and Pax6 loxP/loxP ;α-Cre (E–H) P15 retina cholinergic amacrine labeled by IIF for Isl1 (red), choline acetyltransferase (ChAT, green in A,B,E,F) and Sox2 (green in C,D,G,H) Scale bar in A is 100 µm.
(TIF)
List of differentially expressed genes following Pax6 loss in Pax6loxP/loxP;α-Cre and control mice.
(XLS)
List of differentially expressed genes found in the microarray analysis and validated in situ.
(XLS)
List of primary antibodies used in this study.
(PDF)
Funding Statement
R.A.-P.’s research is supported by the Israel Science Foundation (grant number 610/10), the Israel Ministry of Science (grant number 36494), the Ziegler Foundation, the Binational Science Foundation, the German Israeli Foundation, the Israel Ministry of Health and the Israel-Italy foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Harman AM, Beazley LD (1987) Patterns of cytogenesis in the developing retina of the wallaby Setonix brachyurus. Anat Embryol (Berl) 177: 123-130. doi:10.1007/BF00572536. PubMed: 3434843. [DOI] [PubMed] [Google Scholar]
- 2. Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM (2004) Timing and topography of cell genesis in the rat retina. J Comp Neurol 474: 304-324. doi:10.1002/cne.20134. PubMed: 15164429. [DOI] [PubMed] [Google Scholar]
- 3. Young RW (1985) Cell proliferation during postnatal development of the retina in the mouse. Brain Res 353: 229-239. PubMed: 4041905. [DOI] [PubMed] [Google Scholar]
- 4. Lacomme M, Liaubet L, Pituello F, Bel-Vialar S (2012) NEUROG2 drives cell cycle exit of neuronal precursors by specifically repressing a subset of cyclins acting at the G1 and S phases of the cell cycle. Mol Cell Biol 32: 2596-2607. doi:10.1128/MCB.06745-11. PubMed: 22547683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Das G, Choi Y, Sicinski P, Levine EM (2009) Cyclin D1 fine-tunes the neurogenic output of embryonic retinal progenitor cells. Neural Dev 4: 15. doi:10.1186/1749-8104-4-15. PubMed: 19416500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F et al. (2001) Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105: 43-55. doi:10.1016/S0092-8674(01)00295-1. PubMed: 11301001. [DOI] [PubMed] [Google Scholar]
- 7. Philips GT, Stair CN, Young Lee H, Wroblewski E, Berberoglu MA et al. (2005) Precocious retinal neurons: Pax6 controls timing of differentiation and determination of cell type. Dev Biol 279: 308-321. doi:10.1016/j.ydbio.2004.12.018. PubMed: 15733660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oron-Karni V, Farhy C, Elgart M, Marquardt T, Remizova L et al. (2008) Dual requirement for Pax6 in retinal progenitor cells. Development 135: 4037-4047. doi:10.1242/dev.028308. PubMed: 19004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shaham O, Menuchin Y, Farhy C, Ashery-Padan R (2012) Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res 31: 351-376. doi:10.1016/j.preteyeres.2012.04.002. PubMed: 22561546. [DOI] [PubMed] [Google Scholar]
- 10. Georgala PA, Carr CB, Price DJ (2011) The role of Pax6 in forebrain development. Dev Neurobiol 71: 690-709. doi:10.1002/dneu.20895. PubMed: 21538923. [DOI] [PubMed] [Google Scholar]
- 11. Warren N, Price DJ (1997) Roles of Pax-6 in murine diencephalic development. Development 124: 1573-1582. PubMed: 9108373. [DOI] [PubMed] [Google Scholar]
- 12. Bel-Vialar S, Medevielle F, Pituello F (2007) The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev Biol 305: 659-673. doi:10.1016/j.ydbio.2007.02.012. PubMed: 17399698. [DOI] [PubMed] [Google Scholar]
- 13. Ashery-Padan R, Marquardt T, Zhou X, Gruss P (2000) Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev 14: 2701-2711. doi:10.1101/gad.184000. PubMed: 11069887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S, Mo Z, Yang X, Price SM, Shen MM et al. (2004) Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43: 795-807. doi:10.1016/j.neuron.2004.08.041. PubMed: 15363391. [DOI] [PubMed] [Google Scholar]
- 15. Ma L, Cantrup R, Varrault A, Colak D, Klenin N et al. (2007) Zac1 functions through TGFbetaII to negatively regulate cell number in the developing retina. Neural Dev 2: 11. doi:10.1186/1749-8104-2-11. PubMed: 17559664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Psarros M, Heber S, Sick M, Thoppae G, Harshman K et al. (2005) RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res 33: W638-W643. doi:10.1093/nar/gki490. PubMed: 15980552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. doi:10.1186/gb-2004-5-10-r80. PubMed: 15461798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249-264. doi:10.1093/biostatistics/4.2.249. PubMed: 12925520. [DOI] [PubMed] [Google Scholar]
