Background: TLQP-21 is a bioactive peptide for which the receptor(s) are unknown.
Results: We demonstrate that C3AR1 is a receptor for TLQP-21.
Conclusion: Many of the effects of TLQP-21 can be explained by C3AR1 activation.
Significance: These results provide a bridge linking the regulation of metabolism and the activation of complement in rodents.
Keywords: Cell Migration, Complement System, G Protein-coupled Receptors (GPCRs), Gene Silencing, Metabolism, C3AR1, RNA-Seq, Receptor Identification, TLQP-21
Abstract
TLQP-21, a peptide derived from VGF (non-acronymic) by proteolytic processing, has been shown to modulate energy metabolism, differentiation, and cellular response to stress. Although extensively investigated, the receptor for this endogenous peptide has not previously been described. This study describes the use of a series of studies that show G protein-coupled receptor-mediated biological activity of TLQP-21 signaling in CHO-K1 cells. Unbiased genome-wide sequencing of the transcriptome from responsive CHO-K1 cells identified a prioritized list of possible G protein-coupled receptors bringing about this activity. Further experiments using a series of defined receptor antagonists and siRNAs led to the identification of complement C3a receptor-1 (C3AR1) as a target for TLQP-21 in rodents. We have not been able to demonstrate so far that this finding is translatable to the human receptor. Our results are in line with a large number of physiological observations in rodent models of food intake and metabolic control, where TLQP-21 shows activity. In addition, the sensitivity of TLQP-21 signaling to pertussis toxin is consistent with the known signaling pathway of C3AR1. The binding of TLQP-21 to C3AR1 not only has effects on signaling but also modulates cellular functions, as TLQP-21 was shown to have a role in directing migration of mouse RAW264.7 cells.
Introduction
The vgf (non-acronymic) gene encodes a protein that undergoes multiple processing events resulting in a number of bioactive peptides (1). vgf knock-out mice are hypermetabolic, suggesting that VGF plays a significant role in the control of energy metabolism (2). Among the many peptides studied after this initial observation, TLQP-21 attracted particular attention (3–5). Indeed, intracerebroventricular injection of this peptide induces an increase in resting energy expenditure (3) and prevents high-fat diet-induced weight gain (6). This peptide increases amylase release by rat isolated pancreatic lobule and acinar cells (7). TLQP-21 was also reported to play a role in lipolysis (8). Injection of TLQP-21 decreases gastric acid secretion (9) and gastric emptying (10) in rats. This effect was further explored by Brancia et al. (11), who demonstrated that TLQP-21 is expressed in the ECL (enterochromaffin-like) and somatostatin cells of the stomach, strongly suggesting the existence of a physiological control loop. In addition, TLQP-21 has been shown to promote glucose-stimulated insulin secretion and to protect primary rat pancreatic islet cells from thapsigargin-induced apoptosis (12). TLQP-21 has also been shown to modulate mammotrophic cell differentiation in the GH3 cell line (13). Beyond its contribution to metabolism, TLQP-21 was reported to play a role in stress responses in vivo (14) and in the male reproductive system by stimulating the hypothalamic-pituitary-gonadal axis (15). Therefore, identifying a receptor for TLQP-21 would facilitate the understanding of the regulation of metabolism and might point to novel entry points for pharmacological intervention. Increasing evidence points toward such a membrane receptor for TLQP-21 (8, 13, 16).
RNA-Seq is a recent technique that can be used to analyze changes in gene expression across the entire transcriptome (17, 18). This technology is now being applied to a rapidly increasing number of organisms (19) and presents distinct advantages over microarrays, including greater sensitivity and a much higher dynamic range. Beyond the ability of RNA-Seq to monitor gene expression, it can identify novel transcripts, novel isoforms, alternative splice sites, allele-specific expression, and rare transcripts (18). As RNA-Seq does not require a reference genome to gain useful transcriptomic information, it can be particularly useful in non-model species that have not had their genomes sequenced yet.
In this work, we describes the measurement of a G protein-coupled receptor (GPCR)3-mediated activity for TLQP-21 in two different rodent cell lines. A set of different techniques (including unbiased transcriptome sequencing of the genes expressed in these cell lines, followed by antagonist and siRNA screening to identify the putative receptor, all subsequently supported by recombinant expression of the receptor showing signaling) has been used to demonstrate that the TLQP-21 activity in these two cell lines is mediated by complement C3a receptor-1 (C3AR1). Originally, C3AR1 was thought to be restricted to the innate immune response, having a role in the complement cascade, but its participation has been extended to roles in cancer (20), neurogenesis (21), and hormone release from the pituitary gland (22). Consistent with its role in metabolism, c3ar1 knock-out mice are transiently resistant to diet-induced obesity and are protected against high-fat diet-induced insulin resistance (23). The observations we made describe a novel ligand/receptor association and provide insight into the interconnection between inflammation and metabolism.
EXPERIMENTAL PROCEDURES
Human C3a was purchased from Calbiochem. Rat TLQP-21 was purchased from Tocris, Phoenix, and Bachem, and human TLQP-21 was from Bachem. 125I-Labeled human complement C3a was from PerkinElmer Life Sciences. All cell culture media and reagents were purchased from Invitrogen unless stated otherwise.
Fluo-4 Calcium Assay
CHO-K1 cells were seeded either onto poly-d-lysine-coated 384-well plates (CELLCOAT, Greiner bio-One) for other cells, non-coated 384-well plates (Costar) were used. 24 h before the experiment, cells were seeded at a concentration of 10,000 cells/well for a total of 50 μl. On the day of the experiment, the medium was manually removed and replaced with 40 μl of 1.6 μm Fluo-4/AM (Molecular Probes) in dilution buffer consisting of 20 mm HEPES, Hanks' balanced salt solution, and 0.1% BSA (Calbiochem) and supplemented with 2.5 mm probenecid (Sigma) for 1 h at 37 °C and 5% CO2. Cells were subsequently washed with 20 mm HEPES, Hanks' balanced salt solution, and 2.5 mm probenecid with a BioTek cell washer, leaving 30 μl of buffer covering the cells. Calcium-induced fluorescence was detected using a Hamamatsu FDSS7000 system. Base-line fluorescence was measured, followed by pipetting 15 μl of 3× concentrated compound-containing solution onto the cells. Calcium-induced fluorescence was measured 60 times every 1 s and then 40 times every 2 s. The data were calculated as ΔF/F (maximum of response − base line/base-line signal). For each assay, values were obtained in quadruplicates, and data are expressed as means ± S.D. For the priming protocol, we activated the cells with 100 μm ATP, monitored the signal as indicated above, and let the cells rest for 30 min. The cells were then activated by the ligand as described above. We used a positive control mixture (each at a final concentration of 1 μm) containing ATP (Tocris), lysophosphatidic acid 18:1 (Cayman Chemical), sphingosine 1-phosphate (Tocris), carbachol (Sigma), and bradykinin (Tocris).
