Summary
A key obstacle to understanding neural circuits in the cerebral cortex is that of unraveling the diversity of GABAergic interneurons. This diversity poses general questions for neural circuit analysis: how are these interneuron cell types generated and assembled into stereotyped local circuits and how do they differentially contribute to circuit operations that underlie cortical functions ranging from perception to cognition? Using genetic engineering in mice, we have generated and characterized ~20 Cre and inducible CreER knockin driver lines that reliably target major classes and lineages of GABAergic neurons. More select populations are captured by intersection of Cre and Flp drivers. Genetic targeting allows reliable identification, monitoring, and manipulation of cortical GABAergic neurons, thereby enabling a systematic and comprehensive analysis from cell fate specification, migration, and connectivity, to their functions in network dynamics and behavior. As such, this approach will accelerate the study of GABAergic circuits throughout the mammalian brain.
Introduction
The cerebral cortex is the most recently evolved brain region in vertebrates and supports sophisticated sensory, motor, and cognitive functions in mammals. Despite its large size and functional diversification, the neocortex may have arisen from the duplication of stereotyped local circuits with subtle specializations in different cortical areas and species (Rakic, 2009). A major obstacle to understanding neural circuits in the cerebral cortex is the daunting diversity and heterogeneity of inhibitory interneurons (Markram et al., 2004).
Compared with the more abundant glutamatergic projection neurons, GABAergic interneurons constitute only approximately 20% of cortical neurons, yet these interneurons are crucial in regulating the balance, flexibility, and functional architecture of cortical circuits (Klausberger and Somogyi, 2008; Markram et al., 2004). GABAergic interneurons consist of a rich array of cell types with distinct physiological properties, connectivity patterns, and gene expression profiles. Their diverse intrinsic, synaptic, and dynamic properties allow interneurons to generate a rich repertoire of inhibitory outputs (Jonas et al., 2004). Their distinct connectivity patterns ensure differential recruitment by appropriate inputs as well as strategic distribution of their outputs to stereotyped locations (e.g. specific cellular and subcellular targets) in cortical network (Buzsaki et al., 2004; Somogyi et al., 1998). GABAergic interneurons also play key roles in various forms of network oscillations that provide spatial-temporal frameworks to dynamically organize functional neural ensembles (Bartos et al., 2007; Buzsaki, 2001; Klausberger and Somogyi, 2008).
The assembly of cortical GABAergic circuits begins during mid-gestation and continues well into the postnatal period (Huang et al., 2007). GABAergic neurons are generated mainly in the medial and caudal ganglionic eminences (MGE and CGE) of the basal ganglia and the preoptic area (POA) (Batista-Brito and Fishell, 2009; Gelman and Marin, 2010). MGE and CGE express different sets of transcription factors and give rise to distinct classes of interneurons (Gelman and Marin, 2010). Postmitotic GABAergic neurons navigate towards the developing neocortex through a remarkable process of long distance tangential migration (Marin and Rubenstein, 2001). They subsequently disperse into appropriate cortical areas, settle in appropriate layers, establish specific connectivity patterns, and acquire distinct physiological properties (Huang et al., 2007).
Despite significant progress in past decades, anatomical, physiological, and developmental studies of cortical GABAergic circuits have been hindered by the heterogeneity of cell types. At present, for any given class of interneurons, we often lack comprehensive knowledge of their connectivity patterns, activity during relevant behaviors, and function in cortical information processing. We also have incomplete knowledge as to how they are specified, assemble into circuits, and contribute to activity-dependent maturation and plasticity in cortical networks. This is in part because of the difficulty in tracking the development of interneurons due to the considerable delay between their generation and maturation into potent inhibitory networks, often not complete until early adolescence, depending on cortical areas and species. Individual cell types are the basic units of circuit assembly and function. To achieve a comprehensive understanding of the cortical GABAergic circuits, it is therefore necessary to establish experimental systems that allow precise and reliable identification and manipulation of distinct cell types.
Genetic approaches promise to significantly facilitate the study of the cortical GABAergic circuitry because they engage the intrinsic gene regulatory mechanisms that generate and maintain cell type identity and phenotypes. Using mouse genetic engineering, we have initiated the first round of a systematic effort to genetically target cortical GABAergic neurons. Here we report the generation and characterization of nearly 20 knockin “driver lines” expressing Cre or inducible CreER recombinase. These mouse lines establish reliable experimental access to major classes and lineages of cortical inhibitory neurons. We further demonstrate that more specific subpopulations can be targeted using the intersection of Cre and Flp drivers and by engaging lineage restriction and birth timing mechanisms. These GABA drivers set the stage for a systematic and comprehensive analysis of cortical GABAergic circuits, from cell fate specification,, connectivity, to their functions in network dynamics and behavior. Beyond this, as GABAergic neurons are basic components of neural circuits throughout the mammalian brain, these GABA drivers will also prove useful for analyzing many other brain systems and circuits.
Results
Strategy and resource
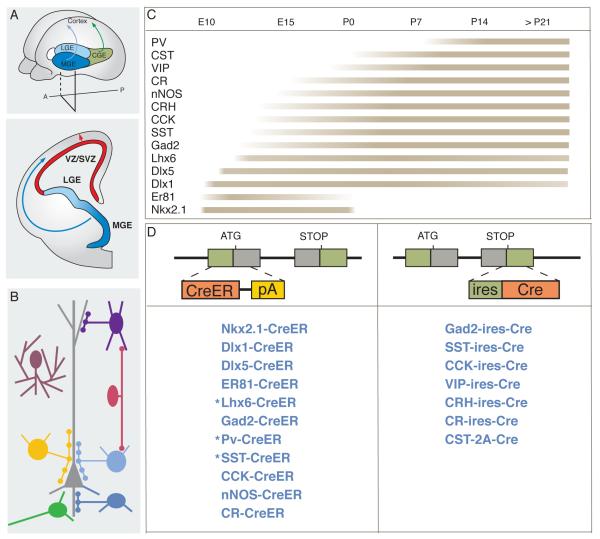
Classification of cortical GABAergic neurons has long been contentious (Ascoli et al., 2008). A useful criterion is the pattern of axon projection along with cellular and subcellular targets of innervation (Figure 1) (Somogyi et al 1998; Markram et al., 2004). For the purpose of genetic targeting, we parse cortical GABAergic populations based on their gene expression. Although gene expression profiles correlate and likely contribute to cell phenotype and identity (Nelson et al., 2006; Sugino et al., 2006), there is often no simple relationship between the expression of a single gene and a morphologically and functionally defined cell type. However, current methods of genetic targeting restrict our approach to cell types based on expression of one or two genes. As a first step, we selected over a dozen genes to target major GABAergic populations and lineages. These included broadly expressed GABA synthetic enzyme and transcription factors, as well as neuropeptides, enzymes, and calcium binding proteins with more restricted expression that correlates with subpopulations (Figure 1).
Figure 1.
Overall scheme on the genetic targeting of GABAergic neurons. (A) Top: Cortical GABAergic neurons in rodents are largely generated from the medial and caudal ganglionic eminence (MGE, CGE), and preoptic area (POA). Bottom, postmitotic GABAergic neurons undergo long distance migration (blue arrow) into the cortex and form circuits with glutamatergic pyramidal neurons that are generated in the dorsal ventricular zone and migrate radially into the cortex (red arrow). (B) Mature cortical GABA neurons can be parsed into several major classes according to their preferential axon innervation of specific cellular and subcellular targets; the expression of several molecular markers tend to correlate, although not exclusively, with these classes. (C) List of genes that were used for genetic targeting in this study and their approximate temporal expression pattern in developing neocortex. (D) Design of CreER and Cre gene targeting strategy and list of knockin lines. Stars (*) indicate lines with low efficiency CreER induction.
We used the Cre/loxP binary gene expression system (Dymecki and Kim, 2007) to target GABAergic neurons. In order to faithfully engage the genetic mechanisms that specify and maintain cell identity, we aimed to generate driver lines in which Cre activity precisely and reliably recapitulates the endogenous gene expression. We therefore used gene targeting in embryonic stem (ES) cells to insert Cre coding cassettes either at the translation initiation codon or immediately after the translation STOP codon of an endogenous gene (Figure 1, Sppl Figure 1, Table 1, SpplTable 1). We used four reporter alleles, all generated at the Rosa26 locus, to assay recombination patterns: 1) RCE-LoxP is a loxP-STOP-loxP-GFP reporter (Miyoshi et al., 2010) , 2) RCE-Frt is an frt-STOP-frt-GFP reporter (Miyoshi et al., 2010), 3) Ai9 is a loxP-STOP-loxP-tdTomado reporter (Madisen et al., 2010), 4) RCE-dual is a loxP-STOP-loxP-frt-STOP-frt-GFP reporter which expresses GFP upon the intersection of Cre and Flp recombination (Miyoshi et al., 2010).
Table 1.