- 19. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116-5121. doi:10.1073/pnas.091062498. PubMed: 11309499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J et al. (2006) Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133: 4439-4450. doi:10.1242/dev.02598. PubMed: 17075007. [DOI] [PubMed] [Google Scholar]
- 21. Jusuf PR, Harris WA (2009) Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev 4: 34. doi:10.1186/1749-8104-4-34. PubMed: 19732413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bassett EA, Pontoriero GF, Feng W, Marquardt T, Fini ME et al. (2007) Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol Cell Biol 27: 7497-7510. doi:10.1128/MCB.00687-07. PubMed: 17724084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng L, Xie X, Joshi PS, Yang Z, Shibasaki K et al. (2006) Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development 133: 4815-4825. doi:10.1242/dev.02664. PubMed: 17092954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B et al. (2007) Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol 502: 1047-1065. doi:10.1002/cne.21368. PubMed: 17444492. [DOI] [PubMed] [Google Scholar]
- 25. Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U et al. (1984) Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 133: 1710-1715. PubMed: 6206131. [PubMed] [Google Scholar]
- 26. Heatley MK (2002) Ki67 protein: the immaculate deception? Histopathology 40: 483. doi:10.1046/j.1365-2559.2002.01390.x. PubMed: 12010370. [DOI] [PubMed] [Google Scholar]
- 27. Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182: 311-322. doi:10.1002/(SICI)1097-4652(200003)182:3. PubMed: 10653597. [DOI] [PubMed] [Google Scholar]
- 28. Barton KM, Levine EM (2008) Expression patterns and cell cycle profiles of PCNA, MCM6, cyclin D1, cyclin A2, cyclin B1, and phosphorylated histone H3 in the developing mouse retina. Dev Dyn 237: 672-682. doi:10.1002/dvdy.21449. PubMed: 18265020. [DOI] [PubMed] [Google Scholar]
- 29. Pacal M, Bremner R (2012) Mapping differentiation kinetics in the mouse retina reveals an extensive period of cell cycle protein expression in post-mitotic newborn neurons. Dev Dyn, 241: 1525–44. PubMed: 22837015. [DOI] [PubMed] [Google Scholar]
- 30. Bäumer N, Marquardt T, Stoykova A, Ashery-Padan R, Chowdhury K et al. (2002) Pax6 is required for establishing naso-temporal and dorsal characteristics of the optic vesicle. Development 129: 4535-4545. PubMed: 12223410. [DOI] [PubMed] [Google Scholar]
- 31. Duparc RH, Boutemmine D, Champagne MP, Tétreault N, Bernier G (2006) Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol 300: 647-655. doi:10.1016/j.ydbio.2006.07.045. PubMed: 16973151. [DOI] [PubMed] [Google Scholar]
- 32. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44-57. PubMed: 19131956. [DOI] [PubMed] [Google Scholar]
- 33. Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1-13. doi:10.1093/nar/gkp505. PubMed: 19033363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rungger-Brändle E, Ripperger JA, Steiner K, Conti A, Stieger A et al. (2010) Retinal patterning by Pax6-dependent cell adhesion molecules. Dev Neurobiol 70: 764-780. doi:10.1002/dneu.20816. PubMed: 20556827. [DOI] [PubMed] [Google Scholar]
- 35. Stoykova A, Götz M, Gruss P, Price J (1997) Pax6-dependent regulation of adhesive patterning, R-cadherin expression and boundary formation in developing forebrain. Development 124: 3765-3777. PubMed: 9367432. [DOI] [PubMed] [Google Scholar]
- 36. Tyas DA, Pearson H, Rashbass P, Price DJ (2003) Pax6 regulates cell adhesion during cortical development. Cereb Cortex 13: 612-619. doi:10.1093/cercor/13.6.612. PubMed: 12764036. [DOI] [PubMed] [Google Scholar]
- 37. Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501-1512. doi:10.1101/gad.13.12.1501. PubMed: 10385618. [DOI] [PubMed] [Google Scholar]
- 38. Bilitou A, Ohnuma S (2010) The role of cell cycle in retinal development: cyclin-dependent kinase inhibitors co-ordinate cell-cycle inhibition, cell-fate determination and differentiation in the developing retina. Dev Dyn 239: 727-736. doi:10.1002/dvdy.22223. PubMed: 20108332. [DOI] [PubMed] [Google Scholar]