RNA-Seq of Cell Lines
Total RNA was isolated from CHO-K1 and CCL39 cells using an RNeasy micro kit (Qiagen). RNA-Seq libraries were prepared using an Illumina RNA prep kit and sequenced using the Illumina HiSeq 2000 platform. A total of 750 million 76-bp end reads were mapped to the Chinese hamster genome (24) using TopHat Aligner (version 1.3.3) (25), a RNA-Seq mapping software specifically designed for detecting splice junctions between exons and based on the fast NGS mapper Bowtie (26). TopHat parameters were set to default. Final transcript levels of all known hamster genes were calculated in fragments/kb of exon/million fragments mapped by counting the number of mapped and spliced reads to exons, normalized by the length of the exons, and averaged over all used exons for each transcript. On average, 87% of the total reads (equivalent to 654 million sequences) were mapped to the genome. The Cuffdiff tool from the Cufflinks package (version 1.1.0) (27) was then used to detect the transcripts whose expression statistically changed between the CHO-K1 and CCL39 cell types. For the correct interpretation of the GPCR expression profiling, this class of genes was re-annotated with a reciprocal best hit approach using the BLASTn algorithm (BLAST e-value cut-off of 10−10) (28) on a total of 433 human non-olfactory GPCR genes collected from Ensembl (version 64, based on Homo sapiens GRCh37). This bioinformatics approach led us to the identification of putative hamster GPCR genes.
Finding the Complete vgf Transcript by RNA-Seq
To characterize the sequence of the hamster vgf gene, transcriptomic data were reanalyzed with an unbiased de novo assembly approach using Trinity software (version r2012-05-18) (29) on a server with 1 TB of RAM. As described previously by Jiménez-Guri et al. (19) and Grabherr et al. (29), Trinity proved to be a reliable approach to reconstruct transcriptomes in organisms with an incomplete genome sequence, in this case, hamster.
PCR
Genomic DNA (isolated from CHO-K1 cells using an DNeasy mini kit (Qiagen)) and cDNA (synthesized from purified total RNA using a high-capacity cDNA synthesis kit (Invitrogen)) were used as templates for PCR amplification of hamster TLQP-21 with sense primer 5′-ACA CTG CAG CCA CCC GCA TCC TCG and reverse primer 5′-GCG CGC AGG TGG CAA CGC GTG. After 35 cycles of amplification, the PCR products were purified and subcloned for sequencing using a Zero Blunt® TOPO® PCR cloning kit (Invitrogen).
siRNA Experiment
Three independent siRNA duplexes for each candidate gene were designed (2 × 21-mer with a 5′–3′-dTdT overhang) and purchased from Microsynth AG (Balgach, Switzerland). Transfection was performed in a 96-well format using Lipofectamine 2000 (Invitrogen). 0.5 μl of Lipofectamine reagent was diluted in 50 μl of Opti-MEM I and added to 50 μl of Opti-MEM I containing 40 pmol of siRNA. After 15 min, 200 μl of a CHO-K1 cell suspension diluted to 2 × 105 cells/ml was added to each well. The transfection mixture was then transferred in four wells for a quadruplicate measurement in a 384-well plate for calcium measurement. Cells were grown for 24 h before the response of the cells to TLQP-21 was monitored using the calcium flux assay.
Screening of GPCR Antagonists
A panel of 110 compounds known to inhibit 90 GPCRs was tested for activity in CHO-K1 cells upon TLQP-21 stimulation. Cells were incubated with 30 μm antagonists for 15 min and then stimulated with 1 μm (EC80) TLQP-21. Compounds causing >70% inhibition of the TLQP-21 response were retested over a range of concentrations to determine their IC50 values.
Cloning
Hamster C3AR1 cDNA was amplified from CHO-K1 cDNA by PCR (forward primer 5′-CACCATGGAGTCTTTCTCTGCTGAC and reverse primer 5′-TACATCTGTACTCAAATTGT) and subcloned into the pcDNA3.1D-TOPO-V5-His vector (Invitrogen). The expression vector was sequenced to confirm integrity of the coding sequence. The same procedure was used for rat C3AR1 (cDNA from O-342 cells used as template with forward primer 5′-CACCATGGAGTCTTTCACTGCTGACACC and reverse primer 5′-CACATCCGTACTCATATGG), for human C3AR1 (forward primer 5′-CACCATGGCGTCTTTCTCTGCT and reverse primer 5′-CACAGTTGTACTATTTCTTTC), and for mouse C3AR1 (cDNA from RAW264.7 cells used as template with forward primer 5′-CACCATGGAGTCTTTCGATGCTGACACC and reverse primer 5′-CACATCTGTACTCATATTG).
Stable Cell Lines
C3AR1 expression vectors were transfected in HEK293 cells (ATCC CRL-1573). 2.5 μg of plasmid was transfected in one well of a subconfluent 6-well plate using Lipofectamine 2000. 24 h after transfection, cells were transferred to a 10-cm dish and grown in selective medium containing 0.4 mg/ml G418 (Invitrogen) until emergence of resistant colonies. A limiting dilution was carried out on this cell population to obtain single clones expressing C3AR1.
Membrane Preparation
CHO-K1 or hamster C3AR1-expressing HEK293 cells were grown to confluence, harvested in PBS containing 5 mm EDTA, and centrifuged at 1000 × g for 5 min at 4 °C before freezing (−80 °C). Frozen pellets were homogenized with a 30-ml Sartorius glass homogenizer vessel in ice-cold Tris buffer (20 mm Tris-HCl (pH 7.7) and 5 mm EDTA) containing Complete protease inhibitor mixture (Roche Applied Science) before centrifugation at 30,000 × g for 30 min at 4 °C. The resulting pellet was washed, and the process was repeated once more before the pellet was resuspended in ice-cold 20 mm HEPES (pH 7.4). The protein content was evaluated (Pierce BCA protein assay kit), and membrane aliquots were kept frozen at −80 °C until used.
Binding Assay
CHO-K1 or hamster C3AR1-expressing HEK293 cell membranes (10–50 μg/ml) were incubated for 60 min at room temperature in 50 mm Tris (pH 7.4), 5 mm MgCl2, 1 mm CaCl2, and 0.5% BSA in a final volume of 200 μl with 125I-labeled C3a and the test compound. Nonspecific binding was defined in the presence of a minimum of 30 nm human C3a. The reaction was stopped by rapid filtration through a UniFilter GF/B presoaked with polyethyleneimine (0.5%), followed by three successive washes with ice-cold buffer. Filters were then counted in a microplate scintillation counter (TopCount NXT, PerkinElmer Life Sciences) with 65% efficiency. Data were analyzed by nonlinear regression using GraphPad Prism 6 software. For displacement studies, rat TLQP-21, human TLQP-21, and human C3a were diluted in water at final concentrations of 10−5–10−12 m and tested against 125I-labeled C3a at 8 pm. Inhibition constants were calculated according to the Cheng-Prusoff equation (using GraphPad Prism 6 software): Ki = IC50/(1 + (L/KD)), where IC50 is the inhibitory concentration 50, L is the ligand concentration, and KD is the dissociation constant of the radioligand.
Migration Assay
Rat TLQP-21 and SB290157 (Calbiochem) dilutions were prepared at the indicated concentrations in migration medium (serum-free RAW264.7 growth medium: RPMI 1640 medium, 1% minimal Eagle's medium nonessential amino acid solution, 1% sodium pyruvate, 0.1% 2-mercaptoethanol, 1% penicillin/streptomycin (Bioconcept), and 10% FCS (Bioconcept)) and distributed in quadruplicates at 160 μl/well to the lower chamber of a CIM-Plate 16 device (ACEA Bioscience). After locking the upper and lower chambers together, 30 μl of migration medium was added to each well. The CIM-Plate 16 device was then loaded onto an xCELLigence RTCA DP analyzer (ACEA Bioscience) and equilibrated for 1 h at 37 °C. In the meantime, ∼70% confluent RAW264.7 cells were washed with PBS and detached with enzyme-free cell dissociation buffer; and after centrifugation, the cells were resuspended in migration medium. Cells were counted and adjusted to 0.5 × 106 cells/ml. Once the equilibration with medium was complete, the background measurement was performed, and 100 μl of cell suspension was added to each well of the upper chamber. To allow the cells to sediment and settle, the CIM-Plate 16 device was left at room temperature for 30 min. After loading the plate again onto the xCELLigence RTCA DP analyzer at 37 °C, the electrical impedance was measured in 5-min intervals for 8 h. The slope of the normalized cell index was then monitored as a measure of cell migration.