| Driver lines | ES cells |
Vector
construction |
5′ homology arm with end primers | 3′ homology arm with end primers |
Targeting
efficiency |
Recomb. Specificity (cortex) |
Recomb. Efficiency (cortex) |
|---|---|---|---|---|---|---|---|
|
Gad2-ires-Cre JAX# 010802 |
V6.5 | recombineering | 2.5 kb 5′end: TGATGTAAGAGATGGTCGCC 3′end: TTACAAATCTTGTCCGAGGC |
4.7 kb 5′end: TCACCTCACTCATCAAACTG 3′end: TGGGAGAGTAGAAGACAATG |
15 % | ~92.2% | ~91% |
|
SOM-ires-Cre JAX# 013044 |
V6.5 | Long PCR | 2.2 kb 5′end: GGAGATTACCTGTTCTATGCC 3′end: CTAACAGGATGTGAATGTCTTC |
5.2 kb 5′end:AATATTGTTGTCCTAGCCAGAC 3′end:TGGACAAGCTCCTAAGTCATC |
1% | ~92.2% | ~93.5% |
|
VIP-ires-Cre JAX# 010908 |
V6.5 | recombineering | 2.3 kb 5′end: CTTAGGAAACATAGCTGAAAG 3′end: GCTCTGCCCAGAGGCCTCTTC |
4.9 kb 5′end: GAAGAGGCCTCTGGGCAGAGC 3′end: GATGCGTGGTGAGGTTCTGTC |
4 % | ~91.5% | ~84.5% |
|
CCK-ires-Cre JAX# 012706 |
V6.5 | recombineering | 5.5 kb 5′end: ATGAGCTACAAAGGCGTCCTG 3′end: CTACGATGGGTATTCGTAGTC |
1.8 kb 5′end:GAGGAGGTGGAATGAGGAAAC 3′end:AGAGGGATACGGTCTACCGTG |
3% | n.d. | n.d. |
|
CR-ires-Cre JAX# 010774 |
Bruce4 | Long PCR | 6.0 Kb 5′end:GCATTCAACCAAAGCACACTTCTTAGCA 3′end: TTACACGGGGGGCTCACTGCAG |
2.0 Kb 5′end: GAAGGGACAGGGGCTGCTTCTG 3′end: CTATTGCTCCAGACGGGCTTC |
6.2% | ~90.8%* | ~71.3%* |
| CRH-ires-Cre JAX# 012704 |
Bruce4 | Long PCR | 4.8 kb 5′end: TGGCTTACATGCTCCATTGTG 3′end: TCATTTCCCGATAATCTCAATC |
2.1 kb 5′end: GCGCTTGGCCAAAACGATTCTG 3′end: TGACTTATCTCACTCATTGAG |
37.5% | n.d. | n.d. |
|
CST-ires-Cre JAX# 010910 |
V6.5 | recombineering | 2.0 kb 5′end: GCTCTTGGTTCATCCTTCCTTG 3′end: CTTGCACGAGGAGAAGGTTTTCCA |
4.2 kb 5′end: CCTATGTAGTTATGAGTGACCTT 3′end: TACACAAATCAAAGGGAAATC |
30% | n.d. | n.d. |
|
Gad2-CreER JAX# 010702 |
V6.5 | recombineering | 5.0 kb 5′end: TGTGATCCCTGCCTGCCAATG 3′end: GGGTTCTGCTAGACTGGCGCT |
2.0 kb 5′end:CTCTGCTCTATGGAGACTCCG 3′end:ACAATGTATCGAGATGATCAG |
8% | ~100% | High |
|
SOM-CreER JAX# 010708 |
Bruce4 | Long PCR | 5.6 kb 5′end: GCTTATGGAAGAGACACAGAC 3′end: CTTCCTTGCCTCAGGCAGCCAA |
~2 kb 5′end: GCTTATGGAAGAGACACAGAC 3′end: CTTCCTTGCCTCAGGCAGCCAA |
0.5% | ~100% | Very low # |
|
PV-CreER JAX# 010777 |
Bruce4 | recombineering | 2.1 kb 5′end: TCAGATGGAGCTGAGAGGTAG 3′end: CCTGCAACTGTTTGAGCGGG |
4.7 kb 5′end: GTGTGTAAAAAGACAAAGCAAC 3′end: TCTTCTTAGTGCTGGGTTGAG |
~10% | ~100% | Very low# |
|
CR-CreER JAX# 013730 |
Bruce4 | recombineering | 4.9 kb 5′end: AGCGTCTTCCCTTGGATAAT 3′end: GGCGAGCCGCTCCGGAGATC |
2.6 kb 5′end: TGACTGCATCCCAGTTCCTGGAAA 3′end: CATCCCCACTGCTAGTGACC |
~ 60% | >70% | Medium |
|
CCK-CreER JAX# 012710 |
Bruce4 | recombineering | ~2.0 kb 5′end:CCTCTCTCCTTCTTGCTTGGTAAG 3′end:GGCTATGGGAAGCAAAGGCGAG |
~5.0 kb 5′end:GTGGCATGGATGGTGACCTCTGG 3′end:ACCGTCTCCTGAGAGTCGGCTTGG |
50% | n.d. | Medium |
|
nNOS-CreER JAX# 014541 |
V6.5 | Long PCR | ~4 kb 5′end: ATGACCTGCTTTGACTGCTT 3′end: GGTATCTGTGTCCTTCAGAAG |
2.1 kb 5′end: TTATCCAAGCCGGCGACATCATTC 3′end: CATGTGGTTGCTGGGGTTTGA |
3.9% | ~100% @ | Medium |
|
Nkx2.1-CreER JAX# 014552 |
V6.5 | recombineering | 2.0 kb 5′end: GAACAGCAGACAAGCAAAGCC 3′end: GATTCGGCGTCGGCTGGAGGAG |
4.6 kb 5′end: GACGTGAGCAAGAACATGGCC 3′end: GCACGGAGCGCTGTCTCCTCG |
2% | n.a. | High |
|
Dlx1-CreER JAX# 014551 |
V6.5 | recombineering | 2.1 kb 5′end: TTCTGCGCACAGCCTCATGCC 3′end: CTCTTCTCGCGGGGTCTGGGT |
5.0 kb 5′end: GTTTCGGATATCAATCTGTGG 3′end: CTCTGTGGACTGAGTCAGATG |
20% | n.a. | High |
|
Dlx5-CreER JAX# 010705 |
Bruce4 | recombineering | 2.0 kb 5′end: GGATCAGTGTCAAGGAACAGC 3′end: AGCTCCCGGGCGGTGGCTGTTC |
4.2 kb 5′end: CGTCTTCCCTGCCCAAGCGTC 3′end: CAGCAGGGAGATGACATCCAC |
1% | n.a. | High |
|
Lhx6-CreER JAX# 010776 |
Bruce4 | recombineering | 2.0 kb 5′end: TGCTTACCATGAGTGGTCACG 3′end: GCTTCCAGTACATGGGCCCGG |
5.0 kb 5′end: TTGCTTCATTAGAGAGACACC 3′end: CTCTGTGCTCTCAATCTCTGG |
20% | n.a. | Very low# |
|
ER81-CreER JAX# 013048 |
Bruce4 | recombineering | 2.0 kb 5′end: GCTCTGATACTGTCCTCTCTC 3′end: TCTGCTCCTCTTCGCAAATAT |
5.0 kb 5′end: AGTCAGCGTGGGAGAAACTG 3′end: CTTGGCTTCAAGGTGTCACTG |
52% | n.a. | Medium |
Specificity and efficiency of Cre recombination were determined from quantification in the neocortex (see Results). Efficiency in CreER lines (High, medium, low) refers to frequency of recombination after tamoxifen induction. 5′ end primer is in the forward direction, 3′ end primer is in the reverse direction.
based on Cre-dependent AAV expression.
<5 cells in cortex in a 50μm sagittal section. n.d. not determined due to lack of specific antibodies.
the nNOS antibody (Zymed) labeled large type I cells but not small type II cells in the cortex; using the RCE reporter, all GFP+ cells were nNOS+; using the Ai9 reporter, the small RFP+ cells were nNOS- (see Results and Figure 8, sppl Figure 7 for details). n.a. not applicable.
Our current characterizations have focused on neocortex and hippocampus, but most GABA driver lines also show Cre activities throughout the brain (Table 2,) from the retina to the spinal cord. A broader characterization of these GABA drivers in the CNS including atlases of Cre-dependent reporter expression is presented at the Cre Driver website http://credrivermice.org. Cre activities in the peripheral nervous system and non-neuronal tissues have not been examined. These GABA driver lines are being distributed by the Jackson Laboratory (http://www.jax.org/).
Table 2.
Overview of Cre recombination patterns in major adult brain regions in selected lines
| OB | CX | HP | AMYG | STR | SC | CB | THAL | HT | BS | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gad2-ires-Cre | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | Reticular nucleus |
++++ | ++++ |
| SST-ires-Cre | ++ | ++ | ++ | ++ | ++ | +++ | + | Reticular nucleus |
++ | ++ |
|
CCK-ires-Cre/
Dlx5-Flp |
++ | ++ | ++ | ++ | − | − | + | − | + | − |
| VIP-ires-Cre | ++ | ++ | ++ | + | − | ++ | + | − | ++ (SCN) | ++ |
| CR-ires-Cre | ++ AOB |
++ | ++ | ++ | ++ | ++ | ++ | − | + | ++ |
| CRH-ires-Cre | ++ AOB |
++ | ++ | ++ | + | ++ | ++ | − | ++ (PVN) |
++ |
| CST-ires-Cre | − | ++ | ++ | − | − | − | − | − | − | − |
|
nNOS-CreER (P21→P30) |
++ AOB |
++ | ++ | ++ | ++ | ++ | ++ | + | ++ | + |
|
Nkx2.1-CreER (E13→P31) |
+ | +++ | +++ | ++ | ++ | − | − | − | − | − |
|
Dlx1-CreER (E12→P22) (P98→P108) |
+++ +++ |
+++ ++ |
++ ++ |
++ ++ |
++ ++ |
+ + |
- - |
- - |
- - |
- - |
|
Dlx5-CreER (E12→P22) (P61→P74) |
+++ +++ |
+++ ++ |
++ ++ |
++ ++ |
++ ++ |
+ - |
- - |
- - |
- - |
- - |
|
Lhx6-CreER (P28→P36) |
− | + | − | − | + | − | − | − | − | − |
|
Ee81-CreER (E13→P28) |
− | + | + | + | + | − | + | − | − | − |
This Table gives an overall impression of the recombination patterns in several brain areas. In each area, recombination often occurs in restricted or highly specific cell populations. In general, recombination pattern tends to recapitulate endogenous mRNA expression assayed by in situ hybridization (e.g. those in the Allen Brain Atlas). Pattern of recombination were determined in three to five animals (>3 weeks) of each line, and were grouped into large categories based on visual observation. “++++,” in most or all GABA neuron; “+++,” in many GABA neuron; “++,” in some GABA neurons; “+,” in a few GABA neurons; “−“ few or no recombination in the sections examined.
The Nkx2.1-CreER driver confers genetic access to MGE progenitors
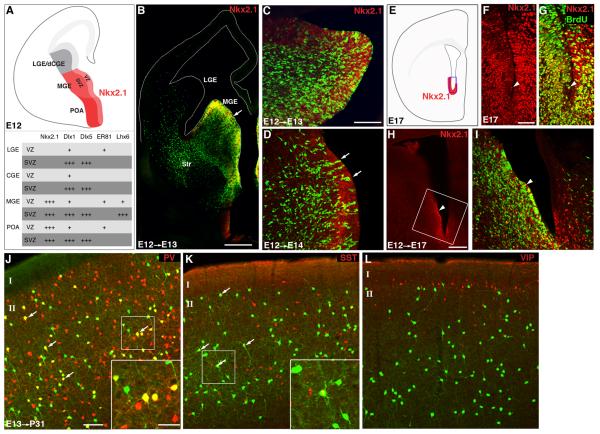
Genetic fate mapping using transcription factors that define progenitor pools should provide insight into the specification and development of GABAergic subtypes. Nkx2.1 is the only known factor specifically expressed within MGE by all progenitors in the ventricular zone (VZ) and SVZ (Flames et al., 2007; Marin and Rubenstein, 2001). In addition, Lhx6 and Er81 are expressed in subdomains of MGE and CGE (Figure 2A) (Flames et al., 2007). We have generated inducible CreER drivers targeting these transcription factor genes (see Table 1, 2).
Figure 2.
The Nkx2.1-CreER driver captures MGE progenitors.