- 39. Duparc RH, Abdouh M, David J, Lépine M, Tétreault N et al. (2007) Pax6 controls the proliferation rate of neuroepithelial progenitors from the mouse optic vesicle. Dev Biol 301: 374-387. doi:10.1016/j.ydbio.2006.11.006. PubMed: 17157287. [DOI] [PubMed] [Google Scholar]
- 40. Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M et al. (2007) Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci 34: 99-119. doi:10.1016/j.mcn.2006.10.008. PubMed: 17158062. [DOI] [PubMed] [Google Scholar]
- 41. Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y et al. (2009) The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLOS Genet 5: e1000511 PubMed: 19521500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carthon BC, Neumann CA, Das M, Pawlyk B, Li T et al. (2005) Genetic replacement of cyclin D1 function in mouse development by cyclin D2. Mol Cell Biol 25: 1081-1088. doi:10.1128/MCB.25.3.1081-1088.2005. PubMed: 15657434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Green ES, Stubbs JL, Levine EM (2003) Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development 130: 539-552. doi:10.1242/dev.00275. PubMed: 12490560. [DOI] [PubMed] [Google Scholar]
- 44. Miyawaki T, Uemura A, Dezawa M, Yu RT, Ide C et al. (2004) Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci 24: 8124-8134. doi:10.1523/JNEUROSCI.2235-04.2004. PubMed: 15371513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM (2006) Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev 20: 1308-1320. doi:10.1101/gad.1413606. PubMed: 16702404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abdollahi A (2007) LOT1 (ZAC1/PLAGL1) and its family members: mechanisms and functions. J Cell Physiol 210: 16-25. doi:10.1002/jcp.20835. PubMed: 17063461. [DOI] [PubMed] [Google Scholar]
- 47. Jadhav AP, Mason HA, Cepko CL (2006) Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133: 913-923. doi:10.1242/dev.02245. PubMed: 16452096. [DOI] [PubMed] [Google Scholar]
- 48. Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R (2006) Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133: 1367-1378. doi:10.1242/dev.02311. PubMed: 16510501. [DOI] [PubMed] [Google Scholar]
- 49. Riesenberg AN, Liu Z, Kopan R, Brown NL (2009) Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci 29: 12865-12877. doi:10.1523/JNEUROSCI.3382-09.2009. PubMed: 19828801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng MH, Shi M, Pei Z, Gao F, Han H et al. (2009) The transcription factor RBP-J is essential for retinal cell differentiation and lamination. Mol Brain 2: 38. doi:10.1186/1756-6606-2-38. PubMed: 20017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sakagami K, Gan L, Yang XJ (2009) Distinct effects of Hedgehog signaling on neuronal fate specification and cell cycle progression in the embryonic mouse retina. J Neurosci 29: 6932-6944. doi:10.1523/JNEUROSCI.0289-09.2009. PubMed: 19474320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA (2005) Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development 132: 5103-5113. doi:10.1242/dev.02096. PubMed: 16236765. [DOI] [PubMed] [Google Scholar]
- 53. Cwinn MA, Mazerolle C, McNeill B, Ringuette R, Thurig S et al. (2011) Suppressor of fused is required to maintain the multipotency of neural progenitor cells in the retina. J Neurosci 31: 5169-5180. doi:10.1523/JNEUROSCI.5495-10.2011. PubMed: 21451052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kenney AM, Rowitch DH (2000) Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol 20: 9055-9067. doi:10.1128/MCB.20.23.9055-9067.2000. PubMed: 11074003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sigulinsky CL, Green ES, Clark AM, Levine EM (2008) Vsx2/Chx10 ensures the correct timing and magnitude of Hedgehog signaling in the mouse retina. Dev Biol 317: 560-575. doi:10.1016/j.ydbio.2008.02.055. PubMed: 18417110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M et al. (1999) Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem 274: 8143-8152. doi:10.1074/jbc.274.12.8143. PubMed: 10075717. [DOI] [PubMed] [Google Scholar]
- 57. Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H (1999) Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126: 3915-3924. PubMed: 10433919. [DOI] [PubMed] [Google Scholar]