RESULTS
Calcium Flux in CHO-K1 and O-342 Cells Reveals a Pertussis Toxin-sensitive TLQP-21 Receptor
Direct activation of CHO-K1 cells by rat or human TLQP-21 did not lead to measurable Fluo-4-based calcium detection (data not shown). This suggests that TLQP-21 does not activate a Gq-coupled receptor in these cells. However, it is possible to redirect non-Gq coupling to a calcium-releasing pathway by presensitization of the cells with a strong stimulus, a process called priming (30). Priming CHO-K1 cells with 100 μm ATP revealed a TLQP-21-driven signal when using Fluo-4 (Fig. 1A). This signal was more potent with rat TLQP-21 than with the human peptide (91 and 276 nm, respectively). The amplitude of this response is significant, as it compares with the endogenous response of these cells to 100 μm ATP (ΔF/F = 2.8 in the same experiment). Such a large signal suggests that the receptor is either highly expressed or efficiently coupled to intracellular signaling pathways. The TLQP-21-mediated increase in intracellular calcium flux could be blocked by pertussis toxin (PTX), indicating that the receptor could be either a Gi- or a Go-coupled receptor (Fig. 1A). To gain a better understanding of the nature of the activated signaling pathways, we screened for additional TLQP-21-responsive cell lines and further interrogated classical GPCR pathways. For example, another hamster cell line (CCL39) was unresponsive to rat TLQP-21, as shown in Fig. 1B. However, we identified another rodent ovary cell line (O-342) as being responsive to TLQP-21 (Fig. 1C). In this rat cell line, TLQP-21 has a slightly higher EC50 of 347 nm.
FIGURE 1.
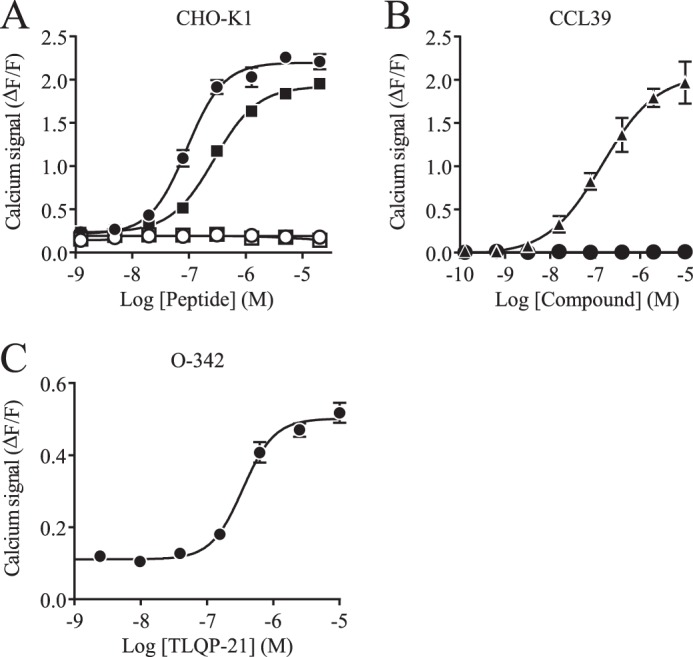
TLQP-21-induced calcium fluxes measured in several cell lines. A, calcium measurements of rat (● and ○) and human (■ and □) TLQP-21 without (● and ■) or with (○ and □) PTX in CHO-K1 cells after ATP priming. B, calcium measurements of positive control (▴; ATP, bradykinin, carbachol, sphingosine 1-phosphate, and lysophosphatidic acid 18:0) and rat (●) TLQP-21-triggered responses after ATP priming in CCL39 cells. C, calcium measurements of rat TLQP-21-triggered responses after ATP priming in O-342 cells. All data are represented as signal − base line/base-line signal (ΔF/F) ± S.D.
The TLQP-21 Receptor Is neither Gs- nor Gi-coupled
TLQP-21-directed signaling mediated by cAMP or inositol phosphate accumulation could not be detected in CHO-K1 cells (data not shown). This indicates that the TLQP-21 receptor endogenously expressed in CHO-K1 cells is not Gs-, Gi-, or Gq-coupled.
RNA-Seq of CHO-K1 Cells
To gain deeper insight into the molecular mechanisms underlying the TLQP-21-induced activity, high-throughput RNA-Seq was performed on CHO-K1 (TLQP-21-responsive) and CCL39 (TLQP-21-nonresponsive) cells. The recently published hamster genome (24) was used to interpret the results produced by RNA-Seq. Expression values (fragments/kb of exon/million fragments mapped) generated by sequencing were further validated by quantitative RT-PCR (data not shown). These values indicate that these two cell lines express different GPCRs. We speculated that the candidate GPCR target for TLQP-21 would show higher expression in CHO-K1 cells compared with CCL39 cells. Table 1 displays the 21 candidates prioritized by this reasoning. The sequence of the hamster vgf gene reported in databases (XM_003500965) is truncated. In particular, the part of the sequence encoding the TLQP-21 peptide is missing. This analysis of the hamster transcriptome enabled the identification of the full transcript sequence (KF309065). To validate this transcript, we chose to amplify part of it (containing TLQP-21). As expected, it was possible to amplify a segment of DNA of the anticipated size (63 bp) from both genomic DNA and cDNA derived from CHO-K1 cells. Subsequent sequencing of these PCR-amplified fragments confirmed the transcript sequence from our RNA-Seq results. Protein sequence alignment (Fig. 2) showed that the hamster, mouse, and rat TLQP-21 peptide sequences are identical to each other but differ from the human sequence.
TABLE 1.
List of the 21 GPCRs overexpressed in CHO-K1 cells compared with CCL39 cells
Expression values are represented in reads/kb/million ± S.D.
| Symbol | RefSeq accession no. | CHO-K1 cells | CCL39 cells |
|---|---|---|---|
| ADORA2A | XM_003508085 | 1.62 ± 0.09 | 0.25 ± 0.08 |
| AGTR2 | XM_003510186 | 0.72 ± 0.12 | 0 ± 0 |
| BAI2 | XM_003498391 | 0.21 ± 0.03 | 0.04 ± 0.02 |
| C3AR1 | XM_003510173 | 16.7 ± 0.49 | 0.08 ± 0.06 |
| CALCR | XM_003496968 | 2.53 ± 0.05 | 0.15 ± 0.15 |
| CCR7 | XM_003510487 | 3.52 ± 0.3 | 0.1 ± 0.02 |
| CD97 | XM_003501912 | 6.45 ± 0.41 | 0.73 ± 0.13 |
| GPR132 | XM_003498768 | 0.33 ± 0.05 | 0.06 ± 0.04 |
| GPR133 | XM_003513353 | 5.48 ± 0.27 | 0.16 ± 0.04 |
| GPR182 | XM_003507483 | 0.7 ± 0.04 | 0.03 ± 0.02 |
| GPR3 | XM_003511439 | 1.18 ± 0.22 | 0.07 ± 0.04 |
| GPR37L1 | XM_003498854 | 0.14 ± 0.07 | 0.02 ± 0.03 |
| GPR56 | XM_003509803 | 23.41 ± 1.1 | 2.46 ± 0.6 |
| GRPR | XM_003507926 | 0.12 ± 0.03 | 0 ± 0 |
| HCRTR1 | XM_003498389 | 0.38 ± 0.02 | 0.04 ± 0.02 |
| OLFML2B | XM_003506879 | 8.86 ± 0.7 | 1.57 ± 0.09 |
| OPN1SW | XM_003497518 | 0.2 ± 0.05 | 0 ± 0 |
| OPRL1 | XM_003512353 | 0.11 ± 0.05 | 0.01 ± 0.02 |
| PTGER4 | XM_003503296 | 1.02 ± 0.02 | 0.06 ± 0.03 |
| TAS2R16 | XM_003504354 | 0.12 ± 0.05 | 0.01 ± 0.02 |
| TMEM185A | XM_003499435 | 13.11 ± 0.73 | 0.74 ± 0.19 |
FIGURE 2.