(A) A schematic of the ventricular (VZ) and subventricular (SVZ) zones of ganglionic eminence at mid-gestation and a summary of transcription factor expression in these regions. (B) GFP expression in an Nkx2.1-CreER::RCE-LoxP brain 1day after induced at E12. MGE progenitors and postmitotic GABA neurons are labeled by GFP; progenitors were colabeled by an NKX2.1 antibody (red) in VZ (arrow). Postmitotic neurons were migrating towards cortex and striatum (Str). (C) High dose tamoxifen induction at E12 labeled many MGE progenitors (colabeled by an NKX2.1 antibody in red) and postmitotic neurons. (D) Low dose tamoxifen induction at E12 labeled sparser MGE progenitors, which showed radial clone-like organization (arrows) in VZ (colabeled by an NKX2.1 antibody). (E) A schematic of NKX2.1 expression in the ventral portion of late embryonic SVZ. (F, G) E17 NKX2.1+ cells (red) in the ventral SVZ were colabeled by a BrdU antibody (green), suggesting that they were mitotic. Four pulses of BrdU were administered every 4 hours at E17. (H, I) MGE progenitors labeled at E12 (green) contributed to NKX2.1+ cells (red) in E17 SVZ. (J-L) NKX2.1 progenitors labeled at E13 gave rise to major types of cortical interneurons including PV (J) and SST (K) cells but not VIP cells (L) at P31. Scale bars: 300 μm in B, 100 μm in C, D, I, 50 μm in F, G, 500 μm in H, 50 μm in J-L, 25 μm in insets of J, K.
Although several Nkx2.1 transgenic lines have been generated expressing a constitutive form of Cre (Fogarty et al., 2007; Xu et al., 2008b), they deviate from the spatiotemporal pattern of endogenous Nkx2.1 to varying degrees, and offer no temporal control over Cre activity. In contrast, our Nkx2.1-CreER driver appeared to precisely recapitulate the endogenous expression and allows temporal regulation of Cre activity, thereby establishing reliable genetic access to the MGE progenitors (Figure 2). E12 tamoxifen induction resulted in robust labeling of the VZ and SVZ progenitors in MGE and POA but not LGE (Figure 2B,C). Low dose tamoxifen induction (0.5 mg/30 g body weight) further revealed radial columns of cells, which likely represent putative progenitor clones in MGE (Figure 2D). Consistent with previous studies (Miyoshi et al., 2007), E12 induction gave rise to cortical GABA neurons expressing parvalbumin (PV), somatostatin (SST), but not vasoactive intestinal peptide (VIP) (Figure 2J-L).
Recent studies demonstrate that Nkx2.1 expression continues beyond mid-gestation and persists in the ventral ridge of SVZ during late embryonic and postnatal ages (Marin et al., 2000; Magno et al., 2009). Indeed, we found Nkx2.1-Cre activity in ventral SVZ beyond E17 (Figure 2E-G), when the characteristic eminence of MGE had already fused with the adjacent LGE. This raised the issue of whether these ventral SVZ cells derived from earlier MGE or acquired Nkx2.1 expression independently. We found that these Nkx2.1+ cells continued to incorporate BrdU labeling at E17 (administered 4 times every 4 hours) and thus retained mitotic competence, which is a key indication of progenitor properties. Using genetic fate mapping, we further demonstrated that Nkx2.1+ progenitors in ventral SVZ derived from earlier progenitors in MGE (e.g. from E12 MGE progenitors; Figure 2H,I) but not the lateral ganglionic eminence (LGE).
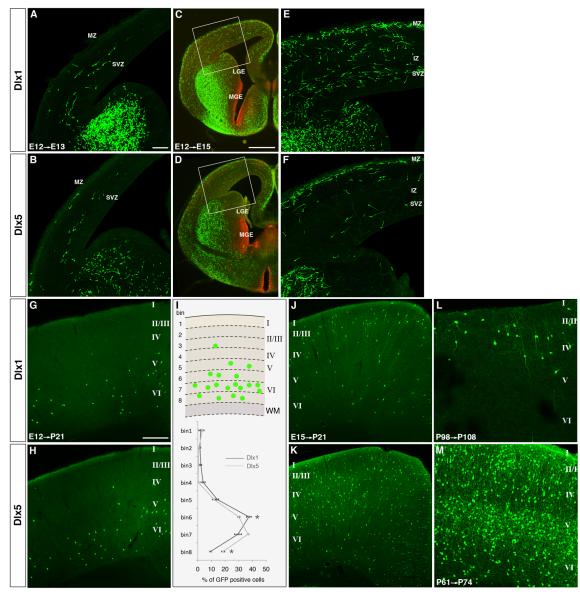
The Dlx1- and Dlx5-CreER drivers show distinct patterns throughout development
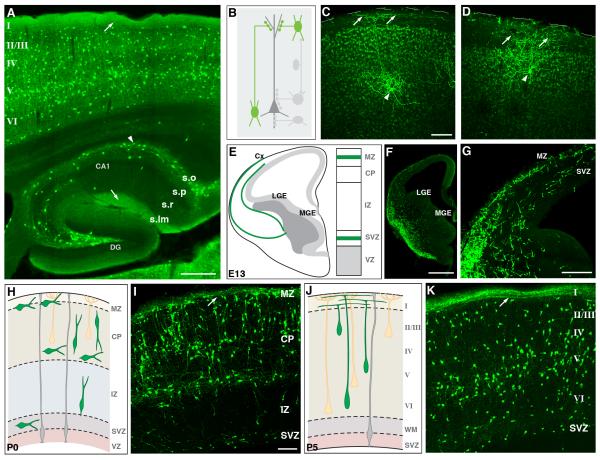
Members of the Dlx family of homeobox transcription factors, Dlx1, Dlx2, Dlx5, and Dlx6, are expressed mainly in the SVZ of embryonic LGE, MGE and CGE (Cobos et al., 2006; Eisenstat et al., 1999). Dlx genes continue to express in subsets of GABAergic neurons in embryonic, postnatal, and mature brains, and have been implicated in regulating their migration, differentiation, survival, and function (Cobos et al., 2007; Cobos et al., 2005; Long et al., 2009). Whether and how different members control the development and function of subpopulations of interneurons is not well understood. Although Dlx5/6-Cre and Dlx1/2-Cre transgenic mice have been generated using short enhancer elements (Potter et al., 2009), these lines do not fully recapitulate endogenous expression and show ectopic Cre recombination. We generated both Dlx1-CreER and Dlx5-CreER knockin drivers, which permit Dlx1+ and Dlx5+ interneurons to be identified and manipulated throughout development. As Dlx1 and Dlx5 are expressed predominantly in the SVZ in putative committed precursors at mid-gestation (i.e. becoming postmitotic after a limited number of cell division) (Eisenstat et al., 1999), CreER induction around this time (e.g. at E12) likely label cohorts of GABA neurons with similar birth dates. Our initial characterization with E12 induction suggests that Dlx1 and Dlx5 may be expressed in at least partially non-overlapping populations of progenitors. During tangential migration at E13, both the E12-induced Dlx1 and Dlx5 cohorts appeared to take similar routes, via the subventricular zone, into the cortex (Figure 3A,B). By E15 however, the two cohorts showed very different patterns of migration. Whereas the Dlx1 cohort migrated throughout the marginal zone (MZ), cortical plate (CP), and intermediate zone (IZ), the Dlx5 cohort migrated predominantly in MZ (Figure 3C-F). In mature cortex (P21), both cohorts settled in deep cortical layers despite their different migration routes, with a larger fraction of Dlx5-CreER labeled neurons situated deeper in layer 6 than Dlx1-CreER labeled neurons (Figure 3G-I).
Figure 3.
The Dlx1- and Dlx5-CreER drivers preferentially target different subsets of developing and mature cortical interneurons.
(A-I) Migration and distribution patterns of interneuorns labeled by the Dlx1-CreER or Dlx5-CreER driver induced at E12. (A, B) At E13, both cohorts of interneuorns entered the cortex through the SVZ. (C-F) At E15, while the Dlx5 cohort migrated predominantly in MZ (D,F), the Dlx1 cohort migrated in both MZ and SVZ and had entered the cortical plate (C,E). (G-I) At P21, both cohorts settled in deep layers of the cortex but with slightly different laminar patterns. Quantification of laminar distribution of GFP positive neurons (schematic in the upper panel of I) in the somatosensory area of Dlx-1-CreER (G) and Dlx5-CreER (H) mice demonstrated that the fraction of Dlx5-CreER labeled neurons occupying the deeper part of layer 6 was higher than that of Dlx1-CreER labeled neurons (I). Results are expressed as the mean±SEM from 3 animals for each genotype. *p<0.01, t-test. (J-M) Distribution of interneuorns at P21 labeled by the Dlx1-CreER or Dlx5-CreER driver induced at E15. While the Dlx1 driver labeled a small subset, the Dlx5 labeled broad population of interneurons.
(L-M) Distribution of interneuorns labeled by the Dlx1- and Dlx5-CreER drivers in adult cortex induced in adult. While the Dlx1 driver labeled a small subset, the Dlx5 labeled broad population of interneurons. Scale bars: 100 μm in A, B, E, F, L, M, 500 μm in C, D, 300 μm in G,H,J,K.
At later embryonic stages (e.g. E15), induction in Dlx1- and Dlx5- drivers gave rise to a very different pattern in the mature cortex (P21, Figure 3J,K). The Dlx1 driver mainly labeled upper layer interneurons. Many of these interneurons showed bipolar morphology and were reminiscent of CGE-derived populations such as VIP or CR positive interneurons. On the other hand, the Dlx5 driver labeled broader populations in all layers, suggesting that induction occurred not only in SVZ progenitors but also in migrating cells that had earlier become postmitotic. This distinction between the Dlx1- and Dlx5- drivers became more evident with adult induction (Figure 3L,M), indicating that in the mature cortex Dlx1 expression is increasingly restricted to a small subset of interneurons, whereas Dlx5 expression is increasingly more ubiquitous among GABAergic neurons. Together, our initial characterization of these two driver lines demonstrated that Dlx1 and Dlx5 are differentially expressed in progenitors and developing interneurons at different developmental stages and thus may play different roles in GABAergic circuit development and function.
The Gad2-ires-Cre and Gad2-CreER: Pan GABA drivers
In mammals, GABA is synthesized by two isoforms of glutamic acid decarboxylases GAD67 and GAD65, encoded by the Gad1 and Gad2 genes, respectively, and co-expressed in most brain regions (Soghomonian and Martin, 1998). To establish genetic access to most or all GABAergic neurons throughout the brain, we generated both Gad2-ires-Cre and Gad2-CreER drivers. In the Gad2-ires-Cre driver, Cre is co-expressed with Gad2 throughout development in GABAergic neurons and in certain non-neuronal cells. Because Cre/loxP recombination converts transient CRE activity to permanent reporter allele activation, reporter expression is a developmental integration of Cre activities up to the time of analysis. In all brain regions examined, Cre-activated RCE reporter expression is almost entirely restricted to GABAergic neurons and includes almost all GABAergic neurons (Sppl Figure 2). In the barrel cortex for example, the fraction of GFP neurons that were GAD67 immuno-fluorescent (i.e. specificity) was 92±2.1% and the fraction of GAD67+ cells expressing GFP (i.e. efficiency) was 91±2.9% (n=300 cells from 3 mice).