- 58. Hsieh YW, Yang XJ (2009) Dynamic Pax6 expression during the neurogenic cell cycle influences proliferation and cell fate choices of retinal progenitors. Neural Dev 4: 32. doi:10.1186/1749-8104-4-32. PubMed: 19686589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Götz M, Stoykova A, Gruss P (1998) Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21: 1031-1044. doi:10.1016/S0896-6273(00)80621-2. PubMed: 9856459. [DOI] [PubMed] [Google Scholar]
- 60. Warren N, Caric D, Pratt T, Clausen JA, Asavaritikrai P et al. (1999) The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cereb Cortex 9: 627-635. doi:10.1093/cercor/9.6.627. PubMed: 10498281. [DOI] [PubMed] [Google Scholar]
- 61. Coutinho P, Pavlou S, Bhatia S, Chalmers KJ, Kleinjan DA et al. (2011) Discovery and assessment of conserved Pax6 target genes and enhancers. Genome Res 21: 1349-1359. doi:10.1101/gr.124115.111. PubMed: 21617155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Skapek SX, Lin SC, Jablonski MM, McKeller RN, Tan M et al. (2001) Persistent expression of cyclin D1 disrupts normal photoreceptor differentiation and retina development. Oncogene 20: 6742-6751. doi:10.1038/sj.onc.1204876. PubMed: 11709709. [DOI] [PubMed] [Google Scholar]
- 63. Rao SS, Kohtz DS (1995) Positive and negative regulation of D-type cyclin expression in skeletal myoblasts by basic fibroblast growth factor and transforming growth factor beta. A role for cyclin D1 in control of myoblast differentiation. J Biol Chem 270: 4093-4100. doi:10.1074/jbc.270.8.4093. PubMed: 7876159. [DOI] [PubMed] [Google Scholar]
- 64. Rao SS, Chu C, Kohtz DS (1994) Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol 14: 5259-5267. PubMed: 8035804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang C, Pattabiraman N, Zhou JN, Fu M, Sakamaki T et al. (2003) Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol 23: 6159-6173. doi:10.1128/MCB.23.17.6159-6173.2003. PubMed: 12917338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tang XM, Beesley JS, Grinspan JB, Seth P, Kamholz J et al. (1999) Cell cycle arrest induced by ectopic expression of p27 is not sufficient to promote oligodendrocyte differentiation. J Cell Biochem 76: 270-279. PubMed: 10618643. [DOI] [PubMed] [Google Scholar]
- 67. Durand B, Gao FB, Raff M (1997) Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation. EMBO J 16: 306-317. doi:10.1093/emboj/16.2.306. PubMed: 9029151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gui H, Li S, Matise MP (2007) A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Dev Biol 301: 14-26. doi:10.1016/j.ydbio.2006.10.035. PubMed: 17123502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Besson A, Hwang HC, Cicero S, Donovan SL, Gurian-West M et al. (2007) Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev 21: 1731-1746. doi:10.1101/gad.1556607. PubMed: 17626791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cvekl A, Kashanchi F, Brady JN, Piatigorsky J (1999) Pax-6 interactions with TATA-box-binding protein and retinoblastoma protein. Invest Ophthalmol Vis Sci 40: 1343-1350. PubMed: 10359315. [PubMed] [Google Scholar]
- 71. Zaccarini R, Cordelières FP, Martin P, Saule S (2007) Pax6p46 binds chromosomes in the pericentromeric region and induces a mitosis defect when overexpressed. Invest Ophthalmol Vis Sci 48: 5408-5419. doi:10.1167/iovs.07-0413. PubMed: 18055787. [DOI] [PubMed] [Google Scholar]
- 72. Asami M, Pilz GA, Ninkovic J, Godinho L, Schroeder T et al. (2011) The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development, 138: 5067–78. PubMed: 22031545. [DOI] [PubMed] [Google Scholar]
- 73. Tamai H, Shinohara H, Miyata T, Saito K, Nishizawa Y et al. (2007) Pax6 transcription factor is required for the interkinetic nuclear movement of neuroepithelial cells. Genes Cells 12: 983-996. doi:10.1111/j.1365-2443.2007.01113.x. PubMed: 17825043. [DOI] [PubMed] [Google Scholar]
- 74. Monaghan AP, Bock D, Gass P, Schwäger A, Wolfer DP et al. (1997) Defective limbic system in mice lacking the tailless gene. Nature 390: 515-517. doi:10.1038/37364. PubMed: 9394001. [DOI] [PubMed] [Google Scholar]