Multiple alignment of VGF proteins from selected species. Shown is the hamster VGF complete coding sequence aligned with human (NP_003369), rat (NP_112259), and mouse (NP_001034474) VGF proteins. The TLQP-21 sequence is indicated by a gray bar.
Antagonist Screen in CHO-K1 Cells
To test the hypothesis that the TLQP-21 receptor could be a known GPCR, we sought to inhibit the TLQP-21 response in CHO-K1 cells with a panel of well characterized antagonists (data not shown). The most effective antagonist was SB290157, a compound described as a C3AR1 antagonist (31). Subsequent concentration-response experiments with the selected candidates confirmed that SB290157 was the most potent antagonist, with an IC50 of 200 nm (Fig. 3A).
FIGURE 3.
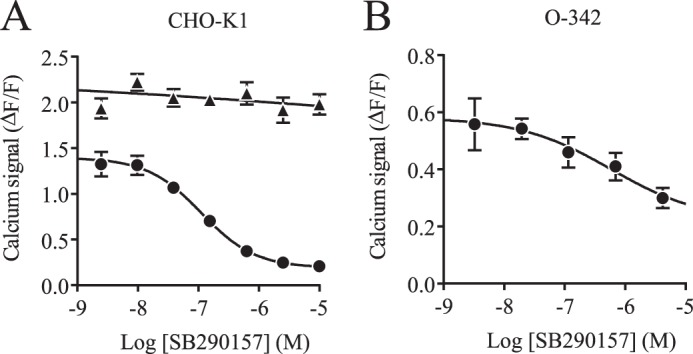
Inhibition of TLQP-21 activity by SB290157. A, calcium measurement after ATP priming of CHO-K1 cells. Shown is the concentration inhibition curve of SB290157 for 1 μm rat TLQP-21 (●) and 1 μm positive control (▴; ATP, bradykinin, carbachol, sphingosine 1-phosphate, and lysophosphatidic acid 18:0). B, concentration inhibition curve of SB290157 for 1 μm rat TLQP-21 generated by Fluo-4-based calcium measurement after ATP priming of O-342 cells. All data are represented as signal − base line/base-line signal (ΔF/F) ± S.D.
TLQP-21 Response in O-342 Cells Is Inhibited by a C3AR1 Antagonist
Because O-342 cells also respond to TLQP-21 after priming with ATP, we speculated that these cells would express the homologous receptor present in CHO-K1 cells. Testing SB290157 in this cell line showed that it also acts as an antagonist of TLQP-21-induced activity (Fig. 3B). Performing quantitative RT-PCR under standard conditions on three biological replicates showed that O-342 cells express C3AR1 with a median Ct of 27.5 (which is significant, being slightly above 10% of the housekeeping transcript hprt1) (data not shown).
siRNA Screen of CHO-K1 Cells
To further test the gene targets suggested by the transcriptomic analysis, we attempted to attenuate the expression of the top candidates using siRNAs. Table 2 lists the siRNAs used. Of the 63 siRNAs tested against the 21 candidate genes, the only gene knockdown that consistently reduced the TLQP-21 mediated response was achieved with siRNAs targeting C3AR1 (Fig. 4).
TABLE 2.
siRNAs used in C3AR1 knockdown (5′ → 3′ direction)
| Name | Sense | Antisense |
|---|---|---|
| C3AR1_1 | GTGTACCAGTATTTGTATAdTdT | TATACAAATACTGGTACACdTdT |
| C3AR1_2 | CTACCAGAAAGCAATTCTAdTdT | TAGAATTGCTTTCTGGTAGdTdT |
| C3AR1_3 | CCGTGACCTGATCACTATAdTdT | TATAGTGATCAGGTCACGGdTdT |
FIGURE 4.
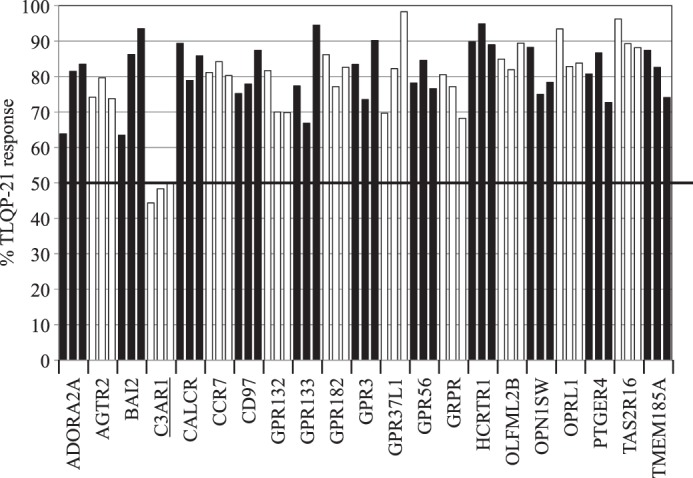
Down-modulation of the TLQP-21 response by selected siRNAs. Shown is the calcium response to rat TLQP-21 in CHO-K1 cells upon siRNA knockdown of the 21 selected GPCR candidates (three siRNAs each). Data are presented as a percentage of the rat TLQP-21 response in untransfected CHO-K1 cells. A number is allocated for each GPCR, which corresponds to the values in Table 1.
Heterologous Expression in HEK293 Cells of Hamster and Rat C3AR1
C3AR1 expression in CHO-K1 and O-342 cells, the potent inhibition of the TLQP-21-directed signal by SB290157, and the signal attenuation by C3AR1 siRNAs all lead to the hypothesis that C3AR1 could be the receptor for TLQP-21. To further test this hypothesis, the hamster and rat C3AR1 genes were cloned and expressed in HEK293 cells. Fig. 5 (B–E) shows that recombinantly expressed C3AR1 was able to elicit a C3a signal in HEK293 cells after priming the cells with ATP. This experiment also demonstrates that TLQP-21 is a ligand for both hamster and rat C3AR1. In contrast, untransfected HEK293 cells did not respond to either human C3a or rat TLQP-21 (data not shown). Untransfected CHO-K1 cells responded to human C3a, as expected (Fig. 5A). As for the TLQP-21 response, the calcium signal could be inhibited by PTX.
FIGURE 5.