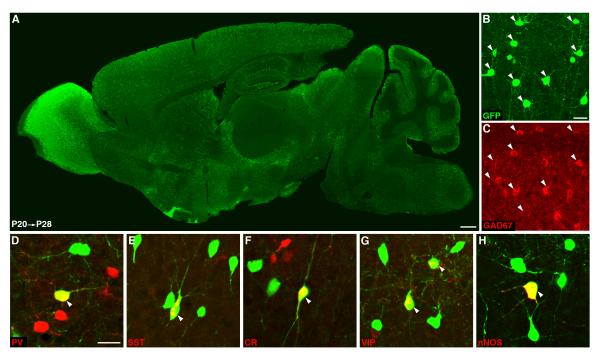
In the Gad2-CreER driver, induction in embryonic or postnatal animals activated reporter expression in GABAergic neurons throughout the brain (Figure 4A). In barrel cortex, reporter expression is entirely restricted to GABAergic neurons and includes all major subpopulations defined by a variety of molecular markers (e.g. PV, SST, Calretinin, VIP, nNOS; Figure 4B-H). Importantly, recombination efficiency can be adjusted by tamoxifen dosage. With low doses, this driver may provide a Golgi-like method by randomly labeling single GABA neurons throughout the brain, and may further allow single neuron genetic manipulation in combination with floxed conditional alleles. With higher doses, this driver allows manipulation of GABA neurons with temporal control. Together, the Gad2-ires-Cre and Gad2-CreER drivers provide robust and flexible genetic tools to manipulate GABAergic neurons throughout the mouse CNS.
Figure 4.
The Gad2-CreER driver. (A) Overview of a P28 brain sagital section with P20 tamoxifen induction. (B,C) GFP expression activated by Cre recombination was restricted to GABAergic neurons (Gad67, red) in cortex. (D-H). GFP labeled neurons in cortex include major populations of interneurons which are positive for parvalbumin (PV), somatostatin (SST), calretinin (CR), vasoactive intestinal peptide (VIP) or nNOS. See SpplFig.1 for recombination patterns of the Gad2-ires-Cre driver. Scale bars: 500 μm in A, 25 μm in B, C, 25 μm in D-H.
The SST-ires-Cre and SST-CreER drivers capture dendrite-targeting interneurons
Somatostatin (SST) is a neuropeptide expressed in a sub-population of dendrite-targeting interneurons derived from the MGE (Miyoshi et al., 2007; Xu et al., 2008a) including: Martinotti cells in neocortex (Wang et al., 2004) and O-LM cells in hippocampus (Sik et al., 1995; Figure 5 diagram). Martinotti cells mediate frequency dependent di-synaptic inhibition among neighboring layer 5 pyramidal neurons and control their synchronous spiking (Berger et al., 2009). O-LM cells modulate pyramidal cell dendrites at distinct phases of hippocampal network oscillation in a brain-state dependent manner (Klausberger et al., 2003). However, the function of these neurons in behaving animals and the mechanism underlying their synaptic specificity are unknown.
Figure 5.
The SST-ires-Cre and SST-CreER drivers. (A) Distribution of SST interneurons in cortex and hippocampus in an adult SST-ire-Cre::RCE-LoxP mouse. Laminar organization in cortex and hippocampus are indicated. Note the prominent axon track in cortical layer 1 (arrow). In hippocampal CA1, O-LM cells (soma indicated by an arrowhead) innervate the distal dendrite of pyramidal cells. O-LM cell axons form a prominent and sharp band in the stratum lacunosum moleculare (arrow) (also see SpplFig.2). (B) A schematic of distal dendrite-targeting interneurons. (C, D) Low frequency recombination in the SST-CreER driver labeled single Martinotti cells (arrow heads) in layer 5 (C) and layer2 (D). Note the characteristic axon arborization in layer1 (arrows). The background green puncta were non-specific signals from GFP antibody staining which were commonly seen when brain sections contained very small number of GFP expressing cells. (E) A schematic showing the migration streams of SST neurons at ~E13. Marginal zone (MZ), cortical plate (CP), intermediate zone (IZ), subventricular zone (SVZ), and ventricular zone (VZ) are indicated. (F, G) At E13, the SST-ires-Cre driver labeled migrating SST neurons soon after they exited MGE. SST neurons entered the cortex mainly through MZ and SVZ. (H) A schematic of P0 cortex depicting SST neurons (green) migrating and entering cortical plate. Pyramidal neurons are in light yellow and radial glia in gray. (I) At P0, while some SST neurons were still migrating in MZ and IZ, a significant portion had entered the cortical plate. Many SST neurons showed vertically oriented neurites, and their layer1 axons (an arrow) became discernable. (J) A schematic of P5 cortex depicting SST neurons (green) which are settling into appropriate cortical layers. (K) At P5, SST neurons were settling into appropriate cortical layers, and their layer1 axons (an arrow) were already prominent. Scale bars: 500 μm in A, 100 μm in C, D, 500 μm in F, 200 μm in G, 100 μm in I, K. stratum oriens (s.o), stratum pyramidale (s.p), stratum radiatum (s.r), stratum lacunosum moleculare (s.lm), dentate gyrus (DG).
The SST-ires-Cre driver provides experimental access to these neurons. In barrel cortex, the fraction of GFP neurons that showed SST immuno-fluorescence (i.e. specificity) was 92±2.08% and the fraction of SST+ cells expressing GFP (i.e. efficiency) was 93.5±3.3% (n=289 cells from 3 mice). The dense axon terminals of Martinotti cells which target the apical tufts of pyramidal cell dendrites are particularly prominent in layer1 (Figure 5A). There is also a notable band of axon fibers in layer 4, which likely target the apical dendrites of layer 6 pyramidal neurons that terminate in this layer (Sppl Figure 3). In the hippocampus, the labeling of all O-LM cell axons revealed their striking subcellular specificity. These axons form a prominent band in stratum lacunosum molecular, which contains the apical tufts of pyramidal neurons with a razor-sharp boarder with stratum radiatum (Figure 5A). The SST-ires-Cre driver provides the first robust in vivo system for manipulating SST interneurons to discover their function and the mechanisms underlying the subcellular specificity of their axons.
Using the Ai9 reporter (Madisen et al., 2010), we imaged cortical SST neurons in live mice with synaptic resolution using 2-photon microscopy (Sppl Figure 3, sppl movie 1). This experimental system allows “on-line” identification of SST interneurons during in vivo physiology and imaging experiments, and longitudinal studies of their development and plasticity.
SST is highly expressed in developing cortical GABA neurons beginning by mid-gestation (Batista-Brito et al., 2008). Since reporter expression remains restricted to SST neurons in the mature cortex, this indicates that Cre activity in SST-ires-Cre driver is specific to the SST population throughout development. SST neurons can be labeled as early as E13, soon after they exit the SVZ of MGE (Figure 5E-F). They reach the developing cortex by E14 and mainly migrate in the marginal zone and subventricular zone (Figure 5E-G). By P0, migrating SST neurons in layer 1 appear to have extended axons and some have entered the cortex trailed by vertically oriental neurites (Figure 5H,I). By P5, most SST cells are in the cortex, and layer 1 axons are already prominent with conspicuous synaptic boutons (Figure 5J,K). Therefore, the SST-ires-Cre driver provides an experimental system to examine the developmental history of SST neurons, including their migration, subcellular synapse targeting, and maturation.
In the SST-CreER line, tamoxifen-induced recombination is also restricted to SST neurons but the efficiency is very low as assayed with both the RCE (Figure 5C,D) and Ai9 reporters (see Discussion). The SST-CreER driver allows imaging and reconstruction of single cortical SST interneurons (Figure 5C,D).
Both SST-ires-Cre and SST-CreER drivers are also active in many other brain regions including: olfactory bulb, striatum, reticular nucleus of the thalamus, superior colliculus, brain stem, as well as in stripes of cerebellar Purkinje cells (Sppl Figure 3, Table 2).
A subtype of perisomatic targeting interneurons is captured by an intersectional strategy
Inhibition directed towards the soma and proximal dendrites of pyramidal neurons controls the gain of summed inputs and thereby the spike discharge. It also regulates the phasing and synchronization of neural ensembles (Freund and Katona, 2007). Perisomatic inhibition is mediated by two broad classes of interneurons that express either the calcium-binding protein parvalbumin (PV) or the neuropeptide cholecystokinin (CCK) (Figure 6A). PV interneurons are characterized by fast and precise intrinsic and synaptic properties and are hypothesized to control the precise timing of cortical network oscillations (Bartos et al., 2007). PV-Cre drivers have been generated and are widely used (Hippenmeyer et al., 2005; Madisen et al., 2010). We generated an inducible PV-CreER driver, which gave low frequency recombination in sparse PV neurons in cerebral cortex and other brain regions (Figure 1).
Figure 6.
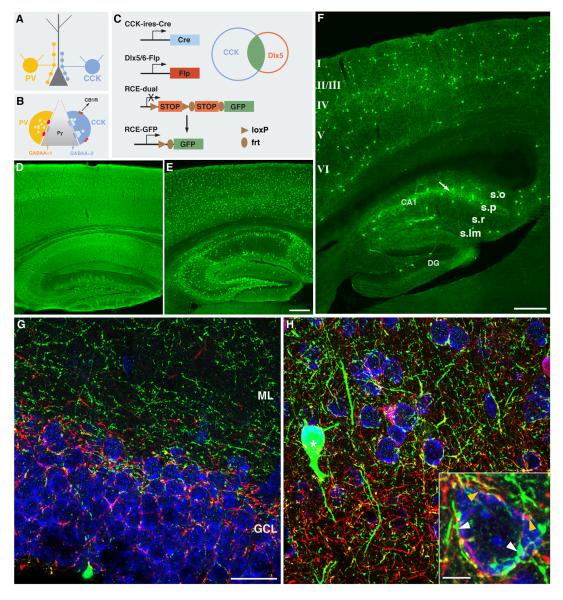
Intersectional strategy captures CCK positive GABA interneurons.