- 75. Roy K, Kuznicki K, Wu Q, Sun Z, Bock D et al. (2004) The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci 24: 8333-8345. doi:10.1523/JNEUROSCI.1148-04.2004. PubMed: 15385616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hollemann T, Bellefroid E, Pieler T (1998) The Xenopus homologue of the Drosophila gene tailless has a function in early eye development. Development 125: 2425-2432. PubMed: 9609825. [DOI] [PubMed] [Google Scholar]
- 77. Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA (2003) Specification of the vertebrate eye by a network of eye field transcription factors. Development 130: 5155-5167. doi:10.1242/dev.00723. PubMed: 12944429. [DOI] [PubMed] [Google Scholar]
- 78. Liu IS, Chen JD, Ploder L, Vidgen D, van der Kooy D et al. (1994) Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron 13: 377-393. doi:10.1016/0896-6273(94)90354-9. PubMed: 7914735. [DOI] [PubMed] [Google Scholar]
- 79. Burmeister M, Novak J, Liang MY, Basu S, Ploder L et al. (1996) Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet 12: 376-384. doi:10.1038/ng0496-376. PubMed: 8630490. [DOI] [PubMed] [Google Scholar]
- 80. Lee J, Platt KA, Censullo P, Ruiz i Altaba A (1997) Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124: 2537-2552 [DOI] [PubMed] [Google Scholar]
- 81. Cayuso J, Ulloa F, Cox B, Briscoe J, Martí E (2006) The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development 133: 517-528. doi:10.1242/dev.02228. PubMed: 16410413. [DOI] [PubMed] [Google Scholar]
- 82. Nishino J, Kim I, Chada K, Morrison SJ (2008) Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135: 227-239. doi:10.1016/j.cell.2008.09.017. PubMed: 18957199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A et al. (2001) Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294: 2186-2189. doi:10.1126/science.1065518. PubMed: 11691952. [DOI] [PubMed] [Google Scholar]
- 84. Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL (2006) Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol 295: 764-778. doi:10.1016/j.ydbio.2006.03.055. PubMed: 16690048. [DOI] [PubMed] [Google Scholar]
- 85. Riesenberg AN, Le TT, Willardsen MI, Blackburn DC, Vetter ML et al. (2009) Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis 47: 175-187. doi:10.1002/dvg.20479. PubMed: 19208436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hufnagel RB, Le TT, Riesenberg AL, Brown NL (2010) Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev Biol 340: 490-503. doi:10.1016/j.ydbio.2010.02.002. PubMed: 20144606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D et al. (2013) Pax6 regulates gene expression in the vertebrate lens through miR-204. PLOS Genet 9: e1003357 PubMed: 23516376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lin YP, Ouchi Y, Satoh S, Watanabe S (2009) Sox2 plays a role in the induction of amacrine and Muller glial cells in mouse retinal progenitor cells. Invest Ophthalmol Vis Sci 50: 68-74. PubMed: 18719084. [DOI] [PubMed] [Google Scholar]
- 89. Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE et al. (2002) Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development 129: 831-842. PubMed: 11861467. [DOI] [PubMed] [Google Scholar]
- 90. Haverkamp S, Wässle H (2000) Immunocytochemical analysis of the mouse retina. J Comp Neurol 424: 1-23. doi:10.1002/1096-9861(20000814)424:1. PubMed: 10888735. [PubMed] [Google Scholar]
- 91. Kay JN, Voinescu PE, Chu MW, Sanes JR (2011) Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nat Neurosci 14: 965-972. doi:10.1038/nn.2859. PubMed: 21743471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reduced expression of amacrine precursor markers in the Pax6loxP/loxP ;α-Cre retina. Expression of amacrine specification and differentiation markers in control (A–D) and Pax6 loxP/loxP ;α-Cre (E–H) OC. IIF was employed for the detection of Pax6 and VC1.1 (E15.5, green and red, respectively, in A,E) Ptf1a (E15.5, green in B,F), syntaxin and Ap2β (E16.5, red and green, respectively, C, G). BarHL2 (E16.5, D,H) was detected using ISH. The recombinant area in the Pax6loxP/loxP;α-Cre retina (marked with dotted line in E–H) was determined by monitoring Pax6 expression by IIF on an adjacent section (E,G adjacent to F,H respectively). Abbreviations: GCL, ganglion cell layer; NBL, neuroblastic layer. Scale bar in A is 100 μm for A,B,E,F. Scale bar in C is 100 µm for C,D ,G,H.