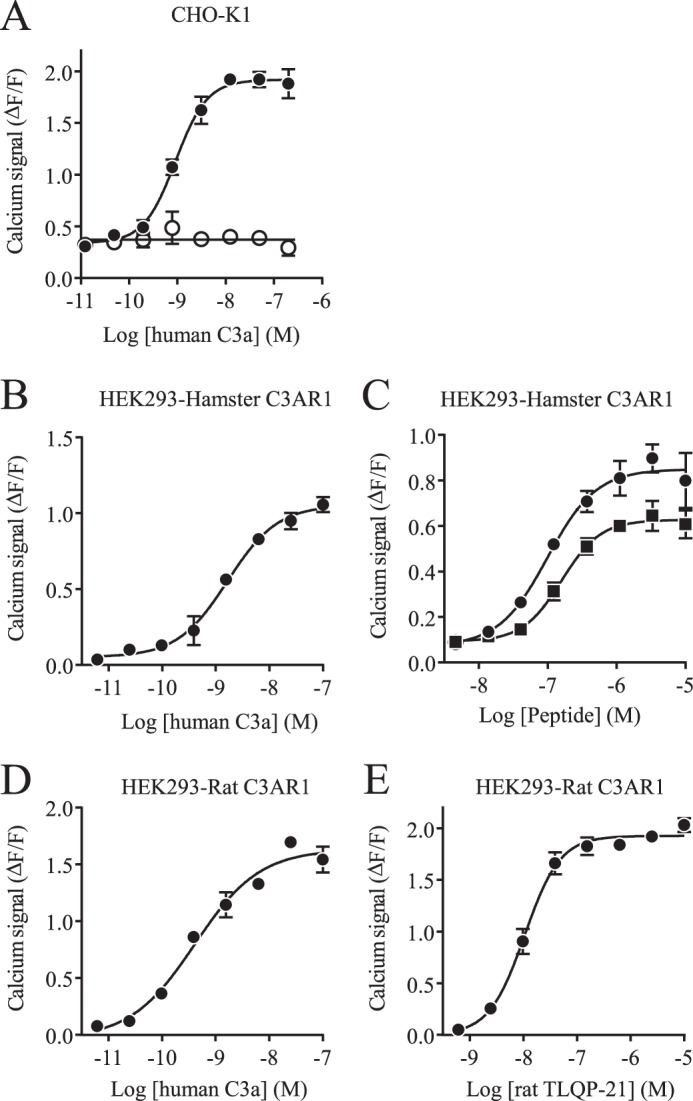
TLQP-21 activity on C3AR1. A, calcium measurement of the human C3a response in CHO-K1 cells after ATP priming in the presence (○) or absence (●) of PTX. B, calcium measurement of the response to human C3a in HEK293 cells expressing hamster C3AR1 after ATP priming. C, calcium measurement in response to rat TLQP-21 (●) and human TLQP-21 (■) in HEK293 cells expressing hamster C3AR1 after ATP priming. D, calcium measurement in response to human C3a in HEK293 cells expressing rat C3AR1 after ATP priming. E, calcium measurement in response to rat TLQP-21 in HEK293 cells expressing rat C3AR1 after ATP priming. All data are represented as signal − base line/base-line signal (ΔF/F) ± S.D.
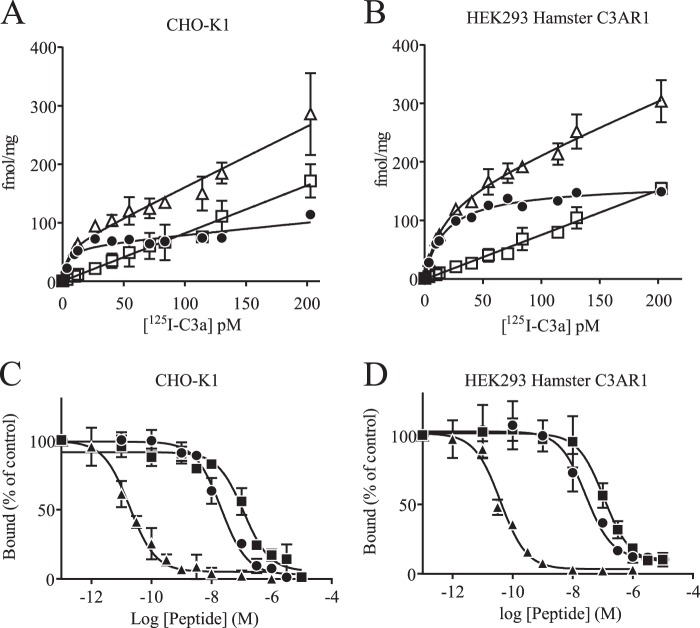
TLQP-21 Binds to Hamster C3AR1
The ability of human 125I-labeled C3a to selectively bind the hamster C3a receptor was evaluated. Dose-dependent, specific, and saturable binding of the radiolabeled material was observed in CHO-K1 membrane preparations (Fig. 6A). Such specific binding was also observed in HEK293 cells expressing recombinant hamster C3AR1 (Fig. 6B). In contrast, no specific binding was observed in untransfected HEK293 cells. Specific binding represented 70–90% of the total binding for a protein concentration of 50 μg/ml, which was then routinely used (data not shown). Characterization of the receptor pharmacology was further assessed in competition studies. As shown in Fig. 6 (C and D), inhibition-binding isotherms of 125I-labeled C3a (8 pm) were monophasic. Human full-length C3a displayed picomolar affinities (Table 3) in both CHO-K1 and HEK293 cells expressing hamster C3AR1. Human and rat TLQP-21 fully displaced 125I-labeled C3a in the high-nanomolar range in CHO-K1 and hamster C3AR1-expressing HEK293 cell membranes with comparable values. Human TLQP-21 was 5-fold less potent than rat TLQP-21 in both assays (Table 3).
FIGURE 6.
TLQP-21 binding studies. A, saturation plot of human 125I-labeled C3a using CHO-K1 membranes. ▵, total; □, nonspecific; ●, specific. B, saturation plot of human 125I-labeled C3a using membranes of HEK293 cells expressing hamster C3AR1. ▵, total; □, nonspecific; ●, specific. C, inhibition-binding isotherms of 125I-labeled C3a in CHO-K1 membrane preparations. D, inhibition-binding isotherms of 125I-labeled C3a in hamster C3AR1-expressing HEK293 membrane preparations. ▴, human C3a; ●, rat TLQP-21; ■, human TLQP-21. Data are represented as means ± S.D.
TABLE 3.
Summary of inhibition concentrations necessary to displace 50% of bound 125I-labeled human C3a
Values are presented as means ± S.D. from three independent experiments.