(A, B) Schematics depict CCK and PV basket interneurons that target pyramidal cell (Py) soma and proximal dendrites (A). CCK interneuron axon terminals show distinct molecular profiles compared to those of PV interneurons, including prominent expression of the CB1 receptor and signaling through distinct postsynaptic GABAA receptors (B). (C) Intersectional strategy using CCK-ires-Cre, Dlx5/6-Flp, and a dual reporter (RCE-dual) to label CCK GABAergic interneurons. (D) GFP reporter expression in the cortex and hippocampus in a CCK-ires-Cre::RCE-LoxP mouse. Note the broad expression presumably including both pyramidal neurons and GABAergic neurons. (E) GFP reporter expression in the cortex and hippocampus in a Dlx5/6-Flp::RCE-Frt mouse. Most of the GABAergic neurons are labeled with GFP. (F) CCK positive GABAergic interneurons in cortex and hippocampus are targeted by the intersectional strategy. Note the dense GABAergic axons (an arrow) in the pyramidal cell layer (stratum pyramidale) in hippocampal CA1. Laminar organization in cortex and hippocampus are as indicated. (G) In hippocampal dentate gyrus, CCK axon terminals (green) segregated from PV axon terminals (red). (H) CCK GABAregic neurons (star) and axons (green) in neocortex. PV axons were labeled with an antibody (red), and all cell somata were labeled with TOTO-3 (blue). Inset: CCK axon terminals (white arrowheads) segregated from PV axon terminals (yellow arrowheads) around the same cell soma. Scale bars: 300 μm in D-F, 25 μm in G, H, 5 μm in inset of H.
In contrast to PV interneurons, CCK-interneurons appear to fine-tune network oscillations and are influenced by subcortical inputs that carry information about motivation, emotion, and autonomic states (Freund and Katona, 2007). CCK interneuron synapses are distinguished from all other inhibitory axon terminals by their specific and high level expression of the cannabinoid type 1 receptors (Katona et al., 1999), which confer a powerful retrograde modulation of GABA release, depending on pyramidal cell activity (Wilson and Nicoll, 2001). We generated both a constitutive CCK-ires-Cre and an inducible CCK-CreER driver but found that they activated the RCE-LoxP reporter in both pyramidal neurons and GABA interneurons in neocortex and hippocampus (Figure 6D, sppl Figure 4A-C). It is likely that CCK or its preprohormone is expressed at low levels in pyramidal neurons or in pyramidal neuron precursors during development.
To selectively target CCK+ GABAergic neurons we used an intersectional strategy that combines two recombinase activities from the CCK-ires-Cre and Dlx5/6-Flp drivers. Dlx5/6-Flp is a transgenic line expressing the Flp recombinase in most cortical GABA neurons (Figure 6E; also see Miyoshi et al 2010). The CCK-ires-Cre and Dlx5/6-Flp intersection was achieved with the RCE-dual reporter, which can only be activated if both Cre and Flp are simultaneously or sequentially expressed in the same cell (Miyoshi et al., 2010). In hippocampus, GABAergic CCK basket cells are located in both stratum pyramidale and stratum radiatum and form a conspicuous band of perisomatic synapses around pyramidal neurons (Figure 6F). In dentate gyrus, CCK basket axons mainly target the proximal dendrites of granule cells in the molecular layer (ML) and segregate from PV basket cell axons, which target granule cell somata (Figure 6G). However, in the neocortex both CCK and PV basket cell axons target the same perisomatic regions of pyramidal neurons. Thus, genetic labeling of CCK+ perisomatic synapses allows them to be distinguished from those formed by PV interneurons around the same pyramidal neuron (Figure 6H). Interestingly, PV+ and CCK+ GABAergic synapses selectively signal through either α1- or α2- containing GABAa receptors, which show fast or slow kinetics, respectively (Nyiri et al., 2001). Genetic access to both PV+ and CCK+ GABAergic synapses may allow study of this exquisite form of synapse specificity. The intersectional strategy can also be used to examine the migration, differentiation, and circuit integration of CCK interneurons.
CCK-ires-Cre and CCK-CreER drivers are also active in other brain regions including olfactory bulb, amygdala, brain stem and a subset of cerebellar Purkinje cells (Sppl Figures 4, Table 2).
The VIP-ires-Cre targets interneurons which selectively innervate other interneurons
Vasoactive intestinal peptide (VIP) is expressed in a subset of cortical GABAergic neurons that are derived from the caudal ganglionic eminence (CGE) and do not overlap with SST or PV interneurons (Miyoshi et al., 2010b). A prominent feature of some VIP interneurons is their preferential innervation of other inhibitory interneurons (David et al., 2007; Somogyi et al., 2003). VIP neurons may also regulate cortical blood flow and metabolism since its receptors appear to localize to blood vessels as well as neurons (Cauli and Hamel, 2010; Cauli et al., 2004).
In the VIP-ires-Cre driver, Cre activity is detected in the neocortex, hippocampus, olfactory bulb, suprachiasmatic nuclei, and other discrete midbrain and brain stem regions (Sppl Figure 5). In the upper layers of barrel cortex, the fraction of GFP neurons that showed VIP immuno-fluorescence was 91.5% and the fraction of VIP+ cells expressing GFP was 84.5% (n=213 cells). Most VIP neurons in layer 2/3 typically extend vertically oriented dendrites (Figure 7A,B and sppl movie 2). In contrast to SST interneurons (Figure 5A), VIP neurons do not elaborate significant axon arbors in layer1 (Figure 7A,B). In hippocampal CA1, VIP neuron axons appear as two distinct bands in stratum pyramidale and stratum orien (Figure 7A).
Figure 7.
The VIP-ires-Cre driver.
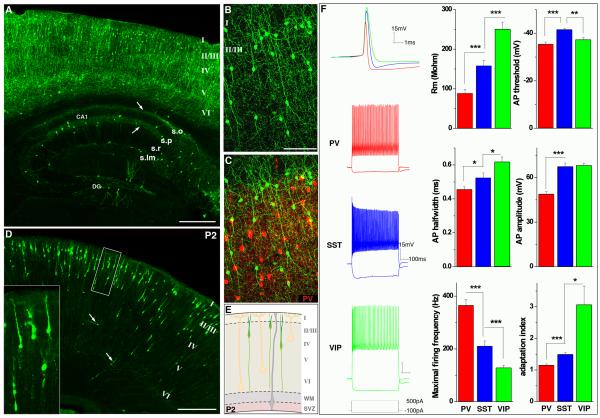
(A) Distribution of VIP interneurons in an adult VIP-ires-cre::Ai9 mouse cortex and hippocampus. Tdtomato signals in A-C were psudocolored green. Laminar organization in cortex and hippocampus are as indicated. Note the prominent vertical orientation of neurites in cortex. In hippocampal CA1, the axons of VIP neurons segregated into distinct lamina in stratum oriens and stratum pyramidale (arrows). (B,C) Higher magnification view of cortical VIP interneurons. A stacked images is available as sppl movie 2. Note that VIP interneurons were completely segregated from PV interneurons (red) in (C). (D) VIP interneurons in a P2 VIP-ires-cre::RCE-LoxP mouse cortex. Young VIP interneurons migrated up into cortical plate from the subventricular zone with strikingly homogeneous and vertically oriented neurites. Inset shows higher magnification view of the boxed region. (E) A schematic depicts VIP neurons in P2 cortex. Pyramidal neurons and radial glias are drawn in light yellow and gray, respectively. (F) Cortical VIP interneurons (green traces) showed distinct intrinsic properties compared with PV (red traces) and SST interneurons (blue trances), including membrane resistance (Rm), action potential (AP) threshold, half width, amplitude, maximum firing frequency, and adaptation. Scale bars: 400 μm in A, 100 μm in B, 300 μm in D. See SpplFig5 for expression pattern in other brain regions.
Using visual cortical slices from VIP-ires-Cre;Ai9, SST-ires-Cre;Ai9, and PV-ires-Cre;Ai9 mice, we compared the intrinsic properties of VIP, SST, and PV interneurons. Consistent with prior studies on the corresponding neurons in the rat (Cauli et al., 2000), VIP cells showed the largest input resistance, broadest action potentials, and exhibited firing patterns with the most accommodation and the lowest maximal frequency among the three cell populations (Figure 7F).
VIP expression, like SST, begins during the neonatal period and thus the VIP-ires-Cre driver can be used to visualize and manipulate developing VIP interneurons. At P0, we found that VIP neurons migrate in the white matter and begin to enter the cortical plate (data not shown). At P2, many VIP neurons have entered the cortex and appeared to strictly migrate radially towards the pia. During this period, they display strikingly homogenous morphology with vertically oriented leading neurites towards the pia and a long trailing process towards the white matter (Figure 7D). These observations suggest that after reaching the cortex, VIP neurons largely disperse within the intermediate zone and attain their laminar positions by radial migration from IZ into the cortex. Such a highly homogeneous mode of migration is in sharp contrast to SST neurons, which enter the cortex in both marginal and intermediate zone and migrate in multiple modes and directions to reach their laminar positions (Figure 5H,I). The VIP-ires-Cre driver therefore provides a reliable genetic handle for studying the development and function of this CGE-derived class of interneuron, including using optogenetic approaches to explore the in vivo release of VIP and its physiological impact.
The nNOS-CreER driver targets neurogliaform cells and long projection GABA neurons
Nitric oxide (NO) is a signaling molecule in the brain synthesized by the neuronal isoform of nitric oxide synthase (nNOS). In cerebral cortex, nNOS is broadly expressed during development (Bredt and Snyder, 1994) and is subsequently restricted to subsets of GABAergic neurons (Kubota et al., 2011). In hippocampus, nNOS+ neurons include neurogliaform cells (NGFCs) and ivy cells (Fuentealba et al., 2008). The most unique feature of NGFCs, including those in the neocortex, is their regulation of local neurons through non-synaptic GABA release and volume transmission (Olah et al., 2009), which may lead to long-lasting network hyperpolarization and widespread suppression in local circuits. NO release from these neurons may also regulate blood vessels and local hemodynamics (Cauli and Hamel, 2010).
In the neocortex, nNOS+ GABA neurons include two types (Kilduff et al., 2011). Whereas Type II cells likely include NGFCs, Type I nNOS+ cells represent another highly unusual population of GABA neurons. First, Type I neurons project long distance axons ipsi- and contralaterally withing cortex, and to subcortical regions, and are conserved from rodent to primate (Higo et al., 2009; Tamamaki and Tomioka; Tomioka et al., 2005). Second, whereas most cortical neurons exhibit reduced firing during slow wave sleep (SWS), Type I neurons are selectively activated during SWS. Thus Type I nNOS+ neurons might be positioned to influence network state across widespread brain areas and may provide a long-sought anatomical link for understanding homeostatic sleep regulation (Kilduff et al., 2011).