(TIF)
Expression of PCNA and Ki67 does not overlap in a subset of Pax6- RPCs. A single pulse of BrdU was administered 24 h prior to sacrifice at E18.5 (A,B) or E15.5 (C,D). Sections of control (Pax6loxP/loxP;A,C) and Pax6 loxP/loxP ;α-Cre (B,D) optic cup were double-stained by IIF with antibodies against BrdU (green in A–D) and either PCNA (red in A, B) or Ki67 (red in C,D). Pax6 expression was detected on adjacent sections and used to identify the recombination area in the Pax6loxP/loxP;α-Cre OC (dotted line in B,D,I–L, N). Coexpression of PCNA and Ki67 (red and green, respectively, in E–L) determined by IIF in control (E–H) and Pax6 loxP/loxP ;α-Cre (I–L) retinas at E12.5 (E,I), E14.5 (F,J), E16,5 (G,K) and P0 (H,L). CyclinB1 expression detected by IIF at E13.5 in control and Pax6loxP/loxP;α-Cre OC (M,N). Abbreviation: NBL, neuroblastic layer. Scale bar in A is 100 µm.
(TIF)
Gene ontology (GO) analysis of genes altered in Pax6loxP/loxP ;α-Cre compared to control RPCs. Histogram depicting average significance of significantly enriched (p<0.05) GO terms as calculated using DAVID Bioinformatics Resources [32,33] clustered into functional and previously reported Pax6 functions.
(TIF)
Aberrant expression of cell-cycle factors in the Pax6 mutant OC. IIF analysis for detection of Pax6 (A,E,I,M) ISH for detection of Ccnd1 transcript (B,F,J,N), IIF for Ccnd1 and Crx (red and green, respectively, C,D,G,H,K,L,O,P) in control (A–D, I–L) and Pax6 loxP/loxP ;α-Cre (E–H, M–P) distal retina. Scale bar in A is 100 µm.
(TIF)
Characterization of components of the Notch signaling pathway during retinogenesis in control and Pax6loxP/loxP ;α-Cre embryos. Expression of Notch-pathway components at E13.5 (A-H) and E16.5 (I-P) in control (A-D, I-L) and Pax6 loxP/loxP ;α-Cre (E-H, M-P) retinas. Pax6 - area was delineated by staining for Pax6 protein on the same or adjacent sections (red in D-H,I,M; dotted line in E–H,M-P). Expression of Notch1 (B,F,J,N), Dll1 (C,G,K,O) and Hes5 (D,H,L,P) detected using fluorescent (A-H) or regular (I-P) ISH. Scale bar in A is 100 µm.
(TIF)
Expression of Gli1 but not of Gli2 or Gli3 is decreased in the Pax6loxP/loxP ;α-Cre retina. Expression of Gli1 (A,D) Gli2 (B,E) and Gli3 (C,F) in Pax6 loxP/loxP control (A–C) and Pax6 loxP/loxP ;α-Cre (D–F) optic cups as detected by ISH at E15.5. Scale bar in A is 100 µm.
(TIF)
Altered expression of amacrine-differentiation-promoting and inhibiting factors in Pax6loxP/loxP ;α-Cre RPCs. Control (A–C) and Pax6 loxP/loxP ;α-Cre (D–F) embryonic retina labeled by IIF for Pax6 (E15, red, A,D,C,F) Sox2 (E15, green, A,D) and by ISH for detection of Six3 (E16, B, E), NeuroD1 (E15, green, C,F). Scale bar in A is 100 µm.
(TIF)
Pax6- amacrines display an abnormal molecular phenotype. Control (A–D) and Pax6 loxP/loxP ;α-Cre (E–H) P15 retina cholinergic amacrine labeled by IIF for Isl1 (red), choline acetyltransferase (ChAT, green in A,B,E,F) and Sox2 (green in C,D,G,H) Scale bar in A is 100 µm.
(TIF)
List of differentially expressed genes following Pax6 loss in Pax6loxP/loxP;α-Cre and control mice.
(XLS)
List of differentially expressed genes found in the microarray analysis and validated in situ.
(XLS)
List of primary antibodies used in this study.
(PDF)