| IC50 ± S.D. |
||
|---|---|---|
| CHO-K1 cells | Hamster C3AR1-expressing HEK293 cells | |
| nm | ||
| Human C3a | 0.048 ± 0.004 | 0.048 ± 0.010 |
| Rat TLQP-21 | 20.42 ± 3.87 | 28.66 ± 9.71 |
| Human TLQP-21 | 98.20 ± 11.98 | 113.9 ± 29.27 |
RAW264.7 Cell Migration
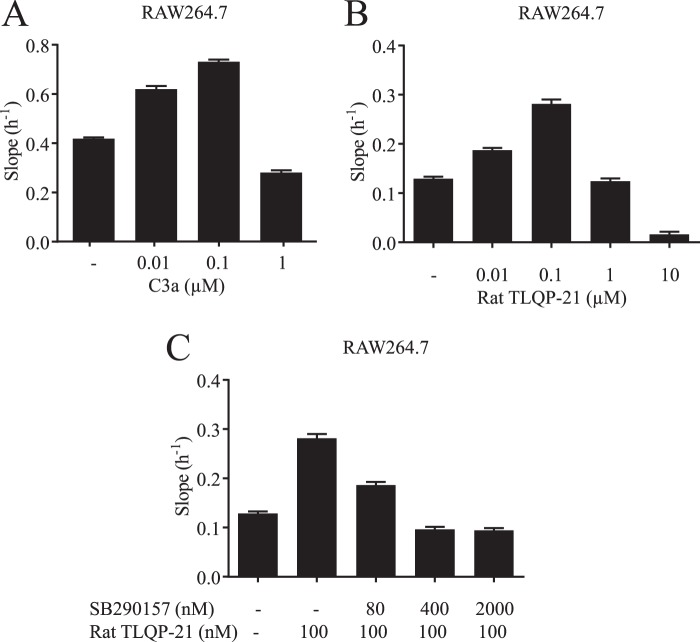
C3AR1 is expressed in immune cells, in particular cells of the monocytic lineage (32). The mouse RAW264.7 cell line is often used to probe various physiological responses, such as TNFα release or migration. First, using real-time PCR, we confirmed that the C3AR1 transcript is highly expressed in these cells (data not shown). The logical consequence of this observation is that RAW264.7 cells would be expected to migrate toward a gradient of C3a (Fig. 7A). Next, the effect of a TLQP-21 gradient on cell migration was tested. Fig. 7B shows that RAW264.7 cells migrated toward a TLQP-21 gradient in a concentration-dependent manner. As is often the case for migration, this effect is bell-shaped, probably due to receptor desensitization or internalization. To assess C3AR1 involvement in the TLQP-21 migration process, we treated the cells with increasing concentrations of SB290157. As demonstrated in Fig. 7C, the C3AR1 antagonist inhibited the TLQP-21-driven migration in a dose-dependent manner.
FIGURE 7.
Effect of TLQP-21 on RAW264.7 cell migration. A, migration of RAW264.7 cells induced by increasing concentrations of human C3a. B, migration of RAW264.7 cells induced by increasing concentrations of rat TLQP-21. C, concentration-dependent inhibition of rat TLQP-21-induced migration by the C3AR1 antagonist SB290157. y axis values correspond to the slope/h.
DISCUSSION
In this study, we have described the identification and characterization of TLQP-21-mediated signaling activity in two rodent cell lines, leading to the discovery of its cognate receptor. In agreement with a recent report (16), we observed that CHO-K1 cells responded to TLQP-21 by an increase in intracellular calcium. However, to obtain a robust signal, this required facilitated coupling using ATP priming, indicating that the TLQP-21 receptor is not a Gq-coupled GPCR. The lack of cAMP modulation, as well as the PTX sensitivity, implied that the G protein mediating these effects was likely to be Go. The second cell line identified in this work to respond to TLQP-21 was the rat cell line O-342. Interestingly, like CHO-K1 cells, O-342 cells are of ovarian origin, suggesting a potential novel role of TLQP-21 in reproduction. This appears to be consistent with a recent publication reporting TLQP-21 effects on reproduction in female rats (33).
To identify the TLQP-21 receptor, the CHO-K1 cell line was chosen because of the amplitude of the calcium response in these cells and the ease with which they can be manipulated. However, one negative aspect of this cell line is that a comprehensive transcriptome study has not previously been performed. Indeed, because hamsters are not commonly used as animal models, most traditional tools for molecular genetics are missing. RNA-Seq is a relatively recent technology that enables the generation of transcriptome-wide expression data without having to design oligonucleotide probes as required for microarray or PCR-based technologies. Another advantage of RNA-Seq, highlighted in this study, is the ability of this method to handle sequences missing in the reference dataset. This is how it was possible to establish, for the first time, the full sequence of the vgf transcript in hamster. The TLQP-21 peptide sequence is identical in hamster, rat, and mouse, whereas the human peptide displays significant differences (Fig. 2). These differences may account for the differences observed between the response of rat and human TLQP-21 in CHO-K1 cells. Utilizing the recently published CHO genome (24) in combination with RNA-Seq enabled us to explore the CHO-K1 transcriptome to identify the GPCRs expressed in this cell line. The CCL39 hamster cell line was chosen as a negative control, as TLQP-21-mediated signaling was not observed in these cells. As we hypothesized that the TLQP-21 receptor is a Go-coupled receptor, we restricted our transcriptional analysis to GPCRs. Comparing the CHO-K1 and CCL39 transcriptomes allowed us to generate a prioritized list of potential GPCRs for TLQP-21. The availability of the genome-wide transcriptome data then allowed us to design RNAi tools for each of the prioritized receptors for knockdown of receptor expression. TLQP-21 signaling was subsequently monitored. This approach was complemented by using pharmacological tool compounds to modulate receptor-mediated signaling. As GPCRs make up one of the largest families of drug targets, there are many compounds known to modulate GPCR activity (34). A set of known GPCR antagonists was assembled and used to probe the TLQP-21 response in CHO-K1 cells. The results from both receptor knockdown and pharmacological approaches are consistent with the receptor mediating this effect being C3AR1. Recombinant receptor expression in HEK293 cells further demonstrated that both the hamster and rat C3AR1 receptors conferred responsiveness to TLQP-21. Finally, direct biochemical evidence showing TLQP-21 binding to membrane preparations containing recombinantly expressed C3AR1 further supports this target/ligand pair. The binding results with recombinantly expressed receptors are consistent with the C3a displacement observed with TLQP-21 in untransfected CHO-K1 cells.
Although surprising at first sight, the fact that TLQP-21 signaling is being mediated by C3AR1 is consistent with many earlier observations. First, C3AR1 was described to mediate Go signaling (35). Second, the knock-out mice for C3AR1 display a metabolic phenotype with a transient resistance to diet-induced obesity and are protected from high-fat diet-induced insulin resistance and liver steatosis (23). This genetic observation was later confirmed by pharmacological intervention (36). The phenotype is also reminiscent of observations with vgf knock-out mice (2, 37). Third, both TLQP-21 and C3a/C3AR1 modulate pituitary gland activity. Indeed, C3a increases prolactin secretion (as well as growth hormone and ACTH) by rat anterior pituitary cells (22). This is in line with the recently reported mammotrophic effect of TLQP-21 in GH3 cells (13). The TLQP-21 effect could indeed be mediated by C3AR1, as it has been demonstrated that this receptor is highly expressed in secretory cells from the pituitary gland and the known cell lines GH3 and AtT20 (22). Although more speculative, it is intriguing to consider that vgf can be induced by various factors, including NGF (38), and that C3a has been described to have some effects on neurogenesis (39, 40).
The fourth argument in favor of a physiological relevance of this newly discovered ligand/receptor pair comes from the study of adipocyte physiology. Indeed, TLQP-21 was described to be able to increase lipolysis in primary adipocytes and to bind to primary adipocytes and 3T3-L1 membranes (8). Mamane et al. (23) have shown that C3AR1 is highly expressed in primary adipocytes and is up-regulated in mice given a high-fat diet. C3AR1 was also reported to be expressed in both differentiated and undifferentiated mouse 3T3-L1 embryonic fibroblast cell lines (36); however, C3a inhibited lipolysis instead of increasing it. At this stage, we can only speculate why TLQP-21 and C3a would induce signaling in these cells but the response would be different. It is possible that these cells express a different TLQP-21 receptor. It would be worth testing the C3AR1 antagonists on the TLQP-21 response in 3T3-L1 cells to dissect this effect further.