In the nNOS-CreER driver, patterns of recombination almost perfectly matched known nNOS neuron profiles throughout the brain. However, the extent of labeling varied in the two reporter lines, as they differ in sensitivity. Whereas the less sensitive RCE reporter labeled only the type I cells (Sppl Figure 7) in cortex, the more sensitive Ai9 reporter labeled both type I and type II cells (Figure 8B). The nNOS cells extend thin, highly profuse axons with notably small boutons throughout cortical layers (Figure 8C,D,F and sppl movie 3), but their terminals avoid the perisomatic regions of pyramidal neurons, which were surrounded by PV+ basket cell axon terminals (Figure 8H). In the hippocampus, nNOS-CreER efficiently labeled neurons whose somata were located in the stratum lacunosum molecular and stratum pyramidale, which likely correspond to NGFCs and ivy cells (Figure 8B,E,G, I and sppl movie 4). These neurons elaborate extremely dense and thin local axons with very small boutons that appear to cover entire volume of stratum oriens and stratum radiatum. Notably, stratum pyramidale and stratum lacunosum moleculare, which were targeted by CCK (Figure 6) or PV (Figure 8I) interneurons and SST (Figure 5) interneurons, respectively, were devoid of nNOS+ axons (Figure 8B,E,G,I). Genetic access to nNOS+ GABAergic projection neurons and NGFCs will facilitate the study of their inputs and outputs, physiological properties, and in vivo functions.
Figure 8.
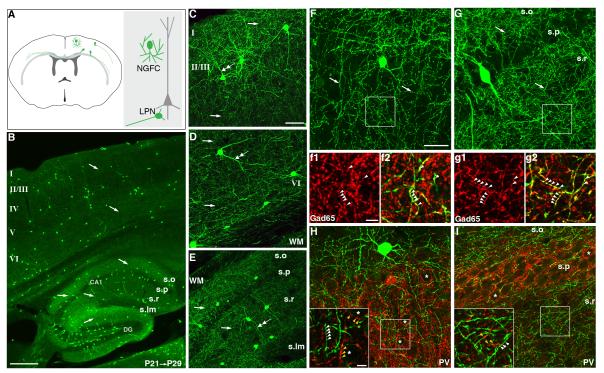
The nNOS-CreER driver
(A) A schematic of nNOS GABAergic neurons depicting the neurogliaform (NGFC) cells and long-range projection neurons (LPN) that extend axons across cortical areas and to the contralateral cortex. (B) Sagittal view of cortex and hippocampus from a P29 nNOS-CreER;Ai9 mouse induced from P21. TdTomato-labeled nNOS neurons were psudocolored green. Cortical nNOS neurons were distributed in all layers and extend highly profuse axons (arrows) throughout the cortex. In hippocampal CA1, NGFC cells elaborate extremely dense axons in stratum oriens and stratum radiatum but were nearly absent in stratum lacunosum moleculare and stratum pyramidale. In the dentate gyrus (DG), some nNOS neurons were located in the area of subgranular zone and hilus. (C, D) Confocal images of superficial (C) and deep (D) cortical layers. nNOS neuron dendrites were indicated by double arrows and axons by arrows. Note the extremely dense axon networks. (E) Confocal image of NGFCs in hippocampal CA1. Note the often vertically oriented dendrites (double arrows) and the extremely dense thin axons (arrows). (F, G) High magnification confocal images of nNOS axons (arrows) in cortical layer 3 (F) and hippocampal CA1 (G). Movie files from the stacked images of F and G are available as sppl movie 3 and 4, respectively. (f.g) Single optical sections in boxed areas from F and G show that nNOS axonal boutons (green) contain GAD65 (red), a GABAergic presynaptic marker. Note the small and closely spaced GAD65 boutons (arrowheads) along green axons. (H, I) Non-overlapping distribution of nNOS and PV axon terminals. In neocortex (H), pyramidal neuron somata (stars) were surrounded by PV axon terminals (red) but were not approached by nNOS axon terminals (green). Inset: higher magnification view of boxed region; white arrowheads indicate nNOS axon terminals, and yellow arrowheads indicate PV axon terminals. In hippocampal CA1 (I), PV basket cell axon terminals heavily innervated the perisomatic regions of pyramidal neurons (stars in stratum pyramidale), whereas nNOS terminals predominantly concentrated around pyramidal cell dendrites in stratum oriens and stratum radiatum. Inset: higher magnification view of the boxed region; white arrowheads indicate nNOS axon terminals, and yellow arrowheads indicate PV axon terminals. Scale bars: 300 μm in B, 50 μm in C-E, 25 μm in F-I, 5 μm in insets of H, I, 5 μm in f1, f2, g1, g2. See SpplFig6 for expression pattern in other brain regions.
The nNOS-CreER driver also efficiently labeled nNOS neurons in olfactory bulb, striatum, amygdala, superioculicullus and hypothalamus (Sppl figure 6, Table 2).
The CRH-ires-Cre driver
Corticotropin releasing hormone (CRH; also known as corticotropin releasing factor-CRF) is best known for mediating neuroendocrine stress response (Korosi and Baram, 2008). CRH and its receptors are widely expressed in the CNS (Korosi and Baram, 2008). CRH modulates a wide range of behaviors including anxiety, arousal, motor function, learning, and memory (Korosi and Baram, 2008), and has been implicated in early life programming (Korosi and Baram, 2009) and depression (Binder and Nemeroff, 2010). In cerebral cortex, CRH neurons constitute a significant fraction of GABA interneurons (Kubota et al., 2011).
The CRH-ires-Cre driver appears to target CRH neurons throughout the brain, including those in the paraventricular nuclei of hypothalamus, bed nucleus of the stria terminalis, locus coeruleus, raphe, and amygdala (Sppl figure 7, Table 2). In superior culicullus, labeled neurons include bottlebrush cells, which project their dendritic terminals in monostritified arrays (“bottlebrush” dendritic endings) and have been implicated in motion processing (Major et al., 2000). In hippocampus and neocortex, the subset of targeted interneurons showed no overlap with PV, SST, and only partial overlap with CR (33±5% (n= 816 cells from 2 mice). The CRH-ires-Cre driver will facilitate studies of the function and development of CRH neurons; it will also allow study of how early life experience and chronic stress alter the connectivity and function of CRH neurons in distributed neural circuits that mediate stress responses in the adult brain.
The CR-ires-Cre and CR-CreER drivers
The calcium binding protein calretinin (CR) is expressed in a subpopulation of GABAergic neurons throughout the brain. In cerebral cortex, CR interneurons include layer 1 GABA neurons and several subpopulations that co-express SST and VIP (Kubota et al., 2011). Labeling mediated by CR-ires-Cre and CR-CreER driver lines largely recapitulate endogenous CR expression (Table 2, Sppl figure 8). The CR-CreER shows high or modest Cre activity, depending on brain regions, upon tamoxifen induction (Sppl figure 8).
The CST-2A-Cre driver
Cortistatin (CST) is a neuropeptide that shares 11 of its 14 amino acids with SST (de Lecea, 2008). CST is predominantly expressed in cerebral cortex, and in subsets of GABA interneurons with partial overlap to SST. In contrast to SST, CST administration in brain ventricles enhances EEG synchronization by selectively promoting slow-wave sleep (de Lecea, 2008). Steady-state levels of CST mRNAs oscillate during the light:dark cycle and are upregulated upon sleep deprivation. The CST-2A-Cre driver appears to selectively target this interneuron population. Cre activity is restricted to a subpopulation of GABA interneurons in cortex and hippocampus and show a partial overlap with SST (37±7.9% (n=568 cells from three sections in one mouse) and PV (15±1.5% (n=573 cells from three sections in one mouse) interneuron populations.
Discussion
Since Cajal’s study of cortical neurons using the Golgi stain more than a century ago (Cajal, 1899), a major obstacle to understanding the organization and function of neural circuits in cerebral cortex has been the lack of methods allowing precise and reliable identification and manipulation of specific cell populations. Genetic targeting is probably the best strategy to systematically establish experimental access to cortical cell types because it engages gene regulatory mechanisms that specify, maintain or correlate with cell types. Combined with modern molecular, optical, and physiological tools, genetic targeting enables labeling of specific cell populations with markers for anatomical analysis, expression of genetically encoded indicators to record their activity, and activation or inactivation of these neurons to examine the consequences in circuit operation and behavior (Luo et al., 2008).
In past decades, genetic approaches have proved increasingly powerful for elucidating a wide array of neural circuits in C. elegans (Macosko et al., 2009), Drosophila (Chiang et al., 2011), zebrafish (McLean and Fetcho, 2008), and mice (Haubensak et al., 2010). For example, genetic analysis of the transcriptional mechanisms that shape neuronal identity and connectivity in the vertebrate spinal cord has provided an entry point into targeting distinct neuronal populations of the central pattern generator networks which control rhythmic movements (Goulding, 2009). However, despite its importance for cognitive function and neuropsychiatric disorders, no coherent effort has been made to systematically apply genetic analysis to neural circuits of the cerebral cortex. Here we have initiated the first round of a systematic genetic targeting of cortical GABAergic neurons by establishing Cre-mediated genetic switches in different cell populations. Reliable genetic access and the combinatorial power of the Cre/loxP binary system will integrate modern physiology, imaging and molecular tools to provide a systematic analysis of GABAergic neurons; they will further enable a comprehensive study of the development, connectivity, function, and plasticity in cortical inhibitory circuitry.
Genetic targeting strategies: transgenesis vs knockin
Two main strategies have been used to target cell types in mice (Huang et al., 2010). In the transgenic approach, including BAC (bacterial artificial chromosome) transgenics (Gong et al., 2003), expression of a transgene is driven by promoter elements contained within the transgenic construct as well as by the gene regulatory elements near the genomic loci of transgene integration. The advantages of transgenesis include relatively easier construction of mouse lines and potentially higher level of transgene expression due to multiple copies. The main limitation of this approach is that transgene expression often does not fully recapitulate that of the endogenous gene and varies among transgenic lines. Given current knowledge about mammalian gene regulation and chromatin biology, this should perhaps come as no surprise. First, cis regulatory elements (enhancers, repressors, insulators) are often very distant from the transcription start site (Kapranov et al., 2007), thus even BAC constructs often do not contain the full complements of regulatory elements of the gene of interest. Second, because cis-regulatory elements can act at a very long distance (Bulger and Groudine, 2011; Heintzman and Ren, 2009), enhancers and repressors near the transgene integration site (but that are unrelated to the promoter elements in the transgene) can influence transcription, leading to ectopic or suppressed expression. Third, different transgenic lines will have different expression patterns due to differential enhancer/repressor influences at different genomic integration sites. Fourth, transgenes inserted into a foreign chromatin environment can be silenced or epigenetically altered in unpredictable ways. Thus the challenges often associated with the transgenic strategy are the uncertainty of the targeted neurons and the effort necessary to ascertain their identity and property.
Here we have used the gene knockin strategy to target GABAergic neurons. Cre cassettes are inserted by homologous recombination at endogenous gene loci, which are embedded in their native chromatin environment with largely intact regulatory elements. The main advantage of this strategy is that Cre expression precisely and reliably recapitulates the targeted endogenous gene. Indeed, after extensive characterization we found that recombination patterns in almost all the GABA Cre drivers often perfectly match the spatial and temporal pattern of the endogenous gene expression. The disadvantages of gene targeting approach include: 1) the possibility of altering the expression of the targeted gene, even when a bicistronic cassette (e.g. ires or T2A) is inserted after the targeted gene (see below); 2) the full expression pattern of a gene may include multiple cell types or brain regions, thus in some cases the partial expression pattern may be more desirable (although this issue can sometimes be addressed by using Cre dependent viral vectors which can be injected to defined brain regions).