It is worth mentioning that, due to the exceptionally large second extracellular loop in C3AR1, the binding and hence the signaling of C3a are not well understood. Indeed, it was established that the docking site also resides in the second extracellular loop but is clearly separable from the activation site, leading to the possibility that other ligands might exist for C3AR1 (41). It is also possible that, in a different cellular context, TLQP-21 could act as a weak partial agonist, whereas C3a would fully activate C3AR1.
Our observations further support the implication of C3AR1 in metabolism, but they also suggest that TLQP-21 could have inflammatory or migration effects on myeloid cells. Here, we have demonstrated that TLQP-21 was able to induce migration of RAW264.7 cells, a mouse monocytic cell line, and that this effect could be blocked by the C3AR1 receptor antagonist SB290157. It may be that TLQP-21 has evolved to allow activation of the chemoattractant function of C3AR1 without the need for activation of the whole of the complement cascade.
It is worth noting that we could not demonstrate TLQP-21 signaling in HEK293 cells expressing human C3AR1, whereas C3a did induce a response comparable to the one observed with hamster and rat C3AR1 (data not shown). The C3AR1 protein sequence contains an exceptionally long and variable second extracellular loop. That, as well as the fact that human TLQP-21 is very different from the rodent version (100% identity between rat, mouse, and hamster), could indicate that the TLQP-21/C3AR1 physiological interaction has been changed during evolution. It is also possible that a different processing leading to a longer peptide is preferred in humans. It is important to note, however, that a previous report described a TLQP-21 effect on human pancreatic islet cells (12). A second hypothesis is of course that there is another receptor other than C3AR1 mediating TLQP-21 effects. Finally, it is also conceivable that human C3AR1 requires a particular cellular context or coreceptor missing in HEK293 cells.
In conclusion, the results presented in this study support the hypothesis that the TLQP-21 peptide is a selective natural agonist of the chemoattractant C3AR1 in rodents. These results could provide new perspectives regarding the physiological role of the TLQP-21 peptide.
Acknowledgment
We thank Steven Charlton for critical reading of the manuscript.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KF309065.
The RNA-Seq data are available in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov) with accession number SRP028606.
- GPCR
- G protein-coupled receptor
- C3AR1
- complement C3a receptor-1
- PTX
- pertussis toxin.
REFERENCES
- 1. Ferri G. L., Noli B., Brancia C., D'Amato F., Cocco C. (2011) VGF: an inducible gene product, precursor of a diverse array of neuro-endocrine peptides and tissue-specific disease biomarkers. J. Chem. Neuroanat. 42, 249–261 [DOI] [PubMed] [Google Scholar]
- 2. Hahm S., Mizuno T. M., Wu T. J., Wisor J. P., Priest C. A., Kozak C. A., Boozer C. N., Peng B., McEvoy R. C., Good P., Kelley K. A., Takahashi J. S., Pintar J. E., Roberts J. L., Mobbs C. V., Salton S. R. (1999) Targeted deletion of the Vgf gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron 23, 537–548 [DOI] [PubMed] [Google Scholar]
- 3. Bartolomucci A., La Corte G., Possenti R., Locatelli V., Rigamonti A. E., Torsello A., Bresciani E., Bulgarelli I., Rizzi R., Pavone F., D'Amato F. R., Severini C., Mignogna G., Giorgi A., Schininà M. E., Elia G., Brancia C., Ferri G. L., Conti R., Ciani B., Pascucci T., Dell'Omo G., Muller E. E., Levi A., Moles A. (2006) TLQP-21, a VGF-derived peptide, increases energy expenditure and prevents the early phase of diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 103, 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartolomucci A., Rigamonti A. E., Bulgarelli I., Torsello A., Locatelli V., Pavone F., Levi A., Possenti R., Muller E. E., Moles A. (2007) Chronic intracerebroventricular TLQP-21 delivery does not modulate the GH/IGF-1-axis and muscle strength in mice. Growth Horm. IGF Res. 17, 342–345 [DOI] [PubMed] [Google Scholar]
- 5. Jethwa P. H., Warner A., Nilaweera K. N., Brameld J. M., Keyte J. W., Carter W. G., Bolton N., Bruggraber M., Morgan P. J., Barrett P., Ebling F. J. (2007) VGF-derived peptide, TLQP-21, regulates food intake and body weight in Siberian hamsters. Endocrinology 148, 4044–4055 [DOI] [PubMed] [Google Scholar]
- 6. Bartolomucci A., Bresciani E., Bulgarelli I., Rigamonti A. E., Pascucci T., Levi A., Possenti R., Torsello A., Locatelli V., Muller E. E., Moles A. (2009) Chronic intracerebroventricular injection of TLQP-21 prevents high fat diet induced weight gain in fast weight-gaining mice. Genes Nutr. 4, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petrella C., Broccardo M., Possenti R., Severini C., Improta G. (2012) TLQP-21, a VGF-derived peptide, stimulates exocrine pancreatic secretion in the rat. Peptides 36, 133–136 [DOI] [PubMed] [Google Scholar]
- 8. Possenti R., Muccioli G., Petrocchi P., Cero C., Cabassi A., Vulchanova L., Riedl M. S., Manieri M., Frontini A., Giordano A., Cinti S., Govoni P., Graiani G., Quaini F., Ghè C., Bresciani E., Bulgarelli I., Torsello A., Locatelli V., Sanghez V., Larsen B. D., Petersen J. S., Palanza P., Parmigiani S., Moles A., Levi A., Bartolomucci A. (2012) Characterization of a novel peripheral pro-lipolytic mechanism in mice: role of VGF-derived peptide TLQP-21. Biochem. J. 441, 511–522 [DOI] [PubMed] [Google Scholar]
- 9. Sibilia V., Pagani F., Bulgarelli I., Tulipano G., Possenti R., Guidobono F. (2012) Characterization of the mechanisms involved in the gastric antisecretory effect of TLQP-21, a vgf-derived peptide, in rats. Amino. Acids 42, 1261–1268 [DOI] [PubMed] [Google Scholar]
- 10. Severini C., La Corte G., Improta G., Broccardo M., Agostini S., Petrella C., Sibilia V., Pagani F., Guidobono F., Bulgarelli I., Ferri G. L., Brancia C., Rinaldi A. M., Levi A., Possenti R. (2009) In vitro and in vivo pharmacological role of TLQP-21, a VGF-derived peptide, in the regulation of rat gastric motor functions. Br. J. Pharmacol. 157, 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brancia C., Cocco C., D'Amato F., Noli B., Sanna F., Possenti R., Argiolas A., Ferri G. L. (2010) Selective expression of TLQP-21 and other VGF peptides in gastric neuroendocrine cells and modulation by feeding. J. Endocrinol. 207, 329–341 [DOI] [PubMed] [Google Scholar]
- 12. Stephens S. B., Schisler J. C., Hohmeier H. E., An J., Sun A. Y., Pitt G. S., Newgard C. B. (2012) A VGF-derived peptide attenuates development of type 2 diabetes via enhancement of islet β-cell survival and function. Cell Metab. 16, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petrocchi Passeri P., Biondini L., Mongiardi M. P., Mordini N., Quaresima S., Frank C., Baratta M., Bartolomucci A., Levi A., Severini C., Possenti R. (2013) Neuropeptide TLQP-21, a VGF internal fragment, modulates hormonal gene expression and secretion in GH3 cell line. Neuroendocrinology 97, 212–224 [DOI] [PubMed] [Google Scholar]
- 14. Razzoli M., Bo E., Pascucci T., Pavone F., D'Amato F. R., Cero C., Sanghez V., Dadomo H., Palanza P., Parmigiani S., Ceresini G., Puglisi-Allegra S., Porta M., Panzica G. C., Moles A., Possenti R., Bartolomucci A. (2012) Implication of the VGF-derived peptide TLQP-21 in mouse acute and chronic stress responses. Behav. Brain Res. 229, 333–339 [DOI] [PubMed] [Google Scholar]
- 15. Pinilla L., Pineda R., Gaytán F., Romero M., García-Galiano D., Sánchez-Garrido M. A., Ruiz-Pino F., Tena-Sempere M., Aguilar E. (2011) Characterization of the reproductive effects of the anorexigenic VGF-derived peptide TLQP-21: in vivo and in vitro studies in male rats. Am. J. Physiol. Endocrinol. Metab 300, E837–E847 [DOI] [PubMed] [Google Scholar]
- 16. Cassina V., Torsello A., Tempestini A., Salerno D., Brogioli D., Tamiazzo L., Bresciani E., Martinez J., Fehrentz J. A., Verdié P., Omeljaniuk R. J., Possenti R., Rizzi L., Locatelli V., Mantegazza F. (2013) Biophysical characterization of a binding site for TLQP-21, a naturally occurring peptide which induces resistance to obesity. Biochim. Biophys. Acta 1828, 455–460 [DOI] [PubMed] [Google Scholar]
- 17. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 18. Wang Z., Gerstein M., Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiménez-Guri E., Huerta-Cepas J., Cozzuto L., Wotton K. R., Kang H., Himmelbauer H., Roma G., Gabaldón T., Jaeger J. (2013) Comparative transcriptomics of early dipteran development. BMC Genomics 14, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Opstal-van Winden A. W., Vermeulen R. C., Peeters P. H., Beijnen J. H., van Gils C. H. (2012) Early diagnostic protein biomarkers for breast cancer: how far have we come? Breast Cancer Res. Treat. 134, 1–12 [DOI] [PubMed] [Google Scholar]
- 21. Klos A., Tenner A. J., Johswich K. O., Ager R. R., Reis E. S., Köhl J. (2009) The role of the anaphylatoxins in health and disease. Mol. Immunol. 46, 2753–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francis K., Lewis B. M., Akatsu H., Monk P. N., Cain S. A., Scanlon M. F., Morgan B. P., Ham J., Gasque P. (2003) Complement C3a receptors in the pituitary gland: a novel pathway by which an innate immune molecule releases hormones involved in the control of inflammation. FASEB J. 17, 2266–2268 [DOI] [PubMed] [Google Scholar]
- 23. Mamane Y., Chung Chan C., Lavallee G., Morin N., Xu L. J., Huang J., Gordon R., Thomas W., Lamb J., Schadt E. E., Kennedy B. P., Mancini J. A. (2009) The C3a anaphylatoxin receptor is a key mediator of insulin resistance and functions by modulating adipose tissue macrophage infiltration and activation. Diabetes 58, 2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu X., Nagarajan H., Lewis N. E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., Andersen M. R., Neff N., Passarelli B., Koh W., Fan H. C., Wang J., Gui Y., Lee K. H., Betenbaugh M. J., Quake S. R., Famili I., Palsson B. O., Wang J. (2011) The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roberts A., Pimentel H., Trapnell C., Pachter L. (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 [DOI] [PubMed] [Google Scholar]
- 28. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 29. Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rosethorne E. M., Leighton-Davies J. R., Beer D., Charlton S. J. (2004) ATP priming of macrophage-derived chemokine responses in CHO cells expressing the CCR4 receptor. Naunyn Schmiedebergs Arch. Pharmacol. 370, 64–70 [DOI] [PubMed] [Google Scholar]
- 31. Ames R. S., Lee D., Foley J. J., Jurewicz A. J., Tornetta M. A., Bautsch W., Settmacher B., Klos A., Erhard K. F., Cousins R. D., Sulpizio A. C., Hieble J. P., McCafferty G., Ward K. W., Adams J. L., Bondinell W. E., Underwood D. C., Osborn R. R., Badger A. M., Sarau H. M. (2001) Identification of a selective nonpeptide antagonist of the anaphylatoxin C3a receptor that demonstrates antiinflammatory activity in animal models. J. Immunol. 166, 6341–6348 [DOI] [PubMed] [Google Scholar]
- 32. Martin U., Bock D., Arseniev L., Tornetta M. A., Ames R. S., Bautsch W., Köhl J., Ganser A., Klos A. (1997) The human C3a receptor is expressed on neutrophils and monocytes, but not on B or T lymphocytes. J. Exp. Med. 186, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aguilar E., Pineda R., Gaytán F., Sánchez-Garrido M. A., Romero M., Romero-Ruiz A., Ruiz-Pino F., Tena-Sempere M., Pinilla L. (2013) Characterization of the reproductive effects of the Vgf -derived peptide TLQP-21 in female rats: in vivo and in vitro studies. Neuroendocrinology 98, 38–50 [DOI] [PubMed] [Google Scholar]
- 34. Overington J. P., Al-Lazikani B., Hopkins A. L. (2006) How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 [DOI] [PubMed] [Google Scholar]
- 35. Settmacher B., Rheinheimer C., Hamacher H., Ames R. S., Wise A., Jenkinson L., Bock D., Schaefer M., Köhl J., Klos A. (2003) Structure-function studies of the C3a-receptor: C-terminal serine and threonine residues which influence receptor internalization and signaling. Eur. J. Immunol. 33, 920–927 [DOI] [PubMed] [Google Scholar]
- 36. Lim J., Iyer A., Suen J. Y., Seow V., Reid R. C., Brown L., Fairlie D. P. (2013) C5aR and C3aR antagonists each inhibit diet-induced obesity, metabolic dysfunction, and adipocyte and macrophage signaling. FASEB J. 27, 822–831 [DOI] [PubMed] [Google Scholar]
- 37. Hahm S., Fekete C., Mizuno T. M., Windsor J., Yan H., Boozer C. N., Lee C., Elmquist J. K., Lechan R. M., Mobbs C. V., Salton S. R. (2002) VGF is required for obesity induced by diet, gold thioglucose treatment, and agouti and is differentially regulated in pro-opiomelanocortin- and neuropeptide Y-containing arcuate neurons in response to fasting. J. Neurosci. 22, 6929–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levi A., Eldridge J. D., Paterson B. M. (1985) Molecular cloning of a gene sequence regulated by nerve growth factor. Science 229, 393–395 [DOI] [PubMed] [Google Scholar]
- 39. Rahpeymai Y., Hietala M. A., Wilhelmsson U., Fotheringham A., Davies I., Nilsson A. K., Zwirner J., Wetsel R. A., Gerard C., Pekny M., Pekna M. (2006) Complement: a novel factor in basal and ischemia-induced neurogenesis. EMBO J. 25, 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bénard M., Raoult E., Vaudry D., Leprince J., Falluel-Morel A., Gonzalez B. J., Galas L., Vaudry H., Fontaine M. (2008) Role of complement anaphylatoxin receptors (C3aR, C5aR) in the development of the rat cerebellum. Mol. Immunol. 45, 3767–3774 [DOI] [PubMed] [Google Scholar]
- 41. Gao J., Choe H., Bota D., Wright P. L., Gerard C., Gerard N. P. (2003) Sulfation of tyrosine 174 in the human C3a receptor is essential for binding of C3a anaphylatoxin. J. Biol. Chem. 278, 37902–37908 [DOI] [PubMed] [Google Scholar]