Constitutive vs inducible GABA Cre drivers
Extensive characterization of eight constitutive drivers indicated that this strategy is highly effective. First, Cre activities appear highly specific and largely match the expression of the targeted genes. In certain lines and brain regions, recombination patterns do deviate from that of the endogenous expression in adult brain (e.g. CCK-ires-Cre, Figure 6D). These most likely result from the fact that recombination patterns reflect the cumulative expression history of a gene throughout development. Second, Cre activity in the bicistronic cassette is quite effective. In nearly all eight lines, reporter allele is activated in over 90% of the targeted cell populations in cortex and hippocampus. On the other hand, we noted that a bicistronic cassette inserted after the STOP codon could still reduce the expression (e.g. translation) of the targeted gene (H.T, Z.J. H., data not shown). This example and others are a reminder that every genetic manipulation is also a genetic lesion to the genome, a fact that must be considered when interpreting results involving genetic targeting.
Our characterization of a dozen inducible drivers confirmed, again, that CreER activities are highly specific and largely matched the expression of the targeted gene. On the other hand, the efficiency of induction varied significantly. While most lines are highly or moderately efficient, 3 lines (PV-, SST-, Lhx6- CreER) were quite inefficient. It is possible that alteration of sequences near the translation initiation codon of these genes reduced transcription levels, leading to low CreER activity. Given the success of the bicistronic strategy, it may be more efficient to insert CreER after the STOP codon of an endogenous gene. The background CreER activity without tamoxifen induction is very low and was only observed occasionally in high efficiency induction lines.
Traditionally, mRNA in situ hybridization and immunohistochemistry have been used as standards for evaluating the specificity of genetic targeting. However, both in situ and immunohistochemistry have intrinsic limitations in specificity and sensitivity, depending on the quality and strength of RNA probes and antibodies. Because Cre knockin often precisely recapitulate endogenous gene expression and Cre-activated reporters amplify expression levels, we suggest that a well-designed Cre knockin line provides an independent and sensitive assay for gene expression and complements mRNA in situ and antibody labeling.
Genetic tracking of interneuron development will facilitate the study of cortical circuit assembly
A remarkable feature of the assembly of cortical inhibitory circuitry is that GABAergic neurons are generated in the embryonic ventral telencephalon and acquire their proper areal and laminar positions through long-distance, multi-modal migration (Marin and Rubenstein, 2001). A major obstacle in studying GABAergic circuit assembly has been that the development of different cell types is prolonged, multi-faceted, and highly intertwined, and there has been no method to track specific cell types from their origin to their circuit integration. The GABA Cre drivers begin to provide genetic tools that allow the tracking of the “life history” of subpopulation of interneurons. Such genetic tracking will link sequential developmental episodes of defined cell types, such as migration, synapse formation and plasticity (which are often studied in separation), within a coherent context of circuit assembly. As cell types are the building blocks of circuit assembly and the units of circuit operation, a cell type-based analysis of GABAergic neuron development will begin to integrate studies of cortical circuit assembly and function.
GABA drivers target inhibitory neurons throughout the mouse brain
It is clear that, in addition to the cerebral cortex, the GABA Cre drivers target diverse and often highly distinct populations of GABA neurons throughout the CNS. Based on our characterization of cortex and hippocampus, it is reasonable to expect that if the mRNA of a gene is detected in a region of interest (e.g. in the Allen Brain Atlas), the corresponding Cre driver will be active in that region (with the exception of the low frequency CreER lines). In addition, Cre activity likely provides a more sensitive means to discern gene expression in regions where in situ analysis is either technically problematic or not sensitive enough to detect the endogenous gene expression. A more thorough characterization of GABA drivers in other brain regions will yield enormously valuable information regarding the organization of the underlying neural circuits and significantly accelerate anatomical, functional, and developmental studies of these circuits.
Conclusion and Perspective
Although the current set of GABA drivers successfully target subpopulations of cortical GABAergic neurons, they have yet to specifically capture individual anatomically and physiologically defined subtypes such as Martinotti and neurogliaform cells. We envision future progress in several areas, following the footsteps of more advanced genetic systems such as Drosophila. First, expression profiling in GABAergic populations targeted by current Driver lines will reveal genes expressed in more restricted populations, providing opportunities for more specific targeting. Second, intersectional strategies using Cre, Flp, or other recombinase drivers will achieve greater specificity not only with respect to cell types but also to brain regions and temporal profiles (Dymecki et al., 2007). Third, lineal and birth timing strategies will be more widely used to target specific cell types (Jensen et al., 2008). Fourth, as more regulatory elements in the mouse genome are annotated by genomic analysis, targeted insertion of enhancers at defined genomic loci might achieve targeting of subtypes with exquisite precision (Pfeiffer et al., 2008).
Current application of genetic analysis to neural circuits has focused on providing experimental access for anatomical and physiological studies (Luo et al., 2008). Beyond experimental access, genetic analysis in past decades has further contributed to discovering the mechanisms and logic underlying biological processes, such as the genetic control of embryonic patterning (Nusslein-Volhard and Wieschaus, 1980). Although the link between genes and cortical circuit function is indirect, it is increasingly evident that gene regulatory programs orchestrate many aspects of circuit assembly, from cell fate specification to synaptic connectivity. Given that the cell type is the basic unit of neural circuit function as well of gene regulation, a cell type-targeted genetic analysis will likely contribute to revealing the logic of cortical circuit assembly and organization. Combined with genetic etiological models in mice, such cell type-based approaches may further contribute to understanding the genetic architecture and pathogenic mechanisms of neurodevelopmental and psychiatric disorders.
Materials and Methods
Knockin vector construction
General scheme
Gene targeting vectors were generated using BAC recombineering (Lee et al., 2001) and, in a few cases, PCR-based cloning approach (Sppl Figure 1). For constitutive Cre lines, either an ires-Cre cassette was inserted immediately after the STOP codon or a 2A-Cre cassette was inserted in frame just before the STOP codon of the targeted gene. For inducible lines, CreER was inserted at the translation initiation site of the targeted gene. If the ATG codon of the targeted gene is in the first coding exon, a CreER-intron-polyA cassette was used; if the ATG codon is not in the first coding exon, a CreER-polyA cassette was used. 2 to 5kb upstream or downstream regions of the targeted loci were cloned into targeting vector as 5′ and 3′ homologous arms, respectively (Table 1). All targeting constructs include an frt-Neo-frt cassette and a tyrosine kinase cassette or Diphtheria toxin cassette for positive and negative selection in ES cells, respectively. Detailed information on targeting constructs for each line is available at http://www.credriver.org.
BAC clones
For each gene of interest, 2 partially overlapping BAC clones from the RPCI-23&24 library (made from C57BL/b mice) were chosen from the Mouse Genome Brower. BAC DNA was transferred from DH10B strain to SW105 strain by electroporation. The identity and integrity of these BAC clones were verified by a panel of PCR primers and restriction digestions.
Building vectors and BAC targeting vectors
We constructed a series of “building vectors” containing the essential elements for different strategies of BAC targeting (sppl Table 1; sppl Figure 1A). These elements were inserted into P451B (gift of Dr. Pentao Liu), a modified version of PL451 without a loxP site (Liu et al., 2003) in front of the frt-Neo-frt cassette. The Neo gene is driven by both the PGK promoter for G418 selection in ES cells and the EM7 promoter for Kan selection in E.coli. A BAC targeting vector was generated for each gene by cloning appropriate 5′ and 3′ homology arms fromthe gene into a building vector, flanking the CreERT2frt-Neo-frt cassette. For targeting to the ATG initiation codon, we typically use 300-500 bp DNA fragments immediate upstream and shortly downstream for 5′ and 3′ homology arms, respectively.
BAC recombineering and generation of knockin constructs
We used the PL253 retrieval vector (Liu et al., 2003) as the backbone of our knockin vectors (sppl Figure 1B). PL253 contains the HSV-TK gene driven by the MC1 promoter for negative selection in ES cells. This cassette is flanked by multi-cloning sites. Knockin cassette was retrieved from the modified BAC clones into PL253 by recombineering. For recombineering (spplFigure 1C), BAC targeting cassettes was excised by restriction digestion, purified to ~100ng/μl, and electroporated into competent SW105 cells containing the BAC clone of interest (Liu et al., 2003). Targeted BAC clones was selected for KanR, and confirmed by a panel of PCR primers and restriction digestions. A correctly targeted BAC clone was used for generating the knockin construct by the BAC retrieval method (Liu et al, 2003). The 5′r and 3′r in the retrieval vector was designed such that between 2-5 kb DNA segment flanking the CreERT2-frt-PGK-EM7-Neo-frt cassette in the BAC clone will be subcloned into PL253. The total length of homology (2-5kb on either side) was sufficient for gene targeting in ES cells. The shorter homology arm was used to design PCR-based screens for targeted ES cells.
Generation of GABA driver lines
Targeting vectors were linearized by NotI or SalI and transfected into either a C57/black6 ES cell line (Bruce4, generously provided by Dr. Collin Stewart) or a 129SVj/B6 F1 hybrid ES cell line (V6.5, Open Biosystems; see Table 1). G418-resistant ES clones were first screened by PCR and then confirmed by Southern blotting using appropriate probes. PCR primers and conditions were first tested on targeted BAC clone, which was used as positive control for ES cell screening. Southern probes were generated by PCR, subcloned, and tested on wildtype genomic DNA and modified BAC DNA to verify that they give clear and expected results. Gene targeting rate varied from ~0.5% to over 60%, depending on the targeted loci (Table 1). For Bruce4 ES cells, positive ES clones were injected into blastocysts from the albino C57BL/6J-Tyrc2j mice to obtain chimeric mice following standard procedures. Chimeric mice were bred with C57BL/6J-Tyrc2j mice to identify germline transmission. For V6.5 ES cells, positive ES cell clones were used for tetraploid complementation to obtain male heterozygous mice following standard procedures. The frt-Neo-frt cassette in the founder line was removed by breeding with Actin-FLPe transgenic mice (gift of Dr. Susan Dymecki). All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of CSHL in accordance with NIH guidelines.
Characterization of Cre driver lines
Cre drivers were bred with the RCE (Myoshi et al., 2010) or Ai9 (Madisen et al., 2010) reporter lines to assay recombination patterns. The offsprings usually contain a mixed C57BL/6 and 129 genetic background carried from the various Cre and reporter lines. For intersectional labeling, mice with triple alleles (CCK-ires-Cre, Dlx5/6-Flp and RCE-dual) were obtained by crossing CCK-ires-Cre::Dlx5/6-Flp with RCE-dual. The RCE-dual allele expresses GFP upon the removal of double STOP cassettes, frt-STOP-frt and loxP-STOP-loxP. 50-μm thick vibratome sections from perfused brains were immunostained and imaged with confocal microscopy or with fluorescent microscopy.
Tamoxifen induction
Tamoxifen was prepared by dissolving in corn oil (20 mg/ml) at 37°C with constant agitation. Appropriate amount of tamoxifen was administered to mice by gavaging or intraperitoneal injection at a dose of 2-5mg/d for 1-5 days, depending on mice age, body weight, and the experiment (e.g. low or high frequency induction). Animals were monitored for adverse effects, and if these became apparent, treatment was stopped. 7-30days after the last induction, mice were processed for analysis.
Immunohistochemistry and confocal microscopy
See Supplemental Methods.
Electrophysiology in cortical slice
See Supplemental Methods.
In vivo 2-photon imaging
See Supplemental Methods.
Supplementary Material
Highlights.
- generation of ~20 Cre lines targeting major classes of cortical interneurons
- intersection of Cre and Flp lines increases specificity of targeting
- enables systematic and comprehensive analysis of cortical interneurons
- accelerates the study of GABAergic circuits throughout the mammalian brain
Acknowledgement
We apologize for not being able to cite more authors due to space limitation. We thank Drs. Hongkui Zeng for generously providing the Ai9 reporter line, Florin Albeanu for help with in vivo 2-photon imaging. This work was supported by NIH grants U01 MH078844-01 and U01 MH078844-02 to ZJH and subaward U01 MH078844-03 to SBN. HT was supported by a NARSAD Postdoctoral Fellowship. ZJH was a McKnight Scholar and a Simons Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, Fairen A, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 2007;8:45–56. doi: 10.1038/nrn2044. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Fishell G. The developmental integration of cortical interneurons into a functional network. Curr Top Dev Biol. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Machold R, Klein C, Fishell G. Gene expression in cortical interneuron precursors is prescient of their mature function. Cereb Cortex. 2008;18:2306–2317. doi: 10.1093/cercor/bhm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger TK, Perin R, Silberberg G, Markram H. Frequency-dependent disynaptic inhibition in the pyramidal network: a ubiquitous pathway in the developing rat neocortex. J Physiol. 2009;587:5411–5425. doi: 10.1113/jphysiol.2009.176552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB, Nemeroff CB. The CRF system, stress, depression and anxiety-insights from human genetic studies. Mol Psychiatry. 2010;15:574–588. doi: 10.1038/mp.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Snyder SH. Transient nitric oxide synthase neurons in embryonic cerebral cortical plate, sensory ganglia, and olfactory epithelium. Neuron. 1994;13:301–313. doi: 10.1016/0896-6273(94)90348-4. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJ, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59:722–732. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G. Hippocampal GABAergic interneurons: a physiological perspective. Neurochem Res. 2001;26:899–905. doi: 10.1023/a:1012324231897. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Geisler C, Henze DA, Wang XJ. Interneuron Diversity series: Circuit complexity and axon wiring economy of cortical interneurons. Trends Neurosci. 2004;27:186–193. doi: 10.1016/j.tins.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Cajal R. La textura del sistema nerviosa del hombre y los vertebrados. 1st edition Moya; Madrid: 1899. [Google Scholar]
- Cauli B, Hamel E. Revisiting the role of neurons in neurovascular coupling. Front Neuroenergetics. 2010;2:9. doi: 10.3389/fnene.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E. Proc Natl Acad Sci U S A. 2000;97(11):6144–6149. doi: 10.1073/pnas.97.11.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, et al. Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- David C, Schleicher A, Zuschratter W, Staiger JF. The innervation of parvalbumin-containing interneurons by VIP-immunopositive interneurons in the primary somatosensory cortex of the adult rat. Eur J Neurosci. 2007;25:2329–2340. doi: 10.1111/j.1460-9568.2007.05496.x. [DOI] [PubMed] [Google Scholar]
- de Lecea L. Cortistatin--functions in the central nervous system. Mol Cell Endocrinol. 2008;286:88–95. doi: 10.1016/j.mce.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Dymecki SM, Kim JC. Molecular neuroanatomy’s “Three Gs”: a primer. Neuron. 2007;54:17–34. doi: 10.1016/j.neuron.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF, Katona I. Perisomatic inhibition. Neuron. 2007;56:33–42. doi: 10.1016/j.neuron.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Fuentealba P, Begum R, Capogna M, Jinno S, Marton LF, Csicsvari J, Thomson A, Somogyi P, Klausberger T. Ivy cells: a population of nitric-oxide-producing, slow-spiking GABAergic neurons and their involvement in hippocampal network activity. Neuron. 2008;57:917–929. doi: 10.1016/j.neuron.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Marin O. Generation of interneuron diversity in the mouse cerebral cortex. Eur J Neurosci. 2010;31:2136–2141. doi: 10.1111/j.1460-9568.2010.07267.x. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Goulding M. Circuits controlling vertebrate locomotion: moving in a new direction. Nat Rev Neurosci. 2009;10:507–518. doi: 10.1038/nrn2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefft S, Jonas P. Asynchronous GABA release generates long-lasting inhibition at a hippocampal interneuron-principal neuron synapse. Nat Neurosci. 2005;8:1319–1328. doi: 10.1038/nn1542. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo S, Akashi K, Sakimura K, Tamamaki N. Subtypes of GABAergic neurons project axons in the neocortex. Front Neuroanat. 2009;3:25. doi: 10.3389/neuro.05.025.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Di Cristo G, Ango F. Development of GABA innervation in the cerebral and cerebellar cortices. Nat Rev Neurosci. 2007;8:673–686. doi: 10.1038/nrn2188. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Taniguchi H, Miao H, Kuhlman S. Imaging in Developmental Biology. A Laboratory Manual Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2010. Genetic labeling of neurons in mouse brain; p. 199. [DOI] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, Dymecki SM. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Bischofberger J, Fricker D, Miles R. Interneuron Diversity series: Fast in, fast out--temporal and spatial signal processing in hippocampal interneurons. Trends Neurosci. 2004;27:30–40. doi: 10.1016/j.tins.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katona I, Sperlagh B, Sik A, Kafalvi A, Vizi ES, Mackie K, Freund TF. Presynaptically located CB1 cannabinoid receptors regulate GABA release from axon terminals of specific hippocampal interneurons. J Neurosci. 1999;19:4544–4558. doi: 10.1523/JNEUROSCI.19-11-04544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilduff TS, Cauli B, Gerashchenko D. Activation of cortical interneurons during sleep: an anatomical link to homeostatic sleep regulation? Trends Neurosci. 2011;34:10–19. doi: 10.1016/j.tins.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Magill PJ, Marton LF, Roberts JD, Cobden PM, Buzsaki G, Somogyi P. Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature. 2003;421:844–848. doi: 10.1038/nature01374. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosi A, Baram TZ. The central corticotropin releasing factor system during development and adulthood. Eur J Pharmacol. 2008;583:204–214. doi: 10.1016/j.ejphar.2007.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosi A, Baram TZ. The pathways from mother’s love to baby’s future. Front Behav Neurosci. 2009;3:27. doi: 10.3389/neuro.08.027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Shigematsu N, Karube F, Sekigawa A, Kato S, Yamaguchi N, Hirai Y, Morishima M, Kawaguchi Y. Selective Coexpression of Multiple Chemical Markers Defines Discrete Populations of Neocortical GABAergic Neurons. Cereb Cortex. 2011 doi: 10.1093/cercor/bhq252. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13(3):476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 2009;19(Suppl 1):i96–106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magno L, Catanzariti V, Nitsch R, Krude H, Naumann T. Ongoing expression of Nkx2.1 in the postnatal mouse forebrain: potential for understanding NKX2.1 haploinsufficiency in humans? Brain Res. 2009;1304:164–186. doi: 10.1016/j.brainres.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Major DE, Luksch H, Karten HJ. Bottlebrush dendritic endings and large dendritic fields: motion-detecting neurons in the mammalian tectum. J Comp Neurol. 2000;423:243–260. [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Using imaging and genetics in zebrafish to study developing spinal circuits in vivo. Dev Neurobiol. 2008;68:817–834. doi: 10.1002/dneu.20617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Hempel C, Sugino K. Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol. 2006;16:571–576. doi: 10.1016/j.conb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Nyiri G, Freund TF, Somogyi P. Input-dependent synaptic targeting of alpha(2)-subunit-containing GABA(A) receptors in synapses of hippocampal pyramidal cells of the rat. Eur J Neurosci. 2001;13:428–442. doi: 10.1046/j.1460-9568.2001.01407.x. [DOI] [PubMed] [Google Scholar]
- Olah S, Fule M, Komlosi G, Varga C, Baldi R, Barzo P, Tamas G. Regulation of cortical microcircuits by unitary GABA-mediated volume transmission. Nature. 2009;461:1278–1281. doi: 10.1038/nature08503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sik A, Penttonen M, Ylinen A, Buzsaki G. Hippocampal CA1 interneurons: an in vivo intracellular labeling study. J Neurosci. 1995;15:6651–6665. doi: 10.1523/JNEUROSCI.15-10-06651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghomonian JJ, Martin DL. Two isoforms of glutamate decarboxylase: why? Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Dalezios Y, Lujan R, Roberts JD, Watanabe M, Shigemoto R. High level of mGluR7 in the presynaptic active zones of select populations of GABAergic terminals innervating interneurons in the rat hippocampus. Eur J Neurosci. 2003;17:2503–2520. doi: 10.1046/j.1460-9568.2003.02697.x. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Tamas G, Lujan R, Buhl EH. Salient features of synaptic organisation in the cerebral cortex. Brain Res Brain Res Rev. 1998;26:113–135. doi: 10.1016/s0165-0173(97)00061-1. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Tomioka R, Okamoto K, Furuta T, Fujiyama F, Iwasato T, Yanagawa Y, Obata K, Kaneko T, Tamamaki N. Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. Eur J Neurosci. 2005;21:1587–1600. doi: 10.1111/j.1460-9568.2005.03989.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Toledo-Rodriguez M, Gupta A, Wu C, Silberberg G, Luo J, Markram H. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 2004;561:65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI, Nicoll RA. Endogenous cannabinoids mediate retrograde signalling at hippocampal synapses. Nature. 2001;410:588–592. doi: 10.1038/35069076. [DOI] [PubMed] [Google Scholar]
- Xu Q, Guo L, Moore H, Waclaw RR, Campbell K, Anderson SA. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron. 2008a;65:328–340. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008b;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Xu XM, Roby KD, Callaway EM. Immunochemical Characterization of Inhibitory Mouse Cortical Neurons: Three Chemically Distinct Classes of Inhibitory Cells. Journal of Comparative Neurology. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.